Abstract
Objective:
To assess the quality of value sets in clinical quality measures, both individually and as a population of value sets.
Materials and methods:
The concepts from a given value set are expected to be rooted by one or few ancestor concepts and the value set is expected to contain all the descendants of its root concepts and only these descendants. (1) We assessed the completeness and correctness of individual value sets by comparison to the extension derived from their roots. (2) We assessed the non-redundancy of value sets for the entire population of value sets (within a given code system) using the Jaccard similarity measure.
Results:
We demonstrated the utility of our approach on some cases of inconsistent value sets and produced a list of 58 potentially duplicate value sets from the current set of clinical quality measures for the 2014 Meaningful Use criteria.
Conclusion:
These metrics are easy to compute and provide compact indicators of the completeness, correctness, and non-redundancy of value sets.
Introduction
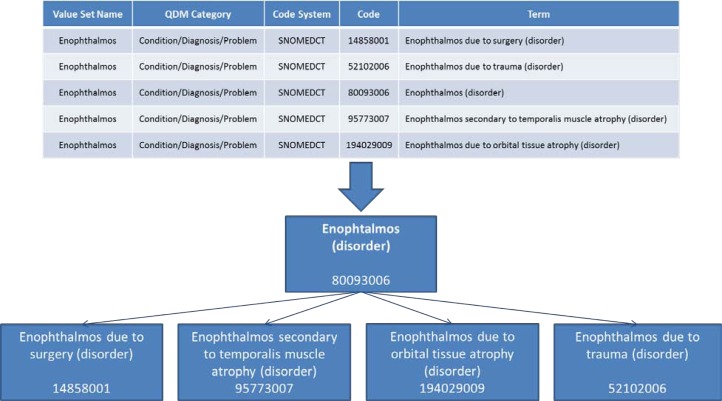
In recent years, there has been an effort to establish quality measures for health care providers, with the objective of improving the quality of health care and comparing performance across institutions. As part of the Meaningful Use incentive program and the certification criteria for electronic health record (EHR) products, the Office of the National Coordinator for Health Information Technology (ONC) and the Centers for Medicare & Medicaid Services (CMS) have selected a set of clinical quality measures (CQMs). For example, one such measure assesses the percentage of pediatric diabetic patients who have been tested for hemoglobin A1c in the past year. The basic information for computing these measures is drawn from EHR data. The implementation of clinical quality measures, i.e., the binding of CQMs to EHR data is realized through sets of codes from standard vocabularies, called value sets, corresponding to specific data elements in the CQM. For example, all procedure codes for Intracranial Neurosurgery in ICD-10 or all diagnosis codes for Enophthalmos in SNOMED CT (see Figure 1, top).
Figure 1.
Value set for the condition Enophthalmos in SNOMED CT, with the value set name (intension) and the list of codes (extension), as well as the hierarchical relations among these codes.
On October 25, 2012, the National Library of Medicine (NLM), in collaboration with ONC and CMS, launched the NLM Value Set Authority Center (VSAC). The VSAC, which is accessible over the web at https://vsac.nlm.nih.gov, provides downloadable access to all official versions of vocabulary value sets contained in the clinical quality measures that are part of the 2014 Meaningful Use criteria. As of December 21, 2012, the VSAC contains 1,520 unique value sets used in 93 clinical quality measures, representing 83,723 unique codes from standard vocabularies including LOINC, RxNorm, SNOMED CT, ICD-9CM, ICD-10-CM, ICD-10-PCS and CPT.
NLM is responsible for both the validation and the delivery of the value sets. All the codes used in the value sets from the clinical quality measures investigated were validated for referential integrity against current versions of the corresponding reference code systems. Types of errors encountered include obsolete codes, errors in codes, and code/description mismatch. Feedback was provided to the measure developers, including suggestions for fixing errors. The value sets were validated iteratively until all errors had been fixed. However, given the short timeframe, limited quality assurance has been performed, beyond ensuring referential integrity and currency of the codes 1.
We define the following quality criteria for value sets in clinical quality measures.
Completeness: A value set should contain all the relevant codes for a particular data element. Moreover, the value set name should also denote this data element. From a terminological perspective, the code corresponding to the data element in the code system should be present in the value set, along with all its descendants. As a consequence, the value set is expected to be rooted by one concept and to contain all the descendants of this root concept.
Correctness: A value set should contain only the relevant codes for a particular data element. From a terminological perspective, the presence in the value set of codes other than the root concept and its descendants might indicate incorrect codes, as they are outside the value domain.
Non-redundancy: A given data element should be represented by one and only one value set (for a given code system). Multiple value sets with the same codes should be harmonized, in order to facilitate maintenance and prevent inconsistency over time. (Duplicate value sets may have been introduced in CQMs at a time when no single repository of value sets existed.)
Figure 1 illustrates an “ideal” value set, for which the name of the value set corresponds to a concept from the underlying terminology, and for which the list of codes provided for the data element (Figure 1, top) exactly includes this concept and all its descendants in the code system (see hierarchical relations, Figure 1, bottom). The name of the data element, Enophthalmos, represents the intension (or intended meaning) of the value set, as would its definition. The list of codes represents the extension of the value set.
The objective of this study is to introduce metrics for assessing the completeness, correctness, and non-redundancy of value sets based on the structure of the underlying terminologies through the UMLS. Focusing on disease value sets, we demonstrate how these metrics can help detect quality issues in value sets.
Background
Quality measure value sets form the basis for guidelines and standards for measuring and reporting on performance regarding preferred practices or measurement frameworks. Each value set is a domain specific list of concepts (codes) derived from standard terminologies, including SNOMED CT®, LOINC®, and RxNorm, used to instantiate data elements from clinical quality measures (e.g., patients with diabetes, clinical visit).
NLM terminology services are used in this study for mapping value set names to terminology concepts. The Unified Medical Language System® (UMLS®) terminology services (UTS) provide exact and normalized match search functions, which identify medical concepts for a given search string. The normalization process is linguistically motivated and involves stripping genitive marks, transforming plural forms into singular, replacing punctuation (including dashes) with spaces, removing stop words, lower-casing each word, breaking a string into its constituent words, and sorting the words in alphabetic order 2. In this study, we use the UTS Java API 2.0 for the normalized name mapping of value set names to terminology concepts. The terminology specific parent/child relations among codes in a value set were extracted from the UMLS and stored in a local database.
Related work. Quality assurance of biomedical terminologies is an active domain of research. In the past few years, quality assurance (QA) of biomedical terminologies and ontologies has become a key issue in the development of standard terminologies, such as SNOMED CT. Approaches to quality assurance include the use of lexical, structural, semantic and statistical techniques applied to particular biomedical terminologies and ontologies, as well as techniques for comparing and contrasting biomedical terminologies and ontologies 3.
In prior work, we have compared sets of concepts from the UMLS, corresponding to the intension and extension of high-level biomedical concepts 4. Other groups have also leveraged intersections of sets for quality assurance purposes 5.
In contrast, little work has been directed at assessing the quality of value sets beyond the validity of their codes. Jiang et al. evaluated value sets from cancer study common data elements with the focus on finding misplaced values in a value set by analyzing UMLS semantic group associations for all values in a value set 6. The same group also provided an approach for context-driven value set extraction from terminologies such as SNOMED CT 7, but did not apply this approach to quality assessment of existing value sets.
The specific contribution of our work is not only to apply quality assurance methods to the value sets, but also to propose specific quality criteria for value sets and to develop operational definitions for the assessment of these quality criteria, in the form of easily computable metrics.
Materials
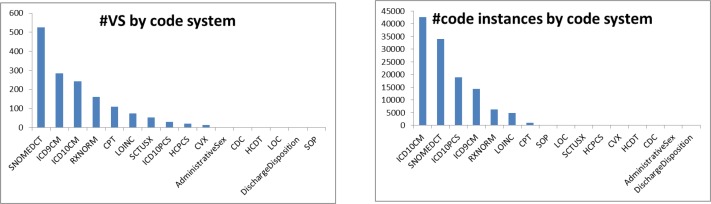
Our study investigates a subset of the 1,520 value sets for the clinical quality measures for the 2014 Meaningful Use criteria, downloaded from the VSAC as Excel files (12/21/2012 release). More specifically, we focus on the 1,054 diagnosis related value sets from SNOMED CT (526), ICD-9-CM (285), and ICD-10-CM (243). As shown in Figure 2, these three code systems cover 86% of all value sets and 74% of all code instances. Information extracted from these value sets includes the value set names, unique identifiers (OIDs), as well as the codes and descriptions (terms). The size of these value sets ranges from one single code up to several thousand codes (median 10), such as the ICD-10-CM Trauma value set with 20,560 codes. SNOMED CT value sets contain between 1 and 3,883 codes (median 11), whereas ICD-9-CM value sets tend to be smaller and range from 1 to 1,213 codes (median 6).
Figure 2.
The diagnosis related value sets from SNOMED CT, ICD-10-CM, and ICD-9-CM account for 1,054 of 1,528 value sets in total (86%). In terms of code instances, these value sets cover 90,937 of all 122,304 code instances (74%).
Methods
Our investigation assesses the quality of the value sets from two different perspectives. On the one hand, we assess the quality of individual value sets (completeness and correctness). On the other hand we examine populations of value sets, with focus on non-redundancy and opportunities for harmonization. As we show later, the population view also provides insights into the quality of individual value sets.
Quality assurance of individual value sets
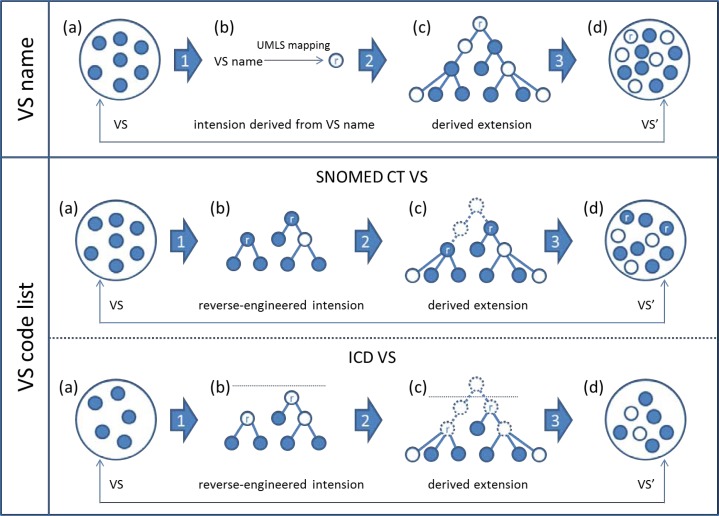
As mentioned earlier, the information currently provided for a value set does not include a detailed, explicit expression of its intension, which is however a prerequisite for assessing a value set in terms of completeness and correctness. As a substitute, we will exploit the information that is available, namely the list of codes and the value set name, of which the latter can be seen as an indirect expression of the intension. Thus, our value set assessment is based on (1) reverse-engineering the intension of a given value set from the value set name and its list of codes, (2) deriving its extension from the reverse-engineered intension, and (3) comparing the actual value set codes with the codes of the derived extension. The overview of our strategy is depicted in Figure 3.
Figure 3.
Quality assurance of individual value sets. Overview of the methods.
(1) Reverse-engineering the value set intension
We propose to reverse-engineer the value set intension from the value set name and from its code list.
Value set name: We leverage the NormalizedString search function of the UTS API 2.0 to establish mappings from the value set names to concepts from a given code system (e.g., SNOMED CT), which is expected to reflect the intension of the value set. Of note, when mapping to concepts from a specific UMLS source vocabulary, the UTS takes advantage of all synonyms for this concept, including synonyms from other source vocabularies.
Code list: We identify as roots those concepts that are not descendants of any other concept inside the value set. For terminologies such as SNOMED CT, where any code can be used for clinical documentation, we look for root concepts within the value set itself. In contrast, coding rules for ICD dictate that only leaf nodes be used in value sets. In this case, we allow aggregation concepts outside a particular value set to be root nodes, if they subsume a maximum of nodes from the value set. However, in order to prevent high-level nodes to be selected as roots for heterogeneous value sets, we set a threshold in the ICD terminology tree, above which aggregation nodes cannot be selected as roots. We mark these root nodes as external roots and distinguish them from the original nodes from the value set. (Of note, multiple parents of the same concept do not interfere with the identification of the root concepts. In SNOMED CT, we identify the root concepts by looking at children, not parents. In ICD-9-CM and ICD-10-CM, concepts do not have multiple parents.)
(2) Deriving the value set extension from the reverse-engineered intension
We compute the full value set extension from the root concepts as identified in step (1) by taking into account the set of all descendants of the roots using the transitive closure of hierarchical relations. We consider the resulting list of codes as the expression of the value set intension.
Value set name: Starting from the concept to which the value set name mapped, we extract all the descendants of this concept in the corresponding code system. The extension consists of the concept mapped to and all its descendants.
Code list: Starting from the root(s) identified from the original value set, we extract all the descendants of these concepts in the corresponding code system. For SNOMED CT, the extension consists of the root concept(s) and all their descendants. For ICD, since only leaf nodes are allowed in the value sets, the extension consists of all the leaf nodes found among the descendants of the root concept(s).
(3) Comparing the actual value set to the derived value set extension
We define a series of metrics to assess the degree to which the value sets conform to the desirable principles of completeness, correctness and non-redundancy, based on the comparison between the original extension of a given value set and the extension derived from reverse-engineering the intension from the name and the roots of the value set.
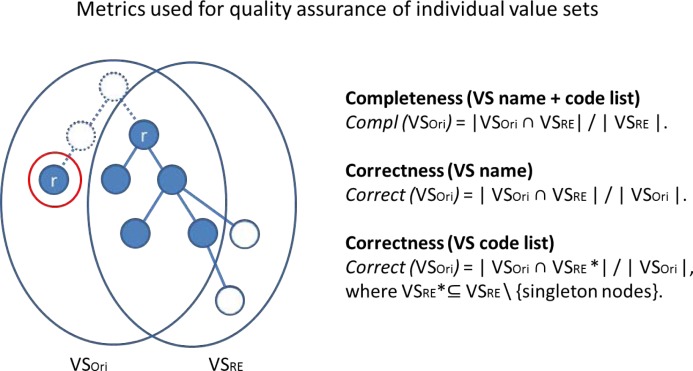
Completeness. Assuming a value set should contain all the codes corresponding to its intension, we compare the cardinality of the original value set extension (VSori) to that of the extension derived from the reverse-engineered intension (VSRE). As a measure of completeness, we use the proportion of the codes from VSRE covered by VSori.
Correctness. Assuming a value set should contain only the codes corresponding to its intension, we compare the cardinality of the reverse-engineered value set extension (VSRE) to that of the original value set (VSori). As a measure of correctness, we use the proportion of the codes from VSori covered by VSRE. This metric works well when using the extension derived from the name of the value set. However, since the extension derived from the roots includes, by design, the entirety of the original value set, a different metric must be defined in this case. Since we assumed that good value sets should have only one or at least few roots, it also means that the codes in a value set should be partitioned into a similar number of large clusters (relatively to the size of the value set). In other words, small disconnected clusters may be indicative of incorrect codes. Intuitively, aggregating the largest clusters together will result in isolating the smaller clusters. If we assume (liberally) that no value set should have more than 10 roots, the corollary is that clusters with less than 10% of the size of the value set are potentially suspicious. The proportion of codes in clusters containing less than 10% of the size of the value set thus provides another metric of correctness. Of note, the presence of a large number of roots and the presence of singleton clusters in the reverse-engineered value set are also indicators of potential quality problems in the value set.
Quality assurance of a population of value sets
In addition to comparing several aspects of a given value set (e.g., intension and extension), it is also possible to compare the value sets to one another in a population of value sets. Under our principle of non-redundancy, there should not be more than one value set for the same intension, and there should not be two value sets with exactly the same extension.
Non-redundancy can be assessed by computing the pairwise similarity of all value sets (within a given code system) in order to identify completely similar (duplicate) or partially similar (redundant) value sets. We use the Jaccard index J to measure the similarity between VSi and VSj based on the codes they contain as follows:
In practice, pairs of VSs with J(VSi, VSj) = 1 are duplicate value sets, as they contain exactly the same codes. Pairs of highly-similar value sets may exhibit redundancy or inconsistency, i.e., redundancy with errors (incorrect or missing codes).
Due to the large number of comparisons and pairwise similarity values generated, we use hierarchical clustering to analyze the results, using a threshold for the minimum distance between value sets to identify redundancy.
Implementation
The methods developed for the analysis of the value sets were implemented in Java 7. We used the 2012AB edition of the UMLS. The mapping of value set names to UMLS concepts were performed using the UMLS Terminology Service (UTS) API 2.0. We computed the transitive closure of hierarchical relations among concepts within a given code system using an RDF (Resource Description Framework) version of the UMLS Metathesaurus in a Virtuoso triple store. Pairwise similarity of value sets and the dendrogram and heat map were generated using the Vegan package of the statistical software tool R.
Results
Quality assurance of individual value sets
(1) Reverse-engineering the value set intension
Value set name: The mapping of value set names to terminology concepts (exact or normalized match to one concept) succeeded for 38% of all value sets. For 214 of the 526 (41%) SNOMED CT value sets, we were able to map the value set name to a SNOMED CT concept. For ICD-9-CM and ICD-10-CM we mapped 108 out of 285 (38%), and 83 out of 243 (34%), respectively.
Code list: Extensions derived from the code lists were computed for all the value sets from any of the three code systems. The number of roots per value set ranged from 1 to 107 with a median of 1 and an average of 6.0 for SNOMED CT, from 1 to 167 (median 2, average 6.7) for ICD-10-CM, and from 1 to 114 (median 1, average 4.7) for ICD-9-CM.
(2) Deriving the value set extension from the reverse-engineered intension
The distribution of the size of the derived extensions (from names or code lists) for each of the three code systems is shown in Table 1.
Table 1.
Distribution of the size of the derived extensions from names or code lists.
| Value set name | Code list | |||||||
|---|---|---|---|---|---|---|---|---|
| # VS | Min | Max | Med | # VS | Min | Max | Med | |
| SNOMED CT | 214 | 1 | 9232 | 9.5 | 526 | 1 | 11793 | 20 |
| ICD-9-CM | 108 | 1 | 1105 | 6 | 285 | 1 | 3629 | 9 |
| ICD-10-CM | 83 | 1 | 234 | 8 | 243 | 1 | 22894 | 16 |
(3) Comparing the actual value set to the derived value set extension
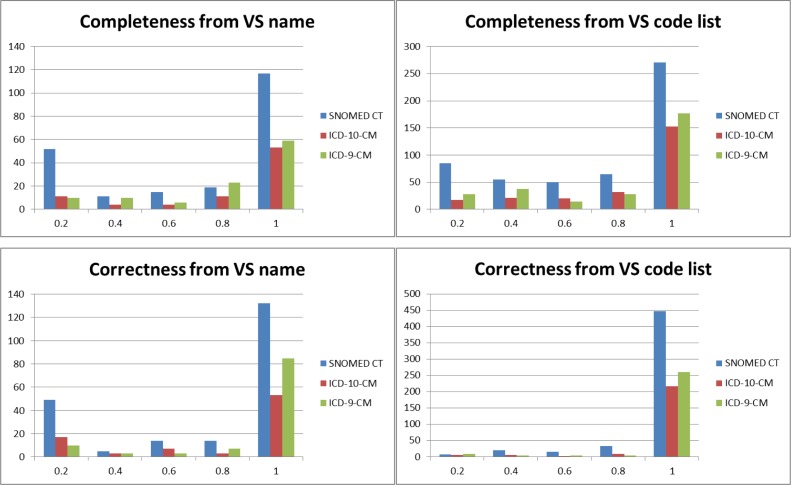
Completeness. Overall, of the 1,054 value sets, 601 (57%) are complete (i.e., have a completeness measure of 1.0) and another 125 (12%) are nearly complete (completeness measure > 0.8). The distribution of completeness measures for all value sets from the three code systems and for each type of reverse engineering (from name and from code list) is shown in Figure 5, top.
Correctness. Overall, of the 1,054 value sets, 927 (88%) are correct (i.e., have a correctness measure of 1.0) and another 48 (5%) are nearly correct (correctness measure > 0.8). The distribution of correctness measures for all value sets from the three code systems and for each type of reverse engineering (from name and from code list) is shown in Figure 5, bottom.
Figure 5.
Completeness and correctness for SNOMED CT, ICD-9-CM, and ICD-10-CM value sets.
Quality assurance of a population of value sets
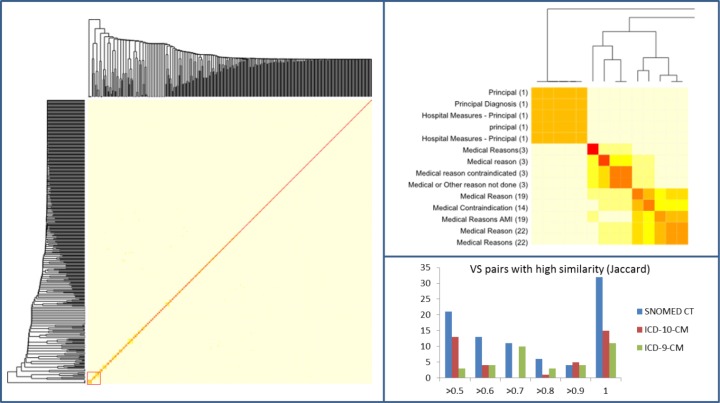
The dendrogram shown in Figure 6 is a visualization artifact for the pairwise similarity matrix computed among the value sets (within a given code system). The left part of the figure provides an overview of the similarity among the value sets. Short branches (as in the bottom left corner) reflect areas of high similarity. In the detailed view, the colors of the heatmap reflect different degrees of similarity. Red (or dark) regions correspond to highly similar value sets (e.g., Medical reason contraindicated and Medical or Other reason not done). The distribution of the pairwise similarity of the value sets is shown in the bottom left portion of the figure. Of note, there are 32 pairs of identical value sets from SNOMED CT.
Figure 6.
Pairwise similarity of SNOMED CT value sets. Left: The heat map shows a low similarity among value sets overall. Right, top: The detailed view reveals local clusters of high similarity for the part marked with a red square in the heat map on the left. Right, bottom: Number of value set pairs with a high similarity for SNOMED CT and ICD value sets.
Discussion
Significance of findings
Our approach was effective in identifying a number of potential errors and inconsistencies in value sets. For example, 58 duplicate value sets were identified, as well as 25 highly-similar value sets. However, the metrics we defined for completeness, correctness and non-redundancy are only indicators provided to help direct the attention of value set developers to areas of the value sets where errors are more likely (e.g., small disconnected clusters of codes within a value set).
Our approach has proved effective on a variety of value sets of different sizes and from code systems with different degrees of granularity (i.e., SNOMED CT vs. versions 9-CM and 10-CM of ICD). They have required little adaptation between code systems. Unlike manual curation, they can easily be reapplied to new versions of the value sets, at no significant cost.
The principles behind the quality criteria (e.g., unique root) have been verified in a large number of cases, where value sets exhibit perfect completeness and correctness. However, these principles may be too strict in some cases and exceptions might apply to some value sets.
Limitations and future work
The metrics that we labeled “completeness” and “correctness” in reference to general quality criteria may simply reflect deviation from these criteria, but not necessarily errors. For example, the SNOMED CT value set for Acute tonsillitis was rooted by the UMLS name mapping approach to SNOMED CT concept Acute tonsillitis (17741008). The completeness for this value set is 1.0 because it indeed contains all descendants of SNOMED CT concept Acute tonsillitis. However, correctness was computed to be 0.92 because the set also contains the concept Acute lingual tonsillitis, which is not a child but a sibling of Acute tonsillitis. Both concepts are descendants of the term Acute pharyngitis, which might be eligible as an alternative value set name. In this case, the perception of an error arises from the lack of a relation in SNOMED CT between Acute lingual tonsillitis and Acute tonsillitis, but the value set actually seems conform to its intension.
Unlike reverse-engineering of the intension of the value set from the code list, reverse-engineering from the name is not always successful, because the name of the value set is not always amenable to mapping to a concept name. Overall, reverse-engineering from the name has only been successful for 38% of all value sets.
Future work
Additional indicators of potential issues in the value sets could be explored further, e.g., the presence of a large number of roots and the presence of roots without expansion. We also would like to compare value sets across code systems (e.g., within a grouping), leveraging equivalence and mapping relations across terminologies. Finally, we plan to generalize this approach to value sets from other code systems, e.g., drug value sets.
Conclusions
We developed operational definitions for the quality assurance of value sets in the form of metrics for completeness, correctness and non-redundancy of value sets, considering the perspective of both individual value sets and value set populations. These metrics are easy to compute and can help direct the attention of value set developers to areas of the value sets where errors are more likely. We recommend that such metrics be integrated into value set authoring systems, such as the NLM Value Set Authority Center.
Figure 4.
Quality assurance of an individual value set based on the derived value set extension. The graph on the right represents the derived extension VSRE for a given VSori. All blue circles represent the six codes from VSori. The two solid white circles represent the codes that were added through the expansion of the root nodes (blue circles marked with r). Completeness: In this case, the completeness, which is the ratio between the common nodes in the intersection (blue) and VSRE, is 5 divided by 7 = 0.71. Correctness (for the value set code list): The ratio between the common nodes in the intersection (blue) and VSori is 5 divided by 6 = 0.83. The blue node in VSori is identified as a singleton and is thus not included in VSRE. The doted nodes and lines indicate the shortest path via the lowest common ancestor at the top between the root nodes.
Acknowledgments
This work was supported in part by the Intramural Research Program of the NIH, National Library of Medicine. This research was supported in part by an appointment to the NLM Research Participation Program.
References
- 1.Winnenburg R, Bodenreider O. Issues in creating and maintaining value sets for clinical quality measures. AMIA Annu Symp Proc; 2012; 2012. pp. 988–96. [PMC free article] [PubMed] [Google Scholar]
- 2.McCray AT, Srinivasan S, Browne AC. Lexical methods for managing variation in biomedical terminologies. Proc Annu Symp Comput Appl Med Care; 1994. pp. 235–9. [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu X, Fan JW, Baorto DM, Weng C, Cimino JJ. A review of auditing methods applied to the content of controlled biomedical terminologies. J Biomed Inform. 2009 Jun;42(3):413–25. doi: 10.1016/j.jbi.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bodenreider O, Burgun A. Aligning knowledge sources in the UMLS: methods, quantitative results, and applications. Stud Health Technol Inform. 2004;107(Pt 1):327–31. [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Y, Gu HH, Perl Y, Geller J, Halper M. Structural group auditing of a UMLS semantic type's extent. J Biomed Inform. 2009 Feb;42(1):41–52. doi: 10.1016/j.jbi.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 6.Jiang G, Solbrig HR, Chute CG. Quality evaluation of cancer study Common Data Elements using the UMLS Semantic Network. J Biomed Inform. 2011 Dec;44(Suppl 1):S78–85. doi: 10.1016/j.jbi.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 7.Pathak J, Jiang G, Dwarkanath SO, Buntrock JD, Chute CG. LexValueSets: an approach for context-driven value sets extraction. AMIA Annu Symp Proc; 2008. pp. 556–60. [PMC free article] [PubMed] [Google Scholar]