Abstract
With antimicrobial resistance increasing worldwide, there is a great need to use automated antimicrobial decision support systems (ADSSs) to lower antimicrobial resistance rates by promoting appropriate antimicrobial use. However, they are infrequently used mostly because of their poor interoperability with different health information technologies. Ontologies can augment portable ADSSs by providing an explicit knowledge representation for biomedical entities and their relationships, helping to standardize and integrate heterogeneous data resources. We developed a bacterial clinical infectious diseases ontology (BCIDO) using Protégé-OWL. BCIDO defines a controlled terminology for clinical infectious diseases along with domain knowledge commonly used in hospital settings for clinical infectious disease treatment decision-making. BCIDO has 599 classes and 2355 object properties. Terms were imported from or mapped to Systematized Nomenclature of Medicine, Unified Medical Language System, RxNorm and National Center for Bitechnology Information Organismal Classification where possible. Domain expert evaluation using the “laddering” technique, ontology visualization, and clinical notes and scenarios, confirmed the correctness and potential usefulness of BCIDO.
INTRODUCTION
Antimicrobial resistance is an increasing problem worldwide and is contributed to by inappropriate antimicrobial prescribing. Incorporating an antibiotic decision support system (ADSS) into clinical decision-making has been shown to be effective at reducing inappropriate antibiotic prescribing and lowering local antimicrobial resistance 1–3. However, despite the apparent benefits of ADSSs, ADSSs are infrequently used in the hospital in-patient setting (Y. Furuya, personal communication, February 8, 2013).
The barriers to widespread adoption and implementation of successful ADSSs include standalone systems that are independent from the electronic health record (EHR) and require interruption of the clinical workflow to use 2,4, a single infectious disease focus (i.e. acute bronchitis) 5, and ADSSs which use their own terminology and cannot be transferred to other EHR systems 5,6. MYCIN was the first ADSS, and though it was hardly used in practice, research indicated that it proposed an acceptable antimicrobial therapy in approximately 69% of cases 4. Interestingly, this was better than the performance of infectious disease experts who were judged using the same criteria, demonstrating the large amount of disagreement that exists between experts about appropriate treatment 7. A number of other automated ADSSs have since been developed for different clinical settings such as intensive care 2,8 and primary care 9. Like MYCIN, some of these systems are standalone 2 requiring data entry by users, while others have been integrated into the local microbiology, prescribing and hospital epidemiology systems 10. Evans et al. developed one of the most successful ADSSs that uses the hospital information system 1,10. However, despite the local success of the system with a reduction in antimicrobial use 11, the system has not been implemented in other hospital systems in the 20 years since it was developed.
Integration and interaction of the ADSS with the EHR is a key requirement for future ADSSs. Successful ADSS have been developed using their own terminologies 5,11, which makes it difficult to transfer the successful system to other institutions or EHR systems. Ontologies can formally represent sharable domain knowledge for key concepts and their semantic relationships 12 and augment clinical decision support systems by providing a standard vocabulary for biomedical entities, helping to standardize and integrate data resources and providing a source of computable domain knowledge that can be exploited for decision support purposes 13–15. To facilitate interoperability and wide spread dissemination of future portable ADSSs, we developed a bacterial clinical infectious diseases ontology (BCIDO). BCIDO serves as an application ontology capturing a controlled terminology for clinical infectious diseases along with domain knowledge commonly used in the hospital in-patient setting. Clinical infectious disease is a broad clinical domain, which not only includes the medical specialty infectious disease, but also has relevance to all other medical and surgical specialties and public health, as clinicians of all disciplines frequently prescribe antimicrobial therapy for their patients. Key roles of an infectious diseases physician include recommending appropriate empiric antimicrobial therapy for an infectious disease in the absence of a definite etiological agent, and selecting the most appropriate antimicrobial therapy and duration when the etiological organism is known. One of the key aspects of selecting an appropriate antibiotic regimen in the absence of bacterial culture results is considering which bacteria may be causing that particular infectious disease, as well as the antibiotics that are effective against those likely bacteria. Therefore, BCIDO encompasses terms and knowledge about common clinical presentations of infections, patient-specific factors that influence differential diagnoses and treatment options, the organisms themselves, and the antimicrobial agents used to treat infections. In the rest of this paper, we present our design and evaluation methods for this ontology.
METHODS
Our method includes four components: (1) determination of the domain and scope of the ontology; (2) review of the literature and related ontologies to evaluate them for reuse; (3) knowledge representation using Protégé OWL; and (4) evaluation of the ontology for its correctness and usefulness.
1. Ontology domain and scope
To determine the functional requirements and clinical scope of the ontology, two meetings lasting one hour were held with two infectious disease physicians, a hospital epidemiologist and an infectious disease pharmacist. The target population for the ADSS to be developed from the ontology is the hospital residents and nurse practitioners, as both groups frequently use the EHR, are comfortable using the EHR, and are responsible for collating microbiology results and presenting treatment options to their attending specialist. Although the ultimate goal is integration of the ADSS throughout the hospital, the medical service will be targeted as the initial user and evaluator of the system. The ontology scope covers all factors relevant to making an appropriate antimicrobial decision in the hospital setting including patient factors and microbiology results, such as gram stain and culture results. Specific antimicrobial treatment recommendations cannot be made in the ontology because they vary widely between clinicians, institutions and countries and are therefore not “universal truths”. However, the factors required for making an antimicrobial treatment decision were included in the ontology so that treatment decisions in an ADSS can be tailored to local preferences. As such, two key common questions in making antimicrobial treatment decisions were identified as the main features to address in the ontology. These questions were used to assess the core competency of the ontology.
What are the potential pathogens for a clinical infectious disease when the causative organism is not known?
What antimicrobials can be used to treat an infection in which the causative organism is known?
The ontology was limited to bacterial infections and excluded Mycobacteria. However, it has been designed so that it can be easily extended to include antimicrobial treatments for mycobacterial, viral, and fungal infections. It can also be extended to include other clinically relevant concepts such as symptoms, anatomical site of infection and infectious disease risk factors. The chosen granularity of the ontology is that at which a diagnosis or treatment recommendation can be made.
2. Review of existing ontologies in the infectious disease domain
In order to serve the purpose of supporting interoperability, ontologies need to be utilized by multiple different groups and applications. Thus, our first step in development was to review existing ontologies to identify those we could re-use. Existing ontologies related to antimicrobials, microorganisms and clinical infectious diseases were evaluated for suitability of content coverage and depth of knowledge, as well as potential to support successful inference. The biomedical literature and ontology repositories such as the NCBO BioPortal (http://bioportal.bioontology.org/), Protégé Wiki (http://protegewiki.stanford.edu/), Swoogle (http://swoogle.umbc.edu/), Ontology Lookup Service from the European Bioinformatics Institute (http://www.ebi.ac.uk/ontology-lookup/) and Open Biomedical Ontology (OBO) Foundry (http://www.obofoundry.org/) were searched to identify potentially relevant ontologies.
The search for relevant ontologies revealed more than 80 vocabulary resources related to the domain of infectious diseases. There were a number of different types of resources ranging from simple taxonomies, glossaries and thesauri through to more formalized clinical vocabularies which are considered to be ontologies. A summary of the key resources is shown in Table 1. The antibiotic prescribing ontology by Bright et al. 16 and the Infectious Disease Ontology (IDO) 17 were identified as the most likely to fulfill the ontology requirements for BCIDO.
Table 1.
The relevance of existing ontologies to the clinical infectious disease decision support domain
| Resource | Type | Clinical infectious disease knowledge content | Suitability of resource for clinical infectious disease decision support domain | Relevance to the BCIDO |
|---|---|---|---|---|
| Medical Subject Headings Controlled Vocabulary (MeSH) | Controlled vocabulary | Comprehensive coverage | Terms appear in hierarchies on the basis of relatedness for document retrieval and are not is-a relationships. Terms are not linked by relations | Terms used in an infectious diseases ontology can be mapped to MeSH |
| International Classification of Diseases – 10 (ICD-10) | Clinical terminology | Reasonable coverage but lacks depth | Disease classification is based on anatomy and pathological structures, however the infectious disease domain does not follow anatomical partitions. Therefore, the hierarchy is not logical for clinical infectious diseases | Terms used in an infectious diseases ontology can be mapped to ICD-10 |
| Systematized Nomenclature of Medicine – Clinical Terms (SNOMED-CT) | Clinical terminology | Comprehensive coverage of clinical terms | Multiple modes of classification limits the capability of automated reasoning The result is the assertion of type-supertype relations that do not hold. There are no natural language definitions. | Terms used in an infectious diseases ontology were mapped to SNOMED-CT |
| Disease Ontology (DO) | Ontology | Limited coverage | Disorganized hierarchy for infectious diseases. Mixes types of infection with types of disease, as well as mixing types based on anatomical location, type of infectious agent or clinical syndrome and properties of infectious agents | Terms used in an infectious diseases ontology can be mapped to DO |
| Antibiotic prescribing ontology | Ontology | Limited to common infections seen in hospital | Suitable, however, the ontology is not publically available. Narrow range of clinical infectious disease syndromes. | Provides proof of principle that an ontology can provide the knowledge base for decision support |
| Infectious Diseases Ontology (IDO) | Ontology | Many of the modules are still in development | Suitable. Before BCIDO, there was no IDO module corresponding to the clinical ID domain. | The core IDO was used as the upper ontology. |
Bright et al. 16 recently developed and evaluated an antibiotic prescribing ontology for guiding appropriate antibiotic prescribing, providing a proof of concept that an infectious disease ontology can provide the knowledge base for an ADSS. The antibiotic prescribing ontology is not implemented in clinical practice, although the following prescribing alerts were successfully generated: (1) antibiotic-microorganism mismatch alert; (2) medication-allergy alert; and (3) non-recommended empiric antibiotic therapy alert 16. The scope of the antibiotic prescribing ontology covers empiric treatment of common clinical infectious diseases and antibiotic choices when a bacterial pathogen is known. In comparison, BCIDO covers a more extensive range of clinical infectious diseases and provides a comprehensive list of differential bacterial diagnoses, which is important if the microbiological identification is pending or no pathogen is identified. The antibiotic prescribing ontology is not publically available and could not be incorporated into BCIDO.
The IDO 17 (http://infectiousdiseaseontology.org/) is a suite of interoperable ontology modules that together aim to cover the entire infectious disease domain. The suite consists of the core IDO, covering terms and relations generally relevant to the infectious disease domain, and a set of domain-specific ontologies developed as extensions from the core 18. To date, disease and pathogen specific extension ontologies have been developed for malaria19, dengue fever 20, influenza, brucellosis 15, and Staphylococcus aureus 21,22. In addition, the Vaccine Ontology (http://www.violinet.org/vaccineontology/index.php) and the Vector Surveillance and Management Ontology 23 are being actively coordinated with IDO development.
The primary purpose of the core IDO is to maximize interoperability between IDO extensions as well as with ontologies outside the IDO suite. To accomplish this, IDO is developed within the framework of the OBO Foundry 18 (http://obofoundry.org/) and adheres to the Foundry’s ontology development guidelines.
To help standardize and integrate data resources, clinical terms and antimicrobials were mapped to Systematized Nomenclature of Medicine – Clinical Terms (SNOMED CT), the reference resource Unified Medical Language System (UMLS) concept unique identifiers) and RxNorm where possible. Bacterial terms were imported from the National Center for Biotechnology Information Organismal Classification (NCBITaxon).
3. Knowledge Representation in Protégé OWL
BCIDO was developed using the core IDO as an upper ontology, and thus the Basic Formal Ontology and Ontology of General Medical Science, which serve as upper ontologies for the IDO suite. BCIDO adheres to the Foundry’s ontology development guidelines and to Cimino’s Desiderata12. Together these include: (1) using Aristotelian definitions with a single mode of classification, (2) using single inheritance hierarchies, (3) using relations with formal, logical definitions based on a distinction between types and instances, and (4) writing definitions and ontology assertions as compositions of ontology terms and relations.
The ontology was represented in the Web Ontology Language (OWL) using the Protégé-OWL editor as a single hierarchical structure. Domain knowledge was obtained from the first author’s experience as an infectious disease physician and supplemented by common clinical infectious disease text books and guidelines 24,25. Expert Protégé users provided regular feedback on the ontology development (M.R.B. and A.G.). The entire IDO core ontology was imported as the upper ontology (http://purl.obolibrary.org/obo/ido.owl). The Basic Formal Ontology was used to assist in designing the structure of our ontology and defining additional ontology classes and properties. Clinical infectious disease terms were mapped to SNOMED-CT and UMLS, and antibiotic terms were mapped to RxNorm, SNOMED-CT and UMLS using the identifier class annotation property defined by the Dublin Core. Bacterial terms were imported from the NCBITaxon using the minimal information to reference an external ontology term (MERIOT) principle using the web-based OntoFox application26 .
4. Ontology Evaluation
The ontology was evaluated from the standpoints of ontology development, domain knowledge, and usability by domain experts. Ontology development was evaluated by checking for adherence to the guidelines and desiderata listed above 12. The ontology was evaluated for domain knowledge and usability using domain expert review and clinical case review. Two domain experts (ID Fellows) evaluated BCIDO for domain accuracy and core competency. The correctness of the domain concepts and the relationships among them was evaluated using the “laddering” technique and visual inspection of the ontology during one written questionnaire and two structured face-to-face evaluation sessions 27. Each evaluation session lasted one hour. An example of a question for a downward probe for the causes relationship is “Can you tell me some bacteria that cause acute meningitis?” Records from the ID expert evaluation sessions and questionnaire were compared to BCIDO. In addition, BCIDO was evaluated using ten de-identified clinical notes selected non-randomly by an ID expert (S.P.), which were reviewed by a clinician specializing in infectious disease research who had authorization to look at these notes. Case notes were reviewed to identify each piece of information needed to make a bacterial differential diagnosis or antibiotic recommendation, and then BCIDO was evaluated to determine whether there was a corresponding term in the ontology. Changes were made to BCIDO after each evaluation event in response to the ID experts’ comments and case notes.
The ID experts (S.P and H.L.P.) who had previously participated in the laddering study assessed the usefulness of BCIDO for knowledge management. One ID expert provided 16 common infectious diseases questions for BCIDO to provide a list of differential diagnoses and possible antimicrobial treatments. Examples of questions are: “What are the bacteria that cause catheter-associated urinary tract infections and are also gram negative bacilli?” and “What antibiotics could be used to treat such an infection?” The evaluator (C.L.G.) verified that the answers were present in BCIDO. The ID experts also interacted with the BCIDO using Protégé with the evaluator’s assistance. During the session, participants were asked to think aloud as they viewed the ontology, as well as completing a structured questionnaire after the session. Verbalizations and written responses were thematically analyzed to assess ID experts’ perceptions of usefulness of the ontology for knowledge management.
RESULTS
Bacterial clinical infectious disease ontology (BCIDO)
The BCIDO was designed focusing on the areas of “infectious disease”, “antibiotic” and “bacteria” and includes 599 classes and 2355 object properties. The “infectious disease” hierarchy contains 255 classes, of which nearly all were mapped to SNOMED-CT on March 8th 2013 either as exact matches or synonyms. Ninety-nine classes were mapped to UMLS CUI. The “antibiotic” hierarchy contains 98 classes, nearly all of which were mapped to SNOMED-CT and UMLS CUI on March 10th 2013, and RxNorm on August 19th, 2013. The “bacteria” hierarchy contains 255 classes, of which all were imported from NCBITaxon on May 16th, 2013. A class “bacterial quality” was created to include the common properties used by clinicians to narrow differential diagnoses of causative bacteria before the final culture result is available. These high-level BCIDO classes are shown in Table 2 along with descriptions, their BFO class type, the corresponding IDO term or parent type in IDO core, and the source ontology for imported terms.
Table 2:
Ontology classes, descriptions, BFO and IDO class types and source ontology
| Class | Description | BFO class type | IDO class type | Source ontology |
|---|---|---|---|---|
| Bacteria | Bacteria are a large group of single-celled prokaryotic organisms, which may have a variety of shapes ranging from spherical, rod-like, comma-shaped to spiral. | Independent Continuant: object | Bacteria | Bacteria is a term in the IDO core imported from NCBITaxon. |
| Infectious disease | A disease whose physical basis is an infectious disorder. | Dependent Continuant: realizable entity | Disease | “Infectious disease” is a term in the IDO core and is a subtype of “disease” which is imported to IDO core from Ontology for General Medical Science. |
| Antibiotic | A chemotherapeutic agent or substance that kills (microbicidal) or inhibits (microbiostatic) the growth of bacteria and treats bacterial infections. | Independent Continuant: object | Antibiotic | “Antibiotic” is a term in IDO core which is linked to the term “antibiotic” in ChEBI. |
| Bacterial quality | The properties of bacteria that allow bacteria to be classified according to phenotypic or morphologic features. These properties assist with narrowing the differential diagnosis before definitive culture results are available. | Dependent Continuant: quality | Quality | “Bacterial quality” is a subtype of “quality” imported to IDO core from BFO. |
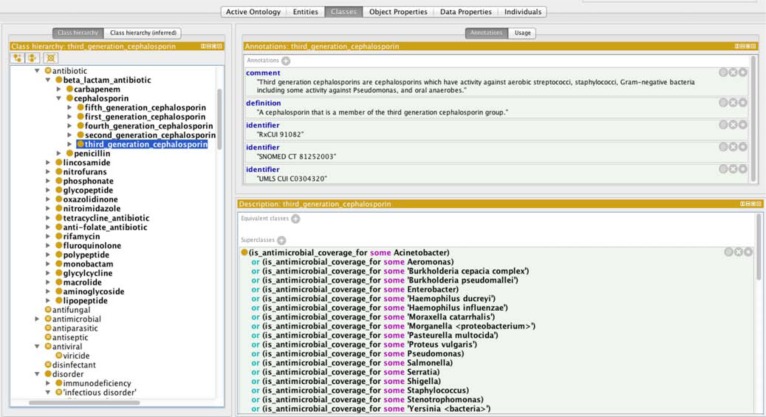
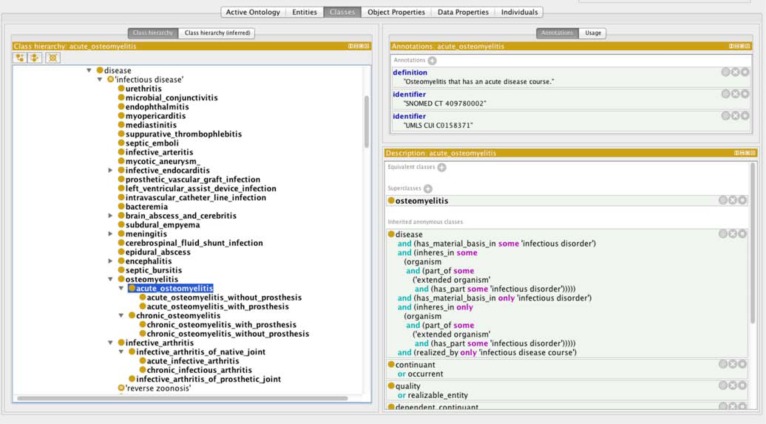
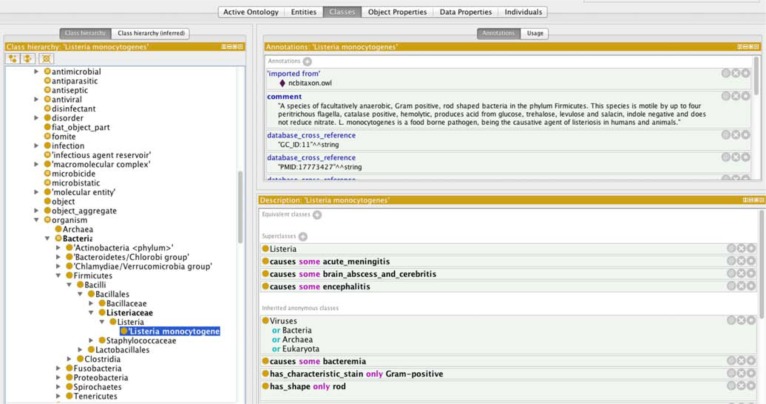
The object properties defined in BCIDO are shown in Table 3. New object properties were created to assert the relations that exist between the three hierarchies. There were 581 object properties between the “bacteria” and “infectious disease” hierarchy; 522 object properties between the “antibiotics” and “bacteria” hierarchies; and 48 object properties between the “bacteria” and “bacterial quality” hierarchies. The object property causes asserts the relation between a “bacteria” and an “infectious disease” and is defined by the existence of a known causative link between the bacteria and the infectious disease. For example, “Neisseria meningitidis causes some meningitis”. The object property is_antimicrobial_coverage_for asserts the relation between an “antibiotic” and a “bacteria” and is asserted when at least some strains of the bacterial type are susceptible to the antimicrobial. For example, “penicillin is_antimicrobial_coverage_for some Treponema pallidum”. The object property has_shape asserts the relation between a “bacteria” and a “bacterial shape” and is defined by the typical shape of the bacteria. For example, “Staphylococcus has_shape spherical”. Figures 1 to 3 demonstrate the three different domain hierarchies and some of their object properties.
Table 3:
Ontology object properties
| Domain class | Object property | Range class |
|---|---|---|
| Bacteria | Causes | Infectious disease |
| Antibiotic | Is_antibiotic_coverage_for | Bacteria |
| Bacteria | Has_characteristic_stain_result | Bacterial quality |
| Bacteria | Has_shape | Bacterial quality |
| Infectious disease | Can_be_associated_with | Infectious disease |
Figure 1:
Domain class of antibiotics showing third generation cephalosporin as an example
Figure 3:
Domain class of infectious disease showing “acute osteomyelitis” as an example
Evaluation Results
1. Correctness of BCIDO
Domain expert review of ontology correctness using the laddering technique and visual review of BCIDO occurred over several weeks and separate sessions were held for each ID expert. Comparison of the ID expert hierarchies using the laddering technique demonstrated agreement with BCIDO class hierarchies, however, the visual review revealed many additional classes to include. BCIDO was refined after each evaluation session for simple changes (e.g.. missing assertion “Mycoplasma genitalium causes some urethritis”) while complex changes were discussed with each ID expert (e.g. location of “sepsis” in “infectious disease” hierarchy). Overall, 8 classes and 35 object properties were added to the “infectious disease” hierarchy; 3 classes and 54 object properties were added to the “bacteria” hierarchy; and 5 classes and 21 object properties were added to the “antibiotic” hierarchy. The majority of the relation assertion additions were between the “infectious disease“ hierarchy and the “bacteria” hierarchy.
In the ten case notes, 100% of antibiotic terms (26/26) and 94% of bacterial terms (15/16) mentioned in the case notes were found in BCIDO. However, 78% of the infectious disease terms were included (18/23). The case notes and ID expert review demonstrated a wide range of synonyms used for the same infectious disease, bacteria and antibiotic. In addition, overlap between infectious disease was not uncommon. For example, a patient with an abdominal wall abscess also had overlying cellulitis. A new object property can_be_associated_with was introduced to link two or more infectious diseases which can commonly occur together.
2. Knowledge management task usefulness of BCIDO
Thematic analysis of the ID expert’s verbal and written responses revealed that both ID experts found that the BCIDO was useful for performing the key knowledge management tasks of guiding differential bacterial diagnoses and initiating antimicrobial treatment. For example, for the question “What are the bacteria that cause catheter-associated urinary tract infections and are also gram negative bacilli?” the options provided by BCIDO were Escherichia coli, Klebsiella, Proteus, Acinetobacter baumannii, Pseudomonas aeruginosa, Stenotrophomonas maltophilia, Enterobacter, Citrobacter, Morganella and Serratia. For the follow-up question “What antibiotics could be used to treat such an infection?” the options provided by BCIDO were aminoglycosides, aztreonam, carbapenem, nitrofurantoin, trimethoprim-sulfamethoxazole and fluroquninolone. ID expert #1 expressed “the ontology was particularly helpful in guiding the differential diagnosis. By including microbiologic data (including appearance of the gram stain, etc.) and grouping disease processes by [infectious disease], I felt that it closely mimicked the thought process I take when evaluating patients. Moving forward, this would be an excellent tool for house staff, as it would guide the differential and assist in informing empiric antibiotic management.” ID expert #2 expressed “I found it interesting and surprisingly quite helpful in the first question of UTI [urinary tract infection], and thought that for the generalist, this … might provide a degree of help in deciding on a treatment regimen.” Both ID experts suggested extensions to BCIDO that may improve its usefulness such as antibiotic resistance knowledge, pharmacologic properties of antibiotics, additional bacterial properties (e.g. coagulase test result for Staphylococci and lactose fermenting result for gram negative bacteria), broadening, narrowing or adding some clinical syndromes and infectious diseases (e.g. fever and rash) and including bacterial prevalence data to prioritize lists of differential bacterial diagnoses. Although ID expert #1 thought BCIDO was “already an excellent tool”, ID expert #2 expressed that future iterations should focus on “increase[ing] user friendliness”.
DISCUSSION
As antimicrobial resistance continues to rise at an alarming rate, solutions such as appropriate antimicrobial prescribing become increasingly important. Although ADSSs have been shown to reduce both antimicrobial prescribing and antibacterial drug resistance, the use of successful ADSS is not widespread. With the exception of the Evans et al. system 1, ADSSs address a single infectious disease or a narrow range of clinical syndromes represented in clinical guidelines and do not comprehensively cover the broad domain of clinical infectious disease. One of the main barriers to dissemination of successful ADSSs is that they tend to use their own terminology. Thus, the system cannot be easily transferred to other EHR systems. Ontologies can improve clinical decision support systems by providing a standard vocabulary for biomedical entities, helping to standardize and integrate data resources. BCIDO serves as an application ontology capturing a controlled terminology for clinical infectious diseases along with domain knowledge commonly used in the hospital in-patient setting with the aim of improving interoperability of portable ADSSs. BCIDO captures much of the knowledge necessary to make clinical decisions about treatment and diagnosis across a broad scope of clinical infectious diseases.
The use of ontologies in decision support is increasing. Members of the Infectious Disease Ontology Consortium have developed and are developing a number of ontologies related to specific infectious diseases such as Brucellosis and Malaria 28,29. The antibiotic prescribing ontology provides the “proof of principle” that an ontology in the infectious diseases domain can successfully enable decision support 16. In comparison to existing decision support ontologies, BCIDO offers comprehensive clinical infectious diseases knowledge required to make clinical decisions before microbiological information is known or in the absence of positive microbial results. This approach accurately reflects the knowledge management tasks of hospital antimicrobial prescribers. BCIDO extends existing ontologies by using the core IDO as the upper ontology, re-using terms and mapping to popular resources such as SNOMED-CT, UMLS and RxNorm. Although not publically available, the antibiotic prescribing ontology could be integrated with BCIDO. Currently, the antibiotic terms in BCIDO are not re-used from an existing ontology. However, the Drug Ontology (DrOn) 30 mediates resources such as Chemical Entities of Biological Interest (ChEBI) and RxNorm and ultimately terms from DrOn will be imported as DrON coverage expands.
Although the current coverage of antibiotics and bacteria in BCIDO is comprehensive, further refinement of the clinical infectious diseases is required as demonstrated by the high number of changes suggested by the ID experts after evaluation. BCIDO also lacks key features of clinical diagnosis such as risk factors for specific infections, symptoms and body location. However, the design of BCIDO is such that these features can be added in future iterations. BCIDO can also be expanded to include viruses, fungi and parasites to create a more comprehensive “organism” hierarchy and relation assertions between the clinical infectious diseases and antimicrobials.
The evaluation of BCIDO has to date been limited. First, the number of domain expert evaluators and number of clinical cases used were small. Thus, our future work will involve continued evaluation using additional domain experts and larger numbers of clinical cases. A second limitation of our evaluation was that it focused only on ontology development best practices and domain knowledge content. Thus, our future work also includes designing an evaluation study to assess how well BCIDO supports clinical decision support.
CONCLUSIONS
BCIDO is a comprehensive application ontology capturing a controlled terminology of bacterial clinical infectious diseases along with domain knowledge commonly used in the hospital in-patient setting. Evaluation by domain experts using several techniques over numerous sessions demonstrated the accuracy and usefulness of BCIDO. BCIDO can improve clinical decision support systems by providing a standard vocabulary for biomedical entities, helping to standardize and integrate data resources to improve portability and interoperability.
Figure 2:
Domain class of bacteria showing Listeria monocytogenes as an example
Acknowledgments
The authors have no conflicts of interest. CLG was supported by a Hutchins Transplant Infectious Diseases Fellowship grant and a Fulbright Scholarship. SP’s contributions were supported by a NIH T32NR013454 grant. LGC's contributions were supported by R01 AI077706 and a Burrough's Wellcome Fund Career Award. AG's contributions were supported by R01 AI077706 to LGC. CW’s contributions were funded under NLM grant R01LM009886. The article’s contents are solely the responsibility of the authors and do not necessarily represent the official view of NIH or BWF..
REFERENCES
- 1.Evans RS, Pestotnik SL, Classen DC, et al. A computer-assisted management program for antibiotics and other antiinfective agents. New England Journal of Medicine. 1998;338:232–8. doi: 10.1056/NEJM199801223380406. [DOI] [PubMed] [Google Scholar]
- 2.Thursky KA, Mahemoff M. User-centered design techniques for a computerised antibiotic decision support system in an intensive care unit. Int J Med Inform. 2007;76:760–8. doi: 10.1016/j.ijmedinf.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 3.Paterson DL. The role of antimicrobial management programs in optimizing antibiotic prescribing within hospitals. Clin Infect Dis. 2006;42(Suppl 2):S90–5. doi: 10.1086/499407. [DOI] [PubMed] [Google Scholar]
- 4.Shortliffe EH, Davis R, Axline SG, et al. Computer-based consultations in clinical therapeutics: explanation and rule acquisition capabilities of the MYCIN system. Comput Biomed Res. 1975;8:303–20. doi: 10.1016/0010-4809(75)90009-9. [DOI] [PubMed] [Google Scholar]
- 5.Gonzales R, Anderer T, McCulloch CE, et al. A cluster randomized trial of decision support strategies for reducing antibiotic use in acute bronchitis. JAMA Intern Med. 2013;173:267–73. doi: 10.1001/jamainternmed.2013.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Linder JA, Schnipper JL, Tsurikova R, et al. Documentation-based clinical decision support to improve antibiotic prescribing for acute respiratory infections in primary care: a cluster randomised controlled trial. Inform Prim Care. 2009;17:231–40. doi: 10.14236/jhi.v17i4.742. [DOI] [PubMed] [Google Scholar]
- 7.Yu VL, Fagan LM, Wraith SM, et al. Antimicrobial selection by a computer. A blinded evaluation by infectious diseases experts. JAMA. 1979;242:1279–82. [PubMed] [Google Scholar]
- 8.Evans RS, Pestotnik SL, Classen DC, et al. Evaluation of a computer-assisted antibiotic-dose monitor. Ann Pharmacother. 1999;33:1026–31. doi: 10.1345/aph.18391. [DOI] [PubMed] [Google Scholar]
- 9.Gulliford MC, van Staa T, McDermott L, et al. Cluster randomised trial in the General Practice Research Database: 1. Electronic decision support to reduce antibiotic prescribing in primary care (eCRT study) Trials. 2011;12:115. doi: 10.1186/1745-6215-12-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans RS, Classen DC, Pestotnik SL, et al. A decision support tool for antibiotic therapy. Proc Annu Symp Comput Appl Med Care. 1995:651–5. [PMC free article] [PubMed] [Google Scholar]
- 11.Evans RS, Pestotnik SL, Classen DC, et al. Development of an automated antibiotic consultant. MD Comput. 1993;10:17–22. [PubMed] [Google Scholar]
- 12.Cimino JJ. Desiderata for controlled medical vocabularies in the twenty-first century. Methods Inf Med. 1998;37:394–403. [PMC free article] [PubMed] [Google Scholar]
- 13.Bodenreider O. Biomedical ontologies in action: role in knowledge management, data integration and decision support. Yearb Med Inform. 2008:67–79. [PMC free article] [PubMed] [Google Scholar]
- 14.Achour SL, Dojat M, Rieux C, et al. A UMLS-based knowledge acquisition tool for rule-based clinical decision support system development. J Am Med Inform Assoc. 2001;8:351–60. doi: 10.1136/jamia.2001.0080351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kashyap V, Morales A, Hongsermeier T. On implementing clinical decision support: achieving scalability and maintainability by combining business rules and ontologies. AMIA Annu Symp Proc. 2006:414–8. [PMC free article] [PubMed] [Google Scholar]
- 16.Bright TJ, Yoko Furuya E, Kuperman GJ, et al. Development and evaluation of an ontology for guiding appropriate antibiotic prescribing. J Biomed Inform. 2012;45:120–8. doi: 10.1016/j.jbi.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cowell L, Smith B. Infectious Diseases Ontology. In: Sintchenko V, editor. Infectious Disease Informatics. New York: Springer; 2010. pp. 373–95. [Google Scholar]
- 18.Smith B, Ashburner M, Rosse C, et al. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat Biotechnol. 2007;25:1251–5. doi: 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Topalis P, Mitraka E, Bujila I, et al. IDOMAL: an ontology for malaria. Malar J. 2010;9:230. doi: 10.1186/1475-2875-9-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitraka E, Topalis P, Dialynas E, et al. IDODEN: An Ontology for Dengue. Proceedings of the International Conference on Biomedical Ontology; 2012; 2012. p. 1. [Google Scholar]
- 21.Goldfain A, Smith B, Cowell LG. Towards an ontological representation of resistance: the case of MRSA. J Biomed Inform. 2011;44:35–41. doi: 10.1016/j.jbi.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goldfain A, Smith B, Cowell LG. Constructing a Lattice of Infectious Disease Ontologies from a Staphylococcus aureus Isolate Repository. Proceedings of the 3rd International Conference on Biomedical Ontology (ICBO 2012); 2012; 2012. pp. 1–5. KR-MED Series; [Google Scholar]
- 23.Lozano-Fuentes S, Bandyopadhyay A, Cowell LG, et al. Ontology for vector surveillance and management. J Med Entomol. 2013;50:1–14. doi: 10.1603/me12169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mandell GL, Bennett JE, Dolin R, Mandell Douglas. Bennett's Principles and Practice of Infectious Diseases. 7th ed. Elsevier; 2010. [Google Scholar]
- 25.Gilbert DN, Moellering RC, Eliopoulos GM, et al. The Sanford Guide to Antimicrobial Therapy 2012. 42nd ed. Antimicrobial Therapy, Inc; 2012. [Google Scholar]
- 26.Xiang Z, Courtot M, Brinkman RR, et al. OntoFox: web-based support for ontology reuse. BMC Res Notes. 2010;3:175. doi: 10.1186/1756-0500-3-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.G R, P M. Laddering. Expert Syst. 1995;12:339–46. [Google Scholar]
- 28.Topalis P, Mitraka E, Bujila I, et al. IDOMAL: an ontology for malaria. Malar J. 2010;9:230. doi: 10.1186/1475-2875-9-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin Y, Xiang Z, He Y. Brucellosis Ontology (IDOBRU) as an extension of the Infectious Disease Ontology. J Biomed Semantics. 2011;2:9. doi: 10.1186/2041-1480-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hogan WR, Hanna J, Joseph E, et al. Towards a Consistent and Scientifically Accurate Drug Ontology. ICBO 2013 Conference Proceedings; 2013; 2013. [PMC free article] [PubMed] [Google Scholar]