Abstract
BioPortal contains over 300 ontologies, for which quality assurance (QA) is critical. Abstraction networks (ANs), compact summarizations of ontology structure and content, have been used in such QA efforts, typically in a “one-off” manner for a single ontology. Ontologies can be characterized—independently of knowledge-content focus—from a structural standpoint leading to the formulation of ontology families. A family is defined as a set of ontologies satisfying some overarching condition regarding their structural features. Seven such families, comprising 186 ontologies, are identified. To increase efficiency, a new family-based QA framework is introduced in which an automated, uniform AN derivation technique and accompanying semi-automated, uniform QA regimen are applicable to the ontologies of a given family. Specifically, across an entire family, the QA efforts exploit family-wide AN features in the characterization of sets of classes that are more likely to harbor errors. The approach is demonstrated on the Cancer Chemoprevention BioPortal ontology.
Introduction
Modern biomedical science is impossible without the management and integration of large data sets. Moreover, the proliferation of interdisciplinary research efforts in the biomedical field is fueling the need to overcome terminological barriers when integrating knowledge from different fields into a unified research project. Thus, biomedical research needs the support of well-developed and well-maintained ontologies that provide structured domain knowledge for data integration, natural language processing, and decision support [1, 2].
The National Center for Biomedical Ontology (NCBO) provides an encyclopedic repository of over 300 ontologies within a uniform development and visualization system covering many different domains. We denote the ontologies hosted in BioPortal as BP ontologies. As BP ontologies underlie various Health Information Systems (HIS), Electronic Health Record (EHR) systems, Health Information Exchanges (HIEs) and healthcare administrative systems, the BioPortal is growing in importance. With the BioPortal framework maturing, the time has come to stress the significance of quality assurance (QA) methodologies for BP ontologies and to further develop them.
Abstraction networks (ANs) are compact networks summarizing the structure and content of ontologies. ANs have been derived in uniquely tailored ways for various individual ontologies. These ANs include: an object-oriented schema for the Medical Entities Dictionary (MED) [3]; the Refined Semantic Network for the UMLS [4]; and various area and partial-area taxonomies for SNOMED CT [5], NCIt [6], Ontology of Clinical Research (OCRe) [7], Sleep Domain Ontology (SDO) [8], and Ontology for Drug Discovery Investigations (DDI) [9]. These ANs were shown to support orientation into the ontologies’ content and structure and have been used to support their QA. However, it would not be practical to derive a unique type of AN for each individual BP ontology. Because the large majority of BP ontologies are released in OWL or OBO formats, many of them share a common underlying structure, such as the usage of domain-defined object properties. We define a family of ontologies as a set of ontologies satisfying some overarching condition regarding their structural features. By structural features, we refer to knowledge elements of classes of an ontology such as kinds of object properties, classes with multiple parents and data properties. Unique combinations of structural features can be used to group BP ontologies into a family.
In this paper, we identify seven families according to combinations of conditions regarding various structural features available in BP ontologies. For example, one family consists of those ontologies with object properties given explicitly defined domain and ranges. Another family contains ontologies with object properties either used as restrictions on classes or given explicitly defined domains and ranges. We collect details and metrics of structural features for 186 BP ontologies and classify each into the proper family.
The organization of ontologies into families serves as the foundation for a new family-based QA framework for ontologies, utilizing a uniform AN derivation technique and uniform AN-based QA regimen for a whole family of ontologies. Such streamlining AN derivation QA process will result in higher efficiency and lower cost of QA. As an illustration of the framework, we apply it to the Cancer Chemoprevention Ontology (CanCo) [10, 11]. The AN for the CanCo is presented. The results of an initial QA review of CanCo based on its AN are given.
Let us note that we do not completely present the new family-based QA framework, although the various aspects of the framework are illustrated using examples. By identifying the structural features defining such families of ontologies, and classifying ontologies into the families, we lay the groundwork for the family-based QA framework. This framework will enable automated AN derivation and semi-automated QA regimens, bringing to bear computer support for the QA of many biomedical ontologies.
Background
The NCBO BioPortal
BioPortal is a repository and uniform development and visualization system for biomedical ontologies provided by the National Center for Biomedical Ontology (NCBO) [12]. BioPortal contains over 300 biomedical ontologies developed in the Web Ontology Language (OWL) [13], Resource Description Framework (RDF) [14], Open Biological and Biomedical Ontologies (OBO) [15] format, Protégé frames, and Rich Release Format. It also provides tools for browsing, developing, editing, and visualizing ontologies to support research in the biomedical sciences.
The NCBO BioPortal has been used in various research projects on biomedical ontologies. Mortensen et al. [16] encoded the Ontology Design Pattern (ODP) from several BP ontologies to facilitate ontology development. Bail et al. [17] examined the justifications from an independently motivated corpus of actively used BP ontologies and exhibited the structural features represented in description logic (DL). Ghazvinian et al. [18] analyzed 53 BP ontologies, identified OBO Foundry candidates and examined their level of term reuse and overlapping.
Abstraction Networks
All but the simplest ontologies are large, complex and heavily interconnected. Thus, diagrammatic displays of ontologies have long been preferred over textual representations. Such diagrams typically take the shape of “node/box and link/arrow” pictures. BioPortal supports a text-based browsing environment, along with a concept-centric diagram display functionality based on FlexViz, a graph based visualization tool [19]. However, when ontologies become large, the advantages of diagrammatic representations disappear, and the graphical representation cannot support ontology orientation and QA efforts. Thus, an alternative compact network, called an abstraction network (AN), summarizing the structure and content of an ontology, can be utilized to make an ontology more comprehensible. An AN consists of nodes connected via child-of hierarchical relationships. Figure 1 demonstrates the general process of deriving an AN from a small ontology of 25 classes (small ovals on the left side). As can be seen on the left, six groups (large ovals) are identified and each is subsequently mapped to and represented by one node on the right side. The derived AN consists of nodes, seen as rectangles, and the child-of links connecting them. The exact nature of the mapping of subsets of the ontology’s classes to AN nodes is separately defined as part of the derivation methodology for each type of AN. By its nature, an AN provides a high-level compact view of the original ontology and can serve as a good entry point for the exploration of its structure and content.
Figure 1.
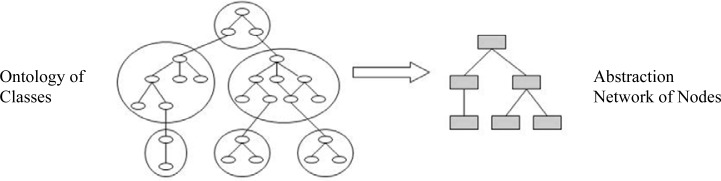
General process of deriving an abstraction network from an ontology
Structural Features of BP Ontologies
In OWL, an object property is an important ontological element, used to relate classes and represent potential relationships between class instances. In ontologies, object properties are utilized in several ways. Object properties can be given explicitly defined domains and ranges, i.e., global limitations on instantiation. An object property’s domain and range can consist of any number of classes from the ontology.
Below is an example in Manchester OWL Syntax of an object property with an explicitly defined domain and range taken from CanCo. In this example the object property is named has disease location and has Disease class as its domain, and Organ class defined as its range. Any instance of has disease location must have a domain that is a disease and a range that is an organ.
ObjectProperty: has_disease_location
Domain: Disease
Range: Organ
Object properties can also be utilized in class restrictions, such as in subclass axioms and class equivalence axioms. Class restrictions are a less strict, local, limitation on the instantiation of object properties. The use of restrictions is more flexible than rigorously defining the domain of every object property and is a common way object properties are utilized (see Results).
The ANs we derived for OCRe [7] and the SDO [8], both available in BioPortal, utilized object properties to create different types of area taxonomies and partial-area taxonomies (taxonomies for short). Taxonomies are a type of dual-level (area and partial-area) AN that group together classes of similar structure and semantics. Taxonomies are used to support orientation and QA of ontologies by highlighting groups of concepts that have a higher likelihood of error. For more details on defining taxonomies see Illustration Section and [5, 7, 8].
For example, the taxonomies derived for OCRe Entity hierarchy utilized only object properties which had explicitly defined domains. For the SDO taxonomies we considered either object properties with explicitly defined domains or object properties used in class restrictions or both to create three different kinds of taxonomies, each of different granularity [8]. A preliminary analysis of the Gene Ontology (GO) (with cross maps to ChEBI) showed that we can derive taxonomies using object properties used in class restrictions on subclass axioms and in class equivalence axioms.
Data properties (attributes) are similar to object properties except the range is a literal value, such as a number or character string. Like object properties, data properties can be given explicitly defined domains or be used in class restrictions. Our previous research has focused on using only object properties to derive taxonomies, but by modifying our derivation methodologies, data properties can potentially be used independent of, or in conjunction with, object properties for deriving new kinds of taxonomies. Below is an example of a data property, has sequence, taken from CanCo, with a domain consisting of two classes, Protein and Nucleic Acid, and a range value defined as a character string.
DataProperty: hasSequence
Domain: Protein, NucleicAcid
Range: xsd:string
Ontologies are organized in a hierarchical structure where the more general classes are at the top and the most specific classes are at the bottom. Ontology hierarchies can be organized either as a directed acyclic graph (DAG), where classes can have multiple superclass, or as strict tree structures where each class can have at most one superclass. Hierarchical relationships can be utilized in deriving abstraction networks as we demonstrated in [20].
Quality Assurance of Biomedical Ontologies using ANs
ANs have typically been used for QA of ontologies in a “one off” manner, one ontology at a time. Previously, ANs were designed individually for SNOMED CT [5], NCIt [6], MED [3], UMLS [4], OCRe [7], SDO [8], and DDI [9]. These ANs were shown to support semi-automated QA of the underlying ontologies by algorithmically identifying sets of classes (or concepts) that are more likely to be erroneous than the general class population. In particular an AN supported the exposure of errors and inconsistencies missed by a DL classifier [21]. Examples of such sets of concepts in SNOMED include small partial-areas, strict-inheritance regions [22], and sets of overlapping concepts [23] corresponding to nodes in a specific kind of AN called the disjoint partial-area taxonomy [20].
Methods
As mentioned, our goal is to create widely applicable uniform AN derivation algorithms and uniform QA methodologies that will work for many ontologies without modification. To accomplish this, we have to group ontologies into families that exhibit similar structural features. For these families, we can then uniformly, algorithmically derive an AN for each ontology of a family. The structural features must be (a) common enough to create families of meaningful sizes; and (b) useful for deriving ANs capable of supporting orientation and QA. The Web Ontology Language (OWL) and the Open Biological and Biomedical Ontologies (OBO) formats are standards based on description logic (DL) that provide a common model for creating ontologies. Most of the ontologies in BioPortal are released in one of these two formats, while some ontologies are released in Rich Release format (RRF). Using the ontological knowledge elements defined for OWL and OBO (e.g. classes, object properties, data properties, subclass axioms, etc.), which we refer to as structural features of an ontology, we can classify ontologies into uniform families of similar structure based on the existence or non-existence of these structural features. Naturally, we focus on features that have been shown in our previous research to support the derivation of ANs, for example, relationships were used for area taxonomies of SNOMED CT [5]. Data properties (as well as relationships) were used to derive an AN for the MED [3]. Hierarchical relationships were used to derive disjoint partial-area taxonomies for SNOMED CT [20]. Concept sets defined by those structural features were shown to exhibit error concentration rates that were statistically significantly higher than error concentration rates for control sample sets.
Ontology Classification
Object properties are widely used in ontology development and introduce a significant amount of knowledge into an ontology. Given a set of ontologies, each ontology can be classified into one (or potentially more) families based on the existence or nonexistence of the previously defined conditions regarding structural features. In this initial study we classify ontologies into seven disjoint families with classification priority given to structural conditions that have been proven useful for deriving ANs. In each case, taxonomies were successfully shown to support orientation and QA of the underlying ontology.
Object properties play a very important role in ontologies since they represent the semantic connections between classes, expressing the domain knowledge of the ontology. The importance of object properties is manifested, for example, in the consideration of classes of ontologies as primitive if they miss some object properties. Thus we have chosen them to initially separate families into two disjoint groups: those with object properties and those without object properties. These high-level groups dictate the type of AN that can be derived for the ontologies of a family. These two groups can be further refined.
Two of the largest and most used ontologies in BioPortal, SNOMED CT and NCIt, share a similar ontological model which is based on description logics. We consider the model of these two ontologies separately from the rest of our sample, and classify them in a separate family since all their relationships are instantiated because each pair of concepts connected by a relationship is concretely linked. In constrast all other BP ontologies’ objet properties are potential connections between classes, with only parts of them instantiated with concrete links. In previous work we derived taxonomies for SNOMED CT [5] and NCIt [6] using both their lateral and hierarchical relationships.
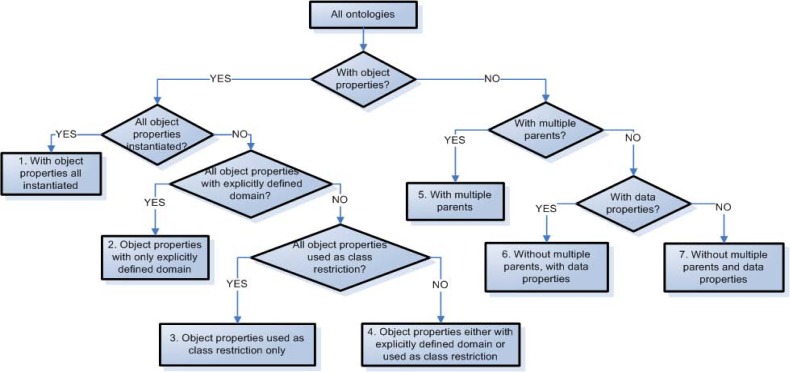
The ontologies that have object properties are further divided into 4 disjoint subgroups: 1. Ontologies that have all their relationships instantiated (e.g. NCIt). The remaining ontologies in this group are further divided as follows. 2. All object properties have only explicitly defined domains. 3. All object properties are only used as class restrictions. 4. The object properties either have explicitly defined domains or are used as class restrictions. The second group of ontologies, which do not have object properties, are divided into three disjoint subgroups. 5. The first subgroup consists of ontologies that have some classes with multiple parents. Ontologies without multiple parents are further divided into 6. ontologies with data properties and 7. ontologies without data properties. In this way, ontologies are grouped into seven disjoint families that exhibit different structural conditions.
Figure 2 illustrates a binary decision tree for classifying ontologies into families. The diamond boxes represent the conditions and the rectangles represent the seven enumerated families of ontologies, plus the starting point.
Figure 2.
A binary decision tree for classifying ontologies into seven disjoint families
Generalizable Design of Abstraction Networks for Families
Previously, AN-based QA was a “one at a time” methodology; the research developing techniques for deriving ANs and developing QA methodologies was done on a per-ontology basis. The process of AN derivation utilizes structural elements from an ontology to algorithmically create a “summary.” Therefore, by deriving ANs using the set of structural features common to a family, ANs can be derived uniformly and automatically for each member of the family.
We will illustrate this generalizable AN-based QA methodology by deriving a partial-area taxonomy for the Cancer Chemoprevention BioPortal Ontology (CanCo) [10, 11]. All of the object properties in CanCo are given explicitly defined domains. Therefore, we can utilize the same taxonomy derivation methodology that we previously developed for OCRe, since both ontologies belong to Family 2. We performed a review of the different partial-areas of the CanCo’s taxonomy, demonstrating how anomalies in the taxonomy design highlight classes with a high likelihood of modeling problems.
With over 300 BP ontologies, and many more biomedical ontologies not hosted in BioPortal, it is necessary to create software for automatically deriving and visualizing ANs for families of ontologies. In previous work, we developed the Biomedical Layout Utility for SNOMED CT (BLUSNO) [24], a tool for automatically deriving and visualizing ANs for SNOMED CT. Our experience with BLUSNO will guide the development of a utility called the Biomedical Layout Utility for the Web Ontology Language (BLUOWL). In an early prototype of BLUOWL, users can select an ontology expressed in OWL from the family of BP ontologies with only object properties with explicitly defined domains (and other similar families), and BLUOWL generates a partial-area taxonomy on the fly. The resulting diagram can be manipulated by the user. The partial-area taxonomy for CanCo (see Figure 3) was derived using the BLUOWL prototype.
Figure 3.
Partial-Area Taxonomy of Cancer Chemoprevention Ontology (CanCo)
Results
Between September 2012 and January 2013 we collected 210 distinct BP ontologies, representing 64% of the 330 BP ontologies available. In addition to SNOMED CT and NCIt, only ontologies released in OWL and OBO formats, were considered. We converted each ontology from the stated view to the inferred view, to utilize all inferable axioms, using the Hermit reasoner [25]. We did not investigate 24 ontologies for various reasons.
Our final sample set consisted of 186 ontologies and included the Gene Ontology (GO), Foundational Model of Anatomy (FMA), Ontology for General Medical Science (OGMS), Ontology of Clinical Research (OCRe), Sleep Domain Ontology (SDO), Vaccine Ontology (VO), Infectious Disease Ontology (IDO), and others. In total, 115 ontologies were in OWL format, 70 were originally in OBO format, and two in flat file format.
Commonality of Structural Conditions
Prior to creating families of ontologies, we had to ensure that enough ontologies exhibited a particular structural condition to meet the criterion that a family is of meaningful size. Table 1 lists the numbers of BP ontologies for each of the structural features that we utilized to analyze the ontologies. For brevity, we use in Table 1 and onward, abbreviated names for those features. For example, object properties with explicitly defined domains are called domain-defined object properties. If used in class restrictions, they are called restriction-defined object properties.
Table 1.
Ontologies in our sample set which exhibited a particular structural condition
| Characteristic | # Ontologies w/Characteristic | % of Sample (n= 186) |
|---|---|---|
| Object properties (total) | 150 | 80.6 |
| Domains defined object properties | 81 | 43.5 |
| Restriction-defined object properties | 131 | 70.4 |
| Data properties (total) | 71 | 38.2 |
| Multiple parents (DAG) | 110 | 59.1 |
| No multiple parents (Tree) | 76 | 40.9 |
From Table 1 one can see that there are some ontologies with both kinds of object properties. In fact, 62 ontologies have some domain-defined object properties and some restriction-defined object properties. Nineteen ontologies have only domain-defined object properties and 69 ontologies have only restriction-defined object properties. Furthermore, 71 out of 186 ontologies have data properties.
For ontologies without object properties, hierarchical structure conditions can potentially be used for AN derivation. There are nine ontologies without object properties having some classes with multiple parents. They are APO, FBSP, HEALTHINDICATORS, HOMHARVARD, HP, IMMDIS, OGMD, PEDTERM and YPO.
Members of Families
Table 2 lists the families of ontologies which have object properties or with instantiated relationships. Since our families were defined as disjoint, the numbers in Table 2 are not coming from Table 1, but from the disjoint partition described above, e.g., there are 19 ontologies with domain-defined object properties.
Table 2.
Families for ontologies that have object properties (relationships)
| Family | Structural Condition | # Ontologies | Samples |
|---|---|---|---|
| 1 | All relationships instantiated | 2 | SNOMED CT, NCIt |
| 2 | With only domain-defined object properties | 19 | Cancer Chemoprevention Ontology (CanCo) International Classification of Functioning, Disability and Health (ICF) Physical Medicine and Rehabilitation (PMR) |
| 3 | With only restriction-defined object properties | 69 | Gene Ontology (GO) Cereal Plant Development (GRO_CPD) Host Pathogen Interactions Ontology (HPIO) |
| 4 | With either domain-defined object properties or restriction-defined object properties | 62 | Sleep Domain Ontology (SDO) Infectious Disease Ontology (IDO) |
Table 3 lists the families of ontologies that have no object properties. As for Table 2, the numbers are computed from the disjoint sets above.
Table 3.
Families of ontologies that have no object properties (relationships)
| Family | Structural Condition | # Ontologies | Samples |
|---|---|---|---|
| 5 | Classes with multiple parents | 9 | Ascomycete phenotype ontology (APO) Human Phenotype Ontology (HP) Ontology of Glucose Metabolism Disorder (OGMD) |
| 6 | Classes without multiple parents and with data properties | 3 | Cell Behavior Ontology (CBO) CareLex |
| 7 | All classes without multiple parents and without data properties | 22 | Ontology for General Medical Science (OGMS) Reproductive trait and phenotype ontology (REPO) Sample processing and separation techniques (SEP) |
Table 4 lists a sample of ontologies in Family 2, i.e., those with only domain-defined object properties.
Table 4.
Sample of ontologies that have only domain-defined object properties
| Ontology Name | # Classes | # Object Properties |
|---|---|---|
| Animal natural history and life history (ADW) | 364 | 16 |
| Biomedical Resource Ontology (BRO) | 487 | 17 |
| Cancer Chemoprevention Ontology (CanCo) | 127 | 37 |
| International Classification of Functioning, Disability and Health (ICF) | 1595 | 41 |
| Physical Medicine and Rehabilitation (PMR) | 137 | 14 |
| RAPID Phenotype Ontology (RPO) | 1544 | 157 |
| Student Health Record (SHR) | 343 | 35 |
| Syndromic Survaillance Ontology (SSO) | 176 | 11 |
| Top-Menelas (Top-Menelas) | 524 | 296 |
In the next section, we illustrate for the CanCo ontology of this family how the taxonomy created automatically by BLUOWL looks and we describe QA work for CanCo, based on the CanCo taxonomy. This example is intended to illustrate the viability of the family-based QA framework.
Illustration for the Cancer Chemoprevention Ontology (CanCo)
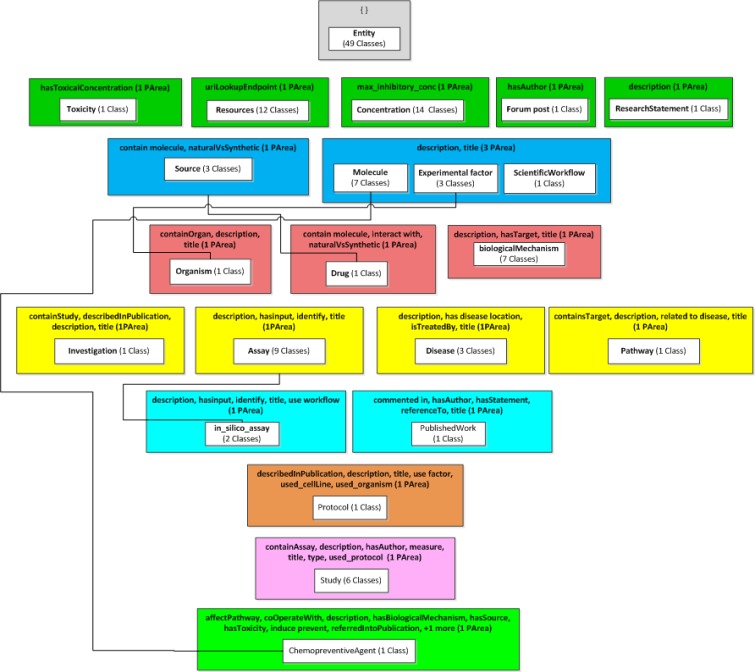
To illustrate the viability of the family-based QA framework, we have chosen the Cancer Chemoprevention Ontology (CanCo) from the domain-defined family, with 127 classes and 37 object properties. The Basic Formal Ontology (BFO), an upper level BP ontology [26], was fully migrated into CanCo for reuse in its design. The BLUOWL prototype is already able to automatically generate and display any domain-defined taxonomy of Family 2. For example, the taxonomy for CanCo appears in Figure 3.
Let us describe the elements and structure of such a taxonomy. An area is defined as the set of all classes that are explicitly defined or inferred as being in exactly the domains of a given set of object properties O. The list of names of the object properties is used to name the area. We define a root of an area as a class that has no parents in the same area. An area may have more than one root. A root of an area defines a partial-area as a set of classes that includes the root and all its descendants in the area. Partial-areas are connected by child-of links derived from the underlying IS-A relationships. Specifically, a partial-area A is child-of partial-area B if a parent of A’s root resides in B. The number of classes (including the root) in each partial-area is shown in parentheses.
In the partial-area taxonomy of CanCo, areas, for every existing set of object properties in the ontology, are organized into color-coded levels based on their numbers of object properties. For example, areas with three object properties are in red. Partial-areas are represented using white boxes within areas’ colored boxes and are labeled using their roots, and the lines are child-of’s. The child-of relationships from most partial-areas are directed to the root Entity partial-area and are not shown to avoid clutter. This indicates that most sets of object properties of areas are disjoint. The only child-of relationships shown are to other partial areas: Source, Experimental Factor, Module and Assay. Except for the area labeled with description and title, with three partial-areas, all areas contain only one partial area. Most partial areas contain only one class, with the exception of three large ones: Entity (49), Concentration (14), and Resources (12). Medium size partial-areas (5–9 classes) include: Assay (9), Module (7), Biological Mechanism (7) and Study (6). These nine partial areas, covering 104 classes, provide an excellent summary of the content and structure of CanCo.
According to our extensive experience with SNOMED CT [5, 22] and NCIt [6] the partial-area taxonomy helped to identify anomalies in the modeling, characterizing sets of concepts with a high likelihood of errors. In our recent QA work on BP ontologies such as OCRe [7] and the Sleep Domain Ontology [8], we found that large partial-areas characterize sets of concepts with a high likelihood of errors. There are three large partial-areas in Figure 3. The second anomaly in the CanCo taxonomy is the unique area (description, title) with three partial-areas: Module (7), Experimental factor (3) and Scientific workflow (1). The third anomaly is defined by the few (four) child-of relationships not directed at Entity.
In the following, we show how these anomalies helped expose modeling problems. First, consider the Entity (49) root partial-area containing all classes with no object properties. When reviewing these classes, we find that 39 out of 49 were migrated from BFO, which is modeled without object properties. Close examination reveals that 20 of them are leaves (classes without children) in CanCo. That means they were not used as the basis for classes in the chemoprevention domain and should not have been migrated to CanCo. The process of “hiding” all 20 such leaves from view would not affect any other CanCo classes. For details on a hiding mechanism for BP ontologies, see [9].
Another modeling problem concerns both large partial-areas Entity (49) and Concentration (14). The class Concentration and all its 13 descendants have the object property max_inhibitory_concentration, but its sibling inhibitory_concentration and the latter’s child Max_inhibitory_concentration do not have this object property and are in Entity (49). Furthermore the last class name is identical to the object property name. Also two subclasses of Concentration, IC and IC50, have related definitions “Maximum inhibitory concentration” and “Half-maximal inhibitory concentration,” respectively.
The two redundant classes inhibitory_concentration and Max_inhibitory_concentration are removed. The object property is removed and replaced by a new data property concentrationValue (domain: Concentration, range: float) defined for Concentration and inherited to its descendants to store the concentration value for all the various types of concentrations.
Two of the child-of relationships not directed at Entity raised questions: Why does it hold that a Drug IS_A Source and why is it true that Organism IS_A Experimental Factor? Source has two children Natural and Synthetic, which should be renamed Natural source and Synthetic source, and Drug IS_A Synthetic source. Regarding the second case, the problem was not resolved yet and is considered future work.
Considering the unique area with three partial-areas, we looked at the seven classes of Molecule. The child of Molecule – Target should be renamed Biological target according to its definition. The five children of Target, e.g., Lipid, Protein and Sugar are macromolecules. Hence a class Macromolecule should be introduced as child of Molecule and become the parent of its current five children. The modeling of the relationships between them and Biological Target will be considered in future work.
There are also issues regarding the three children of Experimental factor, another partial-area in this area (description, title) left for future consideration. The curators of CanCo (co-authors, DZ and KT) have implemented all the above changes, which were incorporated in a new release number 0.3 of CanCo in BioPortal. As was seen, the anomalies found in the CanCo taxonomy helped to detect problems in CanCo’s modeling.
Discussion
The purpose of this paper is to introduce a family-based QA framework for ontologies, which will enable broad applicability and substantial savings by automating part of the QA work. To our knowledge (see, e.g. [8]) current QA techniques for ontologies and taxonomies, typically target a single ontology or terminology. The new framework suggests methods that work uniformly across families of ontologies. This paper provided a proof of concept for the feasibility of such a framework. We discussed the way families are defined and illustrated seven disjoint families consisting of 186 ontologies of the BioPortal repository. The definition of these families together with the classification of the 186 ontologies into them provides a proof of concept for the existence of an groundwork for such a framework. Alternative groupings of families are possible, as described in Future Work.
The two operational aspects of this framework are (1) automatic family-based uniform derivation of abstraction networks and (2) utilization of abstraction networks in characterizations of sets of classes with a high likelihood of errors, recognizable by various aspects and anomalies in the appearance of the abstraction network for a given family of ontologies. Concentrating QA efforts on such sets will increase the yield of QA work in terms of the ratio of problems found and resolved, to the number of classes reviewed.
For each ontology from Families 2–4 (having object properties), our prototype derivation and display tool BLUOWL is able to automatically create an AN. This has been currently demonstrated for CanCo, as well as for OCRe [7] and Top-Manelas [27], all in Family 2. An AN for GO (Family 3) can be found at [27]. ANs for Family 4 members SDO [8] and DDI [9] have been generated. For Family 1, our BLUSNO tool [24] constructs taxonomies for SNOMED hierarchies [5, 20, 23].
The categories of Tables 2 and 3 are intentionally designed to be disjoint since, for ontologies with object properties, the proper taxonomies will typically have sufficient granularity [8] to support QA. These other potential features, e.g. data properties, are not needed for the design of ANs for QA. Let us turn to the families of Table 3 without object properties. An AN can be derived for an ontology with only data properties (Family 6) in a manner similar to that for an ontology with only object properties. Since targets of object properties are not reflected in the taxonomy, classes with the same set of data properties are grouped in an area.
Ontologies having no relationships but having some concepts with multiple parents (Family 5) pose difficulties for AN derivation. Due to the lack of relationships, an area taxonomy cannot be derived. A possible alternative abstraction paradigm might exploit overlapping subhierarchies resulting from concepts with multiple parents. While extensive work is needed for completing the framework, our initial work shows family-based automatic AN derivation is possible. According to our plan, BLUOWL will have a separate module for each family. This tool will be made available for download so that ontology curators can easily derive ANs for their ontologies on demand.
Regarding family-based QA work, we note that for two ontologies of Family 1, SNOMED CT and NCIt, the characterization of small sets represented by nodes of the partial-area taxonomies were shown experimentally to have high likelihoods of errors [5, 6]. For OCRe, SDO, and CanCo a characterization of large partial-areas of the taxonomy was shown to indicate higher concentrations of errors [7, 8]. These examples demonstrate the viability of the QA aspect of the framework introduced in this paper.
Future Work
In this paper, all the families are disjoint, i.e., each ontology is classified into only one family. While we defined families as disjoint for this initial study, an ontology may exhibit several structural features, e.g. domain-defined object properties, a hierarchy with multiple parents, and the existence of data properties, as demonstrated in Table 1. If an ontology has several features, then there are several alternatives how to model it. For example, different types of ANs can be derived, providing additional, independent QA options. If one AN does not work well for QA, others may. If, for example, an ontology has few object properties, yielding too coarse an AN, as was the case for the domain-defined taxonomy for SDO [8], the object properties can be combined with data properties to derive a richer taxonomy. Exploring further families for ontologies will be part of our future research. For example, one can define a refined family for ontologies with restriction-defined object properties and hierarchies with multiple parents. Another example for a refined family is ontologies with few domain-defined object properties and many data properties, e.g., the DermLex BP ontology. In our future research, we will explore such definitions of families. We will also continue to develop the AN derivation and QA groundwork for this family-based QA framework.
Conclusions
In this paper, we analyzed structural features from 186 BP ontologies and identified several structural conditions that enabled the classification of the ontologies into families. Using this family information, we were able to derive abstraction networks for whole families of ontologies, enabling a uniform quality assurance methodology for these similar ontologies. A preliminary QA review of the Cancer Chemoprevention Ontology (CanCo) was used to illustrate the benefits of a uniform family-based QA methodology.
References
- 1.Rubin DL, Shah NH, Noy NF. Biomedical ontologies: a functional perspective. Brief Bioinform. 2008 Jan;9(1):75–90. doi: 10.1093/bib/bbm059. [DOI] [PubMed] [Google Scholar]
- 2.Bodenreider O, Stevens R. Bio-ontologies: current trends and future directions. Brief Bioinform. 2006 Sep;7(3):256–74. doi: 10.1093/bib/bbl027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gu H, Cimino JJ, Halper M, Geller J, Perl Y. Utilizing OODB schema modeling for vocabulary management. Proc AMIA Annu Fall Symp. 1996:274–8. [PMC free article] [PubMed] [Google Scholar]
- 4.Gu H, Perl Y, Geller J, Halper M, Liu LM, Cimino JJ. Representing the UMLS as an object-oriented database: modeling issues and advantages. J Am Med Inform Assoc. 2000;7(1):66–80. doi: 10.1136/jamia.2000.0070066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y, Halper M, Min H, Perl Y, Chen Y, Spackman KA. Structural methodologies for auditing SNOMED. J Biomed Inform. 2007 Oct;40(5):561–81. doi: 10.1016/j.jbi.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 6.Min H, Perl Y, Chen Y, Halper M, Geller J, Wang Y. Auditing as Part of the Terminology Design Life Cycle. J Am Med Inform Assoc. 2006;13(6):676–90. doi: 10.1197/jamia.M2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ochs C, Agrawal A, Perl Y, Halper M, Tu SW, Carini S, Sim I, Noy N, Musen M, Geller J. Deriving an abstraction network to support quality assurance in OCRe. AMIA Annu Symp Proc; 2012; 2012. pp. 681–9. [PMC free article] [PubMed] [Google Scholar]
- 8.Ochs C, He Z, Perl Y, Arabandi S, Halper M, Geller J. Choosing the Granularity of Abstraction Networks for Orientation and Quality Assurance of the Sleep Domain Ontology. the 4th International Conference on Biomedical Ontology; Montreal, QC, Canada. 2013. pp. 84–9. [Google Scholar]
- 9.He Z, Ochs C, Soldatova L, Perl Y, Arabandi S, Geller J. Auditing Redundant Import in Reuse of a Top Level Ontology for the Drug Discovery Investigations Ontology. International Workshop on Vaccine and Drug Ontology Studies; Montreal, QC, Canada. 2013. [Google Scholar]
- 10.BioPortal Cancer Chemoprevention Ontology. 2012. Available from: http://bioportal.bioontology.org/ontologies/3030.
- 11.Zeginis D, Hasnain A, Loutas N, Deus HF, Fox R, Tarabanis K. A collaborative methodology for developing a semantic model for interlinking Cancer Chemoprevention linked-data sources. Semantic Web. 2013 [Google Scholar]
- 12.Whetzel PL, Noy NF, Shah NH, Alexander PR, Nyulas C, Tudorache T, Musen MA. BioPortal: enhanced functionality via new Web services from the National Center for Biomedical Ontology to access and use ontologies in software applications. Nucleic Acids Res. 2011 Jul;39:W541–5. doi: 10.1093/nar/gkr469. (Web Server issue): [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.OWL Web Ontology Language Overview. Available from: http://www.w3.org/TR/owl-features
- 14.Resource Description Framework (RDF) Available from: http://www.w3.org/RDF/
- 15.OBO Foundry Principles Available from: http://www.obofoundry.org/wiki/index.php/Category:Accepted.
- 16.Mortensen JM, Horridge M, Musen MA, Noy NF. Applications of ontology design patterns in biomedical ontologies. AMIA Annu Symp Proc; 2012; 2012. pp. 643–52. [PMC free article] [PubMed] [Google Scholar]
- 17.Bail S, Horridge M, Parsia B, Sattler U. The Justificatory Structure of the NCBO BioPortal Ontologies. International Semantic Web Conference; Bonn, Germany. 2011. pp. 67–82. [Google Scholar]
- 18.Ghazvinian A, Noy NF, Musen MA. How orthogonal are the OBO Foundry ontologies? J Biomed Semantics. 2011;(Suppl 2(S2)) doi: 10.1186/2041-1480-2-S2-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.FlexViz Tool. Available from: http://www.thechiselgroup.org/flexviz
- 20.Wang Y, Halper M, Wei D, Perl Y, Geller J. Abstraction of complex concepts with a refined partial-area taxonomy of SNOMED. J Biomed Inform. 2012 Feb;45(1):15–29. doi: 10.1016/j.jbi.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei D, Bodenreider O. Using the abstraction network in complement to description logics for quality assurance in biomedical terminologies - a case study in SNOMED CT. Studies in health technology and informatics. 2010;160(P2):1070–4. [PMC free article] [PubMed] [Google Scholar]
- 22.Halper M, Wang Y, Min H, Chen Y, Hripcsak G, Perl Y, Spackman KA. Analysis of error concentrations in SNOMED. AMIA Annu Symp Proc. 2007:314–8. [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, Halper M, Wei D, Gu H, Perl Y, Xu J, Elhanan G, Chen Y, Spackman KA, Case JT, Hripcsak G. Auditing complex concepts of SNOMED using a refined hierarchical abstraction network. J Biomed Inform. 2012 Feb;45(1):1–14. doi: 10.1016/j.jbi.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geller J, Ochs C, Perl Y, Xu J. New abstraction networks and a new visualization tool in support of auditing the SNOMED CT content. AMIA Annu Symp Proc; 2012; 2012. pp. 237–46. [PMC free article] [PubMed] [Google Scholar]
- 25.Shearer R, Motik B, Horrocks I. HermiT: a highly-efficient OWL reasoner. Proceedings of the 5th International Workshop on OWL: Experiences and Directions (OWLED 2008); 2008. [Google Scholar]
- 26.BioPortal Basic Formal Ontology. Available from: http://bioportal.bioontology.org/ontologies/1332.
- 27.Figures of the taxonomies of the ontologies Available from: http://cs.njit.edu/~oohvr/SABOC/figures.php