Abstract
Electronic health records are an increasingly important source of data for research, allowing for large-scale longitudinal studies on the same population that is being treated. Unlike in controlled studies, though, these data vary widely in quality, quantity, and structure. In order to know whether algorithms can accurately uncover new knowledge from these records, or whether findings can be extrapolated to new populations, they must be validated. One approach is to conduct the same study in multiple sites and compare results, but it is a challenge to determine whether differences are due to artifacts of the medical process, population differences, or failures of the methods used. In this paper we describe the results of replicating a data-driven experiment to infer possible causes of congestive heart failure and their timing using data from two medical systems and two patient populations. We focus on the difficulties faced in this type of work, lessons learned, and recommendations for future research.
Introduction
Electronic health records (EHRs) have already made vast quantities of data potentially available for research, and new policies (including the PPACA) mean both that more data will be captured electronically and that there will be greater incentives to make use of it. These data are intrinsically heterogeneous, spanning numerical laboratory values, imaging results, and narrative histories, yet there is also heterogeneity in how the same value may be captured by different institutions. Analysis of this data cannot focus on only structured or unstructured data, but will require a broad set of tools and techniques. However, in order to validate methods and determine whether results can be generalized, multisite studies are needed.
As a result, new networks such as the Electronic Medical Records and Genomics (eMERGE) Network [1] and the HMO Research Network (HMORN) aim to make it possible for studies to be conducted in multiple, diverse, populations. One of the key difficulties in trying to replicate studies has been in sharing data (due to HIPAA privacy controls), but interoperability of records for the purpose of research is a growing concern as well. Unlike concerns about interoperability in the context of sharing information about individual patients across multiple medical systems, interoperability for research means being able to apply the same study design and methods in multiple systems, or combine data from multiple systems in one study. Leaving aside issues with access to data, the structural differences in how data are collected and stored across different EHRs make this a substantial challenge. For instance, a study about a disease begins by defining selection criteria for patients, but the same types of data may not be available in all sites. Conversely, reducing the data characteristics used to only those available everywhere in an identical form will reduce the study’s power by ignoring important information. While this work can be tedious and difficult, it is critical if we want to determine whether differing results are due to a failure of the methods used, differences across populations, or issues with data quality and content.
In this paper we discuss the process of replicating a causal inference study in two different medical systems. We focus on describing the challenges faced and lessons learned, before proposing recommendations for future work in designing methods.
Background
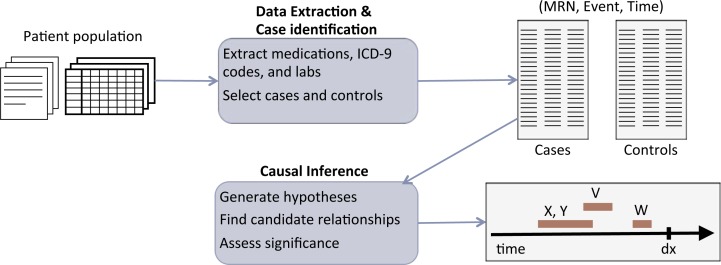
While our work involved conducting a study of causes of one disease in two medical centers, our discussion is based on the broad process of integrating qualitative and quantitative data for research and combining results from multiple medical systems and patient populations. The general process we envision is shown in figure 1, where we begin with structured and unstructured longitudinal data, apply natural language processing (NLP) techniques to extract events from the text data (so they can be combined with the structured data for analysis), separate patients into cases and controls, and use computational approaches for inference on the set of prepared data. This process may be repeated in a number of systems, at which point we then need a way of comparing and interpreting results.
Figure 1.
Overview of inference process. Given raw data for a patient population, ICD-9 codes, medications, and lab measurements are extracted from the structured data and notes when available to generate event triples (patient medical record number, event, and time of event) and used to select cases and controls. Causal inference methods are applied to produce timelines of when significant potential causes occur prior to diagnosis.
We are motivated by an attempt to identify causes of congestive heart failure (CHF) in two different populations: one urban, using data from Columbia University Medical Center (CUMC) in New York City and one rural, using data from Geisinger Health System in Pennsylvania. CHF is a highly disabling progressive disease, but its early detection has remained a challenge, with most cases found too late to have a substantial impact on either reversing cardiac muscle pathology or slowing progression. Thus, improvements in procedures that facilitate early detection may lead to opportunities for delaying onset or slowing the progression of this disease, as earlier interventions have a higher chance of success than late stage ones.
Much work has been carried out on curated longitudinal datasets to identify predictors of heart failure and mortality (e.g., [2,3]). In our scenario, we are instead interested in leveraging raw EHR data for analysis. Due to the challenging nature of EHR data, much of the prior work on analyzing it has focused on applying data mining and machine learning techniques for clustering, learning associations, or finding predictive rules [4–6]. Prior work at Geisinger by Stewart, Roy and Shah [7] applied machine learning methods to predict heart failure 6, 12, and 18 months before diagnosis, but it still remains to determine whether these relationships are causal and whether the findings apply to other populations. While causal inference is a more difficult task, the potential impact of finding causes is substantial, as these allow us to identify targets for intervention in order to prevent or produce particular outcomes.
The causal inference approach used here [8] is based on the idea of causes providing information about the probability of occurrence or expected value of variables that is not contained in other factors. In this approach, relationships are described using logical formulas, so one may construct complex causes involving durations, sequences or conjunctions of factors. There is also a window of time associated with each relationship, so that we find not only that “smoking causes lung cancer,” but rather something like “smoking causes lung cancer in 20–30 years.” Relationships are then inferred from data by calculating the average difference each cause, c, makes to an effect, e, given, pairwise, each of the other potential causes of the same effect (x ∈ X), using:
| (1) |
Other work by Kleinberg [8] has shown how to find the time windows empirically, without any prior knowledge of them. The basic idea is that one can generate a set of candidate time windows (which need only be at the right level of granularity) and then iteratively perturb these (expanding, shifting, shrinking them), while attempting to maximize εavg and this procedure will converge to the correct window.
After computing the average impact of each cause on its effect (εavg) it remains to determine which values are statistically significant. Given that large numbers of hypotheses are being tested, this can be treated as a false discovery rate (FDR) control problem with multiple hypothesis testing [9], and the null hypothesis can be inferred empirically [10]. This allows us to find the relationships that are statistically significant, controlling the FDR, while making few assumptions about the data.
Methods
We aim to learn population-level models of the causes and progression of CHF and aim to do this in a robust way that can be generalized to multiple populations and diseases. Ultimately, the goal is to use the result of these inferences to improve the care of individuals. Within these broad constraints we face a number of challenges in these data, including: heterogeneity, sparsity and missingness, noise, and error. First, data are captured in multiple ways (that may also differ across institutions) so we must incorporate both structured and unstructured information or risk missing important events. Second, the longitudinal nature of EHRs allows us to potentially track the sequence of events leading up to onset of a condition, but also leads to another source of missing data, as there may be large gaps in measurements or variances in timing across patients. We may have data that is entirely absent, but also must disentangle documentation biases from data biases. There is the added challenge of attempting to extract this information from notes, which contain a mixture of current, suspected, and past events in one narrative. Finally, since the data are noisy, we require methods that are robust in the face of uncertainty as well as a large amount of irrelevant information (noise that may overwhelm the signal).
Data and processing
We began with data from subsets of the populations of two medical systems: Geisinger and CUMC, with 32,681 and 13,618 patients respectively. Due to the structured nature of the Geisinger EHR, we used only structured data for that analysis. On the other hand CUMC’s records are in large part text-based, so a mixture of structured and unstructured data was used.
The following processing was carried out on the narrative part of the CUMC EHR data. All notes were collected for the patients in our dataset. Some notes were entirely free text without any pre-determined internal structure (e.g., some primary provider and consult notes, progress notes, signouts), some had some internal structure and well-delimited sections (e.g., radiology reports), and some were templated with a mix of boilerplate text and free text (e.g., structured ambulatory consult note).
The content of all the notes was pre-processed to identify document structure (section boundaries and section headers, lists and paragraph boundaries, and sentence boundaries) [11], shallow syntactic structure (part-of-speech tagging with the GENIA tagger [12] and phrase chunking with the OpenNLP toolkit [13]), and UMLS concept mentions with our in-house named-entity recognizer HealthTermFinder. HealthTermFinder identifies named-entity mentions and maps them against semantic concepts in UMLS [14]. Furthermore, we used a post-processing step to aggregate concepts in the Disorders semantic group whenever possible, so that semantically similar concepts are merged into one whenever the two are used in similar contexts (e.g., if a note uses both concepts “morbidly obese” and “obese”, the two were merged) [15]. At the end of the pipeline, we obtained a list of concept occurrences along with timestamps of the notes in which they are mentioned. While the Geisinger dataset did not contain any notes, patient records contained problem and comorbidities lists defined by ICD-9 codes and relevant dates.
Particular attention was given to mentions of medications in the CUMC notes. Medication names were parsed [16] and were mapped to medication classes and subclasses for a pre-defined set of common and CHF-pertinent classes and subclasses. Similar mapping to classes and subclasses was carried out for medication orders encoded in the CUMC EHR. At the end of this pipeline, we obtained a list of medications (and associated (sub)classes) along with timestamps and source (either note or order). Because of the many open challenges entailed in large-scale, automated medication reconciliation, we did not attempt to reconcile the note-derived and order-derived medication lists. Furthermore, we only kept track of order or mention date, as no information was available about end date for a given medication. There was no indication encoded for the medication either. In contrast, in the Geisinger dataset, we relied on the structured medication fields available in their EHR. As such, Geisinger medications were encoded as the tuple (medication, (sub)classes, indication, start date). The classes and subclasses were the same in both datasets.
Time series of laboratory tests were collected for a total of 75 tests that are either common or pertinent to CHF diagnosis and progression. These were similar across both datasets.
Finally, ICD-9 codes for billing were also collected across both datasets as time series of ICD-9 occurrences for each patient.
Causal inference
Using the inference approach described in the previous section, we tested for causal relationships between all variables in our datasets (medications, lab values, ICD9 codes) and the occurrence of a CHF diagnosis. To use the causal inference method described, all continuous variables were discretized (using standard reference ranges and institution-specific ones when available) and text data was processed into id-event-time triples. We used the date of first diagnosis (using first ICD9 code) instead of all mentions of a condition in our analysis and generated a set of time windows, on the order of months, up to approximately two years. That is, relationships were tested between all variables and CHF diagnosis at 4–6 months, 6–8 months and so on. After finding significant relationships, the candidate windows were refined using the methods described in [8], so that we may begin with a window of 20–24 months but ultimately find that the onset of diabetes leads to CHF in 17–22 months.
Results
Population characteristics
We begin by comparing the composition of the two populations studied. The study population at Geisinger is representative of the population served by Geisinger as a whole and is both stable and homogenous (approximately 95% white, with less than 1% outmigration in most counties served). The structured nature of the EHR and the criteria used to determine cases of CHF and controls made it possible to search for and select controls for each patient. On the other hand, the CUMC population is an ethnically diverse and urban one, with 57% white, 28% black, and 15% asian, and 32% hispanic. There is also substantial in/out migration, gaps in access to healthcare and potentially movement between medical systems in the region. Further, the case selection criteria used (as discussed in the following sections) required both structured and unstructured data. This made matching controls individually to patients infeasible, so we instead focused on a subset of the population at Columbia, patients with at least three visits to the AIM outpatient clinic, excluding AIDS patients (there were a few hundred AIDS patients in the CUMC database and none in the Geisinger data set). This population is relatively stable compared to the CUMC population as a whole and allowed us to select a large group, whose members were then divided into cases and controls. Tables 1 and 2 compare the top 10 most frequent (by patient, rather than total mentions) ICD9 codes and medication subclasses in the two populations. Significant differences are indicated in bold.
Table 1.
Top 10 ICD-9 codes in the two datasets, by raw number of patients and percentage of patients in the dataset.
| CUMC | Geisinger | ||||
|---|---|---|---|---|---|
| 401 | Essential hypertension | 10,669 (79%) | 272 | Disorders of lipoid metabolism | 25,687 (79%) |
| 786 | Symptoms involving respiratory system and other chest symptoms | 7,911 (58%) | 401 | Essential hypertension | 25,065 (77%) |
| 272 | Disorders of lipoid metabolism | 7,908 (58%) | 786 | Symptoms involving respiratory system and other chest symptoms | 19,481(60%) |
| 789 | Other symptoms involving abdomen and pelvis | 7,393 (55%) | 780 | General symptoms | 19,112 (58%) |
| 724 | Other and unspecified disorders of back | 6,518 (48%) | 715 | Osteoarthrosis and allied disorders | 15,974 (49%) |
| 729 | Other disorders of soft tissues | 6,422 (47%) | 724 | Other and unspecified disorders of back | 15,056 (46%) |
| 780 | General symptoms | 6,413 (47%) | 729 | Other disorders of soft tissues | 14,676 (45%) |
| 715 | Osteoarthrosis and allied disorders | 6,048 (45%) | 530 | Diseases of esophagus | 14,016 (43%) |
| 784 | Symptoms involving head and neck | 6,046 (45%) | 719 | Other and unspecified disorders of joint | 13,886 (42%) |
| 250 | Diabetes mellitus | 5,617 (42%) | 788 | Symptoms involving urinary system | 13,529 (41%) |
Table 2.
Top 10 medication sub-classes for the two datasets by raw number of patients and percentage of patients in the dataset.
| CUMC | Geisinger | ||
|---|---|---|---|
| Analgesics Other | 10,230 (76%) | Salicylates | 21,396 (66%) |
| Salicylates | 8,212 (61%) | HMG CoA Reductase Inhibitors | 20,383 (62%) |
| Nonsteroidal Anti-inflammatory Agents (NSAIDs) | 7,903 (58%) | Nonsteroidal Anti-inflammatory Agents (NSAIDs) | 19,878 (61%) |
| Proton Pump Inhibitors | 7,629 (56%) | Opioid Combinations | 19,123 (59%) |
| Non-Barbiturate Hypnotics | 7,351 (54%) | Beta Blockers Cardio-Selective | 18,197 (56%) |
| Opioid Agonists | 7,163 (53%) | ACE Inhibitors | 16,525 (51%) |
| Surfactant Laxatives | 6,875 (51%) | Proton Pump Inhibitors | 16,404 (50%) |
| HMG CoA Reductase Inhibitors | 6,593 (49%) | Glucocorticosteroids | 15,490 (47%) |
| Bulk Chemicals - S’s | 6,274 (46%) | Fluoroquinolones | 14,880 (46%) |
| Calcium | 6,270(46%) | Azithromycin | 13,726 (42%) |
Case and control criteria
One of the biggest challenges for this type of study is replicating the selection criteria for patients. CHF is particularly difficult as there is no single lab value that can objectively determine a CHF diagnosis. This step is critical, though, since mislabeling may lead to substantial false positives and negatives. Further since we aim to determine the timing of risk factors, we have an added challenge in that we must not only determine which patients have CHF, but when. Geisinger developed operational criteria for determining cases of CHF in prior work [7], which formed the basis for the criteria at CUMC. These criteria were reviewed by clinicians and compared against the standard Framingham criteria [17]. The operational criteria used by Geisinger are shown in table 3.
Table 3.
Criteria for establishing cases of CHF.
| CUMC | Geisinger |
|---|---|
|
| |
| 2 ICD9 codes for CHF | 2 medication orders with CHF diagnosis |
| 1 ICD9 code for CHF and 1 mention of CHF in note on same date | 2 outpatient visits with CHF diagnosis |
| 1 ICD9 code for CHF and 1 medication typically indicated for CHF | 1 outpatient CHF diagnosis and 1 medication order with CHF diagnosis |
| CHF on problem list | |
At Geisinger, patients with any indication of CHF (outpatient visit, medication order, CHF on problem list) were considered for further analysis, resulting in 6497 potential cases. Of these, 45% (N=2900) met operational criteria but not Framingham criteria. A much smaller proportion (N=424, 7%) met Framingham criteria but not the operational criteria, and 35% (N=2294) met both. Many of the patients who did not meet the Framingham criteria (2 major, 1 major + 2 minor) had documentation of some of the criteria, though 1322 had none. This is likely due to the fact that most Framingham criteria are documented in notes and difficult to extract with reasonable false positive and negative rates (many mentions involve “suspicion” of a symptom or a “possible” symptom or may refer to a symptom at a prior time). Controls were matched based on age and sex with an average of 7.5 controls for each patient. It was found that a single ICD9 code (whether attached to a visit, or a medication) is a poor indicator for a diagnosis [5, 18], as 20% of the initial pool (N=1303) had only one ICD9 code without CHF also being on the problem list. This also means that there are people with medications for CHF yet no outpatient visits for the condition as well as patients with visits for CHF but no medications for the condition.
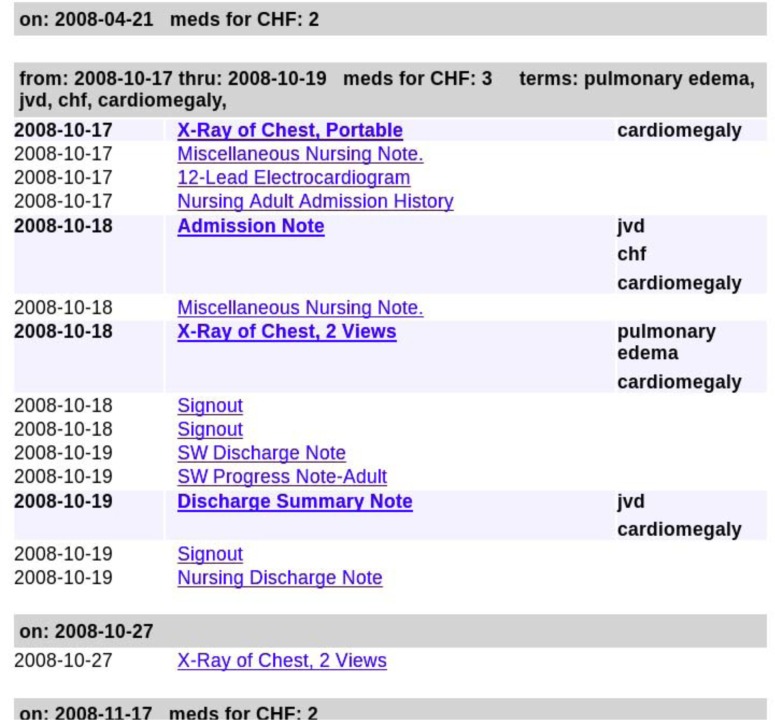
The Geisinger operational criteria depend on a few features of the dataset that were not present in the CUMC data: ICD9 codes associated with each medication, and a structured problem list that is separate from ICD9 codes used for billing. Making use of medications in this way is impossible to replicate without this information, since knowing that a patient was prescribed a beta blocker for their CHF is quite different than simply knowing that a patient is on a beta blocker. As a result of the structural differences between the EHRs, we developed a different set of operational criteria for CUMC, based on the same principle that a single ICD9 code is a noisy indicator of a diagnosis and other evidence is required. Due to the structure of the CUMC EHR system, where a patient’s problem list and medications are at least partly encoded in unstructured notes, we used both structured and unstructured data. Notes were processed to extract positive mentions of CHF as well as Framingham symptoms as follows. The notes’ raw text was indexed and a small set of manual queries were iteratively refined to extract the positive mentions. A dedicated website was created (sample output is shown in figure 2) to allow visualization and review of a patient’s history. The website also enabled us to improve queries by identifying cases where mentions were erroneously classified as positive or missed. Despite our success in getting many of the major Framingham criteria from the notes, we found that it was still not possible to determine when or if a patient is a case based on this criteria alone. Thus we used a mixture of structured and unstructured information.
Figure 2.
View from the system built to visualize CUMC data. Notes with positive mentions of Framingham terms have the terms listed in bold. Visits such as extended hospital stays are consolidated so they can be viewed as a single unit. Dates with CHF medications or ICD9 codes are also indicated.
Overall, the CUMC patient records spanned 12.1 years on average (7.0 years stdev). Together they comprise 2.6 million notes (spread across more than 3,000 note types) corresponding to 210 notes per patient on average (albeit with large variance across patients; 291 notes stdev), 6,180,000 different medications orders, and 4,210,000 different ICD9 codes. The notes are rich in clinical concepts: they contain nearly 4M mentions of UMLS concepts from the Disorder semantic group [19], and 156M UMLS concept mentions overall. Per patient on average, there are more than 1,000 unique UMLS concept and 400 disorder concepts. The patient records for the CUMC CHF cohort spanned 13.3 years (stdev 6.7 years). The Geisinger records had an average span of 7 years (on average 2.2 years pre-diagnosis). There were, on average, 86 encounters per patient, with a total of 1574 unique ICD9 classes, and 558 unique medication subclasses.
Causal inference results
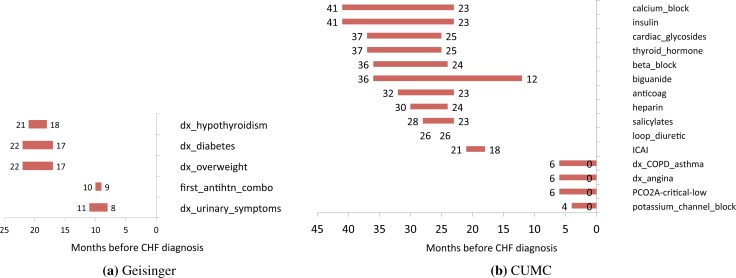
Figures 3a and 3b show timelines of the causal relationships that were inferred with the window of time (in months) prior to CHF diagnosis when these factors occur. In the Geisinger population, we found diagnoses of hypothyroidism, diabetes, overweight and urinary symptoms along with prescription of an antihypertensive medication combination to be causally significant at varying times. Some of these are known risk factors for CHF (diabetes, overweight), but others illustrate the difficulty of interpreting results from EHR data. First, note that our effect is not CHF itself (as we have no way of determining when this progressive illness first begins) but rather clinical diagnosis of the illness.
Figure 3.
Results of causal inference in both populations. Timelines show occurrence of factors prior to CHF diagnosis.
Urinary symptoms such as frequent nighttime urination are a symptom of CHF, suggesting that there is documentation of symptoms prior to a diagnosis (other hypotheses, such as that these were due to diuretics prescribed for CHF were ruled out). Similarly, while hypertension is a risk factor, we did not find this but instead found a medication for hypertension. This is likely due to under-documentation of hypertension, along with errors in the measurement of blood pressure. Note that in all cases this is a new diagnosis of each of these illnesses, as we used the first ICD9 code for each.
In the CUMC data, a larger number of significant relationships were found, yet many of them (aside from COPD asthma and angina) involved medications. We believe that this is due to the noise in billing ICD9 codes (rather than those attached to medications or a problem list), and the text-based nature of the CUMC EHR. In prior work we have extracted clinical information from the free text notes [15], but this work required the timing for each clinical event. Since the notes are comprised of a mix of current and past medical histories, including these may have introduced more noise into the inference [20]. However, if we interpret the medications as indicators for the underlying illness, we see that diabetes (or use of insulin) is found as being a risk factor in both populations, as is thyroid dysfunction (hypothyroidism in Geisinger, thyroid hormone prescription in CUMC). Similarly antihypertensive medications are found in both populations (antihypertensive combination medications in Geisinger, calcium channel blockers and beta blockers in CUMC).
In general, it is not unexpected that we will make fewer findings in a larger population. This is because we have more observations of each individual event. In a smaller population we might, due to coincidence or characteristics of the population, happen to find that people with a particular condition go on to develop CHF. As the population grows, though, we come closer to observing the true underlying probabilities of each event. We also expect that, due to the criteria used and method used for determining dates of diagnosis, in this study the timing of CHF onset will be more accurate in the Geisinger population than in the CUMC one.
Discussion
The use of EHRs to discover actionable knowledge such as causal relationships is a fundamentally new approach to research that will require new methods and considerations. With much of the prior focus on studies of single methods and single hospital systems (aside from that of networks such as eMERGE), we face a similar situation to when genomic sequence data for humans first became available. These data allowed researchers to ask new types of questions, yet researchers initially focused on getting a single sequence at a time, with no real way of determining whether or not the sequence was correct. As genome-wide association studies proliferated1 there too researchers would collect data for one community and attempt to find a set of associations. Only recently have researchers done meta analyses to determine the common findings among these disparate studies [21]. In biomedical informatics replication is critical if we are to know whether a finding is specific to a population (or perhaps due to the idiosyncrasies of an EHR system) or more widely generalizable.
Recommendations
Validation
Studies involving EHRs often face the problem of there being no gold standard against which results can be evaluated. To overcome this experts are often consulted to create labeled datasets [22], but their judgments may be faulty and the time of experts is often costly. Without a gold standard, though, it is challenging to evaluate conflicting results. If we find different causal relationships in different populations, we want to separate out what comes from the data (i.e. as a result of errors and biases), what is from the EHR (artifacts of the healthcare process and documentation practices) and what is from the methods. The computational methods used in this study were validated on simulated data from other domains, since there was none available that mimicked the structure of EHRs. The causal inference methods were validated on simulated financial and neuronal spike train data, and the NLP methods were validated on clinical texts, but the combination of the two were not validated on data with a structure similar to EHRs.
Shared Resources
A primary difficulty in replicating a study in multiple populations is in accessing and sharing these protected data. One of the lessons learned from this project is the importance of having a set of fully de-identified data that can be shared widely. Having such a repository will allow methods to be compared objectively on the same data. Repositories like the MIMIC dataset [23] and the corpora provided through the i2b2 challenges [24] are exciting steps in this direction. More recently, datasets annotated with ground truth labels have started to appear in the informatics community [25].
Simulation
In parallel to the need for annotated de-identified data, there is a need for simulated datasets. We can evaluate whether findings are consistent across populations or with current knowledge, but to determine what computational methods can and cannot do we must have simulated data. A core difficulty in translating computational tools to biomedical informatics is that they are often proven correct theoretically under some assumptions and applied to simulated data, but biomedical data has a radically different structure and unique challenges that are not addressed by generic simulations of networks or data from other fields. While creating realistic data is a challenge, the benefits are substantial and there are no related privacy issues, or concern about whether data is sufficiently de-identified for public release.
Table 4.
Comparison of data for both sites, including availability and format.
| Geisinger | CUMC | |
|---|---|---|
|
| ||
| Cases | 3,838 | 1,853 |
| Controls | 28,843 | 11,765 |
| Lab values | Structured | Structured |
| Medications | Structured orders and reconciliation list | Structured and from notes |
| Diagnoses | Structured comorbidities and problem list | ICD9 codes |
| Framingham symptoms | No | Notes |
| Vital signs | Yes | No |
| Visit types | Out-patient | In-patient and out-patient |
Acknowledgments
This study was supported by NLM contract HHSN276201000024C (NE) and the NSF under Grant #1019343 to the Computing Research Association for the CIFellows project (SK).
Footnotes
As of 7/12/2013, NHGRI’s GWAS catalog contains 1657 publications.
References
- 1.Kho AN, Pacheco JA, Peissig PL, Rasmussen L, Newton KM, Weston N, Crane PK, Pathak J, Chute CG, Bielinski SJ, Kullo IJ, Li R, Manolio TA, Chisholm RL, Denny JC. “Electronic medical records for genetic research: results of the eMERGE consortium,”. Sci Transl Med. 2011;3:79re1. doi: 10.1126/scitranslmed.3001807. 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee D, Austin P, Rouleau J, PP PL, Naimark D, Tu J. “Predicting mortality among patients hospitalized for heart failure: derivation and validation of a clinical model,”. JAMA. 2003;290(19):2581–2587. doi: 10.1001/jama.290.19.2581. [DOI] [PubMed] [Google Scholar]
- 3.Levy W, Mozaffarian D, Linker D, Sutradhar S, Anker S, Cropp A, Anand I, Maggioni A, Burton P, Sullivan M, B BP, Poole-Wilson P, Mann D, Packer M. “The Seattle heart failure model: prediction of survival in heart failure,”. Circulation. 2006;113(11):1424–1433. doi: 10.1161/CIRCULATIONAHA.105.584102. [DOI] [PubMed] [Google Scholar]
- 4.Lee IN, Liao SC, Embrechts M. “Data mining techniques applied to medical information.,”. Medical informatics and the Internet in medicine. 2000;25(2):81. doi: 10.1080/14639230050058275. [DOI] [PubMed] [Google Scholar]
- 5.Wu J, Roy J, Stewart WF. “Prediction modeling using EHR data: challenges, strategies, and a comparison of machine learning approaches,”. Medical care. 2010;48(6):S106. doi: 10.1097/MLR.0b013e3181de9e17. [DOI] [PubMed] [Google Scholar]
- 6.Bellazzi R, Zupan B. “Predictive data mining in clinical medicine: current issues and guidelines,”. International Journal of Medical Informatics. 2008;77(2):81–97. doi: 10.1016/j.ijmedinf.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 7.Shah NR, Roy J, Stewart WF. “Using electronic health record data to predict heart failure diagnosis,”. Clinical Medicine & Research. 2010;8(1):40–40. [Google Scholar]
- 8.Kleinberg S. Causality, probability, and time. Cambridge University Press; 2012. [Google Scholar]
- 9.Benjamini Y, Yekutieli D. “The control of the false discovery rate in multiple testing under dependency,”. Annals of Statistics. 2001;29(4):1165–1188. [Google Scholar]
- 10.Efron B. “Large-scale simultaneous hypothesis testing: The choice of a null hypothesis,”. Journal of the American Statistical Association. 2004;99(465):96–105. [Google Scholar]
- 11.Li Y, Gorman SL, Elhadad N. “Section classification in clinical notes using a supervised hidden markov model,”. Proceedings ACM International Health Informatics Symposium (IHI) 2010:744–750. [Google Scholar]
- 12.Yoshimasa T, Tateishi Y, Jin-Dong K, Tomoko O, McNaught J, Ananiadou S, Junichi T. “Developing a robust part-of-speech tagger for biomedical text,”. Lecture Notes in Computer Science. 2005 [Google Scholar]
- 13.B G, Baldridge J, Morton T. “The OpenNLP maximum entropy package,”. tech. rep., SourceForge. 2002 [Google Scholar]
- 14.Bodenreider O. “The unified medical language system (UMLS): integrating biomedical terminology,”. Nucleic Acids Res. 2004;32(D267) doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pivovarov R, Elhadad N. “A hybrid knowledge-based and data-drien approach to identifying semantically similar concepts,”. Journal of Biomedical Informatics. 2012;45(3):471–481. doi: 10.1016/j.jbi.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gold S, Elhadad N, Zhu X, Cimino J, Hripcsak G. “Extracting structured medication event information from discharge summaries,”. Proceedings AMIA Annual Symposium. 2008:237–241. [PMC free article] [PubMed] [Google Scholar]
- 17.Ho KKL, Pinsky JL, Kannel WB, Levy D. “The epidemiology of heart failure: the Framingham study,”. Journal of the American College of Cardiology. 1993;22(4):A6–A13. doi: 10.1016/0735-1097(93)90455-a. [DOI] [PubMed] [Google Scholar]
- 18.Chase HS, Radhakrishnan J, Shirazian S, Rao MK, Vawdrey DK. “Under-documentation of chronic kidney disease in the electronic health record in outpatients,”. J Am Med Inform Assoc. 2010;17(5):588–594. doi: 10.1136/jamia.2009.001396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bodenreider O, McCray AT. “Exploring semantic groups through visual approaches,”. Journal of Biomedical Informatics. 2003;36(6):414. doi: 10.1016/j.jbi.2003.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hripcsak G, Elhadad N, Chen C, Zhou L, Morrison F. “Using empirical semantic correlation to deduce meaning in temporal assertions in clinical texts,”. J Am Med Inform Assoc. 2009;16:220–227. doi: 10.1197/jamia.M3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heller R, Yekutieli D. “Replicability analysis for genome-wide association studies,”. arXiv preprint arXiv:1209.2829. 2012 doi: 10.1093/bioinformatics/btu434. [DOI] [PubMed] [Google Scholar]
- 22.Hripcsak G, Wilcox A. “Reference standards, judges, and comparison subjects roles for experts in evaluating system performance,”. J Am Med Inform Assoc. 2002;9(1):1–15. doi: 10.1136/jamia.2002.0090001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saeed M, Villarroel M, Reisner AT, Clifford G, Lehman L-W, Moody G, Heldt T, Kyaw TH, Moody B, Mark RG. “Multiparameter intelligent monitoring in intensive care ii (MIMIC-II): A public-access intensive care unit database,”. Critical Care Medicine. 2011 May;39:952–960. doi: 10.1097/CCM.0b013e31820a92c6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uzuner O, South B, Shen S, DuVall S. “2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text,”. J Am Med Inform Assoc. 2011;18(5):552–556. doi: 10.1136/amiajnl-2011-000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Savova G, Chapman W, Elhadad N, Palmer M. “Panel: Shared annotated resources for the clinical domain,”. AMIA Annu Fall Symp. 2011 [Google Scholar]