Abstract
Several recent genome-wide association studies have identified genetic variants associated with breast cancer. However, how much these genetic variants may help advance breast cancer risk prediction based on other clinical features, like mammographic findings, is unknown. We conducted a retrospective case-control study, collecting mammographic findings and high-frequency/low-penetrance genetic variants from an existing personalized medicine data repository. A Bayesian network was developed using Tree Augmented Naive Bayes (TAN) by training on the mammographic findings, with and without the 22 genetic variants collected. We analyzed the predictive performance using the area under the ROC curve, and found that the genetic variants significantly improved breast cancer risk prediction on mammograms. We also identified the interaction effect between the genetic variants and collected mammographic findings in an attempt to link genotype to mammographic phenotype to better understand disease patterns, mechanisms, and/or natural history.
1. Introduction
Large multi-relational databases containing variables that confer disease risk are increasingly available, providing the opportunity for informatics tools to better stratify individuals for appropriate healthcare decisions and explore disease mechanism and behavior. Coincident to this, policy-makers have recommended that interventions, like breast cancer screening with mammography, be increasingly based on individualized risk and shared decision-making1,2. Targeting at risk individuals for intervention after mammographic screening has the potential to decrease recommendations for breast biopsy in women most likely to have an unnecessary procedure for benign findings. Recent large-scale genome-wide association studies have identified 22 susceptibility loci associated with breast cancer (Table 1). In addition, there is a long history of development and codification of features observed by radiologists on mammography that also predict a woman’s risk of breast cancer. However, genetics and mammography abnormality findings have not yet been used together to predict risk. Furthermore, the opportunity to use this data to interpret genotype/phenotype association, explain family aggregation of breast cancer, and shed light on disease mechanism or natural history is just becoming possible.
Table 1:
SNPs evaluated for altered risk of breast cancer.
| SNP ID | Chr | Allele | OR | In Gail (2008,2009) | In Wacholder et al (2010) |
|---|---|---|---|---|---|
| rs11249433 9 | 1 | C | 0.99 | Yes | |
| rs4666451 10 | 2 | A | 0.83 | ||
| rs13387042 11,9 | 2 | G | 0.68 | Yes | Yes |
| rs1045485 12 | 2 | C | 0.86 | Yes | Yes |
| rs17468277 13 | 2 | T | 0.86 | ||
| rs4973768 14 | 3 | T | 0.99 | ||
| rs10941679 15,9 | 5 | G | 1.39 | Proxy of rs7716600 | |
| rs981782 10 | 5 | G | 0.82 | ||
| rs30099 10 | 5 | T | 1.07 | ||
| rs889312 10 | 5 | C | 1.20 | Yes | Yes |
| rs2180341 16 | 6 | G | 1.10 | ||
| rs2046210 17 | 6 | T | 0.91 | ||
| rs13281615 10 | 8 | G | 1.26 | Yes | Yes |
| rs2981582 10,18,16,15,9 | 10 | T | 1.18 | Yes | Yes |
| rs3817198 10,9 | 11 | C | 1.16 | Yes | Yes |
| rs2107425 10 | 11 | T | 1.20 | ||
| rs6220 19,20 | 12 | G | 1.24 | ||
| rs999737 9 | 14 | T | 0.98 | yes | |
| rs3803662 10,11,9 | 16 | T | 1.14 | Yes | Yes |
| rs8051542 10 | 16 | T | 1.29 | ||
| rs12443621 10 | 16 | G | 0.93 | ||
| rs6504950 14 | 17 | A | 0.81 |
Chr: the chromosome the SNP locates.
Allele: the minor allele.
OR: the allelic odds-ratio in the Marshfield Clinic population.
There have been several attempts to incorporate these genetic variants into the Gail model3 which is a standard clinical breast cancer risk model including the number of first-degree relatives with a diagnosis of breast cancer, age at menarche, age at first live birth and the number of previous breast biopsies. Seven associated SNPs, when added to the Gail model, increase the area under the receiver operating characteristic (ROC) curve from 0.607 to 0.6324,5. When ten associated SNPs are added to the Gail model, the area under the ROC curve of the risk model increases from 0.580 to 0.618 on another dataset6. However, the Gail model does not include any mammography features which are clinically used by radiologists. Therefore, it is still unknown how much these genetic variants improve breast cancer diagnosis and clinical decision-making after an abnormal mammogram.
The first purpose of this study is to examine the impact of genetic information on improving breast cancer risk prediction on mammograms. We incorporate genetic polymorphisms with the descriptors that radiologists observe on mammograms while making medical decisions, the American College of Radiology Breast Imaging Reporting and Data System (BI-RADS)7, version 4, including the shape and the margin of masses, the shape and the distribution of microcalcifications, background breast density and other associated findings as defined by this standard lexicon in breast imaging. We also include a small number of predictive variables not included in BI-RADS currently. Specifically, we employ these mammographic findings (49 mammography descriptors) and the 22 genetic variants associated with breast cancer in 404 case subjects and 399 control subjects from a personalized medicine data repository at the Marshfield Clinic. We train a Bayesian network using Tree Augmented Naive Bayes (TAN)8 on the mammographic findings, with and without the 22 genetic variants.
The second purpose of this study is to identify the interaction effect between the genetic variants and the mammographic findings toward risk prediction, in order to understand the genotype/phenotype relationships that may elucidate disease patterns that may not be otherwise evident in this complex multi-relational data. The interaction between the genetic variants and the mammographic findings also sheds light on how the associated SNPs function to increase or decrease the risk of breast cancer. Specifically, we calculate the conditional mutual information between the mammography features and the genetic variants given the class variable on the entire dataset.
2. Materials and Methods
2.1. Data
[Subjects] The Personalized Medicine Research Project21 at the Marshfield Clinic was used as the sampling frame to identify breast cancer cases and controls. The project was reviewed and approved by the Marshfield Clinic IRB. Subjects were selected using clinical data from Marshfield Clinic Cancer Registry and Data Warehouse. We employed a retrospective case-control design. Women with a plasma sample available, a mammogram, and a breast biopsy within 12 months after the mammogram were included in the study. Cases were defined as women having a confirmed diagnosis of breast cancer obtained from the institutional cancer registry. Controls were confirmed through the electronic medical records (and absence from the cancer registry) as never having had a breast cancer diagnosis. In our case cohort, we included both invasive breast cancer (ductal and lobular) as well as ductal carcinoma in situ. In order to construct case and control cohorts that were similar in age distribution, we employed an age matching strategy. Specifically, we selected a control whose age was within five years of the age of each case. Of note, we decided to focus on high-frequency/low-penetrance genes that affect breast cancer risk as opposed to low frequency genes with high penetrance (BRCA1 and BRCA2) or intermediate penetrance (CHEK-2). High-frequency/low-penetrance SNPs generally have frequencies for the rarest allele of > 25% as opposed to the low-frequency, high-penetrance mappings with population frequencies of < 1%. We excluded individuals who had a known high penetrance genetic mutation.
[Genetic Variants] Our study included 22 genetic variants which have been identified by recent large-scale genome-wide association studies. Table 1 summarizes detailed information about the 22 SNPs, including the IDs, the original publications associating them with breast cancer, their chromosomes, the minor alleles and the allelic odds-ratios of the SNPs in the Marshfield Clinic population. The seven SNPs used in Gail study4,5 were also included in our study. Nine of the ten SNPs used in Wacholder et al study6 were included in our study, and the remaining SNP rs7716600 from that study had a proxy rs10941679 in our study. We observed that each SNP only confers a slight increase or decrease in the risk of breast cancer, in accordance with prior literature. Among the 22 associated SNPs, 11 are associated with an increased risk of breast cancer (OR>1.0) and 11 are associated with a decreased risk of breast cancer (OR<1.0). When we built the models with the genetic variants, we coded each genetic variant as whether the subject carries the minor allele, rather than the specific genotype the subject carries.
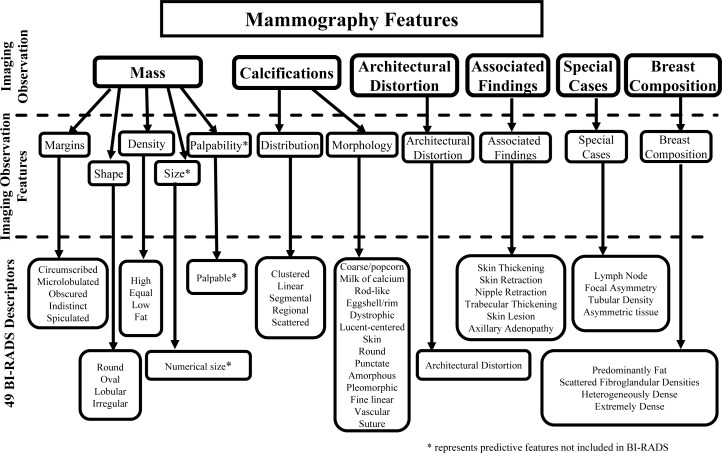
[Mammography Features] The American College of Radiology developed the BI-RADS lexicon7 to homogenize mammographic findings and recommendations. The BI-RADS lexicon consists of a number of mammography descriptors, including the characteristics of masses and microcalcifications, background breast density and other associated findings, which can be organized in a hierarchy as shown in Figure 1. Datasets containing mammography descriptors have been used to build several successful breast cancer risk models and classifiers22,23. Mammography data was originally recorded as free text reports in the Marshfield database, and thus it was difficult to directly access the information contained therein. We used a parser to extract mammography features from the text reports; the parser has been shown to outperform manual extraction24,25. After extraction, every mammography feature takes the value “present” or “not present” except that the variable mass size is discretized into three values, “not present”, “small” and “large”, depending whether there is a reported mass size and whether any dimension of the reported mass size is larger than 30mm.
Figure 1:
Mammography features adopted from the American College of Radiology (BI-RADS lexicon).
Each mammogram also has a BI-RADS category assigned by the radiologist who read the mammogram. The BI-RADS category indicates the radiologist’s opinion of the absence or presence of breast cancer. In our study, the BI-RADS assessment category can take values, with an order of increasing probability of malignancy, of 1, 2, 3, 0, 4a, 4, 4b, 4c and 5. We used the BI-RADS assessment category as the predictions from the radiologists. Our experiment only included diagnostic mammograms, and all the screening mammograms were excluded. Since most of the subjects have multiple diagnostic mammograms in the electronic medical records, we selected one mammogram for each subject as follows, to mimic the scenario of the most important doctor visit before diagnosis. For cases, we selected the mammograms within one year prior to diagnosis. For controls, we selected the mammograms within one year prior to biopsy. If there were still multiple mammograms left for each subject, we selected the mammogram with a more suspicious BI-RADS category, with subsequent tiebreakers being, in order, recency and the number of extracted mammography features.
2.2. Model
We build breast cancer risk models using Bayesian networks, which have been used with mammography data to improve breast cancer diagnosis and clinical decision-making for physicians involved in breast cancer care26,27. Bayesian networks are directed acyclic graphs that allow efficient and effective representation of the joint probability distribution over a set of random variables. Each vertex in the graph represents a random variable, and edges represent conditional independence between the variables. In this paper, we use a special type of Bayesian network model, namely TAN8, which is an effective, provably efficient supervised learning model which captures the strongest pairwise interactions between the features in a compact way. Training a TAN model starts with learning a Naive Bayes model with the case/control output being the class variable and all the other variables being the features. Naive Bayes assumes that all features are conditionally independent of one another given the class28. Because this assumption may be too strong, the TAN learning algorithm next builds a maximum spanning tree over the feature variables with the weight between two variables being the conditional mutual information between two features conditional on the class variable. Eventually the parameters in the model, namely the conditional probability tables, are estimated from the data. In our experiments, we use the TAN implementation in WEKA29.
In total, we construct three TAN models built on different sets of features. The first model is built purely on the 49 mammography features, namely the breast imaging model. The second model is based purely on the 22 associated SNPs, namely the genetic model. The third model is built on the 49 mammography features and the 22 associated SNPs together, namely the combined model. We treat the BI-RADS category scores from the radiologists as the predictions from the radiologists, namely the baseline clinical assessment. We construct ROC curves for each model, and use the area under the curve (AUC) as a measure of performance of the models. We also provide the precision-recall (PR) curves for the models. We evaluate the models in the 10-fold cross-validation fashion. The 404 cases and 399 controls are randomly divided in 10 folds. In each round of the 10-fold cross-validation, we select one fold as the testing data and the remaining nine folds as the training data, so that each fold is used exactly once for testing.
We further evaluate the interaction between the SNPs and the mammography features toward predicting the class label (case or control). Specifically, we calculate the conditional mutual information (CMI) between the 22 SNPs and the 49 mammography features given the class label. We also calculate the 95% confidence intervals for the CMI between each SNP and each mammography feature via bootstrapping. We randomly draw samples with replacement from the 404 cases and the 399 controls, and calculate the conditional mutual information. We bootstrap for 1,000 times and calculate the corresponding 1,000 CMI values. We sort the 1,000 CMI values from the smallest to the largest, and report the 26-th smallest value and the 26-th largest value as the boundaries of the 95% confidence interval.
3. Results
We succeeded in identifying 404 cases for which we could match a mammogram within a year prior to a biopsy. We then identified age-matched controls; however at the end of data collection and verification, 5 of the controls were confirmed to have breast cancer leaving us with 399 controls for which we could match a mammogram within a year prior to a biopsy. Among the 404 cases, there are 401 Caucasian cases, two Asian Hmong cases and one case whose race information is unknown. Among the 399 controls, there are 395 Caucasian controls, three Caucasian/American Indian controls, and one Caucasian/Asian Hmong control. We summarize the distribution of the ages and family breast cancer history of the cases and the controls in Marshfield population in Table 2. There are more young people (age < 50) in the case group than in the control group, and the proportion of elderly people (age ≥ 65) is roughly the same in the case group and in the control group. For the family history of breast cancer, we observe a considerable larger proportion of people with family history in the case group than in the control group, which demonstrates the family aggregation of breast cancer.
Table 2:
The distribution of age at mammogram and family breast cancer history in the cases and the controls.
| Age | Cases | Controls | Family History | Cases | Controls |
|---|---|---|---|---|---|
| < 50 | 96 | 67 | Yes | 178 | 134 |
| ≥ 50, < 65 | 141 | 171 | No | 215 | 251 |
| ≥ 65 | 167 | 161 | Unknown | 11 | 14 |
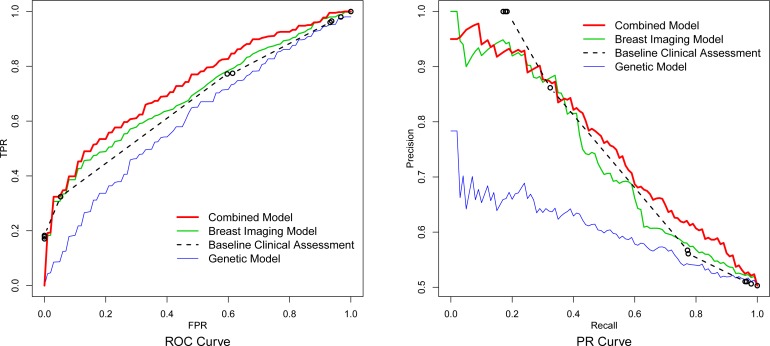
The ROC curves and the PR curves for the baseline clinical assessment, the breast imaging model, the genetic model and the combined model are provided in Figure 2, respectively. For each type of model we vertically average30 its ROC curves from the ten replications of the 10-fold cross-validation to obtain the final curve; we do likewise for the PR curves. The area under the ROC curves for the genetic model, the breast imaging model and the combined model are 0.603, 0.693 and 0.731, respectively. The ROC curve of the combined model almost completely dominates the ROC curve of the breast imaging model, which suggests that adding the 22 genetic variants can help to improve the breast cancer risk prediction based on mammographic findings. We perform a two-sided paired t-test on the area under the ten ROC curves of the breast imaging model and the area under the ten ROC curves of the combined model from the 10-fold cross-validation, and the difference between them is significant with a P-value 0.021. From the PR curves, it is also observed that the combined model dominates the breast imaging model and the baseline clinical assessment at the high recall region (> 0.8) in which we would like to operate, and therefore which we want to optimize.
Figure 2:
The vertical averaged ROC curves and PR curves for the genetic model, the breast imaging model, the combined model and the baseline clinical assessment.
For each SNP, we summarize the mammography feature with the highest conditional mutual information, the conditional mutual information value and the corresponding 95% confidence intervals in Table 3. Most of the interaction effect is moderate with small CMI values. There are four noteworthy interaction pairs between the genetic variants and the mammography features toward breast cancer risk prediction with the conditional mutual information above 0.01. The four interaction pairs are (1) SNP rs1045485 (rs17468277) and pleomorphic calcifications (CMI=0.0141), (2) SNP rs2180341 and dystrophic calcifications (CMI=0.0115), (3) SNP rs2981582 and diffuse calcifications (CMI=0.0112) and (4) SNP rs4666451 and oval masses (CMI=0.0100).
Table 3:
The mammography feature with the highest conditional mutual information (CMI) for each of the 22 SNPs.
| SNP ID | Mammography Feature with Highest CMI | CMI* 95% C.I. | Chr | Associated Gene | OR |
|---|---|---|---|---|---|
| rs1045485 | calcification shape: pleomorphic | 0.0141 (0.006,0.030) | 2q33 | CASP8 | 0.86 |
| rs17468277 | calcification shape: pleomorphic | 0.0141 (0.005,0.032) | 2q33 | CASP8 | 0.86 |
| rs2180341 | calcification shape: dystrophic | 0.0115 (0.006,0.021) | 6q25 | RNF146 | 1.10 |
| rs2981582 | calcification distribution: diffuse | 0.0112 (0.006,0.021) | 10q26 | FGFR2 | 1.18 |
| rs4666451 | mass shape: oval | 0.0100 (0.004,0.017) | 2p | 0.83 | |
| rs11249433 | special case: focal asymmetry | 0.0095 (0.003,0.024) | 1p11 | 0.99 | |
| rs12443621 | calcification shape: dystrophic | 0.0091 (0.004,0.020) | 16q12 | TNRC9/TOX3 | 0.93 |
| rs13281615 | calcification shape: dystrophic | 0.0087 (0.002,0.023) | 8q24 | 1.26 | |
| rs3803662 | calcification distribution: linear | 0.0086 (0.002,0.024) | 16q12 | TNRC9/TOX3 | 1.14 |
| rs2107425 | mass shape: round | 0.0080 (0.003,0.017) | 11p15 | H19 | 1.20 |
| rs889312 | breast composition: extreme | 0.0078 (0.001,0.019) | 5q11 | MAP3K1 | 1.20 |
| rs981782 | breast composition: fat | 0.0076 (0.004,0.015) | 5p12 | HCN1/MRPS30 | 0.82 |
| rs8051542 | calcification distribution: linear | 0.0076 (0.002,0.021) | 16q12 | TNRC9/TOX3 | 1.29 |
| rs3817198 | calcification shape: punctate | 0.0075 (0.002,0.022) | 11p15 | LSP1 | 1.16 |
| rs13387042 | breast composition: extreme | 0.0069 (0.003,0.011) | 2q35 | 0.68 | |
| rs999737 | calcification distribution: linear | 0.0069 (0.001,0.021) | 14q24 | RAD51L1 | 0.98 |
| rs30099 | calcification shape: amorphous | 0.0063 (0.000,0.018) | 5q | 1.07 | |
| rs4973768 | calcification shape: amorphous | 0.0058 (0.003,0.010) | 3p24 | SLC4A7 | 0.99 |
| rs6504950 | mass shape: lobular | 0.0058 (0.001,0.019) | 17q22 | STXBP4 | 0.81 |
| rs2046210 | associated finding: architectural distortion | 0.0053 (0.001,0.018) | 6q25 | C6orf97 | 0.91 |
| rs6220 | calcification shape: amorphous | 0.0050 (0.001,0.014) | 12q23 | IGF-1 | 1.24 |
| rs10941679 | mass shape: oval | 0.0048 (0.000,0.014) | 5p12 | HCN1/MRPS30 | 1.39 |
We order the rows by the CMI values, and the CMI’s above 0.01 are shown in bold.
Chr: the chromosome the SNP locates.
OR: the allelic odds-ratio in the Marshfield Clinic population.
4. Discussion
We found that adding the 22 genetic polymorphisms to the 49 radiologist-reported mammographic findings statistically significantly increased the accuracy (as measured by AUC-ROC) of our Bayesian network model, despite a small sample size. In our preliminary exploration of genotype/phenotype relationships, we identified 4 potentially noteworthy interacting pairs between the genetic variants and the mammographic findings. These observations imply that radiologists may benefit from the availability of patient genotype information when they are making their interpretations of mammogram results.
Statistical models by Gail4,5 and Wacholder et al6 added genetic risk factors to epidemiologic risk factors and found modest improvements in predictive performance. All of these studies used all or a portion of the carefully validated and widely disseminated Gail model (a logistic regression model) as the baseline model. The variables included in the most recent analysis6 were the number of first-degree relatives with a diagnosis of breast cancer, age at menarche, age at first live birth, study entry year, and the number of previous breast biopsies. These investigators added 10 common genetic variants associated with breast cancer in 5,590 case subjects and 5,998 controls. They found that the AUC of the non-genetic model was 0.580, whereas the model with genetic component added (10 SNPs) revealed an AUC of 0.618. Importantly, no model to date has included mammography features describing breast findings. It is not surprising that our discriminative abilities are superior to prior models because we are using highly predictive features from mammography including abnormality descriptors and breast density, which to date, have not been included in previous models. Therefore, we are encouraged by our promising preliminary results.
With the contingency tables in Table 4, we further explore the four interacting pairs from a clinical standpoint:
SNP rs1045485 (rs17468277) and pleomorphic calcifications: The protective minor allele C of SNP rs1045485 was associated with a reduced risk of breast cancer in the genome-wide association study12 with an allelic odds-ratio 0.88, and we observe that the allelic odds-ratio in the Marshfield Clinic population is 0.86. Two earlier studies31,32 found that polymorphisms in CASP8 (rs1045485 and rs1045485/rs17468277) appeared to be specifically associated with a reduced risk of ductal tumors. Generally, pleomorphic calcifications are a malignant descriptor on mammograms indicating ductal carcinoma in situ. We observe 60 cases and 70 controls with pleomorphic calcifications in our data. However, 13 out of the 70 controls carry the minor allele C of rs1045485 whereas only 3 out of the 60 cases carry the minor allele. Therefore, when the person does not carry C at rs1045485 and develops pleomorphic calcification, it is more likely the abnormality is malignant.
SNP rs2180341 and dystrophic calcifications: The risky minor allele G of SNP rs2180341 was identified to be associated with an increased risk of breast cancer in the genome-wide association study16 with an allelic odds-ratio 1.41, and we observe that the allelic odds-ratio in the Marshfield Clinic population is 1.10. Our odds-ratio estimate is consistent with a recent study33 in which the allelic odds-ratio is estimated to be 1.07 in the Cypriot population. Dystrophic calcification is generally a benign descriptor on mammograms although we observe 10 cases and 9 controls with dystrophic calcification in our data. However, none of the cases carry the minor allele G of SNP rs2180341 whereas 3 of the 9 controls carry the minor allele G. Therefore, dystrophic calcifications continue to be a benign feature even if a woman carries the minor allele G of SNP rs2180341.
SNP rs2981582 and diffuse calcifications: SNP rs2981582 lies in intron 2 of FGFR2 (fibroblast growth factor receptor 2), and its risky allele T has been identified to be associated with an increased risk of breast cancer in several studies10,18,16,15,9 with an allelic odds-ratio 1.2610, and we observe that the allelic odds-ratio in the Marshfield Clinic population is 1.18. Diffuse calcifications on mammograms are generally benign. In our dataset, we observe in total 22 people (11 cases and 11 controls) with diffuse calcifications. However, 20 of the 22 people carry the minor allele T of SNP rs2981582, and the 2 people not carrying allele T are both cases.
SNP rs4666451 and oval masses: The protective minor allele A of SNP rs4666451 was associated with a reduced risk of breast cancer in a previous genome-wide association study10 with an allelic odds-ratio 0.97, and we observe the allelic odds-ratio in the Marshfield Clinic population is 0.83. Among the 404 breast cancer cases and 399 controls, there are only 11 subjects (2 cases and 9 controls) whose mammograms exhibit oval masses. However, all the controls carry the minor allele A of rs4666451, whereas neither of two cases carry the minor allele A. Oval masses are generally benign indicators, and we can further strengthen this belief if we know the person also carries minor allele A of SNP rs4666451.
Table 4:
The contingency tables for SNPs rs4666451, rs1045485, rs2180341 and rs2981582 and their interacting mammography features.
| Cases | Controls | |||
|---|---|---|---|---|
| rs1045485 | Carry C | Not Carry C | Carry C | Not Carry C |
|
| ||||
| pleomorphic calcification present | 3 | 57 | 13 | 57 |
| pleomorphic calcification not present | 83 | 261 | 82 | 247 |
|
| ||||
| rs2180341 | Carry G | Not Carry G | Carry G | Not Carry G |
|
| ||||
| dystrophic calcification present | 0 | 10 | 3 | 6 |
| dystrophic calcification not present | 185 | 209 | 168 | 222 |
|
| ||||
| rs2981582 | Carry T | Not Carry T | Carry T | Not Carry T |
|
| ||||
| diffuse calcification present | 9 | 2 | 11 | 0 |
| diffuse calcification not present | 251 | 142 | 235 | 153 |
|
| ||||
| rs4666451 | Carry A | Not Carry A | Carry A | Not Carry A |
|
| ||||
| oval mass present | 0 | 2 | 9 | 0 |
| oval mass not present | 245 | 157 | 257 | 133 |
There are some unavoidable limitations in our study, due to the inherent difficulty of collecting a rich multi-relational dataset. First, the sample size is small compared with large-scale genome-wide association studies10,18,16,15,9. However, other studies do not include mammography features or abnormality data4,5,6. Second, all the mammogram reports in the original database are in free text, rather than structured reports. Although we extract the features with an accurate parser24,25, this extra step introduces noise, and in particular may miss important features for certain subjects. Third, for each SNP we pick the mammography feature with highest CMI value among the 49 mammography features. Although we evaluate the 95% confidence intervals for the CMI’s, the selected mammography features may appear promising by chance. This risk of false positive association generated by this CMI analysis is further exacerbated by small sample size. However, exploring genotype/phenotype relationships is only a secondary goal of this project and we approach this analysis with caution, realizing we need more data and refined methodologies to validate our findings and to eliminate selection bias or the multiple comparison effect.
5. Conclusion
Our study represents the first exploration of breast cancer risk prediction using genetic polymorphisms along with mammography features. The fact that genetic risk factors improve risk prediction to a statistically significant degree raises the possibility that stratification based on these risk factors may provide an opportunity to personalize care. In addition, we plan to further develop the concept of exploring genotype/phenotype relationships to shed light on disease processes that may, in the future, improve diagnosis and treatment. Though we fully realize the necessity of increasing our sample size to validate these promising preliminary results, we are cautiously optimistic of the power of multi-relational databases, like the one we have constructed, both to test risk prediction hypotheses and engage in data-mining that would not otherwise be possible.
Acknowledgments
The authors acknowledge the support of the Wisconsin Genomics Initiative, NCI grant R01CA127379-01 and its ARRA supplement 3R01CA127379-03S1, NIGMS grant R01GM097618-01, NLM grant R01LM011028-01, NIEHS grant 5R01ES017400-03, the UW Institute for Clinical and Translational Research (ICTR) and the UW Carbone Cancer Center.
References
- [1].Nelson HD, Tyne K, Naik A, Bougatsos C, Chan BK, Humphrey L. Screening for breast cancer: an update for the US preventive services task force. Ann Intern Med. 2009;151:727–737. doi: 10.1059/0003-4819-151-10-200911170-00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Schousboe JT, Kerlikowske K, Loh A, Cummings SR. Personalizing mammography by breast density and other risk factors for breast cancer: analysis of health benefits and cost-effectiveness. Ann Intern Med. 2011;155:10–20. doi: 10.7326/0003-4819-155-1-201107050-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Gail MH, Brinton LA, Byar DP, Corle DK, Green SB, Schairer C, et al. Projecting individualized probabilities of developing breast cancer for white females who are being examined annually. J Natl Cancer Inst. 1989;81(24):1879–1886. doi: 10.1093/jnci/81.24.1879. [DOI] [PubMed] [Google Scholar]
- [4].Gail MH. Discriminatory accuracy from single-nucleotide polymorphisms in models to predict breast cancer risk. J Natl Cancer Inst. 2008;100(14):1037–1041. doi: 10.1093/jnci/djn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Gail MH. Value of adding single-nucleotide polymorphism genotypes to a breast cancer risk model. J Natl Cancer Inst. 2009;101(13):959–963. doi: 10.1093/jnci/djp130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Wacholder S, Hartge P, Prentice R, Garcia-Closas M, Feigelson HS, Diver WR, et al. Performance of common genetic variants in breast-cancer risk models. N Engl J Med. 2010;362(11):986–993. doi: 10.1056/NEJMoa0907727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].American College of Radiology and American College of Radiology BI-RADS Committee . Breast imaging reporting and data system. American College of Radiology; 1998. [Google Scholar]
- [8].Friedman N, Geiger D, Goldszmidt M, Provan G, Langley P, Smyth P. Bayesian network classifiers. Machine Learning. 1997:131–163. [Google Scholar]
- [9].Thomas G, Jacobs KB, Kraft P, Yeager M, Wacholder S, Cox DG, et al. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1) Nat Genet. 2009;41(5):579–584. doi: 10.1038/ng.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Easton DF, Pooley KA, Dunning AM, Pharoah PDP, Thompson D, Ballinger DG, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447(17529967):1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Stacey SN, Manolescu A, Sulem P, Rafnar T, Gudmundsson J, Gudjonsson SA, et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2007;39(7):865–869. doi: 10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- [12].Cox A, Dunning AM, Garcia-Closas M, Balasubramanian S, Reed MWR, Pooley KA, et al. A common coding variant in CASP8 is associated with breast cancer risk. Nat Genet. 2007;39(17293864):352–358. doi: 10.1038/ng1981. [DOI] [PubMed] [Google Scholar]
- [13].Odefrey F, Stone J, Gurrin LC, Byrnes GB, Apicella C, Dite GS, et al. Common genetic variants associated with breast cancer and mammographic density measures that predict disease. Cancer Res. 2010;70(20145138):1449–1458. doi: 10.1158/0008-5472.CAN-09-3495. [DOI] [PubMed] [Google Scholar]
- [14].Ahmed S, Thomas G, Ghoussaini M, Healey CS, Humphreys MK, Platte R, et al. Newly discovered breast cancer susceptibility loci on 3p24 and 17q23.2. Nat Genet. 2009;41(19330027):585–590. doi: 10.1038/ng.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Stacey SN, Manolescu A, Sulem P, Thorlacius S, Gudjonsson SA, Jonsson GF, et al. Common variants on chromosome 5p12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2008;40(18438407):703–706. doi: 10.1038/ng.131. [DOI] [PubMed] [Google Scholar]
- [16].Gold B, Kirchhoff T, Stefanov S, Lautenberger J, Viale A, Garber J, et al. Genome-wide association study provides evidence for a breast cancer risk locus at 6q22.33. Proc Natl Acad Sci U S A. 2008;105(18326623):4340–4345. doi: 10.1073/pnas.0800441105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Zheng W, Long J, Gao YT, Li C, Zheng Y, Xiang YB, et al. Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat Genet. 2009;41(19219042):324–328. doi: 10.1038/ng.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hunter DJ, Kraft P, Jacobs KB, Cox DG, Yeager M, Hankinson SE, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39(17529973):870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Kelemen LE, Sellers TA, Vachon CM. Can genes for mammographic density inform cancer aetiology? Nat Rev Cancer. 2008;8(18772892):812–823. doi: 10.1038/nrc2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Biong M, Gram IT, Brill I, Johansen F, Solvang HK, Alnaes GIG, et al. Genotypes and haplotypes in the insulin-like growth factors, their receptors and binding proteins in relation to plasma metabolic levels and mammographic density. BMC Med Genomics. 2010;3(9) doi: 10.1186/1755-8794-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].McCarty C, Wilke R, Giampietro P, Wesbrook S, Caldwell M. Marshfield Clinic Personalized Medicine Research Project (PMRP): design, methods and recruitment for a large population-based biobank. Personalized Med. 2005;2:49–79. doi: 10.1517/17410541.2.1.49. [DOI] [PubMed] [Google Scholar]
- [22].Baker JA, Kornguth PJ, Lo JY, Williford ME, Floyd CE. Breast cancer: prediction with artificial neural network based on BI-RADS standardized lexicon. Radiology. 1995;196(3):817–822. doi: 10.1148/radiology.196.3.7644649. [DOI] [PubMed] [Google Scholar]
- [23].Burnside ES, Davis J, Chhatwal J, Alagoz O, Lindstrom MJ, Geller BM, et al. Probabilistic computer model developed from clinical data in national mammography database format to classify mammographic findings. Radiology. 2009;251(3):663–672. doi: 10.1148/radiol.2513081346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Nassif H, Wood R, Burnside ES, Ayvaci M, Shavlik J, Page D. Information extraction for clinical data mining: a mammography case study. IEEE International Conference on Data Mining (ICDM’09) Workshops; Miami, Florida. 2009. pp. 37–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Percha B, Nassif H, Lipson J, Burnside E, Rubin D. Automatic classification of mammography reports by BI-RADS breast tissue composition class. J Am Med Inform Assn. 2012;19(5):913–916. doi: 10.1136/amiajnl-2011-000607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kahn CE, Jr, Roberts LM, Shaffer KA, Haddawy P. Construction of a Bayesian network for mammographic diagnosis of breast cancer. Comput Biol Med. 1997;27(1):19–29. doi: 10.1016/s0010-4825(96)00039-x. [DOI] [PubMed] [Google Scholar]
- [27].Burnside ES, Rubin DL, Shachter RD. Using a Bayesian network to predict the probability and type of breast cancer represented by microcalcifications on mammography. Stud Health Technol Inform. 2004;107:13–17. [PubMed] [Google Scholar]
- [28].Lowd D, Domingos P. Naive Bayes models for probability estimation; Proceedings of the 22nd international conference on machine learning; 2005. pp. 529–536. [Google Scholar]
- [29].Hall M, Frank E, Holmes G, Pfahringer B, Reutemann P, Witten IH. The WEKA data mining software: an update. SIGKDD Explor Newsl. 2009;11(1):10–18. [Google Scholar]
- [30].Fawcett T. An introduction to ROC analysis. Pattern recognition letters. 2006;27(8):861–874. [Google Scholar]
- [31].MacPherson G, Healey CS, Teare MD, Balasubramanian SP, Reed MW, Pharoah PD, et al. Association of a common variant of the CASP8 gene with reduced risk of breast cancer. J Natl Cancer Inst. 2004;96(24):1866–1869. doi: 10.1093/jnci/dji001. [DOI] [PubMed] [Google Scholar]
- [32].Frank B, Bermejo JL, Hemminki K, Klaes R, Bugert P, Wappenschmidt B, et al. Re: association of a common variant of the CASP8 gene with reduced risk of breast cancer. J Natl Cancer Inst. 2005;97(13):1012. doi: 10.1093/jnci/dji178. [DOI] [PubMed] [Google Scholar]
- [33].Loizidou MA, Hadjisavvas A, Ioannidis JP, Kyriacou K. Replication of genome-wide discovered breast cancer risk loci in the Cypriot population. Breast cancer research and treatment. 2011;128(1):267–272. doi: 10.1007/s10549-010-1319-8. [DOI] [PubMed] [Google Scholar]