Abstract
Deep venous thrombosis and pulmonary embolism are diseases associated with significant morbidity and mortality. Known risk factors are attributed for only slight majority of venous thromboembolic disease (VTE) with the remainder of risk presumably related to unidentified genetic factors. We designed a general purpose Natural Language (NLP) algorithm to retrospectively capture both acute and historical cases of thromboembolic disease in a de-identified electronic health record. Applying the NLP algorithm to a separate evaluation set found a positive predictive value of 84.7% and sensitivity of 95.3% for an F-measure of 0.897, which was similar to the training set of 0.925. Use of the same algorithm on problem lists only in patients without VTE ICD-9s was found to be the best means of capturing historical cases with a PPV of 83%. NLP of VTE ICD-9 positive cases and non-ICD-9 positive problem lists provides an effective means for capture of both acute and historical cases of venous thromboembolic disease.
Introduction
Venous thromboembolism (VTE) includes a spectrum of disease processes from the potentially life threatening pulmonary embolism (PE) to deep venous thrombosis (DVT). Reported incidence of VTE ranges from 7.1 to 11.7 persons per 10,000 person years for community residents.(1–2) VTE risk for post surgical patients can be as much as 20-fold higher within the first three months of surgery.(3) While prophylactic treatment with anticoagulants is routinely utilized for hospitalized patients with known risk factors, current understanding of the genetic components to VTE risk is incomplete.(4) Identification of the VTE phenotype prepares a foundation for larger population studies to identify clinical and genetic risks for thromboembolic disease. In this study we describe a multifilter step algorithm using billing codes and natural language processing (NLP) to capture VTE cases in an electronic health record (EHR).
Defining cases of VTE in an EHR is complex due to the lack of specificity of ICD-9 codes, common presence of VTE prophylaxis for most hospitalized patients, and limited data signal for historical cases (often lacking billing codes, for instance). Research has shown that billing code data is incomplete for capturing VTE events.(5) Even in a well-defined postoperative observation cohort, International Classification of Diseases, Ninth Revision (ICD-9) codes alone had poor positive predictive value (PPV) of 29% and only fair sensitivity (68%) in capturing VTE disease.(6) The AHRQ Patient Safety Indicators, comprise multiple algorithms for VTE identification but ultimately rely on ICD-9 codes. These have been shown to yield slightly better VTE identification results with a PPV (54.5%) and sensitivity (87%).(7) Using an NLP only surveillance process to identify real time Peripherally Inserted Central Catheter line associated DVTs, Evans and colleagues identified an overall incidence of 2.8% with a PPV 98% and sensitivity of 94% for patients with evidence of acute DVTs.(8)
In this study we applied both ICD-9 and NLP methods to identify VTE cases retrospectively in a general academic medical center population, with a goal of identifying both acute and historical VTE. Use of NLP alone was required to identify the smaller subset of patients with no ICD-9s, typically representing previous VTE events and best found in problem list documentation. Given the frequency of VTE “rule out” and prophylaxis language in clinical notes and text-based forms with atypical negation signals, we found general purpose NLP inadequate and required post-processing to achieve sufficient performance.
Methods
Vanderbilt University Medical Center is an 832-bed, tertiary care facility and major teaching center in Nashville, Tennessee. It uses a locally-developed Electronic Health Record (EHR) system, called StarPanel, which has been in use throughout the medical center since the early 1990s. Patient care notes in the form of clinic notes, problem lists, hospital admission and discharges and radiographic data are stored as free text documents. For research use, Vanderbilt has developed the Synthetic Derivative (SD), a de-identified image of StarPanel. Data from the SD includes billing codes, laboratory results, radiographic reports, clinical notes and problem lists. The SD is linked to the Vanderbilt DNA biobank, BioVU.(9) Genetic samples in BioVU are accrued sequentially from discarded blood draws. Approximately the first 10000 patients accrued into BioVU were studied previously and validated for known genetic associations with five common diseases.(10–11) In this study, we used this same set of patients for VTE description and algorithm development. This work represents a preliminary step for further exploration of this phenotype across the SD.
Gold Standard Population:
From the original BioVU set, 590 individuals were identified with at least one VTE ICD-9 code. (See Appendix 1 for ICD-9 codes included). All cases were reviewed in the SD for determination of VTE status by the primary author, a practicing physician board-certified in internal medicine and pediatrics. Author JCD, also a practicing physician board-certified in internal medicine, validated all cases of undetermined status. We defined cases as positive in the SD if radiological procedures reported VTE or if clinical notes or problem lists discussed previous VTE disease. For negative cases, notes and other EHR data were specifically reviewed around date of the ICD-9 to determine whether correlating notes were available to confirm VTE disease. Many of these individuals appear to have ICD-9 codes for secondary processes such as monitoring of coumadin treatment after joint replacement and not for actual VTE disease. A small minority of patients did not have notes with sufficient information to rule in or rule out VTE. These individuals were classified as unknown.
We defined DVT disease as any venous thrombosis associated with a deep vein and with the potential to lead to a pulmonary embolism. Deep veins included internal jugular, super vena cava, inferior venal cava, brachial, radial, ulnar, iliac, femoral, popliteal and profunda femoris veins. Abdominal specific veins splenic, portal, renal and mesenteric were excluded, since these are part of the portal circulation. We also excluded these superficial veins including the external jugular, cephalic, basilica, median cubital, small saphenous and greater saphenous. Finally thrombophlebitis, arterial, tumor or sinus thrombosis and manmade venous conduits were excluded from consideration as VTE disease. Pulmonary embolism positives relied on computed tomography angiography and ventilation/perfusion (V/Q) scans to define positives or documentation supporting historical diagnoses.
Natural Language Processing Definition:
Notes from the Synthetic Derivative for the BioVU subset were processed by the KnowledgeMap Concept Identifier (KMCI), a general purpose NLP program in use at Vanderbilt for the last ten years.(12) For this study, we preprocessed the documents using the SecTag section tagging software to identify subcomponent sections of clinical notes such as History of Present Illness or Physical Exam. Negation detection was completed with a modified version of NegEx.(13–14) KMCI derives lists of concepts (e.g., CUIs) from the Unified Medical Language System (UMLS) with local context of negation attributes and section location. The KMCI output was loaded into a database for secondary processing.
We curated a list of CUI’s for the VTE algorithm, beginning from a sample review of medical school lectures for VTE previously processed in the KnowlegeMap curriculum management system. This system contains 24,000 documents over 10 years of the Vanderbilt School of Medicine curriculum. We expanded the list of related VTE CUIs by manual query of the database for distinct concepts. The final list contained twenty-seven VTE concepts. (See appendix 2 for list.)
Initial database review of the original VTE text strings revealed instances where these CUIs were misidentified (e.g. “PE” for “Physical Exam” and not for Pulmonary Embolism) and note sections with non-patient specific VTE information such as family history. Based upon these findings, all VTE CUIs defined by KMCI as non-negated concepts, without an attribute of “other experiencer”, “risk” or “possible” and not from note sections related to physical exam, family medical history or social history were considered first round positive for VTE. We also excluded medical student notes in general after identifying extensive discussions on VTE disease that did not describe patient specific attributes.
Further study identified two classes of false positives in the training set. The first category of false positives consisted of positive assertions related to discussions of procedural risk, prophylaxis and diagnostic consideration. These assertions represented modifiers other than typical negation, such as a patient being “at risk” for a DVT, a “possible complication” (e.g., from surgery), or a patient needing DVT prophylaxis. This class of false positives was also constrained by a large number of concepts separating the VTE CUI from the negation value.
The second class of false positives derived from overlap of VTE CUIs with non-VTE thrombosis. The second category comprised other thrombosis conditions for which the CUI was not specific enough for VTE disease, such as splenic vein thrombosis, which did not meet our VTE gold standard definition based on venous location.
To further refine our algorithm, we implemented a secondary negation process to remove false positive cases. Since KMCI concepts are broken into subcomponent text strings of their sentences we needed whole sentence level reconstruction to perform a secondary negation. We developed a program to reassemble the VTE containing sentences CUIs then negated concepts with regular expressions for risk, prophylaxis, testing indications and non-VTE thrombosis. For the secondary algorithm, any sentences not negated on this final pass were considered true cases. For this set of negation modifiers, we included the list of status modifiers previously found relevant for colonoscopy reports for presence of procedure.(15)
We iteratively revised the algorithm and compared performance to the gold standard results to determine sensitivity and positive predictive value. We also evaluated patients without ICD-9 codes, which were reviewed as previously described as either true or false for VTE. Documentation was divided into clinical notes and Problem List (PL) only sets. Results were presented for clinical notes only, problem lists only and problem list and notes together.
Evaluation:
Evaluation of the algorithm was divided into two sets. The first replicated application of NLP to an ICD-9 population. A random set of 2275 individuals not a part of the original SD set were processed identically based initially on the presence of a single VTE ICD-9 code. The second evaluation looked at effectiveness of the NLP algorithm for the remaining training set population that did not have a VTE ICD-9. For non-ICD-9 review, the NLP algorithm was applied to the problem lists of the remaining training set individuals identifying a total of 100 non-ICD-9 patients.
Results
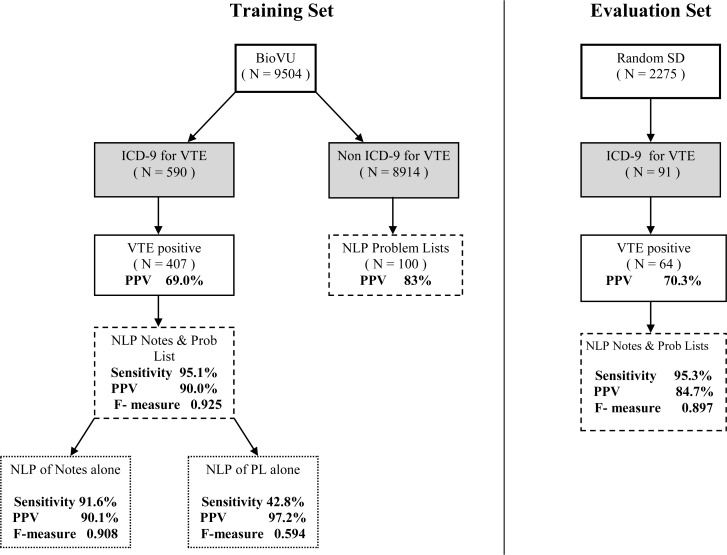
From the 9504 individuals processed from the original BioVU set, 590 individuals had at least one ICD-9 for VTE. Of these patients, 407 were positive for our gold standard VTE definition, giving a positive predictive value (PPV) for billing codes alone of 69.0%. The addition of our NLP algorithm to the notes from ICD-9 positive individuals increased the PPV to 90.1%. and sensitivity to 91.6% with a F-measure of 0.908. The PPV of PL alone was 97.2% with an overall sensitivity 42.8% and a F-measure of 0.594. NLP processed Problem Lists added an addition 14 (3.4%) historical cases that otherwise would not have been captured. The combination of Notes and PL together identified 387 of the original 407 VTE positive cases from the training set giving a Sensitivity of 95.1% and PPV of 90.0% (see Figure 1).
Figure 1.
Flowchart of NLP Algorithm for Training and Evaluation Sets. PL=Problem lists (free-text lists of diagnoses and procedures).
Of the 100 non-ICD-9 Problem List alone identified individuals, manual review of these cases found that 83 were VTE disease positive giving a PPV for these individuals of 83.0%, all historical examples. For the entire training set, 97 individuals or 19.8% of all VTE cases were identified as historical only.
In the evaluation set, 91 individuals were identified with at least one VTE ICD-9 diagnosis. Manual review of cases in the SD identified 64 who met the gold standard VTE definition giving a PPV of 70.3% for billing codes alone. Identical NLP processing of these patients consisted of concept identification by KMCI, followed by primary negation, VTE CUI sentence selection and then positive assertion and thrombosis overlap secondary negation. The PPV in the evaluation set was 84.7%, sensitivity of 95.3 and F-measure of 0.897 with the NLP algorithm finding 61 out of 64 of the true positive patients.
Discussion
Venous thromboembolic disease is a significant cause of morbidity and mortality. The known risk factors for VTE development stasis, injury and hypercoagulability are limited by the unknown risk factors that also contribute to disease development.(16–17) This study presents a general population NLP algorithm utilizing billing codes and problem lists to phenotypically define VTE.
Defining VTE disease separate from procedural risk, hospital prophylaxis or diagnostic consideration is challenging due to the frequency these later concepts are found in the medical record. The significant morbidity risks related to VTE disease drives extensive care processes especially in high-risk populations that leads to extensive documentation in hospital and ED records which are often not related to actual disease in the individual. This documentation often takes the form of a positive assertion that is not accessible to classical negation techniques.
In the algorithm presented here, we applied a stepwise negation technique using classical negation followed by recapitulation of VTE concept sentences treated to another round of negation for positive assertions and overlap of VTE CUIs.
Historical cases accounted for more than 20% of the total true VTE diagnoses. The majority of these did not have a VTE ICD9 associated with them and were primarily identified through the Problem List. For these instances, NLP of our unstructured text Problem List allowed inclusion of these cases that would not have been identified otherwise. The problem list in StarPanel is directly defined by providers for purposes of clinical care only as opposed to ICD9 codes which reflects a more varied needs of billing requirements. As such its PPV was much higher than typically associated with ICD9 codes reflecting it clinical provenance and active ongoing use by providers to help to direct patient care.
In our study here, over 2500 individuals were positive in some form for a VTE concept, whereas only 490 had disease. As such an NLP algorithm provides a method to identify all instances of disease in a large cohort from the medical chart for which manual review would be arduous.
Another challenge in developing the NLP VTE algorithm included overlap of thrombosis concepts with non-VTE disease. This limitation made patient level DVT/PE diagnosis more difficult to confirm. For both problems of frequency of nondisease VTE documentation and overlap of VTE concepts were addressed by a two-step negation process. In the first step, classically defined negated concepts and concepts possibly related to family or social history were removed from consideration. In the second step, the remaining “positive” statements of VTE, were evaluated for other modifying words.
Discussions of procedure risk represents a special case of EHR documentation that is often templated and verbose making correlation of the VTE noun with the risk verb (sometimes 5–6 lines, and >40 concepts, away) more difficult in a traditional limited window approach to connect nouns with verbs. Our sentence-level approach also allowed for negation of this type of risk documentation.
VTE disease identification performed best when derived from the VTE ICD-9 cohort and then refined by further NLP processing of both hospital notes and problem lists together. Problem lists alone showed the best PPV as would be expected for the specificity of this subtype of documentation. Manual review of the non ICD-9 PL notes confirmed that the majority of these concepts were historical.
Causes for failure of the NLP algorithm to remove false positive cases, were often related to overlap of thrombosis concept with nonvenous thrombotic disease and positive assertion sentences that were not negatable. Given the nature of VTE concepts in the EHR, this limitation constrains the maximal effectiveness of our NLP algorithm. Limitations of this study include the possibility over fitting of our secondary negation model by including too many objects and implementation at only one institution. The similar F-measure between the training and evaluation sets suggests a similar consistency of clinical documentation for this problem.
The secondary negation of positive assertion items fell into four categories, concepts related to risk of procedures, prophylaxis, differential diagnosis discussion and overlap of thrombosis concepts. The main components to our VTE NLP algorithm relate to a two-step process of ICD-9 or Problem List for non-ICD-9 initial identification of patients followed by classical negation and then positive assertion negation.
The complex nature of VTE concept presentation within this EHR demonstration constrains this phenotype algorithm to a heuristic model. Consequently a limitation of this model is its dependence on the variability of text formatting of VTE concepts within the EHR.
Conclusion:
This ICD-9 and NLP algorithm provides a method to identify patients retrospectively with venous thromboembolic disease. The use of ICD-9s as a precursor for cohort determination is more effective than standalone NLP for disease refinement. The density of VTE concepts in hospital documentation especially, warrants a two-step negation process for both negative, relatively positive assertions and CUI concept overlap. This work provides a foundation for further larger scale phenotype defined genetic studies.
Appendix 1:
| VTE codes included: | |
| V12.51 | Personal history of venous thrombosis and embolism |
| 453, 453.0, 453.1, 453.2 | |
| 453.4 | Deep vein thrombosis, unspecified |
| 453.40 | Venous embolism and thrombosis of unspecified deep vessels of Lower extremity |
| 453.41 | Venous embolism and thrombosis of deep vessels of proximal lower Extremity |
| 453.42 | Deep Vein thrombosis, distal |
| 453.8 | Embolism and thrombosis of other specified veins |
| 453.81, 453.82, 453.83, 453.84, 453.85, 453.86, 453.87, 453.89 | |
| 453.6, 453.50, 453.75, 453.51, 453.79, 453.77, 453.52, 453.5 | |
| 453.82, 453.7, 453.71, 453.73, 453.74, 453.76 | |
| 415.1 | Pulmonary embolism and infarction |
| 415.11 | Iatrogenic pulmonary embolism and infarction |
| 415.19 | Other pulmonary embolism and infarction |
| Excluding all | |
| 452 | Portal vein thrombosis |
| 451.12 | Septic pulmonary emboli |
Appendix 2: NLP Definition
- Combine CUI definitions with clinically indexed notes processed by KnowledgeMap
- Using CUIs: 34065, 149871, 340708, 856721, 741877, 743004, 877687, 877624, 40038, 40053, 40046, 151946, 42487, 333203, 333204, 392108, 1456118, 151950, 589110, 877618, 1290394, 392108, 87086, 272416, 459853, 38834, 42487
- Excluding clinical sections denoted by Family History or Social History
- Excluding CUI with negation attributes: other_experiencer|negate|negphrase|risk|possible
- Secondary VTE Evaluator
- Reassembles sentence structure
- Negates original string, other non-VTE thrombosis and positive assertion with regular expressions
References
- 1.Snow V, Qaseem A, Barry P, Hornbake ER, Rodnick JE, Tobolic T, Ireland B, Segal J, Bass E, Weiss KB, Green L, Owens DK, The Joint American College Of Physicians/american Academy Of Family Physicians Panel On Deep Venous Thrombosis/pulmonary Embolism Management of Venous Thromboembolism: A Clinical Practice Guideline from the American College of Physicians and the American Academy of Family Physicians. Ann Fam Med. 2007 Jan;5(1):74–80. doi: 10.1370/afm.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spencer FA, Emery C, Joffe SW, Pacifico L, Lessard D, Reed G, Gore JM, Goldberg RJ. Incidence rates, clinical profile, and outcomes of patients with venous thromboembolism. The Worcester VTE study. J. Thromb. Thrombolysis. 2009 Nov;28(4):401–409. doi: 10.1007/s11239-009-0378-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.White RH, Henderson MC. Risk factors for venous thromboembolism after total hip and knee replacement surgery. Curr Opin Pulm Med. 2002 Sep;8(5):365–371. doi: 10.1097/00063198-200209000-00004. [DOI] [PubMed] [Google Scholar]
- 4.Kanaan AO, Silva MA, Donovan JL, Roy T, Al-Homsi AS. Meta-analysis of venous thromboembolism prophylaxis in medically Ill patients. Clin Ther. 2007 Nov;29(11):2395–2405. doi: 10.1016/j.clinthera.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 5.Proctor MS, Wainess RM, Henke PK, Upchurch GR, Wakefield TW. Venous Thromboembolism: Regional Differences in the Nationwide Inpatient Sample, 1993 to 2000. Vascular. 2004 Nov;12(6):374–380. doi: 10.1258/rsmvasc.12.6.374. [DOI] [PubMed] [Google Scholar]
- 6.Zhan C, Battles J, Chiang Y-P, Hunt D. The validity of ICD-9-CM codes in identifying postoperative deep vein thrombosis and pulmonary embolism. Jt Comm J Qual Patient Saf. 2007 Jun;33(6):326–331. doi: 10.1016/s1553-7250(07)33037-7. [DOI] [PubMed] [Google Scholar]
- 7.Henderson KE, Recktenwald A j, Reichley RM, Bailey TC, Waterman BM, Diekemper RL, Storey PE, Ireland BK, Dunagan WC. Clinical validation of the AHRQ postoperative venous thromboembolism patient safety indicator. Jt Comm J Qual Patient Saf. 2009 Jul;35(7):370–376. doi: 10.1016/s1553-7250(09)35052-7. [DOI] [PubMed] [Google Scholar]
- 8.Evans RS, Linford LH, Sharp JH, White G, Lloyd JF, Weaver LK. Computer Identification of Symptomatic Deep Venous Thrombosis Associated with Peripherally Inserted Central Catheters. AMIA Annu Symp Proc; 2007; 2007. pp. 226–230. [PMC free article] [PubMed] [Google Scholar]
- 9.Roden DM, Pulley JM, Basford MA, Bernard GR, Clayton EW, Balser JR, Masys DR. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther. 2008 Sep;84(3):362–369. doi: 10.1038/clpt.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ritchie MD, Denny JC, Crawford DC, Ramirez AH, Weiner JB, Pulley JM, Basford MA, Brown-Gentry K, Balser JR, Masys DR, Haines JL, Roden DM. Robust replication of genotype-phenotype associations across multiple diseases in an electronic medical record. Am J Hum Genet. 2010 Apr;86(4):560–572. doi: 10.1016/j.ajhg.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denny JC, Ritchie MD, Basford MA, Pulley JM, Bastarache L, Brown-Gentry K, Wang D, Masys DR, Roden DM, Crawford DC. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics. 2010 May;26(9):1205–1210. doi: 10.1093/bioinformatics/btq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denny JC, Smithers JD, Miller RA, Spickard A. “Understanding” medical school curriculum content using KnowledgeMap. J Am Med Inform Assoc. 2003 Aug;10(4):351–362. doi: 10.1197/jamia.M1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Denny JC, Spickard A, 3rd, Johnson KB, Peterson NB, Peterson JF, Miller RA. Evaluation of a method to identify and categorize section headers in clinical documents. J Am Med Inform Assoc. 2009 Dec;16(6):806–815. doi: 10.1197/jamia.M3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chapman WW, Bridewell W, Hanbury P, Cooper GF, Buchanan BG. A Simple Algorithm for Identifying Negated Findings and Diseases in Discharge Summaries. Journal of Biomedical Informatics. 2001 Oct;34(5):301–310. doi: 10.1006/jbin.2001.1029. [DOI] [PubMed] [Google Scholar]
- 15.Denny JC, Peterson JF, Choma NN, Xu H, Miller RA, Bastarache L, Peterson NB. Extracting timing and status descriptors for colonoscopy testing from electronic medical records. Journal of the American Medical Informatics Association. 2010 Jul;17(4):383–388. doi: 10.1136/jamia.2010.004804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lijfering WM, Rosendaal FR, Cannegieter SC. Risk factors for venous thrombosis - current understanding from an epidemiological point of view. Br J Haematol. 2010 Jun;149(6):824–833. doi: 10.1111/j.1365-2141.2010.08206.x. [DOI] [PubMed] [Google Scholar]
- 17.Crowther MA, Kelton JG. Congenital thrombophilic states associated with venous thrombosis: a qualitative overview and proposed classification system. Ann Intern Med. 2003 Jan;138(2):128–134. doi: 10.7326/0003-4819-138-2-200301210-00014. [DOI] [PubMed] [Google Scholar]