Abstract
A retrospective clinical evaluation in a cohort of 73 patients receiving stable anticoagulation therapy showed that the addition/elimination of amiodarone resulted in a 6–65% change in warfarin dose requirement. To evaluate the roles of amiodarone and its circulating metabolites in this highly variable inhibitory drug interaction, an LC-ESI+ MS/MS assay was developed for the quantitation of low concentrations of these compounds in human plasma, utilizing newly synthesized deuterated analogs as internal standards. KI’s were determined for the inhibition of (S)-warfarin 7-hydroxylation in human liver microsomes, by parent drug and metabolites, and unbound drug fractions (fu) were measured so that the ratio of unbound plasma concentration to the microsomal KI for unbound drug ([I]u/KI,u) could be calculated. From these ratios, we predict a minor metabolite, N,N-didesethylamiodarone, to be a major contributor to the drug interaction between warfarin and amiodarone.
Keywords: drug-drug interactions, plasma, in vitro, in vivo, pharmacokinetics, warfarin, inhibition, adverse drug reactions, metabolites
Introduction
Amiodarone (AMIO) was originally introduced as an anti-anginal drug over 40 years ago (1), but is now used therapeutically as an effective Class III antiarrhythmic agent (2, 3). AMIO is associated with numerous potential side effects that include pulmonary toxicity (4, 5), hepatotoxicity (6) and thyroid dysfunction (7). Additionally, there have been reports of adverse drug interactions when AMIO is administered together with a wide range of other therapeutic agents including theophylline (8), flecainide (9), cyclosporine A (10, 11) and dextromethorphan (12). However, the most common, as well as potentially the most dangerous drug interaction that AMIO exhibits is its potentiation of the anticoagulant effect of warfarin, which greatly increases the patient’s risk of hemorrhage (8, 13).
AMIO is frequently used for rhythm control in patients with atrial fibrillation (AF). Stroke is a major concern in patients with AF and since warfarin is the preferred treatment for stroke prevention, AMIO is therefore often co-administered with warfarin (14). An interaction always occurs between these two drugs (15, 16) that typically necessitates a dose reduction in warfarin of 25–40% depending on the amiodarone maintenance dose (15, 16).
Early studies by Trager, O’Reilly and co-workers demonstrated that AMIO inhibits the clearance of both (S)- and (R)-warfarin (17) and decreases the formation clearances of (S)-warfarin metabolites to a greater extent than (R)-warfarin metabolites (18). However, AMIO proved to be a relatively weak inhibitor of (S)-warfarin metabolism in human liver microsomes (HLM) (18) and AMIO’s N-dealkylated metabolite, monodesethylamiodarone (MDEA), is 40–90 times more potent an inhibitor of CYP2C9 than the parent drug (19, 20). These observations are significant because CYP2C9 is the major P450 that terminates the pharmacological activity of warfarin (21). Moreover, MDEA is present in human plasma at a concentration approximately equal to that of the parent drug (~1–5 μM) (4, 22). Indeed, there are in vivo data which suggest that plasma MDEA concentration may be a better predictor of the change in INR than plasma AMIO concentration in patients receiving both drugs (23). These observations prompt the hypothesis that a major mechanism of the warfarin-AMIO interaction is inhibition of CYP2C9-mediated (S)-warfarin metabolism by MDEA.
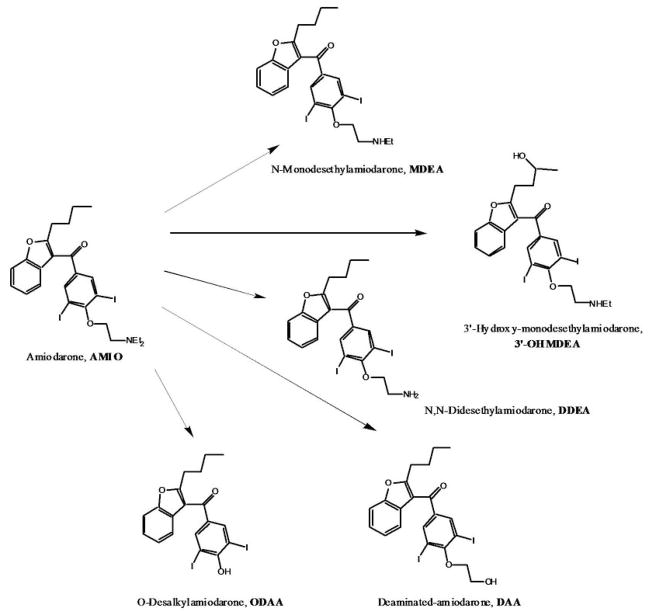
Although MDEA is the major metabolite of AMIO in humans, a number of others have recently been identified in plasma (Figure 1) (24). The most prominent of these metabolites include 3′-hydroxy-N-monodesethylamiodarone (3′-OHMDEA), N,N-didesethylamiodarone (DDEA) and deaminated amiodarone (DAA). Another potential product of AMIO metabolism is the O-desalkylated derivative (ODAA) (25). It is possible that one or more of these minor AMIO metabolites could contribute to the warfarin drug interaction and, in fact, several close analogs of ODAA have been shown to be low nanomolar inhibitors of CYP2C9 (26). Delineation of specific inhibitory AMIO metabolites may also assist in a better understanding of inter-individual variability in the magnitude of this drug-drug interaction (DDI).
Figure 1.
Amiodarone metabolites in human plasma.
The aims of this research were threefold. Firstly, a retrospective study was performed to determine mean and range of warfarin dose change required to maintain an INR of 2.0 – 3.0 after amiodarone was added or discontinued in a cohort of patients undergoing stable warfarin therapy. Next, stabilized plasma concentrations, [I], of AMIO and five of its circulating metabolites were determined in several warfarin patients undergoing combination therapy. In order to measure low metabolite concentrations in vivo, we developed a new, highly sensitive stable-label isotope LC-MS assay that necessitated the synthesis of deuterium-labeled internal standards. Finally, in vitro inhibition experiments were carried out with AMIO and its metabolites to measure the KI against CYP2C9-mediated (S)-warfarin 7-hydroxylation in HLM. To best assess the likelihood of an in vivo drug interaction, we measured the free fraction of AMIO and its various metabolites in HLM and in plasma in order to calculate the respective [I]u/KI,u values for parent drug and circulating metabolites.
Results
In vivo Effect of AMIO on Warfarin Dose Requirement
Over the six year period of the retrospective clinical evaluation, 73 case studies were identified fitting the parameters of our analysis for an out of range INR attributed to a warfarin-AMIO drug interaction which occurred upon either addition or elimination of AMIO therapy to patients undergoing stable warfarin treatment. Results of the analysis are described in Table 1. The mean change in warfarin dose required to maintain an INR of 2–3 was 25.6%. Warfarin dosing change requirements were highly variable, with a range of 5.9% to 65%.
Table 1.
Demographic characteristics of patients, and change in warfarin dose required to maintain INR 2.0 – 3.0 after addition or discontinuation of amiodarone.
| All Episodes | Addition of Amiodarone | Discontinuation of amiodarone | |
|---|---|---|---|
| N | 73 | 46 | 27 |
| Mean Age (range) | 62 (19–92) | 62 (19–92) | 62 (22–88) |
| % male | 78% | 78% | 78% |
| % white | 90% | 89% | 93% |
| Mean warfarin dose prior to addition/discontinuation of amiodarone (range) | 5.1 mg/day (1.9–10) | 3.9 mg/day (1.3–8.2) | |
| Mean warfarin dose after addition/discontinuation of amiodarone (range) | 3.7 mg/day (1.5–8.6) | 4.8 mg/day (1.8–10) | |
| Change in warfarin dose after addition or discontinuation of amiodarone (mean; [range]) | 25.6% (5.9%–65.0%) | 26.9% (5.9%–65.0%) | 24.8% (10.0–50.0%) |
Synthesis of AMIO Metabolite Standards
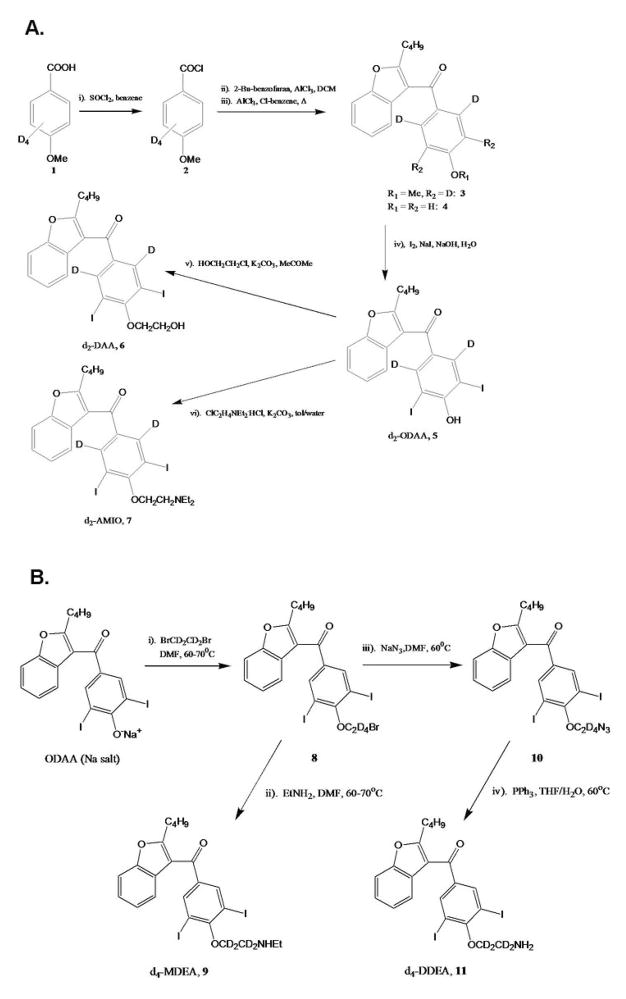
To enable the most rigorous quantitative assessment of AMIO metabolite concentrations in vivo and in vitro, we adopted stable-isotope methodology for all the LC/MS analyses. Literature procedures exist for the synthesis of unlabeled AMIO and many of the drug’s known human plasma metabolites (27–30), but the syntheses of deuterium labeled standards for AMIO, MDEA, DDEA, ODAA and DAA are reported here for the first time. The deuterium source for isotope-labeled AMIO (7), as well as its deaminated (6) and O-desalkylated (5) derivatives, was p-methoxy-(2,3,5,6-d4)-benzoic acid, 1 (Scheme 1A). A slightly different methodology was used in the syntheses of the isotopically-labeled analogs of MDEA and DDEA, 9 and 11, which were both derived from unlabeled ODAA utilizing 1,2-dibromo-d4-ethane as the deuterium source (Scheme 1B). These procedures provided the required stable-isotope internal standards in good yield and purity, and with high deuterium contents (>95%) that reflected those of the labeled starting materials.
Scheme 1.
Synthesis of deuterium-labeled AMIO and circulating AMIO metabolites
Measurement of AMIO Parent Drug and Metabolite Concentrations in Human Plasma
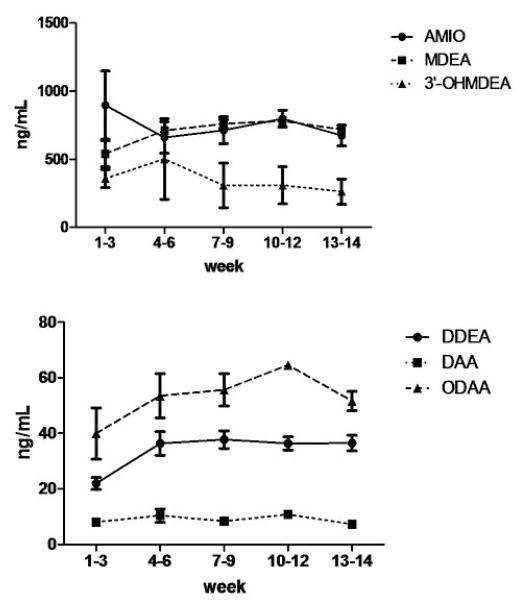
The deuterated standards of AMIO and its metabolites were used to develop an LC-ESI+ MS/MS assay for the quantitation of these compounds in the plasma of clinical patients undergoing concomitant warfarin and AMIO therapy (Figure 2). Blood was drawn from three patients at various time points up to 14 weeks after initiation of AMIO treatment. Parent drug and metabolite plasma concentrations were seen to plateau at around 8 weeks after addition of AMIO therapy to a stabilized warfarin regime (Figure 3), therefore, average drug and metabolite plasma concentrations were calculated for each of the patients between 8 and 14 weeks, and these values were then averaged for the 3 patients to determine the mean steady state plasma concentrations for AMIO, MDEA, DDEA, 3′-OHMDEA, DAA and ODAA (1.09 ± 0.09, 1.16 ± 0.08, 0.059 ± 0.003, 0.42 ± 0.34, 0.015 ± 0.003 and 0.11 ± 0.01 μM, respectively, after converting units from ng/mL, Table 2). The values for AMIO, MDEA, 3′-OHMDEA, DDEA and DAA are similar to those reported in an earlier study by Ha et al., (24). Plasma ODAA concentrations are reported here for the first time.
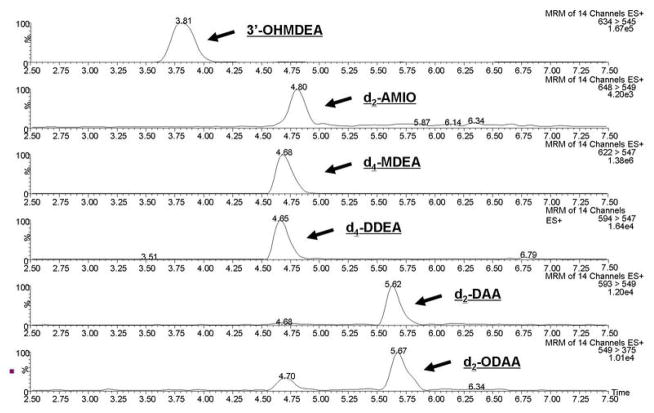
Figure 2.
LC-ESI+ MS/MS trace analysis of a standard mix containing d2-ODAA, d2-DAA, d2-AMIO, d4-MDEA, d4-DDEA and unlabeled 3′-OHMDEA. No overlap of unlabeled material into the channels measuring deuterated standards could be seen above background within the experimental concentration ranges.
Figure 3.
Average plasma concentration vs time curves for AMIO and its circulating metabolites in patients (n = 3), already undergoing warfarin treatment, who were started on AMIO therapy. Error bars denote standard error measurements. AMIO dose varied from 230 to 500 mg/day for each patient with an overall mean of 350 mg/day.
Table 2.
Comparison of [I]/Ki and [I]u/Ki,u values for the inhibition of (S)-warfarin 7-hydroxylation by AMIO and its circulating human metabolites.
| Inhibitor | [I] (μM)a | KI (μM)b | fu(plasma) (%)c | fu(mics) (%)d | [I]/KI | [I]u/KI,ue |
|---|---|---|---|---|---|---|
| AMIO | 1.09 ± 0.09f | 39 ± 14 | 3.3 ± 0.8 | 1.4 ± 0.2 | 0.028 | 0.066 |
| MDEA | 1.16 ± 0.08 | 5.9 ± 0.9 | 2.0 ± 0.4 | 1.2 ± 0.4 | 0.20 | 0.33 |
| DDEA | 0.059 ± 0.003 | 0.053 ± 0.008 | 4.2 ± 0.4 | 1.1 ± 0.3 | 1.1 | 4.30 |
| ODAA | 0.11 ± 0.01 | 0.032 ± 0.006 | 0.13 ± 0.01 | 5.0 ± 2.0 | 3.4 | 0.089 |
| DAA | 0.015 ± 0.003 | 2.0 ± 0.3 | 4.9 ± 1.7 | 1.7 ± 0.1 | 0.008 | 0.022 |
| 3′-OHMDEA | 0.42 ± 0.34 | 5.2 ± 2.0 | 1.8 ± 0.3 | 17 ± 2.8 | 0.081 | 0.009 |
[I] represents mean plateau inhibitor plasma concentration (i.e. at 8 to 14 weeks after initiation of AMIO therapy) and is presented as the mean value derived from n = 3 patients (calculated from Figure 3, with units converted from ng/mL).
KI values were determined in HLM at 0.25mg/mL microsomal protein.
Free fraction measured in human plasma.
Free fraction measured in HLM at 0.25mg/mL microsomal protein.
[I]u = [I] * fu(plasma); KI,u = KI * fu(mics)
Error measurements represent standard deviations of the mean.
Inhibition of (S)-Warfarin 7-Hydroxylation in HLM
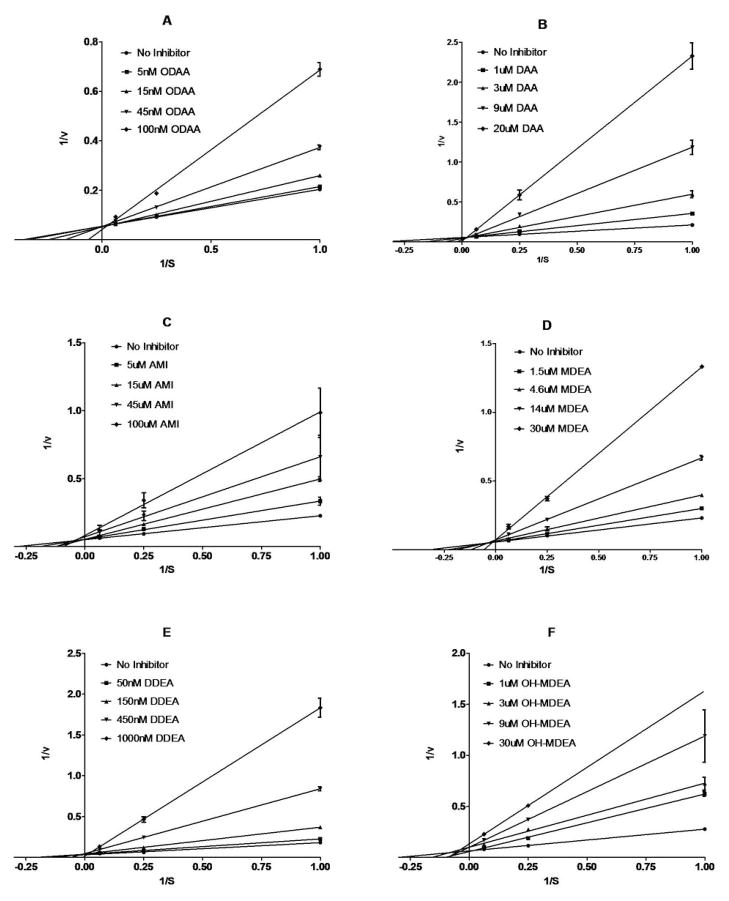
Kinetic studies were carried out to determine the enzymatic mechanism of inhibition and to measure the KI values for the inhibition of (S)-warfarin metabolism by AMIO and its metabolites, in pooled HLM. Both ODAA (Figure 4A) and DAA (Figure 4B) were found to be purely competitive inhibitors of (S)-warfarin 7-hydroxylation, with KI’s of 0.032 ± 0.006 and 2.0 ± 0.3 μM, respectively, while AMIO, MDEA, DDEA and 3′-OHMDEA all appeared to be mixed inhibitors of warfarin metabolism in HLM (Figure 4C–F). The respective KI values for these four nitrogen-bearing compounds, determined from Dixon plots, were calculated to be 39 ± 14, 5.9 ± 0.9, 0.053 ± 0.008 and 5.2 ± 2.0 μM, respectively. Due to extensive non-specific protein binding, IC50 values for the test compounds varied significantly, and linearly, with the amount of microsomal protein that was used in the incubations (data not shown). Therefore, the kinetic estimates presented here are KI (apparent) values and need to be corrected for the free fractions available in metabolic incubations containing 0.25 mg/mL microsomal protein.
Figure 4.
Lineweaver-Burke (L-B) reciprocal plots of the velocity of formation of (S)-7-hydroxywarfarin vs. (S)-warfarin concentration (1, 4 and 16 μM) in the presence of fixed concentrations of A) ODAA, B) DAA, C) AMIO, D) MDEA, E) DDEA or F) 3′-OHMDEA in incubations with HLM at 0.25 mg/mL microsomal protein concentration. Ki’s of 0.032 ± 0.006, 2.0 ± 0.3, 39 ± 14, 5.9 ± 0.9, 0.053 ± 0.008 and 5.2 ± 2.0 μM were determined for A–F, respectively, from replotting the data as the inverse of the velocity of formation of (S)-7-hydroxywarfarin vs. inhibitor concentration (Dixon plots not shown). Data points are mean values derived from triplicate incubations with standard deviations denoted by error bars.
Determination of Fraction Unbound (fu) for Parent Drug and Metabolites
Protein binding experiments demonstrated that AMIO and the majority of its metabolites are very highly bound in both HLM and human plasma. In experiments with HLM, carried out at 0.25 mg/mL microsomal protein, fu values of 1.4 ± 0.2, 1.2 ± 0.4, 1.1 ± 0.3 and 1.7 ± 0.1% were calculated for AMIO, MDEA, DDEA and DAA, while ODAA and 3′-OHMDEA exhibited fu values of 5.0 ± 2.0 and 17 ± 2.8%, respectively. The degree to which most of these compounds bind to plasma protein is slightly less than their protein binding to HLM with calculated plasma fu values of 3.3 ± 0.8, 2.0 ± 0.40, 4.2 ± 0.4 and 4.9 ± 1.7% for AMIO, MDEA, DDEA and DAA, respectively. ODAA and 3′-OHMDEA do not fit this trend as both compounds are much more tightly bound to plasma protein than to microsomal protein, exhibiting plasma fu values of 0.13 ± 0.01 and 1.8 ± 0.3%, respectively (Table 2).
Similar protein binding experiments for these compounds were carried out in the presence of either 4 μM (S)-warfarin, for studies in HLM, or 2 μg/mL rac-warfarin (approximate physiological concentration), for studies in human plasma. Warfarin was found not to affect metabolite protein binding in either HLM or plasma as the fraction unbound values remained essentially unchanged from the numbers reported above, showing less than 2-fold variation in all cases (data not shown).
Determination of [I]/KI and [I]u/KI,u for Parent Drug and Metabolites
[I]/KI, defined here as the total mean plasma drug concentration for the three test subjects after reaching plateau concentrations, i.e. 8–14 weeks after initiation of therapy, divided by the in vitro KI (apparent), measured for the inhibition of (S)-warfarin 7-hydroxylation in HLM, was calculated to be 0.028, 0.20, 1.1, 3.4, 0.0076 and 0.081 for AMIO, MDEA, DDEA, ODAA, DAA and 3′-OHMDEA, respectively (Table 2). [I]u/KI,u is defined as the ratio of unbound plasma drug concentration ([I]u = [I] * fuplasma) to the microsomal KI corrected for unbound microsomal drug concentration (KI,u = KI * fumics). These values were determined as 0.066, 0.33, 4.3, 0.089, 0.022 and 0.0086 for AMIO, MDEA, DDEA, ODAA, DAA and 3′-OHMDEA, respectively (Table 2).
Discussion
The warfarin-AMIO interaction is known to be due to metabolic inhibition of the P450 enzymes responsible for the clearance of warfarin, mainly CYP2C9 (18). In patient studies, we confirmed (i) the wide inter-individual variability in the magnitude of the warfarin-AMIO interaction reported initially by Kerin et al., (15), and (ii) the plasma concentration ranges of AMIO and several metabolites reported earlier by Ha et al. (24). Additionally, in these studies, ODAA was quantified (60.5 ± 5.6 ng/mL, 0.11 ± .01 μM) for the first time in human plasma (although very recently, Deng and coworkers identified ODAA in human bile and as a product of metabolic incubations of AMIO with HLMs (31)). The results of the in vitro inhibition and protein binding experiments with AMIO and its metabolites that we report here serve as a means of elucidating details of the molecular mechanism behind this in vivo interaction.
AMIO is known to be highly protein bound in human plasma and the fu measured in plasma for this study (3.3 ± 0.8%) is in excellent agreement with the value reported previously by Lalloz, et al., (3.7 ± 0.6%) (32). Both values were determined by the ultracentrifugation method for measuring free drug fraction. The plasma free fractions for the metabolites of amiodarone - MDEA, 3′-OHMDEA, DDEA, ODAA and DAA, which varied from ~0.1–5% – have not been previously reported.
Kinetic experiments carried out in HLM, at 0.25 mg/mL microsomal protein, yielded KI (apparent) values of 39, 5.9, 5.2, 0.053, 0.032 and 2.0 μM for the inhibition of (S)-warfarin 7-hydroxylation by AMIO, MDEA, 3′-OHMDEA, DDEA, ODAA and DAA. Both the phenol, ODAA, and the alcohol, DAA, were competitive inhibitors of (S)-warfarin metabolism in HLM while the amine compounds AMIO, MDEA, 3′-OHMDEA and DDEA were mixed inhibitors of (S)-warfarin 7-hydroxylation (Figure 4). Ohyama, et al., previously reported AMIO to be a noncompetitive inhibitor and MDEA to be a mixed inhibitor of (S)-warfarin metabolism in microsomes derived from human B-lymphoblastoid cells co-expressing CYP2C9 and P450 reductase, with KI values of 95 and 2.3 μM (20).
At present, the U.S. Food and Drug Administration guidance for the use of in vitro data to predict in vivo DDIs relies on the comparison of total inhibitor plasma concentration to in vitro inhibition constant, generally measured in microsomes, ([I]/KI) (33). Compounds which exhibit [I]/KI values < 0.1 are considered remote possibilities to cause an interaction, whereas compounds with [I]/KI between 0.1 and 1.0 and [I]/KI > 1.0 are, respectively, considered possible and likely contributors to potential in vivo interactions (33, 34). Using this measure, our data suggest that both DDEA and ODAA are likely to cause a DDI with warfarin, while MDEA is a possible contributor.
There are, however, potential problems with using [I]/KI for prediction of in vivo interactions, especially for compounds that are highly protein-bound in plasma and microsomes. For these compounds, [I]u/KI,u, which corrects for the amount of free drug available in plasma and also normalizes the microsomal inhibition constant for the amount of free drug available in microsomes, is accepted as a more accurate predictor of a drug’s potential to cause an in vivo interaction (35, 36). In comparing the two ratios, [I]/KI appears to both under-predict the potential of DDEA to cause an interaction while greatly over-predicting the potential in vivo interaction of ODAA, which changes from likely to remote when [I]u/KI,u is assessed. By either predictive method, MDEA is seen as a possible contributor, while AMIO, DAA and 3′-OHMDEA would be judged as unlikely to contribute to the potentiation of the warfarin anticoagulant effect. Also, though warfarin binds very strongly to plasma (fu(serum) = 0.008) (37), the anticoagulant was unable to displace AMIO or its metabolites from plasma protein when incubated at a physiological concentration, showing essentially no effect on fu for any of the compounds tested.
The overall change in intrinsic clearance of (S)-warfarin caused by the drug interaction with AMIO can be predicted according to equation 1.
| (Equation 1) |
A 6-fold increase in area under the plasma concentration vs time curve (AUC) is therefore predicted for (S)-warfarin clearance based on the sum of the [I]/KI ratios of the parent drug and circulating metabolites (Table 2). Serendipitously, if [I]u/KI,u ratios are used instead of [I]/KI, the same 6-fold increase in AUC is predicted, however, the prediction using [I]/KI is dominated by the ODAA ratio while the prediction based on [I]u/KI,u values is due almost entirely to DDEA (Table 2).
The actual increase in the mean AUC for (S)-warfarin caused by the drug interaction with AMIO has been reported to range from 27 to 110% in single warfarin dose studies carried out in small groups (n = 5 or 6) of healthy volunteers (17, 18). The data from our retrospective clinical evaluation showing that introduction/discontinuation of AMIO therapy results in a 6 – 65% change in warfarin dose requirement is in good agreement with this clearance data. However, it is also clear that our in vitro data over-predict the inhibitory effect of the overall drug interaction. Tertiary amines are increasingly recognized as mechanism-based inactivators of P450 following sequential metabolism to C-nitroso metabolites whose nitrogen lone pair binds tightly to the reduced heme (38). Since AMIO and its aminated metabolites are mixed inhibitors of CYP2C9, it is possible that one or more of these compounds could be a mechanism-based inhibitor (MBI) of the enzyme. In fact, Mori, et al. (2009) found AMIO and MDEA to be (weak) MBIs of CYP2C9-mediated diclofenac 4′-hydroxylation, both exhibiting KI’s of inactivation of ~100 μM and kinact values of only ~0.1/min (39). However, given that we conducted our kinetic inhibition experiments with AMIO and MDEA at concentrations that were much lower than the KI values reported by Mori et al. (with one exception - for AMIO itself we used a maximum concentration approximately equal to the reported KI), it seems unlikely that time-dependent inhibition of microsomal CYP2C9 is a complicating feature of the kinetic plots obtained under the conditions used here with either the parent drug or its primary metabolite. Nonetheless, studies are underway to evaluate the potential of each of the AMIO metabolites described here to evoke time-dependent P450 inhibition. It is possible that the discrepancy in our prediction of the DDI may be explained by high levels of drug/metabolite efflux resulting in decreased hepatocyte exposure. The recent findings of Deng and coworkers, identifying all of the known circulating AMIO metabolites, (as well as a host of new metabolites) in human bile lends support to this hypothesis (31)
European Medicines Agency guidelines currently recommend that only metabolites of drugs that circulate at unbound or total molar concentrations of ≥ 20% of the parent drug be tested for the potential to cause DDIs (40). Based on our results, DDEA appears to be the most likely metabolite to potentiate the anticoagulant effect when AMIO and warfarin are given in combination, even though AMIO plasma concentrations were roughly 20-fold higher than DDEA concentrations in the patients studied. From our data it would appear that, at least in the specific case of drugs that contain secondary or tertiary amines, the current prediction system might be improved by the inclusion of mono- and especially di-N-desalkyl derivatives – even if these metabolites circulate at very low concentrations – since they are potentially much stronger P450 inhibitors than the parent drug. Studies are underway to determine whether patient variability in formation of DDEA, alone or in combination with other inhibitory metabolites, explains inter-individual variability in the drug interaction between AMIO and warfarin.
Methods
Retrospective Clinical Evaluation of the AMIO/Warfarin Drug-Interaction
From 7/1/2001 to 6/30/2007, a university-affiliated anticoagulation clinic provided routine outpatient anticoagulation management for 3129 patients taking warfarin. During this 6 year period, a total of 230 incidents in which an out-of-range INR was attributed to the warfarin-amiodarone drug interaction were identified. These incidents were associated with 163 episodes in which amiodarone was added or discontinued in patients taking warfarin, or in which the amiodarone dose was changed. Of these 163 episodes, 90 were excluded from analysis due to either unstable warfarin therapy prior to addition of amiodarone (n=40), an interaction due to change in amiodarone dose (n=27); warfarin and amiodarone having been initiated concurrently (n=14), the duration of concurrent warfarin/amiodarone therapy being less than 1 month (n=6) or no records available (n=3). The 73 remaining patients were mainly White (n=66), with Blacks (n = 3) and Asians (n = 4) also represented in the cohort.
Recruitment of Warfarin Patients for Analysis of Amiodarone Plasma Metabolites
Three patients stabilized on warfarin who were to be initiated on concomitant AMIO therapy were recruited through Cardiology and the Anticoagulation Clinic at the University of Washington Medical Center. Briefly, blood was drawn at regular clinic visits for measurement of INR immediately prior to, and over a 14 week period after, addition of AMIO to the treatment regimen. Introduction of AMIO in the three patients resulted in a 33, 40 or 71% reduction in required warfarin dose. All patients gave informed consent for the study which was approved by the University of Washington IRB.
AMIO Metabolite Quantitative LCMS Assay
LCMS analyses were conducted on a Micromass Quattro Premier XE Tandem Quadrupole Mass Spectrometer (Micromass Ltd., Manchester, U.K.) coupled to an ACQUITY Ultra Performance LC™ (UPLC™) System with integral autoinjector (Waters Corp., Milford, MA). The Premier XE was run in ESI+-MS/MS MRM mode at a source temperature of 100 °C and a desolvation temperature of 34 °C. The cone voltage was set at 25 Volts while the collision energy was set to 40 Volts. The following mass transitions were monitored in separate ion channels: m/z 547 to 373 (for d0-ODAA), 549 to 375 (d2-ODAA), 590 to 547 (d0-DDEA), 594 to 547 (d4-DDEA), 591 to 547 (d0-DAA), 593 to 549 (d2-DAA), 618 to 547 (d0-MDEA), 622 to 547 (d4-MDEA), 646 to 547 (d0-AMIO), 648 to 549 (d2-AMIO) and 634 to 545 (3′-OHMDEA). Parent drug and metabolites were separated using only a Nucleosil 7.5×4.6mm 5μC18-guard cartridge as column (Alltech Assoc., Deerfield, IL), with a flow rate of 0.35mL/min, eluted with a binary solvent system consisting of 5mM NH4OAc (solvent A) and 0.5% formic acid in MeOH (solvent B). The solvent was set at 50% B for 2 minutes and increased linearly to 100% B over the following 3.5 min. From 5.5 to 6.5min, the solvent composition was lowered back down to 40% B where it was maintained for one minute (Figure 1).
Data analyses were carried out on Windows XP-based Micromass MassLynxNT®, v 4.1, software.
Chemical Inhibition Experiments
Incubations were carried out using a pool of HLMs consisting of equal quantities of total microsomal protein obtained from eight different liver samples, prepared as described previously (41). All incubation mixtures contained 250μg/mL microsomal protein from the HLM pool, 1 mM NADPH, 1% v/v of a 100x concentrated methanolic inhibitor stock and S-warfarin, added as substrate (at 1, 4 or 16 μM final concentration), made up to 200 μL total volume with 100 mM potassium phosphate buffer, pH 7.4. The concentrations of inhibitor used in the KI studies varied with the compound (Figure 4). All incubations were carried out in triplicate.
HLMs were preincubated with (S)-warfarin, inhibitor and buffer at 37°C and 70 rpm in a water bath for 2.5 min prior to initiation of the reaction with the addition of NADPH. After 30 min incubation the reactions were quenched by the addition of 5 μL of 70% perchloric acid and 3 ng of 7-ethoxycoumarin was added as internal standard. The reaction mixtures were vortexed for 30 sec and centrifuged at 13,400 rpm for 5 min. Supernatants were then analyzed for 7-hydroxywarfarin content by HPLC using fluorescence detection. A time course study revealed that, under the experimental conditions, 7-hydroxywarfarin production was essentially linear out to 30 minutes.
HPLC-Fluorescence Assay for Microsomal CYP2C9 Activity
HPLC was performed on a Shimadzu system equipped with two LC10ADvp pumps, a CBM-20A communication bus module, an RF-10AXL fluorescence detector and an SIL-20AHT autosampler (Shimadzu Scientific Instruments, Inc., Columbia, MD) using a Nucleosil 5μ, 4.6 × 100 mm C18 HPLC column (Macherey-Nagel, Bethlehem, PA), with a flow rate of 1.3mL/min. The analytical method was isocratic, set at 38% solvent B (MeCN, solvent A = 0.5% phosphoric acid) with a run time of 10 min. The fluorescent excitation wavelength was set at 320 nm and the emission was monitored at 415 nm. Data acquisition and analyses were performed on LCsolutions® software (v 1.2, Shimadzu).
Determination of Fraction Unbound (fu) in HLM and in Plasma
Protein binding of AMIO, and its metabolites, was measured in both human plasma and in pooled HLM by ultracentrifugation (42, 43) using a TLA-100 benchtop ultracentrifuge with a TLA-100 rotor (Beckman-Coulter, Palo Alto, CA). AMIO, or one of its metabolites, was spiked (as 100x concentrated methanol stocks), into samples of either HLM (0.25 mg/mL microsomal protein in 100mM KPi buffer, pH 7.4) or blank human plasma. Aliquots of 200 μL were taken from the samples and placed into polycarbonate ultracentrifugation tubes (Beckman-Coulter, no. 343775) which were either centrifuged at 100,000 rpm at 37°C for 2 hours or incubated, without centrifugation, at the same temperature for the same time span. After ultracentrifugation, a 50 μL aliquot was removed from the clear, top layer of the plasma, or the HLM supernatant, and added to the same volume of MeCN. Likewise, after remixing, 50 μL aliquots were taken from the samples which had not been ultracentrifuged and added to MeCN. A standard solution (2.5 μL), containing a mix of d2-AMIO, d4-MDEA, d4-DDEA, d2-ODAA and d2-DAA (80 ng, 80 ng, 2.5 ng, 2.5 ng and 2.5 ng, respectively), in methanol was added to each sample as internal standards. The samples were vortexed then centrifuged (13,400 rpm, 10min), and the supernatants were transferred to vials for LCMS analysis. All reactions were carried out in triplicate. Standard curves were determined for both centrifuged and uncentrifuged samples over a range from 0.010 to 20 μM. The HLM and plasma free drug fractions were calculated as the ratio of the slope of the free concentration curve (determined from the ultracentrifuged samples) to the slope of the total concentration curve (uncentrifuged samples).
Protocols for the synthesis of deuterium-labeled standards and for the quantitation of AMIO and its metabolites in patient plasma samples are included in Supplemental Materials.
Supplementary Material
Acknowledgments
We thank Dr. Nina Isoherranen (Univ. Washington, Seattle), for helpful discussions and Sandy Harris Research Nurse Coordinator at the University of Washington School of Medicine/Division of Cardiology, for her help in providing us with blood samples from patients undergoing warfarin/amiodarone therapy. This work was supported by the National Institutes of Health [Project Program Grant P01GM32165] to AER.
Footnotes
Supplementary Material is linked to the online version of the paper at http://www.nature.com/cpt
Conflict of Interest
The authors declared no conflict of interest.
References
- 1.Vastesaeger E. Fibrin atherosclerosis. Acta Cardiol. 1967;22:581–6. [PubMed] [Google Scholar]
- 2.Julian DG, Camm AJ, Frangin G, Janse MJ, Munoz A, Schwartz PJ, et al. Randomised trial of effect of amiodarone on mortality in patients with left-ventricular dysfunction after recent myocardial infarction: EMIAT. European Myocardial Infarct Amiodarone Trial Investigators. Lancet. 1997;349:667–74. doi: 10.1016/s0140-6736(96)09145-3. [DOI] [PubMed] [Google Scholar]
- 3.Mason JW. Amiodarone. N Engl J Med. 1987;316:455–66. doi: 10.1056/NEJM198702193160807. [DOI] [PubMed] [Google Scholar]
- 4.Heger JJ, Prystowsky EN, Zipes DP. Relationships between amiodarone dosage, drug concentrations, and adverse side effects. Am Heart J. 1983;106:931–5. doi: 10.1016/0002-8703(83)90018-2. [DOI] [PubMed] [Google Scholar]
- 5.Marchlinski FE, Gansler TS, Waxman HL, Josephson ME. Amiodarone pulmonary toxicity. Ann Intern Med. 1982;97:839–45. doi: 10.7326/0003-4819-97-6-839. [DOI] [PubMed] [Google Scholar]
- 6.Rigas B, Rosenfeld LE, Barwick KW, Enriquez R, Helzberg J, Batsford WP, et al. Amiodarone hepatotoxicity. A clinicopathologic study of five patients. Ann Intern Med. 1986;104:348–51. doi: 10.7326/0003-4819-104-3-348. [DOI] [PubMed] [Google Scholar]
- 7.Dickstein G, Amikam S, Riss E, Barzilai D. Thyrotoxicosis induced by amiodarone, a new efficient antiarrhythmic drug with high iodine content. Am J Med Sci. 1984;288:14–7. doi: 10.1097/00000441-198407000-00002. [DOI] [PubMed] [Google Scholar]
- 8.Trujillo TC, Nolan PE. Antiarrhythmic agents - Drug interactions of clinical significance. Drug Safety. 2000;23:509–32. doi: 10.2165/00002018-200023060-00003. [DOI] [PubMed] [Google Scholar]
- 9.Funck-Brentano C, Becquemont L, Kroemer HK, Buhl K, Knebel NG, Eichelbaum M, et al. Variable disposition kinetics and electrocardiographic effects of flecainide during repeated dosing in humans: contribution of genetic factors, dose-dependent clearance, and interaction with amiodarone. Clin Pharmacol Ther. 1994;55:256–69. doi: 10.1038/clpt.1994.26. [DOI] [PubMed] [Google Scholar]
- 10.Chitwood KK, Abdul-Haqq AJ, Heim-Duthoy KL. Cyclosporine-amiodarone interaction. Ann Pharmacother. 1993;27:569–71. doi: 10.1177/106002809302700506. [DOI] [PubMed] [Google Scholar]
- 11.Nicolau DP, Uber WE, Crumbley AJ, 3rd, Strange C. Amiodarone-cyclosporine interaction in a heart transplant patient. J Heart Lung Transplant. 1992;11:564–8. [PubMed] [Google Scholar]
- 12.Funck-Brentano C, Jacqz-Aigrain E, Leenhardt A, Roux A, Poirier JM, Jaillon P. Influence of amiodarone on genetically determined drug metabolism in humans. Clin Pharmacol Ther. 1991;50:259–66. doi: 10.1038/clpt.1991.135. [DOI] [PubMed] [Google Scholar]
- 13.Thi L, Shaw D, Bird J. Warfarin potentiation: a review of the “FAB-4” significant drug interactions. Consult Pharm. 2009;24:227–30. doi: 10.4140/tcp.n.2009.227. [DOI] [PubMed] [Google Scholar]
- 14.Lip GY, Lim HS. Atrial fibrillation and stroke prevention. Lancet Neurol. 2007;6:981–93. doi: 10.1016/S1474-4422(07)70264-8. [DOI] [PubMed] [Google Scholar]
- 15.Kerin NZ, Blevins RD, Goldman L, Faitel K, Rubenfire M. The incidence, magnitude, and time course of the amiodarone-warfarin interaction. Arch Intern Med. 1988;148:1779–81. [PubMed] [Google Scholar]
- 16.Sanoski CA, Bauman JL. Clinical observations with the amiodarone/warfarin interaction: dosing relationships with long-term therapy. Chest. 2002;121:19–23. doi: 10.1378/chest.121.1.19. [DOI] [PubMed] [Google Scholar]
- 17.O’Reilly RA, Trager WF, Rettie AE, Goulart DA. Interaction of amiodarone with racemic warfarin and its separated enantiomorphs in humans. Clin Pharmacol Ther. 1987;42:290–4. doi: 10.1038/clpt.1987.149. [DOI] [PubMed] [Google Scholar]
- 18.Heimark LD, Wienkers L, Kunze K, Gibaldi M, Eddy AC, Trager WF, et al. The mechanism of the interaction between amiodarone and warfarin in humans. Clin Pharmacol Ther. 1992;51:398–407. doi: 10.1038/clpt.1992.39. [DOI] [PubMed] [Google Scholar]
- 19.Kumar V, Locuson CW, Sham YY, Tracy TS. Amiodarone analog-dependent effects on CYP2C9-mediated metabolism and kinetic profiles. Drug Metab Dispos. 2006;34:1688–96. doi: 10.1124/dmd.106.010678. [DOI] [PubMed] [Google Scholar]
- 20.Ohyama K, Nakajima M, Suzuki M, Shimada N, Yamazaki H, Yokoi T. Inhibitory effects of amiodarone and its N-deethylated metabolite on human cytochrome P450 activities: prediction of in vivo drug interactions. Br J Clin Pharmacol. 2000;49:244–53. doi: 10.1046/j.1365-2125.2000.00134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rettie AE, Korzekwa KR, Kunze KL, Lawrence RF, Eddy AC, Aoyama T, et al. Hydroxylation of warfarin by human cDNA-expressed cytochrome P-450: a role for P-4502C9 in the etiology of (S)-warfarin-drug interactions. Chem Res Toxicol. 1992;5:54–9. doi: 10.1021/tx00025a009. [DOI] [PubMed] [Google Scholar]
- 22.Marchiset D, Bruno R, Djiane P, Cano JP, Benichou M, Serradimigni A. Amiodarone and desethylamiodarone elimination kinetics following withdrawal of long-term amiodarone maintenance therapy. Biopharm Drug Dispos. 1985;6:209–15. doi: 10.1002/bdd.2510060211. [DOI] [PubMed] [Google Scholar]
- 23.Naganuma M, Shiga T, Nishikata K, Tsuchiya T, Kasanuki H, Fujii E. Role of desethylamiodarone in the anticoagulant effect of concurrent amiodarone and warfarin therapy. J Cardiovasc Pharmacol Ther. 2001;6:363–7. doi: 10.1177/107424840100600405. [DOI] [PubMed] [Google Scholar]
- 24.Ha HR, Bigler L, Wendt B, Maggiorini M, Follath F. Identification and quantitation of novel metabolites of amiodarone in plasma of treated patients. Eur J Pharm Sci. 2005;24:271–9. doi: 10.1016/j.ejps.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 25.Harris L, Roncucci RR. Amiodarone: pharmacology, pharmacokinetics, toxicology, clinical effects. Médecine et sciences internationales; Paris: 1986. [Google Scholar]
- 26.Locuson CW, 2nd, Wahlstrom JL, Rock DA, Jones JP. A new class of CYP2C9 inhibitors: probing 2C9 specificity with high-affinity benzbromarone derivatives. Drug Metab Dispos. 2003;31:967–71. doi: 10.1124/dmd.31.7.967. [DOI] [PubMed] [Google Scholar]
- 27.Lucas AN, Tanol M, McIntosh MP, Rajewski RA. Preparation and purification of desethylamiodarone hydrochloride. Synthetic Communications. 2006;36:3371–6. [Google Scholar]
- 28.Waldhauser KM, Torok M, Ha HR, Thomet U, Konrad D, Brecht K, et al. Hepatocellular toxicity and pharmacological effect of amiodarone and amiodarone derivatives. J Pharmacol Exp Ther. 2006;319:1413–23. doi: 10.1124/jpet.106.108993. [DOI] [PubMed] [Google Scholar]
- 29.Snead AN, Miyakawa M, Tan ES, Scanlan TS. Trace amine-associated receptor 1 (TAAR1) is activated by amiodarone metabolites. Bioorg Med Chem Lett. 2008;18:5920–2. doi: 10.1016/j.bmcl.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wendt B, Ha HR, Hesse M. Synthesis of two metabolites of the antiarrythmicum amiodarone. Helvetica Chimica Acta. 2002;85:2990–3001. [Google Scholar]
- 31.Deng P, You TG, Chen XY, Yuan T, Huang HH, Zhong DF. Identification of Amiodarone Metabolites in Human Bile by Ultra Performance Liquid Chromatography/Quadrupole Time-of-Flight Mass Spectrometry. Drug Metab Dispos. 2011 doi: 10.1124/dmd.110.037671. [DOI] [PubMed] [Google Scholar]
- 32.Lalloz MR, Byfield PG, Greenwood RM, Himsworth RL. Binding of amiodarone by serum proteins and the effects of drugs, hormones and other interacting ligands. J Pharm Pharmacol. 1984;36:366–72. doi: 10.1111/j.2042-7158.1984.tb04400.x. [DOI] [PubMed] [Google Scholar]
- 33.U.S. Food and Drug Administration. Draft Guidance. Rockville, MD: 2006. Guidance for Industry Drug Interaction Studies - Study Design, Data Analysis, and Implications for Dosing and Labeling. [Google Scholar]
- 34.Yeung CK, Fujioka Y, Hachad H, Levy RH, Isoherranen N. Are circulating metabolites important in drug-drug interactions?: Quantitative analysis of risk prediction and inhibitory potency. Clin Pharmacol Ther. 2011;89:105–13. doi: 10.1038/clpt.2010.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blanchard N, Richert L, Coassolo P, Lave T. Qualitative and quantitative assessment of drug-drug interaction potential in man, based on Ki, IC50 and inhibitor concentration. Curr Drug Metab. 2004;5:147–56. doi: 10.2174/1389200043489072. [DOI] [PubMed] [Google Scholar]
- 36.Ito K, Chiba K, Horikawa M, Ishigami M, Mizuno N, Aoki J, et al. Which concentration of the inhibitor should be used to predict in vivo drug interactions from in vitro data? AAPS PharmSci. 2002;4:E25. doi: 10.1208/ps040425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Obach RS. Nonspecific binding to microsomes: impact on scale-up of in vitro intrinsic clearance to hepatic clearance as assessed through examination of warfarin, imipramine, and propranolol. Drug Metab Dispos. 1997;25:1359–69. [PubMed] [Google Scholar]
- 38.Hanson KL, VandenBrink BM, Babu KN, Allen KE, Nelson WL, Kunze KL. Sequential metabolism of secondary alkyl amines to metabolic-intermediate complexes: opposing roles for the secondary hydroxylamine and primary amine metabolites of desipramine, (s)-fluoxetine, and N-desmethyldiltiazem. Drug Metab Dispos. 2010;38:963–72. doi: 10.1124/dmd.110.032391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mori K, Hashimoto H, Takatsu H, Tsuda-Tsukimoto M, Kume T. Cocktail-substrate assay system for mechanism-based inhibition of CYP2C9, CYP2D6, and CYP3A using human liver microsomes at an early stage of drug development. Xenobiotica. 2009;39:415–22. doi: 10.1080/00498250902822204. [DOI] [PubMed] [Google Scholar]
- 40.European Medicines Agency. Guideline on the Investigation of Drug Interactions. Canary Wharf; London, U.K: 2010. [Google Scholar]
- 41.Sadeque AJ, Eddy AC, Meier GP, Rettie AE. Stereoselective sulfoxidation by human flavin-containing monooxygenase. Evidence for catalytic diversity between hepatic, renal, and fetal forms. Drug Metab Dispos. 1992;20:832–9. [PubMed] [Google Scholar]
- 42.Nakai D, Kumamoto K, Sakikawa C, Kosaka T, Tokui T. Evaluation of the protein binding ratio of drugs by a micro-scale ultracentrifugation method. J Pharm Sci. 2004;93:847–54. doi: 10.1002/jps.20012. [DOI] [PubMed] [Google Scholar]
- 43.Templeton IE, Thummel KE, Kharasch ED, Kunze KL, Hoffer C, Nelson WL, et al. Contribution of itraconazole metabolites to inhibition of CYP3A4 in vivo. Clin Pharmacol Ther. 2008;83:77–85. doi: 10.1038/sj.clpt.6100230. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.