Summary
Chromatin immunoprecipitation studies have mapped protein occupancies at many genomic loci. However, a detailed picture of the complexity of coregulators (CoRs) bound to a defined enhancer along with a transcription factor is missing. To address this, we used biotin-DNA pulldown assays coupled with mass spectrometry-immunoblotting to identify at least 17 CoRs from nuclear extracts bound to 17β-estradiol (E2)-liganded estrogen receptor-α on estrogen response elements (EREs). Unexpectedly, these complexes initially are biochemically stable and contain certain atypical corepressors. Addition of ATP dynamically converts these complexes to an ‘activated’ state by phosphorylation events, primarily mediated by DNA-dependent protein kinase. Importantly, a ‘natural’ ERE-containing enhancer and nucleosomal EREs recruit similar complexes. We further discovered the mechanism whereby H3K4me3 stimulates ERα-mediated transcription as compared with unmodified nucleosomes. H3K4me3 templates promote specific CoR dynamics in the presence of ATP and AcCoA, as manifested by CBP/p300 and SRC-3 dismissal and SAGA and TFIID stabilization/recruitment.
Introduction
Transcriptional coregulators (CoRs) play a central role in the activation (coactivators) or repression (corepressors) of genes and are recruited to DNA by sequence-specific transcription factors (TFs). They contain diverse enzymatic activities essential for regulating transcription processes (reviewed in (Bulynko and O'Malley, 2010)). We recently reported mass spectrometry (MS)-based analyses of 3,290 CoR immunoprecipitations (IPs) from steady-state human cell nuclear extracts (NEs) (Malovannaya et al., 2011). These data revealed that 11,485 unique human gene products were present in CoR complexes, comprising ~50% of the human proteome. Thus, our present study was not aimed at identifying a new coregulator for ERα, rather, it was designed to provide new mechanistic insight(s) into how ERα-CoR complexes initially form and function.
Chromatin immunoprecipitation (ChIP) studies have mapped protein occupancies of TFs and some CoRs to diverse genomic DNA loci (reviewed in (Farnham, 2009)). Additional insight into transcriptional regulation emanated from ‘kinetic’ ChIP experiments in human cell lines that demonstrated that CoRs undergo ‘ordered recruitment’ to different gene promoters in a signal-dependent way (e.g., those estrogen-stimulated (Metivier et al., 2003; Shang et al., 2000)). While these approaches have provided important information about TF and CoR occupancy of target genes as a function of a signal, they are limited by detecting primarily binary interactions and fail to reveal multiple constituents of complexes bound to a particular genetic locus. Alternatively, MS proteomics attempts to reveal CoRs bound with a particular TF on DNA using in vitro ‘immobilized template assays’ have either used artificial TFs and response elements with human cell NE (e.g., GAL-VP16 (Chen et al., 2012)) or used a physiological TF (e.g., E2-liganded ERα), but have revealed only a few bona fide CoRs bound to EREs (e.g., (Nalvarte et al., 2010)).
Here we employed the model ligand-regulated TF, ERα (Gene symbol:ESR1), to mechanistically investigate DNA-bound TF-CoR complexes. There are many CoRs reported to interact with E2-liganded ERα to regulate transcription, but the CoRs in a gene-specific complex are incompletely defined. Major functional players reported to interact with ERα in an E2-dependent manner are steroid receptor coactivators (SRCs), CREB binding protein (CBP/CREBBP) and p300/EP300, and the Mediator complex (reviewed in (McKenna and O'Malley, 2002)). The SRC/p160 family proteins SRC-1/NCoA1, SRC-2/NCoA2, and SRC-3/NCoA3 act as primary coactivators by serving as a scaffold for recruitment of other secondary enzymatic CoRs (e.g., CBP/p300). E2-liganded ERα also recruits the Mediator complex to ERE-containing target genes by interacting with one of its subunits, MED1 (Kang et al., 2002). Besides these ‘classical’ coactivators, certain corepressors have been reported to bind E2-liganded ERα, including RIP140/NRIP1, LCOR, and REA/PHB2 (reviewed in (Gurevich et al., 2007)).
To overcome the limitation of ChIPs on DNA and to provide a more comprehensive picture of the CoR composition nucleated by ERα bound to a defined enhancer, we utilized cell-free biotin-ERE pulldown assays coupled to MS or immunoblotting and identified at least 17 CoRs from HeLa and MCF-7 cells that bind with E2-liganded ERα on canonical EREs. Unexpectedly, these complexes are initially biochemically stable and contain certain corepressors. Addition of ATP dynamically converts these complexes to an ‘activated’ state by phosphorylation events mediated mainly by DNA-dependent protein kinase (DNA-PK). Importantly, we further show that a ‘natural’ ERE from the human GREB1 gene and nucleosomal EREs recruit similar complexes, but also recruit distinct CoRs depending on the histone source. Finally, we have coupled our identifications of CoR-ERα-ERE complexes with a robust in vitro transcription assay to demonstrate how particular CoRs in these complexes and select histone H3 ‘marks’ affect ERα-mediated transcription. Through these cell-free mechanistic studies we reveal two major insights- 1) the dynamic conversion of CoR complexes during the pre-initiation step of ERα-directed transcription by DNA-PK and 2) the identification of CoRs (SAGA/TFIID) important for promoting ERα-driven transcription from H3K4me3-marked templates.
Results
Establishing an ERE DNA Pulldown Assay to Identify Coregulators Bound with E2-Liganded ERα
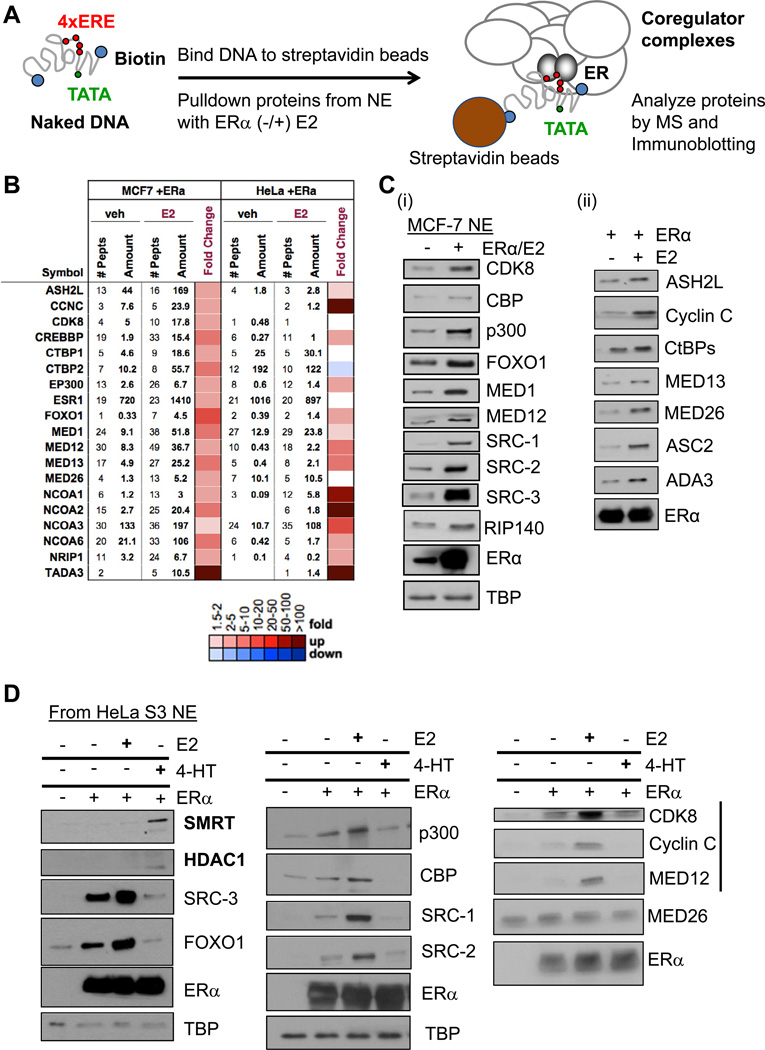
As an extension of our previous CoR IP-MS proteomics analyses (Malovannaya et al., 2011), we set forth to develop a more complete picture of endogenous CoR complexes bound to ERα on its cognate response element, the ERE, by using a biotin-labeled DNA pulldown assay system. Initially, we chose the canonical ERE sequence from the Xenopus Vitellogenin A2 gene promoter (Klein-Hitpass et al., 1986) and immobilized the biotinylated DNA to magnetic beads followed by incubation with HeLa S3 cell nuclear extract (HNE), with/without recombinant ERα, and with/without E2. After rapid washes at a physiological salt concentration along with gentle detergent, bead-bound proteins were identified by either MS or immunoblotting (Figure 1A).
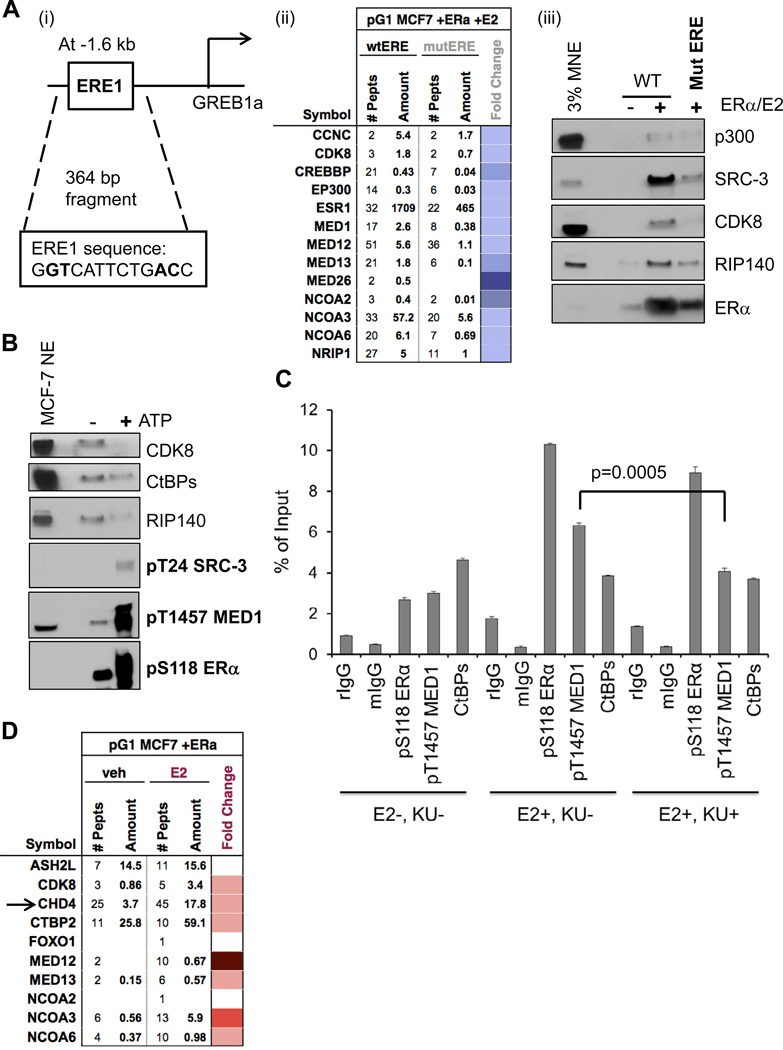
Figure 1. Pulldown Assays Define a ‘Stable’ Set of Coregulators Recruited to E2-Liganded ERα on DNA.
(A) Schematic of the in vitro assay for detection of CoR-ERα complexes on EREs. EREs fused to the Adenovirus E4 gene promoter (4xERE-E4) were immobilized to magnetic beads, incubated with nuclear extract (NE) and recombinant human estrogen receptor-α (ERα), and with/without E2 to allow CoR complex formation. After washes, bound proteins were identified by MS and immunoblotting. See Figure S1 for controls for this system.
(B) MS identifies at least 17 CoRs from MCF-7 and HeLa S3 NEs bound with E2-liganded ERα on 4xERE-E4 and validated by immunoblotting. NCBI gene names are used in this figure, but common protein names are shown for immunoblotting. Number of identified peptides, peptide amount, and fold E2-change (as a heatmap with color scale defined below; same scale is used in later figures) are indicated.
(C) Immunoblotting of 17 MS-identified CoR complex members from MCF-7 NE bound with E2-liganded ERα on 4xERE-E4. (i) Dependency of CoR binding as a function of added ERα. TBP serves a pulldown control, as it binds the TATA box of the E4 promoter independent of ERα. (ii) Dependency of CoR binding as a function of added E2 (100 nM).
(D) Effect of E2 and 4-HT on CoR recruitment to ERE-bound ERα. 1% ethanol served as the vehicle control; 100 nM of each ligand was added. Immunoblotting was performed against a subset of the MS-detected CoRs from HeLa S3 NE. SMRT and HDAC1 (bolded) were recruited to 4-HT-occupied ERα. Also, the CDK8 module of the Mediator complex (indicated by black bar) displayed stronger E2-induced/4-HT-reduced binding than that of MED26. Figure S1D shows data for SRC-3 and RIP140 from MCF-7 NE.
Initially, we performed a series of control experiments to determine the optimal CoR binding conditions on ERE-containing DNA beads. First, we tested how the number of EREs influenced CoR binding by incubating 1x, 2x, 3x, or 4x EREs immobilized on Dynabeads® with HNE and E2-liganded ERα. Consistent with reports that more EREs create a transcriptional ‘synergistic’ response (Klein-Hitpass et al., 1988), we found that four EREs promoted optimal binding of CBP, p300, SRCs, and subunits of the Mediator (MED) complex to E2-liganded ERα (Figure S1A, data not shown). We also tested whether a functional RNA pol II promoter (from the Adenovirus E4 gene) fused to 4xEREs (referred to as 4xERE-E4) would further stabilize CoR complex formation, as it has been used widely in binding and in vitro transcription assays (Acevedo et al., 2004). CBP, p300, MED1, and FOXO1 revealed the best ERα-dependent binding in the presence of the E4 promoter (Figure S1A).
Next, we tested whether the observed CoR recruitment was truly ERE-dependent or was due to non-specific bead binding. We found that the beads themselves without ERE DNA support very little CoR and ERα binding (Figure S1B). Secondly, we tested if four different restriction enzymes that should cleave the ERE DNA away from the beads would ‘release’ CoRs and TFs also. Indeed, incubation of ERE beads with the enzymes released a fraction of p300, SRC-3, MED subunits, ERα, and TBP (Figure S1C).
Defining a Set of CoRs Recruited to E2-Liganded ERα on EREs
After defining 4xERE-E4 as an optimal initial DNA template for investigating CoR recruitment to ERα, we employed unbiased MS (see Experimental Procedures; Table S1) to discover the CoRs bound to EREs. Using both HeLa and MCF-7 NE, we found at least 17 CoRs using this approach (Figure 1B) that were subsequently validated for their binding by immunoblotting (Figure 1C). In this system, recombinant ERα binding to 4xEREs is not E2-dependent (Figure 1Cii; (Brown and Sharp, 1990)). The binding of most CoRs, however, was E2-dependent (Figures 1Cii and 1D). As E2 promotes ERα transcriptional activity in MCF-7 cells, we additionally examined the effects of the antagonist 4-hydroxy-tamoxifen (4-HT) on CoR recruitment to the ERE-E4. Consistent with a prior ChIP study (Shang et al., 2000), we observed a reduction in binding of coactivators to EREs when ERα was occupied with 4-HT, while certain corepressors, such as SMRT and HDAC1, displayed enhanced binding (Figure 1D). However, the RIP140 corepressor is inhibited by 4-HT (Figure S1D; (Cavailles et al., 1995)).
We next investigated the kinetics of CoR-ERE complex formation as a function of added ERα. We found that a subset of CoRs displayed similar temporal binding patterns (Figure S1E, left panel), while others showed delayed kinetic entry (e.g., ASC2, Cyclin C, SRC-2) (right panel). It is interesting that certain corepressors (RIP140/CtBPs (Vo et al., 2001)) displayed binding kinetics similar to the known coactivators p300, SRC-3, and MED26. Importantly, these data substantiated that our standard incubation period allowed for maximal CoR binding.
Besides SRCs, MEDs, CBP, p300, and FOXO1, we also validated the binding of three other reported ERα coactivators to EREs- ASH2L, ASC2, and ADA3. ASH2L is a critical component of mixed-lineage leukemia (MLL) complexes that write the ‘activating’ histone H3K4 trimethyl mark (H3K4me3) (Steward et al., 2006). ASC2 is a component of a steady-state complex containing MLLs and ASH2L (Goo et al., 2003) and certain DNA damage/repair proteins (including DNA-PK (Ju et al., 2006)). Finally, ADA3 is a component of SAGA-type PCAF/GCN5 HAT complexes (Meng et al., 2004).
In addition to assessment of binding, we tested the functional role of some of the identified CoRs by measuring the effect on ERα-driven transcription after their knockdown by RNA interference. A stably integrated ERE-luciferase reporter gene in MCF-7 cells was used for these studies (See Experimental Procedures). We confirmed that SRC-3 and FOXO1 act as coactivators of the ERα-directed reporter gene, while RIP140 acts as a corepressor in this system (Figure S1Fi). In addition, we found that knockdown of the CDK8 module of Mediator reduced luciferase expression, thereby showing that it has coactivation activity in MCF-7 cells (Figure S1Fii), along with its E2-dependent binding.
SRC-3, CBP/p300, and FOXO1 Form an Interaction ‘Hub’ on EREs
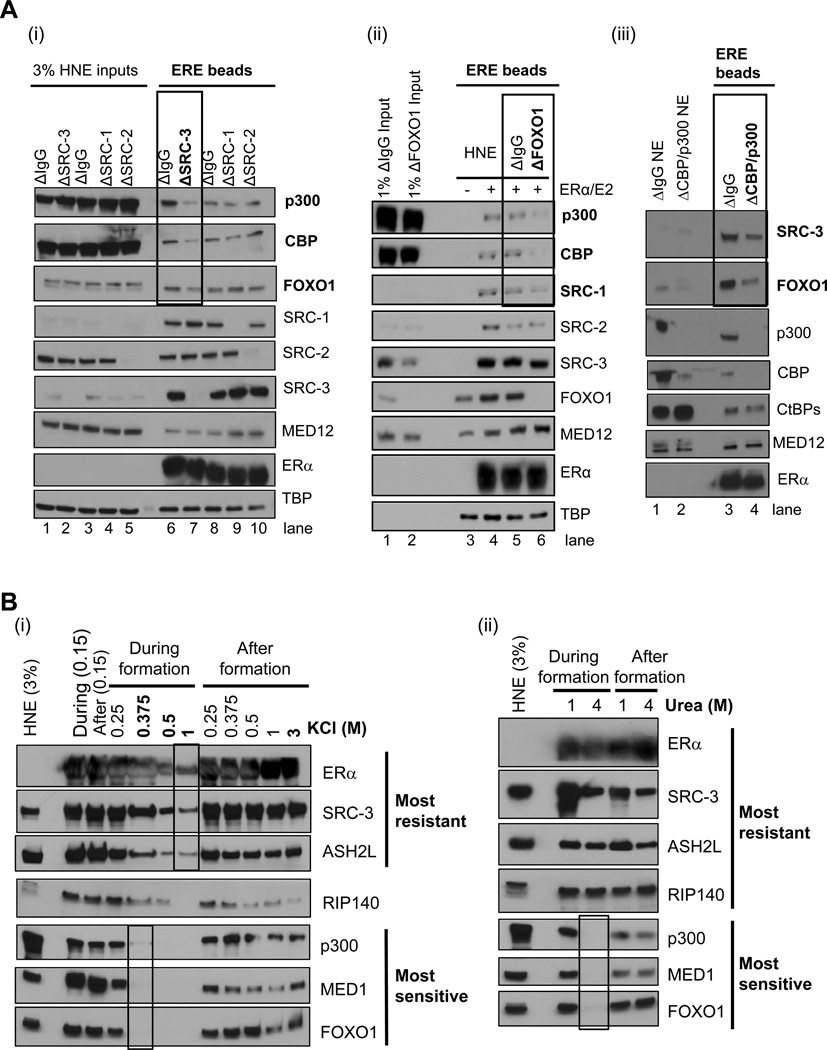
To begin to understand how one CoR might affect the recruitment of another, we decided to immunodeplete a select few and test if their loss affected subsequent CoR binding. We focused these studies on SRC-3, CBP/p300, and FOXO1 as our above data indicated they shared similar kinetics (Figure S1E) and coactivation of ERα (Figure S1Fi). When HNE was cleared of SRC-3 by immunodepletion, p300, CBP, and FOXO1 recruitment to EREs was reduced (compare lanes 6 and 7 in Figure 2Ai), consistent with prior SRC-3-CBP/p300 interaction data (e.g., (Chen et al., 1997)). Similarly, depletion of FOXO1 resulted in a loss of p300, CBP, and SRC-1 (compare lanes 5 and 6 in Figure 2Aii; (Nasrin et al., 2000)). Furthermore, we observed that CBP/p300 depletion reduced SRC-3 and FOXO1 binding to EREs (Figure 2Aiii).
Figure 2. SRC-3, CBP/p300, and FOXO1 Form an Interaction ‘Hub’ that is Resistant to Salt/Urea After Complex Formation.
(A) Effect of immunodepleting SRCs, FOXO1, or CBP/p300 from HeLa NE (HNE) on recruitment of other CoRs to ERα-bound EREs. (i) Depletion of SRC-3, but not other SRCs, reduces CBP/p300 and FOXO1 recruitment to 4xERE-E4. (ii) Immunodepletion of FOXO1 reduces p300/CBP and SRC-1, but not other SRCs, recruitment to 4xERE-E4. DNA pulldowns with regular HNE (−/+) E2-liganded ERα were employed as a positive control. (iii) Immunodepletion of CBP/p300 reduces SRC-3 and FOXO1 recruitment to 4xERE-E4.
(B) Salt/urea added during binding reactions differentially impair CoR complex formation on EREs, yet after formation, salt/urea have minimal effects. (i) Titration of KCl from 150 mM present in HNE to 1M selectively impairs CoR complex binding, with RIP140 displaying intermediate sensitivity. After formation, even 3M KCl cannot completely disrupt these CoR complexes. (ii) Titration of urea from 1M to 4M in HNE selectively impairs CoR complex binding, yet after complex formation, 4M urea cannot effectively disrupt the stable CoR complexes. Figure S2 shows the effect of KCl on a recombinant p300-SRC-3-ERα-ERE complex and on other CoRs when SRC-3 is depleted from HNE.
Coregulators Display Differential Sensitivity to Salt or Urea Only During ‘Formation’ of ERE-ERα Complexes
To further investigate the effect of chemicals disrupting complex formation or stability after formation, various concentrations of KCl or urea were added to the input HNE or added to the pre-formed, washed complexes in an attempt to release them from the DNA. Among the few selected CoRs validated to be present in ERE complexes, at least two classes of CoRs were defined by immunoblotting based on their sensitivities to these treatments (Figure 2B). SRC-3 and ASH2L were the most resistant to increasing KCl in the HNE, RIP140 was intermediate in sensitivity, while p300, MED1, and FOXO1 were the most sensitive (Figure 2Bi). However, after CoR complexes were formed on EREs, even a ‘3M KCl challenge’ did not completely release any of these proteins (although RIP140 and p300 levels did decrease), suggesting that multi-CoR binding dramatically enhances the overall complex stability. Alternatively, complex formation might simply protect proteins from being dissociated by salt by precluding access to their surfaces.
As further tests of these hypotheses, we performed two additional experiments. First, we ‘challenged’ ERα bound to EREs, with and without recombinant SRC-3 or p300, to increasing salt concentrations (Figure S2A). As expected, ERα had reduced DNA binding at 2M KCl, but the addition of SRC-3 protein increased ERα binding (compare lanes 3 and 6). Additionally, when p300 was added, SRC-3 became even more refractory to 2M KCl (compare lanes 6 and 9). Second, we assayed what effect depletion of SRC-3 might have on the salt stability of other CoRs (Figure S2B). While loss of SRC-3 affects the recruitment of interaction ‘hub’ members p300 and FOXO1 (compare lanes 1 and 4) but not MED1 or SRC-1, it is interesting that SRC-3 appears to stabilize p300, but not FOXO1, to the salt challenge (compare lanes 4 and 6).
Given the results of the high salt treatments, we next tested the effect of urea on CoR and ERα retention on EREs. Similar to what was observed with KCl, SRC-3, ASH2L, and RIP140 were the most resistant to addition of urea to the binding reaction, while p300, MED1, and FOXO1 were inhibited when 4M urea was included during complex formation (Figure 2Bii). After complexes were formed, these ‘labile’ proteins became biochemically stable to 4M urea.
Addition of ATP to Stable CoR-ERα-ERE Complexes Results in Rapid Distinct and Dynamic Phosphorylation Events
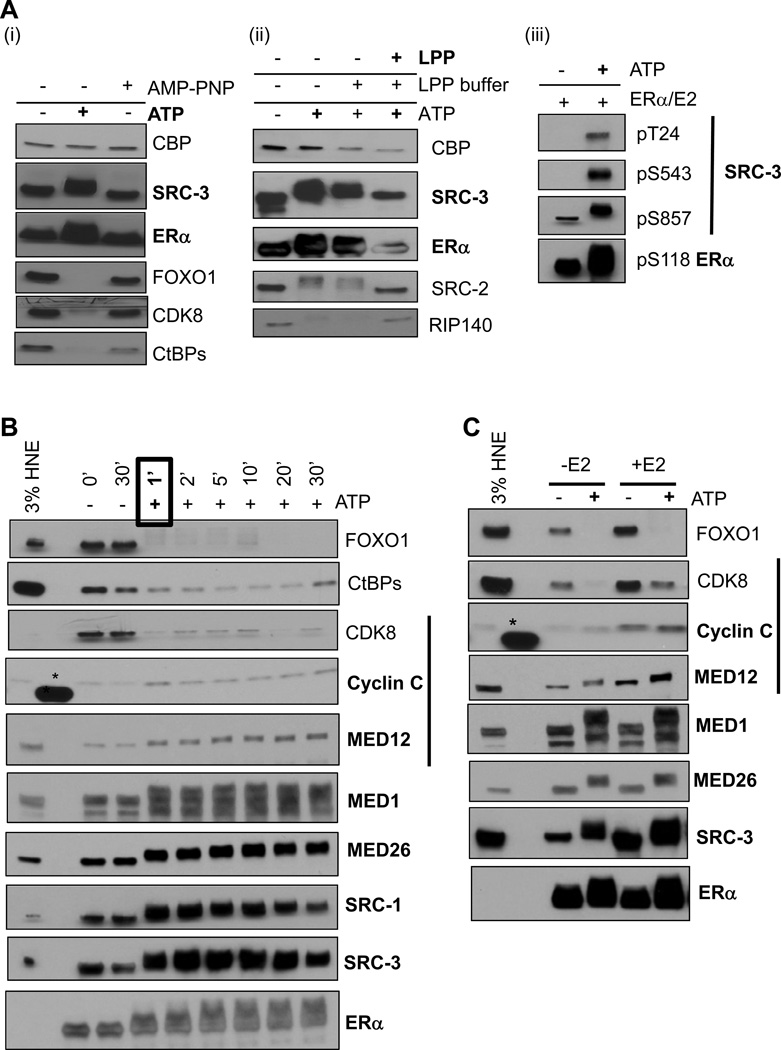
The striking stability of the CoR-ERα-ERE complexes was initially puzzling, especially since ChIP studies in E2-treated MCF-7 cells indicated that ERα and CoRs dynamically occupy the promoters of endogenous target genes (Metivier et al., 2003; Shang et al., 2000). It was possible that our defined stable CoR-ERα-ERE complexes would become more dynamic in the presence of the proper ‘signal’, such as phosphorylation. We asked whether addition of ATP to washed ERE complexes might affect CoR binding. Surprisingly, 0.5 mM ATP treatment of the ERE complexes resulted in the release of five CoRs from the formed complexes (Figures 3Ai and 3Aii). As a control, we tested multiple antibodies to RIP140, CtBPs, CDK8, and FOXO1 and still observed loss of these proteins from EREs in the presence of ATP (data not shown). In contrast, ERα and SRC-3 remained bound but displayed a retarded gel mobility indicating that they might be phosphorylated in the presence of ATP. Importantly, not all CoRs were affected by ATP addition (e.g., CBP). A non-hydrolyzable ATP analog (AMP-PNP) incapable of mediating phosphorylation did not reveal these effects (Figure 3Ai).
Figure 3. Addition of ATP to the Stable CoR-ERα Complexes Results in Dynamic and Distinct Phosphorylation Events.
(A) Addition of 0.5 mM ATP to washed CoR-ERα complexes results in ERα and SRC-3 appearing stabilized on the EREs (bolded), while five other CoRs appear dismissed. (i) ATP, but not AMP-PNP, treatment results in ERα and SRC-3 displaying enhanced gel mobility shifts, while FOXO1, CDK8, and CtBPs appear dismissed. (ii) λ-protein phosphatase (LPP) treatment of ATP-modified CoR-ERα complexes results in reduced gel mobility shifts (bolded) or reappearance in immunoblots. (iii) SRC-3 and ERα are phosphorylated at distinct ‘activating’ residues upon ATP addition to CoR-ERE complexes.
(B) Phosphorylation-mediated effects on CoR-ERE complexes occur with rapid kinetics. After ATP addition, proteins remaining on the beads were assayed by immunoblotting. Within 1 minute of ATP treatment, SRCs, ERα, and MEDs appear stabilized (bolded), while other CoRs are dismissed.
(C) E2 (100 nM) drives CoR-ERα-ERE complex assembly, but not phosphorylation effects. In panels B and C, the black bar indicates the CDK8 Mediator module; *, cross-reaction with protein size standards.
To provide stronger support that phosphorylation of CoRs and ERα was involved in these effects, we carried out the following two experiments. First, we treated washed complexes with ATP and then subsequently treated them with λ-protein phosphatase (LPP) and found that LPP treatment prevented the mobility shifts of SRCs and ERα and reversed dismissal of RIP140 (Figure 3Aii). Second, we tested if three known transcriptionally ‘activating’ phosphorylation sites on SRC-3 (pT24, pS543, and pS857) that we have previously described (Wu et al., 2004) and on ERα (pS118) (Ali et al., 1993) were enhanced with ATP addition to the ERE complexes by immunoblotting with phospho-selective antibodies. All of these tested phosphorylation sites were increased upon ATP addition (Figure 3Aiii).
Given that ATP addition promoted phosphorylation of the receptor-CoR complexes, we next tested the kinetics of these reactions by quenching with SDS-sample buffer after ATP addition at different times. Remarkably, ATP addition to the stable CoR complexes resulted in observable effects within 1 minute (Figure 3B). It was also apparent from this experiment that the CDK8 module of Mediator is not affected the same way by ATP (i.e., MED12 and Cyclin C remain, while CDK8 is dismissed).
Finally, we tested whether similar phosphorylation effects occur on unliganded ERα-ERE-CoR complexes. SRC-3, MED1, MED12, MED26, and ERα displayed mobility shifts and FOXO1 and CDK8 were dismissed without any E2 addition during complex formation, while E2, in general, promoted more CoR recruitment to EREs without ATP (Figure 3C). This data suggests that the role of E2 is to promote CoR complex formation on EREs, and it is not responsible primarily for the subsequent dynamic phosphorylation-mediated events.
DNA-PK Phosphorylates ERα and Some CoRs Within ERE Complexes for Transcriptional Activation
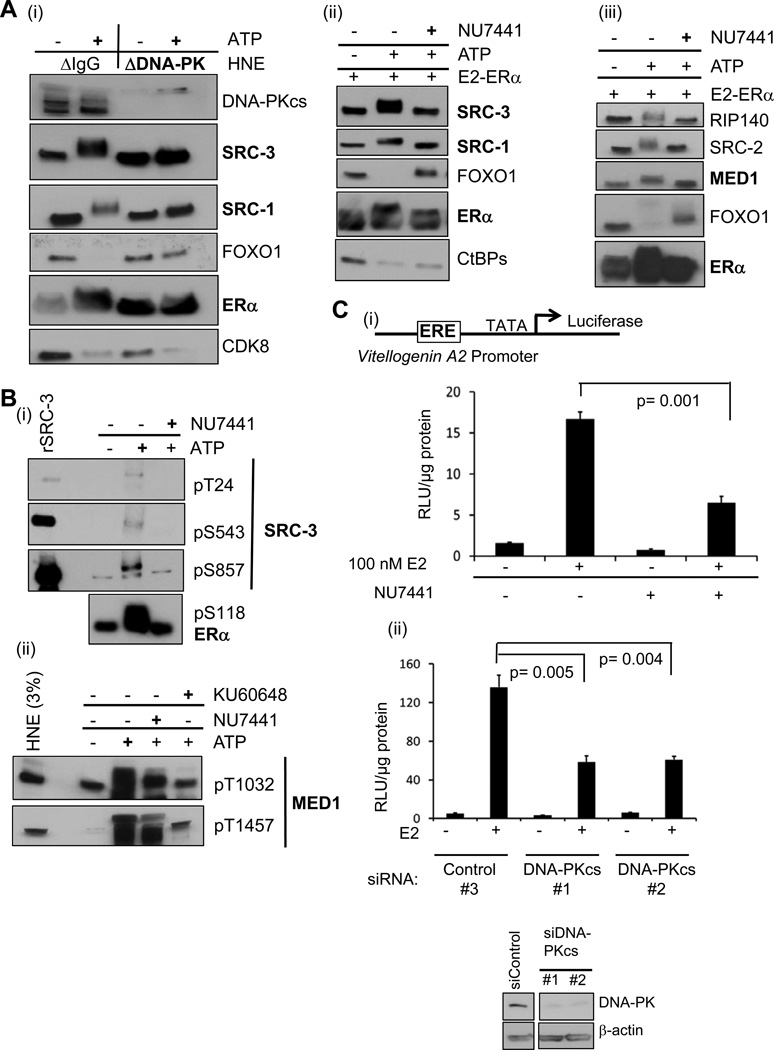
After identifying phosphorylation events occurring with ATP addition to stable ERE complexes, we next attempted to identify a kinase responsible for ‘activating’ these complexes. As a first step, we screened for the presence of 20 kinases from HNE in the ERE complexes. Six kinases were similarly bound without any ERα, and 14 other kinases were not found to be bound to the beads (Table S2). From the group of kinases bound to EREs, we tested how their immunodepletion from HNE would effect CoR phosphorylation, initially focusing on CDK7, CDK8, and DNA-PK catalytic subunit (DNA-PKcs), since CDK7 (Chen et al., 2000) and DNA-PKcs (Medunjanin et al., 2010) can directly phosphorylate ERα at S118, and CDK8 is a member of the stable complexes. While the absence of CDK7 and CDK8 from HNE did not affect the phosphorylation patterns of SRC-3, ERα, or FOXO1 (data not shown), the loss of DNA-PKcs abrogated the ATP-dependent mobility shifts of SRCs and ERα and the dismissal of FOXO1 (but not CDK8) (Figure 4Ai). To confirm our results, we tested the effect of NU7441, a highly selective inhibitor of DNA-PKcs (Leahy et al., 2004); at 9 µM, we observed similar effects as we did for DNA-PKcs depletion (Figure 4Aii). NU7441 also reduced the ATP-mediated MED1 mobility shift and RIP140 and SRC-2 dismissals (Figure 4Aiii), but did not substantially affect CtBPs dismissal (Figure 4Aii).
Figure 4. DNA-PK Phosphorylates ERα and Certain CoRs Producing a Transcriptionally ‘Activated’ State.
(A) Testing the effect of depleting or inhibiting DNA-PKcs on phosphorylation of CoR-ERα complexes. (i) Immunodepletion of DNA-PKcs from HNE reduces ATP-mediated ERα and SRCs gel mobility shifts (bolded) and impairs FOXO1 dismissal. (ii) NU7441 reduces ATP-mediated ERα and SRCs gel mobility shifts and FOXO1 dismissal from EREs. HNE was employed in this experiment. NU7441 (9 µM) was pre-incubated with washed complexes for 5 min before ATP addition. (iii) NU7441 reduces ATP-mediated MED1 and ERα gel mobility shifts (bolded) and reverses other CoRs dismissal from ERE complexes. MCF-7 NE was employed in this experiment.
(B) DNA-PK inhibitors reduce phosphorylation at residues of SRC-3, ERα, and MED1 associated with transcriptional activation. (i) Phosphorylation sites on SRC-3 and ERα (listed) are reduced by 9 µM NU7441. Recombinant SRC-3 protein (rSRC-3) serves as a control for the phospho-selective SRC-3 antibodies. (ii) Two phosphorylation sites on MED1 are reduced by 9 µM NU7441 or KU60648.
(C) Reducing DNA-PKcs activity in MCF-7 MAR-ERE-LUC cells by either inhibition (1 µM NU7441) (i) or siRNA-mediated knockdown (ii) impairs E2-liganded ERα transcription from an ERE-driven promoter (see schematic). (ii) Transfecting siRNA pools (50 nM) targeting DNA-PKcs (#1: Dharmacon; #2: Santa Cruz) reduced ERα activity, as compared to the control siRNA #3 (Dharmacon). The panel below shows efficient knockdown; β-actin serves as a loading control. A few lanes between siControl and siDNA-PKcs were cropped out of the image, as they represented other knockdowns not pertinent to Figure 4Cii. Statistical significance was determined by the Student’s t test from three independent replicates, with mean +/− SEM shown. Figure S3 shows pT1457 MED1 occupancy on the ERE of this luciferase reporter as a function of E2 and DNA-PK activity.
As mentioned above, ATP promotes phosphorylation on SRC-3 and ERα in our ERE complexes at sites known to augment ERα’s transcriptional activity (Figure 3Aiii). MAPK-mediated phosphorylations of MED1 at T1032 and T1457 promote its association with the rest of the Mediator complex, resulting in coactivation of ERα (Belakavadi et al., 2008). We thus decided to test whether NU7441 would inhibit the presence of the transcriptionally activating phosphorylations of SRC-3, MED1, or ERα in the ERE complexes (Figure 4B). Indeed, the DNA-PK inhibitor reduced the phosphorylation of all six sites, although KU60648 (a water-soluble analog of NU7441) was most effective at inhibiting MED1 phosphorylations (Figure 4Bii).
Given that inhibition of DNA-PK activity impaired these activating phosphorylations, we next tested if the loss of DNA-PK activity in MCF-7 ERE-LUC cells would reduce E2-liganded ERα transcriptional activity. When ERE-LUC cells were pre-treated with 1 µM NU7441, we observed a significant loss in E2-liganded ERα activity (Figure 4Ci) that also was seen upon DNA-PKcs knockdown using two different siRNA pools (Figure 4Cii).
ERα and SRC-3 are known to bind the ERE of the Vitellogenin A2 promoter in ERE-LUC cells in an E2-dependent fashion (Peterson et al., 2007), but it was unknown if any DNA-PK mediated phosphorylation events may occur at this ERE within target cells. To address this, we focused on DNA-PK phosphorylation of MED1 at T1457, as it was strongly inhibited by the KU inhibitor. We performed ChIP experiments of ERE-LUC cells that were either hormone-deprived (-E2), treated with E2 for 45 min, or pre-treated with 5 µM KU inhibitor followed by E2 (+E2+KU) comparing ERE occupancy with control or pT1457 MED1 antibodies (Figure S3A). We observed robust pT1457 MED1 ERE occupancy upon E2 treatment that was reduced upon DNA-PK inhibition. Importantly, when another genomic region (CCND1 intron 4) was assayed, pT1457 MED1 occupancy was not reduced, suggesting that the DNA-PK functional targeting of T1457 in MED1 was selective to the ERE and not simply due to reduced MED1 protein level (Figure S3B).
A ‘Natural’ ERE-Containing Enhancer Recruits Similar Stable Complexes That Are Dynamically Phosphorylated
As the ERE-E4 template employed above is artificial, we decided to interrogate CoR-ERα complexes formed on an ERE in its natural environment from the human genome. We chose to assay a 364 base-pair (bp) enhancer region 1.6 kb upstream from the E2- inducible GREB1 gene that has a ‘single’ well-defined ERE (see ERE1 in Figure 5Ai). ERα and SRC-3 are reported to occupy this native ERE in MCF-7 cells and mutations in this ERE reduced E2-induced transcription of a GREB1 promoter-luciferase reporter (Sun et al., 2007). To first discover which CoRs are recruited to E2-liganded ERα bound directly to the ERE1 sequence, we mutated four bp in the GREB1 fragment (Figure 5Ai) and compared ERα/CoR recruitments from MCF-7 NE between wild-type (wt) and mutated (mut) GREB1 fragments. From MS data, we observed that ERα and 12 CoRs that were defined on the ERE-E4 template require an intact ERE1 for optimal binding (Figure 5Aii). We further validated the reduced bindings of ERα and four CoRs on the mutERE by immunoblotting (Figure 5Aiii). Analogous MS data was acquired for GREB1 wt/mut ERE1 using HNE, resulting in the additional appearance of FOXO1 and SRC-1 (Figure S4A). Our data suggest that a similar, but not completely identical, subset of CoRs is bound to the GREB1 ERE1 fragment when compared to the 4xERE-E4. We also asked whether the GREB1 ERE complexes might be phosphorylated. Indeed, when ATP was added to washed CoR-ERα complexes formed on the GREB1 fragment, we observed similar gel mobility shifts for ERα and SRC-3, discrete phosphorylations of ERα, SRC-3, and MED1, and corresponding dismissals of CDK8, CtBPs, RIP140, SRC-2, and FOXO1 (Figures 5B and S4B).
Figure 5. A ‘Natural’ ERE Recruits Similar Dynamic CoR Complexes and DNA-PK Inhibition Reduces pT1457 MED1 Occupancy In Vivo.
(A) The ERE1 sequence in a fragment upstream of the GREB1 gene promoter recruits similar CoR complexes as seen with 4xERE-E4. (i) Schematic of the 364 bp fragment employed in pulldown assays; four base-pairs mutated in ERE1 are bolded. (ii) Mutating the ERE1 sequence impairs formation of stable CoR-ERE complexes, as seen by reduced peptide detection of ERα and CoRs from MCF-7 NE. MS data was presented in a heatmap format for fold change of binding to the mutated ERE (mutERE) over the wild-type ERE (wtERE) indicated. (iii) Immunoblotting shows reduced ERα and four CoRs recruitment to the mutated GREB1 fragment. Figure S4 shows similar data when HNE was employed; Figure S5 further supports this using gel filtration.
(B) Similar CoR phosphorylation events occur on the GREB1 ERE1. After washed complexes were isolated, they were treated with ATP and analyzed by immunoblotting. Like 4xEREs, CDK8, CtBPs, and RIP140 are dismissed, while SRC-3, MED1, and ERα are phosphorylated at distinct sites.
(C) ChIP assays of GREB1 ERE1 occupancy in MCF-7 cells reveal that pS118 ERα and pT1457 MED1 are enhanced with E2 treatment, but only pT1457 MED1 is substantially reduced in cells pre-treated with the DNA-PK inhibitor KU60648 (‘KU’). Normal rabbit IgG (rIgG) or mouse IgG (mIgG) serve as controls. Data is presented as percentage of input chromatin, and statistical significance was determined by the Student’s t test, with mean +/− SEM shown.
(D) MS identification of E2-dependent CoRs enriched on the GREB1 ERE1 fragment with ERα. Unliganded ERα and MCF-7 NE were added to the GREB1 fragment (−/+) 100 nM E2. Ten CoRs were detected as E2-enriched, including the CDK8 module of Mediator (CDK8, MED12, MED13) and CHD4 (indicated by arrow). Fold E2-change is shown as a heatmap. CHD4 and CHD8 are further characterized in Figure S6.
To test if any of the above DNA-PK mediated phosphorylation events may occur at the GREB1 ERE1 inside MCF-7 cells, we performed ChIP assays, specifically testing for enrichment of pS118 ERα, pT1457 MED1, or CtBPs as a function of E2 with/without DNA-PK inhibition (Figure 5C). As expected from the literature (Weitsman et al., 2006), we observed pS118 ERα to be enriched at the GREB1 ERE1 with E2 treatment. Like ERE occupancy of the luciferase reporter, pT1457 MED1 was enriched with E2, while CtBPs occupancy was not. Upon pre-treatment with the KU inhibitor, we observed very minimal reduced pS118 ERα occupancy, hardly any effect on CtBPs, but instead a significant reduction in pT1457 MED1 (a result similar to Figure S3). As multiple kinases can target S118 of ERα in an E2-dependent fashion in cells (Le Romancer et al., 2011), this result was not unexpected. Also, our in vitro data suggested that CtBPs were not targeted by DNA-PK (Figure 4Aii), consistent with the lack of an effect of KU on their occupancy.
To further biochemically support the hypothesis that the GREB1 ERE1 fragment recruits ‘stable’ CoR complexes to ERα, we ran HNE with E2-liganded ERα with and without the ERE DNA on a Superose 6 gel filtration column (Figure S5). ERα added to HNE without ERE DNA can aggregate, leading to its presence in the excluded ‘void’ volume of this column (Figure S5A). However, when GREB1 DNA was included in the binding reaction, it was apparent that a new peak formed on the UV280 chromatogram (Figure S5Ci) that contained DNA-PKcs, p300, SRC-3, MED26, ERα, and ERE1 DNA co-fractionating together (see Fractions 19 and 21 in Figure S5). These data support the existence of this high molecular weight sub-complex as a stable entity.
The GREB1 ERE1 Fragment Additionally Recruits Chromodomain-Helicase DNA Binding Proteins (CHDs)
We next tested the E2-dependency of CoR recruitment to the GREB1 ERE fragment using MCF-7 NE, recombinant ERα, with/without added E2, followed by MS analysis. Ten CoRs observed on 4xERE-E4 appeared E2-enriched on the GREB1 ERE (Figure 5D), including three components of the CDK8 module of Mediator. We validated the E2-dependent binding of three of them (ASH2L, ASC2, SRC-3) as well as for RIP140 (Figure S6). Unexpectedly, we observed chromodomain-helicase-DNA binding protein 4 (CHD4) that is part of the NURD repressive complex (Xue et al., 1998) as being E2-enriched on the GREB1 fragment (Figure 5D) and confirmed this inducible binding by immunoblotting (Figure S6Ai). To test the effect CHD4 might have on E2-liganded ERα transcriptional activity, we knocked down CHD4 with a targeting siRNA pool in ERE-LUC cells (Figure S6Aii) and found that loss of CHD4 reduced E2-dependent transcription, suggesting that it can act as a coactivator for ERα. To further support this hypothesis, we tested whether CHD4 depletion might simply reduce the ERα level in cells, as NURD repression of SNAIL expression may increase ERα expression (Dhasarathy et al., 2007). While CHD4 depletion did increase SNAIL protein, ERα protein levels were not changed (Figure S6Aiii).
We decided also to test CHD8 by immunoblotting, as it is necessary for E2-mediated induction of the cyclin E2 gene (Caldon et al., 2009). Like CHD4, CHD8 displayed E2-enhanced binding to the GREB1 fragment (Figure S6Bi) and its knockdown in ERE-LUC cells reduced E2-dependent transcription (Figure S6Bii). To begin to investigate how CHD8 might act as an ERα coactivator, we reduced its level in MCF-7 NE by immunodepletion (Figure S6Ci) and asked what other CoRs might be affected in recruitment to the GREB1 ERE1 DNA (Figure S6Cii). From the MS data, CHD8 appears to recruit chromatin insulators, modifiers, and remodelers, as we found CTCF, MLL, BPTF, and BAZ1 proteins, respectively, affected by loss of CHD8. We further validated reduced binding of MLL1 and BPTF to the GREB1 ERE1 when CHD8 was depleted (Figure S6Ciii).
Coregulator Recruitment and Transcription Effects Mediated by E2-Liganded ERα on HeLa Core Histone Assembled EREs
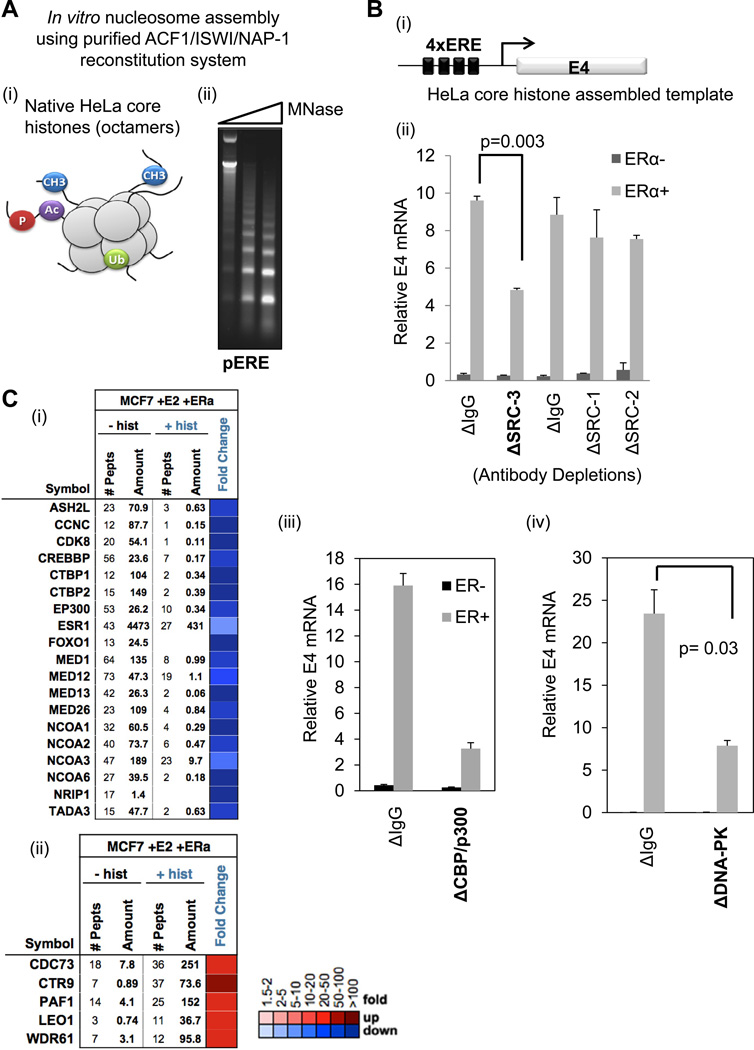
Although protein-free EREs are useful for mechanistic studies, they do not fully represent the natural state of DNA in a living cell. Thus, we tested the effect of assembling native core histone octamers purified from HeLa cells on CoR-ERα-ERE complex formation. These histone octamers contain a variety of post-translational modifications (PTMs) on their histone tails (Figure 6Ai). To couple CoR complex binding data with a functional readout, we again used the ERE-E4 template, as it has been employed for ERα-directed in vitro transcription (Acevedo et al., 2004). We used a recombinant ACF1/ISWI/NAP-1 system for chromatin assembly, and confirmed that this system produces the expected ~146 bp nucleosomal laddering on ERE-E4 plasmid DNA (pERE), upon treatment with micrococcal nuclease (Figure 6Aii). We then assembled HeLa core octamers on the pERE for use in ERα-directed in vitro transcription assays (illustrated in Figure 6Bi). Specifically, we tested the effect of depletion of each SRC, CBP/p300, or DNA-PKcs from HNE on ERα-mediated transcription of the E4 reporter gene. While all three SRCs were efficiently depleted from HNE (Figure 2Ai), only the loss of SRC-3 revealed a twofold loss in ERα-mediated transcription (Figure 6Bii). The data are consistent with SRC-3 acting as the primary recruiting partner for CBP/p300 to ERE-E4 (Figure 2Ai). Likewise, when CBP/p300 were depleted, a four-fold loss in ERα-driven transcription was observed (Figure 6Biii). Similar to cell-based data shown in Figure 4C, we observed a three-fold loss of ERα-mediated reporter gene transcription when DNA-PKcs was depleted from the HNE (Figure 6Biv).
Figure 6. Recruitment of CoRs and Transcription Mediated by ERα Bound to HeLa Core Nucleosomes.
(A) Nucleosomes assembled in vitro on 4xERE-E4 templates. (i) Schematic of HeLa core octamers used for nucleosome assembly; their tails can have various PTMs. (ii) Micrococcal nuclease (MNase) digests reveal nucleosome laddering on plasmid DNA containing 4xERE-E4 (pERE). Increasing amounts of MNase (0.5, 1, and 2 µl) were added to pERE assembled nucleosomes.
(B) Assays with pERE assembled nucleosomes reveal a requirement for SRC-3, CBP/p300, and DNA-PK for optimal ERα-directed transcription. (i) Schematic of nucleosomal DNA template used in transcription reactions. (ii) Cell-free transcription from nucleosomal pERE is reduced when SRC-3 was immunodepleted from HNE. IgG controls (ΔIgG) were either normal goat IgG (for SRC-3) or rabbit IgG (for SRC-1 and SRC-2). (iii) ERα-mediated transcription from nucleosomal pERE is reduced when CBP/p300 were immunodepleted from HNE. Control IgG was normal rabbit IgG. (iv) ERα-mediated transcription from nucleosomal pERE is reduced when DNA-PKcs was immunodepleted from HNE. Control IgG was normal mouse IgG. In all three panels, the relative E4 mRNA level was represented as the mean +/− SEM, and statistical significance was determined by a two-tailed t test from independent duplicates.
(C) HeLa core octamers assembled onto the 4xERE-E4 fragment promote similar and unique CoR recruitments to E2-liganded ERα, as compared to naked EREs. (i) MS data (in heatmap format for fold change in the presence of HeLa core histones) shows that HeLa core nucleosomes (+hist) decrease binding of CoR-ERE complexes, defined in Figure 1, from MCF-7 NE. (ii) MS data showing that HeLa core histones increase the recruitment of the PAF1 complex to 4xERE-E4.
With our in vitro transcription system validated, we next tested the effect of HeLa core histone assembly of the ERE-E4 fragment for CoR recruitment from MCF-7 NE. After MS was performed on nucleosomal (+hist) or nucleosome-free (-hist) ERE-E4 pulldowns, we observed 16 out of 18 CoRs seen on naked EREs to be present on nucleosomal EREs (Figure 6Ci). The reduced CoR peptides identified by MS on HeLa core histones, as compared to naked DNA, was not surprising given that nucleosomes are well-known to inhibit TF DNA binding. However, the HeLa core histone-assembled ERE-E4 template also recruited new CoRs not seen on the naked EREs. Particularly, we observed that the PAF1 RNA pol II elongation complex was enriched with E2-liganded ERα in the presence of the HeLa core histones (Figure 6Cii) - a result further validated by immunoblotting of PAF1 and CDC73 (data not shown).
Recombinant Nucleosomes Containing Different Histone H3 Tail Methylations Display Differences in Transcription and in ‘Reader’ Recruitment to EREs
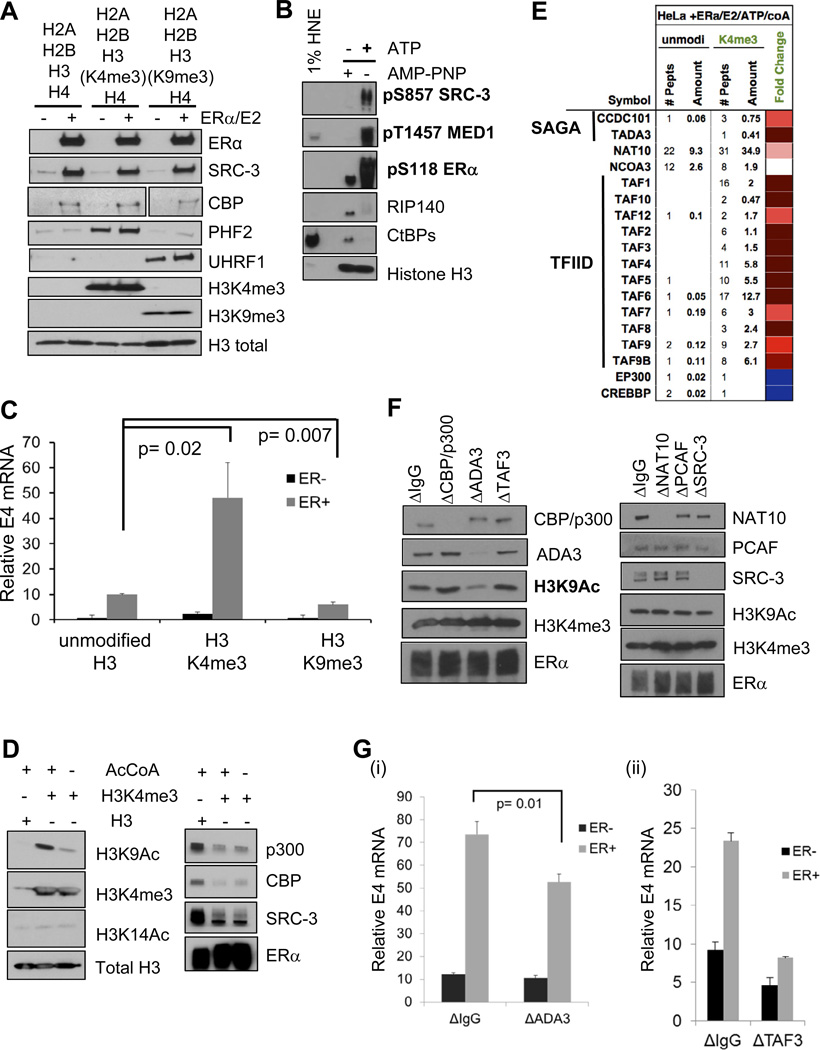
One limitation in the chromatin-based system for ERα function described above is that the arrangement and contribution of any particular mark present on the HeLa core histones cannot be ascertained or controlled during assembly. To solve this problem, we improved the system specificity by the inclusion of recombinant histones H2A, H2B, H3, and H4 and included specific chemically-modified histone H3 to mimic various methylations ((Simon et al., 2007); see Experimental Procedures). We then examined the effect of selected histone modifications on both transcription and CoR recruitment to ERE-E4 templates. We assembled recombinant nucleosomes bearing unmodified H3, H3K4me3, or H3K9me3, with/without added recombinant ERα, E2, and HNE. Micrococcal nuclease digestions showed roughly equal nucleosome laddering on all three H3 proteins (Figure S7A). We showed by immunoblotting after pulldown assays that the differentially marked H3 histones were assembled on the EREs and were competent for ‘reader’ binding (e.g., PHF2 for H3K4me3; UHRF1 for H3K9me3 (Nikolov et al., 2011)) (Figure 7A). Importantly, ERα, SRC-3, and CBP bound to all three H3-assembled nucleosomes equally well.
Figure 7. Differential ‘Reader’ Recruitment and Transcription Mediated by ERα on Recombinant Nucleosomes Bearing H3K4me3 or H3K9me3 Modifications.
(A) Controls for recombinant nucleosomes reconstituted with purified histones (NEB), or those bearing either H3K4me3 or H3K9me3 (Active Motif), assembled onto the 4xERE-E4 fragment and incubated with HNE for pulldown assays. PHF2 and UHRF1 are published H3K4me3 and H3K9me3 ‘readers’, respectively. Total histone H3 levels are also shown. Two lanes between H3K4me3 and H3K9me3 were cropped out of the CBP image, as they represented data not pertinent to Figure 7A.
(B) ATP, but not AMP-PNP, treatment of recombinant unmodified nucleosomal 4xERE-E4 results in ‘activating’ CoR phosphorylations (bolded) and dismissal of corepressors (RIP140, CtBPs) from complexes formed out of HNE with ERα and 100 nM E2.
(C) H3K4me3 present on the recombinant nucleosomal 4xERE-E4 fragment promotes more ERα-directed transcription than unmodified H3, while H3K9me3 further represses transcription. All histones were purchased from Active Motif, except for unmodified H3 (H3.2, NEB). Transcription reactions were performed with 100 nM E2, (−/+) ERα, and acetyl CoA (AcCoA). The bar graph shows the mean +/− SEM of three experiments, with each reaction done in duplicate (N=6). Significance was determined by t test.
(D) AcCoA/ATP addition to H3K4me3-containing nucleosomal templates promotes H3K9 acetylation (H3K9Ac) and CBP/p300 and SRC-3 dismissal. CoR complexes were formed on recombinant nucleosomes, then treated with ATP and AcCoA. AcCoA addition increased H3K9Ac, but not H3K14Ac (left panel). ATP addition triggers CBP/p300 and SRC-3 dismissal from H3K4me3 templates, but not unmodified nucleosomes (right panel).
(E) MS identification (shown as a heatmap) of CoRs/HATs stabilized in the presence of ATP/AcCoA on 4xERE H3K4me3-containing nucleosomes. MS data is shown for fold change of recruitment to H3K4me3 versus unmodified nucleosomes. Two SAGA complex members, 12 TFIID components, and NAT10 appear enriched under these conditions.
(F) Immunodepletion of ADA3 impairs H3K9Ac promoted by H3K4me3 on ERE-containing nucleosomes. ATP/AcCoA were added to H3K4me3 templates and H3K9Ac was assayed by immunoblotting.
(G) ADA3 and TAF3 depletion impairs ERα-driven H3K4me3 transcription from nucleosomal templates in vitro. (i) HNE with a reduced ADA3 level displays a significant reduction in ERα-driven transcription, as compared to control depleted HNE. (ii) TAF3 depletion from HNE has a substantial effect on ERα-driven transcription, as compared to control depleted HNE. The relative E4 mRNA level was represented as the mean +/− SEM, and statistical significance was determined by a two-tailed t test from independent duplicates. See also Figure S7 for additional controls for this figure.
We next tested if phosphorylation effects, similar to that observed with naked EREs, were induced by ATP addition to the unmodified nucleosomal ERE-CoR complexes. Indeed, we found that ATP, but not AMP-PNP, enhanced pS857 on SRC-3, pT1457 on MED1, pS118 on ERα, and led to dismissal of RIP140 and CtBPs (Figure 7B).
With these controls in place, we tested the effect of the different histone H3 tail marks on ERα-mediated in vitro transcription of ERE-E4 nucleosomes. We focused on H3K4me3 and H3K9me3, as these two ‘marks’ present at promoters correlate with transcriptionally active or repressed genes, respectively (e.g., (Li et al., 2007)). Consistent with these observations, H3K4me3-containing nucleosomes increased E2-liganded ERα-mediated transcription roughly five-fold over the unmodified H3, while H3K9me3 significantly inhibited the transcriptional activity (Figure 7C).
To address possible mechanisms to explain why H3K4me3 promotes more transcription directed by E2-liganded ERα versus that mediated by unmodified H3 or H3K9me3, we subjected the specific marked H3 recombinant nucleosomes on ERE-E4 to DNA pulldown-MS analyses. We first tested whether our previously defined CoR-ERα-ERE complexes seen on naked EREs were differentially enriched on one H3 mark versus the other. Similar CoRs were enriched with E2-liganded ERα on all three recombinant histone H3-assembled ERE templates (data not shown).
The above data suggested that the recruitment of distinct epigenomic ‘readers’, selectively enriched on H3K4me3 or H3K9me3 in the presence of E2-liganded ERα, could provide the mechanism. Indeed, we found at least six known H3K4me3 readers, DIDO1, SPIN1, CHD1, PHF8, PHF2, and ING1 (Bartke et al., 2010; Nikolov et al., 2011) associated with H3K4me3-assembled complexes (Figure S7Bi). H3K9me3, unlike H3K4me3, appeared to promote the binding of distinct ‘readers’, such as UHFR1, SFPQ, and NONO (Bartke et al., 2010; Nikolov et al., 2011) (Figure S7Bii). We further validated select CoRs (BRD2, CNOT3, and SMARCAD1) from the MS data, showing their preferential binding to H3K4me3 and H3K9me3-marked nucleosomes, respectively (Figure S7Biii). We then tested whether immunodepletion of CHD1 or BRD2 from HNE (Figure S7Ci) would affect ERα-directed transcription from H3K4me3 templates. However, neither CHD1 nor BRD2 loss had a significant effect on transcription (Figure S7Cii).
At this point, we considered that our binding assays might be missing an important ingredient. A clue came from performing ERα-driven transcription reactions with/without acetyl CoA (AcCoA), the donor substrate for protein acetylation. We found that the addition of AcCoA greatly stimulated transcription from H3K4me3 marked nucleosomes (Figure S7D). Based on this data, we tested whether H3K4me3 templates might influence subsequent histone acetylation events by treating washed ERE-CoR- nucleosomal complexes with AcCoA and ATP (to mimic transcription conditions), followed by immunoblotting for different histone H3 acetylations. Indeed, we found that H3K4me3 promoted acetylation of H3 at K9 (H3K9Ac), but not at other sites, including H3K14, K18, and K27 (Figure 7D, left panel; data not shown). Furthermore, CBP/p300 and SRC-3 now displayed reduced binding to H3K4me3 templates as compared to unmodified ones under these conditions (Figure 7D, right panel).
To find the relevant ‘readers’ under these transcription-mimic conditions, we subjected unmodified or H3K4me3-bearing nucleosomes to ERE DNA pulldowns, followed by AcCoA/ATP treatment, and MS analysis. Consistent with our immunoblotting data, CBP/p300 and SRC-3 were reduced on H3K4me3 as compared with unmodified H3 nucleosomes (Figure 7E). In addition, we observed two components of SAGA HAT complexes, NAT10 (a GCN5-like HAT), and 12 TAFs (part of the general factor TFIID) to have increased binding. We further validated that NAT10 and two TAFs displayed H3K4me3-preferential binding with just ATP addition (Figure S7E). We next tested whether immunodepletion of each of these CoRs might affect production of H3K9Ac on H3K4me3 templates (Figure 7F). Loss of CBP/p300, TAF3, NAT10, or SRC-3 had no effect on the H3K9Ac that occurred in response to prior H3K4me3. However, ADA3 depletion severely inhibited H3K9Ac, consistent with the loss of GCN5, PCAF, and CCDC101 (Figure S7F, left panel). As CCDC101 and TAF3 are known H3K4me3 ‘readers’ (Vermeulen et al., 2010; Vermeulen et al., 2007), we finally tested what effect their immunodepletion may have on ERα-driven transcription from H3K4me3 templates. While loss of ADA3 produced a small but significant reduction in H3K4me3 transcription (Figure 7Gi), we found that TAF3 depletion (Figure S7F, right panel) severely impaired ERα-driven transcription from H3K4me3 (Figure 7Gii).
Discussion
Our present study represents the most comprehensive identification of bona fide ERE-dependent CoR complexes, and our MS datasets represent a useful unbiased catalog of a variety of other potential interacting proteins for future studies. Here, we identify at least 17 endogenous CoRs enriched from MCF-7 cells with E2-liganded ERα on the nucleosome-free ERE-E4 template (Figure 1C). Consistent with our in vitro data, ERα was recently found associated with both coactivators (SRC-3, CBP/p300, and MED15) and corepressors (CtBPs, RIP140) in formaldehyde crosslinked MCF-7 cells (Mohammed et al., 2013). However, we provide a great deal of new mechanistic information concerning SRCs and components of ‘natural’ and nucleosomal ERE complexes. We found that SRC-3-CBP/p300-FOXO1 form a functional interaction ‘hub’ (Figures 2, S1Fi, 6B) and our kinetic data suggest, but not absolutely prove, coactivators and atypical corepressors may be present in the same complex (Figure S1E). A similar subset of stably-bound CoRs on a ‘natural’ single ERE-containing fragment from the human GREB1 gene was observed (Figures 5, S4, and S5), thereby substantiating the physiological relevance of our findings with artificial EREs. Nucleosomal ERE templates also recruited distinct CoRs, depending on the source of histones and in particular, the H3 ‘marks’ and whether ATP/AcCoA was present (Figures 6Cii, S7B, and 7E). Our study therefore provides a systematic definition of CoR complexes formed on different specific naked and nucleosomal EREs. Furthermore, we define mechanisms regulating CoR in vitro binding dynamics on these EREs.
Existence of Uniquely Stable CoR Complexes Bound to EREs
A completely unpredicted finding of our study is that once CoR-ERα-ERE complexes form from NE, they remain biochemically stable until ‘activating’ phosphorylation events ensue. This initial enhanced stability provides additional insight into multi-protein CoR complexes, in which multiple CoRs transform protein-protein ‘synergy’ into transcriptional ‘synergy’. We show that CoRs bound to E2-liganded ERα on EREs may exist in ‘stable sub-complexes’ based on the following data: 1) kinetics shows a subset of CoRs bind together (Figure S1E), 2) sensitivity to increasing salt and urea concentrations occurs during complex formation (Figure 2B), and 3) DNA-PKcs, p300, SRC-3, and MED26 co-fractionate with an ERE on a gel filtration column (Figure S5). Perhaps most remarkably, after complexes are formed, formerly ‘labile’ CoRs remain stable to challenge by harsh dissociation conditions. We provide additional data indicating that even an ‘individual’ CoR protein can have stabilizing effects on the overall complex on DNA. For example, full-length SRC-3 stabilizes the ERα-ERE interaction and p300 recruitment, while p300 stabilizes SRC-3 bound to ERα on EREs (Figure S2). Consider the enhancement in affinity for members of such binary complexes when the numbers of CoRs reach 10 or greater, a concept that makes the stability of CoR complexes to high salt and urea understandable. Also, DNA binding can affect receptor structure and CoR recruitment (e.g., SRCs and ERs (Wood et al., 2001)). Taken together, our data suggest that stable CoR complex formation is what occurs initially in cells and drives transcriptional synergy due to the cooperative energetics of multiple CoR-CoR, CoR-ERα, and ERα-ERE interactions.
Intriguingly, biochemically stable CoR-ERE complexes do not contain just coactivators, but also, contain certain ‘atypical’ corepressors. Thus, ‘activation’ of coactivators by phosphorylation may not be entirely responsible for robust signal-regulated transcription, but ‘transient’ corepressors also may need to be dismissed from an initial temporarily repressed and inactive complex. In particular, we see that RIP140 and CtBPs are recruited with E2-liganded ERα to EREs. In our system, E2 promoted the binding of stable CoR complexes, but it was only the addition of ATP and the act of phosphorylation that drove dismissal of these ‘transient’ corepressors from naked (Figures 3 and 5B) or nucleosomal EREs (Figure 7B).
Role of DNA-PK in ERα-Mediated Transcription- Dynamic ‘Activation’ of CoR Complexes
DNA-PK is a DNA-activated serine/threonine-directed protein kinase, consisting of a ~460 kDa catalytic subunit (DNA-PKcs) and two autoimmune Ku antigens, Ku70/Ku80 (Kong et al., 2011). Unlike its roles in DNA repair and V(D)J recombination, how DNA-PK may affect transcriptional regulation is less well understood. In our study, we found that DNA-PK mediates phosphorylation events to dynamically ‘activate’ CoR-ERα-ERE complexes for E2-mediated transcription (Figure 4).
When DNA-PKcs activity was reduced in vitro, we found that phosphorylation of many of the CoRs bound with ERα on EREs was affected (Figures 4A and 4B). It is formally possible that DNA-PK may not directly phosphorylate all of these CoRs, but rather some downstream kinase that is activated by DNA-PK participates, such as AKT (Feng et al., 2004). However, we saw very little effect on CoR phosphorylation when an AKT inhibitor was added to HeLa NE (data not shown). The net result of the DNA-PK dependent phosphorylation events is to produce ‘activation PTMs’ on ERα, MED1, and SRC-3, at residues in each protein that previously have been published to enhance transcription in cells (Ali et al., 1993; Belakavadi et al., 2008; Wu et al., 2004), and to dismiss RIP140. Consistent with this model, reduction of DNA-PKcs activity in cells (Figure 4C) or depletion from HNE reduced E2-mediated reporter gene transcription (Figure 6Biv). To validate that an event of DNA-PK phosphorylation that we detected in vitro (Figure 4) may occur inside cells, we performed ChIP experiments in MCF-7 cells and found that MED1 is phosphorylated at T1457 by DNA-PK at two different EREs (Figures S3 and 5C). Whether other CoRs are indeed bona fide DNA-PK targets at EREs inside cells is an important question for future studies.
Although DNA-PK association with and phosphorylation of ERα was previously reported (Ju et al., 2006; Medunjanin et al., 2010), how phosphorylation activated transcription remained unclear. Our present in vitro findings suggest that DNA-PK phosphorylates certain CoRs (SRCs and MED1) for their activation and the RIP140 corepressor for its dismissal, thereby providing missing mechanistic insight as to how DNA-PK can enhance ERα-mediated transcription.
Mechanism for ERα-Driven Transcription from Nucleosomal EREs Bearing H3K4me3
We found that H3K4me3 enhanced ERα-mediated transcription in vitro, as compared with unmodified H3, by promoting enhanced H3K9Ac and recruitment/stabilization of TFIID (Figures 7 and S7). Our data extend previous MS-based studies that identified H3K4me3-binding ‘readers’ on different DNA templates (Bartke et al., 2010; Nikolov et al., 2011). Unlike these studies, we found that unmodified H3, H3K4me3, and H3K9me3-containing ERE nucleosomes equally bound ERα, SRC-3, and CBP (Figure 7A), until the addition of ATP/AcCoA that revealed dynamic CoR complex changes. Namely, we found that as CBP/p300 and SRC-3 leave ERα-bound H3K4me3 EREs (Figures 7D and 7E), two distinct coactivating ‘readers’ of the H3K4me3 mark (CCDC101 and TAF3) appear, allowing for the subsequent production of H3K9Ac and efficient TFIID complex binding for enhanced initiation, respectively (Figures 7F, 7G, and S7E). While it was known that H3K4me3 and H3K9Ac co-localize at ERα target gene promoters in E2-treated MCF-7 cells (e.g., (Kwon et al., 2007)), which mark may be ‘dominant’ and occur first was not known. Here, we extend these studies and conclude that H3K4me3 is ‘dominant’ for the creation of H3K9Ac by CCDC101 recruitment of SAGA HAT complexes. In terms of the enhanced TFIID binding we observed on H3K4me3-bearing EREs, it was recently reported that p53-driven transcription of H3K4me3-marked nucleosomes is dependent on TAF3 by regulating preinitiation complex assembly (Lauberth et al., 2013). Our data with ERα also show a strong role for TAF3 in promoting transcription from H3K4me3-bearing EREs, but we suggest that CBP/p300 and SRC-3 subsequently may need to leave for SAGA and TFIID entrance and initiation to ensue. Future experiments will test this hypothesis.
Experimental Procedures
Human Cell Lines and Nuclear Extract (NE) Preparation
HeLa S3 and MCF-7 cells were obtained from the National Cell Culture Center (Minneapolis, MN) and the BCM Tissue Culture Core, respectively. Details on NE preparation are found in Supplemental Experimental Procedures.
Immunodepletions from NE
Specific details for particular antigen depletions are found in Supplemental Experimental Procedures.
Hormones, Chemicals, and Pharmacological Inhibitors
Sources of ligands and inhibitors are given in Supplemental Experimental Procedures.
ERE-DNA Pulldown Assays
Detailed ERE-DNA pulldown reaction conditions and how biotinylated EREs were made are listed in Supplemental Experimental Procedures.
Protein Identification by Mass Spectrometry (MS)
For MS analysis, the entire ERE pulldown reaction was scaled up two-fold and was separated on 1D SDS-PAGE. Additional details are found in Supplemental Experimental Procedures. All MS datasets described in this study are available online at http://www.epicome.org/ under msProjects/ERE Resource.
In Vitro Phosphorylation of CoR-ERα-ERE Complexes
After washing, complexes on magnetic beads were resuspended in 30 µl kinase reaction buffer and then incubated at 30°C for various time points. After the incubation period, final beads were resuspended in 2x SDS sample buffer for immunoblotting analysis. See Supplemental Experimental Procedures for further detail.
In Vitro Acetylation of Histone H3 in Nucleosomal EREs
After nucleosomal EREs were immobilized to Dynabeads, they were incubated with HNE and E2-liganded ERα and washed as above. Beads were resuspended in 30 µl kinase reaction buffer, 0.5 mM ATP, and 9 µM AcCoA and incubated at 30°C for 1 hour. Final beads were resuspended in 20–30 µl 2x SDS sample buffer for immunoblotting analysis.
siRNA Transient Transfections and ERE-Luciferase Reporter Analysis
Sources of siRNAs, controls checking CoR knockdown level, and details of how luciferase assays were performed in MCF-7 MAR-ERE-LUC cells are listed in Supplemental Experimental Procedures.
Chromatin Immunoprecipitation (ChIP)
ChIP assays were performed from MCF-7 cells incubated in stripped media for at least two days (as per (Metivier et al., 2003; Shang et al., 2000)), with additional specific details given in Supplemental Experimental Procedures.
Chromatin Assembly, Histones, and Micrococcal Nuclease (MNase) Analysis
Chromatin was assembled onto pERE or the biotinylated 921 bp 4xERE-E4 fragment using a recombinant Drosophila ACF1/ISWI/NAP-1 assembly system and assayed by MNase partial digestion (see Supplemental Experimental Procedures for more detail).
In Vitro Transcription
Detailed procedures for ERα-activated cell-free transcription reactions from ERE-containing nucleosomal templates are given in Supplemental Experimental Procedures.
Supplementary Material
Highlights.
-
➢
Proteomics of DNA-bound E2-liganded ERα defines an initial set of stably bound CoRs
-
➢
DNA-dependent protein kinase dynamically ‘activates’ CoR-ERE complexes
-
➢
An ERE from the GREB1 gene recruits similar CoR complexes as does consensus EREs
-
➢
Histone mark-dependent ‘readers’ produce direct effects on ERα-driven transcription
Acknowledgements
We thank W. Lee Kraus (UT Southwestern) for providing pERE, Steffi Oesterreich (University of Pittsburgh) for MCF-7 MAR-ERE-LUC cells, Malcolm Parker (Imperial College London) for RIP140 antibodies, Xiaoting Zhang (University of Cincinnati) for the pT1032 MED1 antibody, and the following colleagues at BCM: Judy Roscoe (MCF-7 cells); Nancy Weigel (AKT inhibitor); the Monoclonal Antibody/Recombinant Protein Shared Resource funded by NCI Cancer Center Support Grant 5 P30 CA125123 (recombinant protein expression); and Rainer Lanz, Giedre Krenciute, and Margaret S.Y. Fung (preparing data for epicome). This work was supported by funds from the National Institutes of Health (5 R01 HD008188 and 2 P01 DK059820 to B.W.O., and 5 K01 DK084209 and P30DK079638PJ4 to Q.F.) and the Welch Foundation (Q-152) to B.W.O.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Acevedo ML, Lee KC, Stender JD, Katzenellenbogen BS, Kraus WL. Selective recognition of distinct classes of coactivators by a ligand-inducible activation domain. Mol Cell. 2004;13:725–738. doi: 10.1016/s1097-2765(04)00121-2. [DOI] [PubMed] [Google Scholar]
- Ali S, Metzger D, Bornert JM, Chambon P. Modulation of transcriptional activation by ligand-dependent phosphorylation of the human oestrogen receptor A/B region. EMBO J. 1993;12:1153–1160. doi: 10.1002/j.1460-2075.1993.tb05756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartke T, Vermeulen M, Xhemalce B, Robson SC, Mann M, Kouzarides T. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell. 2010;143:470–484. doi: 10.1016/j.cell.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belakavadi M, Pandey PK, Vijayvargia R, Fondell JD. MED1 phosphorylation promotes its association with mediator: implications for nuclear receptor signaling. Mol Cell Biol. 2008;28:3932–3942. doi: 10.1128/MCB.02191-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M, Sharp PA. Human estrogen receptor forms multiple protein-DNA complexes. J Biol Chem. 1990;265:11238–11243. [PubMed] [Google Scholar]
- Bulynko YA, O'Malley BW. Nuclear Receptor Coactivators: Structural and Functional Biochemistry. Biochemistry. 2010 doi: 10.1021/bi101762x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldon CE, Sergio CM, Schutte J, Boersma MN, Sutherland RL, Carroll J, Musgrove EA. Estrogen regulation of cyclin E2 requires cyclin D1 but not c-Myc. Mol Cell Biol. 2009;29:4623–4639. doi: 10.1128/MCB.00269-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavailles V, Dauvois S, L'Horset F, Lopez G, Hoare S, Kushner PJ, Parker MG. Nuclear factor RIP140 modulates transcriptional activation by the estrogen receptor. EMBO J. 1995;14:3741–3751. doi: 10.1002/j.1460-2075.1995.tb00044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Riedl T, Washbrook E, Pace PE, Coombes RC, Egly JM, Ali S. Activation of estrogen receptor alpha by S118 phosphorylation involves a ligand-dependent interaction with TFIIH and participation of CDK7. Mol Cell. 2000;6:127–137. [PubMed] [Google Scholar]
- Chen H, Lin RJ, Schiltz RL, Chakravarti D, Nash A, Nagy L, Privalsky ML, Nakatani Y, Evans RM. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- Chen XF, Lehmann L, Lin JJ, Vashisht A, Schmidt R, Ferrari R, Huang C, McKee R, Mosley A, Plath K, et al. Mediator and SAGA have distinct roles in Pol II preinitiation complex assembly and function. Cell reports. 2012;2:1061–1067. doi: 10.1016/j.celrep.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhasarathy A, Kajita M, Wade PA. The transcription factor snail mediates epithelial to mesenchymal transitions by repression of estrogen receptor-alpha. Mol Endocrinol. 2007;21:2907–2918. doi: 10.1210/me.2007-0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnham PJ. Insights from genomic profiling of transcription factors. Nat Rev Genet. 2009;10:605–616. doi: 10.1038/nrg2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Park J, Cron P, Hess D, Hemmings BA. Identification of a PKB/Akt hydrophobic motif Ser-473 kinase as DNA-dependent protein kinase. J Biol Chem. 2004;279:41189–41196. doi: 10.1074/jbc.M406731200. [DOI] [PubMed] [Google Scholar]
- Goo YH, Sohn YC, Kim DH, Kim SW, Kang MJ, Jung DJ, Kwak E, Barlev NA, Berger SL, Chow VT, et al. Activating signal cointegrator 2 belongs to a novel steady-state complex that contains a subset of trithorax group proteins. Mol Cell Biol. 2003;23:140–149. doi: 10.1128/MCB.23.1.140-149.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich I, Flores AM, Aneskievich BJ. Corepressors of agonist-bound nuclear receptors. Toxicol Appl Pharmacol. 2007;223:288–298. doi: 10.1016/j.taap.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science. 2006;312:1798–1802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- Kang YK, Guermah M, Yuan CX, Roeder RG. The TRAP/Mediator coactivator complex interacts directly with estrogen receptors alpha and beta through the TRAP220 subunit and directly enhances estrogen receptor function in vitro. Proc Natl Acad Sci U S A. 2002;99:2642–2647. doi: 10.1073/pnas.261715899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein-Hitpass L, Kaling M, Ryffel GU. Synergism of closely adjacent estrogen-responsive elements increases their regulatory potential. J Mol Biol. 1988;201:537–544. doi: 10.1016/0022-2836(88)90635-3. [DOI] [PubMed] [Google Scholar]
- Klein-Hitpass L, Schorpp M, Wagner U, Ryffel GU. An estrogen-responsive element derived from the 5' flanking region of the Xenopus vitellogenin A2 gene functions in transfected human cells. Cell. 1986;46:1053–1061. doi: 10.1016/0092-8674(86)90705-1. [DOI] [PubMed] [Google Scholar]
- Kong X, Shen Y, Jiang N, Fei X, Mi J. Emerging roles of DNA-PK besides DNA repair. Cellular signalling. 2011;23:1273–1280. doi: 10.1016/j.cellsig.2011.04.005. [DOI] [PubMed] [Google Scholar]
- Kwon YS, Garcia-Bassets I, Hutt KR, Cheng CS, Jin M, Liu D, Benner C, Wang D, Ye Z, Bibikova M, et al. Sensitive ChIP-DSL technology reveals an extensive estrogen receptor alpha-binding program on human gene promoters. Proc Natl Acad Sci U S A. 2007;104:4852–4857. doi: 10.1073/pnas.0700715104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauberth SM, Nakayama T, Wu X, Ferris AL, Tang Z, Hughes SH, Roeder RG. H3K4me3 Interactions with TAF3 Regulate Preinitiation Complex Assembly and Selective Gene Activation. Cell. 2013;152:1021–1036. doi: 10.1016/j.cell.2013.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Romancer M, Poulard C, Cohen P, Sentis S, Renoir JM, Corbo L. Cracking the estrogen receptor's posttranslational code in breast tumors. Endocr Rev. 2011;32:597–622. doi: 10.1210/er.2010-0016. [DOI] [PubMed] [Google Scholar]
- Leahy JJ, Golding BT, Griffin RJ, Hardcastle IR, Richardson C, Rigoreau L, Smith GC. Identification of a highly potent and selective DNA-dependent protein kinase (DNA-PK) inhibitor (NU7441) by screening of chromenone libraries. Bioorg Med Chem Lett. 2004;14:6083–6087. doi: 10.1016/j.bmcl.2004.09.060. [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Malovannaya A, Lanz RB, Jung SY, Bulynko Y, Le NT, Chan DW, Ding C, Shi Y, Yucer N, Krenciute G, et al. Analysis of the human endogenous coregulator complexome. Cell. 2011;145:787–799. doi: 10.1016/j.cell.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna NJ, O'Malley BW. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell. 2002;108:465–474. doi: 10.1016/s0092-8674(02)00641-4. [DOI] [PubMed] [Google Scholar]
- Medunjanin S, Weinert S, Schmeisser A, Mayer D, Braun-Dullaeus RC. Interaction of the double-strand break repair kinase DNA-PK and estrogen receptor-alpha. Molecular biology of the cell. 2010;21:1620–1628. doi: 10.1091/mbc.E09-08-0724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng G, Zhao Y, Nag A, Zeng M, Dimri G, Gao Q, Wazer DE, Kumar R, Band H, Band V. Human ADA3 binds to estrogen receptor (ER) and functions as a coactivator for ER-mediated transactivation. J Biol Chem. 2004;279:54230–54240. doi: 10.1074/jbc.M404482200. [DOI] [PubMed] [Google Scholar]
- Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, Gannon F. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- Mohammed H, D'Santos C, Serandour AA, Ali HR, Brown GD, Atkins A, Rueda OM, Holmes KA, Theodorou V, Robinson JL, et al. Endogenous Purification Reveals GREB1 as a Key Estrogen Receptor Regulatory Factor. Cell reports. 2013;3:342–349. doi: 10.1016/j.celrep.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalvarte I, Schwend T, Gustafsson JA. Proteomics analysis of the estrogen receptor alpha receptosome. Molecular & cellular proteomics : MCP. 2010;9:1411–1422. doi: 10.1074/mcp.M900457-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasrin N, Ogg S, Cahill CM, Biggs W, Nui S, Dore J, Calvo D, Shi Y, Ruvkun G, Alexander-Bridges MC. DAF-16 recruits the CREB-binding protein coactivator complex to the insulin-like growth factor binding protein 1 promoter in HepG2 cells. Proc Natl Acad Sci U S A. 2000;97:10412–10417. doi: 10.1073/pnas.190326997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolov M, Stützer A, Mosch K, Krasauskas A, Soeroes S, Stark H, Urlaub H, Fischle W. Chromatin affinity purification and quantitative mass spectrometry defining the interactome of histone modification patterns. Molecular & cellular proteomics : MCP. 2011;10 doi: 10.1074/mcp.M110.005371. M110.005371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson TJ, Karmakar S, Pace MC, Gao T, Smith CL. The silencing mediator of retinoic acid and thyroid hormone receptor (SMRT) corepressor is required for full estrogen receptor alpha transcriptional activity. Mol Cell Biol. 2007;27:5933–5948. doi: 10.1128/MCB.00237-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–852. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- Simon MD, Chu F, Racki LR, de la Cruz CC, Burlingame AL, Panning B, Narlikar GJ, Shokat KM. The site-specific installation of methyl-lysine analogs into recombinant histones. Cell. 2007;128:1003–1012. doi: 10.1016/j.cell.2006.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward MM, Lee JS, O'Donovan A, Wyatt M, Bernstein BE, Shilatifard A. Molecular regulation of H3K4 trimethylation by ASH2L, a shared subunit of MLL complexes. Nat Struct Mol Biol. 2006;13:852–854. doi: 10.1038/nsmb1131. [DOI] [PubMed] [Google Scholar]
- Sun J, Nawaz Z, Slingerland JM. Long-range activation of GREB1 by estrogen receptor via three distal consensus estrogen-responsive elements in breast cancer cells. Mol Endocrinol. 2007;21:2651–2662. doi: 10.1210/me.2007-0082. [DOI] [PubMed] [Google Scholar]
- Vermeulen M, Eberl HC, Matarese F, Marks H, Denissov S, Butter F, Lee KK, Olsen JV, Hyman AA, Stunnenberg HG, et al. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell. 2010;142:967–980. doi: 10.1016/j.cell.2010.08.020. [DOI] [PubMed] [Google Scholar]
- Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA, Baltissen MP, Stunnenberg HG, Mann M, Timmers HT. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131:58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- Vo N, Fjeld C, Goodman RH. Acetylation of nuclear hormone receptor-interacting protein RIP140 regulates binding of the transcriptional corepressor CtBP. Mol Cell Biol. 2001;21:6181–6188. doi: 10.1128/MCB.21.18.6181-6188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitsman GE, Li L, Skliris GP, Davie JR, Ung K, Niu Y, Curtis-Snell L, Tomes L, Watson PH, Murphy LC. Estrogen receptor-alpha phosphorylated at Ser118 is present at the promoters of estrogen-regulated genes and is not altered due to HER-2 overexpression. Cancer Res. 2006;66:10162–10170. doi: 10.1158/0008-5472.CAN-05-4111. [DOI] [PubMed] [Google Scholar]
- Wood JR, Likhite VS, Loven MA, Nardulli AM. Allosteric modulation of estrogen receptor conformation by different estrogen response elements. Mol Endocrinol. 2001;15:1114–1126. doi: 10.1210/mend.15.7.0671. [DOI] [PubMed] [Google Scholar]
- Wu RC, Qin J, Yi P, Wong J, Tsai SY, Tsai MJ, O'Malley BW. Selective phosphorylations of the SRC-3/AIB1 coactivator integrate genomic reponses to multiple cellular signaling pathways. Mol Cell. 2004;15:937–949. doi: 10.1016/j.molcel.2004.08.019. [DOI] [PubMed] [Google Scholar]
- Xue Y, Wong J, Moreno GT, Young MK, Cote J, Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.