1. Overview, history and trajectory
Multi-subunit RNA polymerases (RNAPs) are ornate molecular machines that translocate on a DNA template as they generate a complementary RNA chain. RNAPs are highly conserved in evolution among eukarya, eubacteria, archaea and some viruses. As such, multi-subunit RNAPs appear to be an irreplaceable advance in the evolution of complex life on earth. Because of their stepwise movement on DNA, RNAPs are considered to be molecular motors, and, because RNAPs catalyze a templated polymerization reaction, they are central to biological information flow. Ubiquitous transcription of intergenic regions in eukaryotes has emerged as a process crucial for regulation of chromatin structure and synthesis of regulatory RNA molecules, and, in animals with complex body plans, promoter-proximal pausing of RNAP II is recognized as a common feature of gene transcription control. RNAPs synthesize very long polymers, so their exceptional processivity, accuracy, reaction to DNA lesions, response to accessory factors, termination and recycling are of significance. The carboxy terminal domain (CTD) of eukaryotic RNAP II is an unusual heptapeptide repeat (i.e. 52 nearly identical repeats in human RNAP II) that integrates regulated and dynamic interactions with initiation, capping, elongation, chromatin modification, termination, and RNA export factors. As the initial step in expression of genes, eukaryotic RNAPs are highly regulated with central roles in human AIDS, cancer, viral infection, leukodystrophies4 and normal development. Eubacterial RNAPs are targets for antibiotics.
The history of RNAP discovery and investigation of its mechanism and regulation appear as complex as RNAP itself. The central role of templated RNA synthesis in biological information flow was predicted by Jacob and Monod, and the enzymatic activity promoting formation of RNA polymers was reported in eubacteria and eukaryotes at that time by several groups. However, the DNA-dependent RNA polymerase proved elusive until 1960 when it was independently identified in bacteria by Hurwitz and Stevens and in plants by the Bonner group5. Later, investigations focused on mechanisms of RNA polymerization established that synthesis was not monotonous. Observations of distinct pauses by E. coli RNAP followed by isolation and characterization of individual stalled elongation complexes by the Chamberlin group6 lead to the “inchworming” model of RNAP movement along the template7 suggesting that RNAP translocation is driven by major conformational changes. The alternative mechanism, which explained RNAP pausing by backward sliding (backtracking) of RNAP along the DNA, accompanied by extrusion of the 3′-end of the nascent transcript, was proposed by investigators from the Goldfarb laboratory8. The backtracking mechanism was largely proposed from investigation of the thermodynamics of the transcription elongation complex by von Hippel9. Later, RNAP backtracking was observed in vivo in bacteria10 and confirmed by crystallographic studies11, and eukaryotic RNAP II undergoes backtracking during promoter-proximal pausing12. Transcriptional pausing by bacterial RNAP can also occur without backtracking, often in a hairpin-dependent manner13. Beyond the thermodynamic dimension, promoting backtracking from weaker to stronger RNA-DNA hybrids9, and the allosteric component, which is dependent on RNA hairpin interactions with the RNA exit channel14, the exact mechanisms underlying pausing remain poorly understood. Detailed analyses of the nucleotide addition cycle (NAC), including pre-steady-state kinetic methods15 and mutagenesis guided by computational approaches similar to the studies described in this issue by Weinzierl and by Wang, Feig, Cukier and Burton are required to understand the fine regulation of catalysis by RNAPs. An epochal advance in the understanding of RNAP structure-function was the solution of x-ray crystal structures for these large and dynamic enzymes both without and with associated nucleic acid scaffolds2,16. In 2006, Roger Kornberg was awarded the Nobel Prize in recognition in part of his success in determining yeast RNAP II structures.
Shortly after its discovery and in parallel with investigations of RNAP translocation along template, regulation by external factors emerged as an important mechanism of gene expression, starting with the bacterial sigma factor17. Building a detailed understanding of higher order structures, such as TFIID18, TFIIH19, SAGA20, mediator21 and the decorated CTD (Jeronimo, Bataille and Robert; Geyer and Eick; Cordon), is ongoing. Enhancer and silencer structure and function and their interaction with promoters remain incompletely understood22. Chromatin and histone transaction and modification factors have multiple dynamic and regulated interactions with RNAP and its accessory factors23. It is clear that gene expression and interacting epigenetics are at the very core of human development, viral infections, cancer and other disease, and in the future, the connection between gene expression, the transcription apparatus and human health will result in development of innovative and effective therapeutics24.
2. Genesis according to multi-subunit RNAPs
Multi-subunit RNAPs are a fascinating feature of evolution of complex life on earth. With minor gaps in the fossil and evolutionary record, multi-subunit RNAPs appear rooted the RNA world1. The initial templated RNAP was probably a ribozyme. In time, proteinaceous RNA-dependent RNAPs (RDRPs), which have living representatives in many eukaryotes, probably invaded and then replaced a primordial ribozyme precursor. Multi-subunit RNAPs share a core structure with these RDRPs (termed a “double-psi beta barrel (DPBB)” (Figure 1). Remarkably, DPBBs now found in the two largest subunits of multi-subunit RNAPs were generated from a primordial RDRP dimer. Connected to the DPBB is the defining DXDGD Mg2+ binding active site motif of eukaryotic RDRPs that corresponds to the nearly invariant NADFDGD Mg2+ binding site in RNAPs1a. The active site, buried deep within the structure and wedged between the two DPBBs, is a composite of from two (i.e. human, bacterial) to four (some archaea) subunits encoded by separate genes. Remarkably, the three domains of life, eukarya, eubacteria and archaea are fully dependent on multi-subunit RNAPs. As described in the Werner and Weinzierl reviews, eukarya have RNAPs I, II and III and some plants have RNAPs IV and V. Core catalytic function was entrenched and retained in evolution, and regulatory mechanisms have developed around interactions with RNAP surfaces far from the active center. Werner also describes regulation of elongation and factors that support high processivity as a probable evolutionary precursor to initiation control. For instance, high processivity is essential for end-to-end RNA genome replication.
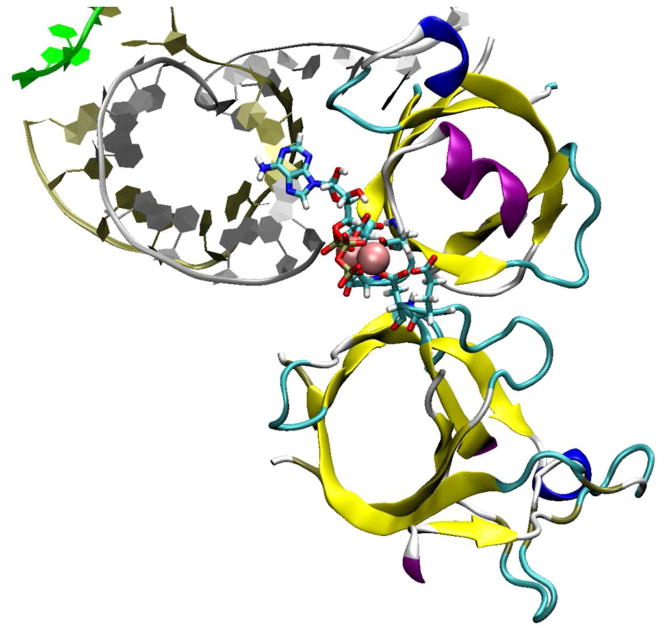
Figure 1.
Duplication and divergence of two double-psi β barrels (yellow), one from the β and one from the β′ subunit, describes evolution of the multi-subunit RNAP active site and may indicate a mechanism for high processivity and stepwise translocation1. An ATP substrate is shown with catalytic Mg2+ (magenta). RNA is silver; DNA template is gold; DNA non-template is green. The image is from pdb 205J2, prepared using visual molecular dynamics3.
Building organisms of increasing complexity and differentiation has divided RNAP I, II and III functions in eukarya and layered fascinating regulatory schemes on RNAP II, including the CTD (the RNAP II Rpb1 subunit carboxy terminal domain), the mediator, TFIID, TFIIH and SAGA and promoter-proximal pausing (regulation by DSIF-NELF involving the CTD and P-TEFb), discussed below. In evolution of modern animals and plants, these CTD-mediated innovations in control of gene expression very likely made possible complex differentiation of cell types. The evolution of the CTD is highlighted by Corden, and the roles of the CTD in driving evolution of eukarya are discussed by Eick and Geyer.
3. Getting the NAC: catalysis, translocation and fidelity
RNAPs have complex functions as targets of regulators, but the mechanism by which these enzymes acquire substrates, polymerize RNA and translocate on template are also of significant interest and importance. Encoded life is built on templated polymerization mechanisms including DNA replication, transcription and translation. Transcription (or RNA synthesis) involves RNAP translocating on DNA, reading a DNA template strand to polymerize a complementary RNA strand. The nucleotide addition cycle (NAC) is the process by which RNAP elongates a RNA polymer from a DNA template that involves NTP binding, NTP sequestration, NMP incorporation, pyrophosphate release and translocation (see the Werner review). Because living systems are fully dependent on templated polymerization reactions, transmission of the genomic code/cipher into gene products is fundamental. Because templated polymerization requires multiple similar substrates, errors in polymerization occur in replication, transcription and translation. Maintenance of adequate fidelity, therefore, is an essential feature of complex life. On the other hand, in order to evolve, living systems must cope with some coding errors. RNAP fidelity is discussed by Wang et al. and Weinzierl.
3.1. The RNAP factory (in vitro)
Molecular motions of RNAP domains during transcription elongation have been addressed using a combination of site-directed mutagenesis and biochemical assays. A high-throughput example of this approach (The RNAP “factory”) is described by Weinzierl. Archaeal RNAP is highly amenable to a factory analysis because, in some archaea, the two largest RNAP subunits found in eubacteria and eukarya are each divided into two polypeptide chains encoded by separate genes. This allows more efficient assembly of these archaeal RNAPs from recombinant proteins produced in E. coli, facilitating mutagenesis. Systematic substitution of the catalytic subunits of archaeal RNAP and subsequent high-throughput functional analyses of the mutants revealed flexible hinges in the bridge helix domain, which are important for transcription elongation. Weinzierl champions the use of all-atom molecular dynamics analyses to help interpret high-throughput mutagenic data.
3.2. Biophysics and translocation: RNAP as a motor and nanobot
Single molecule studies provide unique insights into the dynamics and functioning of complex macromolecular machines. As reviewed by Michaelis and Treutlein, transcription initiation, elongation dynamics, effects of nucleosome packaging of the template, and possible consequences of transcription errors have been addressed using single molecule approaches. Details of the initiation and elongation complex architecture can be viewed using nanopositioning systems. Optical tweezers are employed to analyze RNAP elongation and encounters with nucleosomes and terminators. RNAP-nucleosome transactions have also been viewed by atomic force microscopy. Sophisticated probes of transcription initiation and elongation are developed for in vivo studies resulting in observation of transcriptional bursting, initiation, elongation and factor recruitment and cycling.
Shimamoto considers RNAP as a complex nanoscale machine with diverse modes and functions. His review seeks to elucidate the biological roles of heterogeneity in RNAP locating a promoter, initiating transcription and commencing elongation. Investigations of molecular motions during transcription, including RNAP translocation, using single molecule approaches are also discussed in his review. The energetics and kinetics of RNAP translocation via a thermal ratchet or a power stroke are compared. The application of single molecule techniques to understand heterogeneity in initiation and elongation complexes is discussed. Despite enthusiasm for new methods, caution is recommended in interpreting results from some single molecule approaches including fluorescence resonance energy transfer.
3.3. Computational approaches
All-atom molecular dynamics simulation techniques and related approaches have been applied to multi-subunit RNAPs, reviewed by Wang et al. Because RNAP and DNAP mechanisms are so similar, both are considered. Quantum methods to understand the core RNAP and DNAP mechanisms are discussed. Molecular dynamics is computationally expensive and has limitations in making and breaking covalent bonds. Quantum approaches potentially solve this problem but are limited to a small collection of selected atoms and are difficult to correlate with experiment. An interesting discussion of basic residues termed “histidine and arginine micro-switches” that may interact with the NTP, RNA and DNA and drive conformational changes during each NAC is offered.
4. Regulation
Multi-subunit RNAPs are key enzymes in gene expression. RNAPs are subject to complex regulation by a multitude of protein factors during transcription initiation, elongation and termination. In eukaryotes, RNAP passage along the DNA leads to changes in chromatin packaging and histone modifications. Investigation of this regulation provides insights into regulation, RNA processing and export, and is essential for promoting understanding of cell division, differentiation and function.
4.1. The CTD
The Rpb1 CTD is a humbling topic, giving transcription in higher eukaryotes a distinctly Rube Goldberg look and flavor. The CTD, NELF (negative elongation factor), P-TEFb, promoter-proximal pausing, chromatin, epigenetics and interacting and modifying factors comprise a seemingly endless set of layered regulatory add-ons and compensations. The evolution of complex organisms from the single-celled appears to have been driven by the conflicting restrictions of the need for innovation and differentiation on a scaffold that will only tolerate limited stress to core essential functions. As a result, the final regulatory scheme seems overly intricate with layers upon layers of positive, anti-negative and negative regulation.
The RNAP II CTD is a repeat of heptapeptide units of varying length in different organisms. In humans, 52 near perfect repeats of the amino acid sequence 1-YSPTSPS-7 are found at the C-terminal end of the Rpb1 subunit. The repeat is heavily modified, for instance, by cyclin-dependent kinases, including TFIIH and P-TEFb. The CTD appears to function as an integrator to track the progress of RNAP II through the transcription cycle from initiation to elongation to termination, mRNA export and recycling. During initiation, the CTD is largely unmodified for interaction with enhancer-binding and promoter-binding regulators. The complex mediator, which envelops RNAP II, is a target of the unmodified CTD. During initiation, the CTD is heavily phosphorylated by TFIIH at SP serine 5. S5-phosphorylation appears to signal dissociation from mediator and promoter factors and entry into early elongation and is a precursor to 5′-end mRNA capping. Productive elongation is associated with a decrease in S5-phosphorylation and an increase in S2-phosphorylation, catalyzed by P-TEFb and related cyclin-dependent kinases. Modification of the CTD is important in recruitment of specific elongation, chromatin-modification, termination and recycling factors. Of course, modification marks must be placed, regulated and removed, and CTD reader factors must bind and dissociate. The CTD appears, therefore, to be an interaction scaffold for factors that regulate position and progression through the transcription cycle. Progression involves checkpoints regulated by cyclin-dependent kinases, phosphatases, ubiquitin ligases, protein degradation, acetylation/deacetylation and methylation/demethylation. Thus, the transcription cycle by RNAP II resembles a cell cycle in its inception and regulation. Three reviews in this issue relate to CTD evolution, readers, writers and control (Cordon; Jeronimo et al.; and Eick and Geyer).
Reviews in this issue focus on various aspects of the CTD complex contacts and roles. Evolution, composition and function of the CTD addressed by structural, biochemical and genetic approaches are reviewed by Corden. Eick and Geyer discuss the role of the CTD in evolution of eukaryotic organisms specifically emphasizing its essential role in splicing, which, in turn, increases the coding capacity of the eukaryotic genome. Nino, Herissant, Babour and Dargemont address the ornate machinery for mRNA export through the nuclear pore. Initial recruitment of export factors occurs co-transcriptionally and depends on the CTD and specific chromatin modifications (i.e. H2B ubiquitinylation). Jeronimo et al. focus on the CTD modification machinery and discuss the mechanisms of recruitment of chromatin modification factors and histone chaperons to the CTD. Lu, Li and Zhou focus on the role of CTD phosphorylation by P-TEFb in human AIDS, which leads to an increase in RNAP II processivity on pro-viral DNA, activating HIV-1 transcription.
4.2. mRNA nuclear export
In eukarya, transcription is decoupled from translation by the nuclear envelope and nuclear pore, but mRNA synthesis, processing, decoration, export and tagging for translation are coupled processes. The RNAP II CTD couples transcription to capping, splicing, 3′ end cleavage and polyadenylation. RNA synthesis is strongly coupled to export-competent mRNP transport through the nuclear pore. One purpose of coupling transcription, processing and handoff factors is to ensure the accuracy of mRNA processing and mRNP completion. After mRNA export to the cytoplasm, many transport factors dissociate to reenter the nucleus and restart the export cycle. Higher order cellular organization in eukaryotes, therefore, is coupled to mRNP production, fidelity and targeted mRNA decay.
4.3. Promoter-proximal pausing
In higher animals (including Drosophila), promoter proximal pausing of RNAP II has recently been recognized as an important mechanism of gene control. The essential evolutionary advance supporting this pausing mechanism appears to be the multi-subunit NELF (negative elongation factor) that cooperates with DSIF and P-TEFb. Organisms with NELF (i.e. Drosophila) demonstrate promoter-proximal RNAP II pausing, and organisms lacking NELF (i.e. C. elegans) do not. P-TEFb and related factors are found in virtually all eukaryotes. Two reviews in this issue deal with promoter proximal pausing. One from the Price laboratory stresses the importance of P-TEFb, NELF and Gdown1 in pausing control. Gdown1 is a factor that communicates to the mediator complex, making Gdown1 a link between initiation and elongation of RNAP II transcription. Protein factors involved in eukaryotic RNAP II initiation, elongation and termination are also reviewed by Guo and Price. They summarize the mechanisms of action of elongation factors identified to date; some, including TFIIS, TFIIF, ELL, Gdown1, GNAF, NELF and DSIF bind to core RNAP II. These factors affect the efficiency of promoter-proximal pausing observed in complex animals (i.e. Drosophila but not C. elegans).
A paper from the Zhou laboratory focuses on the regulation of HIV-1 (human immunodeficiency virus-type 1) transcription. The deadly human disease AIDS (auto-immune deficiency syndrome) is caused by HIV-1 infection. Attack by this virus involves regulation of early RNAP II elongation control mediated by the transcriptional regulator Tat, encoded by the virus, and a TAR RNA sequence just downstream of the viral promoter within the long terminal repeat (LTR). Transcription from the LTR regulates viral latency versus virus production, and clinical strategies versus AIDS involve suppression of latency to allow more complete and effective HIV-1 clearing using anti-viral agents. Positive Tat/TAR regulation involves host P-TEFb recruitment and binding of a host Super Elongation Complex (SEC), which is also recruited inappropriately in human lymphomas and leukemias. Inappropriate readthrough of an early RNAP II elongation checkpoint (promoter-proximal pausing), therefore, is an important issue in human development and disease.
4.4. Coupling of transcription and translation and the universal elongation factor
Function and conformational transitions of NusG, a bacterial analogue of metazoan DSIF (Spt5-Spt4; DRB-sensitivity inducing factor) and archaeal (and yeast) Spt5, are discussed by Tomar and Artsimovitch. Potential mechanisms of RNAP pause suppression by NusG and its paralog RfaH are considered: these proteins prevent transitions to an elemental pause state and might promote translocation by binding to RNAP and stabilizing the closed position of the downstream DNA-binding clamp. It remains to be established whether archaeal Spt5 and eukaryotic DSIF promote transcription elongation by a similar mechanism.
Prokaryotic NusG is related by evolution to the Spt5 subunit of DSIF/Spt5-Spt4, making this factor universal in transcription elongation control. In eubacteria, NusG couples transcription and translation. In Archaea, Spt5 is the NusG homolog. In eukarya, DSIF/Spt5-Spt4 takes on variable responsibility for elongation, depending on organismal complexity. In yeast, Spt5-Spt4 regulates elongation, and Spt5 is a major target of cyclin-dependent kinases and phosphatases that regulate the transcription cycle. In animals with more complex body structures, DSIF, NELF and P-TEFb cooperate to regulate promoter-proximal pausing of RNAP II. Artsimovitch describes eubacterial NusG and related factors. RfaH is a striking example, termed a “transformer” protein, that radically rearranges the secondary and tertiary structure of a domain to participate in transcriptional-translational coupling for a select set of bacterial genes.
5. RNAP and the genomic DNA
5.1. What could go wrong?
What could possibly go wrong in initiating on and transcribing across a complex, congested and dynamic genome and how could transcription and DNA structure anomalies be subverted in gene rearrangement and regulation? Two reviews address aspects of these broad questions. Belotserkovskii, Mirkin and Hanawalt summarize the effects of unusual DNA conformations on transcription, outlining several possible mechanisms of RNAP stalling, including guanosine-quadruplexes, triplex DNA, Z-DNA, H-DNA, cruciforms, (GAA)n repeats, RNA hairpins and R-loops. In immunoglobulin gene recombination switching, formation of R-loops exposes single-stranded DNA that can form G-quadruplexes. Telomere repeat-containing RNA (TERRA) also generates G-quadruplexes, indicating that these features of single-stranded RNA and DNA play key regulatory roles. YY1 binds extended DNA/RNA, as in R-loops, and supports X-chromosome silencing. Collisions between RNAPs or RNAPs and DNA polymerases can be highly consequential for generation of nucleic acid anomalies.
Thus, transcriptional processes may lead to genome modifications, including transcription-coupled DNA repair, as well as transcription-induced mutagenesis and recombination, reviewed in detail by Aguilera, Gaillard and Herrera-Moyano, who emphasize the impact of transcription on genome (in)stability and discuss various biological impacts of transcription-driven events leading to DNA modifications and damage. Ongoing transcription of genes is associated with recombination and mutation (TAM/TAR=transcription-associated mutation/recombination). TAM+TAR=TAGIN (transcription-associated genomic instability). TC-NER=transcription-coupled nucleotide excision repair. In many systems there is a strong bias for development of mutations in the non-template DNA strand for transcription versus the template DNA strand within a gene. TC-NER in bacteria is mediated by Mfd (mutation frequency decline) and Cockayne Syndrome B in complex eukaryotes and its homologue Rad26 in yeast. Interestingly, hyperactive NER can result in spontaneous mutations, indicating that RNAP helps direct and restrain NER targeting. Replication fork arrest due to collision between RNAP and DNAP can also enhance mutagenesis, recombination and generate chromosome instability. Biogenesis and export of mRNP through the eukaryotic nuclear pore is also coupled to TAR, possibly through generation of R loops (extended RNA/DNA hybrids displacing and exposing a long and fragile single stranded DNA).
5.2. The bacterial nucleoid
The role of transcription in organization of the bacterial nucleoid is elucidated by Jin, Cagliero and Zhou. Dramatic changes occur in genome compaction under different growth conditions that directly correlate with RNAP activity and distribution. They also suggest the concept of macromolecular crowding and discuss how extremely high concentrations of macromolecules in the bacterial cytoplasm might affect the conformation of DNA in the nucleoid. Because of its large size and fluid nature, probing nucleoid structure-function presents challenges. Gene amplification of rRNA genes via accelerated replication initiation from proximal oriC provides one mechanism to organize nucleoid dynamics. Studies of global transcription and gene regulation in eubacteria indicate that the genome is arranged as an evolved unit to support the optimal functioning of the organism during diverse growth conditions, sequestering and/or providing RNAP and cellular resources to match the needs of a cell to its changing environment. Transcription foci of dedicated RNAP (mostly for rRNA transcription) modulate nucleoid compaction. A model is developed to explain the interaction of RNAP with establishment and maintenance of the bacterial nucleoid structure, which is similar to the generation of the nucleolus around eukaryotic RNAP I foci.
5.3. Transcription factories (in vivo)
Papantonis and Cook propose a RNAP-centered model of genome and nucleus organization. They review experimental evidence indicating that, in the nucleus, RNAPs become immobilized on the matrix together with associated factors, and the DNA template is threaded through this macromolecular assembly. Higher organization of the nucleus (including the nucleolus) develops around RNAP factories. This model explains how the topological problem of RNA and DNA separation is resolved without intertwining during transcription, and serves as a foundation for a general model of gene regulation, proposed in the review. The concept of transcription factories has long been considered, but, with the development of new techniques, becomes easier to analyze and understand. The factory concept extends to many processes involving DNA, including replication, repair, chromosome maintenance, telomerase and recombination. The nucleolus is the prototypic factory for transcription by RNAP I, but RNAP II and III transcription units also organize and compress into factories building higher order units within the nucleus. Regulated bunching of RNAP II transcription units can begin to describe regulation of transcription by enhancers and silencers. Gene looping and mRNA export with linkage to the nuclear pore also support the transcription factory concept.
Biographies

Maria Kireeva started her research career as an undergraduate student of Moscow University in the Institute of Protein Research in Pushchino, Russia, participating in investigations of molecular mechanisms of bacterial translation and protein folding under the supervision of Dr. Anatoly Gudkov. She continued her education in the U.S., receiving a Ph.D. from the University of Illinois at Chicago in 1997 for characterization of mammalian growth factor-inducible immediate early genes in the laboratory of Dr. Lester Lau, and joining Dr. Mikhail Kashlev’s group at the NIH as a postdoc to study molecular mechanisms of eukaryotic transcription. Because of her unwavering passion to experimental research, Maria stayed in the same group as a staff scientist; she is currently interested in mechanisms of RNAP translocation and transcription fidelity, as well as RNA applications in nanomedicine and development of anti-viral therapeutics targeting viral RNA synthesis.

Dr. Mikhail Kashlev received his Ph.D. in molecular biology from Moscow Institute of Molecular Genetics in 1990. He was a postdoctoral fellow in the Department of Microbiology at Columbia University from 1991 to 1992 and a research associate at the Public Health Research Institute from 1993 to 1996. In 1996, he joined the ABL-Basic Research Program at the National Cancer Institute - Frederick and established the Molecular Mechanisms of Transcription Section. In 1999, he joined the Center for Cancer Research, NCI-Frederick where he is currently serving as a Senior Principal Investigator. Kashlev’s laboratory has been conducting biochemical and genetic studies of the mechanisms regulating transcription elongation and termination in prokaryotes and eukaryotes using E. coli and yeast S. cerevisiae as model organisms. His current research involves biochemical and genetic analysis of the mechanisms of transcription fidelity and impact of transcription errors to cell physiology, aging, and cancer and role of RNA polymerase in transcription coupled DNA repair.

Zachary Burton received his Ph.D. from UCLA in 1980, working in the laboratory of Dr. David Eisenberg. He was a postdoc in Dr. Richard Burgess’ laboratory until 1983 and a postdoc with Dr. Jack Greenblatt until 1987. Since then, he has been a professor at Michigan State University. Current research interests include RNAP structure, function, dynamics and fidelity.
References
- 1.(a) Iyer LM, Koonin EV, Aravind L. BMC Struct Biol. 2003;3:1. doi: 10.1186/1472-6807-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Iyer LM, Aravind L. J Struct Biol. 2012;179:299. doi: 10.1016/j.jsb.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, Artsimovitch I. Nature. 2007;448:157. doi: 10.1038/nature05932. [DOI] [PubMed] [Google Scholar]
- 3.Humphrey W, Dalke A, Schulten K. J Mol Graph. 1996;14:33. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 4.(a) Bernard G, Chouery E, Putorti ML, Tetreault M, Takanohashi A, Carosso G, Clement I, Boespflug-Tanguy O, Rodriguez D, Delague V, Abou Ghoch J, Jalkh N, Dorboz I, Fribourg S, Teichmann M, Megarbane A, Schiffmann R, Vanderver A, Brais B. Am J Hum Gen. 2011;89:415. doi: 10.1016/j.ajhg.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Tetreault M, Choquet K, Orcesi S, Tonduti D, Balottin U, Teichmann M, Fribourg S, Schiffmann R, Brais B, Vanderver A, Bernard G. Am J Hum Gen. 2011;89:652. doi: 10.1016/j.ajhg.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hurwitz J. J Biol Chem. 2005;280:42477. doi: 10.1074/jbc.X500006200. [DOI] [PubMed] [Google Scholar]
- 6.Krummel B, Chamberlin MJ. J Mol Biol. 1992;225:221. doi: 10.1016/0022-2836(92)90917-9. [DOI] [PubMed] [Google Scholar]
- 7.Uptain SM, Kane CM, Chamberlin MJ. Ann Rev Biochem. 1997;66:117. doi: 10.1146/annurev.biochem.66.1.117. [DOI] [PubMed] [Google Scholar]
- 8.(a) Komissarova N, Kashlev M. J Biol Chem. 1997;272:15329. doi: 10.1074/jbc.272.24.15329. [DOI] [PubMed] [Google Scholar]; (b) Nudler E, Mustaev A, Lukhtanov E, Goldfarb A. Cell. 1997;89:33. doi: 10.1016/s0092-8674(00)80180-4. [DOI] [PubMed] [Google Scholar]
- 9.von Hippel PH. Science. 1998;281:660. doi: 10.1126/science.281.5377.660. [DOI] [PubMed] [Google Scholar]
- 10.Toulme F, Mosrin-Huaman C, Artsimovitch I, Rahmouni AR. J Mol Biol. 2005;351:39. doi: 10.1016/j.jmb.2005.05.052. [DOI] [PubMed] [Google Scholar]
- 11.Cheung AC, Cramer P. Nature. 2011;471:249. doi: 10.1038/nature09785. [DOI] [PubMed] [Google Scholar]
- 12.(a) Nechaev S, Adelman K. BBA. 2011;1809:34. doi: 10.1016/j.bbagrm.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Nechaev S, Fargo DC, dos Santos G, Liu L, Gao Y, Adelman K. Science. 2010;327:335. doi: 10.1126/science.1181421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Landick R. Biochem Soc Trans. 2006;34:1062. doi: 10.1042/BST0341062. [DOI] [PubMed] [Google Scholar]
- 14.Artsimovitch I, Landick R. PNAS USA. 2000;97:7090. doi: 10.1073/pnas.97.13.7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kireeva M, Nedialkov YA, Gong XQ, Zhang C, Xiong Y, Moon W, Burton ZF, Kashlev M. Methods. 2009;48:333. doi: 10.1016/j.ymeth.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.(a) Cheung AC, Cramer P. Cell. 2012;149:1431. doi: 10.1016/j.cell.2012.06.006. [DOI] [PubMed] [Google Scholar]; (b) Wang D, Bushnell DA, Westover KD, Kaplan CD, Kornberg RD. Cell. 2006;127:941. doi: 10.1016/j.cell.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Vassylyev DG, Vassylyeva MN, Zhang J, Palangat M, Artsimovitch I, Landick R. Nature. 2007;448:163. doi: 10.1038/nature05931. [DOI] [PubMed] [Google Scholar]; (d) Zhang Y, Feng Y, Chatterjee S, Tuske S, Ho MX, Arnold E, Ebright RH. Science. 2012;338:1076. doi: 10.1126/science.1227786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burgess RR, Travers AA, Dunn JJ, Bautz EK. Nature. 1969;221:43. doi: 10.1038/221043a0. [DOI] [PubMed] [Google Scholar]
- 18.Bieniossek C, Papai G, Schaffitzel C, Garzoni F, Chaillet M, Scheer E, Papadopoulos P, Tora L, Schultz P, Berger I. Nature. 2013;493:699. doi: 10.1038/nature11791. [DOI] [PubMed] [Google Scholar]
- 19.(a) Compe E, Egly JM. Nat Rev Mol Cell Biol. 2012;13:343. doi: 10.1038/nrm3350. [DOI] [PubMed] [Google Scholar]; (b) Egly JM, Coin F. DNA repair. 2011;10:714. doi: 10.1016/j.dnarep.2011.04.021. [DOI] [PubMed] [Google Scholar]
- 20.Spedale G, Timmers HT, Pijnappel WW. Genes & Dev. 2012;26:527. doi: 10.1101/gad.184705.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.(a) Conaway RC, Conaway JW. BBA. 2013;1829:69. doi: 10.1016/j.bbagrm.2012.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Malik S, Roeder RG. Nature Rev Gen. 2010;11:761. doi: 10.1038/nrg2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roy AL, Sen R, Roeder RG. Trends in Immunol. 2011;32:532. doi: 10.1016/j.it.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.(a) Tomson BN, Arndt KM. BBA. 2013;1829:116. doi: 10.1016/j.bbagrm.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Smolle M, Workman JL. BBA. 2013;1829:84. doi: 10.1016/j.bbagrm.2012.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.(a) Smith E, Shilatifard A. Genes & Dev. 2013;27:1079. doi: 10.1101/gad.215137.113. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Beck DB, Oda H, Shen SS, Reinberg D. Genes & Dev. 2012;26:325. doi: 10.1101/gad.177444.111. [DOI] [PMC free article] [PubMed] [Google Scholar]