Abstract
We have explored the adaptation of the cytochromes P450 (P450) of deep-sea bacteria to high hydrostatic pressures. Strict conservation of the protein fold and functional importance of protein-bound water make P450 a unique subject for the studies of high pressure adaptation. Earlier we expressed and purified a fatty-acid binding P450 from the deep-sea bacteria Photobacterium profundum SS9 (CYP261C1). Here we report purification and initial characterization of its mesophilic ortholog from the shallow-water P. profundum 3TCK (CYP261C2), as well as another piezophilic enzyme, CYP261D1 from deep-see Moritella sp. PE36. Comparison of the three enzymes revealed a striking peculiarity of the piezophilic enzymes. Both CYP261C1 and CYP261D1 possess an apparent pressure-induced conformational toggle actuated at the pressures commensurate with the physiological pressure of habitation of the host bacteria. Furthermore, in contrast to CYP261C2, the piezophilic CYP261 enzymes may be chromatographically separated into two fractions with different properties, and different thermodynamic parameters of spin equilibrium in particular. According to our concept, the changes in the energy landscape that evolved in pressure-tolerant enzymes must stabilize the less-hydrated, closed conformers, which may be transient in the catalytic mechanisms of non-piezophilic enzymes. The studies of enzymes of piezophiles should help unravel the mechanisms that control water access during the catalytic cycle.
Keywords: pressure-tolerant enzymes, piezophiles, conformational heterogeneity, cytochrome, P450, monooxygenases, molecular mechanism
Cytochromes P450, the heme-thiolate enzymes that catalyze oxidation of a wide variety of endogenous and exogenous lipophilic compounds, play diverse physiological functions varying from the synthesis of steroid hormones, prostaglandins and second messengers to oxidation of exogenous compounds, such as drugs and other xenobiotics. Monooxygenation reactions require the input of two electrons (supplied by flavoprotein or ferredoxin partners) used to activate the dioxygen molecule. One oxygen atom is then inserted into the substrate, while the second atom is reduced and released in the form of a water molecule. However, an important fraction of the activated oxygen may be released without substrate oxidation through the “leaky” shortcuts in the catalytic cycle (see (1) for a review). This leakage, which results in a futile cycling and generation of reactive oxygen species, such as the superoxide anion-radical and hydrogen peroxide, is closely related to the degree of active site hydration. The mechanisms of conformational gating of the heme pocket that minimize futile cycling through regulation of its solvent accessibility, prevention of aberrant proton delivery and efficient expulsion of the water molecule formed in catalysis represents one of the most important challenges for the current mechanistic enzymology of cytochromes P450. In our search for new approaches to study the conformational dynamics during the P450 catalytic cycle, we turned our attention to the enzymes from the deep-sea organisms (piezophiles). This premise is that studies of the evolutionary adaptation of pressure-tolerant enzymes may provide indispensable information about functionally important conformational dynamics in the enzymes.
Studies of piezophiles have revealed a profound adaptation of their biochemical systems for function at elevated hydrostatic pressure (2-4). The most extensive studies have been devoted to the search for pressure-sensing systems and pressure-regulated genes, as well as to exploration of the basics of high pressure adaptation in biological membranes. However, the adaptation of the enzymes of piezophiles has received less attention and remains unexplored.
The basis of pressure effects on proteins is the change in system volume that accompanies biochemical processes. According to the Le Chatellier’s principle, increased pressure enhances those processes that are accompanied by a decrease in system volume, and conversely, inhibits processes occurring with a volume increase. The most important part of the volume changes in protein transitions is attributable to protein interactions with solvent (5-8). These include electrostriction of water on solvent-exposed charges, changes in the solvent packing around exposed nonpolar groups, and water penetration into the protein cavities (8-10). Pressure increase results in enhancement of protein hydration, which therefore constitutes the core of pressure-induced protein transitions.
Due to the fundamental role that the changes in protein hydration play in enzymatic mechanisms, the sensitivity of protein-solvent interactions to hydrostatic pressure constitutes the most important challenge for evolutionary adaptation in pressure-tolerant enzymes (“piezozymes”). In particular, this structural adaptation must prevent excessive protein hydration at critical steps of the catalytic cycle, while maintaining the functionality of water channeling to the active site when required. Therefore, this adaptation must change the energy landscape of the protein through redistribution of the populations of the enzyme conformers, so that the conformations that are only transient in the catalytic mechanisms of mesophilic enzymes may become stabilized in their piezophilic orthologs. Therefore, comparative studies of enzymes with different degree of high-pressure adaptation may provide a clue to functionally important conformational dynamics that are common to both piezozymes and their mesophilic orthologs.
According to our basic hypothesis, the role of structural adaptation to high pressure is especially important in enzymes with catalytic cycles that involve controlled expulsion and/or penetration of water molecule(s) from/into the active site. Perhaps the most illustrative example of such an enzyme system is represented by cytochromes P450. These enzymes, where the catalytic efficiency is largely determined by the degree of hydration and water accessibility of the active site, provide unparalleled opportunities for the studies of the mechanisms of high pressure adaptation. A comparative study with a series of homologous cytochromes P450 with different degree of piezophilic adaptation may provide a clue to understanding the mechanisms that regulate water accessibility of the active site during the catalytic cycle of the enzyme.
We recently reported expression and purification of the first P450 enzyme from a piezophilic bacterium, CYP261C1 or P450-SS9 from Photobacterium profundum SS9, and characterized its basic properties. Even the very first studies revealed a well-pronounced effect of high-pressure adaptation. Using pressure-perturbation absorbance spectroscopy we were able to demonstrate that this piezozyme possesses a pressure-actuated conformational toggle, which controls the solvent accessibility of the heme pocket and prevents the pressure-induced hydration of the active site (11-12).
In this article we report expression and purification of two other enzymes from the same P450 family - the mesophilic CYP261C2 from the shallow-water strain Photobacterium profundum 3TCK and the piezophilic CYP261D1 from the deep-sea bacterium Moritella sp. PE36. Comparative studies with the group of closely related P450 enzymes with different degree of piezophilic adaptation demonstrate the power of our strategy for elucidation of the functionally-important conformational dynamics in cytochromes P450.
Materials and methods
P450 cloning
Bacterial cytochromes P450 were cloned by add-on PCR as C-terminal hexa-histidine (His6) tag fusions into the Pharmacia plasmid pKK233-2. (GE Healthcare Life Sciences, Uppsala, Sweden).
The Moritella sp
PE36 CYP261D1 gene (NCBI reference sequence ZP_01899073.1) was PCR-amplified using the bacterial chromosomal DNA as a template. The PCR product was digested with BspHI and HindIII and digested PCR product was ligated to the vector digested with NcoI and HindIII, resulting in plasmid pKK233-P450-PE36.
The genes encoding CYP261C1 and CYP261C2 were obtained by amplification of the YP_133374.1 gene from the chromosomal DNA of P. profundum SS9 and the ZP_01217946.1 gene from the DNA of P. profundum 3TCK respectively. The corresponding PCR products were digested with BspHI and NsiI restriction enzymes and inserted into pKK233-2 vector digested with NcoI and PstI. The resulting plasmids are termed pKK233-P450-SS9 and pKK233-P450-3TCK. The nucleotide sequence of the amplified genes was verified by DNA sequencing and found to be identical to the genomic sequence.
Expression and initial purification of CYP261C1, CYP261C2 and CYP261D1
The recombinant proteins were expressed in E. coli TOPP3 cells harboring the corresponding plasmids. Four liters of bacterial cultures were grown to mid-log phase (OD600≈0.6) and supplemented with 80 μg/ml of γ-aminolevulinic acid (γ-ALA). Protein expression was induced by the addition of isopropyl β-D-1-thiogalactopyranoside (IPTG) to a final concentration of 0.5 mM. Cells were grown for an additional 48 h at 30 °C and harvested by centrifugation.
Crude extracts were prepared by resuspending the cell pellet in 5 volumes of 100 mM Hepes, pH 7.4, containing 500 mM KCl and 10% glycerol (buffer A) following by sonication of the cell suspension. The extracts were clarified from cell debris by centrifugation for 1 h at 100,000 g at 4 °C.. The extracts were clarified from cell debris by centrifugation for 1 h at 100,000 g at 4 °C and loaded onto a nickel-NTA resin (Qiagen, USA) column (1 ml of the gel per 60 nmols of P450). The column containing the protein-bound resin was washed consequently with 5 volumes of Buffer A containing 20 mM imidazole and 15 volumes of the same buffer with no imidazole added. Proteins were eluted with the same buffer containing 250 mM imidazole.
Separation of (+) and (−) forms of CYP261C1 and CYP261D1
Separation of CYP261C1 and CYP261D1 into two stable forms termed here as (+) and (−) conformers was achieved with two sequential ion-exchange chromatography steps using anion- and cation-exchange resins. The crude preparation obtained as described above was diluted with 10 volumes of 10% glycerol in deionized water and 10 volumes of 10 mM Na-Hepes buffer (pH 7.4) containing 10% glycerol. The diluted solution was applied to a column of Macro-Prep High S Support resin (Bio-Rad Laboratories, Hercules, CA; 1 ml of the gel per 40 nmol P450) equilibrated with 10 mM Na-Hepes buffer pH 7.4 containing 10% glycerol. Application to the column resulted in binding of ~50% of the applied P450 protein. Unbound protein was applied to a column of Toyopearl DEAE-650S resin (Tosoh Bioscience, Japan; 1ml of the gel per 100 nmol P450 ).
The Macro-Prep High-S column with the bound (+) form of the protein was washed with 10 volumes of 100 mM Na-Hepes buffer containing 10% glycerol. The protein was eluted with a gradient of 0 – 300 mM of KCl in100 mM Na-Hepes buffer containing 10% glycerol. The fractions containing P450, which elutes at 180-250 mM KCl, where pooled, concentrated to a protein concentration of 150-300 μM, and dialyzed against 100 mM Na-Hepes buffer, pH 7.4, containing 10% glycerol, 150 mM KCl and 3 mM TCEP.
The column of Toyopearl DEAE-650S with bound − form of the protein was washed with 10 volumes of 100 mM Na-Hepes buffer containing 10% glycerol, and the protein was eluted with the same buffer containing 300 mM KCl. The protein was concentrated and dialyzed as described above for the (+) forms of the proteins.
Determination of molecular masses
of purified preparations of CYP261D1 was done by matrix-assisted laser desorption/ionization time-of-flight spectroscopy (MALDI-TOF) with the use of a Voyager DE-STR mass spectrometer (Applied Biosystems, Foster City, CA). Precise positions of the peaks in the mass spectra were determined by their approximation with a combination of Gaussian and Lorenzian distributions with the use of GRAMS AI spectroscopy software as provided in GRAMS 8.0 suite (ThermoFisher Scientific, Philadelphia, PA).
Absorbance spectra
were recorded with a MC2000-2 CCD rapid scanning spectrometer (Ocean Optics Inc., Dunedin, FL). In the experiments at ambient pressure this instrument was equipped with a custom-made thermostated cell chamber with a magnetic stirrer and a L7893 UV-Vis fiber optics light source (Hamamatsu Photonics K.K., Japan). All experiments were performed at 25 °C in 0.1 M Na-Hepes, pH 7.4, containing 0.5mM EDTA.
Pressure-perturbation studies
were performed as described (13) using a custom-built high-pressure optical cell (14) connected to a manual pressure generator (High Pressure Equipment, Erie, PA) capable of generating a pressure of up to 6000 bar.
Analysis of series of spectra
obtained in absorbance spectroscopy experiments was done using a principal component analysis (PCA) method, which is also known as singular value decomposition (SVD) technique, as described earlier (15-16). To interpret the spectral transitions in terms of the changes in the concentration of P450 species, we used a least-squares fitting of the spectra of principal components by the set of the spectral standards of pure high-spin, low-spin and P420 species of the heme protein (15-17). In the case of pressure-perturbation studies the series of spectra were corrected for solvent compression as described (18).
Fitting of the titration curves
obtained in our absorbance titration and pressure-perturbation experiments was done with the use of the equation for the isotherm of bimolecular association ((19) p 73, eq. II-53), which is also known as “tight binding” or “square root equation”
Approximation of the ionic strength dependencies
of the spin state of cytochromes P450 was performed quantitatively with a semi-empirical relationship similar to that used by Peyser and co-authors to analyze the effect of ionic strength on the conformation of myosin (20). Following this approach the fraction of the high-spin heme protein in the enzyme-substrate complex (Fh) is represented as a function of the degree of occupation of a putative ion-binding site in the protein (20):
| (1) |
where [I] is the ion concentration (which is formally equivalent to the ionic strength in this context), , , and are the high-spin fraction of P450 at ion concentration [I], and at zero and infinite ionic strength respectively, and Kion is the equilibrium constant of ion binding to a putative ion binding site in the enzyme. Despite the evident oversimplifications in this model, equation (1) provides a necessary quantitative measure for comparing CYP261 enzymes in terms of responsiveness to ionic strength changes.
Results and Discussion
CYP261 family: a group of putative fatty acid hydroxylases with different degrees of high-pressure adaptation
Our search through genomes of deep-sea bacteria revealed sequences encoding cytochrome P450 proteins in Moritella sp. strain PE36 and Photobacterium profundum strain SS9 (11). Both strains have evolved to live at pressures of 250 – 500 bar (21). The proteins from Moritella and Photobacterium display 46% sequence identity… This comparative study also required closely related mesophilic proteins as reference points. The genome of the shallow water strain of Photobacterium, P. profundum 3TCK also encodes a P450 protein, which is 97% identical to the piezozyme from the strain SS9. Very recently we also found a close mesophilic ortholog of the piezozyme from Moritella in the partially sequenced genome of the marine bacterium Moritella viscosa, a causative agent of winter ulcer in atlantic salmon (22). This putative P450 enzyme is 87% similar to the enzyme from Moritella sp. PE36. Although we have not yet expressed and purified this enzyme, we use its sequence in the comparative analysis of amino acid substitutions involved in high-pressure adaptation.
The four proteins belong to the P450 family 261. The proteins from P. profundum SS9 and 3TCK constitute subfamily 261C and are designated P450 261C1 (CYP261C1) and P450 261C2 (CYP261C2) respectively. The enzymes from Moritella sp. P36 and Moritella viscosa belong to the subfamily 261D. They are referred as CYP261D1 and CYP261D2 respectively. Alignment of the sequences of these four enzymes may be found in the supporting material to this publication.
A BLAST search with CYP261 sequences through the genome database showed that all four proteins have important homology to P450s known as hydroxylases of fatty acid and their derivatives, such as CYP4V, CYP4F and CYP4X enzymes, or P450BM-3. Consistent with these findings, the screening of potential substrates of CYP261C1 showed that this enzyme has the highest affinity for saturated fatty acids with medium chain length (such as myristic acid) (11-12).
Expression, and initial purification of CYP261 enzymes
We recently reported expression and purification of the first CYP261 enzyme, CYP261C1 from piezophilic Photobacterium profundum SS9 (P450-SS9), and characterized its basic properties (11-12). For this study we changed the expression system for this enzyme. We cloned CYP261C1 into the pKK233-2 vector (Pharmacia) and expressed this protein in E. coli strain TOPP-3. The use of this expression system allowed us to increase the yield of the protein from 200 to ~500 nmol/l. A similar approach was used with two other members of the same family – CYP261C2 and CYP261D1. We amplified the genes encoding these P450 proteins, incorporated a hexa-histidine tag at the C-termini and cloned them into the pKK233-2 vector (Pharmacia). In the expression system with TOPP-3 the yield of CYP261D1 (P450-PE36) was as high as ~1200 nmol/liter of culture. Although the level of expression of CYP261C2 (P450-3TCK) was considerably lower (~200 nmol/liter), it was high enough to provide a sufficient amount of purified protein. Initial purification of the proteins was performed with the use of Ni-NTA agarose, similar to that described for CYP261C1 (11-12).
All three purified CYP261 enzymes are characterized by a significant displacement of the spin equilibrium toward the high-spin state (60-90%) and exhibit a maximum of the Soret band of the ferrous carbonyl complexes at 445-446 nm, similar to CYP261C1 (11-12).
Pressure-induced transitions in CYP261 enzymes reveal a contrast between the piezophilic and mesophilic orthologs
The formalism behind the use of pressure in studies of protein transitions is based on the general relationship that defines the dependence of chemical equilibrium on temperature (T) and pressure (P):
| (2) |
where ΔG, ΔH, ΔU, ΔV and ΔS are the changes in free energy, enthalpy, internal energy, volume, and entropy; R is the gas constant and Keq is the equilibrium constant. According to this relationship, the molar volume change of a chemical transition can be determined from the pressure dependence of ΔG at constant temperature:
In practice, the effect of pressure on ΔU and ΔS (i.e. the pressure dependence of the changes in isothermal compressibility, ΔβT) in protein transitions is small, and the plots of ln(Keq) versus pressure are usually linear, at least in the pressure range below 2 kbar (23-25). In good agreement with this analysis, the pressure dependencies of the logarithm of constant of spin equilibrium (Kh) in most cytochromes P450 studied to date are linear (13, 26-29).
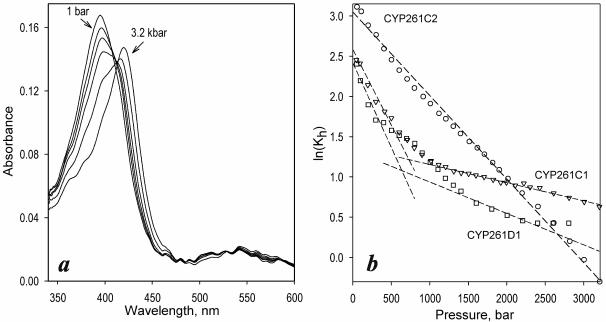
At first glance, the effect of pressure on the absorbance of CYP261 enzymes is quite similar to that observed with other cytochromes P450. At the initial steps of pressure increase, the Soret band of the heme protein gradually shifts from 393 to 416 nm, which is indicative of a pressure-induced spin transition (Fig 1a). At pressures above 1500 bar this reversible process was complemented with a pressure-induced conversion into the inactive P420 state. Similar to that observed earlier with CYP3A4 (17), this pressure-induced inactivation involves an irreversible slow phase (τ>10 min), so that the content of the P420 state gradually increases upon incubation at pressures >1500 bar. In contrast, the reversible pressure-induced spin shift was very rapid. No time-dependent changes in the spin state of the enzyme were detected upon incubation for up to 30 minutes at high pressures (data not shown).
Figure 1.
Effect of hydrostatic pressure on CYP261 proteins. (a) a series of spectra of absorbance of CYP261C2 measured at the pressures of 1 bar and 1, 1.4, 1.6, 1.8, 2.4 and 3.2 kbar. (b): Pressure dependencies of ln(Kh) for CYP261C1 (triangles), CYP261D1 (circles) and CYP261D1 (squares). Dashed lines show linear approximations of the initial and final parts of the plots.
Detailed analysis of pressure induced transitions in these enzymes reveals a striking difference between the piezophilic CYP261C1 and CYP261D1 and the mesophilic CYP261C2. As we reported earlier, increasing pressure fails to displace the spin equilibrium in CYP261C1 completely to the low-spin state, in contrast to other P450s. The pressure dependencies of the logarithm of the constant of spin equilibrium (Kh) in this enzyme revealed a well-pronounced break (Fig. 1b, triangles). Such nonlinearity of the plots of ln(Keq) versus pressure are often observed in systems involving several interlinked equilibria with different thermodynamic parameters (25). Consequently, we interpret the break in the pressure dependencies of ln(Kh) observed with CYP261C1 as an indication of a conformational splitting of the enzyme between two states with different thermodynamic parameters of spin equilibrium (11). The behavior of CYP1261C1 versus pressure is consistent with a pressure-induced conformational transition with P½ in the range of 400-600 bar which is close to the physiological pressure of habitation of P. profundum (11-12).
Our experiments with CYP261D1 revealed very similar behavior (Fig 1b, squares). In contrast, the pressure dependence ln(Kh) in CYP261C2, a mesophilic ortholog of CYP261C1, was linear in the whole pressure range, similar to other mesophilic P450s, such as P450cam or P450BM-3. The positive value of 18.0 ± 3.6 ml/mol found for ΔVspin from a series of four individual measurements with CYP261C2 is also very close to published ΔVspin values for the substrate-free forms of other cytochromes P450, such as P450cam (20 ml/mol), P450BM-3 (23 ml/mol), CYP2B4 (21 ml/mol) or CYP3A4 (14 ml/mol) (27-28).
These results suggest strongly that the break in the pressure dependence of ln(Kh) in CYP261C1 and CYP261D1 represents a consequence of the high-pressure adaptation in these enzymes. The piezophilic P450 enzymes appear to possess a pressure-actuated conformational toggle, which controls the solvent accessibility of the heme pocket and prevents the pressure-induced hydration of the active site.
Conformational splitting in piezophilic enzymes revealed in purification trials: ambiguous folding of pressure-adapted proteins at ambient pressure?
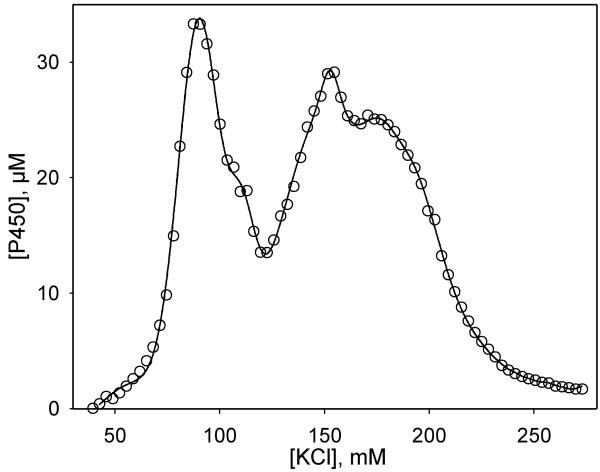
In order to better purify the CYP261 proteins for crystallization trials we attempted to use cation-exchange chromatography, as applied to other P450s. To our surprise, only about 50% of the P450 pool of the piezophilic CYP261C1 and CYP261D1 is retained by Macro-Prep High-S cation-exchange resin. In contrast, retention of the proteins by the anion-exchanger (Toyopearl DEAE-650S) approaches 100%. However, elution of the proteins from the DEAE column with a KCl gradient revealed two separate P450 fractions (Fig. 2). The spectra of the ferrous carboxycomplexes revealed no measurable content of the P420 state in the purified preparations of both forms of either CYP261C1 or CYP261D1 and exhibited Soret bands positioned at 445.5 nm, similar to our earlier report on the crude preparation of CYP261C1 (11-12). This unusual heterogeneity was characteristic of both CYP261D1 and CYP261C1, but absent in mesophilic CYP261C2, which binds equally well to the cation- and anion-exchangers and elutes as a single band from both resins. This contrast between CYP261C1 and CYP261C2 is especially stunning in view of the 97% sequence similarity between these proteins, which differ by 11 amino acid residues only (see Fig. S1 in Supporting Information).
Figure 2.
Profile of elution of CYP261D1 from Toyopearl DEAE-650S resin.
In our view, this unusual conformational splitting may reveal a consequence of high-pressure adaptation in CYP261C1 and CYP261D1, which have evolved to fold and function at elevated pressure. Transfer of these proteins from the physiological 250-500 bar to ambient pressure results in an important change in the energetic landscape of the protein and therefore causes an ambiguity in folding when the proteins are expressed in E. coli grown at ambient pressure. Stabilization of two dissimilar conformers with very different thermodynamic parameters of the spin shift in CYP261D1 and CYP261C1 (but not in the mesophilic CYP261C2) may provide an important insight into the mechanism of the conformational control of spin equilibrium and its relationship to protein hydration.
The (+) and − conformers exhibit pronounced differences in the position of spin equilibrium and its dependence on ionic strength
In order to explore the conformational heterogeneity further we developed a 3-column procedure for purification of each of the two stable conformers of CYP261C1 and CYP261D1 (See Materials and Methods). These two forms of the piezophilic proteins we designate as (+) and − conformers (for the fraction binding to the cation-exchanger and the one retained by DEAE-resin only, respectively). Analyses of the fractions of CYP261D1 with MALDI-TOF mass spectroscopy showed that both preparations are homogenous and have similar molecular masses. The masses of the (+) and − forms were 55,222 and 55,224 Daltons respectively, which is in a good agreement with the value of 55,220 Daltons calculated for the hexahistidine-tagged CYP261D1 with a cleaved N-terminal methionine. These results suggest that the primary structure of the two conformers is identical, so that the divergence of the piezozyme into two stable fractions cannot be explained by a posttranslational modification.
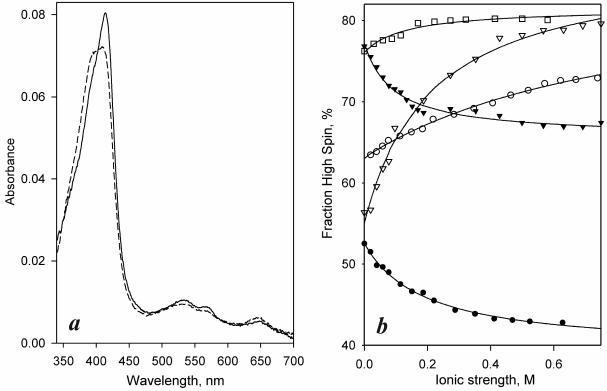
Despite similarity in their primary structure the (+) and − conformers exhibit clear differences in the position of spin equilibrium and its dependence on ionic strength (Fig 3, Table 1). In the (+) form of CYP261D1 the spin equilibrium is considerably more shifted toward the high-spin state then in CYP261D1− (Fig. 3a). Interestingly, both conformers of this enzyme exhibit an unusual response to increasing ionic strength, which displaces the spin equilibrium toward the low spin state (Fig 3b, filled symbols). This direction of changes is opposite to that observed with other cytochromes P450 studied up to date (30-36).
Figure 3.
Differences in position of spin equilibrium and their dependencies on ionic strength among different forms of CYP261 enzymes. (a) Absorbance spectra of purified CYP261D1− and CYP261D1(+) conformers (solid and dashed lines respectively). (b) Effect of ionic strength on the spin state of CYP261C2 (open squares) and the (+) (triangles) and − (circles) conformers of CYP261C1 (open symbols) and CYP261D1 (closed symbols). Solid lines represent the approximations of the data sets with a hyperbolic equation.
Table 1. Effect of ionic strength on the position of spin equilibrium in CYP261 species*.
| P450 species |
Fh(0), %a | ΔFh, %b | Kion, mMc |
|---|---|---|---|
|
| |||
| 261C1(−) | 62.6 ± 1.9 | 19.4 ± 5.9 | 832 ± 89 |
| 261C1(+) | 61.2 ± 2.8 | 23.1 ± 8.7 | 150 ± 89 |
| 261C2 | 74.7 ± 10.8 | 6.0 ± 4.2 | 89 ± 49 |
| 261D1(−) | 58.0 ± 10.6 | −15.0 ± 3.4 | 163 ± 52 |
| 261D1(+) | 72.6 ± 2.6 | −13.2 ± 4.1 | 138 ± 42 |
The values given in the table represent the averages of 2-4 individual measurements, and the “±” values show the confidence interval calculated for p = 0.05.
P450 high-spin content in cytochrome P450 extrapolated to zero ionic strength (see equation (1)).
Maximal amplitude of the change in the content of the P450 high spin state induced by an increase in ionic strength.
Apparent equilibrium constant of ion binding to a putative ion binding site found by approximation of the ionic strength dependencies with equation (1).
In contrast to CYP261D1, the effect of ionic strength on spin equilibrium in both conformers of CYP261C1 (Fig 3b, open symbols) is similar to that observed with other cytochromes P450. The difference in the spin state of (+) and − conformers at low ionic strength is also opposite to that of CYP261D1. However, the effect of ionic strength on the spin equilibrium is considerably more pronounced in CYP261C1(+) than in CYP261C1− (Fig. 3b, open triangles). Accordingly, at an ionic strength above 200 mM the (+) form of the enzyme becomes more enriched in the high-spin state than the − conformer, similar to CYP261D1.
Importantly, the mesophilic CYP261C2 (Fig 3b, dotted symbols) exhibits a considerably higher content of the high-spin state than either of the two conformers of its piezophilic ortholog. Furthermore, the magnitude of the ionic-strength-induced changes in CYP261C2 is considerably lower than that exhibited by either CYP261C1(+) or CYP261−. These differences in the effect of ionic strengths on the piezozyme and its mesophilic ortholog may indicate that the evolutionary adaptation of CYP261 enzymes to high hydrostatic pressure involves an important rearrangement of intramolecular electrostatic interactions.
Interactions of CYP261 enzymes with fatty acids
The parameters of interactions of (+) and − forms of CYP261 enzymes with fatty acids are summarized in Table 2. Similar to earlier findings with a CYP261C1 preparation containing both (+) and − forms (11-12), interactions of all forms of CYP261 enzymes with myristic acid results in a displacement of the spin equilibrium to the high spin state, whereas the binding of the unsaturated arachidonic acid results in the opposite direction of the spin shift. As seen from Table 2, the (+) and − conformers of CYP261C1 and CYP261D1 exhibit some modest differences in the affinities for both substrates and distinct differences in the amplitudes of the substrate-induced spin shift. However, most important is the contrast between the mesophilic CYP261C2 and the piezozymes in their affinity for arachidonic acid. As seen from Table 2, the KD of the complex of CYP261C2 with arachidonate is an order of magnitude higher than the values for either CYP261C1 or CYP261D1. The difference between CYP261C1 and CYP261C2 is especially interesting in view of very high sequence identity between these proteins (11 amino acid residue differences; see Fig. S1 in Supporting Information). Importantly, analysis of the position of these dissimilar residues shows no differences between CYP261C1 and CYP261C2 in the regions of putative substrate recognition (SRS) defined by analogy with other cytochromes P450 (Fig S1 in Supporting information). Therefore, decreased affinity of CYP261C2 for arachidonic acid seems to reflect some indirect effect of a conformational difference between CYP261C2 and CYP261C1 caused by amino acid substitutions outside of the substrate binding pocket.
Table 2. Interactions of CYP261C and CYP261D with fatty acids*.
| P450 species |
Arachidonic acid | Myristic acid | ||
|---|---|---|---|---|
|
| ||||
| ΔFh, %a | KD, μM | ΔFh, %a | KD, μM | |
|
| ||||
| 261C1(−) | −6.3 ± 2.7 | 9.7 ± 4.4 | 27.3 ± 1.4 | 8.1 ± 2.9 |
| 261C1(+) | −11.2 ± 4.3 | 10.4 ± 2.4 | 18.4 ± 3.7 | 3.6 ± 0.9 |
| 261C2 | −15.4 ± 7.9 | 89.0 ± 7.4 | 12.2 ± 4.3 | 1.4 ± 0.3 |
| 261D1(−) | −2.9 ± 1.3 | 8.9 ± 2.2 | 11.3 ± 5.8 | 3.0 ± 1.0 |
| 261D1(+) | −5.9 ± 1.6 | 3.1 ± 0.6 | 3.7 ± 2.0 | 2.4 ± 0.7 |
The values given in the table represent the averages of 2-4 individual measurements and the “±” values show the confidence interval calculated for p = 0.05.
Maximal amplitude of the ligand-induced change in the content of the P450 high spin state.
Effect of pressure on thermodynamic parameters of spin transitions in CYP261D1
As discussed above, non-linearity in pressure dependencies of spin equilibrium in crude preparations of CYP261C1 and CYP261D2 (Fig. 1b) was interpreted as an indication of a divergence of the piezophilic enzymes between two conformers with different thermodynamic parameters (ΔS and ΔV) of spin equilibrium. Partitioning between these conformers is controlled by a pressure dependent equilibrium with P½ of 400 – 600 bar.
We next studied the effect of pressure on the temperature dependence of spin equilibria in purified (+) and − conformers. Comparison of thermodynamic parameters (ΔH and ΔS) of equilibrium determined at different pressures is an attractive alternative to probing the linearity of the pressure dependence of ln(Keq). Comparison of ΔH and ΔS values obtained at different pressures assesses directly the effect of pressure on ΔS and ΔV and may be used to reveal non-linear behavior of the system and pressure-dependent conformational heterogeneity. Furthermore, in the case of a “linear” system where the value of ΔS remains constant irrespective of pressure, the molar volume change may be calculated from the enthalpy changes determined at pressures p1 and p2 using a simple relationship:
| (3) |
The studies of temperature dependencies at different pressures have been successfully used by Balny and Hooper to demonstrate pressure-dependent conformational transition in hydroxylamine oxidoreductase (37).
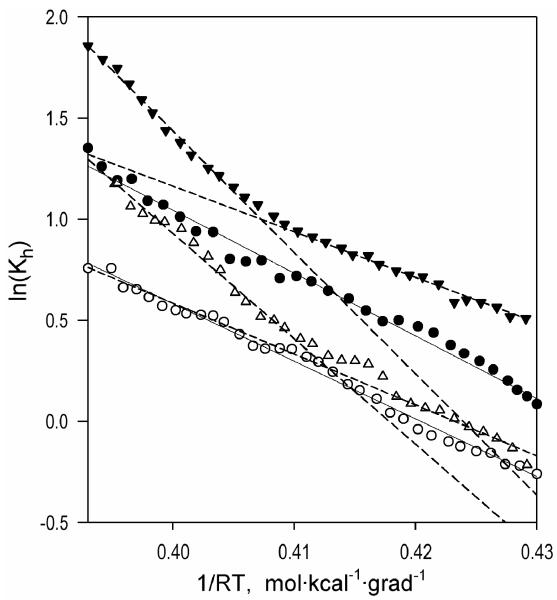
The temperature dependencies of (+) and − conformers of CYP261D1 obtained at two different pressures are illustrated in Figure 4, which shows the Van’t Hoff plots for the constant of spin equilibrium (Kh) in CYP261D1(+) (triangles) and CYP261D1− (circles) at ambient pressure (closed symbols) and a pressure of 500 bar (open symbols). In CYP261D1(+) these dependencies exhibit a break at approximately 18-22 °C, after which the ΔHspin increases considerably. In contrast, the Van’t Hoff plots for the − form of the enzyme are linear, so that the value of ΔHspin remains constant in the whole temperature range studied (+2 − +32 °C).
Figure 4.
Van’t Hoff plots for the temperature dependencies of the spin equilibrium in CYP261D1− (circles, solid lines) and CYP261D1(+) (triangles, dashed lines) placed at the pressures of 1 and 500 bar (filled and empty circles, respectively).
In further analysis we focused on the low-temperature (<20°C) portions of these plots (below the break observed in (+) conformer). As seen from the values of the thermodynamic parameters determined in this range of temperatures (Table 3), the behavior exhibited by CYP261(+) is consistent with a simple reversible pressure-dependent equilibrium. Here pressure exerts no appreciable effect on either ΔSspin or ΔVspin (which is defined as the molar change in the system volume in a transition from the low-spin to the high-spin state) calculated according to equation (3). The latter is equal to +18.3 ml/mol, which is similar to value of +18.0 ± 3.6 ml/mol determined from the pressure dependencies ln(Keq) in CYP261C2 in this study and commensurate with the values of 14 -23 ml/mol obtained with the substrate-free forms of other cytochromes P450 (27-28).
Table 3. Effect of pressure on thermodynamic parameters of spin transitions in CYP261D1.
| P450 species |
Pressure, bar |
ΔH, kcal/mol | ΔS, cal/mol·grad | ΔV, ml/mola |
|---|---|---|---|---|
| 261D1(−) | 1 500 |
32.5 ±3.9 22.7 ±6.3 |
118.8 ±12.5 62.2 ±6.9 |
−46.8 |
| 261D1(+) | 1 500 |
21.7 ±1.4 25.5 ±8.8 |
81.7 ±5.5 89.0 ±33.1 |
+18.3 |
The values given in the table represent the averages of 2-4 individual measurements, and the “±” values show the confidence interval calculated for p = 0.05.
Apparent value of molar volume change in low-to-high spin transition calculated according to equation (2).
In contrast, the CYP261D1− conformer reveals a fundamental deviation from the canonical behavior. Here increasing pressure results in a decrease in the ΔHspin, so that the value of ΔVspin determined according to (2) appears to be negative. This sign of ΔVspin suggests a pressure-induced transition to the high-spin state, which is not consistent with our experimental observations. This contradiction is caused by the fact that the requirement of pressure-insensitivity of ΔS, which is necessary for equation (3) to be applicable, does not hold in this case. The ΔSspin in this conformer at ambient pressure is considerably higher than that observed in CYP261D1(+) and exhibits a pronounced decrease with increasing pressure. This analysis reveals a pronounced non-linearity in the pressure-related behavior of the − conformer and suggests an involvement of pressure-induced changes in the partitioning of two different substates of CYP261D1− that differ in the thermodynamic parameters of spin transitions.
Notably, the increase in pressure to 500 bar, which is commensurate with the physiological pressure of habitation of the host bacteria, diminishes the contrast between the conformers by rendering the thermodynamic parameters of spin transitions nearly similar (Fig. 4, empty symbols). This convergence of the properties at increased pressure indicates that the conformational difference between the (+) and − forms of the enzyme is closely related to the difference between the hypothetical “relaxed” and “pressure-promoted” conformations, which is fundamental to the mechanism of the putative pressure-actuated conformational toggle (11). According to the above analysis, the − conformer exhibits a pressure-dependent equilibrium between two substates with different parameters of spin equilibrium, whereas the (+) conformer appears to be stabilized in the conformation nearly similar to that of the pressure-promoted state of the − conformer.
“Hot spots” in the structure of CYP261 enzymes: analysis of the amino acid substitutions evolved in piezozymes
Marked differences between the piezophilic CYP261C1 and CYP261D1 and mesophilic CYP261C2 in the interactions with ion-exchange resins and in the effect of ionic strength on the position of spin equilibrium suggests that the mechanism of high-pressure adaptation must involve a change in surface exposure of charged amino acid residues and a rearrangement of a network of intramolecular salt bridges in the enzyme. Therefore, our analysis of amino acid substitutions evolved in piezozymes was based on a search for cassettes of spatially-adjacent substitutions that involve appearance, elimination or substitution of charged residues (D,E,K,R).
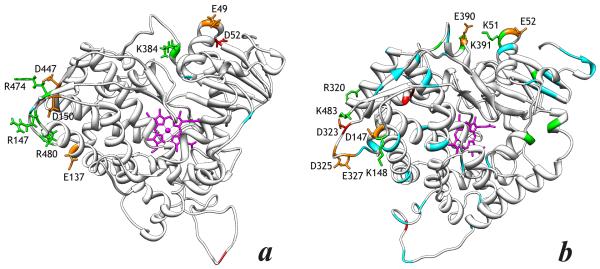
The positions of dissimilar amino acid residues in each of the 261C1/C2 and 261D1/D2 pairs are illustrated in the alignment shown in Fig. S1 in the Supplementary Information. In order to evaluate spatial relationship between the individual substitutions we used SWISS-MODEL Workspace (38) to build homology models of the two piezophilic proteins based on the known X-ray structure of the I401P mutant of the heme domain of P450BM-3 (39), which shares 25%-26% sequence identity with CYP261 proteins. Positions of the individual substitutions mapped on these models are illustrated in Fig. 5.
Figure 5.
Homology model of the structucture of piezophilic CYP261C1 (a) and CYP261D1 (b). The positions of the substitutions that result in appearance, elimination or substitution of charged residues (D,E,K,R) are highlighted in green (Arg and Lys residues) and red (Glu and Asp). Positions of other substitutions are shown in cyan. The substitutions presumably involved in the mechanism of high-pressure adaptation are highlighted by displaying their side chains. The side chains of potentially important Glu and Asp residues involved in charge-pairing interactions, which are conserved within each of the CYP261C1/C2 and CYP261D1/D2 pairs, are highlighted in orange.
In the case of CYP261C1 and 261C2, which have only 11 dissimilar residues, a pair of N55D and P384K substitutions (the substitutions are termed here using the CYP261C2 sequence as a base) may be the most important, as it may create a new salt bridge between the α-helices A and K in CYP261C1. Electrostatic interactions in this region may also involve the spatially-adjacent Glu-59 (which is present in both CYP261C1 and CYP261C2). Interestingly, the CYP261D1/D2 pair also display a group of substitutions in the same regions, namely R51K, S391K, and K394A, which are likely to modify the electrostatic interactions between the helices A and K in the piezozyme. Here the electrostatic interactions may also involve Glu-390 and Glu-52, which are conserved between 261D1 and 261D2 proteins.
Another group of structurally-important substitutions in the CYP261C1/C2 pair may include K147R in the C/D loop and a cassette of Q474R, T477P and K480R in the C-terminal portion of the protein. These four substitutions are spatially adjacent and may affect the conformation and mobility of the C/D loop, which is important for the heme pocket accessibility in P450 enzymes. The network of salt bridges in this region is likely to involve adjacent Glu-137, Asp-150 and Asp-447. Here, again the CYP261D1/D2 pair reveals the closely related differences located in the same region of the protein. Here the list of substitutions include the C-terminal residue K428 which is absent in 261D2, Q148K in α-helix D and N323D and Q320R in the loop between the helices I and J. Similar to the suggestion about the CYP261C pair, these substitutions between CYP261D1 and D2 are like to modify the interactions in the region of the C-terminal loop of the protein.
Therefore, the analysis of the amino acid substitutions in CYP261C1/C2 and CYP261D1/D2 reveals similar patterns in both pairs of a piezozyme with its mesophilic ortholog. The most probable intramolecular interactions involved in the conformational adaptation to high hydrostatic pressure in CYP261C1 and CYP261D1 include the interactions between α-helices A and K and the salt bridges between the C-terminal loop, I/J loop and α-helix D.
The conformational splitting revealed in piezozymes may provide a clue to general mechanisms of conformational gating of the active site in cytochromes P450
The conformational splitting that we discovered in pressure-adapted CYP261 enzymes has an important parallel with the apparent conformational heterogeneity revealed in such microsomal cytochromes P450 as CYP2B4, CYP3A4, and P450scc. The early evidence of such heterogeneity was obtained in studies of the kinetics of P450 reduction by artificial donors, such as dithionite (40) or eosin radical (41) and the kinetics of CO-recombination with P450(Fe2+) (42-43). An important indication of divergence of the pool of microsomal cytochromes P450 into two non-interconverting sub-populations is the differences in the kinetics of NADPH- and dithionite-dependent reduction between the high- and low-spin states of the heme protein, which are otherwise expected to exist in rapid equilibrium. This striking inconsistency demonstrated for CYP2C11 (44), CYP2B4 (40), and most recently for CYP3A4 (13, 45), suggests that the term “spin equilibrium” is inapplicable to the P450 pool taken as a whole, but that there are several slowly-interconverting sub-populations of the enzyme with different position of spin equilibrium. Further evidence includes the heterogeneity in the resonance Raman spectral properties (46) and sensitivity to high hydrostatic pressures (15, 18, 47-48). Most of these experiments were done with CYP2B4 and CYP3A4, although some of the studies involve cytochrome P450scc (46, 48), and CYP2E1 (47).
Divergence of pressure-adapted CYP261 heme proteins into two forms with different thermodynamic parameters of spin transitions which may be separated chromatographically appears to be closely related to above indications of conformational heterogeneity in microsomal P450 enzymes. Furthermore, pressure-induced conformational switching observed in CYP261D1 and CYP261C1 closely resembles the situation with microsomal CYP3A4 in the complexes with allosteric substrates, where hydrostatic pressure could not completely displace the spin equilibrium toward the low-spin state (28). In our opinion, the transition to a state with confined water accessibility seen in CYP261C1 and CYP261D1 may represent a common feature of cytochromes P450. This state may be transient in the P450 catalytic and serve to coordinate the heme pocket hydration with substrate binding, interactions with redox partners and the redox state of the enzyme. We propose that alteration of the conformational equilibria that distinguish CYP261C1 and CYP261D1 from CYP261C2 has evolved in the piezophilic proteins to allow this conformational gating mechanism to function at pressures of habitation of their host bacteria. Hence, characterization of these enzymes at ambient pressure will provide insight into functionally-important conformational transition in the P450 family.
Concluding remarks
Our comparison of CYP261 enzymes from organisms with different degree of high-pressure adaptation suggests strongly that the unique pressure-related behavior of CYP261C1 and CYP261D1 represent a consequence of the high-pressure adaptation in these enzymes. These results demonstrate an exceptional potential of the studies of conformational adaptation of piezozymes to their function at high hydrostatic pressure. Merging molecular evolution with biochemical thermodynamics allows a new strategy for exploration of protein conformational mobility through pressure-perturbation studies in a series of enzymes with different degree of evolutionary adaptation to high pressure.
Besides their importance for molecular enzymology and protein biophysics, the studies of high-pressure adaptation have a high value for biotechnology and bioengineering. The concept of changing the direction of enzymatic reactions and increasing the efficiency of catalysis by high hydrostatic pressure is widely discussed in modern literature (49-51). An increase in hydrostatic pressure often provides a potent means to change the substrate specificity and stereoselectivity of enzymatic catalysis (52). Elevated pressures are known to increase temperature stability of proteins (10, 50), and the combination of high-pressures with increased temperatures enables an increase in enzyme turnover by over an order of magnitude (10, 51). However, at present the application of these pressure-based approaches in biotechnology is restricted to a limited number of enzymes, the function of which is not obstructed by increased hydration. Functional and structural comparison of piezozymes with their mesophilic counterparts will facilitate elaboration of the universal principles of engineering piezotolerant enzymes, thus extending the applicability of pressure-based approaches in biotechnology.
Supplementary Material
Acknowledgements
Authors are grateful to Dr. Majid Gassemian (UC San Diego Biomolecular/Proteomics Mass Spectrometry Facility, UC San Diego) for his assistance with MALDI-TOF assays. This work was supported in part by NIH grant GM054995.
List of abbreviations
- CYP261C1
cytochrome P450 261C1 from Photobacterium profundum SS9
- CYP261C2
cytochrome P450 261C2 from P. profundum 3TCK
- CYP261D1
cytochrome P450 261D1 from Moritella sp. PE36
- CYP261D2
putative cytochrome P450 from M. viscosa.
References
- [1].Zangar RC, Davydov DR, Verma S. Mechanisms that regulate production of reactive oxygen species by cytochrome P450. Toxicol. Appl. Pharmacol. 2004;199:316–331. doi: 10.1016/j.taap.2004.01.018. [DOI] [PubMed] [Google Scholar]
- [2].Abe F. Exploration of the effects of high hydrostatic presure on microbial growth, physiology and survival: perspectives from piezophysiology. Biosc. Biotechnol. Biochem. 2007;71:2347–2357. doi: 10.1271/bbb.70015. [DOI] [PubMed] [Google Scholar]
- [3].Bartlett DH. Microbial life at high pressures. Sci. Progress. 1992;76:479–496. [PubMed] [Google Scholar]
- [4].Kato C, Bartlett DH. The molecular biology of barophilic bacteria. Extremophiles. 1997;1:111–116. doi: 10.1007/s007920050023. [DOI] [PubMed] [Google Scholar]
- [5].Cioni P, Gabellieri E. Protein dynamics and pressure: What can high pressure tell us about protein structural flexibility? Biochim. Biophys. Acta. 2011;1814:934–941. doi: 10.1016/j.bbapap.2010.09.017. [DOI] [PubMed] [Google Scholar]
- [6].Kornblatt JA, Kornblatt MJ. The effects of osmotic and hydrostatic pressures on macromolecular systems. Biochim. Biophys. Acta. 2002;1595:30–47. doi: 10.1016/s0167-4838(01)00333-8. [DOI] [PubMed] [Google Scholar]
- [7].Low PS, Somero GN. Protein hydration changes during catalysis - new mechanism of enzymic rate-enchancement and ion activation inhibition of catalysis. Proc. Natl. Acad. USA. 1975;72:3305–3309. doi: 10.1073/pnas.72.9.3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Low PS, Somero GN. Activation volumes in enzymic catalysis - their sources and modification by low-molecular-weight solutes. Proc. Natl. Acad. Sci. USA. 1975;72:3014–3018. doi: 10.1073/pnas.72.8.3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Royer CA. Revisiting volume changes in pressure-induced protein unfolding. Biochim. Biophys. Acta. 2002;1595:201–209. doi: 10.1016/s0167-4838(01)00344-2. [DOI] [PubMed] [Google Scholar]
- [10].Mozhaev VV, Lange R, Kudryashova EV, Balny C. Application of high hydrostatic pressure for increasing activity and stability of enzymes. Biotechnol. Bioeng. 1996;52:320–331. doi: 10.1002/(SICI)1097-0290(19961020)52:2<320::AID-BIT12>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- [11].Sineva EV, Davydov DR. Cytochrome P450 from Photobacterium profundum SS9, a piezophilic bacterium, exhibits a tightened control of water access to the active site. Biochemistry. 2010;49:10636–10646. doi: 10.1021/bi101466y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Sineva EV, Davydov DR. Constrained water access to the active site of cytochrome P450 from the piezophilic bacterium Photobacterium profundum. High Pressure Res. 2010;30:466–474. doi: 10.1080/08957959.2010.535208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Davydov DR, Davydova NY, Halpert JR. Allosteric transitions in cytochrome P450eryF explored with pressure-perturbation spectroscopy, lifetime FRET, and a novel fluorescent substrate, Fluorol-7GA. Biochemistry. 2008;47:11348–11359. doi: 10.1021/bi8011803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hui Bon Hoa G, Marden MC. The pressure dependence of the spin equilibrium in camphor-bound ferric cytochrome P-450. Eur. J. Biochem. 1982;124:311–315. doi: 10.1111/j.1432-1033.1982.tb06593.x. [DOI] [PubMed] [Google Scholar]
- [15].Davydov DR, Deprez E, Hui Bon Hoa G, Knyushko TV, et al. High-pressure-induced transitions in microsomal cytochrome P450 2B4 in solution: evidence for conformational inhomogeneity in the oligomers. Arch. Biochem. Biophys. 1995;320:330–344. doi: 10.1016/0003-9861(95)90017-9. [DOI] [PubMed] [Google Scholar]
- [16].Renaud JP, Davydov DR, Heirwegh KP, Mansuy D, Hui Bon Hoa GH. Thermodynamic studies of substrate binding and spin transitions in human cytochrome P-450 3A4 expressed in yeast microsomes. Biochem. J. 1996;319(Pt 3):675–681. doi: 10.1042/bj3190675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Davydov DR, Halpert JR, Renaud JP, Hui Bon Hoa G. Conformational heterogeneity of cytochrome P450 3A4 revealed by high pressure spectroscopy. Biochem. Biophys. Res. Commun. 2003;312:121–130. doi: 10.1016/j.bbrc.2003.09.247. [DOI] [PubMed] [Google Scholar]
- [18].Davydov DR, Knyushko TV, Hui Bon Hoa G. High pressure induced inactivation of ferrous cytochrome P-450 LM2 (IIB4) CO complex: evidence for the presence of two conformers in the oligomer. Biochem. Biophys. Res. Commun. 1992;188:216–221. doi: 10.1016/0006-291x(92)92372-5. [DOI] [PubMed] [Google Scholar]
- [19].Segel IH. Behavior and Analysis of Rapid Equilibrium and Steady-State Systems. Wiley, New York: 1975. Enzyme Kinetics. [Google Scholar]
- [20].Peyser YM, Ajtai K, Burghardt TP, Muhlrad A. Effect of ionic strength on the conformation of myosin subfragment 1-nucleotide complexes. Biophys. J. 2001;81:1101–1114. doi: 10.1016/s0006-3495(01)75767-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Yayanos AA. Evolutional and ecological implications of the properties of deep-sea barophilic bacteria. Proc. Natl. Acad. Sci. USA. 1986;83:9542–9546. doi: 10.1073/pnas.83.24.9542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Bjornsdottir B, Hjerde E, Bragason BT, Gudmundsdottir T, et al. Identification of type VI secretion systems in Moritella viscosa. Vet. Microbiol. 2012;158:436–442. doi: 10.1016/j.vetmic.2012.02.030. [DOI] [PubMed] [Google Scholar]
- [23].Bruins ME, Janssen AEM, Boom RM. Equilibrium shifts in enzyme reactions at high pressure. J. Mol. Catalysis B. 2006;39:124–127. [Google Scholar]
- [24].Akasaka K. Probing conformational fluctuation of proteins by pressure perturbation. Chem. Rev. 2006;106:1814–1835. doi: 10.1021/cr040440z. [DOI] [PubMed] [Google Scholar]
- [25].Masson P, Balny C. Linear and non-linear pressure dependence of enzyme catalytic parameters. Biochim. Biophys. Acta. 2005;1724:440–450. doi: 10.1016/j.bbagen.2005.05.003. [DOI] [PubMed] [Google Scholar]
- [26].Hui Bon Hoa G, McLean MA, Sligar SG. High pressure, a tool for exploring heme protein active sites. Biochim. Biophys. Acta. 2002;1595:297–308. doi: 10.1016/s0167-4838(01)00352-1. [DOI] [PubMed] [Google Scholar]
- [27].Davydov DR, Hui Bon Hoa G, Peterson JA. Dynamics of protein-bound water in the heme domain of P450BM3 studied by high-pressure spectroscopy: comparison with P450cam and P450 2B4. Biochemistry. 1999;38:751–761. doi: 10.1021/bi981397a. [DOI] [PubMed] [Google Scholar]
- [28].Davydov DR, Baas BJ, Sligar SG, Halpert JR. Allosteric mechanisms in cytochrome P450 3A4 studied by high-pressure spectroscopy: pivotal role of substrate-induced changes in the accessibility and degree of hydration of the heme pocket. Biochemistry. 2007;46:7852–7864. doi: 10.1021/bi602400y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Fisher MT, Scarlata SF, Sligar SG. High-pressure investigations of cytochrome P-450 spin and substrate binding equilibria. Arch. Biochem. Biophys. 1985;240:456–463. doi: 10.1016/0003-9861(85)90050-5. [DOI] [PubMed] [Google Scholar]
- [30].Lange R, Bonfils C, Debey P. Low-spin reversible high-spin transition of camphor-bound cytochrome-p-450 - effects of medium and temperature on equilibrium data. Eur. J. Biochem. 1977;79:623–628. doi: 10.1111/j.1432-1033.1977.tb11847.x. [DOI] [PubMed] [Google Scholar]
- [31].Davydov DR, Botchkareva AE, Kumar S, He YQ, Halpert JR. An Electrostatically Driven Conformational Transition Is Involved in = the Mechanisms of Substrate Binding and Cooperativity in Cytochrome = P450eryF. Biochemistry. 2004;43:6475–6485. doi: 10.1021/bi036260l. [DOI] [PubMed] [Google Scholar]
- [32].Deprez E, Gerber NC, Di Primo C, Douzou P, et al. Electrostatic control of the substrate access channel in cytochrome P-450cam. Biochemistry. 1994;33:14464–14468. doi: 10.1021/bi00252a012. [DOI] [PubMed] [Google Scholar]
- [33].Deprez E, Di Primo C, Hui Bon Hoa G, Douzou P. Effects of monovalent cations on cytochrome-P-450 camphor - evidence for preferential binding of potassium. FEBS Lett. 1994;347:207–210. doi: 10.1016/0014-5793(94)00545-1. [DOI] [PubMed] [Google Scholar]
- [34].Mansuy D, Carlier M, Bertrand JC, Azoulay E. Spectral characterization of cytochrome P-450 of a strain of Candida tropicalis grown on tetradecane. Eur. J. Biochem. 1980;109:103–108. doi: 10.1111/j.1432-1033.1980.tb04773.x. [DOI] [PubMed] [Google Scholar]
- [35].Yun CH, Song M, Kim H. Conformational change of cytochrome P450 1A2 induced by phospholipids and detergents. J. Biol. Chem. 1997;272:19725–19730. doi: 10.1074/jbc.272.32.19725. [DOI] [PubMed] [Google Scholar]
- [36].Yun CH, Ahn T, Guengerich FP. Conformational change and activation of cytochrome P450 2B1 induced by salt and phospholipid. Arch. Biochem. Biophys. 1998;356:229–238. doi: 10.1006/abbi.1998.0759. [DOI] [PubMed] [Google Scholar]
- [37].Balny C, Hooper AB. Effect of solvent, pressure and temperature on reaction-rates of the multiheme hydroxylamine oxidoreductase - evidence for conformational change. Eur. J. Biochem. 1988;176:273–279. doi: 10.1111/j.1432-1033.1988.tb14278.x. [DOI] [PubMed] [Google Scholar]
- [38].Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- [39].Whitehouse CJC, Bell SG, Yang W, Yorke JA, et al. A Highly Active Single-Mutation Variant of P450(BM3) (CYP102A1) ChemBioChem. 2009;10:1654–1656. doi: 10.1002/cbic.200900279. [DOI] [PubMed] [Google Scholar]
- [40].Davydov DR, Karyakin AV, Binas B, Kurganov BI, Archakov AI. Kinetic studies on reduction of cytochromes P-450 and b5 by dithionite. Eur. J. Biochem. 1985;150:155–159. doi: 10.1111/j.1432-1033.1985.tb09001.x. [DOI] [PubMed] [Google Scholar]
- [41].Ledenev AN, Tverdokhlebov EN, Davydov RM. Reduction of Ferricytochrome P-450 with Eosin Photoradical. Biofizika. 1984;29:730–732. [PubMed] [Google Scholar]
- [42].Davydov RM, Khanina OY, Iagofarov S, Uvarov VY, Archakov AI. Effect of lipids and substrates on the kinetics of interactions of ferrocytochrome P-450 with CO. Biokhimiia. 1986;51:125–129. [PubMed] [Google Scholar]
- [43].Koley AP, Buters JT, Robinson RC, Markowitz A, Friedman FK. CO binding kinetics of human cytochrome P450 3A4. Specific interaction of substrates with kinetically distinguishable conformers. J. Biol. Chem. 1995;270:5014–5018. doi: 10.1074/jbc.270.10.5014. [DOI] [PubMed] [Google Scholar]
- [44].Backes WL, Tamburini PP, Jansson I, Gibson GG, et al. Kinetics of cytochrome P-450 reduction: evidence for faster reduction of the high-spin ferric state. Biochemistry. 1985;24:5130–5136. doi: 10.1021/bi00340a026. [DOI] [PubMed] [Google Scholar]
- [45].Davydov DR, Botchkareva AE, Davydova NE, Halpert JR. Resolution of two substrate-binding sites in an engineered cytochrome P450eryF bearing a fluorescent probe. Biophys. J. 2005;89:418–432. doi: 10.1529/biophysj.104.058479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Hildebrandt P, Heibel G, Anzenbacher P, Lange R, et al. Conformational analysis of mitochondrial and microsomal cytochrome P-450 by resonance Raman spectroscopy. Biochemistry. 1994;33:12920–12929. doi: 10.1021/bi00209a024. [DOI] [PubMed] [Google Scholar]
- [47].Anzenbacherova E, Hudecek J, Murgida D, Hildebrandt P, et al. Active sites of two orthologous cytochromes P450 2E1: Differences revealed by spectroscopic methods. Biochem. Biophys. Res. Commun. 2005;338:477–482. doi: 10.1016/j.bbrc.2005.08.063. [DOI] [PubMed] [Google Scholar]
- [48].Bancel F, Bec N, Ebel C, Lange R. A central role for water in the control of the spin state of cytochrome P-450scc. Eur. J. Biochem. 1997;250:276–285. doi: 10.1111/j.1432-1033.1997.0276a.x. [DOI] [PubMed] [Google Scholar]
- [49].Masson P, Tonello C. Potential applications of high pressures in pharmaceutical science and medicine. High Pressure Res. 2000;19:613–621. [Google Scholar]
- [50].Eisenmenger MJ, Reyes-de-Corcuera JI. High pressure enhancement of enzymes: A review. Enzyme Microbial Technol. 2009;45:331–347. [Google Scholar]
- [51].Aertsen A, Meersman F, Hendrickx MEG, Vogel RF, Michiels CW. Biotechnology under high pressure: applications and implications. Trends Biotechnol. 2009;27:434–441. doi: 10.1016/j.tibtech.2009.04.001. [DOI] [PubMed] [Google Scholar]
- [52].Mombelli E, Shehi E, Fusi P, Tortora P. Exploring hyperthermophilic proteins under pressure: theoretical aspects and experimental findings. Biochim. Biophys. Acta. 2002;1595:392–396. doi: 10.1016/s0167-4838(01)00361-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.