Abstract
The DNA sequences found repeated in opposite orientation at the ends of insertion (IS) elements are thought to contain sites at which transposase proteins act during transposition. Many elements have repeats of at least 15 base pairs (bp). Those of IS50 are quite short, however: just 8 of the first 9 bp. Functional tests had indicated that one end of IS50 is more effective in transposition than the other end and suggested that at least one of the recognition sites of IS50 extends beyond the common 8/9 bp. To determine the lengths of recognition sites of IS50 we mutagenized IS50 in vitro and tested the transposition proficiency of the resulting mutants. Our results show that the recognition sites at each end of IS50 are about 19 bp long. These findings suggest models for the evolution of IS elements from simpler immobile gene complexes.
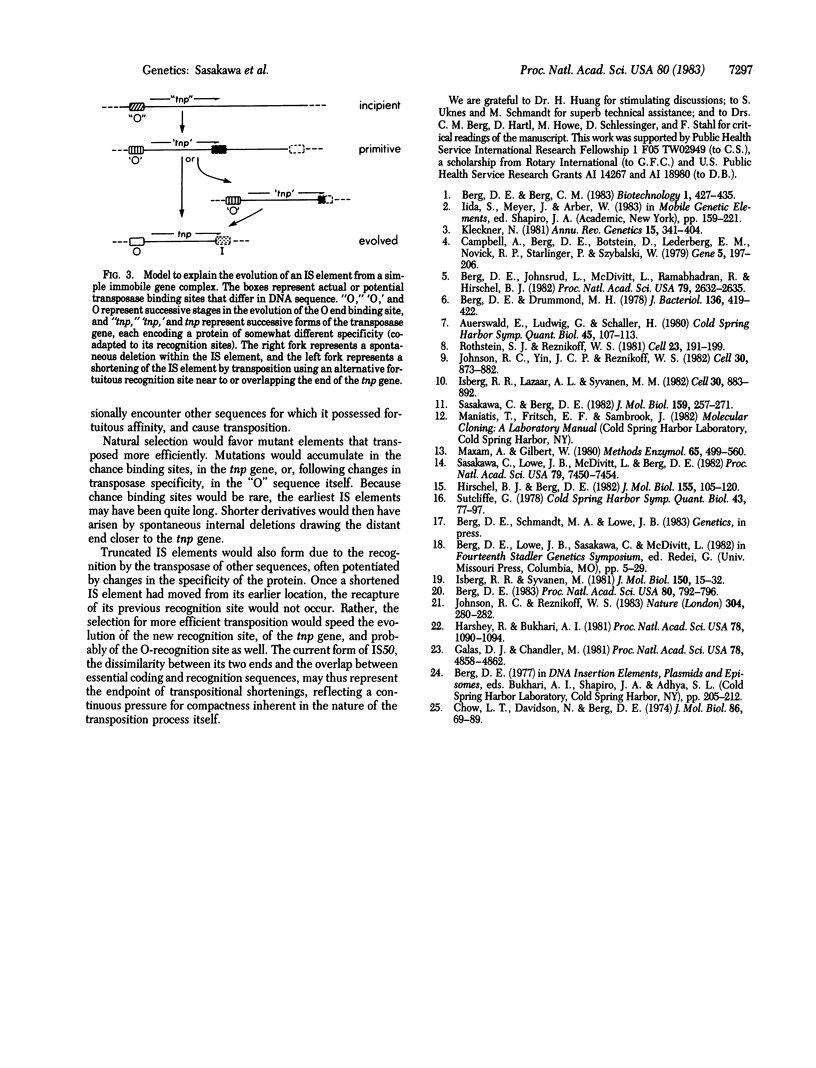
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Auerswald E. A., Ludwig G., Schaller H. Structural analysis of Tn5. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):107–113. doi: 10.1101/sqb.1981.045.01.019. [DOI] [PubMed] [Google Scholar]
- Berg D. E., Drummond M. Absence of DNA sequences homologous to transposable element Tn5 (Kan) in the chromosome of Escherichia coli K-12. J Bacteriol. 1978 Oct;136(1):419–422. doi: 10.1128/jb.136.1.419-422.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg D. E., Johnsrud L., McDivitt L., Ramabhadran R., Hirschel B. J. Inverted repeats of Tn5 are transposable elements. Proc Natl Acad Sci U S A. 1982 Apr;79(8):2632–2635. doi: 10.1073/pnas.79.8.2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg D. E. Structural requirement for IS50-mediated gene transposition. Proc Natl Acad Sci U S A. 1983 Feb;80(3):792–796. doi: 10.1073/pnas.80.3.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell A., Berg D. E., Botstein D., Lederberg E. M., Novick R. P., Starlinger P., Szybalski W. Nomenclature of transposable elements in prokaryotes. Gene. 1979 Mar;5(3):197–206. doi: 10.1016/0378-1119(79)90078-7. [DOI] [PubMed] [Google Scholar]
- Chow L. T., Davidson N. Electron microscope study of the structures of lambdadv DNAs. J Mol Biol. 1974 Jun 15;86(1):69–89. doi: 10.1016/s0022-2836(74)80008-2. [DOI] [PubMed] [Google Scholar]
- Galas D. J., Chandler M. On the molecular mechanisms of transposition. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4858–4862. doi: 10.1073/pnas.78.8.4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harshey R. M., Bukhari A. I. A mechanism of DNA transposition. Proc Natl Acad Sci U S A. 1981 Feb;78(2):1090–1094. doi: 10.1073/pnas.78.2.1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschel B. J., Berg D. E. A derivative of Tn5 with direct terminal repeats can transpose. J Mol Biol. 1982 Feb 25;155(2):105–120. doi: 10.1016/0022-2836(82)90439-9. [DOI] [PubMed] [Google Scholar]
- Isberg R. R., Lazaar A. L., Syvanen M. Regulation of Tn5 by the right-repeat proteins: control at the level of the transposition reaction? Cell. 1982 Oct;30(3):883–892. doi: 10.1016/0092-8674(82)90293-8. [DOI] [PubMed] [Google Scholar]
- Isberg R. R., Syvanen M. Replicon fusions promoted by the inverted repeats of Tn5. The right repeat is an insertion sequence. J Mol Biol. 1981 Jul 25;150(1):15–32. doi: 10.1016/0022-2836(81)90322-3. [DOI] [PubMed] [Google Scholar]
- Johnson R. C., Reznikoff W. S. DNA sequences at the ends of transposon Tn5 required for transposition. Nature. 1983 Jul 21;304(5923):280–282. doi: 10.1038/304280a0. [DOI] [PubMed] [Google Scholar]
- Johnson R. C., Yin J. C., Reznikoff W. S. Control of Tn5 transposition in Escherichia coli is mediated by protein from the right repeat. Cell. 1982 Oct;30(3):873–882. doi: 10.1016/0092-8674(82)90292-6. [DOI] [PubMed] [Google Scholar]
- Kleckner N. Transposable elements in prokaryotes. Annu Rev Genet. 1981;15:341–404. doi: 10.1146/annurev.ge.15.120181.002013. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Rothstein S. J., Reznikoff W. S. The functional differences in the inverted repeats of Tn5 are caused by a single base pair nonhomology. Cell. 1981 Jan;23(1):191–199. doi: 10.1016/0092-8674(81)90284-1. [DOI] [PubMed] [Google Scholar]
- Sasakawa C., Berg D. E. IS50-mediated inverse transposition. Discrimination between the two ends of an IS element. J Mol Biol. 1982 Aug 5;159(2):257–271. doi: 10.1016/0022-2836(82)90495-8. [DOI] [PubMed] [Google Scholar]
- Sasakawa C., Lowe J. B., McDivitt L., Berg D. E. Control of transposon Tn5 transposition in Escherichia coli. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7450–7454. doi: 10.1073/pnas.79.23.7450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]



