Abstract
Staphylococcal strains (CoNS) were speciated in this study. Digests of proteins released from whole cells were converted to tryptic peptides for analysis. Liquid chromatography electrospray ionization tandem mass spectrometry (LC-ESI MS/MS, Orbitrap) was employed for peptide analysis. Data analysis was performed employing the open-source software X!Tandem which uses sequenced genomes to generate a virtual peptide database for comparison to experimental data. The search database was modified to include the genomes of the 11 Staphylococcus species most commonly isolated from man. The number of total peptides matching each protein along with the number of peptides specifically matching to the homologue (or homologues) for strains of the same species were assesed. Any peptides not matching to the species examined were considered conflict peptides. The proteins typically identified with the largest percentage of sequence coverage, number of matched peptides and number of peptides corresponding to only the correct species were elongation factor Tu (EF Tu) and enolase (Enol). Additional proteins with consistently observed peptides as well as peptides matching only homologues from the same species were citrate synthase (CS) and 1-pyrroline-5-carboxylate dehydrogenase (1P5CD). Protein markers, previously identified from gel slices, (aconitate hydratase and oxoglutarate dehydrogenase) were found to provide low confidence scores when employing whole cell digests. The methodological approach described here provides a simple yet elegant way of identification of staphylococci. However, perhaps more importantly the technology should be applicable universally for identification of any bacterial species.
Introduction
In bacteriology, proteomics is primarily utilized in identifying proteins expressed by an organism under specific growth conditions, not for chemotaxonomic characterization. In contrast MALDI-TOF MS (matrix assisted time of flight/deionization mass spectrometry) protein profiling Cain, Lubman, and Weber. 1994; Holland et al., 1996; Krishnamurphy, Ross and Rajamani, 1996) has become an established technique for identification of bacteria particularly with relevance to clinical microbiology (Seng, et al., 2009; Sauer and Kliem, 2010.). MALDI-TOF MS is used for rapid determination of a mass pattern of proteins for determination of species identity using a data base of mass profiles of previously characterized species; these proteins are generally not identified (Intelicato-Young and Fox, 2013). Alternatively, there have been a handful of reports identifying bacteria with potentially greater confidence utilizing sequence differences among peptides released by tryptic digestion (“tryptic peptides”). Experimental spectra of tryptic peptides (generated using liquid chromatography-electrospray ionization tandem mass spectrometry [LC-ESI MS/MS] analysis are compared to virtual spectra generated from genomic data bases. Custom software has been used successfully primarily by one group (Jabbour et al., 2005generated using liquid chromatography-electrospray ionization tandem mass spectrometry [LC-ESI MS/MS] analysis are compared to virtual spectra generated from genomic data bases. Custom software has been used successfully primarily by one group (Jabbour et al., 2010a, 2010b). However more recently standard proteomics software has been also used (Dobryan et al., 2013).
Microbiological testing in clinical settings is still largely based on biochemical characteristics. Many of these tests are routinely used for accurate identification of many pathogenic or opportunistic species but for less studied species the results are often less than optimal (Morgan et al., 2009). Numerous variants of the polymerase chain reaction (PCR) and/or restriction enzymes are commonly used in more advanced reference laboratories in species identification. However in developing new genetic markers two conserved genetic regions (surrounding a variable region) are required for primers to provide initiation for amplification and for the variable regions to provide the information for discriminating closely related species; selection of a gene for assessing sequence variation can be somewhat arbitrary. Whole genome sequencing and annotation is still a technically demanding and expensive alternative (Ivanova, et al., 2003), although this situation is changing rapidly. Accordingly genes that are present universally in bacteria, most commonly16S rDNA, are still widely employed. However, it has become clear over the years that 16S rDNA sequences are too conserved amongst many closely related species and only genus-level identification is provided (Stackebrandt and Goebel, 1994).
Our group has studied members of the genera Staphylococcus including coagulase negative staphylococci (CoNS) and Micrococcus (Fox et al., 2010; Fox et al., 2011, Kooken et al., 2013a). Commercial biochemical tests do not provide adequate identification of CoNS. Variation in sequences of 16S rRNA, but not sodA sequence, correlates with MALDI TOF MS (Dubois et al., 2010) suggesting that genus level identification is being achieved. SodA sequence is the current gold standard for identification of CoNS. Others have also found MALDI TOF MS inadequate for CoNS identification (Carbonnelle et al., 2012). However, in a companion study to the current work excellent correlation with variation in sodA sequence and peptide marker sequences were found (Kooken et al., 2013a). MALDI TOF MS or LC-MS/MS analysis of gel bands identified aconitate hydratase and oxoglutarate dehydrogenase as the dominant proteins present in these gel bands. LC-MS/MS analysis of tryptic peptides, released from whole cells, also provided speciation, correlating with sodA sequence. However, different marker proteins (including elongation factor Tu) were found in whole cell digests. The current work further explores differences in the utility of the different marker proteins.
Materials an Methods
Strains analyzed
S. aureus ATCC 12598, S. capitis ATCC 27841, S. chonii ATCC 29972, S. chromogenes ATCC 43764, S. epidermidis ATCC12228, S. lugdunensis ATCC 49576, CNS 1, CNS5, CNS18, CNS20, ASO2 C21, ASO15 C28, ASO15C 40Y, ASO5 C106, cow924RR, cow970RR, MUS5949, MUS5951, 09-304-034, 900-200-150, 990RR, ASO1 C8, ASO2 C53, ASO2 C63, ASO3 C6, ASO3 C59W, ASO15 C64, ASO15 C84, ASUNO2-15 C6Y and ASUNO2-15 C11Y.
Culture conditions and sample preparation
Bacteria were grown on nutrient agar plates at 37 °C for 24–48 h. The isolates were identified as staphylococci by Gram stain morphology and fermentation characteristics. Colonies were harvested from plates using 0.1 M NaCl, 50 mM Tris HCl, 0.5 mM phenylmethylsulfonyl fluoride [PMSF] and placed in a FastPrep®-24 (MP Biomedicals, Solon, OH) for 6 ms × 30 sec with 5 min on ice between each cycle for a total of 6 cycles. The samples were then centrifuged at 4 °C for 1 h at 10,000 g. The supernatants were removed and placed at −70 °C for two freeze-thaw cycles to eliminate DNA. Each strain was generally analyzed once. Reproducibility was assessed for several species by analyzing multiple strains. For any aberrant results, the analysis was repeated again.
Protein separation, tryptic digestion and MS analysis
Bacteria were harvested (after growing as confluent lawns) from plates using 0.1 M NaCl, 50 mM Tris HCl, 0.5 mM phenylmethylsulfonyl fluoride [PMSF] and placed in a FastPrep®-24 bead beater (MP Biomedicals, Solon, OH) for 6 ms × 30 sec with 5 min on ice between each cycle for a total of 6 cycles to release proteins. The samples were then centrifuged at 4 °C for 1 h at 10,000 g. The supernatants were removed and placed at −70 °C for two freeze-thaw cycles to eliminate DNA.
Peptide Preparation
Bacteria samples were thawed, vortexed briefly to re-suspend. Fif+ty μl of supernatant was transferred to labeled low protein binding 1.5ml microfuge tubes. 50 μl of freshly made 8M urea, 1μl of β-mercapto ethanol, 24 ul of water, and 25 μl of 200 mM Tris-HCl pH 8.0 were added tubes were vortexed briefly and incubated at 60°C for one hour in a Thermomixer (ThermoFisher Scientific) shaking at 300RPM. The tubes were centrifuged briefly after the incubation to collect the evaporate on top of the lids. 800ul of 50mM ammonium bicarbonate was added to each tube to reduce the urea concentration to below 1M. 2 μl of trypsin gold at a concentration of 2 μg/μl was added to each tube and briefly vortexed to mix. The samples were then incubated at 37°C for 15 hours in a Thermomixer shaking at 300RPM.
Purification of peptides
Solid phase extraction (SPE) was performed with a vacuum manifold using Strata C-18 T solid phase extraction columns (Phenomenex, Torrence, CA) and following the manufacturer’s protocol. Briefly, 1 ml of 100% methanol was added to activate the resin, followed by a conditioning step of 1ml 0.1% TFA water, then addition of the samples. The samples are washed with 5% acetonitrile in 0.1% TFA water, and finally elution of the samples with 80% acetonitrile in 0.1% TFA water into labeled clean low protein binding 1.5ml microfuge tubes. The desired flow rate for vacuuming steps is 0.5ml/min with the vacuum seal released after each solution. Samples were dried down to near completeness (5–10 μl remaining) with a Thermo speed vac. 25ul of 0.5% formic acid was added to each sample, using the pipettor gently wash the sides of the tube to recover as much of the sample as possible. The samples were then transferred to labeled HPLC vials with 200 μl inert glass inserts and capped with screw caps.
MS-MS analysis of the peptides
Peptides were separated using an Agilent 1200 HPLC with a 40cm long 0.15 mm ID fused silica column packed with Jupiter 5μm C-18 resin. Column was made in house. A 50 min gradient was established by changing the relative concentrations of a two solvent systems where “A” is 5% acetonitrile, 0.1% formic acid in H2O and “B” is 95% acetonitrile, 0.1% formic acid in H2O at a flow rate of 2 μl per minute. The separation had a 10 min isocratic step at 5% B and a gradient from 5% to 60% B over 50 minutes. Eluate from the HPLC was directly transferred into an LTQFT Orbitrap system (Thermo electron, Billercia MA). The electrospray conditions used were: 2.5kV spray voltage, 200°C and 10V ion transfer tube voltage. T he ion injection time was set for automatic gain control with a maximum injection time of 200ms for 5X10e7 charges in the trap. The MS parent scan was performed using the Orbitrap mass analyzer using a resolution setting of 30,000. Dynamic parent ion selection was performed where the top five most abundant ions were selected for MS-MS in the linear quadrupole ion trap using a 3 m/z mass window.
Database searches
Searches employing MS/MS data were performed using the open source software X!Tandem (www.thegpm.org/tandem) (Craig et al., 2004). Raw files from the Orbitrap (MS-MS data) were converted into the mascot general format (mgf) files) using the program Proteowizard (proteowizard.sourceforge.net). The data base was modified by downloading staphylococcal genomes including eleven species representing the 29 strains in this study. Fasta files were downloaded from (Uniprot, www.uniprot.org) and then converted to FastaPro files for use with X!Tandem. The Fastaprofiles were placed in the fasta section of the software the pro_species.jl (controls program-interface for bacterial species) and taxonomy.xml files (lists each file pathway for each fastaPro file) of the X!Tandem software. Trypsin-specific enzymatic digestion rules were selected and two missed cleavages. The parent mass accuracy was specified at +/− 10 parts per million (ppm) (data acquired with Orbitrap mass spectrometer) and fragment mass accuracy of +/− 300 ppm for MS-MS scans acquired with the LTQ system. maximum parent charge set to 5, and no modifications selected for these samples. The X!Tandem output was also processed into a file format compatible with the mass spectrometry generating function (MSGF) for secondary validation (Kim et al., 2008). Peptide spectrum matches with Log expectation values of −2 or smaller (more significant) by X!Tandem were validated using the MSGF. Peptides with a score of 1 × 10−9 or larger (less significant) were excluded from consideration. All peptide to spectrum matches pass these criteria were captured into a SQLite database for sorting of matches.
Species and strains in the custom X!Tandem database
Staphylococcus aureus O46, Staphylococcus aureus strain Newman, Staphylococcus aureus subsp aureus CIG1242, Staphylococcus capitis, Staphylococcus capitis SK14, Staphylococcus capitis subsp capitis, Staphylococcus capitis subsp urealyticus, Staphylococcus capitis VCU116, Staphylococcus carnosus, Staphylococcus carnosus strain TM300, Staphylococcus carnosus subsp carnosus, Staphylococcus carnosus subsp utilis, Staphylococcus chromogenes, Staphylococcus cohnii, Staphylococcus cohnii subsp cohnii, Staphylococcus cohnii subsp urealyticus, Staphylococcus epidermidis strain ATCC 12228, Staphylococcus epidermidis strain ATCC 35984 RP62A, Staphylococcus epidermidis VCU127, Staphylococcus haemolyticus, Staphylococcus haemolyticus strain JCSC1435, Staphylococcus hominis, Staphylococcus hominis SK119, Staphylococcus hominis subsp hominis, Staphylococcus hominis subsp hominis C80, Staphylococcus hominis subsp novobiosepticus, Staphylococcus hominis VCU122, Staphylococcus lugdunensis, Staphylococcus lugdunensis ACS-027 V Sch2, Staphylococcus lugdunensis M23590, Staphylococcus lugdunensis strain HKU09-01, Staphylococcus lugdunensis strain N920143, Staphylococcus lugdunensis VCU139, Staphylococcus saprophyticus, Staphylococcus saprophyticus subsp bovis, Staphylococcus saprophyticus subsp saprophyticus, Staphylococcus saprophyticus subsp saprophyticus KACC 16562, Staphylococcus saprophyticus subsp saprophyticus MS1146, Staphylococcus saprophyticus subsp saprophyticus strain ATCC 15305 DSM 20229, Staphylococcus simulans ACS-120-V-Sch1, Staphylococcus warneri, Staphylococcus warneri L37603, Staphylococcus warneri VCU121
Results and Discussion
Our previous method work employing MS and MS/MS for bacterial identification employed SDS-PAGE gels for protein separation and extraction of the protein from the gel slice (Fox et al., 2011, Kooken, Fox and Fox 2012, Kooken et al, 2013a) focused primarily on aconitate hydratase or oxoglutarate reductase proteins. Overall analysis takes several days. By utilizing trypsin digests of whole cell extracts, sample preparation time was decreased to less than 24 hr. Furthermore when using whole cell digests numerous proteins are identified leading to greater flexibility in species identification.
MS scans were performed on the Orbitrap, where the 5 most abundant peaks per scan were then sent on for MS-MS analysis in the ion trap. This setup allows for high resolution and fast initial MS scans, followed by a MS-MS scan that have a more focused MW range and fast scan times. Digestion of a whole cell preparation produces a complex mixture of peptides detected among >1500 spectra per run. The spectra are related to several hundred proteins present within the annotated database.
MS/MS results were analyzed using the X!Tandem search engine in which genomic sequence information gathered from the UNIPROT data base for the 11 most common Staphylococcus species associated with humans was uploaded. The X!Tandem program allows for additions of genomic sequences to that provided in the standard database so a more complete evaluation of spectra is possible. Thus a customized library was employed using genomic sequences stored in the UNIPROT database. To prevent oversampling of one species, such as S. aureus, only the genomic sequences for the highest reported, and fully annotated strains were downloaded into X!Tandem. With all Staphylococcus species selected the advance settings in the program allowed for several adjustments to be made. The Log of the E score is typically reported with the smaller the score (larger negative exponent) the more significant the peptide to sequence matches and larger number of peptides corresponding to a given protein sequence (Craig and Beavis 2004, Bioinformatics 20, 1466.). Values with a negative exponent larger than -100 are considered highly significant with an excess of 4 peptides supporting the identification.
Selected ATCC type strains were run as controls to verify proper identification. For example, the top results for S. aureus and S. lugdunensis, for ten commonly identified proteins, are shown in Table 1 which lists proteins identified with high confidence and peptide coverage including elongation factor Tu (EF Tu) and enolase (Enol). The number of total peptides matching each protein along with the number of peptides specifically matching to the homologue (or homologues) for strains of the same species being examined is listed. Any peptides not matching to the species examined were considered conflict peptides. The Log of the protein expectation value and next suggested species for S. aureus and S. lugdunensis is also listed.
Table 1.
The peptides identified for ten protein markers detected in datasets from ATCC strains of S. aureus and S. lugdunensis. The number of peptides that match to alternative species are listed as conflict peptides. The Log protein expectation value is provided for each protein in the S. aureus and S. lugdunensis datasets.
| Protein | Staphylococcus lugdunensis ATCC 49576 | S. aureus ATCC 12598 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Total | Specific | Conflict | Log Epro/Diff | % seq cover | Total | Specific | Conflict | Log Epro/Diff | % seq cover | |
| 10 kDa Chaperonin | 4 | 2 | 0 | (−32/22) | 48% | 1 | 1 | 0 | - | - |
| 1-pyrroline-5-carboxylate dehydrogenase | 18 | 14 | 0 | (−205/140) | 75% | 14 | 9 | 0 | (−145/22) | 58% |
| 2-oxoglutarate dehydrogenase | 14 | 2 | 0 | (−135/100) | 36% | 0 | 0 | 0 | - | - |
| 60 kDa Chapernonin | 14 | 5 | 0 | (−154/85) | 58% | 2 | 1 | 0 | - | - |
| Aconitate hydratase | 22 | 7 | 2 | (−212/82) | 35% | 1 | 0 | 0 | - | - |
| Catalase | 4 | 2 | 0 | (−24/9) | 16% | 4 | 3 | 0 | (−30/−30) | 20% |
| Citrate Synthase | 26 | 16 | 0 | (−297/220) | 72% | 4 | 2 | 0 | (−31/26) | 19% |
| Elongation factor Tu | 32 | 6 | 0 | (−405/85) | 79% | 19 | 3 | 0 | (−262/85) | 88% |
| Enolase | 26 | 6 | 0 | (−341/162) | 69% | 21 | 11 | 0 | (−235/130) | 68% |
| Glyceraldehyde-3-PO4 dehydrogenase | 20 | 12 | 1 | (−227/140) | 67% | 6 | 3 | 0 | (−98/70) | 44% |
The total number of peptides identified for ten commonly observed proteins in each of the ATCC strains for S. simulans (ATCC), S. chromogenes (ATCC 43764), and S. epidermidis (ATCC 12228) are listed in Table 2. The proteins typically identified with the largest percentage of sequence coverage, number of matched peptides and number of peptides corresponding to only the correct species was elongation factor Tu (EF Tu) and enolase (Enol). Additional proteins with consistently observed peptides as well as peptides matching only homologues from the same species were citrate synthase (CS) and 1-pyrroline-5-carboxylate dehydrogenase (1P5CD). The previous work by Kooken et al (Kooken et al., 2013a) had used aconitate hydratase (AH) and 2-oxoglutarate dehydrogenase (2-OD) as markers. The proteins were not always observed with large numbers of identified peptides in cell lysates from all of the ATCC strains examined.
Table 2.
The total numbers of peptides identified for S. chromogenes, S. simulans and S. epidermidis are listed along with the number of peptides specific for the correct species as well as the conflict peptides not matched.
| Protein | S. simulans ACS-120-V-Sch1 | S. chromogenes ATCC 43764 | S. epidermidis ATCC 12228 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Total | Specific | Conflict | Total | Specific | Conflict | Total | Specific | Conflict | |
| 10 kDa Chaperonin | 4 | 3 | 0 | 0 | 0 | 0 | 2 | 0 | 0 |
| 1-pyrroline-5-carboxylate dehydrogenase | 29 | 22 | 0 | 5 | 0 | 5 | 2 | 1 | 0 |
| 2-oxoglutarate dehydrogenase | 16 | 10 | 0 | 0 | 0 | 0 | 41 | 5 | 6 |
| 60 kDa Chapernonin | 12 | 6 | 0 | 2 | 0 | 2 | 13 | 4 | 0 |
| Aconitate hydratase | 17 | 10 | 0 | 2 | 0 | 2 | 5 | 5 | 0 |
| Catalase | 9 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Citrate Synthase | 19 | 15 | 0 | 2 | 0 | 2 | 11 | 7 | 1 |
| Elongation factor Tu | 41 | 10 | 0 | 26 | 11 | 3 | 11 | 2 | 1 |
| Enolase | 19 | 3 | 0 | 8 | 0 | 8 | 8 | 5 | 0 |
| Glyceraldehyde-3-phosphate dehydrogenase | 22 | 6 | 0 | 8 | 7 | 1 | 9 | 5 | 0 |
The available databases were not complete for all the species of interest. For example, the S. chromogenes sample had 26 and 8 peptides identified for EF Tu and G3PD respectively. For each protein, a significant proportion of the peptides matched only to the S. chromogenes homologue within the database. By contrast, Enol, CS and 1P5CD were all identified with all peptides matching only to other staphylococcal species, indicating that the S. chromogenes sequences for each were missing from the Uniprot database. A BLAST search for Enol sequences in the Uniprot database revealed a number of staphylococcal Enol homologues but none for S. chromogenes. All the peptides identified for that protein matched to sequences from other species. The other seven proteins appeared to have peptides matched specifically to the S. simulans homologue in each case, indicating their presence in the database.
The detected peptides from EF Tu and Enol protein markers were then investigated as a means to identify 28 clinical, veterinary and environmental isolates (Table 3). These strains had also been examined by Kooken et al (2013a). Of the 28 strains, 23 of the isolates examined indicated a specific species by both markers. In each case the number of species specific peptides was much larger than any conflict peptides identified. The identifications corresponded with the identification results obtained by sodA and MALDI-MS analyses. The five strains that did not have a specific species selected from the database were CNS5, aso15c28, aso15c106, aso3c59 and Cow924RR.
Table 3.
The total number of peptides identified for the protein markers indicated is given for each isolate for EF Tu and Enol protein markers. The species identified using the LC-MS-MS datasets are indicated. The number of peptides specific for the indicated species given along with the number of conflict peptides is also given. Isolates that had multiple conflicting species indicated using specific peptides are shaded in grey. All were considered conflict peptides.
| Elongation factor Tu (EF Tu) | Enolase (Enol) | ||||||
|---|---|---|---|---|---|---|---|
| Isolate Name | Species Indicated | Total | Specific | Conflict | Total | Specific | Conflict |
| 9304034 | S. saprophyticus | 23 | 5 | 1 | 19 | 6 | 1 |
| 900-200-150 | S. warneri | 30 | 4 | 0 | 9 | 2 | 0 |
| aso15c64 | S. aureus | 20 | 4 | 0 | 18 | 8 | 0 |
| aso15c84 | S. aureus | 18 | 2 | 0 | 17 | 9 | 0 |
| aso1c8 | S. aureus | 21 | 3 | 0 | 15 | 7 | 0 |
| aso2c21 | S. warneri | 38 | 6 | 1 | 10 | 3 | 0 |
| aso2c44 | S. simulans | 36 | 6 | 0 | 16 | 7 | 0 |
| aso2c53 | S. epidermidis | 30 | 6 | 0 | 12 | 4 | 0 |
| aso3c22 | S. saprophyticus | 36 | 10 | 1 | 29 | 13 | 1 |
| aso3c6 | S. aureus | 19 | 3 | 0 | 16 | 6 | 1 |
| asuno15c6Y | S. aureus | 19 | 3 | 0 | 21 | 12 | 0 |
| asuno215c6Y | S. aureus | 12 | 3 | 0 | 15 | 6 | 0 |
| CNS 10 | S. simulans | 48 | 15 | 1 | 26 | 8 | 1 |
| CNS 18 | S. aureus | 22 | 6 | 1 | 14 | 5 | 2 |
| CNS 20 | S. aureus | 23 | 2 | 0 | 18 | 9 | 0 |
| CNS 6 | S. lugdunensis | 35 | 8 | 1 | 28 | 6 | 1 |
| CNS 7 | S. simulans | 43 | 9 | 1 | 18 | 2 | 1 |
| CNS 8 | S. lugdunensis | 39 | 9 | 3 | 26 | 7 | 1 |
| Cow970 RR | S. saprophyticus | 29 | 7 | 0 | 15 | 6 | 0 |
| aso2 c53 | S. epidermidis | 29 | 7 | 0 | 6 | 0 | 1 |
| mus 5949 | S. chromogenes | 39 | 19 | 13 | 9 | 0 | 9 |
| CNS1 | S. simulans | 21 | 8 | 0 | 2 | 0 | 0 |
| 990RR | S. simulans | 21 | 5 | 2 | 6 | 1 | 0 |
| aso15c106 | S. lugdunensis/S. aureus/S. saprophyticus/S. capitis/S.warneri | 29 | 0 | 3 | 9 | 0 | 4 |
| aso15c28 | S. aureus/S. lugdunensis | 26 | 0 | 0 | 12 | 0 | 4 |
| Cow924RR | S. warnerii/S. capitis/S. haemolyticus/S. Saprophyticus | 11 | 0 | 1 | 5 | 0 | 3 |
| aso3c59w | S. lugdunensis/S. cohnii/S. saprophyticus | 31 | 0 | 7 | 10 | 0 | 2 |
| CNS5 | S. lugdunensis/S. cohnii/S. saprophyticus | 45 | 0 | 22 | 6 | 0 | 0 |
Additional protein markers CS and AH were used as supplementary markers to aid in species identification (Table 4) The peptides identified for these two protein markers supported the results for EF Tu and Enol when species specific peptides were identified. The peptide identification results for CS and AH for the five strains not specifically associated with a species using EF Tu and Enol were able to specify species from the database. For strains aso15c28 and aso15c106, S. haemolyticus was selected from the database which supports the findings from Kooken et al. (2013) using other techniques. The Cow924 RR strain had inconclusive results using the EF Tu, Enol and AH data while CS marker indicated S. saprophyticus. Further information on full sequencing of the sodA gene identified the strain as a closely related species, S. equorum, that was not represented in the protein sequence database (Nováková et al. 2006.). The CNS5 and aso3c59 strains had S. simulans and S. warneri indicated respectively using the CS marker. However, additional data on the sequence of sodA for strain aso3c59 indicated that it was actually an isolate of S. pastueri, a species with no representatives in our protein sequence database and only 14 total protein sequences present within the Uniprot database (Chesneau et al., 1993).
Table 4.
The total number of peptides identified for the protein markers indicated is given for each isolate for citrate synthase (CS) and aconitate hydratase (AH) protein markers. The species identified using the LC-MS-MS datasets are indicated. The number of peptides specific for the indicated species given along with the number of conflict peptides. Isolates that had no dominant species indicated by EF Tu or Enol are indicated in grey along with the species indicated by CS or AH.
| Citrate synthase | Aconitate hydratase | ||||||
|---|---|---|---|---|---|---|---|
| Isolate Name | Species Indicated | Total | Specific | Conflict | Total | Specific | Conflict |
| 9304034 | S. saprophyticus | 0 | 0 | 0 | 7 | 3 | 1 |
| 900-200-150 | S. warneri | 4 | 4 | 0 | 1 | 0 | 0 |
| aso15c64 | S. aureus | 3 | 2 | 0 | 1 | 0 | 0 |
| aso15c84 | S. aureus | 4 | 3 | 0 | 3 | 1 | 0 |
| aso1c8 | S. aureus | 5 | 4 | 0 | 2 | 1 | 0 |
| aso2c21 | S. warneri | 9 | 5 | 0 | 5 | 1 | 1 |
| aso2c44 | S. simulans | 18 | 13 | 0 | 22 | 13 | 0 |
| aso2C53 | S. epidermidis | 2 | 2 | 0 | 2 | 0 | 0 |
| aso3C22 | S. saprophyticus | 14 | 10 | 0 | 22 | 13 | 0 |
| aso3c6 | S. aureus | 5 | 3 | 0 | 3 | 1 | 0 |
| asuno15c6y | S. aureus | 5 | 4 | 0 | 4 | 2 | 0 |
| asuno2-15c6y | S. aureus | 1 | 0 | 0 | 3 | 2 | 0 |
| CNS10 | S. simulans | 14 | 11 | 0 | 24 | 14 | 0 |
| CNS18 | S. aureus | 5 | 4 | 0 | 3 | 1 | 0 |
| CNS20 | S. aureus | 3 | 1 | 0 | 3 | 1 | 0 |
| CNS6 | S. lugdunensis | 18 | 12 | 0 | 27 | 8 | 3 |
| CNS7 | S. simulans | 16 | 13 | 0 | 26 | 14 | 1 |
| CNS8 | S. lugdunensis | 9 | 6 | 0 | 15 | 6 | 0 |
| Cow970RR | S. saprophyticus | 1 | 1 | 0 | 6 | 2 | 0 |
| aso2c53 | S. epidermidis | 2 | 2 | 0 | 2 | 0 | 0 |
| mus5949 | S. chromogenes | 2 | 0 | 0 | 2 | 0 | 0 |
| CNS1 | S. simulans | 4 | 2 | 0 | 2 | 0 | 0 |
| 990rr | S. simulans | 10 | 8 | 0 | 0 | 0 | 0 |
| aso15c106 | S. haemolyticus | 6 | 3 | 0 | 2 | 0 | 0 |
| aso15c28 | S. haemolyticus | 5 | 1 | 0 | 2 | 0 | 0 |
| Cow924RR | S. saprophyticus | 0 | 0 | 0 | 7 | 4 | 0 |
| aso3c59w | S. warneri | 4 | 3 | 0 | 2 | 0 | 0 |
| CNS5 | S. simulans | 4 | 2 | 1 | 6 | 2 | 0 |
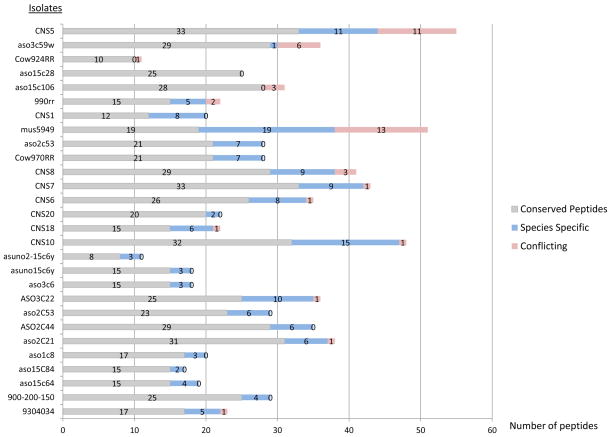
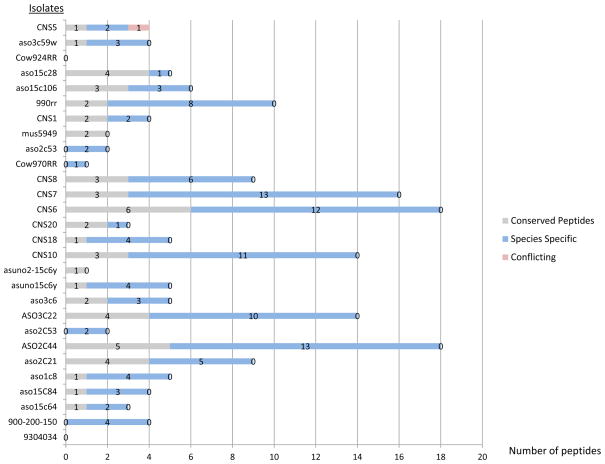
The EF Tu and Enol were proteins generally identified with the largest number of peptides. The profile of EF Tu peptides identified across the 28 strains is given in Figure 1. For seven of the isolates, there was no species selectivity or an ambiguous mixture of species indicated by similar numbers of specific and conflict peptides. The CS marker was the only one that consistently had species identifying peptides when present and almost no peptides matching to conflicting species (Figure 2). Two of the strains did not have any CS peptides identified while two strains had one or two conserved peptides identified, providing no species selective information.
Figure 1.
The total number of Elongation factor Tu identified peptides for 28 clinical and environmental isolates. Among the total peptides identified, the number of conserved peptides are indicated in grey while the number of specific peptides is indicated in blue and the number of conflict peptides in red.
Figure 2.
Cysteine synthase peptide coverage observed for the clinical and environmental isolates. Among the total peptides identified, the number of conserved peptides are indicated in grey while the number of specific peptides is indicated in blue and the number of conflict peptides in red.
Conclusions
Our previous work involved gel separation of intact proteins, followed by extraction and generation of tryptic peptides (Fox et al. 2011, Kooken et al., 2012; Kooken et al., 2013a). Using MALDI TOF MS and MS/MS aconitate hydratase and oxoglutarate dehydrogenase were identified as marker proteins. These markers were not consistently identified with sufficient peptide coverage to provide species identification on LC-MS/MS analysis of tryptic peptides from whole cell supernatants. Additional protein markers that were more consistently observed to contribute peptides in the digest were used for species identification. EF Tu, and Enol provided much greater peptide coverage and confidence in speciation in most samples. Furthermore the available peptide coverage for CS consistently had a significant number of species-specific peptides and no conflicting peptides when detected. The identification results could be supported by AH when detected as well.
The strength of this approach is its ability to utilize multiple abundant proteins and available sequence databases. While the database composition is constantly changing as new genomes are sequenced, it cannot be considered to be complete and comprehensive for all species of interest. The species that have received more attention and have larger numbers of strains with sequenced genomes (e.g. S. aureus) have more of the potential sequence diversity represented. Conversely, the species that have a few or only a single sequenced strain, far less of the sequence diversity for that species has been captured within the database. For this reason, the total number of peptide identifications matching any given species is often uneven and so this number is not useful for species identification. Instead the utility of individual protein markers have been evaluated for their taxonomic specificity. Similar to using DNA sequence databases, it is vital to understand the composition of the database and which species and strains are represented as well as which species are underrepresented or missing.
It is hypothesized that only the most abundant proteins should provide almost complete coverage on LC-MS-MS and many peptides derived from less abundant proteins will be buried in the complex chromatograms. This methodology is simple to perform but the software requires further development for routine use. Furthermore, LC-ESI MS-MS technology is currently too complex for implementation in a clinical microbiology laboratory where the emphasis is not analytical chemistry. However, we are optimistic that changes will be implemented in the not too distant future. Indeed it has taken almost 20 years for MALDI TOF MS profiling to be fully developed and accepted by the clinical microbiology community (Intelicato and Fox, 2013). In the mean-time in research settings, LC-MS-MS may help expand the knowledge-base of proteins (with unique variable regions) derived from bacterial species allowing their identification or detection in simple or complex matrices.
These observations highlight the need to verify when proteins for a species of interest are missing from a database if protein identification is to be used for species identification. A failure to find peptides corresponding to a protein homologue from a species of interest may indicate incomplete sequence information rather than excluding that species as the source of the sample. An additional consideration is that in some cases, only a fragment of a protein may be present in the database causing some peptides to match specifically to a homologue while other peptides (matching the missing portion of the protein sequence fragment) match only to protein homologues from the wrong species. Finally, the isolate being analyzed may have significant sequence variation from the available sequences in the database.
Acknowledgments
Support for this work was provided by the National Science Foundation (# 0959427, J. Rose, P.I, K. Fox Co-P.I). Jennifer Kooken received pre-doctoral support from the Sloan Foundation and NIH R25GM076277 (B. Ely, P.I. and R. Hunt, Co-P.I.). The clinical strains were kindly provided by Dr. Gustavo Medino obtained from 2 hospitals in Valdavia, Chile. The veterinary strains were obtained from Drs. George Stewart and John Middleton, College of Veterinary Medicine, University of Missouri, Columbia, MO 65211. Thanks to Aaron Robinson for assistance in data analysis. Battelle Memorial Institute operates Pacific Northwest National Laboratory for the U.S. DOE under Contract DE-AC06-76RLO.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Jennifer Kooken, Department of Pathology, Microbiology and Immunology, School of Medicine, University of South Carolina, Columbia, SC 29208, US.
Karen Fox, Department of Pathology, Microbiology and Immunology, School of Medicine, University of South Carolina, Columbia, SC 29208, US.
Alvin Fox, Department of Pathology, Microbiology and Immunology, School of Medicine, University of South Carolina, Columbia, SC 29208, US.
David Wunschel, Chemical and Biological Signature Sciences, Pacific Northwest National Laboratory, PO Box 999 MS P7-50, Richland WA, 99354, US.
References
- Cain T, Lubman D, Weber J. Differentiation of bacteria using protein profiles from matrix assisted laser desorption/ionization time of flight mass spectrometry. Rapid Commun Mass Spectrom. 1994;8:1026–1030. [Google Scholar]
- Carbonnelle E, Jacquier GH, Day N, Tenza S, Dewailly A, Vissouarn O, Rottman M, Herrmann J-L, Podglajen I, Raskine L. Robustness of two MALDI-TOF mass spectrometry systems for bacterial identification. J Microbiol Meth. 2012;89:133–136. doi: 10.1016/j.mimet.2012.03.003. [DOI] [PubMed] [Google Scholar]
- Chesneau O, Morvan A, Grimont F, Labischinski H, el Solh N. Staphylococcus pasteuri sp nov., isolated from human, animal, and food specimens. IJSB. 1993;43:237–244. doi: 10.1099/00207713-43-2-237. [DOI] [PubMed] [Google Scholar]
- Claydon M, Davey S, Edwards-Jones V, Gordon D. The rapid identification of intact microorganisms using mass spectrometry. Nature Biotechnol. 1996;14:1584–1586. doi: 10.1038/nbt1196-1584. [DOI] [PubMed] [Google Scholar]
- Craig R, Cortens JP, Beavis RC. Open source system for analyzing, validating, and storing protein identification data. J Proteome Res. 2004;3:1234–42. doi: 10.1021/pr049882h. [DOI] [PubMed] [Google Scholar]
- Dworzanski JP, Snyder AP. Classification and identification of bacteria using mass spectrometry-based proteomics. Expert Rev Proteomics. 2005;2:863–878. doi: 10.1586/14789450.2.6.863. [DOI] [PubMed] [Google Scholar]
- Dobryan MT, McCorrister S, Chong PM, Lee DM, Corbett CR, Westmacott GR. A simple shotgun proteomics method for rapid bacterial identification. J Microbiol Meth. doi: 10.1016/j.mimet.2013.04.008. (in press) [DOI] [PubMed] [Google Scholar]
- Dubois, et al. Identification of a variety of Staphylococcus species by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol. 2010;48:941–945. doi: 10.1128/JCM.00413-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nováková D, 1, Sedláček I, 1, Pantůček R, Štĕtina V, Švec P, Petráš P. Staphylococcus equorum and Staphylococcus succinus isolated from human clinical specimens. J Med Microbiol. 2006;55:523–528. doi: 10.1099/jmm.0.46246-0. [DOI] [PubMed] [Google Scholar]
- Fox K, Fox A, Elssner T, Feigley C, Salzberg D. MALDI-TOF mass spectrometry speciation of staphylococci and their discrimination from micrococci isolated from indoor air of schoolrooms. J Environ Monit. 2010;12:917–23. doi: 10.1039/b925250a. [DOI] [PubMed] [Google Scholar]
- Fox K, Fox A, Rose J, Walla M. Speciation of coagulase negative staphylococci, isolated from indoor air, using SDS PAGE gel bands of expressed proteins followed by MALDI TOF MS and MALDI TOF-TOF MS-MS analysis of tryptic peptides. J Microbiol Meth. 2011;84:243–50. doi: 10.1016/j.mimet.2010.12.007. [DOI] [PubMed] [Google Scholar]
- Holland RD, Wilkes G, Rafii F, Sutherland JB, Persons CC, Voorhees KJ, Lay JO., Jr Rapid Identification of intact whole bacteria based on spectral patterns using matrix-assisted laser desorption/ionization with time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. 1996;10:1227–1232. doi: 10.1002/(SICI)1097-0231(19960731)10:10<1227::AID-RCM659>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Intelicato-Young J, Fox A. Mass Spectrometry and tandem mass spectrometry characterization of protein patterns, protein markers and whole proteomes for pathogenic bacteria. J Microbiol Meth. 2013;92:381–386. doi: 10.1016/j.mimet.2013.01.004. [DOI] [PubMed] [Google Scholar]
- Ivanova N, Sorokin A, Anderson I, Galleron N, Candelon B, Kapatral V, Bhattacharyya A, Reznik G, Mikhailova N, Lapidus A, Chu L, Mazur M, Goltsman E, Larsen N, D’Souza M, Walunas T, Grechkin Y, Pusch G, Haselkorn R, Fonstein M, Ehrlich S, Overbeek R, Kyrpides N. Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature. 2003;423:87–89. doi: 10.1038/nature01582. [DOI] [PubMed] [Google Scholar]
- Jabbour RE, Wade MM, Deshpande SV, Stanford MF, Wick CH, Zulich AW, Snyder AP. Identification of Yersinia pestis and Escherichia coli strains by whole cell and outer membrane protein extracts with mass spectrometry-based proteomics. J Proteome Res. 2010;9:3647–3655. doi: 10.1021/pr100402y. [DOI] [PubMed] [Google Scholar]
- Jabbour RE, et al. Double-blind characterization of non-genome-sequenced bacteria by mass spectrometry-based proteomics. Appl Environ Microbiol. 2010;76:3637–44. doi: 10.1128/AEM.00055-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy T, Ross PL, Rajamani U. Detection of pathogenic and non- pathogenic bacteria by matrix-assister laser desorption/ionization time of flight mass spectrometry. Rapid Commun Mass Spectrom. 1996;10:883–888. doi: 10.1002/(SICI)1097-0231(19960610)10:8<883::AID-RCM594>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Kooken JM, Fox KF, Fox A. Characterization of Micrococcus strains isolated from indoor air. Mol Cell Probes. 2012;26:1–5. doi: 10.1016/j.mcp.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooken J, Fox Karen, Fox Alvin, Altomare Diego, Creek Kim, Wunschel David. Identification of staphylococcal species based on variations in protein sequence (tandem mass spectrometry) and DNA sequence (microarray) 2013. 2013a. [DOI] [PubMed] [Google Scholar]
- Morgan M, Boyette M, Goforth C, Sperry K, Greene S. Comparison of the Biolog OmniLog identification system and 16S ribosomal RNA gene sequencing for accuracy in identification of atypical bacteria of clinical origin. J Microbiol Meth. 2009;79:336–343. doi: 10.1016/j.mimet.2009.10.005. [DOI] [PubMed] [Google Scholar]
- Stackebrandt E, Goebel BM. Taxonomic note: A place for DNA-DNA reassociation and16s rRNA sequence analysis in the present species definition in Bacteriology. Int J System Bacteriol. 1994;4:846–849. [Google Scholar]
- Sauer S, Kliem M. Mass spectrometry tools for the classification and Identification of bacteria. Nat Rev Microbiol. 2010;8:74–82. doi: 10.1038/nrmicro2243. [DOI] [PubMed] [Google Scholar]
- Seng P, Drancourt M, Gouriet F, La SB, Fournier PE, Rlain JM, Raoult D. Ongoing revolution in bacteriology: routine identification of bacteria by matrix assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis. 2009;49:543–551. doi: 10.1086/600885. [DOI] [PubMed] [Google Scholar]