Abstract
Pheromone biosynthesis activating neuropeptide (PBAN) promotes synthesis and release of sex pheromones in moths. We have identified and functionally expressed a PBAN receptor from Heliothis virescens (HevPBANR) and elucidated structure-activity relationships of PBAN analogs. Screening of a larval CNS cDNA library revealed three putative receptor subtypes and nucleotide sequence comparisons suggest they are produced through alternative splicing at the 3’-end. RT-PCR amplified preferentially HevPBANR-C from female pheromone glands. CHO cells expressing HevPBANR-C are highly sensitive to PBAN and related analogs, especially those sharing the C-terminal pentapeptide core, FXPRLamide (X = T, S or V). Alanine replacements in the C-terminal hexapeptide (YFTPRLamide) revealed the relative importance of each residue in the active core as follows, R5>L6>F2≫P4>T3≫Y1. This study provides a framework for the rational design of PBANR-specific agonists and/or antagonists that could be exploited for disruption of reproductive function in agriculturally important insect pests.
1. Introduction
Sex pheromones provide important olfactory signals for species-specific courtship and mating. In some butterfly and moth species, synthesis and release of pheromones are regulated by pheromone biosynthesis activating neuropeptides (PBANs). The first PBANs identified from brain and subesophageal ganglia contain 33 or 34 amino acids, amidated at the C-terminus [5, 14, 16, 24, 27]. Despite significant advances over the last decades [4, 10], many questions regarding the precise functional roles of PBANs remain. Recent efforts have focused on identification and tissue-specific expression of PBAN receptors.
Analysis of the Drosophila melanogaster genome led to a prediction that 44 G protein-coupled receptors GPCRs mediate peptide hormone signaling [7]. We and others showed previously that GPCRs CG8795 and CG8784 are highly sensitive to D. melanogaster pyrokinin-2 (DrmPK-2), a neuropeptide with high C-terminal sequence similarity to PBAN [21, 26]. These findings suggested that the moth counterpart of CG8795 would likely encode a functional PBAN receptor. Indeed, moth orthologs of CG8795 have been identified as functional PBAN receptors in the moth Helicoverpa zea [3] and Bombyx mori [9].
Mating disruption by field application of synthetic pheromones has become a viable strategy for integrated pest management (IPM) of agriculturally important Lepidopteran pests. It therefore seems likely that chemicals interfering with PBAN signaling could become novel insect control agents of the future. Consistent with this suggestion, a bromo-fluorene analog of PBAN is toxic to H. virescens in a manner consistent with pyrokinin receptor activation [29]. Molecular identification of the PBAN receptor would provide a key target for development of agrochemical reagents, as well as provide further insights into possible broader physiological roles for PBAN.
Here we describe the cloning and functional expression of a PBAN receptor from H. virescens. The expressed receptor is sensitive and selective for PBAN and its analogs. Through systematic alanine substitution, we identify the active ligand core, and characterize critical chemical information within, that interacts with the receptor.
2. Materials and Methods
2.1. Experimental Animals
Heliothis virescens larvae were reared on a modified pinto bean diet [28]. Eggs and larvae of both insect species were kept at 27 °C on a 16 h:8 h L:D cycle.
2.2. RNA isolation and cDNA library construction
Total RNA was isolated from the brain and ventral nerve cord of 1-3 day old fifth instar Heliothis virescens larvae using Trizol and precipitated by isopropanol. Poly(A)+ RNA was purified by two passages of total RNA through an oligo(dT) column, and then used for the construction of cDNA using an oligo-(dT) primer containing a NotI cut site as previously described [17]. Approximately 1.5μg of cDNA was then separated by size exclusion chromatography on a Superose 6 column using a SMART system (Pharmacia LKB Biotechnology, Piscataway, NJ). cDNA fragments >2kb were used for the library construction in pSport1 vector (Invitrogen) and electroporated into DH10B cells.
2.3. Cloning of HevPBAN Receptor
Recombinant DNA techniques used for cloning of GPCRs were described previously [9, 22]. Briefly, the partial sequence of HevPBANR (Heliothis virescens PBAN receptor) was obtained from the larval CNS cDNA by performing PCRs using a set of degenerate primers. Degenerate primers were designed and synthesized according to protein sequences of highly conserved regions of Drosophila melanogaster GPCRs, CG8795 (GenBank accession number, AF522190), CG8784 (AF522189), Anopheles gambiae ortholog of CG8795 (Genbank accession number, XP_312761), Caenorhabditis elegans receptor (NP_509515) and rat neuromedin U receptor 1 (NMUR 1, AB038649); forward primer (5’-ACXGCXACNAAYTTYTAYYT; X is inosine) and reverse primer (5’-TGRAANGGNGCCCARCADAT). PCRs were performed with Taq (Invitrogen), and PCR products of the expected size (~636 bps) were cloned and sequenced.
Three subtypes of HevPBANR (HevPBANR-A, -B and –C; Fig. 1) were isolated from a H. virescens CNS cDNA library using primers based on the partial sequence. Since the cDNA library was prepared using pSPORT vector (Invitrogen), the 5’-sequence was amplified in PCRs using pairs of gene-specific reverse primers (PBANR-R1 and -R2) and vector specific primers (pSPORT-D69, -L29, -A83 –A82 and –B6); PBANR-R1 (5’-CAACGAGCATTCTGATGACT), PBANR-R2 (5’-CCAGGTCTCTCGTTACTCTC), PSPORT-D69 (5’- GTCGCATGCACGCGTACGTAAGCTTGG), PSPORT-L29 (5’- GGATCCTCTAGAGCGGCCGC), PSPORT-A83 (5’-GGTACCGGTCCGGAATTCCCGGG), PSPORT-A82 (5’-GGGAAAGCTGGTACGCCTGCAGG), PSPORT-B6 (5’- CCAAGCTCTAATACGACTCACTAT). Likewise, the 3’-sequence was obtained with the nested sets of two forward primers from the gene (PBANR-F1 and PBANR-F2) and primers from the vector (pSPORT-D69, -L29, -A83 –A82 and –B6); PBANR-F1 (5’- GTGGTCTGCCATTTGAAGTA), PBANR-F2 (5’-AGCTAACGCGACAGTATTGA). After obtaining entire cDNA sequences of all three PBANR subtypes, ORFs of each subtype were amplified from either the larval CNS cDNA (for HevPBANR-A and –B) or the female adult pheromone gland cDNA (for HevPBANR-C) using a mixture of pfu (Stratagen) and Taq (Invitrogen) (0.5U:0.5U) with the following sets of primers; for HevPBANR-A, PBANR-F3 (5’- GTGCTAGTGGTGAAGTTACG) and PBANR-R3 (5’-GGTTTGATTCCCGTGATGTC); for HevPBANR-B and –C, PBANR-F3 and PBANR-R4 (5’- CGTGGTCACTGTCGCTTACA). The ORFs of HevPBANR-A, -B or -C subtypes were subcloned into the pcDNA3.1 vector (Invitrogen) for subsequent Chinese hamster ovary (CHO) cell expression.
Fig. 1.
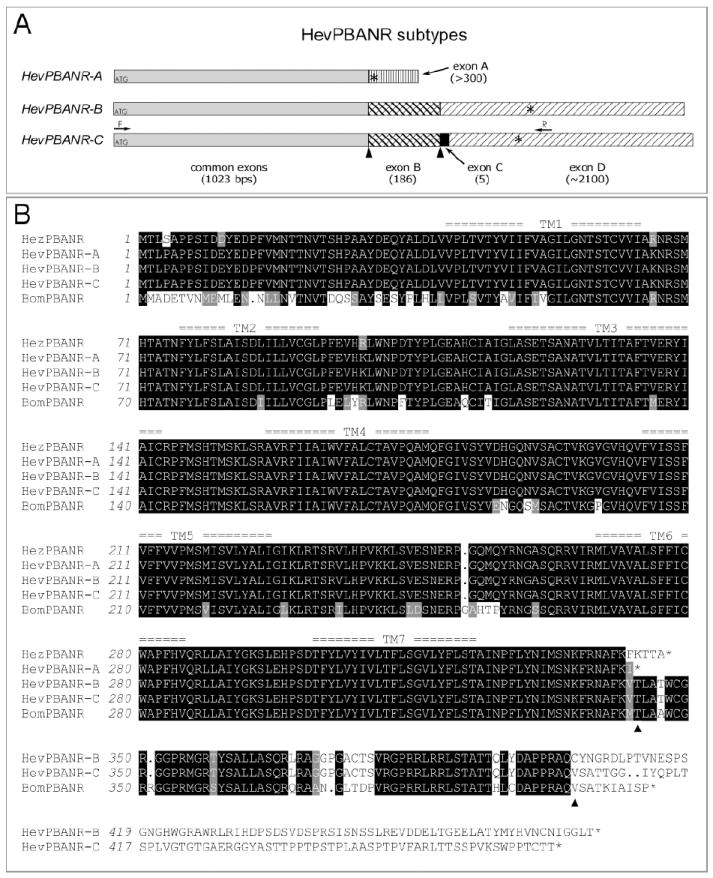
Three subtypes of HevPBANR.
(A) Schematic diagram showing the organization of putative exons in three HevPBANR subtypes. cDNA sequence comparisons between subtypes suggest the cDNA of each receptor subtype consists of a common region (gray bar) and four additional exons (variously patterned bars; exon A, -B, -C and -D), which are arranged in subtype-specific configurations. Numbers in parentheses indicate exon lengths. Translation initiation and termination sites are indicated by ATGs and asterisks (*) respectively. Arrows labeled with either F or R indicate primer binding sites used to amplify the entire open reading frame of HevPBANR-C from pheromone glands. (B) Protein sequence alignment of three HevPBANR subtypes described in this study. Helicoverpa zea PBANR (HezPBANR, GenBank accession number, AY319852) and Bombyx mori PBANR (BomPBANR, AB181298). Note that C-terminal region of HevPBANR subtype-A is quite similar to that of HezPBANR, while subtypes-B, and C are similar to BomPBANR. Triangles indicate junctions between exons corresponding to those labeled with triangles in (A). Seven transmembrane domains are numbered as TM1- TM7. Identical amino acids are highlighted in black shadow and conservative amino acids are in light gray. Dots are gaps introduced for alignment.
2.4. Phylogenetic Analysis
For the PBANR phylogram, ClustalX [30] was used to align amino acid sequences of HevPBANR-C (this study), BomPBANR (GenBank accession number, AB181298), CG8795 (AF522190), CG8784 (AF522189), CG9918 (AF522191), CG14575 (AF522193), NMUR1 (AB038649) and NMUR2 (NM_022275). Neurotensin receptor (NTR, JH0164) and growth hormone secretagogue receptor (GHSR, NM_032075) were included as outgroups. The rooted tree was produced from aligned sequences of transmembrane domains 1 to 7 by PAUP (V4.0b8a) using bootstrap with heuristic search and 1000 replicates and distance was used as the optimality criterion.
2.5. CHO Cell Expression
HevPBANRs were expressed and assayed in two CHO cell lines. One line (WTA11-CHO) exhibited stable expression of aequorin and Gα16 (Euroscreen), and the other was a wild type line (CHO-K1). WTA11-CHO cells were transiently transfected with ORFs of HevPBANR-A, -B, or –C [22], whereas CHO-K1 cells were transfected with two plasmid containing ORFs of HevPBANR-C and codon-optimized aequorin as described [12, 31]. Procedures used for CHO cell transfection and receptor assay were described previously [12, 13, 22].
Amino acid sequences of peptides used in this study are shown in Fig. 3B and Fig. 4. HezPBAN was kindly provided by Dr. Timothy Kingan. Manduca sexta diapause hormone (MasDH) was synthesized at the UC Riverside Genomics Institute. α-Subesophageal neuropeptide (α-SG-NP), β-SG-NP, γ-SG-NP from M. sexta were synthesized by Synthetic Biomolecules (San Diego). The natural members of the pyrokinin/PBAN family, truncated PBAN fragments, and the PBAN C-terminal hexapeptide Ala-series of analogs listed in Fig. 4 were synthesized by the solid-phase method on an ABI 433A Peptide Synthesizer with modified FastMoc0.25 procedure (0.25mM scale, prolonged double coupling), using the Fmoc-strategy, HPLC purified and quantified via amino acid analysis as previously described [20].
Fig. 3.
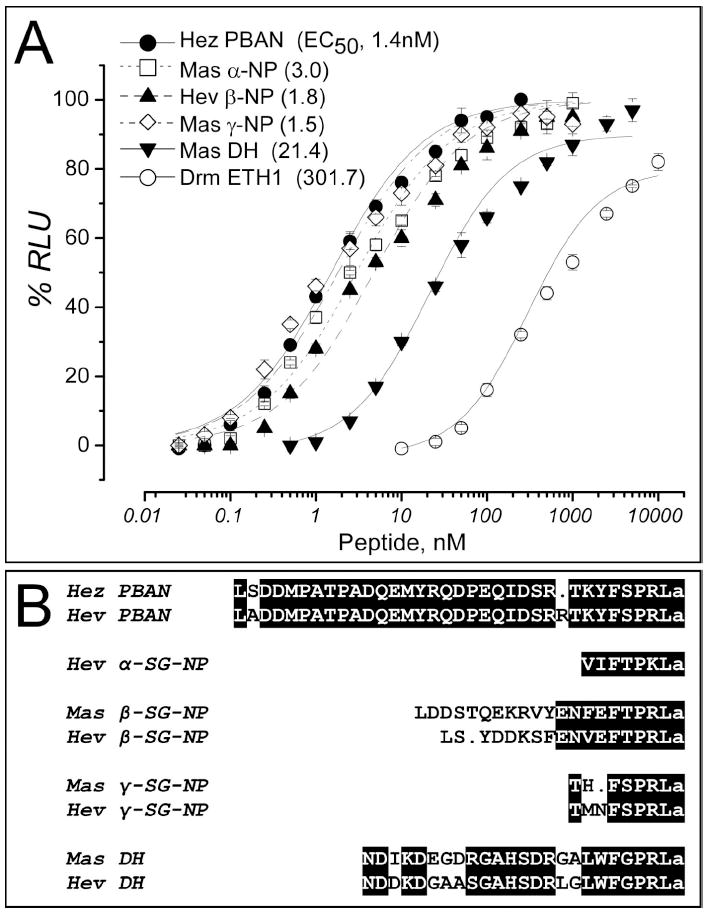
Pharmacological profile of HevPBANR.
(A) Luminescence responses of CHO cells co-expressing HevPBANR-C, aequorin, and Gα16 are plotted as a function of ligand concentration. The PBAN precursor encodes five mature peptides; diapause hormone (DH), α-subesophageal neuropeptide (α-SG-NP), β-SG-NP, PBAN and γ-SGNP. Each point is a mean value ± S.D. for percent of maximum relative luminescence unit (RLU). Number in parenthesis indicates EC50 value of each ligand.
(B) Protein sequence alignments of peptide ligands tested in (A) and their counterparts originated from HevPBAN precursor. ‘Hez’ and ‘Mas’ indicate Helicoverpa zea and Manduca sexta, respectively. Identical amino acids are highlighted in black shadow and conservative amino acids are in light gray. Dots are gaps introduced for alignment.
Fig. 4.
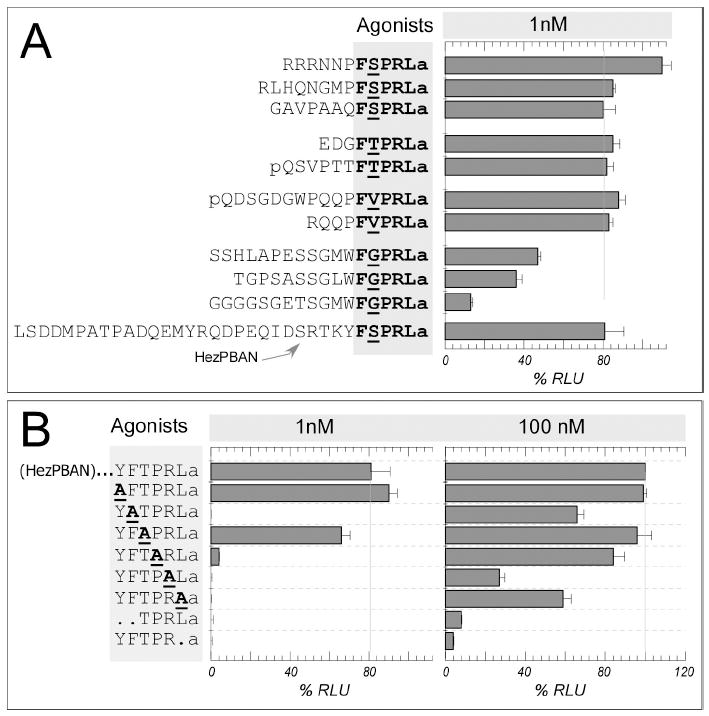
Structure-activity relationships of PBAN receptor agonists assayed on the HevPBANR.
3. RESULTS
3.1. Molecular Identification of Three HevPBANR Subtypes
In order to identify a PBAN receptor from Heliothis virescens, we synthesized degenerate PCR primers based on previously published sequences of Drosophila melanogaster GPCRs, CG8795 and CG8784, both of which are highly sensitive to DrmPK-2, a likely homolog of PBAN [21, 26]. PCRs using nested sets of degenerate primers yielded a ~636 bp cDNA fragment from the CNS of 4th instar larvae. BLAST analysis using the Drosophila local database (http://www.flybase.org/blast/) showed this fragment to retain high sequence similarity to CG8795 and CG8784 among all annotated proteins in the Drosophila genome. Using primers based on the partial cDNA sequence, we identified three distinctive cDNAs that share a common 5’-sequence (-188~+1211), but diverge at the 3’-end (>+1212), suggesting they originate from alternative splicing of 3’-exons (Fig. 1A). These receptor subtypes were named HevPBANR-A, -B and –C, respectively (Fig. 1; GenBank accession numbers, EU000525, EU000526, EU000527). Using RT-PCR, the complete ORF of HevPBANR-C was amplified from pheromone gland cDNA, while ORFs of HevPBANR-A and –B were amplified from larval CNS cDNA.
ORFs of HevPBANR subtype-A, -B and -C encode 342, 476 and 469 amino acid polypeptides, respectively. The predicted protein for each subtype contains seven putative transmembrane (TM) domains according to the TM prediction algorithm [8]; http://www.ch.embnet.org/software/TMPRED_form.html). The 5’ sequence shared by all subtypes encodes the major portion of the predicted GPCR extending from the N-terminus to the end of TM7, while subtype-specific 3’ sequences encode a C-terminal intracellular tail of varying size and sequence (Fig. 1). Comparisons of C-terminal intracellular regions of GPCRs indicate that PBANRs reported previously from Helicoverpa zea [3] and Bombyx mori [9] were highly similar to HevPBANR-A and HevPBANR-C, respectively (Fig. 1B).
3.2. Phylogenetic relationships of HevPBANR and other related GPCRs
Phylogenetic analysis revealed that HevPBANR occurs in a monophyletic clade with PBANRs of the moths Helicoverpa zea (HezPBANR) and Bombyx mori (BomPBANR), four Drosophila GPCRs CG8784, CG8795, CG9918, CG14575, and vertebrate neuromedin U receptors (NMURs) (Fig. 2). In the distance tree, three moth PBANRs (HevPBANR, HezPBANR and BomPBANR) are grouped with Drosophila GPCRs CG8784 and CG8795 to produce the monophyletic ‘PBANR clade’ with a 97% bootstrap value. The PBANR-clade is further supported by pharmacological profiles shared by its members: All receptors in this clade show marked sensitivity to pyrokinins/PBAN, which feature the FXPRLa C-terminal amino acid motif [3, 9, 21, 26]. The PBANR clade is grouped with CG9918, mainly sensitive to CAP2b-3 and a diapause hormone (DH) analog that contains a C-terminal WFGPRLa motif [2, 21]. Finally, the PBANR clade and CG9918 are grouped with CG14575, which is highly sensitive and selective to CAP2b-1 and CAP2b-2 peptides featuring the C-terminal sequence motif FPRVa (Fig. 2; [22]).
Fig. 2.
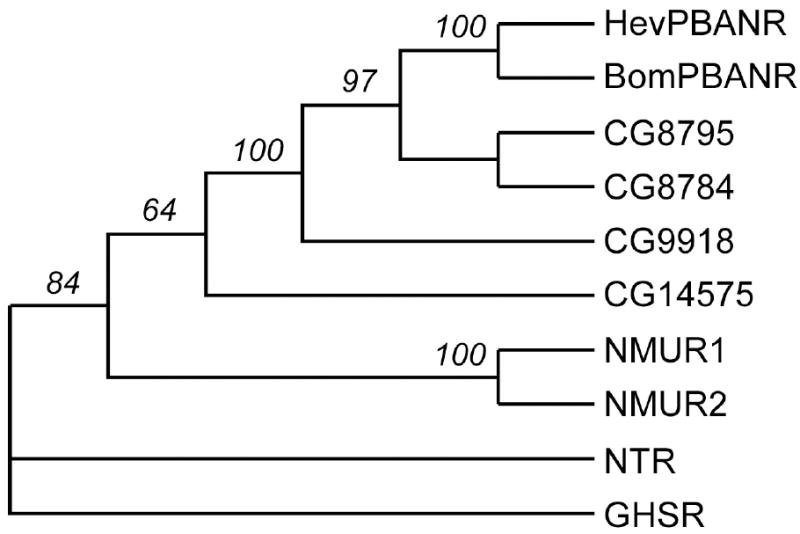
Phylogenetic tree of HevPBANR and its related G-protein coupled receptors. The percentage of 1000 bootstrap replicates supporting each node is indicated. NTR (neurotensin receptor) and GHSR (growth hormone secretogogue receptor) were analyzed as an outgroup. For details of the molecules analyzed, see the Materials and Methods section.
3.3. Functional Expression and Pharmacological Characterization of HevPBANR
Pharmacological profiles of the HevPBANR were investigated in two different CHO cell lines (CHO-WTA11 and CHO-K1) expressing aequorin and HevPBANR. In this assay, ligand-mediated activation of the GPCR mobilizes calcium from intracellular stores, resulting in a luminescent flash through oxidation of an aequorin-coelantrazine complex [9, 15, 22]. Expressed in CHO-WTA11, HevPBANR-C generated robust luminescence responses to PBAN and related peptides (Fig. 3), while HevPBANR-A and -B showed little or no Ca2+ responses to PBAN (not shown). HevPBANR-C showed slightly higher sensitivity when expressed in the CHO-K1 cell line (Fig. 4).
Luminescence responses of CHO cells expressing HevPBANR-C were highly sensitive to all peptides processed from the PBAN precursor, including PBAN, α-SG-NP, β-SG-NP, γ-SG-NP and diapause hormone (Fig. 3). Among these peptides, PBAN and γ-SG-NP were almost equally potent and the most active. Ca2+ responses were evident at 0.5 nM and reached peak levels at 50 nM, with an EC50 value of ~1.5 nM. α-SG-NP and β-SG-NP were slightly less active, with EC50 values of ~2-3 nM. Diapause hormone (DH) was least active, showing 10-fold lower activity than PBAN (Fig. 3A; see below).
3.4. Structure-Activity Relationships of PBAN-Related Agonists on the Expressed PBANR-C
We wanted to define the functional core of the PBAN peptide as well as amino acids in the C-terminal FXPRLa motif that are critical for PBANR activation. Accordingly, we assayed a panel of PBAN peptide analogs on CHO-K1 cells expressing HevPBANR-C (Fig. 4). Those natural analogs, many of which are native to a variety of insect species and share the common C-terminal FXPRLa (X=S/T/V) ending, elicit strong responses (Fig. 4A), while peptides carrying truncations of the C-terminal pentapeptide (i.e., TPRLa and YFTPRa) fail to do so (Fig. 4B). This result clearly demonstrates the C-terminal pentapeptide is the active core domain of PBAN, critical for interaction with the HevPBANR, in agreement with pheromonotropic bioassays in Heliothine insects [6, 11, 25].
We further evaluated the functional role of each residue in the active core, by testing a series of PBAN analogs carrying an Ala-replacement in the C-terminal hexapeptide YFTPRLa (Fig. 4B), which had demonstrated far greater activity than the minimal C-terminal pentapeptide core in pheromonotropic bioassays [11]. Ala features a simple side-chain that is intermediate on the hydrophilic/hydrophobic scale and lacks both branched chain character and a functional group. At 1 nM, analogs carrying the replacement of Y (AFTPRLa) or T (YFAPRLa) retain significant activity. Interestingly, DH analogs native to other insect species, subsets of pyrokinins that share C-terminal WFGPRLa, demonstrate markedly reduced activity in comparison with other pyrokinins (Fig. 4A). The aromatic W residue may be considered to be similar to the Y residue in the analogous position of PBAN, but a larger difference may be found in the variable residue of position 2 of the C-terminal pentapeptide, occupied by G. Apparently, the receptor demonstrates less compatibility with G, which features only a hydrogen at the alpha carbon, the smallest and most flexible of the amino acid sidechains. On the other hand, replacement of the positively charged R within the core by Ala leads to an analog (YFTPALa) with little or no activity even at 100 nM (Fig. 4B), suggesting that this residue is most critical for interaction of pyrokinin/PBAN ligands with HevPBANR. Replacement of F (YATPRLa) and the C-terminal L with Ala (YFTPRAa) leads to analogs that fail to elicit activity at 1 nM, but show measurable activity at 100 nM, an indication that the importance of these residues is secondary only to R. The aromatic sidechain of F and the branched-chain character of the hydrophobic residue L provide important contributions to the interaction of the ligand with the receptor. Finally, the P residue appear to be of moderate importance, as the Ala replacement analog YFTARLa shows markedly reduced activity at 1 nM compared with the parent hexapeptide YFTPRLa (Fig. 4B).
4. DISCUSSION
4.1. Multiple Subtypes of PBANR
We have reported here three subtypes of HevPBANR, which are likely produced by alternative splicing at the 3’-end of the receptor gene. One of these subtypes (HevPBANR-C) was preferentially amplified from the pheromone gland. All subtypes share the sequence extending from the N-terminus to the end of transmembrane domain 7 (TM 7), but not the C-terminal intracellular loop. Recently, two independent groups reported PBAN receptors from Helicoverpa zea (HezPBANR) [3] and Bombyx mori (BomPBANR) [9], respectively. Strikingly, we found that the HevPBANR-C subtype described here shares a considerable portion of the C-terminal intracellular loop with the BomPBANR, while the HezPBANR was quite similar to HevPBANR-A subtype (Fig. 1). Moreover, homologous multiple subtypes were also identified in a distantly related moth species, Manduca sexta (Kim et al., unpublished). Taken together, these findings suggest that alternative splicing of the gene encoding PBANR is common in moths, generating multiple receptor subtypes.
Each of the three PBANR subtypes described here differs only at the C-terminus. Therefore, we hypothesize that each subtype couples to different intracellular signaling pathways, which in turn carry out distinct physiological functions. In the current work, we found that not all subtypes were functional when expressed in CHO cells. Only the HevPBANR-C responded to PBAN under the conditions of our particular assay system. Intriguingly, the H. zea counterpart of HevPBANR-A known as HezPBANR (GenBank Accession number, AY319852) showed strong sensitivity to PBAN and its related peptides when expressed in Sf9 cells, a cell line derived from the Noctuid moth Spodoptera frugiperda [3], suggesting that subtype specific domains are involved in coupling receptors with their downstream intracellular signaling partners.
4.2. Pharmacological Profile Supports a Role as the Physiological PBAN Receptor
The pentameric C-terminal ending (FSPRLa) of PBAN is conserved, and many peptides with the same or highly similar C-terminal pentapeptide (FXPRLa), collectively referred to as members of the pyrokinin family [23], were isolated from other insect species, including non-Lepidopteran insects. The C-terminal pentapeptide fragment of PBAN-related peptides is critical for myotropic activity [18] and subsequently for their pheromonotropic function [6, 25]. This is shown in the current study to be equally true for the PBANR receptor from H. virescens. Some PBAN analogs were tested for pheromonotropic activity in H. zea [1]. This study showed that Locusta migratoria myotropin II (Lom-MT-II; EGDFTPRLa) is equally or even more potent than PBAN, while B. mori diapause hormone (BomDH; -WFGPRLa) was about 10 times less active than PBAN. Likewise, the PBANR reported in this study showed almost undistinguishable sensitivity to both Lom-MT-II (EGDFTPRLa) and PBAN, while it was about 10 times less sensitive to BomDH. Taken together, our results strongly implicate that the PBANR described here mediates the pheromonotropic activity of PBAN in H. virescens.
4.3. The PBAN Receptor as a Target for Novel Peptidomimetics
The current study is the first to address structure-activity relationships directly with an expressed PBAN receptor, and the first to use an Ala-scan to assess the relative importance of amino acid residues within the C-terminal active core region to either the successful interaction with the receptor or to the pheromonotropic response. The results indicate that the positively charged, basic R5 residue is the most critical within the hexapeptide core region for receptor interaction, followed by the branched chain L6 and aromatic F2. The residue P4 is of intermediate importance, perhaps an indication that the conformation adopted by the core PBAN sequence is important for successful receptor interaction. The proline residue is often associated with the promotion of a turn or bend structure in peptides [19]. Residues T3 and Y1 are of less importance. Overall, a ranking of residues according to relative importance to receptor-ligand interaction can be represented as follows, R5>L6>F2≫P4>T3≫Y1. This structure-activity relationship data represents an important step towards the development of potent pseudopeptide and/or non-peptide peptidomimetic agonists and/or antagonists of the PBAN receptor with enhanced biostability and bioavailability with the potential to disrupt the reproductive process in insect pests of agricultural importance. Future studies with this expressed receptor will be undertaken to identify additional aspects of conformation and chemical structure critical to the binding and activation of PBAN with the active site. Information on the identities of critical chemical and conformational structures to PBAN/receptor interaction in this and future studies can be incorporated into the developmental process of rationally designed non-peptide libraries, which can in turn be evaluated on the Hev-PBANR receptor. The availability of an expressed Hev-PBANR receptor, and the initial structure-activity relationships identified in this study, can accelerate the development of peptidomimetics with potential utility in the future management of pest insects in a selective and environmentally sound fashion.
Acknowledgments
We thank Timothy Kingan and Yoonseong Park for sage advice, and Vincent Dupriez, Euroscreen, Belgium for providing the WTA11-CHO cell line. This work was supported by a grant from the National Institutes of Health (GM 067310).
Abbreviations footnote
- CHO
Chinese hamster ovary
- EC50
cells, effective concentration for 50% response
- GPCR
G protein-coupled receptor
Footnotes
Data-deposition footnote: The nucleotide sequences and translations reported in this paper have been deposited in the GenBank database and have the following accession numbers: HevPBANR-A (EU000525), HevPBANR-B (EU000526), HevPBANR-C (EU000527).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Abernathy RL, Teal PEA, Meredith JA, Nachman RJ. Induction of pheromone production in a moth by topical application of a pseudopeptide mimic of a pheromonotropic neuropeptide. Proc Nat Acad Sci U S A. 1996;93:12621–5. doi: 10.1073/pnas.93.22.12621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cazzamali G, Torp M, Hauser F, Williamson M, Grimmelikhuijzen CJ. The Drosophila gene CG9918 codes for a pyrokinin-1 receptor. Biochem Biophys Res Commun. 2005;335:14–9. doi: 10.1016/j.bbrc.2005.07.038. [DOI] [PubMed] [Google Scholar]
- 3.Choi MY, Fuerst EJ, Rafaeli A, Jurenka R. Identification of a G protein-coupled receptor for pheromone biosynthesis activating neuropeptide from pheromone glands of the moth Helicoverpa zea. Proc Natl Acad Sci U S A. 2003;100:9721–6. doi: 10.1073/pnas.1632485100. Epub 2003 Jul 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christensen TA, Itagaki H, Teal PE, Jasensky RD, Tumlinson JH, Hildebrand JG. Innervation and neural regulation of the sex pheromone gland in female Heliothis moths. Proc Natl Acad Sci U S A. 1991;88:4971–5. doi: 10.1073/pnas.88.11.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis MT, Vakharia VN, Henry J, Kempe TG, Raina AK. Molecular cloning of the pheromone biosynthesis-activating neuropeptide in Helicoverpa zea. Proc Natl Acad Sci U S A. 1992;89:142–6. doi: 10.1073/pnas.89.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gazit Y, Dunkelbaum E, Benichus M, Altstein M. Effect of synthetic PBAN and derived peptides on sex pheromone biosynthesis in Heliothis peltigera. Insect Biochem. 1990;20:853–8. [Google Scholar]
- 7.Hewes RS, Taghert PH. Neuropeptides and neuropeptide receptors in the Drosophila melanogaster genome. Genome Res. 2001;11:1126–42. doi: 10.1101/gr.169901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hofmann K, Stoffel W. TMbase - A database of membrane spanning proteins segments. Biol Chem Hoppe-Seyler. 1993;374:166. [Google Scholar]
- 9.Hull JJ, Ohnishi A, Moto K, Kawasaki Y, Kurata R, Suzuki MG, et al. Cloning and characterization of the pheromone biosynthesis activating neuropeptide receptor from the silkmoth, Bombyx mori. Significance of the carboxyl terminus in receptor internalization. J Biol Chem. 2004;279:51500–7. doi: 10.1074/jbc.M408142200. [DOI] [PubMed] [Google Scholar]
- 10.Jurenka RA, Jacquin E, Roelofs WL. Stimulation of pheromone biosynthesis in the moth Helicoverpa zea: action of a brain hormone on pheromone glands involves Ca2+ and cAMP as second messengers. Proc Natl Acad Sci U S A. 1991;88:8621–5. doi: 10.1073/pnas.88.19.8621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kempe TG, Raina AK, Menn JJ. Structure-activity studies of pheromone-biosynthesisactivating neuropeptide PBAN. Pest Sci. 1990;30:436–8. [Google Scholar]
- 12.Kim YJ, Spalovska-Valachova I, Cho KH, Zitnanova I, Park Y, Adams ME, et al. Corazonin receptor signaling in ecdysis initiation. Proc Natl Acad Sci U S A. 2004;101:6704–9. doi: 10.1073/pnas.0305291101. Epub 2004 Apr 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim YJ, Zitnan D, Cho KH, Schooley DA, Mizoguchi A, Adams ME. Central peptidergic ensembles associated with organization of an innate behavior. Proc Natl Acad Sci U S A. 2006;103:14211–6. doi: 10.1073/pnas.0603459103. Epub 2006 Sep 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kitamura A, Nagasawa H, Kataoka H, Inoue T, Matsumoto S, Ando T, et al. Amino acid sequence of pheromone-biosynthesis-activating neuropeptide (PBAN) of the silkworm, Bombyx mori. Biochem Biophys Res Commun. 1989;163:520–6. doi: 10.1016/0006-291x(89)92168-2. [DOI] [PubMed] [Google Scholar]
- 15.Le Poul E, Hisada S, Mizuguchi Y, Dupriez VJ, Burgeon E, Detheux M. Adaptation of aequorin functional assay to high throughput screening. J Biomol Screen. 2002;7:57–65. doi: 10.1177/108705710200700108. [DOI] [PubMed] [Google Scholar]
- 16.Ma PW, Knipple DC, Roelofs WL. Structural organization of the Helicoverpa zea gene encoding the precursor protein for pheromone biosynthesis-activating neuropeptide and other neuropeptides. Proc Natl Acad Sci U S A. 1994;91:6506–10. doi: 10.1073/pnas.91.14.6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mbungu D, Ross LS, Gill SS. Cloning, functional expression, and pharmacology of a GABA transporter from Manduca sexta. Arch Biochem Biophys. 1995;318:489–97. doi: 10.1006/abbi.1995.1258. [DOI] [PubMed] [Google Scholar]
- 18.Nachman RJ, Holman GM, Cook BJ. Active fragments and analogs of the neuropeptide leucopyrokinin: Structure-function studies. Biochem Biophys Res Commun. 1986;137:936–42. doi: 10.1016/0006-291x(86)90315-3. [DOI] [PubMed] [Google Scholar]
- 19.Nachman RJ, Roberts VA, Dyson HJ, Holman GM, Tainer JA. Active conformation of an insect neuropeptide family. Proc Natl Acad Sci U S A. 1991;88:4518–22. doi: 10.1073/pnas.88.10.4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nachman RJ, Strey A, Isaac E, Pryor N, Lopez JD, Deng JG, et al. Enhanced in vivo activity of peptidase-resistant analogs of the insect kinin neuropeptide family. Peptides. 2002;23:735–45. doi: 10.1016/s0196-9781(01)00654-4. [DOI] [PubMed] [Google Scholar]
- 21.Park Y, Kim YJ, Adams ME. Identification of G protein-coupled receptors for Drosophila PRXamide peptides, CCAP, corazonin, and AKH supports a theory of ligandreceptor coevolution. Proc Natl Acad Sci U S A. 2002;99:11423–8. doi: 10.1073/pnas.162276199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park Y, Kim YJ, Dupriez V, Adams ME. Two subtypes of ecdysis-triggering hormone receptor in Drosophila melanogaster. J Biol Chem. 2003;278:17710–5. doi: 10.1074/jbc.M301119200. [DOI] [PubMed] [Google Scholar]
- 23.Predel R, Nachman RJ. The FXPRLamide (Pyrokinin/PBAN) peptide family. In: Kastin A, editor. Handbook of Biologically Active Peptides. Elsevier; 2006. pp. 207–13. [Google Scholar]
- 24.Raina AK, Jaffe H, Kempe TG, Keim P, Blacher RW, Fales HM, et al. Identification of a neuropeptide hormone that regulates sex pheromone production in female moths. Science. 1989;244:796–8. doi: 10.1126/science.244.4906.796. [DOI] [PubMed] [Google Scholar]
- 25.Raina AK, Kempe TG. A pentapeptide of the C-terminal sequence of PBAN with pheromonotropic activity. Insect Biochem. 1990;20:849–51. [Google Scholar]
- 26.Rosenkilde C, Cazzamali G, Williamson M, Hauser F, Sondergaard L, DeLotto R, et al. Molecular cloning, functional expression, and gene silencing of two Drosophila receptors for the Drosophila neuropeptide pyrokinin-2. Biochem Biophys Res Commun. 2003;309:485–94. doi: 10.1016/j.bbrc.2003.08.022. [DOI] [PubMed] [Google Scholar]
- 27.Sato Y, Oguchi M, Menjo N, Imai K, Saito H, Ikeda M, et al. Precursor polyprotein for multiple neuropeptides secreted from the suboesophageal ganglion of the silkworm Bombyx mori: characterization of the cDNA encoding the diapause hormone precursor and identification of additional peptides. Proc Natl Acad Sci U S A. 1993;90:3251–5. doi: 10.1073/pnas.90.8.3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shorey HH, Hale RL. Mass-rearing of the larvae of nine noctuid species on a simple artificial medium. J Econ Entomol. 1965;58:522–4. [Google Scholar]
- 29.Teal PE, Nachman RJ. A brominated-fluorene insect neuropeptide analog exhibits pyrokinin/PBAN-specific toxicity for adult females of the tobacco budworm moth. Peptides. 2002;23:801–6. doi: 10.1016/s0196-9781(01)00656-8. [DOI] [PubMed] [Google Scholar]
- 30.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–82. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vernon WI, Printen JA. Assay for intracellular calcium using a codon-optimized aequorin. Biotechniques. 2002;33:730–732. doi: 10.2144/02334bm02. [DOI] [PubMed] [Google Scholar]


