Abstract
Human pseudouridine (Ψ) synthase Pus1 (hPus1) modifies specific uridine residues in several non-coding RNAs; tRNA, U2 spliceosomal RNA and steroid receptor activator RNA. We report three structures of the catalytic core domain of hPus1 from two crystal forms, at 1.8 Å resolution. The structures are the first of a mammalian Ψ synthase from the set of five Ψ synthase families common to all kingdoms of life. hPus1 adopts a fold similar to bacterial Ψ synthases, with a central antiparallel ß-sheet flanked by helices and loops. A flexible hinge at the base of the sheet allows the enzyme to open and close around an electropositive active site cleft. In one crystal form a molecule of MES mimics the target uridine of an RNA substrate. A positively charged electrostatic surface extends from the active site towards the N-terminus of the catalytic domain suggesting an extensive binding site specific for target RNAs. Two alpha helices C-terminal to the core domain, but unique to hPus1, extend along the back and top of the central ß-sheet and form the walls of the RNA binding surface. Docking of tRNA to hPus1 in a productive orientation requires only minor conformational changes to enzyme and tRNA. The docked tRNA is bound by the electropositive surface of the protein employing a completely different binding mode than that seen for the tRNA complex of the E. coli homolog TruA.
Keywords: RNA modifying enzyme, isomerase, pseudouridine, X-ray crystallography, tRNA
Introduction
Pseudouridine synthases (Ψ synthases) are a class of RNA modifying enzymes that catalyze the conversion of uridine to Ψ in a variety of RNA substrates (rRNA, snRNA, tRNA, snoRNA), without any cofactor. Pseudouridine (Ψ), the C5-glycosyl isomer of uridine, is the most common modified nucleotide found in RNA throughout all kingdoms of life.1; 2 Pseudouridylation generates an extra hydrogen bond donor at the new N1 position, and brings stability to tertiary structure of RNA, for maintaining accuracy and efficiency of translation, and for function of the spliceosome.1; 2; 3; 4 Multiple Ψs need to be present for proper formation and function of the ribosome and spliceosome in yeast or E. coli.
Based on sequence homology Ψ synthases are grouped into six families: TruA, TruB, TruD, RluA and RsuA are named after their bacterial representative; the sixth family, the Pus10 family is present in human, eukaryotes and archaea and does not have significant sequence homology to the other five families.5 Despite low sequence homology between families, these Ψ synthases have catalytic domains with a common fold containing conserved motifs required for catalytic activity.5; 6 Some Ψ synthases possess additional RNA-binding domains N- or C-terminal to the catalytic core such as S4 or PUA domains that contribute to RNA binding.7 In eukaryotes pseudouridylation is carried out by site-specific Ψ synthases (e.g. Pus1 and Pus10), or by Cbf5 (dyskerin in human), which functions as part of a H/ACA small nucleolar ribonucleoprotein (snoRNP) complex with a guide RNA conferring site-specificity.5; 8
Human pseudouridine synthase 1 (hPus1) is a member of the TruA family and modifies several specific RNA substrates. It acts on mature and intron-containing tRNAs, the U2 snRNA and the steroid receptor RNA activator (SRA).9 Pseudouridylation is critical for SRAs function as a co-activator of the nuclear receptor estrogen receptor α.10; 11
The rare genetic disease ‘mitochondrial myopathy and sideroblastic anemia’ (MLASA) is associated with absent or greatly reduced Pus1 activity because tRNA from MLASA patients lacks uridine modifications at positions normally modified by Pus1. Two Pus1 mutations, an Arg144-to-Trp† mutation in the active site of the enzyme and a mutation of Glu220, which leads to C-terminally truncated protein, have been identified independently and probably form the genetic basis for the reduced activity.12, 13
Yeast and mammalian Pus1 proteins have been studied in vitro and in vivo and display multiple overlapping but not identical modification sites. The enzymes have been shown to modify positions 27/28 in the anticodon stem-loop of many tRNAs, positions 34/36 in intron-containing tRNAs and position 1 in tRNAArg.14; 15 Mouse and human Pus1 also modify position 30 in some tRNAs.15 Interestingly, modification of U2 snRNA at U44 by mouse Pus1 has only been seen when the protein is expressed in a pus1 deleted yeast strain, whereas yeast Pus1 can modify U2 snRNA in vitro and in vivo.15; 16 All tRNA modification sites and the target nucleotide in SRA lie in base-paired regions of stem-loops whereas the target nucleotide in U2 snRNA is base-unpaired. In a recent study Sibert and Patton analyzed the structural and sequence requirements of human Pus1.17 A minimal substrate was identified that consists of the ASL and the TΨC-loop of tRNASer. While base pair interactions 3′ to the modification site in the ASL are critical for activity; hPus1 does not display specific sequence requirements in the TΨC-loop.
Our own and other structures of several bacterial and archaeal Ψ synthases in complex with RNA substrates reveal important aspects of how these enzymes recognize their RNA substrates and achieve site specific modification.7; 18 The only eukaryotic pseudouridine structure solved to date is that of Pus10 that represents a previously uncharacterized Ψ synthase family. To gain insight into how a single Ψ synthase recognizes specific sites in several different RNA tertiary structures, we determined high-resolution crystal structures of the core domain of hPus1, the first structure of a eukaryotic Ψ synthase from one of the five families conserved in all kingdoms of life, and the second structure of a eukaryotic Ψ synthase.
Results and Discussion
Structure determination and overall structure of hPus1
hPus1 is a 427 amino acid protein in the TruA family of Ψ synthases. Protein disorder prediction analysis (using the RONN19 server) indicated residues N-and C-terminal to the putative catalytic domain to be disordered. Since we did not obtain crystals of the full-length protein, we designed a six-histidine tagged candidate construct encompassing residues 79-408 that could be expressed in E. coli, and purified to homogeneity using Ni-NTA affinity- and size exclusion chromatography. This construct comprises the complete catalytic core, and forms a complex with tRNA, and with the recently identified hPus1 minimal tRNA substrate that encompasses nucleotides 26-65 of human tRNASer.
Truncated hPus1 (79-408) shows comparable activity (99% after 2h of incubation) to full-length hPus1 in tritium release assays, using the 3H-UTP radiolabeled minimal tRNA substrate.20 This result indicates that the predicted disordered N- and C-terminal residues are not essential for hPus1' activity.
The minimal tRNA-protein complex was purified using size exclusion chromatography, subjected to crystallization trials, and led to two different crystal forms, hexagonal plates and rod shaped crystals, under different crystallization conditions.
The structures of both crystal forms were determined using molecular replacement with the apo TruA structure (PDB ID: 1DJ0) as a search model. Although we used a minimal-tRNA to prepare a protein complex for crystallization, we did not observe electron density corresponding to RNA in either crystal form. We determined two structures of the hexagonal plate form (form I), and one structure of the rod form (form II). A MES (2-(N-morpholino)ethane sulfonic acid) molecule and two sulfate ions, derived from the crystallization condition, were bound in the active site in one form I structure. The second form I structure, which was obtained by soaking form I crystals in a MES-free uridine solution, had only water in the active site. We refer to these structures as “form I MES” and “form I apo”.
The form I MES structure was in space group P6522 with one molecule in the asymmetric unit. The structure was refined to 1.85Å to an Rwork of 18.2% and an Rfree of 22.6%. The final structure includes 296 out of the 336 residues. Residues at the N- and C-terminus and three loop regions could not be interpreted due to poor electron density (residues 79-80, 103-107, 192-195, 341-354 and 400-408). The form II structure was in space group C2 with one molecule in the asymmetric unit. The structure was refined to 1.8Å resolution to give an Rwork of 18.6% and an Rfree of 22.7%. For form II, 313 out of 336 residues were interpreted (missing residues: 79-80, 104-108, 346-354 and 408). The form I and form II crystal structures are identical except for small conformational differences, to be discussed below. The form II structure is used for the description of the hPus1 protein fold and for comparison to other Ψ synthases because it is more complete.
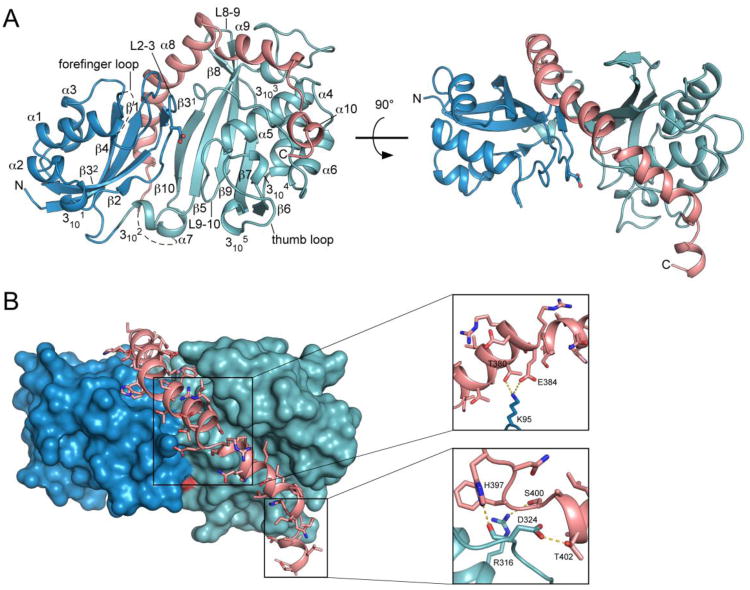
The Pus1 catalytic domain adopts a mixed α/β fold with an extended central antiparallel ß-sheet decorated by helices and loops as found in other Ψ synthases (Figure 1A).7 The ß-sheet consists of a four-stranded N-terminal sheet (ß4, ß1, ß31 and ß32, ß2) that packs against a six-stranded C-terminal sheet (ß10, ß5, ß9, ß8, ß7, ß6), with hydrogen bonds at the N-terminal ends of strands ß31 and ß10 joining the two sheets together. Helices α1-α3 flank the N-terminal sheet and helices α4-α6 decorate the C-terminal sheet. Catalytic Asp146 is located in the center of a cleft between the N- and C-terminal β-sheets that is about 8 Å wide (distance CA/Asp146 to CA/Met291) (Figure 1A, 2B). The walls of the cleft are formed by the N-terminal domain loops L1-2 and L2-3 and the C-terminal domain loops L6-7 and L9-10. Loops L1-2 and L6-7 are commonly called forefinger and thumb loop, respectively.
Figure 1.
Overall structure of the Pus1 catalytic domain. (A) Cartoon representation of the Pus1 crystal structure (form II) in two different orientations, rotated by 90°, with the N-terminal domain colored dark-blue and the C-terminal domain colored green. Catalytic Asp146, rendered in ball-and-stick form, is located in the center of the cleft. The C-terminal extension helices α8, α9 and α10 unique to Pus1 are colored salmon (B) Interactions between amino acid residues of helices α8 and α9, with the surface of the Ψ synthase core shown in dark-blue and green. C-terminal helices are colored salmon with amino acid side chains shown as sticks. Pus1 is shown in the same orientation as in Figure 1 A right panel. Right: Close-up views of two main regions of interactions between residues of α8 and α9 with the Ψ synthase core. Hydrogen bonds are depicted as yellow dashes.
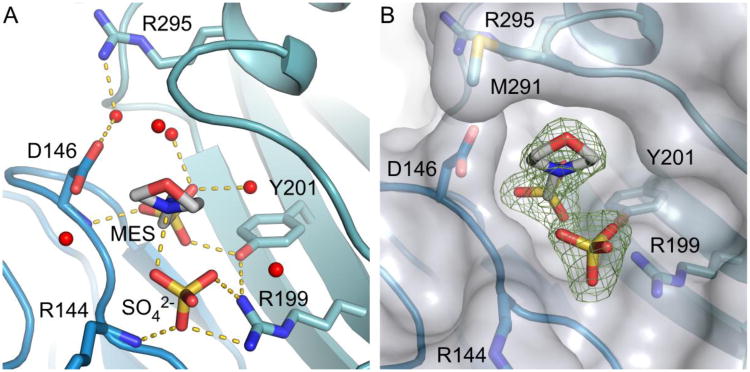
Figure 2.
Active site cleft and polar contacts between MES, SO42- and water molecules and residues at the active site of Pus1. (A) Active site of crystal form I depicting interactions between MES, SO4 and residues in the active site as yellow dashed lines. Waters lining the cleft are represented as red spheres. (B) Surface representation of Pus1 superimposed on (Fo-Fc) omitmap electron density (4σ) showing the width of the active site cleft. The distance between Cαs of Asp146 and Met291 is about 8 Å.
Unique to Pus1, three C-terminal helices, α8, α9 and α10 (α8: Ala359-Glu384, α9: Ser386-Ile396, α10: Ser400-Thr405) border the substrate-binding cleft and protrude from the conserved Ψ synthase core. Two orthogonal views of Pus1 in Figure 1 illustrate that these helices are integrated into the catalytic domain, intimately associated with both the N-terminal (blue) and C-terminal (green) domains of the conserved core. α8 is positioned at the convex site of the N-and C-terminal ß-sheets and extends up to the beginning of the ß9 strand, and α9 wraps around the C-terminal domain. Residues 369-373 in the middle of α8 form a π-helix.21 Major interactions between the C-terminal helices and the Ψ core include hydrogen bonds between Lys364 and Leu181; Lys95 and Thr380 as well as Glu384; His397 and the carbonyl oxygen of Asp324; Ser400 and Arg316 and Thr402 and Asp324 (Figure 1B). We were unable to obtain soluble protein of a construct Pus1 (79-345), in which helices α8-α10 have been deleted, suggesting that the unique C-terminal helices are essential for protein structure and stability.
Mimicking the substrate nucleotide and phosphate, a MES molecule and sulfate ion are bound in the active site cleft of Pus1 in crystal form I MES (Figure 2). When the Pus1 structure is superposed on the structure of TruA-tRNA (PDB ID: 2NR0) the morpholine ring of MES and the ribose of the flipped-out nucleotide in the TruA active site overlap and the sulfate ion aligns with the phosphate group of the RNA (Figure S2). Catalytic Asp146 points toward the sulfonic acid group of the MES molecule and interacts through a water-mediated hydrogen bond with Arg295 from α5 on the opposite site of the cleft. Because MES mimics the substrate nucleotide it borrows residues from the uridine-binding site, specifically interacting with Asp146, with conserved Tyr201 from ß5, and with a neighboring sulfate molecule at the backbone phosphate-binding site. O3 from the sulfonic acid group of MES makes a hydrogen bond to the main-chain N-H of Asp146, O1 hydrogen bonds to the hydroxyl of Tyr201, and O2 hydrogen bonds to two water molecules. N4 of MES interacts with a sulfate ion, which in turn is coordinated by Arg199 and the main-chain N-H of Arg144 (Figure 2A).
To exchange the MES with a substrate mimic more similar to the natural substrate RNA, we soaked the form I crystals in a uridine solution that did not include MES. The soaked crystals were isomorphous to crystal form I, and the structure was solved and refined to a resolution of 1.75Å with Rfree of 23.9% and Rwork of 19.9%. However, no electron density for either MES or uridine was visible in the active site. We refer to this structure as form I apo. The form I apo- and the MES-bound structures align with an rms deviation of 0.3 Å (rigid superposition over 296 Cα). In the apo-structure L9-10 moved towards the active site loop that contains Asp 146, L2-3, by 0.95 Å, resulting in a narrower active site cleft. The small but distinct shift of L9-10 is clearly seen in an (|Fobs|-|Fobs_apo|)αcalc difference map (Figure S3), which more sensitively indicates differences between the form I MES and form I apo structures than does comparison of their refined coordinates. The MES-induced opening of the RNA-binding cleft suggests an induced-fit substrate-binding mechanism similar to that observed in other Ψ synthases, where the N-and C-terminal domains adjust to enclose the RNA bound to the cleft.22; 23 Without MES, the side chain orientation of Asp146 is no longer restrained and Asp146 adopts an alternative rotamer conformation pointing away from the cleft into the solvent.
The absence of ligand in the active site of the soaked crystals is in line with observations made with RluE and Pus10 where soaking with uracil and UMP in the case of RluE, and uridine and UMP in case of Pus10 did not lead to nucleotide-bound structures.24; 25 The fact that we were unable to soak uridine into the crystals suggests that the binding affinity of hPus1 for uridine alone is low.
Conformational flexibility is indicated by differences among Pus1 crystal structures
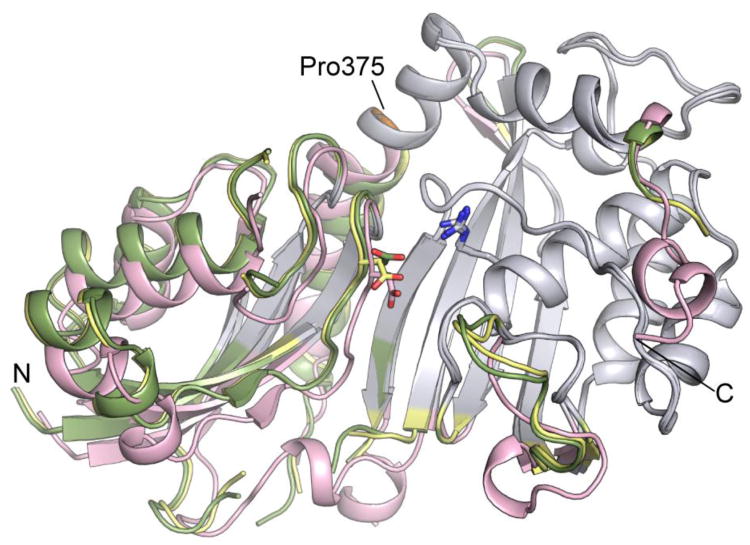
The conformational differences observed in the three crystal structures reported here allow us to identify flexible regions of the protein that are potentially important for induced fit substrate recognition. Pairwise comparison of the form II structure with the form I structures using the program Rapido26 identifies two structurally conserved ‘rigid bodies’. Separately, the conformations of these rigid bodies are conserved, but their relative positions are different in each crystal structure. The larger rigid body encompasses residues in the C-terminal domain and 16 residues from the N-terminal ß-sheet. The smaller rigid body includes residues in the N-terminal domain and residues in the C-terminal helix α8 up to Pro375 that forms a hinge point. In Figure 3 the three structures are aligned on the C-terminal rigid bodies (gray) to show the differences in the relative positions of the N-terminal rigid bodies. Form II structure has a more closed active site than the form I structures, and indicates that Pus1 is hinged to open and close to bind substrate as observed in other Ψ synthases.22, 23 Interestingly, residues C-terminal to the π-helix at Pro375 are part of the larger ‘rigid body domain’ suggesting a role for the π-helix in the hinge motion. In addition to conformational changes by movement of the rigid bodies, structural differences are seen in several loop regions indicating their flexibility. These include the loop regions L8-9, L9-10 and the thumb loop, which in TruA interacts with the major groove of the anti codon stem loop in the tRNA.
Figure 3.
Conformational changes between Pus1 form I (MES, apo) and form II (apo) crystal structures. The conserved cores of the C-terminal domains of the three Pus1 structures were superposed (colored grey). Form I (MES) is colored in yellow. Form I (apo) in green shows a narrowed active site cleft, and form II (apo) in salmon shows a crystal-form dependent rigid body shift in the N-terminal domain. Catalytic Asp146 and Arg295 are represented as sticks. Pro375 is colored orange. The orientation shown corresponds to the orientation shown in Figure 1 A left panel.
There are two disulfide bonds in the open (form I) structures: an intramolecular disulfide bond between residues Cys142 and Cys196 and an intermolecular disulfide bond between Cys260 and Cys260′ from a symmetry related molecule. Cys260 is not conserved in TruA and the low interface surface of 325 Å2 indicates that the Pus1 interface containing these residues is not likely to be biologically relevant. The intramolecular disulfide bond is associated with a shift of residues 195-197 closer to L2-3, where the catalytic aspartate resides. However, no major conformational changes were induced by disulfide bond formation in either form I structure. We suggest that disulfide bond formation is not physiological and simply stems from the loss of reducing potential in the crystallization solution as crystals grew over a period of 2-3 weeks. However reducing agents are generally included for maximum activity in Ψ synthase assays.27
Pus1 and TruA share a common fold
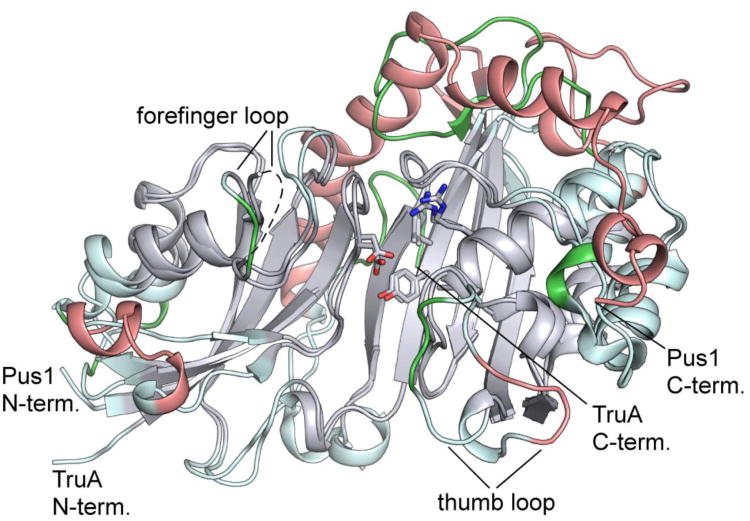
Despite their low overall sequence identity (24%), the core regions of E. coli TruA (PDB ID: 1DJ0) and human Pus1 have the same basic topology and very similar folds, justifying their assignment to the same Ψ synthase family. The rmsd for 225 Cαs of TruA aligned with form II Pus1 is 2.15 Å . When the alignment is performed on the conserved core identified by difference-distance analysis in Rapido, the rmsd is 0.8Å (over 146 Cα residues). Thus the catalytic cores of the bacterial and human homologues have a highly conserved conformation. Superposition of Pus1 and TruA on their catalytic cores (Figure 4) shows that inserts and nonconserved loops in the two molecules are arranged around the periphery of the core.
Figure 4.
Comparison of the structures of apo-Pus1 (form II) and apo-TruA. Superposition of the apo-Pus1 structure (form II) with the structure of apo-TruA (PDB: 3DJ0) aligned based on the most structurally conserved regions of their common cores, colored grey. Regions of the core not used in the alignment are colored light cyan. Regions outside the core are colored salmon for Pus1 and green for TruA. The five signature Ψ synthase residues, including catalytic Asp, are superimposable, represented as sticks. The orientation shown corresponds to the orientation shown in Figure 1 A left panel.
The five signature residues of Ψ synthase in the active site, Asp146, Tyr201, Arg295, Ile294 (Val in TruA) and Leu333, align closely in the superposed structures, indicating that the target uridine is bound in a similar orientation in the two homologues. The structural equivalence of these signature residues in TruA and hPus1 was thus correctly assumed by Sibert et al. (2008) in interpretation of site-directed mutagenesis experiments with hPus1.9 They mutated Tyr201 and Arg295 to several other amino acids and found that many of the variants had significant activity. These results indicate Tyr201 and Arg295 do not have essential roles in catalysis, which is consistent with the fact that neither residue is invariant among all Ψ synthases.
The target uridine of the substrate tRNA has to be flipped out of the RNA structure into the active site of the enzyme. We have shown how Arg58 in TruA, which is conserved in the RluA, RsuA and TruA families but not in the TruB and TruD families, serves to eject the base to reach the flipped state.28 Using MD simulations we identified a transition pathway of the target uridine nucleotide in which Arg58 stacks against the uridine and tracks its movement out of the anticodon loop into the active site. Mutation of the arginine did not affect the affinity of the enzyme for the substrate tRNA but led to a complete loss of enzymatic activity. In RluA, RluF and RluB the corresponding Arg is proposed to drive base-flipping by intercalating into the RNA stem at the location of the target base.29 In hPus1 the equivalent Arg144 is also positioned to intercalate into an RNA stem to replace the target base. Mutation of Arg 144 in hPus1 to tryptophan has been identified as the genetic basis for the rare autosomal disease MLASA.13 Presumably the bulky tryptophan can't intercalate into the stem and thus interferes with enzymatic activity. On the other hand, R144A, R144K and R144S variants show near wild-type activity at positions 27 and 28 of tRNA, but reduced or absent activity at other modification sites.9 These results suggest that while Arg144 probably substitutes for the target uridine when the target flips into the hPus1 active site, it isn't essential for base-flipping in hPus1, at least during modification of U27 or U28 of tRNA. However the Arg 144 may be required for stabilizing the RNA conformation required for catalysis at the other modification sites.
Major differences between Pus1 and TruA are seen in the length and orientation of the helices and loop regions surrounding the core. The density shows that the forefinger loop in Pus1 is flexible. In TruA this loop is shorter by three residues and is ordered in both the apo and RNA-bound structures. Pus1 and TruA structures also differ by a nine-residue insert that forms two short helices (α2 and 3101) between helix α1 and Asp146 in Pus1. In TruA, Loop L6-7, also called the thumb loop, participates in RNA recognition. In Pus1, the thumb loop points away from the cleft, generating a wider opening than in TruA. The higher than average B-factors of the loop indicate flexibility that may be important for RNA binding. Flexibility of the loop regions has also been seen in other Ψ synthases: In TruB the thumb loop is highly flexible in the apo protein and becomes ordered only upon RNA binding.23 Some of these loops must be key determinants of RNA specificity.
Pus1 has three additional helices C-terminal to the catalytic domain (α8: Ala359-Glu384, α9: Ser386-Ile396 and α10: Ser400-Thr405), comprising an insert found in all species of Pus1, which has not been reported for any other Ψ synthase to date. α8 covers the convex side of the 10-stranded ß-sheet while α9 runs orthogonal to α5 (Figure 1). In contrast in TruA, the C-terminal 12 residues form a non-helical and proline rich structure that occupies an area covered by residues N-terminal to the π-helix of α8 in Pus1. α9 in Pus1 restrains the orientation of the loop between ß5 and α4 while the equivalent loop in TruA is pointed towards the active site cleft and defines the walls of the tRNA binding site. The loop between ß8 and ß9 is six residues longer in Pus1 and interacts with α8 (Arg274 hydrogen bonds to Glu370). The C-terminal helices together with other inserts in Pus1 enlarge the area and change the topography of the protein surface relative to that of TruA.
Size exclusion chromatography of full-length hPus1 and hPus1 (79-408) combined with ‘tetra detector’ analyses that allows the determination of the molecular mass of full-length and truncated Pus1 show single, monodisperse peaks, which correspond to a monomeric state of the enzyme. Analysis of the interfaces between symmetry related Pus1 molecules in both form I and form II crystals using the program PISA30 identifies an interface area of 1084 Å2 in the form I crystals as the largest one between Pus1 molecules. This area equals 8% of the total accessible Pus1 surface area, and approximately one third of the buried surface area in the TruA dimer interface. Though the value lies within the range of biological significant interfaces, the Pus1 interface does not have the characteristics of an oligomeric interface that would be stable in solution (as analyzed by PISA). The interface is composed of mainly polar residues, while interfaces involved in the formation of multimers usually contain hydrophobic residues. The interface also includes residues close to the active site (Lys147). Together these observations indicate that the Pus1 interface in the form I crystals is rather formed by crystal packing than reflecting a biological dimer. In contrast E. coli TruA behaves as a dimer in solution and both apo-TruA and tRNA-bound TruA crystallized as dimers (Figure 5).28, 31
Figure 5.
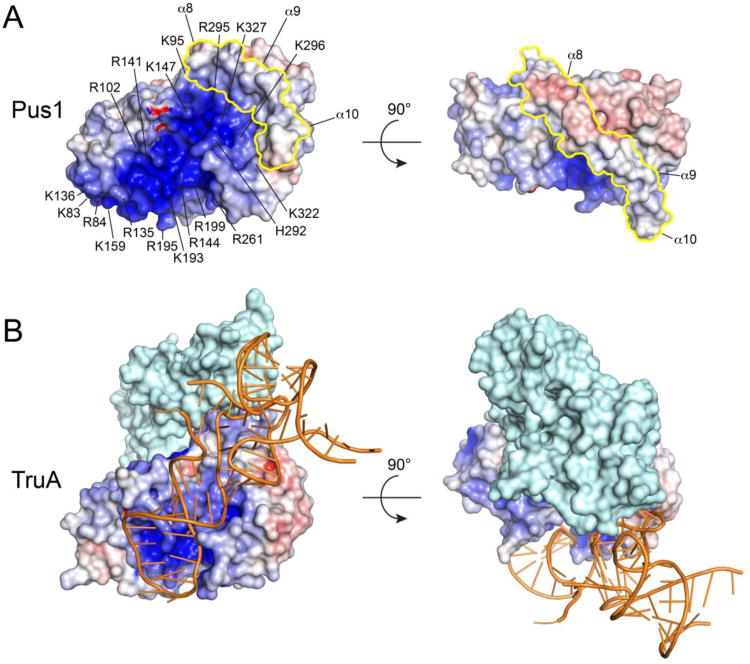
Electrostatic surface potentials of apo-Pus1 (form II) (A) and TruA from the tRNA-TruA complex (PDB ID: 2NR0) (B). The Pus1 C-terminal insert is colored in yellow. Positive electrostatic potential is colored blue (+10 kT/e), negative electrostatic potential red (-10 kT/e). Electrostatic potentials were calculated with APBS46 Pus1 and TruA were superposed on their catalytic cores and the two orientations shown correspond to the orientations in Figure 1A. The surface of a dimer of TruA is shown with only one protomer TruA colored according to its electrostatic surface potential and the other protomer colored in light cyan. Bound tRNA is shown in orange cartoon format.
We were not able to conclusively determine the stoichiometry of the Pus1-RNA complex we used for crystallization (see Material and Methods, Protein expression and purification). Using ultracentrifugation analysis Arluison et al. show that yeast Pus1 binds to tRNA as a monomer32, and residues in α8 are conserved between human and yeast Pus1. Though human Pus1 is more similar to the yeast enzyme than to its bacterial homologue TruA, large structural inserts in the yeast enzyme and the fact that it depends on Zn2+ to bind tRNA, make simple predictions on Pus1 RNA binding properties difficult by this approach. Future analysis of the stoichiometry of a hPus1-RNA complex using full-length tRNA should be more conclusive.
Pseudouridine formation in humans: Comparison between Pus1 and Pus10 crystal structures
The Pus10 structure is the only other X-ray structure of a human Ψ synthase25 and because it lacks sequence similarity to the five bacterial Ψ synthase families, it is classified as the first and so far the only member of a sixth family of Ψ synthases. Although not proven, Pus10 most likely catalyzes the formation of the conserved Ψ55 in tRNAs.33 In addition to its catalytic domain Pus10 has an N-terminal THUMP (thiouridine synthases, methylases and PSUSs) domain, an RNA-binding domain that has not been observed in structures of other Ψ synthases. The catalytic domains of Pus1 and Pus10 share the central ß-sheet structure but differ in decorating secondary structure elements giving rise to an rms deviation of 4.2 Å (over 184 Cα). It is clear from the overlap of Pus 1 (form II) and the catalytic domain of Pus10 (Figure S4) that these enzymes are much less conserved in structure than are Pus1 and TruA (Figure 4). However, the conserved signature Ψ synthase motifs, shown as sticks in Figure S4, closely align, suggesting a conserved catalytic mechanism. Docking experiments with tRNA and Pus10 performed by McCleverty et al. suggest that the THUMP domain of Pus10 is involved in binding the 3′-acceptor stem of the tRNA.25 Pus1 does not have an additional RNA binding domain suggesting that the basis for RNA recognition differs between Pus1 and Pus10.
Zinc ions are not involved in the human Pus1 complexes. The N-terminal THUMP domain of Pus10 incorporates Zn2+, which is coordinated by four cysteine residues and is probably essential for maintaining the structural integrity of the domain. Arluison et al. have shown that yeast Pus1 binds one Zn2+ ion and that Zn2+-depleted Pus1 is unable to bind tRNA.34 Two zinc binding motifs with a total of six cysteines and histidines have been proposed for yeast Pus1. However, out of the six proposed Zn2+-binding residues, only Cys196 and His291 are conserved in hPus1 and the latter residue is located in the active site cleft. Further, hPus1 binds to RNA in the absence of Zn2+, and our crystal structures of the catalytic domain of hPus1 do not contain Zn2+. No other Zn-binding motifs are identified in hPus1, suggesting it doesn't bind Zn2+. Thus the evidence is that Pus 1, and Pus 10 are both related to the bacterial structures but target different bases in tRNA, using different domains for recognition.
RNA binding
Using a 5′-FAM labeled minimal tRNASer (nt. 26-65), we measured the change of its fluorescence polarization in presence of increasing amounts of truncated hPus1D146N (79-408) and full-length hPus1D146N (Figure S5). The affinities of both proteins for the minimal RNA substrate are in the low nanomolar range (Kd, Pus1 (79-408) < 14 nM and Kd (Pus1 full-length) = 53 nM) suggesting that the RNA binding determinants are present in the core protein. Deletion of the predicted disordered N-and C-terminal residues thus does not impair RNA binding. These binding constants are similar to previously reported Kds of Pus 1 binding RNAs. Sibert and Patton determined a Kd of 240 nM for the interaction between human Pus1 and tRNASer.17 Affinities for the interaction between S. cerevisiae Pus1 to substrate tRNAs varied from 15 nM to 150 nM.35
The calculated surface electrostatic potential of Pus1 shows a patch of positive charge lining the active site cleft and spanning the length of the protein towards the N-terminus of the catalytic domain (Figure 5). The cleft is wide enough to accommodate an RNA stem. Helix 9, 3101 and the forefinger and thumb loop form the rim of this positive surface. Arg135 and Lys136 of 3101 are solvent exposed and possibly involved in RNA binding. Lys83, Arg84, Arg102, Lys159 and Arg195 contribute to the basic surface at the N-terminal domain and Arg141, Arg144 and Lys193 and Arg199 line the active site cleft leading to Asp146. The C-terminal sheet residues His292, Arg295, Lys296 and Lys327 further contribute to the basic surface. The C-terminal region of α8 together with the N-terminal residues of α9 are composed of acidic residues. In particular residues Asp383, Glu384 and Glu389 constitute a negatively charged surface patch that demarcates the electropositive surface. In contrast to Pus1, TruA does not have a continuous positive surface that extends along the length of the enzyme but is rather restricted to the active site cleft.
Pus1 helices α8, α9 and α10 alter the topology and electrostatics of the Ψ synthase core surface in such a way as to prevent tRNA from binding to Pus1 in the same orientation as in TruA. In the TruA-tRNA product complexes reported the tRNA is bound to a composite binding site contributed by both protomers of the obligate homodimer.28 This is shown in a surface rendition of the TruA dimer with the bound tRNA plotted in cartoon format (Figure 5B). In order to distinguish the two protomers in Figure 5, only the primary one is colored by electrostatic potential. The anticodon stem loop binds along the RNA-binding groove of the primary protomer with the anticodon stem loop adjacent to the electropositive active site, while the elbow between the D- and T-loops packs against two helices (corresponding to helices α 1 and α3 in Pus1) in the second protomer.
The TruA dimer interface is formed primarily by a long loop between β5 and α3 (β5 and α4 in Pus1). The β5-α3 loops from the two protomers are juxtaposed at the interface, making extensive contacts with each other and with the N-terminal β -sheet of the opposite protomer. In Pus1, α8 occupies the space where both loops would need to bind in order to form a dimer analogous to the TruA dimer. The other unique Pus1 helix, α9, sits in the space occupied by the tRNA D-stem in the TruA-tRNA complexes.28 Thus the Pus1 α8/α9 insert prevents tRNA from binding in the same manner as in TruA in two ways. It prevents dimer formation, so the composite binding site cannot form, and it blocks binding to the tRNA D-stem. We propose that these restrictions on tRNA binding mode select against modification at sites 38, 39, and 40 in the anticodon loop. The α8, α9 and α10 helices may similarly sterically and electrostatically restrict the binding modes of other large RNAs, thus contributing to substrate selectivity.
tRNA docking to Pus1
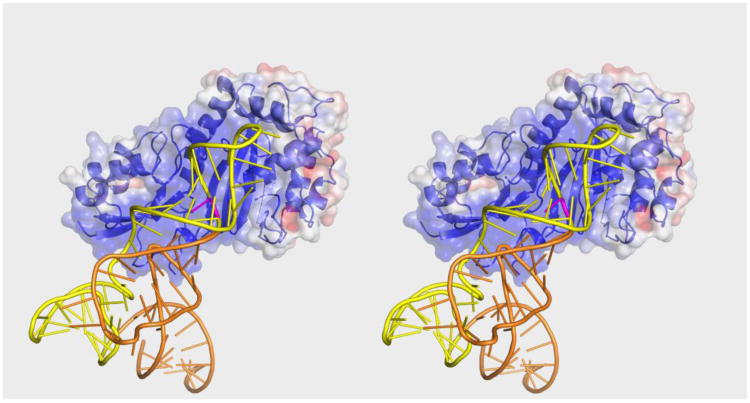
We next attempted to predict a binding mode that would allow Pus1 to pseudouridylate tRNA at known target sites in the anticodon stem (sites 27, 28, 30). Substrate-bound structures of four Ψ synthases, TruB, RluA, RluF, and RluB, reveal that the target uridine and the two nucleotides 5′ to the target bind in a conserved manner to the RNA-binding cleft.29; 36; 22 In the RluA, RluF and RluB complexes the binding mode for the nucleotide 3′ to the target is also conserved. We thus manually docked tRNAPhe as a rigid body into the active site of Pus 1 (form I) with the sole constraint that the nucleotides 3′ and 5′ to the target uridine, U27, bind in this conserved orientation in the cleft (Figure 6). Energy minimization of this complex relieved the few collisions between tRNA and protein without major conformational changes to either protein or tRNA. The minimal adjustment required suggests that Pus1 can bind tRNA in its canonical cloverleaf conformation. The archaeosine tRNA-guanine transglycosylase, in contrast, must bind tRNA in an alternative (‘lambda’) conformation in order to access its target base.37 The tRNA binds to the large positive surface, with the anticodon stem loop and TΨC loop making most of the contacts with the protein. This binding mode is consistent with the finding that an RNA construct comprised only of the anticodon stem loop linked to the TΨC loop of tRNASer is a good hPus1 substrate.
Figure 6.
Stereo Plot of tRNAPhe docked to apo-Pus1 (form I). The surface representation of Pus1 is colored according to its electrostatic potential (-5 kT/e to 5 kT/e). tRNAPhe is colored orange with nucleotides corresponding to the Pus1 minimal substrate in yellow. The substrate base is colored purple. The orientation of Pus1 is the same as the orientation of TruA in Figure 5.
The anticodon loop is at the top of the binding groove abutting C-terminal α9 (the upper rim of the positive surface) at the edge of the positive patch. Given the flexibility of the anticodon loop it is conceivable it would refold on binding to Pus1 to more fully occupy the positive patch. Three flexible loops on the protein, which are disordered in the form I crystal structures, surround the docked tRNA and may refold upon complex formation. These include the forefinger and thumb loops, both poised to bind to the major groove of the anticodon stem loop and the residues connecting β10 and α8, which could interact with the acceptor stem. If docking is performed with the form II structure severe clashes between RNA and protein exist, indicating that form I resembles a conformation more similar to the substrate bound form than the apo-form. Our docking results suggest that Pus1 binds tRNA in a fundamentally different manner than TruA, which has a composite tRNA binding site comprised of residues from both protomers.28 The docked tRNA binds to a monomer of Pus1 such that the RNA axis is fully ∼180° rotated versus tRNA in TruA.
Almost all the other tRNA and pre-tRNAIle Pus1 sites are at the ends of the anticodon stem, the only exception being position 1 in the acceptor stem. The Pus1 sites in the anticodon stem are all on the same side of the stem, thus we predict that for modification of all of these, Pus1 would bind tRNA in the docked orientation shown in Figure 6, that is, with the RNA axis in the opposite direction to tRNA in TruA complexes. Pus1 site 30 of tRNA, for example, can be aligned with the enzyme active site with few steric clashes when tRNA is docked in this orientation. This orientation would position the 62-nt intron of pre-tRNAIle at the predicted binding site for the anticodon loop of tRNAPhe seen in Fig. 6. How Pus1accommodates this large RNA domain is difficult to predict in the absence of the pre-tRNA tertiary structure. The fact that Pus1 modifies the anticodon stem at several positions, and also modifies substrates unrelated to tRNA, suggests that there are few specific Pus1-RNA interactions with the targeted stem loop (outside of the active site) and substrate recognition includes shape and electrostatic complementarity between Pus1 and more remote regions of the RNA. This would be consistent with the finding that like TruA, hPus1 is not active against isolated stem loops.
Conclusion
We determined three crystal structures of the catalytic domain of the human Ψ synthase Pus1. Pus1 forms complexes both with tRNA and the minimal tRNA substrate. The catalytic domain of Pus1 binds the minimal tRNA substrate however no co-crystals were produced. Unlike its E. coli homolog, TruA, which exclusively modifies anticodon stemloops of many tRNAs, Pus1 has greater promiscuity and pseudouridylates several types of ncRNAs. Pus1 and TruA share the same protein fold and contain conserved sequence motifs at the active site cleft, indicating a common catalytic mechanism; however, Pus1 has a unique C-terminal insert comprised of two long α-helices that does not allow most orientations of tRNA, including that seen in TruA. This insert demarcates a large electropositive surface in Pus1 to which a molecule of substrate tRNA can be docked with the target uridine oriented for catalysis, but rotated ∼180° with respect to the orientation in TruA. The large positive surface in Pus1 contrasts with the much smaller positive surface surrounding the active site of TruA, reflecting a less tightly constrained target selectivity.
Materials and Methods
Protein expression and purification
hPus1 (79-408) was subcloned into a modified pET47 vector containing an N-terminal hexahistidine tag and a 3C protease cleavage site. For protein expression cells were grown at 30°C in LB-medium with 50 μg/ml kanamycin. Protein expression was induced at 20°C after addition of 0.1 mM isopropyl-β-D-1-thio-galactopyranoside at OD600 0.6 and cells were further grown for 16 hours.
For purification cells were lysed in 50 mM Hepes pH 7.0, 500 mM NaCl, 5 mM β-mercaptoethanol using an EmulsiFlex-C5 homogenizer (Avestin) and the lysate was cleared by centrifugation for 30 min at 32.000g. The supernatant was incubated with nickel-nitrilotriacetic acid (Ni-NTA) (GE Healthcare) resin, the resin was washed with lysis buffer containing 20 mM imidazole and proteins were eluted using lysis buffer containing 250 mM imidazole as final buffer. Protein containing fractions were combined and dialyzed over night at 4°C against 50 mM Hepes pH 7.0, 100 mM NaCl, 0.5 mM TCEP in the presence of 3 C protease for cleavage of the His-tag. The protein solution was then concentrated and loaded on a Superdex S200(10/300) (GE Healthcare) size exclusion column equilibrated in 50 mM Hepes pH 7.0, 100 mM NaCl, 0.5 mM TCEP. Peak fractions were analyzed by SDS-PAGE, pooled, concentrated, flash frozen in liquid nitrogen, and stored at -80°C. Numbering of Pus1 residues is based on UniProt accession number Q9Y606-1. Our full-length Pus1 encodes residues 29-427 and lacks the predicted N-terminal mitochondrial-targeting motif.
The minimal hPus1 tRNA substrate17, encompassing the wild type anticodon stem-loop and the TΨC stem-loop of human cytoplasmic tRNASer was purchased from Dharmacon (ThermoScientific) and deprotected according to the manufacturer's protocol. For refolding, RNA was diluted to 30 μM in water, heated in a metal block to 96°C for 2 minutes followed by slow hybridization to room temperature. RNA-protein complexes were formed by incubating RNA with Pus1 (79-408) at equimolar concentrations for 15 min at room temperature. The RNA-protein solution was applied to a Superdex S200 (10/300) column equilibrated with 50 mM Hepes pH 7.0, 100 mM NaCl, 0.5 mM TCEP. The elution profile showed two overlapping peaks, both containing protein and RNA, that were pooled independently and concentrated to 2 mg/ml. Crystals were obtained using protein from both peaks. We assume that the peak at lower elution volume presents a complex consisting of two Pus1 molecules and possibly one minimal tRNA, whereas the peak eluting at higher volume presents a one to one complex of Pus1 and RNA.
Crystallization and Data Collection
Crystallization experiments were carried out at 16°C using hanging-drop vapor diffusion by mixing 1 μl protein with 1 μl reservoir solution (500 μl reservoir solution). The best diffracting hexagonal crystals grew within 2-3 weeks in 18% PEG8000, 0.1 M MES pH 6.1, 200 mM AmSO4 using the protein/RNA complex corresponding to the higher elution volume size exclusion peak. Diffraction data were collected on beamline 23ID-D of the Advanced Photon Source (Argonne National Laboratory, USA). Soaking was performed by exchanging the drop solution of a crystal grown in 18% PEG8000, 0.1 M MES pH 6.5, 0.3 M AmSO4 with a soaking solution containing 18% PEG8000, 0.1 M Bis-Tris pH 6.5, 0.1M AmSO4 and 89.9 mM uridine three times before incubating the crystal with soaking solution for 45 minutes. Crystals were cryoprotected in reservoir solution containing 20% ethyleneglycol. Rod shaped crystals originally appeared in 96-well sitting drop format in 30% PEG4000, 0.1 M Tris-HCl pH 8.5, 0.22 M MgCl2 and 1 mM spermine. Crystals were optimized in 15 well plates but did not give usable diffraction. Seeding improved crystal morphology and produced well diffracting crystals in 28% PEG4000, 0.1 M Tris-HCl pH 8.5, 0.2 M MgCl2 and 1 mM spermine. Diffraction data for crystals derived from soaking and seeding experiments were collected on beamline 8.3.1 of the Advanced Light Source (Berkeley, USA). Crystals required the minimal-RNA substrate to be present but excluded it from the protein, resulting in apo-protein crystals.
Structure Determination
Diffraction data was processed with XDS.38 Rmeas represents the redundancy independent R-factor as given in Diederichs and Karplus.39 The structure of MES-bound Pus1 (form I) was solved by molecular replacement with AutoMR40 from the PHENIX41 package using the structure of apo-TruA (PDB ID: 1DJ0) as a search model. Form I apo and form II structures were solved by molecular replacement with Phaser40, using the MES-bound Pus1 structure as search model. Refinement was performed in PHENIX, COOT42 was used for model building and visualization. Hydrogens in riding positions were added in the last cycles of refinement of the form I apo structure. TLS refinement using two TLS groups43 was carried out in the final cycles of refinement of the MES-bound Pus1 structure. Data collection and refinement statistics are shown in Table 1. Molecular presentations were prepared with PyMol.44
Table 1. Data collection and refinement statistics.
| Form I (MES) | Form I (apo) | Form II | |
|---|---|---|---|
| Data collection | |||
| X-ray source | APS 23ID-D | ALS 8.3.1 | ALS 8.3.1 |
| X-ray wavelength (Å) | 0.9793 | 1.115869 | 1.115869 |
| Space group | P6522 | P6522 | C2 |
| Cell dimensions | |||
| a, b, c (Å) | 51.9, 51.9, 446.2 | 51.9, 51.9, 445.5 | 118.6, 42.3, 71.8 |
| β(°) | 117.1 | ||
| Resolution (Å) a | 50-1.85 (1.90-1.85) | 50-1.75 (1.80-1.75) | 20-1.80 (1.85-1.80) |
| No. of observed/unique reflections | 366287/32146 | 427599/37347 | 90367/29280 |
| Rmeas (%) b | 7.0 (76) | 11.1 (152) | 8.3 (98) |
| <I/σI> | 24.7 (3.4) | 19.2 (1.33) | 15.6 (1.75) |
| Completeness (%) | 99.5 (97.8) | 98.4 (89.8) | 98.3 (99.1) |
| <Redundancy> | 11.5 (11.5) | 11.5 (4.8) | 3.1 (3.0) |
| Refinement | |||
| Resolution | 45-1.85 | 45-1.75 | 20-1.80 |
| Rwork/Rfree (%) | 18.2/22.6 | 19.9/23.9 | 18.6/22.7 |
| No. of non hydrogen | 2557 | 2602 | 2669 |
| atoms in AU | |||
| No. of water molecules | 154 | 184 | 149 |
| <B-factor (Å2)> | 37 | 25 | 19 |
| r.m.s. deviations | |||
| Bond length (Å) | 0.014 | 0.012 | 0.007 |
| Bond angles (°) | 1.539 | 1.231 | 1.050 |
| Ramachandran Plot (%) | |||
| Favored regions | 98.6 | 98 | 98.1 |
| Allowed regions | 1.4 | 1.7 | 1.9 |
|
| |||
| PDB ID | 4ITS | 4J37 | 4IQM |
Structure analysis
For the purpose of comparing homologous structures, difference distance matrices were calculated using Rapido26 and used to identify common structural domains that could be used for alignment. Superposition of structures by alignment of their common cores was done with LSQAB from the CCP445 suite of programs. In order to construct a model of tRNAPhe bound to Pus1, the RluB-stem-loop structure (unpublished) was first superimposed on the Pus1 form I crystal structure. Then two nucleotides on either side of the Pus1 target in tRNAPhe (U27) were aligned using LSQAB with two nucleotides on either side of the RluB target (U2605). The geometry of the resulting hPus1-tRNAPhe complex was regularized with Phenix PDB tools using Ramachandran restraints.
RNA synthesis and tritium release assays
The minimal [5-3H]-tRNASer substrate (nt. 26-65) was in vitro transcribed using the MEGAshortscript Kit (Ambion) and an oligodeoxynucleotide template in the presence of 0.3 mM cold UTP, 0.1 mM [5-3H]-UTP (23.9 Ci/mmol, Moravek Biochemicals) and 3.75 mM ATP, GTP and CTP. For template generation the T7 primer (5′ TAATACGACTCACTATAG) was annealed with the following oligodeoxynucleotide (5′ GCAGGATTCGAACCTGCGCCAATGG ATTTCAAGTCCATCTATAGTGAGTCGTATTA).
After 2 hours of incubation the in vitro transcription reaction was treated with DNAse I and nucleotides and protein were subsequently removed by phenol/chloroform extraction. RNA was EtOH precipitated and purified by DEAE Sepharose chromatography (GE Healthcare) using a NaCl gradient to elute the RNA. RNA containing fractions were collected at 0.4 M and 0.6 M NaCl, EtOH precipitated and resuspended in water. Before performing the tritium release assays the RNA was refolded by heating to 78°C for 2 min and slow equilibration to room temperature.
Activity assays were carried out at room temperature in 25 mM Tris-HCl (pH 7.5), 50 mM ammonium chloride, 5 mM DTT and 1 mM MgCl2 in a reaction containing 0.1 μM Pus1 and 0.3 μM minimal tRNA substrate. After 2 hours the reaction was quenched with 5% (w/v) Norit A in 0.1 N HCl, the sample was centrifuged (5 min, 5000g) and the supernatant was again treated with Norit A, followed by centrifugation. The supernatant was filtered through Ultrafree-MC centrifugal filters (Millipore) to remove residual Norit A. The filtrate was mixed with Aquasol-2 (Perkin Elmer) and released 3H was counted.
Fluorescence Anisotropy Measurements
The affinity of hPus1D146N full-length and hPus1D146N (79-408) to a 5′-fluorescein amidite (FAM)-labeled minimal tRNASer (nt. 26-65) substrate was determined by fluorescence anisotropy measurements using a SpectraMax M5 plate reader (Molecular Devices). Measurements were carried out at room temperature in a buffer containing 50 mM Hepes (pH 7.5), 100 mM NaCl, 1 mM DTT, 1 mM MgCl2 and 0.2 mg/ml bovine serum albumin. Pus1 proteins were added at increasing concentrations to 20 nM FAM-labeled RNA, and changes in fluorescent polarization were determined at an excitation wavelength of 495 nm and emission wavelength of 525 nM. Data were fitted to a quadratic binding equation.
Supplementary Material
Figure S1. Sequence alignment of Pus1 proteins, human Pus3 and E. coli TruA. Sequence alignment of human Pus1 (UniProt accession number Q9Y606), mouse Pus1 (accession number Q9WU56), Saccharomyces cerevisiae Pus1 (UniProt accession number Q12211), Candida glabrata Pus1 (UniProt accession number Q6FV05), human Pus3 (UniProt accession number Q9BZE2) and E. coli TruA (UniProt accession number P07649). The alignment was constructed with T-Coffee and manually edited so that inserts in the hPus1 and TruA sequences corresponded to the inserts identified in their three-dimensional structures. The secondary structure organization of human Pus1 and TruA is depicted next to the respective sequences. Residues are colored according to their degree of conservation with red representing residues that are conserved in all species. The five signature Ψ synthase residues are highlighted in bold. The program ALINE was used for visualization of the alignment.
Figure S2. MES and SO42- mimic the substrate base and phosphate of tRNA in the structure of TruA-tRNA. The structures of Pus1 (form I MES) and TruA-tRNA (PDB ID: 2NR0) were superposed. The base from one of the TruA target sites (grey sticks) is flipped into the active site of TruA. MES and active site residues of Pus1 are represented as sticks with green carbon atoms.
Figure S3. Binding to MES induces opening of the active site cleft of Pus1. The isomorphous form I structures are shown as grey (form I) and blue (form I apo) ribbons. MES and SO42- are depicted as grey sticks. The (|Fobs|-|Fobs_apo|)αcalc difference map, contoured at 4 sigma illustrates the opening of the active site cleft. The distance between Cαs positions of Met291 in both structures is 0.95 Å.
Figures S4. The structures of Pus1 and Pus10 share the central ß-sheet. Superposition of the structures of Pus1 (form II), shown as green cartoon, and the catalytic domain of Pus10 (PDB: 2V9K, magenta). The five signature Ψ synthase residues, depicted in stick representation, align closely. The orientations shown correspond to the perspectives shown in Figure 1(A).
Figures S5. Binding of hPus1D146N full-length and hPus1D146N (79-408) to the Pus1 minimal-RNA substrate. The change in polarization of 5′FAM-labeled tRNA minimal-substrate upon addition of increasing concentrations of Pus1 proteins is shown. Kd values (Kd, Pus1 (79-408) < 14 nM and Kd (Pus1 full-length) = 53 nM) were obtained by fitting the fluorescence polarization change to a quadratic binding equation.
Highlights.
Pus1 is a pseudouridine synthase with a diverse range of RNA substrates (tRNA, pre-tRNA, U2 snRNA, steroid receptor RNA activator)
Three crystal structures of the catalytic core of human Pus1 from two different crystal forms are reported
The structure of Pus1 is the first structure of a eukaryotic pseudouridine synthase from the five families conserved in all kingdoms of life
Two helices C-terminal to the catalytic core of the enzyme are unique to Pus1 and demarcate a large electropositive surface that might be key to the Pus1′s broadened specificity
tRNA can be docked to Pus1 and employs a different binding mode as seen for the bacterial homologue TruA
Acknowledgments
We thank the staff of beamline 8.3.1 at the Advanced Light Source (Berkeley, USA) and the staff of GM/CA CAT at the Advanced Photon Source (Argonne National Laboratory) for excellent facilities and assistance with data collection. John Pak is thanked for collecting the form I apo dataset. Data were collected during the 2012 CCP4/APS school, and we thank the presenters for advice on structure solution. Rebecca Robbins is thanked for expert technical assistance with the tetra detector analysis. Pat Greene is gratefully acknowledged for critical reading of the manuscript. N.C was supported by a fellowship from the Deutsche Forschungsgemeinschaft. Research was supported by GM51232 to RMS.
Footnotes
Numbering of Pus1 residues is based on UniProt accession number Q9Y606-1.
Accession Numbers: Coordinates and structure factors have been deposited in the Protein Data Bank with accession number 4IQM, 4ITS and 4J37.
Supplementary Data Supplemental data include five figures.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Charette M, Gray MW. Pseudouridine in RNA: what, where, how, and why. IUBMB Life. 2000;49:341–51. doi: 10.1080/152165400410182. [DOI] [PubMed] [Google Scholar]
- 2.Agris PF. Decoding the genome: a modified view. Nucleic Acids Res. 2004;32:223–38. doi: 10.1093/nar/gkh185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang C, McPheeters DS, Yu YT. Psi35 in the branch site recognition region of U2 small nuclear RNA is important for pre-mRNA splicing in Saccharomyces cerevisiae. J Biol Chem. 2005;280:6655–62. doi: 10.1074/jbc.M413288200. [DOI] [PubMed] [Google Scholar]
- 4.Yu YT, Shu MD, Steitz JA. Modifications of U2 snRNA are required for snRNP assembly and pre-mRNA splicing. EMBO J. 1998;17:5783–95. doi: 10.1093/emboj/17.19.5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamma T, Ferré-D'Amaré AR. Pseudouridine synthases. Chem Biol. 2006;13:1125–35. doi: 10.1016/j.chembiol.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 6.Del Campo M, Ofengand J, Malhotra A. Crystal structure of the catalytic domain of RluD, the only rRNA pseudouridine synthase required for normal growth of Escherichia coli. RNA. 2004;10:231–9. doi: 10.1261/rna.5187404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hur S, Stroud R, Finer-Moore J. Substrate recognition by RNA 5-methyluridine methyltransferases and pseudouridine synthases: a structural perspective. J Biol Chem. 2006;281:38969–73. doi: 10.1074/jbc.R600034200. [DOI] [PubMed] [Google Scholar]
- 8.Hamma T, Ferré-D'Amaré A. The box H/ACA ribonucleoprotein complex: interplay of RNA and protein structures in post-transcriptional RNA modification. J Biol Chem. 2010;285:805–9. doi: 10.1074/jbc.R109.076893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sibert BS, Fischel-Ghodsian N, Patton JR. Partial activity is seen with many substitutions of highly conserved active site residues in human Pseudouridine synthase 1. RNA. 2008;14:1895–906. doi: 10.1261/rna.984508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao X, Patton JR, Davis SL, Florence B, Ames SJ, Spanjaard RA. Regulation of nuclear receptor activity by a pseudouridine synthase through posttranscriptional modification of steroid receptor RNA activator. Mol Cell. 2004;15:549–58. doi: 10.1016/j.molcel.2004.06.044. [DOI] [PubMed] [Google Scholar]
- 11.Leygue E. Steroid receptor RNA activator (SRA1): unusual bifaceted gene products with suspected relevance to breast cancer. Nucl Recept Signal. 2007;5:e006. doi: 10.1621/nrs.05006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patton JR, Bykhovskaya Y, Mengesha E, Bertolotto C, Fischel-Ghodsian N. Mitochondrial myopathy and sideroblastic anemia (MLASA): missense mutation in the pseudouridine synthase 1 (PUS1) gene is associated with the loss of tRNA pseudouridylation. J Biol Chem. 2005;280:19823–8. doi: 10.1074/jbc.M500216200. [DOI] [PubMed] [Google Scholar]
- 13.Bykhovskaya Y, Casas K, Mengesha E, Inbal A, Fischel-Ghodsian N. Missense mutation in pseudouridine synthase 1 (PUS1) causes mitochondrial myopathy and sideroblastic anemia (MLASA) Am J Hum Genet. 2004;74:1303–8. doi: 10.1086/421530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen J, Patton JR. Cloning and characterization of a mammalian pseudouridine synthase. RNA. 1999;5:409–19. doi: 10.1017/s1355838299981591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Behm-Ansmant I, Massenet S, Immel F, Patton JR, Motorin Y, Branlant C. A previously unidentified activity of yeast and mouse RNA:pseudouridine synthases 1 (Pus1p) on tRNAs. RNA. 2006;12:1583–93. doi: 10.1261/rna.100806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Massenet S, Motorin Y, Lafontaine DL, Hurt EC, Grosjean H, Branlant C. Pseudouridine mapping in the Saccharomyces cerevisiae spliceosomal U small nuclear RNAs (snRNAs) reveals that pseudouridine synthase pus1p exhibits a dual substrate specificity for U2 snRNA and tRNA. Mol Cell Biol. 1999;19:2142–54. doi: 10.1128/mcb.19.3.2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sibert BS, Patton JR. Pseudouridine synthase 1: a site-specific synthase without strict sequence recognition requirements. Nucleic Acids Res. 2012;40:2107–18. doi: 10.1093/nar/gkr1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guelorget A, Golinelli-Pimpaneau B. Mechanism-Based Strategies for Trapping and Crystallizing Complexes of RNA-Modifying Enzymes. Structure. 2011;19:282–91. doi: 10.1016/j.str.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 19.Yang ZR, Thomson R, McNeil P, Esnouf RM. RONN: the bio-basis function neural network technique applied to the detection of natively disordered regions in proteins. Bioinformatics. 2005;21:3369–76. doi: 10.1093/bioinformatics/bti534. [DOI] [PubMed] [Google Scholar]
- 20.Huang L, Pookanjanatavip M, Gu X, Santi DV. A conserved aspartate of tRNA pseudouridine synthase is essential for activity and a probable nucleophilic catalyst. Biochemistry. 1998;37:344–51. doi: 10.1021/bi971874+. [DOI] [PubMed] [Google Scholar]
- 21.Cooley RB, Arp DJ, Karplus PA. Evolutionary origin of a secondary structure: π-helices as cryptic but widespread insertional variations of α-helices that enhance protein functionality. J Mol Biol. 2010;404:232–46. doi: 10.1016/j.jmb.2010.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alian A, DeGiovanni A, Griner SL, Finer-Moore JS, Stroud RM. Crystal structure of an RluF-RNA complex: a base-pair rearrangement is the key to selectivity of RluF for U2604 of the ribosome. J Mol Biol. 2009;388:785–800. doi: 10.1016/j.jmb.2009.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pan H, Agarwalla S, Moustakas DT, Finer-Moore J, Stroud RM. Structure of tRNA pseudouridine synthase TruB and its RNA complex: RNA recognition through a combination of rigid docking and induced fit. Proc Natl Acad Sci U S A. 2003;100:12648–53. doi: 10.1073/pnas.2135585100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pan H, Ho JD, Stroud RM, Finer-Moore J. The crystal structure of E. coli rRNA pseudouridine synthase RluE. J Mol Biol. 2007;367:1459–70. doi: 10.1016/j.jmb.2007.01.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCleverty CJ, Hornsby M, Spraggon G, Kreusch A. Crystal structure of human Pus10, a novel pseudouridine synthase. J Mol Biol. 2007;373:1243–54. doi: 10.1016/j.jmb.2007.08.053. [DOI] [PubMed] [Google Scholar]
- 26.Mosca R, Schneider TR. RAPIDO: a web server for the alignment of protein structures in the presence of conformational changes. Nucleic Acids Res. 2008;36:W42–6. doi: 10.1093/nar/gkn197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nurse K, Wrzesinski J, Bakin A, Lane BG, Ofengand J. Purification, cloning, and properties of the tRNA psi 55 synthase from Escherichia coli. RNA. 1995;1:102–12. [PMC free article] [PubMed] [Google Scholar]
- 28.Hur S, Stroud R. How U38, 39, and 40 of many tRNAs become the targets for pseudouridylation by TruA. Mol Cell. 2007;26:189–203. doi: 10.1016/j.molcel.2007.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoang C, Ferré-D'Amaré AR. Cocrystal structure of a tRNA Psi55 pseudouridine synthase: nucleotide flipping by an RNA-modifying enzyme. Cell. 2001;107:929–39. doi: 10.1016/s0092-8674(01)00618-3. [DOI] [PubMed] [Google Scholar]
- 30.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–97. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 31.Foster PG, Huang L, Santi DV, Stroud RM. The structural basis for tRNA recognition and pseudouridine formation by pseudouridine synthase I. Nat Struct Biol. 2000;7:23–7. doi: 10.1038/71219. [DOI] [PubMed] [Google Scholar]
- 32.Arluison V, Batelier G, Riès-Kautt M, Grosjean H. RNA:pseudouridine synthetase Pus1 from Saccharomyces cerevisiae: oligomerization property and stoichiometry of the complex with yeast tRNA(Phe) Biochimie. 1999;81:751–6. doi: 10.1016/s0300-9084(99)80133-3. [DOI] [PubMed] [Google Scholar]
- 33.Roovers M, Hale C, Tricot C, Terns MP, Terns RM, Grosjean H, Droogmans L. Formation of the conserved pseudouridine at position 55 in archaeal tRNA. Nucleic Acids Res. 2006;34:4293–301. doi: 10.1093/nar/gkl530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arluison V, Hountondji C, Robert B, Grosjean H. Transfer RNA-pseudouridine synthetase Pus1 of Saccharomyces cerevisiae contains one atom of zinc essential for its native conformation and tRNA recognition. Biochemistry. 1998;37:7268–76. doi: 10.1021/bi972671o. [DOI] [PubMed] [Google Scholar]
- 35.Arluison V, Buckle M, Grosjean H. Pseudouridine synthetase Pus1 of Saccharomyces cerevisiae: kinetic characterisation, tRNA structural requirement and real-time analysis of its complex with tRNA. J Mol Biol. 1999;289:491–502. doi: 10.1006/jmbi.1999.2789. [DOI] [PubMed] [Google Scholar]
- 36.Hoang C, Chen J, Vizthum CA, Kandel JM, Hamilton CS, Mueller EG, Ferré-D'Amaré AR. Crystal structure of pseudouridine synthase RluA: indirect sequence readout through protein-induced RNA structure. Mol Cell. 2006;24:535–45. doi: 10.1016/j.molcel.2006.09.017. [DOI] [PubMed] [Google Scholar]
- 37.Ishitani R, Nureki O, Nameki N, Okada N, Nishimura S, Yokoyama S. Alternative tertiary structure of tRNA for recognition by a posttranscriptional modification enzyme. Cell. 2003;113:383–94. doi: 10.1016/s0092-8674(03)00280-0. [DOI] [PubMed] [Google Scholar]
- 38.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–32. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Diederichs K, Karplus PA. Improved R-factors for diffraction data analysis in macromolecular crystallography. Nat Struct Biol. 1997;4:269–75. doi: 10.1038/nsb0497-269. [DOI] [PubMed] [Google Scholar]
- 40.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–21. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 43.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D Biol Crystallogr. 2006;62:439–50. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 44.Schrodinger LLC. The PyMOL Molecular Graphics System. 2010 Version 1.5.0.4. [Google Scholar]
- 45.Collaborative Computational Project, N. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–3. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 46.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–41. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Sequence alignment of Pus1 proteins, human Pus3 and E. coli TruA. Sequence alignment of human Pus1 (UniProt accession number Q9Y606), mouse Pus1 (accession number Q9WU56), Saccharomyces cerevisiae Pus1 (UniProt accession number Q12211), Candida glabrata Pus1 (UniProt accession number Q6FV05), human Pus3 (UniProt accession number Q9BZE2) and E. coli TruA (UniProt accession number P07649). The alignment was constructed with T-Coffee and manually edited so that inserts in the hPus1 and TruA sequences corresponded to the inserts identified in their three-dimensional structures. The secondary structure organization of human Pus1 and TruA is depicted next to the respective sequences. Residues are colored according to their degree of conservation with red representing residues that are conserved in all species. The five signature Ψ synthase residues are highlighted in bold. The program ALINE was used for visualization of the alignment.
Figure S2. MES and SO42- mimic the substrate base and phosphate of tRNA in the structure of TruA-tRNA. The structures of Pus1 (form I MES) and TruA-tRNA (PDB ID: 2NR0) were superposed. The base from one of the TruA target sites (grey sticks) is flipped into the active site of TruA. MES and active site residues of Pus1 are represented as sticks with green carbon atoms.
Figure S3. Binding to MES induces opening of the active site cleft of Pus1. The isomorphous form I structures are shown as grey (form I) and blue (form I apo) ribbons. MES and SO42- are depicted as grey sticks. The (|Fobs|-|Fobs_apo|)αcalc difference map, contoured at 4 sigma illustrates the opening of the active site cleft. The distance between Cαs positions of Met291 in both structures is 0.95 Å.
Figures S4. The structures of Pus1 and Pus10 share the central ß-sheet. Superposition of the structures of Pus1 (form II), shown as green cartoon, and the catalytic domain of Pus10 (PDB: 2V9K, magenta). The five signature Ψ synthase residues, depicted in stick representation, align closely. The orientations shown correspond to the perspectives shown in Figure 1(A).
Figures S5. Binding of hPus1D146N full-length and hPus1D146N (79-408) to the Pus1 minimal-RNA substrate. The change in polarization of 5′FAM-labeled tRNA minimal-substrate upon addition of increasing concentrations of Pus1 proteins is shown. Kd values (Kd, Pus1 (79-408) < 14 nM and Kd (Pus1 full-length) = 53 nM) were obtained by fitting the fluorescence polarization change to a quadratic binding equation.