Abstract
In China, brucellosis is an endemic disease typically caused by Brucella melitensis infection (biovars 1 and 3). Brucella canis infection in dogs has not traditionally recognized as a major problem. In recent years however, brucellosis resulting from Brucella canis infection has also been reported, suggesting that infections from this species may be increasing. Data concerning the epidemiology of brucellosis resulting from Brucella canis infection is limited. Therefore, the purpose of this study was to assess the diversity among Chinese Brucella canis strains for epidemiological purposes. First, we employed a 16-marker VNTR assay (Brucella MLVA-16) to assess the diversity and epidemiological relationship of 29 Brucella canis isolates from diverse locations throughout China with 38 isolates from other countries. MLVA-16 analysis separated the 67 Brucella canis isolates into 57 genotypes that grouped into five clusters with genetic similarity coefficients ranging from 67.73 to 100%. Moreover, this analysis revealed a new genotype (2-3-9-11-3-1-5-1:118), which was present in two isolates recovered from Guangxi in 1986 and 1987. Second, multiplex PCR and sequencing analysis were used to determine whether the 29 Chinese Brucella canis isolates had the characteristic BMEI1435 gene deletion. Only two isolates had this deletion. Third, amplification of the omp25 gene revealed that 26 isolates from China had a T545C mutation. Collectively, this study reveals that considerable diversity exists among Brucella canis isolates in China and provides resources for studying the genetic variation and microevolution of Brucella.
Introduction
Brucellosis, caused by various species of the Gram-negative bacterium Brucella, continues to be one of the most serious zoonotic diseases for humans and animals throughout the world [1]. In recent years, the number of published reports describing brucellosis resulting from B. canis infection has increased [2]–[6]. Although, B. canis infection has not been the major cause of brucellosis in China [7]–[10], recent outbreaks in Beijing and other provinces suggest that this infection may be on the rise [8]. In order to effectively prevent this disease, it is important to identify what strain of Brucella has caused the infection. However, most molecular subtyping tools and “classical biotyping” methods lack sufficient discriminatory power for epidemiological investigations. Thus, effective molecular subtyping tools capable of reproducibly distinguishing differences between strains must be implemented.
The Brucella genus is genetically conserved [11]–[13], making rapid and accurate identification of species in the genus difficult. For instance, distinguishing between the species B. suis and B. canis has been particularly challenging [14]–[20]. This difficulty is due to the fact that few genetic polymorphisms discriminate between these two species at the molecular level as well as B. canis being a clonal lineage that arose from B. suis [21]. Several studies have optimized PCR based tests to address this issue [22]. For example, three multiplex PCR assays, a Bruce-ladder, a 19-primer multiplex PCR, and a new Bruce-ladder multiplex PCR assay called “v2.0 multiplex PCR” have been developed to characterize Brucella isolates at the species level [21], [23], [24]. More recently, multi-locus variable-number tandem-repeat analysis (MLVA) has been confirmed as a useful tool for identifying and genotyping Brucella isolates, and the data have been used for epidemiological investigations [25]–[28].
Bruce-ladder analysis generally performs well and has been recommended by the World Organization for Animal Health (OIE) as a rapid and simple one-step molecular test for identification and typing of Brucella species [24]. The 19-primer multiplex PCR assay was used to differentiate isolates of the genus Brucella at the species and biovar levels [21]. However, routine and widespread use of these two multiplex PCR typing tools has revealed that they misidentify a substantial proportion of B. canis isolates as B. suis [21], [29], [30]. The primary reason for this misclassification is the selection of the BMEI1435 gene as a marker for B. canis. This gene was originally chosen because it was thought to be naturally deleted in isolates of B. canis and would thus yield a smaller PCR product than other Brucella species containing this gene [31]; however, the BMEI1435 gene is not deleted in all B. canis isolates. Therefore, a substantial proportion of B. canis isolates cannot be correctly identified using this approach.
In this report, we present both Brucella MLVA-16 and PCR data from Chinese B. canis isolates recovered from brucellosis outbreaks from different geographical origins. The aims of this study were to twofold. Firstly, we sought to assess the performance of MLVA-16 molecular typing assay applied to Chinese B. canis isolates and assess the diversity among B. canis strains for epidemiological purposes. Secondly, we wanted to evaluate the genetic relationship between Chinese B. canis strains and exogenous strains. We compared the results of Chinese B. canis isolates identified by deletion of the BMEI1435 gene, 19-primer multiplex PCR, Bruce-ladder PCR, and Bruce-ladder v2.0. In addition, because the omp25 gene can be used to distinguish B. canis from other Brucella species [11], [32]–[34], we employed SNP-based typing using the omp25 gene as an alternative target.
Materials and Methods
Strains
A collection of B. canis isolates from infected dogs was obtained from various locations in China (including ten outbreak isolates from Beijing in 2011) and characterized using classical biotyping [35] (Table 1). Growth and harvesting of Brucella cells and bacterial DNA extraction were performed as previously described [23], [36]. All isolates were further examined using SNP typing, Bruce-ladder v2.0, 19-primer multiplex PCR (B. canis specific primers only) and Bruce-ladder. SNP-based typing to identify Brucella isolates at the species level was carried out as described by Gopaul et al. [32]. Bruce-ladder v2.0 was performed according to the method of Lopez-Goni et al. [24]. The 19-primer multiplex PCR was performed as described by Huber et al. [21]. Bruce-ladder was performed as described by Garcia-Yoldi et al. [23].
Table 1. Brucella canis isolates and characterization using different molecular typing methods.
| Isolate1 , 2 | Biotype | SNP typing | Bruce-ladder v2.0 | 19-primer multiplex PCRa | Bruce-ladder | Deletionb | Mutationc | Year | Place |
| B. canis RM6/661 | B. canis | B. canis | B. canis | B. canis | B. canis | Yes | T | 1966 | USA |
| B. canis 11241 | B. canis | B. canis | B. canis | B. canis | B. canis | Yes | T | 2010 | Inner Mongolia |
| B. canis 2331 | B. canis | B. canis | B. canis | B. canis | B. canis | Yes | T | 1986 | Xinjiang |
| B. canis 2311 | B. canis | B. canis | B. canis | Atypical B. canis | B. suis | No | T | 1983 | Shanghai |
| B. canis 2321 | B. canis | B. canis | B. canis | Atypical B. canis | B. suis | No | C | 1986 | Jiangsu |
| B. canis 2351 | B. canis | B. canis | B. canis | Atypical B. canis | B. suis | No | C | 1986 | Guangxi |
| B. canis 2361 | B. canis | B. canis | B. canis | Atypical B. canis | B. suis | No | C | 1986 | Zhejiang |
| B. canis 2371 | B. canis | B. canis | B. canis | Atypical B. canis | B. suis | No | C | 1986 | Guangxi |
| B. canis 2391 | B. canis | B. canis | B. canis | Atypical B. canis | B. suis | No | C | 1987 | Henan |
| B. canis 2401 | B. canis | B. canis | B. canis | Atypical B. canis | B. suis | No | C | 1987 | Jiangxi |
| B. canis 2411 | B. canis | B. canis | B. canis | Atypical B. canis | B. suis | No | C | 1987 | Shandong |
| B. canis 2431 | B. canis | B. canis | B. canis | Atypical B. canis | B. suis | No | C | 1987 | Anhui |
| B. canis 2441 | B. canis | B. canis | B. canis | Atypical B. canis | B. suis | No | C | 1987 | Guangxi |
| B. canis 2451 | B. canis | B. canis | B. canis | Atypical B. canis | B. suis | No | C | 1988 | Hubei |
| B. canis 2471 | B. canis | B. canis | B. canis | Atypical B. canis | B. suis | No | C | 1988 | Jiangxi |
| B. canis 2491 | B. canis | B. canis | B. canis | Atypical B. canis | B. suis | No | C | 1988 | Liaoning |
| B. canis 2511 | B. canis | B. canis | B. canis | Atypical B. canis | B. suis | No | C | 1989 | Hebei |
| B. canis RU1 | B. canis | B. canis | B. canis | Atypical B. canis | B. suis | No | C | 2007 | Jiangsu |
| B. canis XUE11 | B. canis | B. canis | B. canis | Atypical B. canis | B. suis | No | C | 2010 | Inner Mongolia |
| B. canis LI2 | B. canis | B. canis | B. canis | Atypical B. canis | B. suis | No | C | 2010 | Liaoning |
| B. canis BJ-031 | B. canis | B. canis | B. canis | Atypical B. canis | B. suis | No | C | 2011 | Beijing |
| B. canis BJ-101 | B. canis | B. canis | B. canis | Atypical B. canis | B. suis | No | C | 2011 | Beijing |
| B. canis BJ-131 | B. canis | B. canis | B. canis | Atypical B. canis | B. suis | No | C | 2011 | Beijing |
| B. canis BJ-151 | B. canis | B. canis | B. canis | Atypical B. canis | B. suis | No | C | 2011 | Beijing |
| B. canis BJ-181 | B. canis | B. canis | B. canis | Atypical B. canis | B. suis | No | C | 2011 | Beijing |
| B. canis BJ-191 | B. canis | B. canis | B. canis | Atypical B. canis | B. suis | No | C | 2011 | Beijing |
| B. canis BJ-381 | B. canis | B. canis | B. canis | Atypical B. canis | B. suis | No | C | 2011 | Beijing |
| B. canis BJ-721 | B. canis | B. canis | B. canis | Atypical B. canis | B. suis | No | C | 2011 | Beijing |
| B. canis BJ-731 | B. canis | B. canis | B. canis | Atypical B. canis | B. suis | No | C | 2011 | Beijing |
| B. canis BJ-891 | B. canis | B. canis | B. canis | Atypical B. canis | B. suis | No | C | 2011 | Beijing |
: B.canis specific primers only;
: Characterization of B. canis BMEI1435 gene deletion (related to the genome of B. melitensis 16M, AE008917).
: The mutation of omp25 gene at position 545 (SNP position related to the genome of B. abortus 9-941, GenBank ID: AE017223).
: isolated from dog.
: isolated from human.
Characterization of B. canis BMEI1435 gene deletion
In order to characterize deletion of the BMEI1435 gene, primers BMEI 1434F (5′-GCCAGCCACAGGATCAGGTGAT-3′) and BMEI 1436R (5′- GGATCCGTTCGTTTCGCTCG-3′) [31] were used to amplify the BMEI1435 gene. The length of the product was 1674 bp (containing the BMEI1435 gene) or 607 bp (the BMEI1435 gene deletion). Products were then separated by agarose gel electrophoresis and sequenced.
Characterization of B. canis in omp25 gene
Primers were designed based on the sequence of the omp25 gene from B. melitensis 16M (omp25-F: CATGGGCGGTTTACTC; omp25-R: CGGCCAGATCATAGTTC). The omp25 gene of 29 Chinese B. canis isolates was amplified using primers omp25-F and omp25-R. The length of the product was 652 bp, and the products were amplified and sequenced by the Sanger method. These omp25 gene sequences were aligned with B. canis RM6/66 and 13 other B. canis omp25 gene sequences obtained from GenBank.
Characterization of B. canis by MLVA genotyping
MLVA was performed as described previously with the following modifications [37], [38]. The 16 primer pairs were divided into three groups: MLVA-8 (eight loci including bruce06, bruce08, bruce11, bruce12, bruce42, bruce43, bruce45 and bruce55), panel 2A (three loci including bruce18, bruce19 and bruce21), and panel 2B (five loci including bruce04, bruce07, bruce09, bruce16 and bruce30). Forward primers of the panel 2A and 2B loci were labeled with one of four 5′-fluorescent labels (6-FAM, ROX, HEX, or TAMRA). These primers were obtained from Shenggong Biosciences, Inc., (Shanghai, China). After an initial denaturation at 94°C for 3 min, the PCR conditions were as follows: 30 cycles of denaturation at 94°C for 30 seconds, annealing at 60°C for 30 seconds, and extension at 72°C for 50 seconds. Five microliters of the panel one loci amplification products were loaded in to 2% agarose gels containing ethidium bromide (0.5 µg/ml), visualized under UV light, and photographed. To determine the number of repeats from the sample products, PCR products were purified and directly sequenced using an ABI Prism Big Dye Terminator (v3.1) cycle sequencing ready reaction kit (v5.0; Applied Biosystems, Foster City, CA, USA). The PCR products of these samples were then sequenced and compared to the sequence of B. melitensis 16M. PCR products of panel 2A and 2B loci were denatured and resolved by capillary electrophoresis on an ABI Prism 3130 automated fluorescent capillary DNA sequencer (Applied Biosystems). Fragments were sized by comparison to a ROX (carboxy-X-rhodamine)-labeled molecular ladder (MapMaker 1000; BioVentures Inc., Murfreesboro, TN, USA) with GeneMapper software version 4.0 (Applied Biosystems). Appropriate VNTR designations of the fragments were assigned based on size calling through internal software binning capabilities and the corresponding repeat copy numbers of B. melitensis 16M.
Analysis of MLVA data
All data were analyzed using BioNumerics software version 5.1 (Applied Maths, Sint-Martins-Latem, Belgium). Cluster analysis was based on the categorical coefficient and the unweighted pair group method using arithmetic averages (UPGMA). The genotypes were then compared using the web-based Brucella 2012 MLVA database (http://mlva.u-psud.fr/). The genotyping data can be found in the supplementary data (Table S1).
Results
Molecular typing
Given that routine multiplex PCR typing tools often misidentify a substantial proportion of B. canis isolates as B. suis, 29 Chinese B. canis isolates were identified using classical biotyping method, and then further examined them by SNP typing, Bruce-ladder v2.0, 19-primer multiplex PCR (B. canis specific primers only) and Bruce-ladder. Consistent with phenotypic typing, SNP genotyping and Bruce-ladder v2.0 identified all 29 isolates as B. canis. However, only two of the 29 B. canis isolates (1124 and 233) were identified as B. canis by Bruce-ladder and 19-primer multiplex PCR. The other 27 isolates were identified as B. suis (Table 1). In addition, using the B. canis specific primers from the 19-primer multiplex PCR assay reported by Huber et al. (2009) [21], we amplified products of 836 and 1903 base pairs. These two products represented B. canis isolates lacking and containing the BMEI1435 gene, respectively. To assess the correlation between B. canis and BMEI1435 gene deletion, these 29 isolates were then analyzed for the presence of the BMEI1435 gene by PCR amplification. The BMEI1435 gene was only deleted in isolates 1124 and 233, the same two isolates identified as B. canis by Bruce ladder and 19-primer multiplex (Table 1). These data suggest that SNP typing and Bruce-ladder v2.0 assay are more effective methods for identifying B. canis and that not all B. canis strains have a deletion of the BMEI1435 gene.
Characterization of the omp25 gene in B. canis
In addition to the BMEI1435 marker, the omp25 gene has also been used to distinguish between species of Brucella. Therefore, the omp25 gene of the 29 Chinese B. canis isolates was amplified a 625 bp product. Twenty-six of 29 B. canis isolates had an omp25 T545C mutation, while the other three had no mutations (Table 1).
MLVA analysis of 29 Chinese B. canis isolates
We next sought to identify a more effective molecular tool for detecting the genetic variation of Brucella isolates. Thus, we employed a Brucella MLVA-16 assay to assess the diversity and epidemiological relationship between 29 B. canis isolates from diverse locations throughout China. This method provided a high discriminatory power (HGDI of 0.956) with a genetic similarity coefficient ranging from 69.46 to 100% (Table 2). The HGDI values were determined at seven loci: Bruce 04, 07, 09, 11, 16, 18 and 55 (Table 2). Bruce09 was identified as having the highest diversity overall. In addition, MLVA-16 analysis distributed the 29 Chinese B. canis isolates into three MLVA-8 genotypes (2-3-9-11-3-1-5-2, 2-3-8-11-3-1-5-2 and 2-3-9-11-3-1-5-1) and 21 MLVA-16 genotypes (Table 2). A vast majority of Chinese B. canis isolates belonged to MLVA-8 genotype 3 (2-3-9-11-3-1-5-2); however, a new genotype 118 (2-3-9-11-3-1-5-1) was detected in two isolates from Guangxi recovered in 1986 and 1987.
Table 2. Hunter-Gaston Diversity Index (HGDI) for the 67 Brucella canis isolates.
| 29 Chinese B. canis isolates | 67 B. canis strains | |||||
| Locus | No. of alleles | HGDIa | CI 95%b | No. of alleles | HGDIa | CI 95%b |
| MLVA-16 | 21 | 0.956 | 0.906–1.000 | 57 | 0.991 | 0.981–1.000 |
| MLVA-8 | 3 | 0.310 | 0.107–0.513 | 4 | 0.197 | 0.071–0.323 |
| Bruce06 | 1 | 0.000 | 0.000–0.210 | 1 | 0.000 | 0.000–0.101 |
| Bruce08 | 1 | 0.000 | 0.000–0.210 | 1 | 0.000 | 0.000–0.101 |
| Bruce11 | 2 | 0.192 | 0.016–0.368 | 3 | 0.143 | 0.031–0.255 |
| Bruce12 | 1 | 0.000 | 0.000–0.210 | 1 | 0.000 | 0.000–0.101 |
| Bruce42 | 1 | 0.000 | 0.000–0.210 | 1 | 0.000 | 0.000–0.101 |
| Bruce43 | 1 | 0.000 | 0.000–0.210 | 1 | 0.000 | 0.000–0.101 |
| Bruce45 | 1 | 0.000 | 0.000–0.210 | 1 | 0.000 | 0.000–0.101 |
| Bruce55 | 2 | 0.133 | 0.000–0.292 | 2 | 0.059 | 0.000–0.135 |
| Panel 2A | 3 | 0.616 | 0.519–0.712 | 4 | 0.532 | 0.423–0.641 |
| Bruce18 | 3 | 0.616 | 0.519–0.712 | 3 | 0.454 | 0.343–0.565 |
| Bruce19 | 1 | 0.000 | 0.000–0.210 | 2 | 0.114 | 0.014–0.214 |
| Bruce21 | 1 | 0.000 | 0.000–0.210 | 1 | 0.000 | 0.000–0.101 |
| Panel 2B | 21 | 0.956 | 0.906–1.000 | 56 | 0.991 | 0.980–1.000 |
| Bruce04 | 5 | 0.547 | 0.362–0.732 | 6 | 0.697 | 0.626–0.768 |
| Bruce07 | 7 | 0.714 | 0.587–0.841 | 8 | 0.624 | 0.528–0.720 |
| Bruce09 | 10 | 0.860 | 0.790–0.929 | 12 | 0.887 | 0.861–0.913 |
| Bruce16 | 7 | 0.825 | 0.747–0.903 | 7 | 0.842 | 0.818–0.866 |
| Bruce30 | 1 | 0.000 | 0.000–0.210 | 1 | 0.000 | 0.000–0.101 |
: Hunter and Gaston index.
: Precision of the diversity index, expressed as 95% upper and lower boundaries.
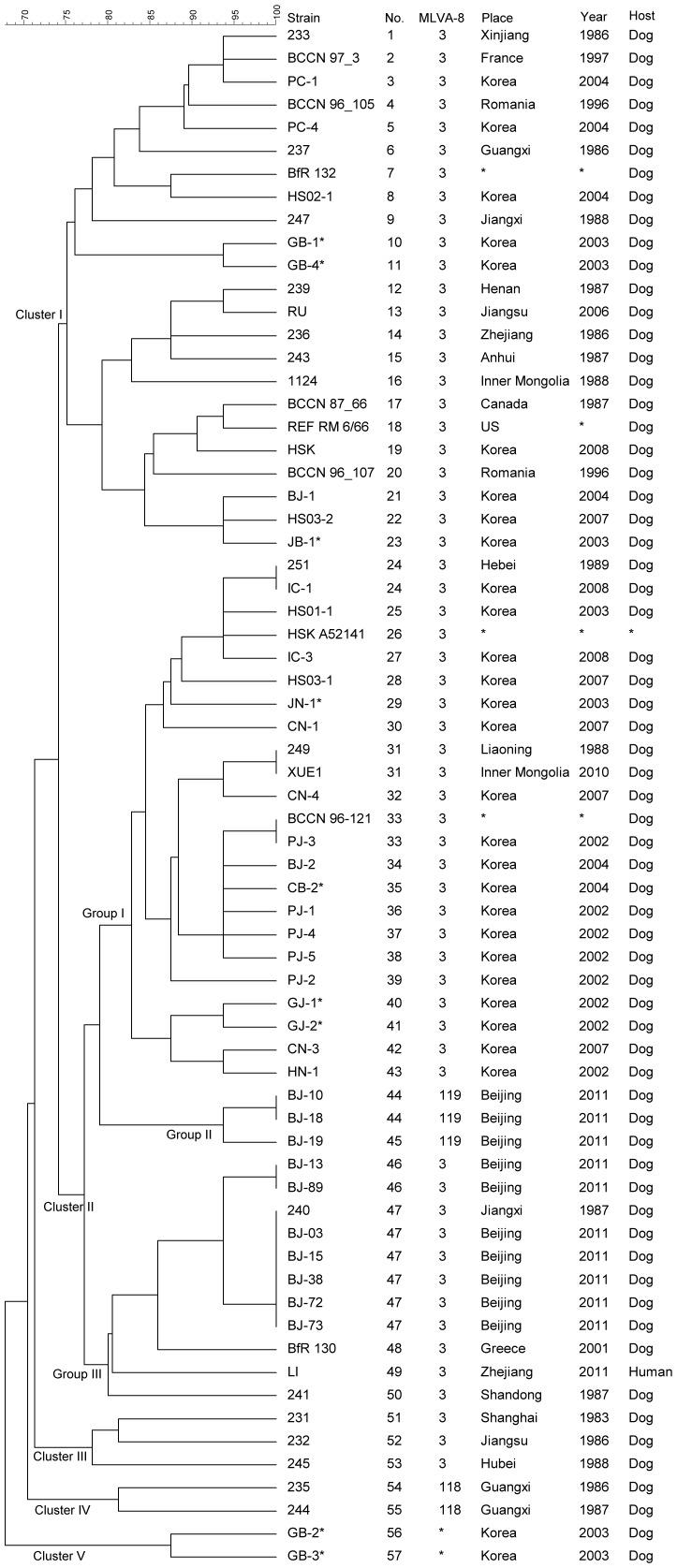
We next sought to extend this analysis to isolates from regions outside of China. MLVA-16 analysis of 67 B. canis strains (29 from China, and 38 from the Brucella2012MLVA database and a report by Kang, SI et al. in 2011) provided a high discriminatory power (HGDI of 0.991) with a genetic similarity coefficient ranging from 67.73 to 100% (Figure 1). These 67 B. canis strains were distributed into four MLVA-8 genotypes and 57 MLVA-16 genotypes (Table 2), and separated into five major clusters. Cluster I covered 23 MLVA-16 genotypes comprising eight isolates recovered from China (Xinjiang, Guangxi, Jiangxi, Henan, Jiangsu, Zhejiang, Anhui and Inner Mongolia), 15 from other countries (US, Canada, Romania, France, Greece, Korea and one unknown source) and the RM6/66 reference strain. All Cluster I isolates shared the MLVA-8 genotype 3.
Figure 1. Dendrogram derived from the MLVA-16 genotyping assay.
This dendrogram illustrates the various relationships among the 29 Chinese B. canis isolates and the 38 non-Chinese B. canis strains. In the columns, the following data for isolates are indicated: Strain: laboratory identifier of isolate in which the DNA extraction was performed, No. genotype numbering, Place/Year: country and year of isolation (when known), MLVA-8: genotype numbers associated with the genotypes corresponding to each isolates in the database.
Cluster II contained 37 B. canis isolates (16 from China, 18 from Korea, one from Greece and two from unknown sources) grouped in 27 MLVA-16 genotypes. All Cluster II isolates shared the MLVA-8 genotype 3, with only three exceptions, corresponding to the MLVA-8 genotypes 119. In addition, the 37 isolates of Cluster II were separated into three groups. Group I contained 23 isolates, with only six of the Chinese B. canis isolates included. Interestingly, three pairs of isolates with the same MLVA-16 pattern (Hebei [1989] and Korea [2008]; Inner Mongolia [2010] and Liaoning [1988]; and Korea [2002] and one unknown source) were clustered into three MLVA-16 genotypes (24, 31 and 33) in Cluster II Group I. Group II consisted of only three outbreak isolates recovered from Beijing, and separated into two MLVA-16 genotypes (44 and 45). The other five Beijing outbreak isolates (2011) and one of the Jiangxi isolates (1987) clustered to one MLVA-16 genotype (47) in Group III. In addition, two other isolates from a Beijing outbreak (2011) grouped in one MLVA-16 genotype (46) in Group III.
Cluster III contained three isolates from Shanghai, Jiangsu and Hebei. Cluster IV contained two Guangxi isolates (MLVA-8: 118). Cluster V was comprised of two Korea isolates.
Discussion
B. canis was first isolated in China from domestic and imported beagles in 1984 [7]; however, the epidemiological characteristics of B. canis infection are limited. Although B. canis infection has not been a major national concern, recent reports of B. canis infections in Beijing and other provinces highlight the significance of defining the epidemiological characteristics of this organism [8]. Currently, the MLVA-16 assay is being used to analyze epidemiological correlations between various strains. This method can also be used to track the geographic origin by comparing genetic patterns of endogenous strains with foreign isolates [2], [27], [28]. In this study, 29 Chinese B. canis isolates from different locations were analyzed by MLVA-16 assay, and compared with 38 B. canis isolates from other countries.
This study revealed that MLVA-16 genotype 31 (249 from Liaoning in 1988 and XUE1 from Inner Mongolia in 2010) was detected in different provinces with over twenty years between outbreaks. In addition, five of ten Beijing B. canis outbreak isolates (in 2011) and the Jiangxi isolate (in 1987) belonged to MLVA-16 genotype 47 [8]. These data suggest that B. canis strains might spread throughout China. Similarly, when compared with foreign strains, the MLVA-16 pattern of the domestic B. canis isolate 251 was identical with a Korean strain (IC-1). This suggests that poor importation quarantine may account for a subset of B. canis infections. Together, these data suggest that the MLVA-16 assay can be applied to long-term surveillance, and investigation of B. canis origins and epidemiological relatedness. This information will be valuable to establish strategies for a nationwide survey of B. canis infections and potential human exposure risks.
Another layer of B. canis diversity is the presence or absence of the BMEI1435 gene. Garcia-Yoldi et al. reported that 11 B. canis isolates (from USA, Mexico, Argentina, Germany, South Africa and Japan) contained the BMEI1435 gene; however this gene was not present in 13 different B. canis isolates (from USA, Germany, Peru and United Kingdom) [29]. Similarly, Huber et al. reported that nine B. canis isolates contained the BMEI1435 gene, but this gene was absent in six B. canis isolates (isolated from dogs and one unknown source) [21]. In the present study, sequence analysis of the BMEI1435 gene region was performed after amplification using the primers BMEI1434F and BMEI1436R. Compared to the reference train RM6/66, the BMEI1435 gene was absent in only two of the 29 B. canis isolates (1124 and 233) analyzed. The remaining 27 isolates contained the BMEI1435 gene, which was identical to B. canis HSK A52141 (CP003174). These data reveal that B. canis isolates with and without the BMEI1435 gene exist in China.
Huber et al. also reported that a PCR product of 887 bp (B. canis isolates lacking the BMEI1435 gene, including B. canis reference strain RM6/66) or a faint amplicon of 1863 bp (B. canis isolates containing the BMEI1435 gene) were amplified using B. canis specific primers [21]. In this study, using the B. canis specific primers reported by Huber et al. [21], PCR products of 836 bp and 1903 bp were obtained. To account for this discrepancy, we performed sequence analysis. Alignment of the 1903 bp product with B. canis HSK A52141 (CP003174) revealed 100% sequence identity, and alignment of the 1903 bp product with the sequence of the B. canis reference strain RM6/66 revealed that strain RM6/66 had the expected deletion (1067 bp). A representative sequence was submitted to GenBank (Accession number KC572141). A similar analysis of the 836 bp product revealed 100% identity with the B. canis reference sequence from strain RM6/66. Sequences of this product have also been submitted to GenBank (Accession number KC572142).
We also sought to identify various B. canis strains by sequencing the omp25 gene. Gene sequences from the 29 Chinese B. canis isolates were aligned with omp25 gene sequences from B. canis RM6/66 and other strains obtained from GenBank. This analysis revealed that the omp25 gene sequences of 26 Chinese B. canis isolates have the omp25 T545C mutation, and this mutation is consistent with three B. canis isolates isolated from Germany and South Africa (AM695188, AM695179 and AM695170). Like B. canis RM6/66, the omp25 gene position T545 of the remaining three isolates (1124, 233 and 231) was not mutated. The omp25 gene sequence also correlated to the presence of the BMEI1435 gene. Two of the B. canis isolates without the omp25 T545C mutation (1124 and 233) lacked the BMEI1435 gene like B. canis RM6/66. Twenty-six of the 27 remaining isolates were strains with the omp25 T545C mutation also had the BMEI1435 gene. These data revealed that the base at position 545 of the omp25 gene correlated well with the presence of the BMEI1435 gene. One exception was the isolate 231. This isolate did not have the typical B. canis deletion of BMEI1435 gene, but had the omp25 T545C mutation. Further experimentation is required to fully elucidate this relationship.
Conclusions
SNP genotyping and Bruce-ladder 2.0 assay were able to sufficiently resolve B. canis and B. suis species. B. canis isolates with and without the BMEI1435 gene were present in China. In addition, a point mutation in the omp25 gene position 545 correlated with the presence of the BMEI1435 gene. This study reveals that the MLVA-16 assay is an effective molecular tool for detecting genotype distribution of Brucella isolates from endemic and non-endemic areas, and that considerable diversity among B. canis isolates in China.
Supporting Information
MLVA-16 genotypes for 67 B. canis strains.
(XLS)
Acknowledgments
We thank John Klena for editorial and scientific input.
Funding Statement
This study was funded by the National Natural Science Foundation of China (81271900), the Science and Technology Basic Work Program (2012FY111000), MOST (Ministry of Science and Technology of the People's Republic of China) (2012ZX10004-209), and the NIH. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Bricker BJ (2002) PCR as a diagnostic tool for brucellosis. Vet Microbiol 90: 435–446. [DOI] [PubMed] [Google Scholar]
- 2. Kang SI, Heo EJ, Cho D, Kim JW, Kim JY, et al. (2011) Genetic comparison of Brucella canis isolates by the MLVA assay in South Korea. J Vet Med Sci 73: 779–786. [DOI] [PubMed] [Google Scholar]
- 3. Nomura A, Imaoka K, Imanishi H, Shimizu H, Nagura F, et al. (2010) Human Brucella canis infections diagnosed by blood culture. Emerg Infect Dis 16: 1183–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sayan M, Erdenlig S, Stack J, Kilic S, Guducuoglu H, et al. (2011) A serological diagnostic survey for Brucella canis infection in Turkish patients with Brucellosis-like symptoms. Jpn J Infect Dis 64: 516–519. [PubMed] [Google Scholar]
- 5. Kulakov I, Tsirel'Son LE, Zheludkov MM (2012) Molecular-genetic characterization of canine and rangiferine Brucella isolates from different regions of Russia. Mol Gen Mikrobiol Virusol 28–33. [PubMed] [Google Scholar]
- 6. Holst BS, Lofqvist K, Ernholm L, Eld K, Cedersmyg M, et al. (2012) The first case of Brucella canis in Sweden: background, case report and recommendations from a northern European perspective. Acta Vet Scand 54: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Deqiu S, Donglou X, Jiming Y (2002) Epidemiology and control of brucellosis in China. Vet Microbiol 90: 165–182. [DOI] [PubMed] [Google Scholar]
- 8. Jiang H, Mao LL, Zhao HY, Li LY, Piao DR, et al. (2012) Reemergence and genetic comparison of Brucella canis in China, using a multiple-locus variable-number tandem-repeat assay. Vet Microbiol 154: 419–421. [DOI] [PubMed] [Google Scholar]
- 9. Jiang H, Wang H, Xu L, Hu G, Ma J, et al. (2013) MLVA Genotyping of Brucella melitensis and Brucella abortus Isolates from Different Animal Species and Humans and Identification of Brucella suis Vaccine Strain S2 from Cattle in China. PLoS One 8: e76332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jiang H, Fan M, Chen J, Mi J, Yu R, et al. (2011) MLVA genotyping of Chinese human Brucella melitensis biovar 1, 2 and 3 isolates. BMC Microbiol 11: 256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Whatmore AM, Perrett LL, MacMillan AP (2007) Characterisation of the genetic diversity of Brucella by multilocus sequencing. BMC Microbiol 7: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Halling SM, Peterson-Burch BD, Bricker BJ, Zuerner RL, Qing Z, et al. (2005) Completion of the genome sequence of Brucella abortus and comparison to the highly similar genomes of Brucella melitensis and Brucella suis . J Bacteriol 187: 2715–2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vizcaino N, Cloeckaert A, Verger J, Grayon M, Fernandez-Lago L (2000) DNA polymorphism in the genus Brucella . Microbes Infect 2: 1089–1100. [DOI] [PubMed] [Google Scholar]
- 14. Whatmore AM, Murphy TJ, Shankster S, Young E, Cutler SJ, et al. (2005) Use of amplified fragment length polymorphism to identify and type Brucella isolates of medical and veterinary interest. J Clin Microbiol 43: 761–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Moreno E, Cloeckaert A, Moriyon I (2002) Brucella evolution and taxonomy. Vet Microbiol 90: 209–227. [DOI] [PubMed] [Google Scholar]
- 16. Gandara B, Merino AL, Rogel MA, Martinez-Romero E (2001) Limited genetic diversity of Brucella spp. J Clin Microbiol 39: 235–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Michaux-Charachon S, Bourg G, Jumas-Bilak E, Guigue-Talet P, Allardet-Servent A, et al. (1997) Genome structure and phylogeny in the genus Brucella . J Bacteriol 179: 3244–3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cloeckaert A, Verger JM, Grayon M, Vizcaino N (1996) Molecular and immunological characterization of the major outer membrane proteins of Brucella . FEMS Microbiol Lett 145: 1–8. [DOI] [PubMed] [Google Scholar]
- 19. Cloeckaert A, Verger JM, Grayon M, Grepinet O (1995) Restriction site polymorphism of the genes encoding the major 25 kDa and 36 kDa outer-membrane proteins of Brucella . Microbiology 141 (Pt 9): 2111–2121. [DOI] [PubMed] [Google Scholar]
- 20. Ouahrani S, Michaux S, Sri WJ, Bourg G, Tournebize R, et al. (1993) Identification and sequence analysis of IS6501, an insertion sequence in Brucella spp.: relationship between genomic structure and the number of IS6501 copies. J Gen Microbiol 139: 3265–3273. [DOI] [PubMed] [Google Scholar]
- 21. Huber B, Scholz HC, Lucero N, Busse HJ (2009) Development of a PCR assay for typing and subtyping of Brucella species. Int J Med Microbiol 299: 563–573. [DOI] [PubMed] [Google Scholar]
- 22. Whatmore AM (2009) Current understanding of the genetic diversity of Brucella, an expanding genus of zoonotic pathogens. Infect Genet Evol 9: 1168–1184. [DOI] [PubMed] [Google Scholar]
- 23. Garcia-Yoldi D, Marin CM, de Miguel MJ, Munoz PM, Vizmanos JL, et al. (2006) Multiplex PCR assay for the identification and differentiation of all Brucella species and the vaccine strains Brucella abortus S19 and RB51 and Brucella melitensis Rev1. Clin Chem 52: 779–781. [DOI] [PubMed] [Google Scholar]
- 24. Lopez-Goni I, Garcia-Yoldi D, Marin CM, de Miguel MJ, Barquero-Calvo E, et al. (2011) New Bruce-ladder multiplex PCR assay for the biovar typing of Brucella suis and the discrimination of Brucella suis and Brucella canis . Vet Microbiol 154: 152–155. [DOI] [PubMed] [Google Scholar]
- 25. Valdezate S, Navarro A, Villalon P, Carrasco G, Saez-Nieto JA (2010) Epidemiological and phylogenetic analysis of Spanish human Brucella melitensis strains by multiple-locus variable-number tandem-repeat typing, hypervariable octameric oligonucleotide fingerprinting, and rpoB typing. J Clin Microbiol 48: 2734–2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Marianelli C, Petrucca A, Pasquali P, Ciuchini F, Papadopoulou S, et al. (2008) Use of MLVA-16 typing to trace the source of a laboratory-acquired Brucella infection. J Hosp Infect 68: 274–276. [DOI] [PubMed] [Google Scholar]
- 27. Kattar MM, Jaafar RF, Araj GF, Le FP, Matar GM, et al. (2008) Evaluation of a multilocus variable-number tandem-repeat analysis scheme for typing human Brucella isolates in a region of brucellosis endemicity. J Clin Microbiol 46: 3935–3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ferreira AC, Chambel L, Tenreiro T, Cardoso R, Flor L, et al. (2012) MLVA16 typing of Portuguese human and animal Brucella melitensis and Brucella abortus isolates. PLoS One 7: e42514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Koylass MS, King AC, Edwards-Smallbone J, Gopaul KK, Perrett LL, et al. (2010) Comparative performance of SNP typing and ‘Bruce-ladder’ in the discrimination of Brucella suis and Brucella canis . Vet Microbiol 142: 450–454. [DOI] [PubMed] [Google Scholar]
- 30. Lopez-Goni I, Garcia-Yoldi D, Marin CM, de Miguel MJ, Munoz PM, et al. (2008) Evaluation of a multiplex PCR assay (Bruce-ladder) for molecular typing of all Brucella species, including the vaccine strains. J Clin Microbiol 46: 3484–3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rajashekara G, Glasner JD, Glover DA, Splitter GA (2004) Comparative whole-genome hybridization reveals genomic islands in Brucella species. J Bacteriol 186: 5040–5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gopaul KK, Koylass MS, Smith CJ, Whatmore AM (2008) Rapid identification of Brucella isolates to the species level by real time PCR based single nucleotide polymorphism (SNP) analysis. BMC Microbiol 8: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Foster JT, Okinaka RT, Svensson R, Shaw K, De BK, et al. (2008) Real-time PCR assays of single-nucleotide polymorphisms defining the major Brucella clades. J Clin Microbiol 46: 296–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Scott JC, Koylass MS, Stubberfield MR, Whatmore AM (2007) Multiplex assay based on single-nucleotide polymorphisms for rapid identification of Brucella isolates at the species level. Appl Environ Microbiol 73: 7331–7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Alton GG, Jones LM, Pietz DE (1975) Laboratory techniques in brucellosis. Monogr Ser World Health Organ 1–163. [PubMed] [Google Scholar]
- 36. Garcia-Yoldi D, Le FP, De Miguel MJ, Munoz PM, Blasco JM, et al. (2007) Comparison of multiple-locus variable-number tandem-repeat analysis with other PCR-based methods for typing Brucella suis isolates. J Clin Microbiol 45: 4070–4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Le Flèche P, Jacques I, Grayon M, Al Dahouk S, Bouchon P, et al. (2006) Evaluation and selection of tandem repeat loci for a Brucella MLVA typing assay. BMC Microbiol 6: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Al Dahouk S, Flèche PL, Nockler K, Jacques I, Grayon M, et al. (2007) Evaluation of Brucella MLVA typing for human brucellosis. J Microbiol Methods 69: 137–145. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
MLVA-16 genotypes for 67 B. canis strains.
(XLS)