Abstract
In the past decade, a number of case–control studies have been carried out to investigate the relationship between the CTLA4 gene polymorphisms and type 1 diabetes (T1D). However, these studies have yielded contradictory results. To investigate this inconsistency, we performed a meta-analysis of all available studies dealing with the relationship between the CTLA4 polymorphism and T1D. In total, 58 association studies on two CTLA4 polymorphisms (G49A and C60T) and risk of T1D, including a total of 30,723 T1D cases and 45,254 controls were included. In a combined analysis, the summary per-allele odds ratio (OR) for T1D of the G49A and C60T polymorphism was 1.42 [95% confidence interval (CI): 1.31–1.53, P<10−5] and 1.23 (95% CI: 1.18–1.29, P<10−5), respectively. Significant results were also observed using dominant or recessive genetic model. In the subgroup analysis by ethnicity and sample size, significantly increased risks were also found for these polymorphisms. This meta-analysis demonstrated that the G49A and C60T polymorphism of CTLA4 is a risk factor associated with increased T1D susceptibility, but these associations vary in different ethnic populations.
Introduction
Type 1 diabetes (T1D) is an autoimmune disease characterized by destruction of the insulin-producing β-cells in the pancreatic islets. Although its etiology is not yet understood, strong genetic and environmental components appear to modulate individual disease susceptibility in patients and in animal models [1]. The cytotoxic T lymphocyte antigen-4 gene (CTLA4) and the gene encoding CD28 have been mapped to chromosome 2q33. CTLA4 is a glycoprotein receptor expressed on activated T cells and CD28 is involved in the regulation process of the activation of T cells by antigen-presenting cells and subsequent cellular immunity [2]. Based on its role in the regulation of the activation of T cells and T cell and B cell interactions [3], CTLA4 has been considered to be a permissive candidate gene involved in the etiology of autoimmune diseases. A number of common polymorphisms have been reported both in the coding and promoter regions of the CTLA4 gene. Among them, one common polymorphism in the coding region, which leads to a alanine→threonine substitution at exon 1 (G49A, rs231775) and one located at 3′-UTR (G6230A, C60T, rs3087243) were studied widely for their association with T1D susceptibility.
In the past decade, several association studies have investigated the associations between the CTLA4 gene and T1D susceptibility. However, these studies yielded conflicting results. Genetic association studies can be problematic to reproduce due to inadequate statistical power, multiple hypothesis testing, population stratification, publication bias, and phenotypic heterogeneity. In addition, with the increased studies in recent years among Caucasian, Asian, and other populations, there is a need to reconcile these data. Therefore, we performed a systematic meta-analysis of published studies to clarify the relationship between CTLA4 and T1D.
Materials and Methods
Literature search strategy and inclusion criteria
The literature included in our analysis was selected from PubMed, EMBASE, ISI web of science and Chinese National Knowledge Infrastructure with keywords relating to the relevant genes (e.g. ‘cytotoxic T lymphocyte antigen-4’ or ‘CTLA4’) in combination with words related to T1D (e.g. ‘Type 1 diabetes’ or ‘insulin dependent diabetes mellitus’) and ‘polymorphism’ or ‘variation’. Genetic association studies published before the 31 Jan. 2013 on T1D and polymorphisms in the CTLA4 gene described above were retrieved, and their references were checked to identify other relevant publications. The search was supplemented by reviews of reference lists for all relevant studies and review articles. The major inclusion criteria were (a) original papers containing independent data, (b) case–control or cohort studies and (c) available genotype distribution information or odds ratio (OR) with its 95% confidence interval (OR) with its 95% confidence interval (CI) and P-value. The major reasons for exclusion of studies were (a) overlapping data and (b) case-only studies and review articles.
Data extraction
Data extraction was performed independently by two reviewers, and differences were resolved by further discussion among all authors. For each included study, the following information was extracted according to a fixed protocol: first author's surname, publication year, definition and numbers of cases and controls, diagnostic criterion, frequency of genotypes, source of controls, gender, age at onset, Hardy–Weinberg equilibrium (HWE) status, ethnicity and genotyping method.
Statistical methods
The strength of association between polymorphisms of CTLA4 and T1D risk was assessed by OR with the corresponding 95% CI. The per-allele OR of the risk allele was compared between cases and controls. Then, we examined the association between risk genotype of polymorphisms and T1D susceptibility using dominant and recessive genetic models.
Heterogeneity across individual studies was calculated using the Cochran chi-square Q test followed by subsidiary analysis or by random-effects regression models with restricted maximum likelihood estimation [4]–[6]. Random-effects and fixed-effect summary measures were calculated as inverse variance-weighted average of the log OR. The results of random-effects summary were reported in the text because they take into account the variation between studies. In addition, we investigated potential sources of identified heterogeneity among studies by stratifying by ethnic group and the number of cases (≥300 and <300). Ethnic group was defined as East Asians, Caucasians (i.e. people of European origin) and Middle Eastern (e.g. Iran, Egyptian and Lebanon), Indian and African. The Z test was used to determine the significance of the pooled OR.
We assessed publication bias by using an ancillary procedure attributed to Egger et al. [7], which uses a linear regression approach to measure funnel plot asymmetry on the natural logarithm of the OR. The larger the deviation from the funnel curve of each study, the more pronounced the asymmetry will be. The results from small studies tend to scatter widely at the bottom of the graph, with the spread narrowing among larger studies. The significance of the intercept is evaluated using the t test. Sensitivity analysis was performed by removing each individual study in turn from the total and re-analyzing the remainder. This procedure was used to ensure that no individual study was entirely responsible for the combined results. All statistical analyses were carried out with the Stata software version 10.0 (Stata Corporation, College Station, TX, USA). The type I error rate was set at 0.05. All the P-values were for two-sided analysis.
Results
Characteristics of included studies
The combined search yielded 193 references. 135 articles were excluded because they did not meet the criteria or reported overlapping data (Figure S1). Finally, a total of 58 case–control studies were retrieved based on the search criteria for T1D susceptibility related to the CTLA4 polymorphisms [8]–[65]. The main study characteristics were summarized in Table 1. There are 51 studies with 10,969 T1D cases and 14,111 controls concerning G49A polymorphism and 15 studies with 22,437 T1D cases and 34,599 controls concerning C60T variation. These two polymorphisms were found to occur in frequencies consistent with HWE in the control populations of the vast majority of the published studies.
Table 1. Characteristics of the studies included in the meta-analysis.
| Study | Year | Ethnicity | Case | Control | No. of case/control | Genotyping method | Mean age at onset |
| Nistico [8] | 1996 | Belgian | T1D per NDDG criteria | Non-diabetic participants | 483/529 | allele-specific PCR | NA |
| Donner [9] | 1997 | American | T1D patients | Healthy | 293/325 | SSCP | 17.9 |
| Van der Auwera [10] | 1997 | Belgian | T1D per NDDG criteria | Healthy | 425/530 | RFLP | 20.0 |
| Awata [11] | 1998 | Japanese | T1D patients | Healthy | 173/425 | NA | 24.5 |
| Djilali-Saiah [12] | 1998 | French | T1D patients | Healthy | 112/100 | NA | 24.9 |
| Krokowski [13] | 1998 | Polish | T1D patients | Healthy | 192/136 | allele-specific PCR | 9.5 |
| Abe [14] | 1999 | Japanese | T1D per NDDG criteria | Healthy | 111/445 | RFLP | NA |
| Hayashi [15] | 1999 | Japanese | T1D per ADA criteria | Healthy | 117/141 | RFLP | 34.0 |
| Yanagawa [16] | 1999 | Japanese | T1D patients | Non-diabetic participants | 110/200 | RFLP | 25.9 |
| Lee [17] | 2000 | Chinese | T1D per NDDG criteria | Non-diabetic participants | 253/91 | RFLP | 7.1 |
| Takara [18] | 2000 | Japanese | T1D patients | Healthy | 74/107 | RFLP | 21.8 |
| Ihara [19] | 2001 | Japanese | T1D per NDDG criteria | Non-diabetic participants | 160/200 | SSCP | 7.9 |
| Kamoun Abid [20] | 2001 | Tunisian | T1D patients | Healthy | 74/49 | RFLP | 10.3 |
| Kikuoka [21] | 2001 | Japanese | T1D per WHO criteria | Non-diabetic participants | 125/200 | RFLP | NA |
| McCormack [22] | 2001 | Irish | T1D patients | Healthy | 130/307 | NA | NA |
| Osei-Hyiaman [23] | 2001 | Chinese, African | T1D per NDDG criteria | Healthy | 532/621 | SSCP | NA |
| Cinek [24] | 2002 | Czech | T1D per WHO criteria | Non-diabetic participants | 305/289 | allele-specific PCR | 7.6 |
| Cosentino [25] | 2002 | Italian | T1D patients | Healthy | 80/85 | RFLP | NA |
| Fajardy [26] | 2002 | French | T1D per WHO criteria | Non-diabetic participants | 134/273 | RFLP | 17.0 |
| Klitz [27] | 2002 | Philippine | T1D per ADA criteria | Non-diabetic participants | 90/94 | allele-specific PCR | NA |
| Ma [28] | 2002 | Chinese | T1D per ADA criteria | Healthy | 31/36 | RFLP | NA |
| Ongagna [29] | 2002 | French | T1D per WHO criteria | Non-diabetic participants | 62/84 | RFLP | 13.3 |
| Wood [30] | 2002 | German | T1D patients | Non-diabetic participants | 176/220 | RFLP | NA |
| Bouqbis [31] | 2003 | Moroccan | T1D patients | Healthy | 118/114 | SNaPshot | NA |
| Mochizuki [32] | 2003 | Japanese | T1D per ADA criteria | Non-diabetic participants | 97/60 | RFLP | NA |
| Haller [33] | 2004 | Estonian | T1D per ECDC criteria | Healthy | 69/158 | RFLP | NA |
| Ide [34] | 2004 | Japanese | T1D per ADA criteria | Healthy | 116/114 | RFLP | 22.0 |
| Liang [35] | 2004 | Japanese | T1D per ADA criteria | Normal glucose tolerance | 29/40 | RFLP | 25.3 |
| Zalloua [36] | 2004 | Lebanese | T1D patients | Healthy | 190/96 | allele-specific PCR | 8.9 |
| Caputo [37] | 2005 | Argentinean | T1D per WHO criteria | Healthy | 123/168 | RFLP | 15.0 |
| Mojtahedi [38] | 2005 | Iranian | T1D per NDDG criteria | Healthy | 109/331 | SSCP | 16.4 |
| Zhernakova [39] | 2005 | Dutch | IS-PAD | Healthy | 350/900 | TaqMan | 17.0 |
| Ahmedov [40] | 2006 | Azeri | T1D per WHO criteria | Non-diabetic participants | 160/271 | SSCP | 9.1 |
| Baniasadi [41] | 2006 | Indian | T1D per ADA criteria | Healthy | 130/180 | RFLP | 15.4 |
| Kanazawa [42] | 2006 | Japanese | T1D patients | Normal glucose tolerance | 71/39 | RFLP | 35.4 |
| Ikegami [43] | 2006 | Japanese | T1D patients | Non-diabetic participants | 769/723 | Invader | 27.3 |
| Haller [44] | 2007 | Estonian | T1D per ECDCD criteria | Healthy | 70/252 | RFLP | 24.3 |
| Howson [45] | 2007 | British | T1D patients | Non-diabetic participants | 4066/6866 | TaqMan | 7.5 |
| Butty [46] | 2008 | American | T1D patients | Normoglycemic participants | 224/343 | TaqMan | NA |
| Kawasaki [47] | 2008 | Japanese | T1D per WHO criteria | Healthy | 91/369 | RFLP | NA |
| Saleh [48] | 2008 | Egyptian | T1D patients | Healthy | 396/396 | SSCP | 6.7 |
| Smyth [49] | 2008 | British | T1D patients | Healthy | 5253/9161 | TaqMan | 7.5 |
| Balic [50] | 2009 | Chilean | T1D per ADA criteria | Healthy | 300/310 | RFLP | 8.9 |
| Douroudis [51] | 2009 | Estonian, Finnish | T1D per WHO criteria | Healthy | 574/955 | TaqMan | NA |
| Jin [52] | 2009 | Chinese | T1D per WHO criteria | Healthy | 413/476 | RFLP | 17.0 |
| Jung [53] | 2009 | Korean | T1D per WHO criteria | Healthy | 176/90 | RFLP | 7.5 |
| Korolija [54] | 2009 | Croatian | T1D patients | Healthy | 102/193 | RFLP | 11.5 |
| Lemos [55] | 2009 | Portuguese | T1D patients | Healthy | 207/249 | RFLP | 16.1 |
| Momin [56] | 2009 | Chilean | T1D per ADA criteria | Healthy | 261/280 | RFLP | 8.2 |
| Benmansour [57] | 2010 | Tunisian | T1D patients | Normal glucose tolerance | 228/193 | RFLP | 15.7 |
| Klinker [58] | 2010 | Finnish | T1D patients | Normoglycemic participants | 591/1538 | TaqMan | 26.0 |
| Howson [59] | 2011 | British | T1D per WHO criteria | Normoglycemic participants | 928/2043 | TaqMan | 33.3 |
| Philip [60] | 2011 | Indian | T1D patients | Healthy | 53/53 | RFLP | NA |
| Plagnol [61] | 2011 | British | T1D patients | Healthy | 8506/10596 | Affymetrix chip | 8.0 |
| Reddy [62] | 2011 | American | T1D per ADA criteria | Healthy | 1434/1864 | TaqMan | NA |
| Wafai [63] | 2011 | Lebanese | T1D per ADA criteria | Healthy | 39/46 | RFLP | 8.9 |
| Horie [64] | 2012 | Japanese | T1D patients | Normoglycemic participants | 134/222 | RFLP | NA |
| Mosaad [65] | 2012 | Egyptian | T1D per ADA criteria | Healthy | 104/78 | RFLP | 8.2 |
NA: Not Available, WHO: World Health Organization, ADA: American Diabetes Association, ECDC: Expert Committee on the Diagnosis and Classification of Diabetes Mellitus, IS-PAD: International Society of Paediatric and Adolescent Diabetes.
Association of CTLA4 G49A polymorphism and T1D
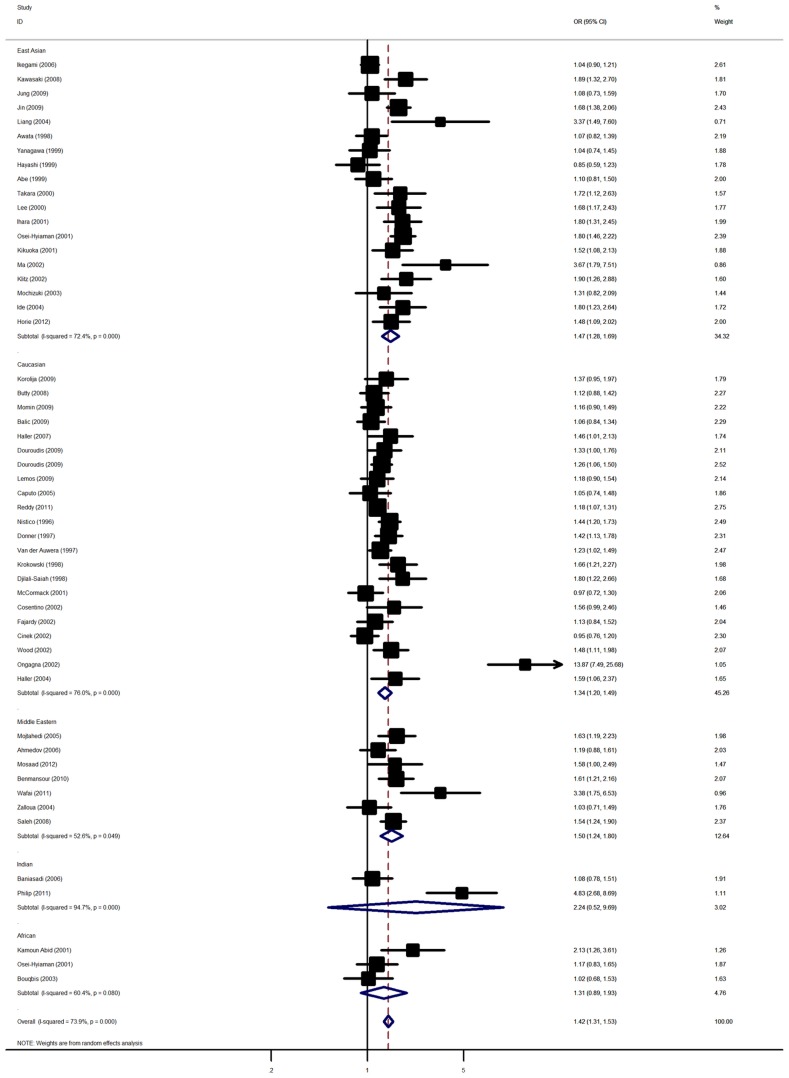
Overall, there was evidence of an association between the increased risk of T1D and the variant in different genetic models when all the eligible studies were pooled into the meta-analysis. Using random effect model, the summary per-allele OR of the G variant for T1D was 1.42 [95% CI: 1.31–1.53; P(Z)<10−5; P(Q)<10−5; Figure 1], with corresponding results under dominant and recessive genetic models of 1.48 [95% CI: 1.31–1.66; P(Z)<10−5; P(Q)<10−5 ] and 1.68 [95% CI: 1.47–1.91; P(Z)<10−5; P(Q)<10−5], respectively.
Figure 1. Forest plot from the meta-analysis of type 1 diabetes risk and CTLA4 G49A polymorphism.
In the stratified analysis by ethnicity, significantly increased risks were found among East Asian populations [G allele: OR = 1.47, 95% CI: 1.28–1.69; dominant model: OR = 1.65, 95% CI: 1.29–2.11; recessive model: OR = 1.65, 95% CI: 1.35–2.02] and Caucasian populations [G allele: OR = 1.23, 95% CI: 1.20–1.49; dominant model: OR = 1.31, 95% CI: 1.14–1.49; recessive model: OR = 1.68, 95% CI: 1.37–2.06]. Similar significant associations were also observed for Middle Eastern population [G allele: OR = 1.50, 95% CI: 1.24–1.80; dominant model: OR = 1.62, 95% CI: 1.15–2.29; recessive model: OR = 1.93, 95% CI: 1.26–2.96]. However, no significant associations were detected among Indian and African populations (Table 2). Subsidiary analyses of sample size yielded a per-allele OR for small studies of 1.52 (95% CI: 1.34–1.72) and for large studies of 1.30 (95% CI: 1.19–1.42).
Table 2. Meta-analysis of the CTLA-4 G49A polymorphism on type 1 diabetes risk.
| Sub-group analysis | No. of cases/controls | G allele vs. A allele | Dominant model | Recessive model | ||||||
| OR (95%CI) | P(Z) | P(Q) | OR (95%CI) | P(Z) | P(Q) | OR (95%CI) | P(Z) | P(Q) | ||
| Total | 10969/14111 | 1.42 (1.31–1.53) | <10−5 | <10−5 | 1.48 (1.31–1.66) | <10−5 | <10−5 | 1.68 (1.47–1.91) | <10−5 | <10−5 |
| Ethnicity | ||||||||||
| East Asians | 3430/4453 | 1.47 (1.28–1.69) | <10−5 | <10−5 | 1.65 (1.29–2.11) | <10−4 | 0.008 | 1.66 (1.35–2.02) | <10−5 | 0.0009 |
| Caucasians | 5756/7650 | 1.23 (1.20–1.49) | <10−5 | <10−5 | 1.31 (1.14–1.49) | <10−4 | 0.002 | 1.68 (1.37–2.06) | <10−5 | <10−5 |
| Middle Eastern | 1226/1411 | 1.50 (1.24–1.80) | <10−4 | 0.05 | 1.62 (1.15–2.29) | 0.006 | 0.0007 | 1.93 (1.26–2.96) | 0.003 | 0.07 |
| African | 374/364 | 1.31 (0.89–1.93) | 0.17 | 0.001 | 1.43 (0.84–2.44) | 0.18 | 0.11 | 1.31 (0.61–2.82) | 0.49 | 0.12 |
| Indian | 183/233 | 2.24 (0.52–9.69) | 0.28 | 0.08 | 3.94 (0.33–47.54) | 0.28 | <10−4 | 1.89 (0.48–7.41) | 0.36 | 0.02 |
| Sample size | ||||||||||
| Small | 4224/6368 | 1.52 (1.34–1.72) | <10−5 | <10−4 | 1.58 (1.32–1.88) | <10−5 | <10−5 | 1.77 (1.46–2.16) | <10−5 | <10−5 |
| Large | 6745/7743 | 1.30 (1.19–1.42) | <10−5 | <10−5 | 1.39 (1.20–1.61) | <10−4 | 0.001 | 1.58 (1.38–1.82) | <10−5 | 0.09 |
Association of CTLA4 C60T polymorphism and T1D
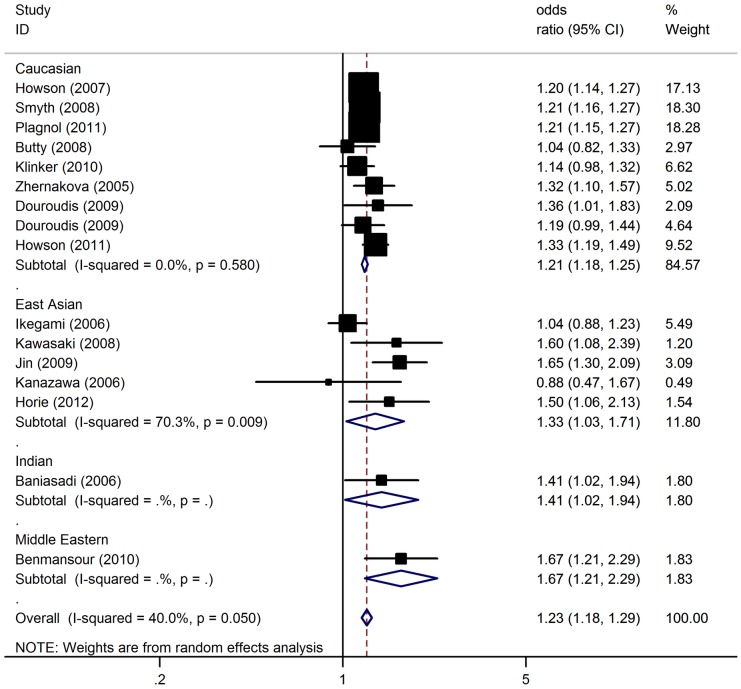
In the overall analysis, the C60T polymorphism of CTLA4 was significantly associated with elevated T1D risk with a per-allele OR of 1.23 [95% CI: 1.18–1.29; P(Z)<10−5; P(Q) = 0.05; Figure 2]. Significant associations were also found under dominant [OR = 1.31; 95% CI: 1.16–1.47; P(Z)<10−5; P(Q) = 0.11] and recessive [OR = 1.32; 95% CI: 1.19–1.43; P(Z)<10−5; P(Q) = 0.06] genetic model.
Figure 2. Forest plot from the meta-analysis of type 1 diabetes risk and CTLA4 C60T polymorphism.
When studies were stratified for ethnicity, significant risks were found among Caucasians in all genetic models [C allele: OR = 1.21, 95% CI: 1.18–1.25; dominant model: OR = 1.31, 95% CI: 1.20–1.44; recessive model: OR = 1.24, 95% CI: 1.18–1.31]. Similar results were also found in the Middle Eastern populations and Indians with a per-allele OR of 1.67 (95% CI: 1.21–2.29) and 1.41 (95% CI: 1.02–1.94), respectively. Only marginal significant results were detected for East Asians with per-allele OR of 1.33 (95% CI: 1.03–1.71). In the stratified analysis by sample size, significant associations were detected in both large and small studies (Table 3).
Table 3. Meta-analysis of the CTLA-4 C60T polymorphism on type 1 diabetes risk.
| Sub-group analysis | No. of cases/controls | C allele vs. T allele | Dominant model | Recessive model | ||||||
| OR (95%CI) | P(Z) | P(Q) | OR (95%CI) | P(Z) | P(Q) | OR (95%CI) | P(Z) | P(Q) | ||
| Total | 22437/34599 | 1.23 (1.18–1.29) | <10−5 | 0.05 | 1.31 (1.16–1.47) | <10−5 | 0.11 | 1.32 (1.19–1.43) | <10−5 | 0.06 |
| Ethnicity | ||||||||||
| East Asians | 1487/1821 | 1.33 (1.03–1.71) | 0.03 | 0.009 | 1.18 (0.58–2.43) | 0.65 | 0.06 | 1.44 (1.00–2.03) | 0.05 | 0.02 |
| Caucasians | 20592/32405 | 1.21 (1.18–1.25) | <10−5 | 0.58 | 1.31 (1.20–1.44) | <10−5 | 0.26 | 1.24 (1.18–1.31) | <10−5 | 0.43 |
| Middle Eastern | 228/193 | 1.67 (1.21–2.29) | 0.002 | NA | 1.63 (1.10–2.42) | 0.01 | NA | 2.37 (1.18–4.76) | 0.01 | NA |
| Indian | 130/180 | 1.41 (1.02–1.94) | 0.04 | NA | 1.74 (1.06–2.87) | 0.03 | NA | 1.34 (0.78–2.33) | 0.29 | NA |
| Sample size | ||||||||||
| Small | 2226/3680 | 1.35 (1.21–1.50) | <10−5 | 0.15 | 1.39 (1.04–1.86) | 0.02 | 0.09 | 1.51 (1.30–1.75) | <10−5 | 0.34 |
| Large | 20211/30919 | 1.21 (1.17–1.25) | <10−5 | 0.25 | 1.34 (1.23–1.46) | <10−5 | 0.21 | 1.22 (1.16–1.29) | <10−5 | 0.47 |
NA: not available.
Haplotype analysis
The linkage disequilibrium (LD) analysis revealed a tight LD between the G49A and C60T sites with D′ score of 0.95. Haplotype analyses between 49G>A, and C60T polymorphisms were performed in the 4 studies, involving 2242 cases and 2581 controls. Three prevalent haplotypes (Table S1), which represent more than 95% of the haplotypes among those studied at the CTLA4 loci (49G>A, C60T), were detected both in affected and unaffected subjects. The AT haplotype was significantly associated with decreased diabetes risk in the overall analysis (OR = 0.87, 95% CI: 0.77–0.98, P = 0.03). In addition, the frequency of GC haplotype (OR = 1.12, 95% CI: 0.98–1.28, P = 0.10) was also higher in diabetes patients compared with controls.
Sensitivity analyses and publication bias
Sensitivity analysis indicated that no single study influenced the pooled OR qualitatively, suggesting that the results of this meta-analysis are stable (data not shown). A funnel plot of these included studies suggested a possibility of the preferential publication of positive findings in smaller studies for G49A polymorphism of CTLA4 (Begg test, P = 0.001; Egger test, P = 0.0002; Figure S2). The Duval and Tweedie nonparametric “trim and fill” method was used to adjust for publication bias. Meta-analysis with and without “trim and fill” method did not draw different conclusion (data not shown), indicating that our results were statistically robust. The shape of the funnel plots seemed symmetrical for CTLA4 C60T polymorphism, suggesting that no bias from selected studies have been included (Figure S3). The statistical results still did not show publication bias (Begg test, P = 0.32; Egger test, P = 0.18).
Discussion
Large sample and unbiased epidemiological studies of predisposition genes polymorphisms could provide insight to etiology of diseases. This is the most comprehensive meta-analysis examined the CTLA4 polymorphisms and the relationship to susceptibility for T1D. Its strength was based on the accumulation of published data giving greater information to detect significant differences. In total, the meta-analysis involved 58 studies for T1D which provided 30,734 cases and 40,754 controls.
Our results demonstrated that the G49A and C60T polymorphism of CTLA4 is a risk factor for developing T1D. In the stratified analysis by ethnicity, significant associations were found in Caucasians and Middle Eastern population for the two polymorphisms in all genetic models. Significant associations were detected among East Asians for G49A polymorphism; while no associations were found for C60T polymorphism. Among Indian population, only marginal significant associations were detected for C60T polymorphism. No associations were found in Africans. The reasons that the same polymorphism plays a different role in different ethnic populations or across different studies may arise from many aspects. Firstly, T1D is a complex disease and genetic heterogeneity exists in different populations. Whole genome linkage studies on T1D have confirmed this genetic heterogeneity [66]. Secondly, clinical heterogeneity may also explain the discrepancy. Potential contribution of differences in patient populations (e.g., age and years from onset, female proportion, disease severity…) might cause different results. Thirdly, population structure difference may also contribute to the discrepancy. Different populations often have different LD patterns. The same polymorphism plays a different role in disease susceptibility in different ethnic populations, implicating that this polymorphism might not be a causal variant. The fact is that this polymorphism may be in LD with a nearby causal variant in one ethnic population but not in another. Moreover, the difference might come from type I error. Therefore, additional studies are needed to further validate ethnic difference of the effect of these polymorphisms on T1D risk Therefore, additional studies are warranted to further validate ethnic difference in the effect of these polymorphisms on T1D risk.
An important source of bias in every meta-analysis is related to the studies that have been published and thus can be included in the analysis. Nevertheless in our meta-analysis, we included many studies with negative findings. Although the funnel plot for G49A polymorphism is not symmetric, the overall results of different ethnic groups are concordant, indicating that this bias cannot affect the final result. On the other hand, funnel plot asymmetry is not always caused by publication bias. True heterogeneity may also lead to funnel plot asymmetry. For example, significant difference may be seen only in high-risk individuals, and these high-risk people are usually more likely to be included in small studies. This is particularly true in our meta-analysis because the majority of the significant associations have been observed among the studies with small sample size. Language bias or citation bias also could be an important source in this group of studies, meaning that the studies without significant findings are preferentially published in languages other than English and less likely to be cited in other articles. Finally, it is possible that an asymmetrical funnel plot arises simply by chance.
The heterogeneity of OR is high in our data, especially in the studies for African and Indian populations, based on a small number of individuals. Nevertheless, the total number of subjects included in this part of the analysis comprises the largest sample size so far. Future studies including larger numbers of Africans and Indians are necessary to clarify the consistency of findings across ethnic groups. Another possible source of heterogeneity is difference in age at onset of T1D: early or late onset. Unfortunately, it was not possible to tease out this association because the breakdown of the two types was not consistently reported.
A number of factors predict T1D, however, detailed pathogenesis mechanisms of T1D remain a matter of speculation. Polymorphisms of the CTLA4 gene have been shown to confer susceptibility to several autoimmune diseases, due to its role in the down-regulation of the activated immune response [67]. The 49 G/G genotype of the CTLA4 gene was associated with reduced inhibitory function of cytotoxic T-lymphocyte antigen 4 [67]. In addition, it is proposed that a reduced function of CTLA4 associated with the C allele of C60T polymorphism allows T cells to be more hyperactive and to respond to peripheral antigens to a greater degree than individuals carrying the T/T genotype, which is associated with autoimmune disease protection and increased peripheral tolerance [68], [69]. Association studies and functional data along with our meta-analysis suggest that G49A and C60T polymorphisms of CTLA4 are risk factors for developing T1D.
Several meta-analyses addressing the same theme have been recently published [70]–[72]. However, Chen et al. and Si et al. mainly focused on the G49A polymorphism without assessing the relationship between CTLA4 C60T polymorphism and T1D [70], [71]. Furthermore, the results reported by Tang et al. were believed to be not entirely credible for insufficient literature identification and overlapping samples [72], [73]. Compared to those previous meta-analyses, the present study has considered more studies from the literature. In addition, we also investigated the two common variants on CTLA4 gene and genetic susceptibility to T1D. Furthermore, we also explored whether the CTLA4 gene haplotypes were associated with T1D risk. Our results also suggest the importance of including a haplotype-based approach to assess genetic associations. Haplotype-based case–control studies are warranted to confirm our findings in the future.
In summary, this meta-analysis showed that the CTLA4 G49A and C60T polymorphism was significantly associated with increased risk of T1D, particularly in Caucasian and Middle Eastern population. While the CTLA4 G49A and C60T polymorphisms are significantly associated with increased risk of T1D, larger cohorts of Indian and African subjects are needed to test the effect of these SNPs in these populations.
Supporting Information
(DOC)
Flow chart of literature search for studies examining CTLA4 gene polymorphism and risk of T1D.
(TIF)
Funnel plot of studies of the G49A polymorphism of CTLA4 and T1D showing a possible excess of smaller studies with strikingly positive findings beyond the 95% CI.
(TIF)
Funnel plot for the association between CTLA4 C60T polymorphism and T1D risk.
(TIF)
Meta-analysis of haplotype combinations between G49A, and C60T polymorphisms of CTLA4 gene and T1D risk.
(DOCX)
Funding Statement
The authors have no support or funding to report.
References
- 1. Eisenbarth GS (2007) Update in type 1 diabetes. J Clin Endocrinol Metab 92: 2403–2407. [DOI] [PubMed] [Google Scholar]
- 2. Walunas TL, Bakker CY, Bluestone JA (1996) CTLA4 ligation blocks CD28-dependent T cell activation. J Exp Med 183: 2541–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lane P (1997) Regulation of T and B cell responses by modulating interactions between CD28/CTLA4 and their ligands, CD80 and CD86. Ann NY Acad Sci 815: 392–400. [DOI] [PubMed] [Google Scholar]
- 4. Cochran WG (1954) The combination of estimates from different experiments. Biometrics 10: 101–129. [Google Scholar]
- 5. Thompson SG, Shar SJ (1999) Explaining heterogeneity in meta-analysis: a comparison of methods. Stat Med 18: 2693–2708. [DOI] [PubMed] [Google Scholar]
- 6. DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Trials 7: 177–188. [DOI] [PubMed] [Google Scholar]
- 7. Egger M, Davey Smith G, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315: 629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nisticò L, Buzzetti R, Pritchard LE, Van der Auwera B, Giovannini C, et al. (1996) The CTLA-4 gene region of chromosome 2q33 is linked to, and associated with, type 1 diabetes. Belgian Diabetes Registry. Hum Mol Genet 5: 1075–1080. [DOI] [PubMed] [Google Scholar]
- 9. Donner H, Rau H, Walfish PG, Braun J, Siegmund T, et al. (1997) CTLA4 alanine-17 confers genetic susceptibility to Graves' disease and to type 1 diabetes mellitus. J Clin Endocrinol Metab 82: 143–146. [DOI] [PubMed] [Google Scholar]
- 10. Van der Auwera BJ, Vandewalle CL, Schuit FC, Winnock F, De Leeuw IH, et al. (1997) CTLA-4 gene polymorphism confers susceptibility to insulin-dependent diabetes mellitus (IDDM) independently from age and from other genetic or immune disease markers. The Belgian Diabetes Registry. Clin Exp Immunol 110: 98–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Awata T, Kurihara S, Iitaka M, Takei S, Inoue I, et al. (1998) Association of CTLA-4 gene A-G polymorphism (IDDM12 locus) with acute-onset and insulin-depleted IDDM as well as autoimmune thyroid disease (Graves' disease and Hashimoto's thyroiditis) in the Japanese population. Diabetes 47: 128–129. [DOI] [PubMed] [Google Scholar]
- 12. Djilali-Saiah I, Larger E, Harfouch-Hammoud E, Timsit J, Clerc J, et al. (1998) No major role for the CTLA-4 gene in the association of autoimmune thyroid disease with IDDM. Diabetes 47: 125–127. [DOI] [PubMed] [Google Scholar]
- 13. Krokowski M, Bodalski J, Bratek A, Machejko P, Caillat-Zucman S (1998) CTLA-4 gene polymorphism is associated with predisposition to IDDM in a population from central Poland. Diabetes Metab 24: 241–243. [PubMed] [Google Scholar]
- 14. Abe T, Takino H, Yamasaki H, Ozaki M, Sera Y, et al. (1999) CTLA4 gene polymorphism correlates with the mode of onset and presence of ICA512 Ab in Japanese type 1 diabetes. Diabetes Res Clin Pract 46: 169–175. [DOI] [PubMed] [Google Scholar]
- 15. Hayashi H, Kusaka I, Nagasaka S, Kawakami A, Rokkaku K, et al. (1999) Association of CTLA-4 polymorphism with positive anti-GAD antibody in Japanese subjects with type 1 diabetes mellitus. Clin Endocrinol (Oxf) 51: 793–799. [DOI] [PubMed] [Google Scholar]
- 16. Yanagawa T, Maruyama T, Gomi K, Taniyama M, Kasuga A, et al. (1999) Lack of association between CTLA-4 gene polymorphism and IDDM in Japanese subjects. Autoimmunity 29: 53–56. [DOI] [PubMed] [Google Scholar]
- 17. Lee YJ, Huang FY, Lo FS, Wang WC, Hsu CH, et al. (2000) Association of CTLA4 gene A-G polymorphism with type 1 diabetes in Chinese children. Clin Endocrinol (Oxf) 52: 153–157. [DOI] [PubMed] [Google Scholar]
- 18. Takara M, Komiya I, Kinjo Y, Tomoyose T, Yamashiro S, et al. (2000) Association of CTLA-4 gene A/G polymorphism in Japanese type 1 diabetic patients with younger age of onset and autoimmune thyroid disease. Diabetes Care 23: 975–978. [DOI] [PubMed] [Google Scholar]
- 19. Ihara K, Ahmed S, Nakao F, Kinukawa N, Kuromaru R, et al. (2001) Association studies of CTLA-4, CD28, and ICOS gene polymorphisms with type 1 diabetes in the Japanese population. Immunogenetics 53: 447–454. [DOI] [PubMed] [Google Scholar]
- 20. Kamoun Abid H, Hmida S, Smaoui N, Kaabi H, Abid A, et al. (2001) Association between type 1 diabetes and polymorphism of the CTLA-4 gene in a Tunisian population. Pathol Biol (Paris) 49: 794–798. [DOI] [PubMed] [Google Scholar]
- 21. Kikuoka N, Sugihara S, Yanagawa T, Ikezaki A, Kim HS, et al. (2001) Cytotoxic T lymphocyte antigen 4 gene polymorphism confers susceptibility to type 1 diabetes in Japanese children: analysis of association with HLA genotypes and autoantibodies. Clin Endocrinol (Oxf) 55: 597–603. [DOI] [PubMed] [Google Scholar]
- 22. McCormack RM, Maxwell AP, Carson D, Patterson CC, Bingham A, et al. (2001) Possible association between CTLA4 DNA polymorphisms and early onset type 1 diabetes in a UK population. Genes Immun 2: 233–235. [DOI] [PubMed] [Google Scholar]
- 23. Osei-Hyiaman D, Hou L, Zhiyin R, Zhiming Z, Yu H, et al. (2001) Association of a novel point mutation (C159G) of the CTLA4 gene with type 1 diabetes in West Africans but not in Chinese. Diabetes 50: 2169–2171. [DOI] [PubMed] [Google Scholar]
- 24. Cinek O, Drevínek P, Sumník Z, Bendlová B, Kolousková S, et al. (2002) The CTLA4 +49 A/G dimorphism is not associated with type 1 diabetes in Czech children. Eur J Immunogenet 29: 219–222. [DOI] [PubMed] [Google Scholar]
- 25. Cosentino A, Gambelunghe G, Tortoioli C, Falorni A (2002) CTLA-4 gene polymorphism contributes to the genetic risk for latent autoimmune diabetes in adults. Ann N Y Acad Sci 958: 337–340. [DOI] [PubMed] [Google Scholar]
- 26. Fajardy I, Vambergue A, Stuckens C, Weill J, Danze PM, et al. (2002) CTLA-4 49 A/G dimorphism and type 1 diabetes susceptibility: a French case-control study and segregation analysis. Evidence of a maternal effect. Eur J Immunogenet 29: 251–257. [DOI] [PubMed] [Google Scholar]
- 27. Klitz W, Bugawan TL, Panelo A, Solfelix CM, Buzzetti R, et al. (2002) Association of CTLA-4 variation with type I diabetes in Filipinos. Immunogenetics 54: 310–313. [DOI] [PubMed] [Google Scholar]
- 28. Ma Y, Tang X, Chang W, Gao L, Li M, et al. (2002) CTLA-4 gene A/G polymorphism associated with diabetes mellitus in Han Chinese. Chin Med J (Engl) 115: 1248–1250. [PubMed] [Google Scholar]
- 29. Ongagna JC, Sapin R, Pinget M, Belcourt A (2002) Markers for risk of type 1 diabetes in relatives of Alsacian patients with type 1 diabetes. Int J Exp Diabetes Res 3: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wood JP, Pani MA, Bieda K, Meyer G, Usadel KH, et al. (2002) A recently described polymorphism in the CD28 gene on chromosome 2q33 is not associated with susceptibility to type 1 diabetes. Eur J Immunogenet 29: 347–349. [DOI] [PubMed] [Google Scholar]
- 31. Bouqbis L, Izaabel H, Akhayat O, Pérez-Lezaun A, Calafell F, et al. (2003) Association of the CTLA4 promoter region (−1661G allele) with type 1 diabetes in the South Moroccan population. Genes Immun 4: 132–137. [DOI] [PubMed] [Google Scholar]
- 32. Mochizuki M, Amemiya S, Kobayashi K, Kobayashi K, Shimura Y, et al. (2003) Association of the CTLA-4 gene 49 A/G polymorphism with type 1 diabetes and autoimmune thyroid disease in Japanese children. Diabetes Care 26: 843–847. [DOI] [PubMed] [Google Scholar]
- 33. Haller K, Kisand K, Nemvalts V, Laine AP, Ilonen J, et al. (2004) Type 1 diabetes is insulin −2221 MspI and CTLA-4 +49 A/G polymorphism dependent. Eur J Clin Invest 34: 543–548. [DOI] [PubMed] [Google Scholar]
- 34. Ide A, Kawasaki E, Abiru N, Sun F, Kobayashi M, et al. (2004) Association between IL-18 gene promoter polymorphisms and CTLA-4 gene 49A/G polymorphism in Japanese patients with type 1 diabetes. J Autoimmun 22: 73–78. [DOI] [PubMed] [Google Scholar]
- 35. Liang H, Yagi K, Asano A, Kobayashi J, Mabuchi H (2004) Association between CTLA-4 +49 A/G polymorphism and type 1B diabetes in Japanese population. Endocrine 25: 105–109. [DOI] [PubMed] [Google Scholar]
- 36. Zalloua PA, Abchee A, Shbaklo H, Zreik TG, Terwedow H, et al. (2004) Patients with early onset of type 1 diabetes have significantly higher GG genotype at position 49 of the CTLA4 gene. Hum Immunol 65: 719–724. [DOI] [PubMed] [Google Scholar]
- 37. Caputo M, Cerrone GE, López AP, Villalba A, Krochik GA, et al. (2005) Cytotoxic T lymphocyte antigen 4 heterozygous codon 49 A/G dimorphism is associated to latent autoimmune diabetes in adults (LADA). Autoimmunity 38: 277–281. [DOI] [PubMed] [Google Scholar]
- 38. Mojtahedi Z, Omrani GR, Doroudchi M, Ghaderi A (2005) CTLA-4 +49 A/G polymorphism is associated with predisposition to type 1 diabetes in Iranians. Diabetes Res Clin Pract 68: 111–116. [DOI] [PubMed] [Google Scholar]
- 39. Zhernakova A, Eerligh P, Barrera P, Wesoly JZ, Huizinga TW, et al. (2005) CTLA4 is differentially associated with autoimmune diseases in the Dutch population. Hum Genet 118: 58–66. [DOI] [PubMed] [Google Scholar]
- 40. Ahmedov G, Ahmedova L, Sedlakova P, Cinek O (2006) Genetic association of type 1 diabetes in an Azerbaijanian population: the HLA-DQ, -DRB1*04, the insulin gene, and CTLA4. Pediatr Diabetes 7: 88–93. [DOI] [PubMed] [Google Scholar]
- 41. Baniasadi V, Narain N, Goswami R, Das SN (2006) Promoter region −318 C/T and −1661 A/G CTLA-4 single nucleotide polymorphisms and type 1 diabetes in North Indians. Tissue Antigens 67: 383–389. [DOI] [PubMed] [Google Scholar]
- 42. Kanazawa Y, Motohashi Y, Yamada S, Oikawa Y, Shigihara T, et al. (2006) Frequency of CTLA-4 gene CT60 polymorphism may not be affected by vitamin D receptor gene Bsm I polymorphism or HLA DR9 in autoimmune-related type 1 diabetes in the Japanese. Ann N Y Acad Sci 1079: 251–256. [DOI] [PubMed] [Google Scholar]
- 43. Ikegami H, Awata T, Kawasaki E, Kobayashi T, Maruyama T, et al. (2006) The association of CTLA4 polymorphism with type 1 diabetes is concentrated in patients complicated with autoimmune thyroid disease: a multicenter collaborative study in Japan. J Clin Endocrinol Metab 91: 1087–1092. [DOI] [PubMed] [Google Scholar]
- 44. Haller K, Kisand K, Pisarev H, Salur L, Laisk T, et al. (2007) Insulin gene VNTR, CTLA-4 +49A/G and HLA-DQB1 alleles distinguish latent autoimmune diabetes in adults from type 1 diabetes and from type 2 diabetes group. Tissue Antigens 69: 121–127. [DOI] [PubMed] [Google Scholar]
- 45. Howson JM, Dunger DB, Nutland S, Stevens H, Wicker LS, et al. (2007) A type 1 diabetes subgroup with a female bias is characterised by failure in tolerance to thyroid peroxidase at an early age and a strong association with the cytotoxic T-lymphocyte-associated antigen-4 gene. Diabetologia 50: 741–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Butty V, Campbell C, Mathis D, Benoist C (2008) Impact of diabetes susceptibility loci on progression from pre-diabetes to diabetes in at-risk individuals of the diabetes prevention trial-type 1 (DPT-1). Diabetes 57: 2348–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kawasaki E, Imagawa A, Makino H, Uga M, Abiru N, et al. (2008) Differences in the contribution of the CTLA4 gene to susceptibility to fulminant and type 1A diabetes in Japanese patients. Diabetes Care 31: 1608–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Saleh HM, Rohowsky N, Leski M (2008) The CTLA4 −819 C/T and +49 A/G dimorphisms are associated with Type 1 diabetes in Egyptian children. Indian J Hum Genet 14: 92–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Smyth DJ, Plagnol V, Walker NM, Cooper JD, Downes K, et al. (2008) Shared and distinct genetic variants in type 1 diabetes and celiac disease. N Engl J Med 359: 2767–2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Balic I, Angel B, Codner E, Carrasco E, Perez-Bravo F (2009) Association of CTLA-4 polymorphisms and clinical-immunologic characteristics at onset of type 1 diabetes mellitus in children. Hum Immunol 70: 116–120. [DOI] [PubMed] [Google Scholar]
- 51. Douroudis K, Laine AP, Heinonen M, Hermann R, Lipponen K, et al. (2009) Association of CTLA4 but not ICOS polymorphisms with type 1 diabetes in two populations with different disease rates. Hum Immunol 70: 536–539. [DOI] [PubMed] [Google Scholar]
- 52. Jin P, Xiang B, Lin J, Huang G, Zhou WD, et al. (2009) Association of CTLA-4 + 49A/G and CT60 gene polymorphism with type 1 diabetes and thyroid autoimmunity. Zhonghua Yi Xue Za Zhi 89: 1246–1249. [PubMed] [Google Scholar]
- 53. Jung MH, Yu J, Shin CH, Suh BK, Yang SW, et al. (2009) Association of cytotoxic T lymphocyte antigen-4 gene polymorphisms and HLA class II alleles with the development of type 1 diabetes in Korean children and adolescents. J Korean Med Sci 24: 1004–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Korolija M, Renar IP, Hadzija M, Medvidović EP, Pavković P, et al. (2009) Association of PTPN22 C1858T and CTLA-4 A49G polymorphisms with Type 1 Diabetes in Croatians. Diabetes Res Clin Pract 86: e54–57. [DOI] [PubMed] [Google Scholar]
- 55. Lemos MC, Coutinho E, Gomes L, Bastos M, Fagulha A, et al. (2009) The CTLA4 +49 A/G polymorphism is not associated with susceptibility to type 1 diabetes mellitus in the Portuguese population. Int J Immunogenet 36: 193–195. [DOI] [PubMed] [Google Scholar]
- 56. Momin S, Flores S, Angel BB, Codner DE, Carrasco PE, et al. (2009) Interactions between programmed death 1 (PD-1) and cytotoxic T lymphocyte antigen 4 (CTLA-4) gene polymorphisms in type 1 diabetes. Diabetes Res Clin Pract 83: 289–294. [DOI] [PubMed] [Google Scholar]
- 57. Benmansour J, Stayoussef M, Al-Jenaidi FA, Rajab MH, Rayana CB, et al. (2010) Association of single nucleotide polymorphisms in cytotoxic T-lymphocyte antigen 4 and susceptibility to autoimmune type 1 diabetes in Tunisians. Clin Vaccine Immunol 17: 1473–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Klinker MW, Schiller JJ, Magnuson VL, Wang T, Basken J, et al. (2010) Single-nucleotide polymorphisms in the IL2RA gene are associated with age at diagnosis in late-onset Finnish type 1 diabetes subjects. Immunogenetics 62: 101–107. [DOI] [PubMed] [Google Scholar]
- 59. Howson JM, Rosinger S, Smyth DJ, Boehm BO, Todd JA (2011) Genetic analysis of adult-onset autoimmune diabetes. Diabetes 60: 2645–2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Philip B, Isabel W (2011) Association of cytotoxic T lymphocyte-associated antigen 4 gene single nucleotide polymorphism with type 1 diabetes mellitus in Madurai population of Southern India. Indian J Hum Genet 17: 85–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Plagnol V, Howson JM, Smyth DJ, Walker N, Hafler JP, et al. (2011) Genome-wide association analysis of autoantibody positivity in type 1 diabetes cases. PLoS Genet 7: e1002216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Reddy MV, Wang H, Liu S, Bode B, Reed JC, et al. (2011) Association between type 1 diabetes and GWAS SNPs in the southeast US Caucasian population. Genes Immun 12: 208–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ei Wafai RJ, Chmaisse HN, Makki RF, Fakhoury H (2011) Association of HLA class II alleles and CTLA-4 polymorphism with type 1 diabetes. Saudi J Kidney Dis Transpl 22: 273–281. [PubMed] [Google Scholar]
- 64. Horie I, Kawasaki E, Ando T, Kuwahara H, Abiru N, et al. (2012) Clinical and genetic characteristics of autoimmune polyglandular syndrome type 3 variant in the Japanese population. J Clin Endocrinol Metab 97: E1043–1050. [DOI] [PubMed] [Google Scholar]
- 65. Mosaad YM, Elsharkawy AA, El-Deek BS (2012) Association of CTLA-4 (+49A/G) Gene Polymorphism with Type 1 Diabetes Mellitus in Egyptian Children. Immunol Invest 41: 28–37. [DOI] [PubMed] [Google Scholar]
- 66. Villano MJ, Huber AK, Greenberg DA, Golden BK, Concepcion E, et al. (2009) Autoimmune thyroiditis and diabetes: dissecting the joint genetic susceptibility in a large cohort of multiplex families. J Clin Endocrinol Metab 94: 1458–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kouki T, Sawai Y, Gardine CA, Fisfalen ME, Alegre ML, et al. (2000) CTLA-4 gene polymorphism at position 49 in exon 1 reduces the inhibitory function of CTLA-4 and contributes to the pathogenesis of Graves disease. J Immunol 165: 6606–6611. [DOI] [PubMed] [Google Scholar]
- 68. Ueda H, Howson JM, Esposito L, Heward J, Snook H, et al. (2003) Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature 423: 506–511. [DOI] [PubMed] [Google Scholar]
- 69. Atabani SF, Thio CL, Divanovic S, Trompette A, Belkaid Y, et al. (2005) Association of CTLA4 polymorphism with regulatory T cell frequency. Eur J Immunol 35: 2157–2162. [DOI] [PubMed] [Google Scholar]
- 70. Chen Z, Fei M, Fu D, Zhang L, Ma Y, et al. (2013) Association between cytotoxic T lymphocyte antigen-4 polymorphism and type 1 diabetes: a meta-analysis. Gene 516: 263–70. [DOI] [PubMed] [Google Scholar]
- 71. Si X, Zhang X, Luo Y, Tang W (2012) Association between the CTLA-4 +49A/G polymorphism and type 1 diabetes: a meta-analysis. Genet Test Mol Biomarkers 16: 1336–42. [DOI] [PubMed] [Google Scholar]
- 72. Tang ST, Tang HQ, Zhang Q, Wang CJ, Wang YM, et al. (2012) Association of cytotoxic T-lymphocyte associated antigen 4 gene polymorphism with type 1 diabetes mellitus: a meta-analysis. Gene 508: 165–87. [DOI] [PubMed] [Google Scholar]
- 73. Chang WW, Zhang L, Su H, Jin YL, Chen Y, et al. (2012) Need for clarification of data in the recent meta-analysis about cytotoxic T-lymphocyte associated antigen 4 gene polymorphism and type 1 diabetes mellitus. Gene doi:10.1016/j.gene.2012.12.035 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
Flow chart of literature search for studies examining CTLA4 gene polymorphism and risk of T1D.
(TIF)
Funnel plot of studies of the G49A polymorphism of CTLA4 and T1D showing a possible excess of smaller studies with strikingly positive findings beyond the 95% CI.
(TIF)
Funnel plot for the association between CTLA4 C60T polymorphism and T1D risk.
(TIF)
Meta-analysis of haplotype combinations between G49A, and C60T polymorphisms of CTLA4 gene and T1D risk.
(DOCX)