Abstract
Background
Metabolic syndrome is a cluster of metabolic risk factors including dyslipidemia, impaired glucose tolerance, hypertension and central obesity. BDNF (Brain-derived neurotrophic factor) and leptin have been implied in the energy homeostasis. The purposes of this study were to examine concentrations of leptin, BDNF and biochemical parameters in metabolic-syndrome subjects and healthy controls, and also to search for associations of leptin gene (LEP) G2548A, leptin receptor gene (LEPR) Gln223Arg, and BDNF gene (BDNF) Val66Met polymorphisms with leptin levels, BDNF levels and metabolic syndrome among Thais.
Methods
The case-controlled design was performed using 322 Thai volunteers (160 metabolic-syndrome subjects; 162 controls) during the health screening program. Metabolic syndrome was assessed by using the modified National Cholesterol Education Program, Adult Treatment Panel III criteria. The levels of leptin, BDNF, insulin, glucose and lipids were measured in samples. Genotyping of LEP G2548A, LEPR Gln223Arg and BDNF Val66Met was carried out using polymerase chain reaction-restriction fragment length polymorphism technique.
Results
Serum leptin levels were significantly higher in the metabolic-syndrome group than the control group (p < 0.01), but the BDNF difference between them was not significant. Significant associations of LEPR Gln223Arg polymorphism were found with leptin and glucose levels (p < 0.05), after adjusting for potential covariates. This LEPR polymorphism in the metabolic-syndrome group was also significantly more frequent than in the control group (p < 0.05). However, other gene polymorphisms, LEP G2548A and BDNF Val66Met, showed no significant relationship with leptin levels, BDNF levels or metabolic syndrome.
Conclusion
These findings suggest leptin levels are linked with metabolic syndrome. LEPR Gln223Arg polymorphism impacted leptin concentrations, and this gene polymorphism may influence susceptibility to metabolic syndrome among Thais.
Keywords: Leptin, BDNF, Gene polymorphisms, Metabolic syndrome
Introduction
Metabolic syndrome (MS), which involves abdominal obesity, dyslipidemia, hypertension, and glucose tolerance represents one of the world’s most challenging health problems, and is an important risk factor for type 2 diabetes and cardiovascular diseases [1]. Leptin, an adipocytokine, is a key metabolic regulator and acts to reduce food intake, increase energy expenditure, and modulate immune and inflammatory responses by binding and activating the leptin receptor [2]. Serum leptin is proportional to body adiposity [3]. There are several studies indicating links of leptin gene (LEP) and leptin receptor gene (LEPR) variations with obesity [4], insulin resistance [5], and type 2 diabetes [6]. The gene polymorphisms of LEP G2548A (rs7799039: guanine > adenine) and LEPR Gln223Arg (rs1137101: glutamine > arginine) are studied widely because LEP G2548A has been associated with leptin production and secretion [7], and LEPR Gln223Arg has been associated with impaired signaling capacity of the leptin receptor [8]. However, evidence for an association of LEP G2548A and LEPR Gln223Arg with leptin concentrations and MS is inconsistent [9-11]. Moreover, the relationship of LEP G2548A and LEPR Gln223Arg gene polymorphisms with MS among Thais has not been investigated to date.
Brain-derived neurotrophic factor (BDNF) is a member of the neurotrophic factor family and is one of the molecules that mediate the function in the control of energy balance. The associations of BDNF and leptin with functions involved in energy homeostasis have been studied [12]. Previous studies have shown links between BDNF concentrations and obesity and diabetes [13,14]. The most common BDNF gene (BDNF) polymorphism, Val66Met (rs6265: valine > methionine), affects intracellular trafficking of pro-BDNF, and also decreases secretion and production of the mature BDNF with the Met allele [15]. However, previous studies have shown inconsistent results in relation to BDNF concentrations in MS subjects [16,17], and information on BDNF Val66Met gene polymorphism and MS is scarce. Therefore, the aims of this study were to examine concentrations of leptin, BDNF and biochemical parameters in MS subjects and healthy controls, and to search for links of LEP G2548A, LEPR Gln223Arg and BDNF Val66Met gene polymorphisms with leptin levels, BDNF levels and MS among Thais.
Methods
Following approval by the Ethics Committee of Rangsit University (RSEC No.016/53), Thailand and in accordance with the Declaration of Helsinki, this case-control study enrolled 322 Thai volunteers aged between 24 to 64 years and living in urban and suburban residential areas of Bangkok, Thailand. Among them, 162 healthy controls (86 male, 76 female) and 160 MS subjects (73 male, 87 female) were chosen during the health screening program among check-up subjects in March 2011- February 2012 and the statistical power in our sample size calculation was 80% at alpha =0.05. A physical examination and medical history check on all subjects was performed. Subjects with a history of liver, kidney and cardiovascular diseases were excluded from the study. Metabolic syndrome was defined using the modified National Cholesterol Education Program/Adult Treatment Panel III (NCEP/ATP III) criteria [18]. The new cut-off for waist circumference in the Asia-Pacific Region, instead of the original cut-off in the ATP III criteria, was used. Adoption of the new cut-off point for fasting plasma glucose criterion has already been reported (> 100 mg/dl). The modified NCEP/ATP III definition required at least three of the following: raised triglyceride (TG) levels: > 150 mg/dl, reduced high-density lipoprotein cholesterol (HDL-C): < 40 mg/dl in men, and < 50 mg/dl in women, raised fasting plasma glucose: > 100 mg/dl, raised waist circumference (WC): > 90 cm in Asian men and > 80 cm in Asian women or raised blood pressure: systolic blood pressure > 130 mmHg or diastolic blood pressure > 85.
Anthropometric measurements
The body weight of each subject was measured using a carefully calibrated beam balance (Detecto®, Cardinal Detecto Scale Manufacturing, MO). Height was measured using a vertical measuring rod. Body mass index (BMI) was conventionally expressed as weight (kg)/height (m2). Waist and hip circumferences were measured. Waist and hip circumferences were calculated for waist and hip circumference ratio. Furthermore, this study had also determined triceps skin-fold thickness (TSF) and arm circumference (AC) because TSF was suggested as the simple predictor of body density and hence percentage total body fat [19] and AC measurement could reflect adult nutritional status as defined by BMI [20]. The upper arm length midpoint mark was used to measure TSF and AC. The TSF was measured by using the Holtain Skinfold Caliper. Blood pressure (BP) was measured by a nurse after 5 to 10 minutes’ rest in the sitting position.
Laboratory measurements
10 ml of venous blood was taken from subjects in the morning after overnight fast. Glucose, TG, HDL-C levels were measured using enzymatic methods by DADE Dimension AR®. Leptin and insulin levels were measured using a radioimmunoassay kit from Linco Research (St Louis, USA). Serum BDNF levels were determined using an ELISA protocol, according to the manufacturer’s instructions (DBD00; R & D Systems, Europe).
Polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) technique
DNA was extracted from EDTA-treated whole blood by FlexiGene DNA kit (Qiagen, Hilden, Germany). Genotyping of BDNF Val66Met, LEP G2548A and LEPR Gln223Arg was carried out using PCR-RFLP assay. DNA fragments were amplified by PCR (PE Applied Biosystems). For analysis of the BDNF Val66Met polymorphism, the following primers were used:
Forward primer – 5′-ATCCGAGGACAAGGTGGC-3′
Reverse primer – 5′-CCTCATGGACATGTTTGCAG-3′
A 50 μl PCR reaction was performed according to the protocol described by Matsushita et al.[21]. For analysis of the LEP G2548A polymorphism, the following primers were used:
Forward primer – 5′-TTTCTGTAATTTTCCCGTGAG-3′
Reverse primer – 5′-AAAGCAAAGACAGGCATA AAAA-3′
A 50 μl PCR reaction was conducted, according to the protocol described by Boumaiza et al.[10]. For analysis of the LEPR Gln223Arg polymorphism, the following primers were used:
Forward primer – 5′-ACCCTTTAAGCTGGGTGTCCCAAATAG-3′
Reverse primer – 5′-AGCTAGCAAATATTTTTGTAAGCAATT-3′
A 50 μl PCR reaction was conducted, according to the protocol described by Duarte et al.[4]. PCR products (304 bp for BDNF, 242 bp for LEP and 421 bp for LEPR) were detected on 2% agarose gel containing ethidium bromide. Aliquots of the PCR products were digested with 8 U PmlI, HhaI and MspI restriction enzymes for the BDNF Val66Met, the LEP G2548A, and the LEPR Gln223Arg polymorphisms, respectively. Alleles were visualized as fragments by electrophoresis through an ethidium bromide-stained 2.5% agarose gel. Distribution patterns of the BDNF Val66Met, the LEP G2548A, and the LEPR Gln223Arg polymorphisms were shown in Figures 1, 2, 3.
Figure 1.
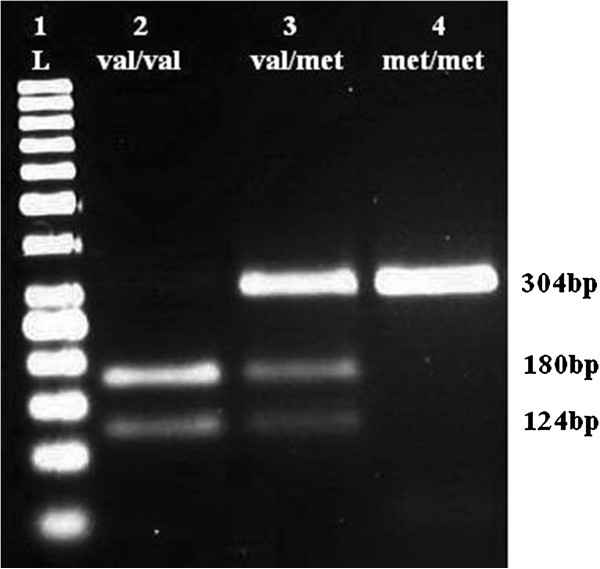
Restriction fragments length polymorphism (RFLP) distribution pattern of BDNF Val66Met polymorphism. Lane 1 = ladder (50 bp), Lane 2 = Val/Val (124 and 180 bp), Lane 3 = Val/Met (304, 180 and 124 bp), Lane 4 = Met/Met (304 bp).
Figure 2.
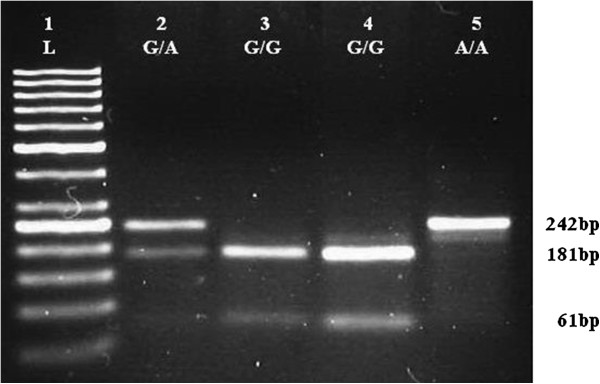
Restriction fragments length polymorphism (RFLP) distribution pattern of LEP G2548A polymorphism. Lane 1 = ladder (50 bp), Lane 2 = G/A (242,181, and 61 bp), Lane 3-4 = G/G (181 and 61 bp), Lane 5 = A/A (242 bp).
Figure 3.
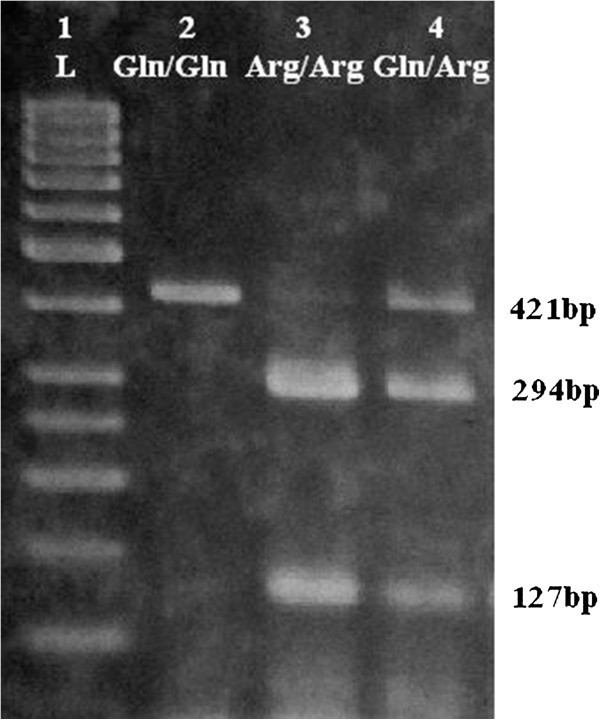
Restriction fragments length polymorphism (RFLP) distribution pattern of LEPR Gln223Arg polymorphism. Lane 1 = ladder (50 bp), Lane 2 = Gln/Gln (421 bp), Lane 3 = Arg/Arg (294 and 127 bp), Lane 4 = Gln/Arg (421, 294 and 127 bp).
Statistical analysis
Statistical analysis was performed using SPSS for Windows version 11.5 (SPSS, Chicago, IL). Median, range and 95% confidence intervals (CI) were calculated. The difference between the two groups was compared by Mann-Whitney U/Wilcoxon Rank Sum W test. The differences in genotypic and allelic frequencies of the two groups were assessed by Chi-square test. The Minitab statistical computer program was used to calculate the odds ratio (OR). Spearman rank was used to calculate correlations among the variables. To assess links between MS as a dependent variable and other potential factors, logistic regression was applied. A p-value < 0.05 was considered statistically significant. Goodness-of-fit of the logistic regression models was established by Hosmer-Lemeshow test.
Results
Anthropometric and biochemical data of the MS and control groups are shown in Table 1. Serum leptin levels were significantly higher in the MS group than the control group (p < 0.01), while lower HDL-C levels were observed in MS group as compared to the control group (p < 0.01). Meanwhile, there were significant differences between the MS and control groups in glucose, insulin, TG, systolic BP, diastolic BP, BMI, waist/hip ratio, WC, AC and TSF (p < 0.01). The BDNF difference between the MS and control groups was not significant. Spearman’s Rank correlation test results are shown in Table 2. Leptin level was positively correlated with age, insulin, TG, systolic BP, diastolic BP, BMI, waist/hip ratio, WC, AC and TSF, but negatively correlated with HDL-C level (p < 0.05). Moreover, BDNF level correlated significantly with age, AC and TSF, as shown in Table 2.
Table 1.
Comparison of anthropometric, biochemical variables and age between individuals with and without MS
|
Variables |
Control (n = 162) |
MS (n = 160) |
p-value |
|---|---|---|---|
| Median (range) | Median (range) | ||
| Age (years) |
46.0 (24.0-64.0) |
48.0 (28.0-63.0) |
0.241 |
| Leptin (ng/mL) |
6.2 (2.2-32.3) |
11.4 (2.9-36.8) |
<0.001** |
| BDNF (ng/mL) |
11.7 (2.8-29.2) |
11.5 (2.7-27.8) |
0.946 |
| Glucose (mg/dL) |
84.0 (66.0-113.0) |
95.5 (70.0-134.0) |
<0.001** |
| Insulin (μU/mL) |
11.7 (4.5-30.7) |
15.8 (5.6-37.3) |
<0.001** |
| TG (mg/dL) |
84.0 (58.0-270.0) |
169.0 (63.0-302.0) |
<0.001** |
| HDL-C (mg/dL) |
50.0 (28.0-77.0) |
40.0 (27.0-71.0) |
<0.001** |
| Systolic BP (mmHg) |
120.0 (95.0-160.0) |
130.0 (100.0-170.0) |
<0.001** |
| Diastolic BP (mmHg) |
74.0 (60.0-95.0) |
80.0 (64.0-100.0) |
<0.001** |
| BMI (kg/m2) |
22.8 (18.9-35.5) |
26.5 (19.5-37.9) |
<0.001** |
| Waist/Hip ratio |
0.82 (0.69-0.97) |
0.88 (0.72-0.99) |
<0.001** |
| WC (cm) |
77.0 (61.5-101.3) |
91.0 (66.5-106.0) |
<0.001** |
| AC (cm) |
27.5 (21.8-34.0) |
30.2 (22.9-38.5) |
<0.001** |
| TSF (mm) | 17.9 (6.5-37.5) | 26.0 (8.5-40.9) | <0.001** |
Abbreviation: BDNF Brain-derived neurotrophic factor, TG triglyceride, HDL-C high-density lipoprotein cholesterol.
BMI Body mass index, WC waist circumference, AC arm circumference, TSF triceps skin-fold thickness.
Significant level;** = p < 0.01 by Mann-Whitney U / Wilcoxon Rank Sum W test (2-tailed).
Table 2.
Correlation coefficients of leptin as well as BDNF with other parameters, in all subjects
| Variables | Leptin | BDNF |
|---|---|---|
| Leptin |
1 |
0.021 |
| BDNF |
0.021 |
1 |
| Age |
0.233** |
-0.280** |
| Glucose |
0.097 |
-0.059 |
| Insulin |
0.152* |
-0.127 |
| TG |
0.236** |
0.127 |
| HDL-C |
-0.214** |
-0.057 |
| Systolic BP |
0.358** |
-0.018 |
| Diastolic BP |
0.199** |
-0.015 |
| BMI |
0.383** |
0.036 |
| Waist/Hip ratio |
0.301** |
0.027 |
| WC |
0.293** |
0.052 |
| AC |
0.408** |
0.314** |
| TSF | 0.516** | 0.224** |
Abbreviation: BDNF Brain-derived neurotrophic factor, TG triglyceride,
HDL-C high-density lipoprotein cholesterol, BMI Body mass index, WC waist circumference, AC arm circumference, TSF triceps skin-fold thickness.
Significant levels;* = p < 0.05 by Spearman’s rank correlation (2-tailed).
**= p < 0.01 by Spearman’s rank correlation (2-tailed).
To evaluate the associations of single-nucleotide polymorphisms (SNPs), LEPR Gln223Arg, LEP G2548A and BDNF Val66Met, with leptin level, BDNF level and metabolic features we performed multivariate analysis. Significant associations of LEPR Gln223Arg polymorphism were found with leptin and glucose levels, after adjusting for potential covariates including age, sex, weight and smoking status; the results are shown in Table 3. There were no significant associations of LEP G2548A and BDNF Val66Met polymorphisms with leptin level, BDNF level and metabolic features (p > 0.05); the results are shown in Table 3. The genotypes of SNPs LEPR Gln223Arg, LEP G2548A and BDNF Val66Met were in Hardy-Weinberg equilibrium (HWE) (p > 0.05) and the values of chi-square and degrees of freedom were shown in Table 4. Moreover, the results in genotypic and allelic frequencies of LEPR Gln233Arg, LEP G2548A, and BDNF Val66Met polymorphisms in MS and control subjects are also shown in Table 4. There were significant differences in genotypic and allelic frequencies of LEPR Gln233Arg polymorphism between MS and control groups (p < 0.05), but genotypic and allelic distributions of LEP G2548A and BDNF Val66Met polymorphisms between these two groups were not significantly different (p > 0.05).
Table 3.
Adjusted odds ratio for LEPR Gln223Arg, LEP G2548A and BDNF Val66Met polymorphisms with leptin level, BDNF level and metabolic features
|
Variables |
SNP
LEPR
Gln223Arg |
p-value |
SNP
LEP
G2548A |
p-value |
SNP
BDNF
Val66Met |
p-value |
|---|---|---|---|---|---|---|
| OR (95% CI) | OR (95% CI) | OR (95% CI) | ||||
| Systolic BP |
0.989 (0.952-1.027) |
0.568 |
1.030 (0.982-1.090) |
0.318 |
1.022 (0.978-1.068) |
0.331 |
| Diastolic BP |
1.019 (0.968-1.072) |
0.474 |
1.000 (0.932-1.073) |
0.999 |
1.017 (0.972-1.075) |
0.447 |
| WC |
0.991 (0.972-1.046) |
0.437 |
1.044 (0.966-1.129) |
0.270 |
0.968 (0.899-1.043) |
0.410 |
| TG |
0.999 (0.993-1.004) |
0.635 |
1.002 (0.994-1.009) |
0.636 |
0.998 (0.991-1.007) |
0.378 |
| HDL-C |
1.000 (0.974-1.038) |
0.994 |
0.992 (0.945-1.041) |
0.745 |
1.002 (0.965-1.029) |
0.722 |
| Glucose |
1.047 (1.004-1.092) |
0.033* |
1.021 (0.989-1.065) |
0.340 |
0.989 (0.965-1.019) |
0.354 |
| Insulin |
1.016 (0.997-1.056) |
0.099 |
0.985 (0.944-1.027) |
0.473 |
1.009 (0.972-1.050) |
0.600 |
| Leptin |
1.192 (1.038-1.379) |
0.009** |
0.924 (0.816-1.039) |
0.209 |
0.963 (0.873-1.050) |
0.903 |
| BDNF | 0.997 (0.950-1.045) | 0.737 | 0.986 (0.925-1.051) | 0.576 | 0.980 (0.943-1.045) | 0.909 |
Abbreviation: BDNF Brain-derived neurotrophic factor, TG triglyceride, HDL-C high-density lipoprotein cholesterol.
WC waist circumference, LEP leptin gene, LEPR leptin receptor gene, BDNF Brain-derived neurotrophic factor gene.
Significant levels;* = p < 0.05, **= p < 0.01.
Table 4.
Genotypic and allelic distribution of LEPR Gln233Arg, LEP G2548A, and BDNF Val66Met polymorphisms in MS and control subjects
|
MS n (%) |
HWE of MS p-value a | Control n (%) | HWE of control p-value* | Genotypic or allelic p-value b | |
|---|---|---|---|---|---|
|
LEPR
Gln233Arg | |||||
| Genotype | |||||
| Arg/Arg |
20 (12.5) |
0.399 |
17 (10.5) |
0.295 |
0.044* |
| Arg/Gln |
80 (50.0) |
(df = 1) |
62 (38.3) |
(df = 1) |
|
| Gln/Gln |
60 (37.5) |
(χ2 = 0.71) |
83 (51.2) |
(χ2 = 1.09) |
|
| Allele | |||||
| Arg |
120 (0.375) |
|
96 (0.296) |
|
0.034* |
| Gln |
200 (0.625) |
|
228 (0.704) |
|
|
|
LEP
G2548A | |||||
| Genotype | |||||
| A/A |
58 (36.2) |
0.638 |
55 (34.0) |
0.483 |
0.439 |
| A/G |
79 (49.4) |
(df = 1) |
75 (46.3) |
(df = 1) |
|
| G/G |
23 (14.4) |
(χ2 = 0.22) |
32 (19.7) |
(χ2 = 0.49) |
|
| Allele | |||||
| A |
195 (0.609) |
|
185 (0.571) |
|
0.321 |
| G |
125 (0.391) |
|
139 (0.429) |
|
|
|
BDNF
Val66Met | |||||
| Genetype | |||||
| Met/Met |
32 (20.0) |
0.479 |
30 (18.5) |
0.544 |
0.327 |
| Met/Val |
84 (52.5) |
(df = 1) |
75 (46.3) |
(df = 1) |
|
| Val/Val |
44 (27.5) |
(χ2 = 0.50) |
57 (35.2) |
(χ2 = 0.37) |
|
| Allele | |||||
| Met |
148 (0.463) |
|
135 (0.417) |
|
0.241 |
| Val | 172 (0.537) | 189 (0.583) | |||
Abbreviation: HWE Hardy-Weinberg equilibrium, LEP leptin gene, LEPR leptin receptor gene, BDNF Brain-derived neurotrophic factor gene.
Significant level;* = p < 0.05.
aBased on the results of chi-square test.
bBased on the results of chi-square test for the comparison between MS and control groups.
Association between LEPR Gln233Arg, LEP G2548A and BDNF Val66Met polymorphisms and MS among study subjects are shown in Table 5. Polymorphism of LEPR Gln223Arg showed a significant link to MS (OR = 1.8, p < 0.05), and Gln/Arg with Arg/Arg genotype frequencies in the MS and control groups were 62.5% and 48.8%, respectively. In contrast, LEP G2548A and BDNF Val66Met polymorphisms were not related to MS (p > 0.05).
Table 5.
Association between LEPR Gln233Arg, LEP G2548A and BDNF Val66Met polymorphisms and MS
|
SNPs |
Genotype frequencies |
OR (95% CI) |
p-value
a
|
|
|---|---|---|---|---|
| MS n (%) | Control n (%) | |||
|
LEPR
Gln223Arg |
|
|
|
|
| Genotype |
|
|
|
|
| Arg/Arg + Arg/Gln |
100 (62.5) |
79 (48.8) |
1.8 (1.2-2.7) |
0.013* |
| Gln/Gln |
60 (37.5) |
83 (51.2) |
|
|
|
LEP
G2548A |
|
|
|
|
| Genotype |
|
|
|
|
| A/A + A/G |
137 (85.6) |
130 (80.2) |
1.5 (0.8-2.6) |
0.200 |
| G/G |
23 (14.4) |
32 (19.8) |
|
|
|
BDNF
Val66Met |
|
|
|
|
| Genotype |
|
|
|
|
| Met/Met + Met/Val |
116 (72.5) |
105 (64.8) |
1.4 (0.8-2.5) |
0.230 |
| Val/Val | 44 (27.5) | 57 (35.2) | ||
Abbreviation: LEP leptin gene, LEPR leptin receptor gene, BDNF Brain-derived neurotrophic factor gene.
Significant level;* = p < 0.05.
aBased on the results of chi-square test.
Logistic regression results for possible associations between MS and age, sex, TSF, waist/hip ratio, insulin, leptin, LEP G2548A, LEPR Gln223Arg and BDNF Val66Met, are shown in Table 6. Of these variables, leptin, LEPR Gln223Arg, TSF and waist/hip ratio were significantly related to MS (p < 0.05). The Hosmer-Lemeshow Goodness-of-Fit test (χ2 = 0.896, p > 0.05) was not statistically significant; the fit between the predictive model and the data was acceptable.
Table 6.
Logistic regression analysis when MS was used as a dependent variable, and age, sex, TSF, waist/hip ratio, insulin, leptin, LEPR Gln223Arg, LEP G2548A and BDNF Val66Met, were taken as independent variables
| Variables | β | p-value | OR | (95% CI) |
|---|---|---|---|---|
| Leptin |
0.46 |
0.007* |
1.91 |
1.25-3.03 |
|
LEPR Gln223Arg |
2.37 |
0.033* |
9.07 |
1.22-37.14 |
| Waist/hip ratio |
5.58 |
<0.001* |
22.04 |
2.42-91.36 |
| TSF |
0.17 |
0.029* |
1.31 |
1.03-1.95 |
| Sex |
-2.34 |
0.335 |
0.20 |
0.02-6.26 |
| Age |
0.03 |
0.625 |
0.97 |
0.89-1.14 |
| Insulin |
0.07 |
0.101 |
1.10 |
0.98-1.21 |
|
BDNF Val66Met |
0.88 |
0.179 |
2.39 |
0.74-8.54 |
| LEP G2548A | 1.84 | 0.124 | 6.03 | 0.80-26.11 |
Abbreviation: LEP leptin gene, LEPR leptin receptor gene, BDNF Brain-derived neurotrophic factor gene, TSF triceps skin-fold thickness.
Significant level; *= p < 0.05.
Discussion
To the best of our knowledge, this is the first study to examine the links of leptin level, BDNF level, and polymorphisms of LEPR Gln223Arg, LEP G2548A and BDNF Val66Met with MS among Thais. The key observations of this study were that leptin levels were significantly higher in the MS group than in the control group, and that the LEPR Gln223Arg polymorphism showed a significant link to leptin concentrations and MS among Thais. In contrast, LEP G2548A and BDNF Val66Met polymorphisms were not related to MS.
MS is a cluster of risk factors predictive of future cardiovascular diseases and type 2 diabetes mellitus [1]. Factors which appear to promote the development of MS include genetics, obesity, physical inactivity and an unhealthy diet. Mechanisms causing the onset of this syndrome, however, are still not clearly understood. Leptin is an adipocyte-derived signaling factor that has an important role in metabolic control, such as in stimulating glucose uptake, fatty acid oxidation, and reducing food intake [2,22]. Moreover, circulating leptin levels are closely related to stored adipose tissues [3] and metabolic function [22]. However, most previous studies have focused on the relationship between leptin and obesity, and each component of MS, instead of a cluster of MS components. Therefore, the present study investigated links between leptin and MS among Thais, especially. Our results were in accordance with previous studies in Korean populations [23] and Lebanese volunteers [24], in that serum leptin was associated with MS. Bremer et al. also showed increased amounts of subcutaneous adipose tissue secretion and circulating leptin concentrations in subjects with nascent MS [25]. Increased leptin concentration in MS may represent a form of leptin resistance, as in obese subjects [26]; obesity is one of the risk factors for MS. Therefore, alternation of leptin levels may be a part of the etiology of MS.
Leptin exerts a number of significant influences, by binding and activating specific leptin receptors in the hypothalamus and other organs including pancreatic β cells, adipose tissue, and muscle. Previous studies reported LEPR polymorphisms were associated with glucose metabolism [27] and insulin resistance [5], possibly leading to a predisposition towards greater MS risk. LEPR Gln223Arg polymorphism has been associated with impaired signaling capacity of the leptin receptor [8]. LEPR Gln223Arg polymorphism has also been linked with impaired glucose tolerance and conversion to type 2 diabetes [27]. Our study was therefore interested in this position of the LEPR gene polymorphism. Present data points to a relationship between LEPR Gln223Arg polymorphism and increased leptin and glucose levels in Thai subjects, similar to results in Dutch adults [28] and Romanian subjects [29]. In contrast to our results, however, LEPR Gln223Arg polymorphism demonstrated no relationship with leptin levels in Turkish children [11]. Moreover, few studies have determined a correlation between LEPR gene polymorphism and MS. Our findings among Thais were in accordance with results from a sample of elderly Brazilian subjects [9], which showed links between LEPR Gln223Arg polymorphism and MS. On the other hand, the study in Turkish children did not find any relation between LEPR Gln223Arg polymorphism and MS [11]. Therefore, our study demonstrated that LEPR Gln223Arg polymorphism in Thai subjects was associated with increased MS risk, which may be explained by increasing glucose and leptin levels. Although the pathogenesis of MS and each of its components is complex and not well understood, our investigation proposes that LEPR Gln223Arg polymorphism associated with increased leptin levels may represent a modified functional leptin receptor and may take part in the development of MS. The previous studies also suggested that LEPR Gln223Arg polymorphism has been associated with insulin resistance capacity and an altered leptin-binding activity [30] and the leptin can modulates insulin secretion and action via leptin receptors in pancreatic β cells [31].
Although many researches indicate a role for genetic susceptibility to the MS and our results supporting the relationship between the LEPR Gln223Arg polymorphism and increased MS risk among Thais, lifestyle factors have also found to be affecting factors in the pathogenesis and progression of MS. Thailand has importantly experienced economic transition and its structure has gradually changed from the traditional agricultural surroundings to an industrialized structure. The dietary intake pattern in Thais has changed from traditional high-carbohydrate diets, which rely heavily on rice and vegetables, to diets high in fat and sugar; physical activity has progressively declined [32]. These transitions have consequently resulted in increasing the prevalence of MS in Thailand [33].Therefore, lifestyle interventions including eating healthy diet and improving physical activity should be regarded as important management for reducing the development of MS among Thais, especially in SNP LEPR Gln223Arg subjects. With regard to links between LEP G2548A gene polymorphism and leptin levels, the previous study reported a common gene polymorphism in the promoter LEP G2548A influencing leptin expression and adipose tissue secretion of leptin [7]. However, studies yielded inconsistent results; data in Egyptian subjects [34] and Romanian subjects [29] points to a relationship between LEP 2548GG variants and serum leptin levels, meanwhile studies in Melanesian and Micronesian Solomon Islanders [35] found no relationship between LEP G2548A polymorphism and leptin levels, consistent with our results among Thais. In investigating the links between LEP G2548A polymorphism and MS, LEP G2548A polymorphism was shown to be significantly associated with MS in Tunisian volunteers [10] and Egyptian subjects [34], while no links were found in our study. Investigations among Romanian subjects also found no association of the LEP G2548A polymorphism with common obesity-related variables and metabolic traits [29]. It is possible that inconsistencies among the results of the LEP and LEPR gene polymorphisms in these studies are the result of the different genetic backgrounds or environmental conditions of the populations studied.
BDNF is involved in regulating energy homoeostasis, blood glucose levels, and eating behaviors, and it appears to function downstream of the leptin-melanocortin signaling pathway [13,36]. BDNF is expressed at high levels in the ventromedial hypothalamus (VMH) where its expression is regulated by nutritional state and by melanocortin-4 receptor (MC4R) signaling [36]. Moreover, Komori et al. demonstrated that administration of leptin causes an increase in expression of BDNF mRNA and protein in the VMH [37]. The precise mechanism that mediates BDNF expression by leptin remains to be elucidated and there are two evidences that have been proposed. The first possibility is that the LEP- LEPR interaction directly triggers a signal transduction cascade that induces BDNF in the VMH. The second option involves the leptin-mediated production of the melanocortin precursor α-melanocyte stimulating hormone (α-MSH) in the arcuate nucleus, which activates BDNF in the VMH via MC4R [13]. However, the mechanistic implication of the association between leptin, BDNF and the LEPR gene in MS is unclear. Our results may imply that Thai MS subjects have related to LEPR Gln223Arg polymorphism and increased leptin levels representing a form of leptin resistant. These defects may be involved in the LEP-LEPR interaction in mechanisms induced BDNF expression by leptin signaling. Although previous studies have investigated a correlation between BDNF levels and MS, these investigations have shown inconsistent results of BDNF levels in subjects with MS [16,17,38,39]. In examining links between BDNF concentrations and BDNF Val66Met gene polymorphisms with MS among Thais, the present study found neither BDNF concentrations nor BDNF gene polymorphism relates to MS. Our results were in line with studies in Spanish and Taiwanese subjects, in that, BDNF concentrations did not correlate with MS and any metabolic syndrome component [16,38]. In contrast, other reports have found BDNF levels to be linked with MS, and with several features of MS [17,39]. However, data on BDNF Val66Met gene polymorphism with MS is scarcely available. Indeed, ours is the first publication to determine these associations among Thais. Previous studies found that BDNF Val66Met gene polymorphism influenced BDNF expression and/or activity [15], and was related to a higher risk of becoming obese [40]. One study among Caucasian subjects found no significant association between Val66Met gene polymorphism and MS [41]. Similarly, our results confirm this polymorphism not to be linked with MS or with features of MS. Therefore, our findings suggest that BDNF concentrations and BDNF Val66Met polymorphism are not involved in the pathophysiology of MS among Thais. However, the relationship between BDNF levels and gene polymorphisms with MS is still not clearly understood, and further studies should be undertaken investigating different ethnicities, as well as other points of the BDNF gene.
There are a few limitations of our study. We did not analyze the whole gene of LEP, LEPR and BDNF with MS and further studies should be replicated our findings in larger sample numbers.
In conclusion, the present study found leptin concentrations of MS subjects to be higher than the controls. Although no correlations between MS with BDNF level or BDNF Val66Met were observed, there was an association between LEPR Gln223Arg polymorphism and leptin level and MS. Therefore, genetic polymorphism in the LEPR Gln223Arg appeared to affect the leptin concentration and the susceptibility to MS and these findings might play a significant function in the etiology of MS among the Thai population. Moreover, we believe further studies on MS genetics will help to develop preventive strategies and the role of other genetic and environmental factors should be studied in the other populations.
Competing interests
The authors have no conflicts of interest to declare.
Authors’ contributions
KS contributed to the conception and design of the study, collected specimens, carried out laboratory analysis and prepared the manuscript. KT carried out laboratory analysis and helped to draft the manuscript. RT collected specimens and interpretation of the data. All authors read and approved the final manuscript.
Contributor Information
Kanjana Suriyaprom, Email: kanjana.su@rsu.ac.th.
Rungsunn Tungtrongchitr, Email: rungsunn.tun@mahidol.ac.th.
Kittisak Thawnasom, Email: kittisak.th@rsu.ac.th.
Acknowledgments
The authors wish to express their sincere thanks to all volunteers and staff of the Faculty of Medical Technology, Rangsit University, as well as staff of the Department of Tropical Nutrition & Food Science, Faculty of Tropical Medicine, Mahidol University for their assistance in carrying out this research. Also, our thanks to Mr. Gary Hutton and Mr. Paul Adams for proofreading this manuscript. The project was supported by funds from Rangsit University and grant through the Center for Biopharmaceutical Development and Innovative Therapy, Mahidol University, Thailand.
References
- Di Chiara T, Argano C, Corrao S, Scaglione R, Licata G. Hypoadiponectinemia: a link between visceral obesity and metabolic syndrome. J Nutr Metab. 2012;2012:175245. doi: 10.1155/2012/175245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poeggeler B, Schulz C, Pappolla MA, Bodó E, Tiede S, Lehnert H, Paus R. Leptin and the skin: a new frontier. Exp Dermatol. 2010;19:12–18. doi: 10.1111/j.1600-0625.2009.00930.x. [DOI] [PubMed] [Google Scholar]
- Martins M do C, Lima Faleiro L, Fonseca A. Relationship between leptin and body mass and metabolic syndrome in an adult population. Rev Port Cardiol. 2012;31:711–719. doi: 10.1016/j.repc.2012.08.002. [DOI] [PubMed] [Google Scholar]
- Duarte SFP, Francischetti EA, Genelhu-Abreu V, Barroso SG, Braga JU, Cabello PH, Pimentel MM. p.Q223R leptin receptor polymorphism associated with obesity in Brazilian multiethnic subjects. Am J Hum Biol. 2006;18:448–453. doi: 10.1002/ajhb.20519. [DOI] [PubMed] [Google Scholar]
- Chiu KC, Chu A, Chuang L-M, Saad MF. Association of leptin receptor polymorphism with insulin resistance. Eur J Endocrinol. 2004;150:725–729. doi: 10.1530/eje.0.1500725. [DOI] [PubMed] [Google Scholar]
- Han HR, Ryu HJ, Cha HS, Go MJ, Ahn Y, Koo BK, Cho YM, Lee HK, Cho NH, Shin C, Shin HD, Kimm K, Kim HL, Oh B, Park KS. Genetic variations in the leptin and leptin receptor genes are associated with type 2 diabetes mellitus and metabolic traits in the Korean female population. Clin Genet. 2008;74:105–115. doi: 10.1111/j.1399-0004.2008.01033.x. [DOI] [PubMed] [Google Scholar]
- Hoffstedt J, Eriksson P, Mottagui-Tabar S, Arner P. A polymorphism in the leptin promoter region (-2548 G/A) influences gene expression and adipose tissue secretion of leptin. Horm Metab Res. 2002;34:355–359. doi: 10.1055/s-2002-33466. [DOI] [PubMed] [Google Scholar]
- Yiannakouris N, Yannakoulia M, Melistas L, Chan JL, Klimis-Zacas D, Mantzoros CS. The Q223R polymorphism of the leptin receptor gene is significantly associated with obesity and predicts a small percentage of body weight and body composition variability. J Clin Endocrinol Metab. 2001;86:4434–4439. doi: 10.1210/jcem.86.9.7842. [DOI] [PubMed] [Google Scholar]
- Gottlieb MG, Bodanese LC, Leite LE, Schwanke CH, Piccoli Jda C, da Rocha MI, da Cruz IB. Association between the Gln223Arg polymorphism of the leptin receptor and metabolic syndrome in free-living community elderly. Metab Syndr Relat Disord. 2009;7:341–348. doi: 10.1089/met.2008.0029. [DOI] [PubMed] [Google Scholar]
- Boumaiza I, Omezzine A, Rejeb J, Rebhi L, Ouedrani A, Ben Rejeb N, Nabli N, Ben Abdelaziz A, Bouslama A. Relationship between leptin G2548A and leptin receptor Q223R gene polymorphisms and obesity and metabolic syndrome risk in Tunisian volunteers. Genet Test Mol Biomarkers. 2012;16:726–733. doi: 10.1089/gtmb.2011.0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komşu-Ornek Z, Demirel F, Dursun A, Ermiş B, Pişkin E, Bideci A. Leptin receptor gene Gln223Arg polymorphism is not associated with obesity and metabolic syndrome in Turkish children. Turk J Pediatr. 2012;54:20–24. [PubMed] [Google Scholar]
- Tapia-Arancibia L, Rage F, Givalois L, Arancibia S. Physiology of BDNF: focus on hypothalamic function. Front Neuroendocrinol. 2004;25:77–107. doi: 10.1016/j.yfrne.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Rosas-Vargas H, Martínez-Ezquerro JD, Bienvenu T. Brain-derived neurotrophic factor, food intake regulation, and obesity. Arch Med Res. 2011;42:482–494. doi: 10.1016/j.arcmed.2011.09.005. [DOI] [PubMed] [Google Scholar]
- Krabbe KS, Nielsen AR, Krogh-Madsen R, Plomgaard P, Rasmussen P, Erikstrup C, Fischer CP, Lindegaard B, Petersen AM, Taudorf S, Secher NH, Pilegaard H, Bruunsgaard H, Pedersen BK. Brain-derived neurotrophic factor (BDNF) and type 2 diabetes. Diabetologia. 2007;50:431–438. doi: 10.1007/s00125-006-0537-4. [DOI] [PubMed] [Google Scholar]
- Chen Z-Y, Patel PD, Sant G, Meng C-X, Teng KK, Hempstead BL, Lee FS. Variant brain-derived neurotrophic factor (BDNF) (Met66) alters the intracellular trafficking and activity-dependent secretion of wild-type BDNF in neurosecretory cells and cortical neurons. J Neurosci. 2004;24:4401–4411. doi: 10.1523/JNEUROSCI.0348-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee IT, Lee WJ, Tsai IC, Liang KW, Lin SY, Wan CJ, Fu CP, Sheu WH. Brain-derived neurotrophic factor not associated with metabolic syndrome but inversely correlated with vascular cell adhesion molecule-1 in men without diabetes. Clin Chim Acta. 2012;413:944–948. doi: 10.1016/j.cca.2012.02.013. [DOI] [PubMed] [Google Scholar]
- Chaldakov GN, Fiore M, Stankulov IS, Hristova M, Antonelli A, Manni L, Ghenev PI, Angelucci F, Aloe L. NGF, BDNF, leptin, and mast cells in human coronary atherosclerosis and metabolic syndrome. Arch Physiol Biochem. 2001;09:357–360. doi: 10.1076/apab.109.4.357.4249. [DOI] [PubMed] [Google Scholar]
- Jesmin S, Islam MR, Islam AM, Mia MS, Sultana SN, Zaedi S, Yamaguchi N, Iwashima Y, Hiroe M, Watanabe T. Comprehensive assessment of metabolic syndrome among rural Bangladeshi women. BMC Public Health. 2012;12:49. doi: 10.1186/1471-2458-12-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seltzer CC, Goldman RF, Mayer J. The triceps skinfold as a predictive measure of body density and body fat in obese adolescent girls. Pediatrics. 1965;36:212–218. [PubMed] [Google Scholar]
- Collins S. Using middle upper arm circumference to assess severe adult malnutrition during famine. JAMA. 1996;276:391–395. doi: 10.1001/jama.1996.03540050051023. [DOI] [PubMed] [Google Scholar]
- Matsushita S, Kimura M, Miyakawa T, Yoshino A, Murayama M, Masaki T, Higuchi S. Association study of brain-derived neurotrophic factor gene polymorphism and alcoholism. Alcohol Clin Exp Res. 2004;28:1609–1612. doi: 10.1097/01.ALC.0000145697.81741.D2. [DOI] [PubMed] [Google Scholar]
- Wauters M, Considine RV, Van Gaal LF. Human leptin: from an adipocyte hormone to an endocrine mediator. Eur J Endocrinol. 2000;143:293–311. doi: 10.1530/eje.0.1430293. [DOI] [PubMed] [Google Scholar]
- Yun JE, Kimm H, Jo J, Jee SH. Serum leptin is associated with metabolic syndrome in obese and nonobese Korean populations. Metabolism. 2010;59:424–429. doi: 10.1016/j.metabol.2009.08.012. [DOI] [PubMed] [Google Scholar]
- Gannagé-Yared M-H, Khalife S, Semaan M, Fares F, Jambart S, Halaby G. Serum adiponectin and leptin levels in relation to the metabolic syndrome, androgenic profile and somatotropic axis in healthy non-diabetic elderly men. Eur J Endocrinol. 2006;155:167–176. doi: 10.1530/eje.1.02175. [DOI] [PubMed] [Google Scholar]
- Bremer AA, Jialal I. Adipose tissue dysfunction in nascent metabolic syndrome. J Obes. 2013;2013:393192. doi: 10.1155/2013/393192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpace PJ, Tümer N. Peripheral and hypothalamic leptin resistance with age-related obesity. Physiol Behav. 2001;74:721–727. doi: 10.1016/S0031-9384(01)00616-3. [DOI] [PubMed] [Google Scholar]
- Salopuro T, Pulkkinen L, Lindström J, Eriksson JG, Valle TT, Hämäläinen H, Ilanne-Parikka P, Keinänen-Kiukaanniemi S, Tuomilehto J, Laakso M, Uusitupa M. Genetic variation in leptin receptor gene is associated with type 2 diabetes and body weight: the finnish diabetes prevention study. Int J Obes. 2005;29:1245–1251. doi: 10.1038/sj.ijo.0803024. [DOI] [PubMed] [Google Scholar]
- Van Rossum CTM, Hoebee B, van Baak MA, Mars M, Saris WHM, Seidell JC. Genetic variation in the leptin receptor gene, leptin, and weight gain in young Dutch adults. Obes Res. 2003;11:377–386. doi: 10.1038/oby.2003.51. [DOI] [PubMed] [Google Scholar]
- Constantin A, Costache G, Sima AV, Glavce CS, Vladica M, Popov DL. Leptin G-2548A and leptin receptor Q223R gene polymorphisms are not associated with obesity in Romanian subjects. Biochem Biophys Res Commun. 2010;391:282–286. doi: 10.1016/j.bbrc.2009.11.050. [DOI] [PubMed] [Google Scholar]
- Utsunomiya K, Shinkai T, Sakata S, Hwang R, Yamada K, Chen HI, Fukunaka Y, Ohmori O, Nakamura J. Lack of association between the leptin receptor gene (LEPR) Gln223Arg polymorphism and late-onset Alzheimer disease. Alzheimer Dis Assoc Disor. 2010;24:101–103. doi: 10.1097/WAD.0b013e3181b982dc. [DOI] [PubMed] [Google Scholar]
- Seufert J, Kieffer TJ, Leech CA, Holz GG, Moritz W, Ricordi C, Habener JF. Leptin suppression of insulin secretion and gene expression in human pancreatic islets: implications for the development of adipogenic diabetes mellitus. J Clin Endocrinol Metab. 1999;84:670–676. doi: 10.1210/jcem.84.2.5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popkin BM. Urbanization, lifestyle changes and the nutrition transition. World Dev. 1999;27:1905–1916. doi: 10.1016/S0305-750X(99)00094-7. [DOI] [Google Scholar]
- Santibhavank P. Prevalence of metabolic syndrome in Nakhon Sawan population. J Med Assoc Thai. 2007;90:1109–1115. [PubMed] [Google Scholar]
- Abdel Hay RM, Rashed LA. Association between the leptin gene 2548G/A polymorphism, the plasma leptin and the metabolic syndrome with psoriasis. Exp Dermatol. 2011;20:715–719. doi: 10.1111/j.1600-0625.2011.01299.x. [DOI] [PubMed] [Google Scholar]
- Furusawa T, Naka I, Yamauchi T, Natsuhara K, Kimura R, Nakazawa M, Ishida T, Nishida N, Eddie R, Ohtsuka R, Ohashi J. The serum leptin level and body mass index in Melanesian and Micronesian Solomon Islanders: focus on genetic factors and urbanization. Am J Hum Biol. 2011;23:435–444. doi: 10.1002/ajhb.21124. [DOI] [PubMed] [Google Scholar]
- Xu B, Goulding EH, Zang K, Cepoi D, Cone RD, Jones KR, Tecott LH, Reichardt LF. Brain-derived neurotrophic factor regulates energy balance downstream of melanocortin-4 receptor. Nat Neurosci. 2003;6:736–742. doi: 10.1038/nn1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori T, Morikawa Y, Nanjo K, Senba E. Induction of brain-derived neurotrophic factor by leptin in the ventromedial hypothalamus. Neuroscience. 2006;139:1107–1115. doi: 10.1016/j.neuroscience.2005.12.066. [DOI] [PubMed] [Google Scholar]
- Corripio R, Gónzalez-Clemente JM, Jacobo PS, Silvia N, Lluis G, Joan V, Assumpta C. Plasma brain-derived neurotrophic factor in prepubertal obese children: results from a 2-year lifestyle intervention programme. Clin Endocrinol (Oxf) 2012;77:715–720. doi: 10.1111/j.1365-2265.2012.04431.x. [DOI] [PubMed] [Google Scholar]
- Golden E, Emiliano A, Maudsley S, Windham BG, Carlson OD, Egan JM, Driscoll I, Ferrucci L, Martin B, Mattson MP. Plasma brain-derived neurotrophic factor in prepubertal obese children: results from a 2-year lifestyle intervention programme. PLoS One. 2010;5:e10099. doi: 10.1371/journal.pone.0010099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckers S, Peeters A, Zegers D, Mertens I, Van Gaal L, Van Hul W. Association of the BDNF Val66Met variation with obesity in women. Mol Genet Metab. 2008;95:110–112. doi: 10.1016/j.ymgme.2008.06.008. [DOI] [PubMed] [Google Scholar]
- Zeman M, Jáchymová M, Jirák R, Vecka M, Tvrzická E, Stanková B, Zák A. Polymorphisms of genes for brain-derived neurotrophic factor, methylenetetrahydrofolate reductase, tyrosine hydroxylase, and endothelial nitric oxide synthase in depression and metabolic syndrome. Folia Biol (Praha) 2010;56:19–26. [PubMed] [Google Scholar]


