Abstract
The degree of substitution and valency of bioconjugate reaction products are often poorly judged or require multiple time- and product- consuming chemical characterization methods. These aspects become critical when analyzing and optimizing the potency of costly polyvalent bioactive conjugates. In this study, size-exclusion chromatography with multi-angle laser light scattering was paired with refractive index detection and ultraviolet spectroscopy (SEC-MALS-RI-UV) to characterize the reaction efficiency, degree of substitution, and valency of the products of conjugation of either peptides or proteins to a biopolymer scaffold, i.e., hyaluronic acid (HyA). Molecular characterization was more complete compared to estimates from a protein quantification assay, and exploitation of this method led to more accurate deduction of the molecular structures of polymer bioconjugates. Information obtained using this technique can improve macromolecular engineering design principles and better understand multivalent macromolecular interactions in biological systems.
Introduction
Bioconjugation reaction methods have become increasingly important in biomaterial, biochemistry, pharmaceutical, and medical endeavors. They have been employed in diverse research fields for material synthesis1, macromolecular engineering2, biochemical modification3,4, and surface treatment5,6. A theme widely explored, for various medical applications, is the conjugation of a therapeutic biomolecule to either a synthetic or natural polymer or protein ‘scaffold’ to create a decorated ‘carrier’ for the bioactive agent. Knowing the true reaction efficiency, degree of substitution on the scaffold (i.e., the average number of ligands per potential reaction site on the molecular scaffold), conjugate valency (i.e., the average number of ligands per scaffold molecule), and macromolecule structure and size are crucial to understanding and controlling the chemical and biological performance of the conjugated scaffold.
Many extracellular proteins of mammalian tissue are multivalent and multifunctional, and this biochemical information directs cell behavior and tissue dynamics through receptor-ligand engagement and down-stream signaling7,8. Therefore, multivalent bioactive conjugates have the potential to greatly improve pharmaceutical and biomaterial potency, and reduce the need for inefficient and potentially harmful doses of expensive therapeutics. They may also alter receptor-ligand dynamics (e.g. increased ligand localization and avidity, inhibited complex internalization and degradation), which could lead to improved potency of peptide and protein conjugates6,9. Furthermore, multivalent ligands have been known to increase activation of intracellular signaling when membrane-bound protein complexes are formed10,11, and natural physiological roles of soluble extracellular signaling aggregates have been recognized12. However, in many applications of bioconjugate reactions, the products are incompletely characterized in terms of molecular weight, degree of substitution, conjugate valency, and conjugate structure.
Conjugate valency is often poorly judged using common characterization techniques and can easily deviate from the expected. For example, for fixed weight concentrations of monomer and scaffold, the valency of a conjugate can vary considerably depending on the molecular weight of the scaffold. In addition, it is difficult to obtain information not only about the average valency, but the distribution of valencies within a preparation. Degradation, particularly of naturally derived or degradable polymer backbones (e.g., hyaluronic acid), can occur under the chemical conditions necessary for conjugation. Characterization of degrees of substitution and molecular weights therefore become critical when measuring the potency of various scaffolds, as well as in optimizing these multivalent biomimetic polymers for specific applications2,13.
Many chemical characterization techniques provide an incomplete picture of bioconjugate molecular structure, and therefore several are often used in conjunction14. Mass spectrometry is highly sensitive, but requires the ability to deconvolute molecular fragments and is more appropriate for lower molecular weight derivatized proteins and conjugates15-17. Protein quantification methods (e.g. bicinchoninic acid assay, Coomassie staining, ultraviolet light absorbance) can be used to estimate the average mass fraction of peptide or protein2; however, in the absence of accompanying measurements of scaffold concentration and molecular weight, these measurements do not provide direct information on the average or the distribution of conjugate valencies. Also, they generally require highly accurate protein standards that are difficult and time-consuming to prepare and validate. Fourier transform infrared spectroscopy (FTIR)18, UV-VIS spectroscopy19, and nuclear magnetic resonance (NMR)20 provide molecular composition information, but end group identification or tagging is required for molecular weight characterization. These methods, along with biochemical, molecular biology, and fluorescence18 assays, provide only an average ligand content or total molecular weight with no indication of their distribution. While traditional chromatographic methods can be used to obtain relative physicochemical properties and chemical composition16,17, light scattering methods can provide absolute molecular weight measurements without the use of standards21.
In this work we sought to develop a technique and workflow that would characterize the distributions of molecular masses and ligand valencies of an entire macromolecule population within a conjugate reaction product, using a single measurement system with a reasonable sample size (< 1.5 mg). An online three-detector method was used with continuous monitoring of differential refractive index (RI), ultraviolet light absorbance (UV), and multi-angle laser light scattering (MALS) after size-exclusion chromatography (SEC)22,23. We explored the natural biopolymer hyaluronic acid as a model scaffold, since it has been studied for numerous medical and pharmaceutical applications2,24-28. The reaction products of either peptides or proteins conjugated to a hyaluronic acid backbone were analyzed using this technique. The mass fractions and degrees of substitution determined were used to calculate valencies for comparison to those estimated using a standard protein quantification assay. We observed that reaction efficiency varied with the type and concentration of ligand conjugated. Furthermore, under some conditions the scaffold molecular weight decreased due to fragmentation of the backbone during the bioconjugation reaction. The triple detection method additionally provided novel information about ligand valency during this molecular weight change.
Materials and methods
Hyaluronic acid sodium salts (HyA) of 0.5 and 1.0 MDa MW served as the scaffolds for the conjugation reactions (Genzyme,Cambridge, Mass.). Conjugation reagents, including 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC), N-hydroxysulfosuccinimide (sulfoNHS), and [N-ε-maleimidocaproic acid] hydrazide trifluoroacetic acid (EMCH), were purchased from Thermo Scientific Pierce. Tris(2-carboxyethyl) phosphine (TCEP), Dulbecco's phosphate buffered saline (DPBS), 2-(N-morpholino)ethanesulfonic acid (MES), and 10 kDa MW cutoff dialysis tubing (SnakeSkin™) were also obtained from Pierce. All dry buffers were constituted in ultra-pure ASTM type I reagent grade water (<18.2 MΩ•cm, pyrogen free, UPW). An Arg-Gly-Asp (RGD)-containing 15-mer peptide derived from bone sialoprotein (Ac-CGGNGEPRGDTYRAY-NH2: bsp-RGD(15))29 was synthesized (American Peptide Co., Sunnyvale, CA) with two glycine spacer amino acids and an acylated cysteine amino-terminus for coupling via thiol reaction with maleimide groups of an activated HyA backbone. Recombinant modified Sonic hedgehog (Shh) protein was produced as described below. All reactions were performed under aseptic conditions using sterile-filtered solutions and autoclaved containers and instruments.
Coupling of peptide to hyaluronic acid
Hyaluronic acid was first activated with maleimide groups by carbodiimide coupling of EMCH. Hyaluronic acid sodium salt 0.5 MDa (50 mg) was added to 8 mL of MES buffer (0.1 M, pH 6.5) and stirred under low shear until dissolved. Conjugation reagents (110 mg EDC, 30.8 mg sulfoNHS, and 13.2 mg EMCH) were dissolved in 2.2 mL MES buffer. Two mL of the conjugation reaction mixture was added through sterile-filter to the HyA solution and stirred. The final concentrations of each reagent for EMCH activation of HyA were: 5 mg/mL HyA, 10 mg/mL EDC, 2.8 mg/mL sulfoNHS, and 1.2 mg/mL EMCH. The reaction was allowed to proceed for 2 hrs with stirring at room temperature in the dark. The solution was then thoroughly dialyzed through a 10 kDa MWCO membrane against MES buffer (0.1 M, pH 6.5) and transferred to another reaction vessel.
The peptide was then reduced and added to the activated HyA-EMCH solution for conjugation via reaction of the maleimide moieties with the thiol group of the peptide. In order to reduce any potential disulfide bonds formed by the peptide via spontaneous oxidation, 3 mg of NaOH and 5.7 mg of TCEP were added to 1 mL UPW. The bsp-RGD peptide (4.4 mg) was added to the alkaline reducing solution, allowed to react for 30 min at 4 °C, and added through a sterile-filter to the activated HyA-EMCH solution. The final solution was reacted overnight at 4°C in the dark. The conjugate solution was then dialyzed three times for 2 hrs through a 10 kDa MWCO membrane against UPW. After dialysis, the solution was transferred to a 50 mL Steriflip tube (BD Bioscience) and lyophilized through the sterile-filter cap. The dry product was stored at -20 °C until characterization. The final chemical structure of the bioconjugate is depicted in Figure 1.
Figure 1.

Hyaluronic acid-peptide conjugate chemical structure. A heterobifunctional linker was used to anchor peptide and protein via their terminal thiol-functional cysteine residues to the carboxylic acid groups of the linear hyaluronic acid polymer.
Recombinant production of modified Sonic hedgehog protein
A recombinant cysteine-modified N-terminal Shh was produced as previously described2,31. A sequence encoding a cysteine residue and a 6xHis tag were added via PCR to 3' end of cDNA encoding the N-terminal signaling domain of rat Shh to allow for sulfhydryl-based reactions and protein purification, respectively. The conjugation site was chosen because it is distant from the active receptor-binding domain. In addition, valine and isoleucine residues were introduced to the N-terminus to increase potency by mimicking the hydrophobic palmitic acid modification of endogenous Shh. The modified Shh PCR product was inserted into a pBAD-HisA plasmid (Invitrogen), sequenced for confirmation, and expressed in BL21(DE3)pLysE E. coli upon arabinose induction. The cells were lysed, and the resulting expressed Shh was purified using NiNTA binding through the BioRad DuoFlow System and elution with imidazole. The isolated protein was dialyzed into pH 7.4 PBS containing 10% glycerol, 2 mM EDTA, and 50 μM ZnSO4 and stored at -20°C until conjugation.
Conjugation of Sonic hedgehog protein to hyaluronic acid
The modified Shh protein was conjugated to 1.0 MDa HyA through a similar two-step reaction process to the bsp-RGD (HyA-bsp-RGD(15)), using carbodiimide chemistry at the carboxylate group of the HyA and maleimide reaction at the protein C-terminal cysteine2. Hyaluronic acid was activated at 3 mg/mL in MES (0.1 M pH 5.0) with 3.9 mg/mL EDC, 1.1 mg/mL sulfoNHS, and 0.5 mg/mL EMCH. Activated HyA was purified by sequential dilution and centrifugation in 100 kDa MWCO centrifuge filters (Pall Gellman). The activated HyA-EMCH was reacted with the modified Shh at varying stoichiometric feed ratios to produce conjugates with different conjugate valency. The reaction was performed at 4 °C overnight in MES buffer (0.1 M pH 6.5) with 50 μM TCEP to maintain cysteine reduction. Following the reaction, the remaining maleimide groups were quenched by the addition of 0.5 mM dithiothreitol and incubation at 4 °C for 1 hr. The HyA-Shh conjugation product was dialyzed into pH 7.4 PBS containing 2 mM EDTA, and 50 μM ZnSO4 and stored at -20 °C until characterization.
SEC-MALS-RI-UV analysis of bioconjugates
The SEC-MALS-RI-UV setup consisted of an Agilent HPLC 1100 (including degasser, quaternary pump, autosampler, column holder, and UV-VIS diode array detector) in line with a DAWN-EOS multi-angle laser light scattering detector and Optilab-DSP relative refractive interferometer (Wyatt Technology, Santa Barbara, CA). Refractive index change was measured differentially with a GaAs laser at a wavelength of 690 nm, and UV absorbance was measured with the diode array detector at 280 nm. A PolySep-GFC-P Linear column was used for the HPLC (Phenomenex). HyA-peptide and HyA-protein conjugates were injected at 5 mg/mL and 3 mg/mL conjugate, respectively. The temperatures of the DSP and column holder were both maintained at 45 °C. Agilent software was used to control the HPLC, and Wyatt Astra V software was used for data collection and analysis. Normalization of the multi-angle detectors was performed with 300 μL of 5 mg/mL bovine serum albumin (BSA) (Aldrich) in DPBS at a flow rate of 0.8 mL/min (used throughout unless otherwise noted) with DPBS as the mobile phase. Peak alignment and band broadening correction between the UV, MALS, and RI detectors were performed using Astra software algorithms. Analysis was restricted to detectors at moderate angles due to high noise at very low and very high angle detectors.
The specific refractive index values of the peptide, protein, HyA, and HyA-EMCH were confirmed by a 100% mass recovery method at a wavelength of 690 nm (the same as used in online detection). The UV absorbance coefficient values for each conjugate component were measured using an off-line UV-VIS spectrophotometer at 280 nm wavelength. These values were confirmed using a 100% mass recovery method to corroborate UV and RI concentration source detectors. Zimm plots were created for 0.5 MDa HyA scaffold and a conjugate of 0.5 MDa HyA with bsp-RGD(15) at a 1 to 25 molar ratio (1:25, HyA:bsp-RGD(15)) using 200 μL injections of 1, 2, 3, and 4 mg/mL in DPBS at 45 °C with DPBS as the mobile phase.
Results and discussion
Online SEC-MALS-RI-UV analysis of hyaluronic acid bioconjugates
Measurement of RI, UV, and MALS signals allowed simultaneous online measurement of concentration, conjugate molar mass, and ligand mass fraction distribution amongst the modified polymer chains. Table 1 lists the physicochemical parameters of the conjugate components employed in this study. The weight-averaged molar mass (Mw) and z-averaged radius of gyration (RG,z) were determined for each slice along the chromatogram (Figure 2a) using the Rayleigh-Gans-Debye equation:
Table 1.
Specific refractive indices (∂ n/ ∂ c) and 280 nm absorbance coefficients (ε abs) for hyaluronic acid, EMCH-activated hyaluronic acid, peptide, and proteins.
| Component | ∂ n/ ∂ c (mL/g) | ε abs (mL/g cm) | MWw | Sequence |
|---|---|---|---|---|
| HyA | 0.167 | 22 | 0.5 and 1.0 MDa | |
| HyA-EMCH | 0.167 | 22 | ||
| bspRGD | 0.185 | 1,840 | 1,657 | Ac-CGGNGEPRGDTYRAY-NH2 |
| Shh | 0.185 | 974 | 21,376 | See reference (Wall et al. Bioconjug Chem, 2008) |
Figure 2.

Representative chromatogram (a) with 90° light scattering (LS), refractive index (RI), and ultraviolet light absorbance (UV) signals and Debye plot (b) calculated from mulit-angle laser light scattering (MALS) data at slices along the size-exclusion chromatogram for HyA-bsp-RGD(15) (1:25, HyA:bsp-RGD(15) molar reaction ratio). A Zimm plot (c) created from injectons of varying total conjugate mass for HyA-bsp-RGD(15) (1:25) used to calculate weight-averaged molecular mass (Mw), z-averaged radius of gyration, (RG,z,) and second virial coefficient, (A2).
where c is the concentration of polymer in solution, R(θ) is the excess Rayleigh scattering ratio, A2 is the second virial coefficient, A3 is the third virial coefficient, θ is the scattering angle, and is the specific refractive index increment of the macromolecule in solution. Debye plots were created for each slice along the chromatogram (Figure 2b). Zimm plots were also constructed by injecting a series of concentrations through the SEC-MALS system, which gave the A2, RG,z, and Mw values of the backbone polymer and a representative conjugate (Figure 2c and Table 2).
Table 2.
Weight-averaged molecular mass (Mw), z-averaged radius of gyration (RG,z), and second virial coefficient (A2) for HyA and HyA-bsp-RGD(15) (1:25) as determined by Zimm plot construction (as measured at 45 °C in phosphate buffer).
| Sample | MWw (105 g/mol) | RG,z (nm) | A2 (10-3 mol mL/g2) |
|---|---|---|---|
| HyA, 0.5 MDa | 4.9±0.24 | 55±3.5 | 2.4±0.76 |
| HyA-RGD (1:25) | 2.1±0.74 | 70±15.8 | 3±1.6 |
The use of two concentration detectors (RI and UV) in combination with SEC-MALS allowed characterization of the molecular weight distribution and the relative composition of conjugated macromolecules. This was possible since the measured values of specific refractive index and UV absorption coefficient (εabs) along the chromatogram are weight-averaged values for each component individually:
Knowing ∂ n/ ∂ c and εabs for each component, these equations can be solved simultaneously to determine the mass fractions of each component of the conjugate. Valency information was obtained in this manner by dividing the total average ligand molar mass of the conjugate (mass protein or peptide per mole conjugate) by the individual ligand molecular weight.
Analysis of hyaluronic acid conjugate synthesis products
Conjugates were made with HyA and Shh at several reaction ratios, and the products were characterized by SEC-MALS-RI-UV. Signals from the concentration detectors combined with MALS data were used to calculate the HyA and ligand molecular masses, as depicted in Figure 3 for HyA-Shh (1:40, HyA:Shh molar reaction ratio) conjugates made with 1.0 MDa HyA. These values for each conjugate were used to determine protein fraction distributions along the chromatograms, as presented in Figure 4. The quality of the data collected depended on the overall concentration of the conjugate as well as its degree of substitution. The cumulative and differential mass fractions were plotted against molar mass in Figure 5, depicting molecular weight distributions. The reaction and final ligand concentrations along with HyA and total ligand molecular weights of conjugate products are listed in Table 3 and Table 4.
Figure 3.

Detector signals (90° light scattering, LS, refractive index, RI, and ultraviolet light absorbance, UV) (a) collected during size-exclusion chromatography (SEC) combined with MALS data allowed calculation of hyaluronic acid and protein molecular weight distributions (b) of the HyA-Shh (1:40, HyA:Shh molar reaction ratio) conjugate.
Figure 4.

Protein fraction (dark line) and 90° light scattering intensity (light line) of HyA-bsp-RGD(15) (1:25) (a), HyA-Shh (1:20) (b), and HyA-Shh (1:40) (c) following size-exclusion chromatography. Light scattering instensity depends on concentration and total molar mass, with higher molar masses eluted first. Conjugation of bsp-RGD was more efficient than Shh, with smaller input reaction ratios achieving a higher final protein fraction due to the small size and ease of accessibility to the bsp-RGD reaction site when compared to Shh.
Figure 5.
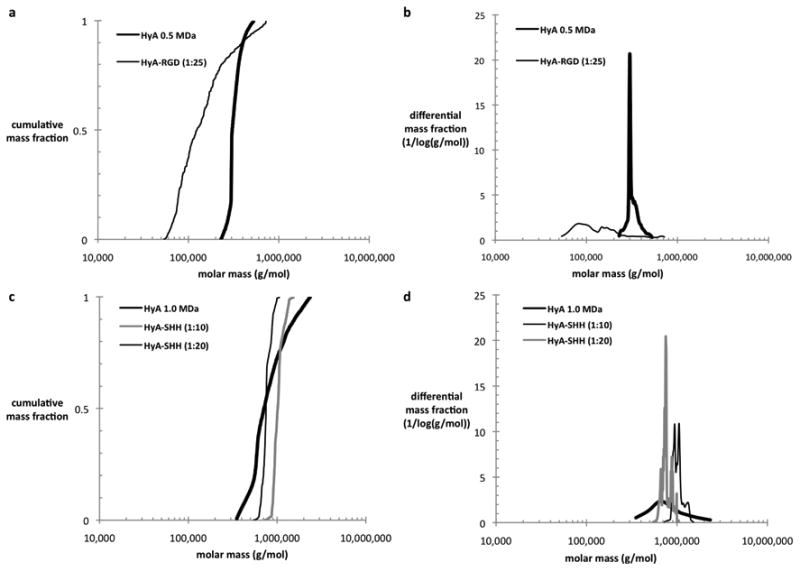
Cumulative (a, c) and differential (b, d) mass fractions versus molar mass of 0.5 MDa HyA and HyA-bsp-RGD(15) (1:25) (a, b) along with 1.0 MDa HyA, HyA-Shh (1:20), and HyA-Shh (1:40) (c, d).
Table 3.
Bioconjugate characterization of HyA-peptide products by SEC-MALS-RI-UV.
| reaction molar ratio | reaction mass fraction | output ligand mass fraction | MWw HyA (kDa) | MWw ligand (kDa) | conjugate valency | |
|---|---|---|---|---|---|---|
| HyA, 0.5 MDa | 491 | |||||
| HyA-bspRGD 1:10 | 10 | 0.04 | 0.09 | 413 | 19 | 11.5 |
| HyA-bspRGD 1:25 | 25 | 0.11 | 0.33 | 141 | 72 | 43.4 |
Table 4.
Bioconjugate characterization of HyA-Shh products by SEC-MALS-RI-UV.
| reaction molar ratio | reaction mass fraction | output ligand mass fraction | MWw HyA (kDa) | MWw ligand (kDa) | conjugate valency | |
|---|---|---|---|---|---|---|
| HyA, 1.0 MDa | 508±335 | |||||
| HyA-EMCH | 909±398 | 0 | 0.0 | |||
| HyA-Shh 1:10 | 10 | 0.22 | 0.06±.01 | 496±403 | 36±22 | 1.7±1.0 |
| HyA-Shh 1:20 | 20 | 0.43 | 0.23±.04 | 468±118 | 128±49 | 6.0±2.3 |
| HyA-Shh 1:40 | 40 | 0.87 | 0.27±.10 | 578±252 | 187±33 | 8.7±1.6 |
The molecular weights of the HyA after Shh conjugation varied from batch to batch (Figure 6). It is known that HyA is susceptible to hydrolytic degradation, particularly under shear (e.g. in a stirred reaction, upon vortexing, or during sonication) or catalytic chemical conditions (e.g. acidic, alkaline, reductive, or oxidative). A reducing agent was used in the conjugation reaction, and the conjugate was exposed to shear in several steps of the reaction (stirring, pipetting). It is believed that the reducing conditions and shear led to molecular weight degradation of the HyA bioconjugates. The large variation in molecular weights is likely further due to differences in the way that the conjugates were handled, such as differences in stirring times or shear rates.
Figure 6.
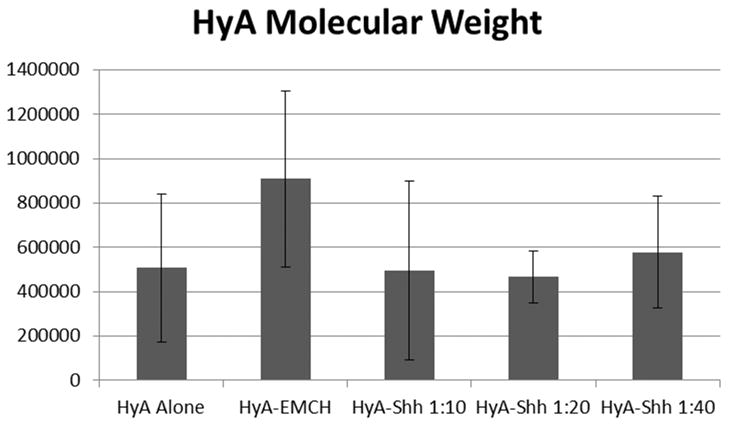
Weight-averaged molecular weights of hyaluronic acid in conjugates of varying reaction ratio. The error bars represent the standard deviation (N=3).
The input and output degrees of substitution (DS) for several HyA-Shh conjugates are plotted in Figure 7. Input and output DS were calculated according to:
Figure 7.

Input and output degrees of substitution (DS) values for HyA-Shh conjugates. The line represents correspondence of 1:1.
*based on nominal HyA MW.
In these equations, primed quantities refer to the molecular weight of a single peptide or protein molecule. MWw refers to the weight average molecular weight. Hyaluronic acid molecular weight was normalized by the dimer molecular weight as there is one reactive carboxylate group per dimer.
Differences in reaction efficiency between bsp-RGD(15) and Shh protein can be attributed to the higher molecular weight and steric hindrance of Shh protein. Higher conjugation efficiency of Shh to HyA was achieved when the reactions were performed at lower total reagent concentration (data not shown).
Comparison SEC-MALS-RI-UV results with input ratios and BCA results
Protein fractions measured by SEC-MALS-RI-UV corresponded well with results determined by BCA protein measurements for a given batch of conjugate. The final conjugate valency differed from that calculated through the BCA assay, however (Figure 8). Differences in calculated valency between SEC-MALS-RI-UV and BCA assay were due largely to fragmentation of the HyA backbone during the conjugate synthesis, and subsequently accounting for changes in the molecular weight. Valency determined by SEC-MALS-RI-UV was calculated based on the total weight-averaged ligand molecular weight in each conjugate divided by the molecular weight of the individual ligand, whereas in the BCA assay determination, the mass ratio of ligand to HyA was used to calculate the molar ratio based on the ligand and original HyA molecular weights. Using the valency data collected from the BCA measurements leads to a misinterpretation of multivalent macromolecular values, whereas that calculated using the SEC-MALS-RI-UV does not.
Figure 8.
Comparison of input and output molar ratios as determined by SEC-MALS-RI-UV and BCA assay methods. Error bars represent the standard deviation (N=3).
Conclusion
SEC-MALS with RI and UV detection is valuable for determining the molecular weight and protein fraction distributions of polymer bioconjugates. The amount of material needed for this method depends upon instrument sensitivity, molecular weight, and reagent physiochemical parameters, but is significantly less than standard methods (i.e., BCA). Accurate measurement of protein fraction requires that the bioconjugate components have differing refractive index increments and UV absorbance coefficients. These values can be determined using online or offline analysis of un-conjugated reagents. SEC-MALS with triple detection (MALS, RI, UV) can also be used to accurately measure bioconjugation reaction efficiency and provide detailed molecular structural information along with precise distribution of substitution and valency data. In contrast, with the large variation in actual HyA molecular weight after the conjugation reaction, accurate measurement of the valency of the bioconjugates is impossible using more conventional batch techniques such as a BCA assay. Moreover, the capabilities of this method could be expanded by using the diode array detector (DAD) on a high-performance liquid chromatography (HPLC) instrument to measure UV absorbance at multiple wavelengths in order to simultaneously identify conjugate components with different absorbance spectra. In either case, more accurate deduction of the molecular structures of polymer bioconjugates can be used to improve macromolecular engineering design principles and better understand multivalent macromolecular interactions (e.g. receptor-ligand dynamics and enzyme kinetics). Furthermore, this analysis method can be used for quality assurance and to optimize reactor design and operations for large-scale production of designer bioconjugate products.
Acknowledgments
This work was supported in part by the American Heart Association #20082098, National Science Foundation (NSF Graduate Fellowship to JFP), the University of California at Berkeley (Berkeley Graduate Fellowship to JFP), the California Institute for Regenerative Medicine (Postdoctoral Fellowship to RSA), NIH R21EB007295, NIH NHLBI R01HL096525. The authors thank Melissa Velasquez for her assistance in HyA-peptide synthesis.
References
- 1.Elbert D, Hubbell J. Conjugate addition reactions combined with free-radical cross-linking for the design of materials for tissue engineering. Biomacromolecules. 2001;2:430–441. doi: 10.1021/bm0056299. [DOI] [PubMed] [Google Scholar]
- 2.Wall ST, Saha K, Ashton RS, Kam KR, Schaffer DV, Healy KE. Multivalency of Sonic hedgehog conjugated to linear polymer chains modulates protein potency. Bioconjug Chem. 2008;19:806–12. doi: 10.1021/bc700265k. [DOI] [PubMed] [Google Scholar]
- 3.Drumheller P, Elbert D, Hubbell J. Multifunctional poly (ethylene glycol) semi-interpenetrating polymer networks as highly selective adhesive substrates for bioadhesive peptide grafting. Biotechnology and bioengineering. 1994;43:772–780. doi: 10.1002/bit.260430812. [DOI] [PubMed] [Google Scholar]
- 4.Stile RA, Healy KE. Thermo-responsive peptide-modified hydrogels for tissue regeneration. Biomacromolecules. 2001;2:185–94. doi: 10.1021/bm0000945. [DOI] [PubMed] [Google Scholar]
- 5.Ho M, Wang D, Hsieh H, Liu H, Hsien T, Lai J, Hou L. Preparation and characterization of RGD-immobilized chitosan scaffolds. Biomaterials. 2005;26:3197–3206. doi: 10.1016/j.biomaterials.2004.08.032. [DOI] [PubMed] [Google Scholar]
- 6.Ito Y. Covalently immobilized biosignal molecule materials for tissue engineering. Soft Matter. 2008;4:46–56. doi: 10.1039/b708359a. [DOI] [PubMed] [Google Scholar]
- 7.Sacchettini JC, Baum LG, Brewer CF. Multivalent protein-carbohydrate interactions. A new paradigm for supermolecular assembly and signal transduction. Biochemistry. 2001;40:3009–15. doi: 10.1021/bi002544j. [DOI] [PubMed] [Google Scholar]
- 8.Kiessling LL, Gestwicki JE, Strong LE. Synthetic multivalent ligands in the exploration of cell-surface interactions. Current Opinion in Chemical Biology. 2000;4:696–703. doi: 10.1016/s1367-5931(00)00153-8. [DOI] [PubMed] [Google Scholar]
- 9.Schamel WWA, Reth M. Clustering models. Adv Exp Med Biol. 2008;640:64–73. doi: 10.1007/978-0-387-09789-3_6. [DOI] [PubMed] [Google Scholar]
- 10.Geiger B, Bershadsky A, Pankov R, Yamada KM. Transmembrane crosstalk between the extracellular matrix--cytoskeleton crosstalk. Nat Rev Mol Cell Biol. 2001;2:793–805. doi: 10.1038/35099066. [DOI] [PubMed] [Google Scholar]
- 11.Huveneers S, Danen EHJ. Adhesion signaling - crosstalk between integrins, Src and Rho. Journal of Cell Science. 2009;122:1059–69. doi: 10.1242/jcs.039446. [DOI] [PubMed] [Google Scholar]
- 12.McLellan JS, Zheng X, Hauk G, Ghirlando R, Beachy PA, Leahy DJ. The mode of Hedgehog binding to Ihog homologues is not conserved across different phyla. Nature. 2008;455:979–83. doi: 10.1038/nature07358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saha K, Pollock JF, Schaffer DV, Healy KE. Designing synthetic materials to control stem cell phenotype. Current Opinion in Chemical Biology. 2007;11:381–7. doi: 10.1016/j.cbpa.2007.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bi X, Shi X, Baker JR. Synthesis, characterization and stability of a luteinizing hormone-releasing hormone (LHRH)-functionalized poly(amidoamine) dendrimer conjugate. Journal of biomaterials science Polymer edition. 2008;19:131–42. doi: 10.1163/156856208783227686. [DOI] [PubMed] [Google Scholar]
- 15.Wang L, Amphlett G, Blättler WA, Lambert JM, Zhang W. Structural characterization of the maytansinoid-monoclonal antibody immunoconjugate, huN901-DM1, by mass spectrometry. Protein Sci. 2005;14:2436–46. doi: 10.1110/ps.051478705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng W, Satyanarayanajois S, Lim LY. Aqueous-soluble, non-reversible lipid conjugate of salmon calcitonin: synthesis, characterization and in vivo activity. Pharm Res. 2007;24:99–110. doi: 10.1007/s11095-006-9128-9. [DOI] [PubMed] [Google Scholar]
- 17.Tzokova N, Fernyhough CM, Butler MF, Armes SP, Ryan AJ, Topham PD, Adams DJ. The effect of PEO length on the self-assembly of poly(ethylene oxide)-tetrapeptide conjugates prepared by “Click” chemistry. Langmuir : the ACS journal of surfaces and colloids. 2009;25:11082–9. doi: 10.1021/la901413n. [DOI] [PubMed] [Google Scholar]
- 18.Mi FL. Synthesis and characterization of a novel chitosan-gelatin bioconjugate with fluorescence emission. Biomacromolecules. 2005;6:975–87. doi: 10.1021/bm049335p. [DOI] [PubMed] [Google Scholar]
- 19.Demolliens A, Boucher C, Durocher Y, Jolicoeur M, Buschmann MD, De Crescenzo G. Tyrosinase-catalyzed synthesis of a universal coil-chitosan bioconjugate for protein immobilization. Bioconjug Chem. 2008;19:1849–54. doi: 10.1021/bc800066b. [DOI] [PubMed] [Google Scholar]
- 20.Oh EJ, Park K, Choi JS, Joo CK, Hahn SK. Synthesis, characterization, and preliminary assessment of anti-Flt1 peptide-hyaluronate conjugate for the treatment of corneal neovascularization. Biomaterials. 2009;30:6026–34. doi: 10.1016/j.biomaterials.2009.07.024. [DOI] [PubMed] [Google Scholar]
- 21.Wyatt P. Light scattering and the absolute characterization of macromolecules. Analytica Chimica Acta 1993 [Google Scholar]
- 22.Wen J, Arakawa T, Philo JS. Size-exclusion chromatography with on-line light-scattering, absorbance, and refractive index detectors for studying proteins and their interactions. Anal Biochem. 1996;240:155–66. doi: 10.1006/abio.1996.0345. [DOI] [PubMed] [Google Scholar]
- 23.Kendrick BS, Kerwin BA, Chang BS, Philo JS. Online Size-Exclusion High-Performance Liquid Chromatography Light Scattering and Differential Refractometry Methods to Determine Degree of Polymer Conjugation to Proteins and Protein–Protein or Protein–Ligand Association States. Anal Biochem. 2001;299:136–146. doi: 10.1006/abio.2001.5411. [DOI] [PubMed] [Google Scholar]
- 24.Ghosh K, Shu XZ, Mou R, Lombardi J, Prestwich GD, Rafailovich MH, Clark RA. Rheological characterization of in situ cross-linkable hyaluronan hydrogels. Biomacromolecules. 2005;6:2857–65. doi: 10.1021/bm050361c. [DOI] [PubMed] [Google Scholar]
- 25.Leach JB, Bivens KA, Collins CN, Schmidt CE. Development of photocrosslinkable hyaluronic acid-polyethylene glycol-peptide composite hydrogels for soft tissue engineering. J Biomed Mater Res A. 2004;70:74–82. doi: 10.1002/jbm.a.30063. [DOI] [PubMed] [Google Scholar]
- 26.Pouyani T, Prestwich GD. Functionalized derivatives of hyaluronic acid oligosaccharides: drug carriers and novel biomaterials. Bioconjug Chem. 1994;5:339–47. doi: 10.1021/bc00028a010. [DOI] [PubMed] [Google Scholar]
- 27.Park YD, Tirelli N, Hubbell JA. Photopolymerized hyaluronic acid-based hydrogels and interpenetrating networks. Biomaterials. 2003;24:893–900. doi: 10.1016/s0142-9612(02)00420-9. [DOI] [PubMed] [Google Scholar]
- 28.Peattie RA, Nayate AP, Firpo MA, Shelby J, Fisher RJ, Prestwich GD. Stimulation of in vivo angiogenesis by cytokine-loaded hyaluronic acid hydrogel implants. Biomaterials. 2004;25:2789–98. doi: 10.1016/j.biomaterials.2003.09.054. [DOI] [PubMed] [Google Scholar]
- 29.Rezania A, Healy K. The effect of peptide surface density on mineralization of a matrix deposited by osteogenic cells. Journal of biomedical materials research. 2000;52:595–600. doi: 10.1002/1097-4636(20001215)52:4<595::aid-jbm3>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 30.Nomizu M, Kim W, Yamamura K, Utani A, Song S, Otaka A, Roller P, Kleinman H, Yamada Y. Identification of Cell Binding Sites in the Laminin alpha1 Chain Carboxyl-terminal Globular Domain by Systematic Screening of Synthetic Peptides. Journal of Biological Chemistry. 1995;270:20583–90. doi: 10.1074/jbc.270.35.20583. [DOI] [PubMed] [Google Scholar]
- 31.Ho J, Chung E, Wall S, Schaffer D, Healy K. Journal of Biomedical Materials Research Part A. 2007;83:1200–1208. doi: 10.1002/jbm.a.31355. [DOI] [PubMed] [Google Scholar]



