Abstract
The Arabidopsis SUPERMAN (SUP) gene encodes a C2H2 type zinc finger protein that is required for maintaining the boundaries between stamens and carpels, and for regulating development of ovule outer integument. Orthologs of SUP have been characterized in bisexual flowers as well as dioecious species, but it remains elusive in monoecious plants with unisexual flowers on the same individual. Here we isolate the SUP ortholog in Cucumis sativus L (CsSUP), a monoecious vegetable. CsSUP is predominantly expressed in female specific organs: the female flower buds and ovules. Ectopic expression of CsSUP in Arabidopsis can partially complement the fruit development in sup-5 mutant, and its over-expression in wide-type leads to reduced silique length, suppressed stamen development and distorted petal patterning. Our data suggest that CsSUP plays conserved as well as distinct roles during flower and fruit development, and it may function in the boundaries and ovules to balance petal patterning, stamen and ovule development in Arabidopsis.
Introduction
In flowering plants, approximately 90% species produce bisexual flowers with specialized male and female organs in the same flower, while 6% species are dioecious with separate male and female plants, and the remaining species are monoecious, producing male and female flowers on the same individual [1], [2]. Arabidopsis thaliana is a model species for bisexual plant, whose flowers consist of four types of floral organs arranged into concentric whorls, specifically sepals in outermost (whorl 1), petals in whorl 2, stamens in whorl 3 and carpels in innermost (whorl 4). The patterning of floral organs in different whorls is controlled by the combinatorial interactions between three classes of homeotic genes, designated A, B, and C class of genes [3], [4]. Sepal identity is determined by the function of A genes APETALA1 and APETALA2, petal identity is specified by the simultaneous function of A and B genes (which include APETALA3 and PISTILLATA), stamen identity is determined by combinatorial action of B and C (provided by the AGAMOUS gene), whereas carpel identity is specified by C function alone [3]–[5]. Recently, D class of genes, FLORAL BINDING PROTEIN7 (FBP7) and FLORAL BINDING PROTEIN11 (FBP11), have been shown to be involved in carpel and ovule development, whereas E class of genes (provided by SEPALLATA 1-4) are required for all the floral organ development, thus the classical ABC model has been modified into ABCDE model for floral patterning [6]–[10].
In Arabidopsis, the cadastral gene SUPERMAN (SUP) has shown to be a negative regulator of B class of genes at the boundaries between stamens and carpels. Loss of function of AtSUP leads to extra stamen production in the fourth whorl at the expense of carpel development, and expansion of the B class of genes APETALA3 and PISTILLATA from the second and third whorls into the forth whorl [11], [12]. AtSUP encodes a C2H2 type zinc finger transcription factor that is expressed in the subdomain of third whorl adjacent to the forth whorl, and may function at the boundaries to balance cell proliferation in whorl 3 and 4 [12], [13]. The DLELRL hexapeptide in the AtSUP carboxy terminal domain confers the transcriptional repression activity and is required for the normal flower development [14], [15]. In addition, AtSUP also function in cell proliferation suppression of outer integument on the adaxial side of the ovule [16]. In sup mutants, outer integument grows evenly and leads to production of infertile ovules that are radially symmetrical [16]. Consistent with the function in ovule, AtSUP is expressed in the developing ovules and then limited to the stalks of ovules [16].
Numerous studies have explored the function of AtSUP upon ectopic expression in various species. Overexpression of AtSUP under APETALA1 promoter leads to reduced size as well as decreased number of all four types of floral organs in Arabidopsis [17]. Similarly, ectopic expression of AtSUP using FLORAL BINDING PROTEIN1 (FBP1) promoter results in size reduction in petals and stamens in petunia or tobacco [18]. However, when overexpressed under constitutive promoter in dicotyledonous species such as Arabidopsis or tobacco, AtSUP causes dwarfness at the whole plant level, and restores stamen development and produces functional pollen in an alloplasmic CMS tobacco plant [14], [19], [20]. While overexpressed in monocotyledon such as rice, high level of AtSUP expression leads to juvenile death, and low level of AtSUP expression results in expansion of ventral carpel and decreased number of stamen [21]. Recently, several orthologs of SUP have been cloned, including NtSUP in Nicotiana tabacum, PhSUP1 in Petunia hybrida and SlSUP in Silene latifolia, and their functions are generally conserved, but the expression patterns are quite divergent [19], [20], [22].
Despite the extensive studies of AtSUP and SUP orthologs, no study has been performed in monoecious plant with unisexual flowers on the same individual. Cucumber (Cucumis sativus L.) is a typical monoecious species with male flowers produce at the bottom and female flowers form at the top. Therefore, we cloned the SUP ortholog in cucumber designated as CsSUP, and the expression pattern of CsSUP was characterized by both qRT-PCR and in situ hybridization. CsSUP is predominantly expressed in female organs: female flower buds and ovules of fruit. Ectopic expression of CsSUP in Arabidopsis can partially complement the fruit development in sup-5 mutant, and its over-expression in wide-type leads to reduced silique length, suppressed stamen development, and disorganized petal patterning. Our data suggested that CsSUP possesses conserved functions in stamen and fruit development as well as a distinct role in floral patterning.
Results
Identification of CsSUP gene from C. sativus L
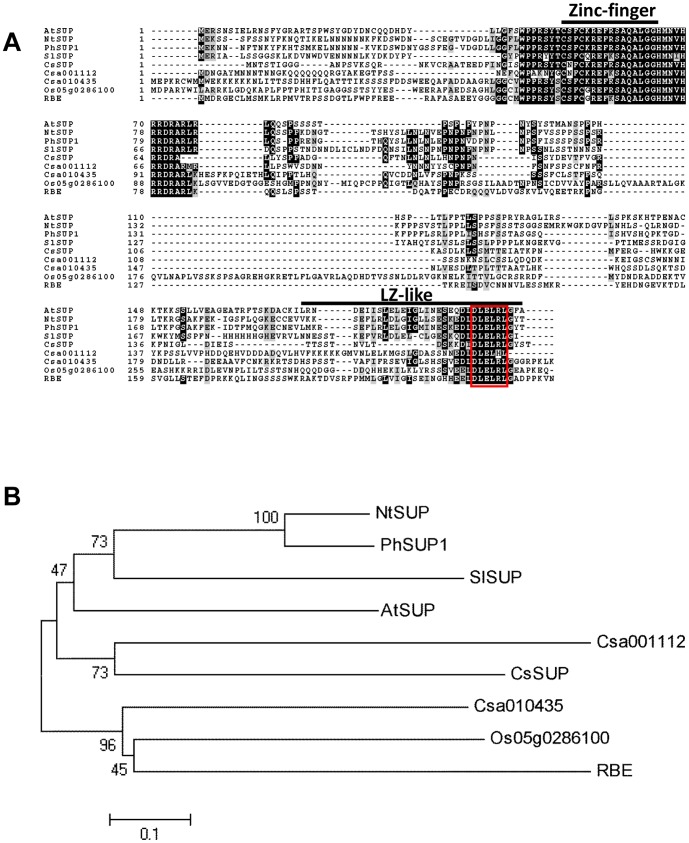
To isolate the potential SUP-like genes in cucumber, we performed BLAST analysis against Cucumber Genome Database [23] using the sequence information of AtSUP in Arabidopsis. Three SUP-like genes were identified in cucumber, Csa000134, Csa001112 and Cas010435, in which Csa000134 displays the lowest e-value with AtSUP using both cDNA (3e-16) and deduced protein sequence (2e-17), thus was designated as CsSUP in this study. The cDNA of CsSUP were cloned from the female buds. Consistent with known SUP orthologs [12], [20], [22], [24], CsSUP has no intron, and the full length CsSUP encodes a protein of 171 amino acids. ClustalW was used to align the amino acid sequence of CsSUP with known SUP-like genes, including AtSUP and RABBIT EARS (RBE) in Arabidopsis, NtSUP in tobacco, PhSUP in petunia, SlSUP in white campion, and Os05g0286100 in rice [25] (Figure 1A). Despite CsSUP shows only 28.78%, 33.62%, 29.78% and 30.36% identity with AtSUP,NtSUP,PhSUP and SlSUP respectively, the zinc-finger domains and the leucine zipper (LZ)-like domains are highly conserved (Figure 1A). The DLELRL domains, which are required for transcriptional repression during cell proliferation in Arabidopsis and petunia [14], [15], are identical over all the proteins we aligned except for Csa001112. Phylogenetic analysis was performed with the entire amino acid sequences using the neighbor-joining (NJ) method [26]. As shown in Figure 1B, CsSUP and Csa001112 belong to the same clade with known SUP orthologs, which are distinct from the clade consisting of SUP-like genes RBE, Os05g0286100 and Cas010435 [25], suggesting that CsSUP and Csa001112 maybe the SUP homologues in cucumber.
Figure 1. Alignment and phylogenetic analysis of SUP-like genes.
(A) Alignment of CsSUP and SUP-like genes. The amino acid sequences of CsSUP, Csa001112 and Cas010435 in cucumber, AtSUP and RBE in Arabidopsis, NtSUP in tobacco, PhSUP in petunia, SlSUP in white campion and Os05g0286100 in rice were aligned using ClustalW in the MEGA5 software package. The black and gray areas indicate identical and similar amino acid, respectively. Zinc-finger and leucine zipper (LZ)-like domains were indicated in black lines. The DLELRL domain [15] was showed in red box. (B) An unrooted phylogenetic tree constructed using the amino acid sequences of CsSUP, Csa001112, Cas010435, AtSUP, RBE, NtSUP, PhSUP, SlSUP and Os05g0286100 based on the neighbor joining method. Branch length is proportional to evolutionary distance.
Expression pattern of CsSUP in Cucumber
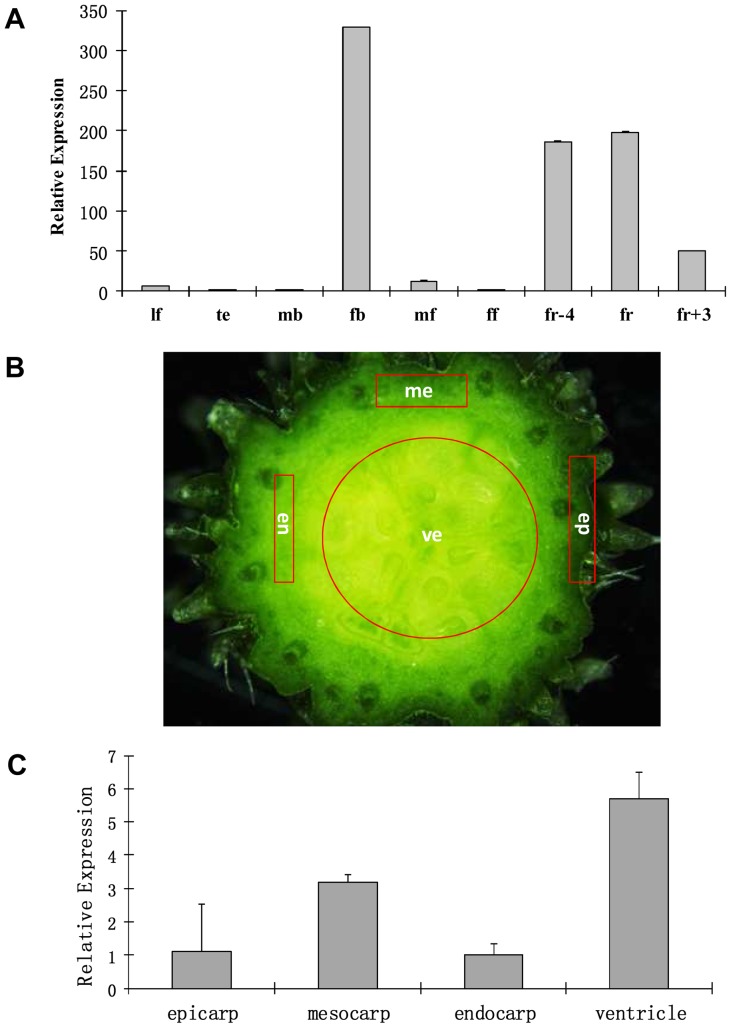
Next, we examined the transcription of CsSUP and Csa001112 in different organs of cucumber through qRT-PCR (Figure 2A). Total RNA was isolated from leaves (lf), tendril (te), male flower buds (mb), female flower buds (fb), male open flowers (mf), female open flowers (ff), and fruits at three different developmental stages. Our results showed that CsSUP and Csa001112 display similar expression patterns, which are highly enriched in female specific tissues such as female flower buds and developing fruits, whereas there were very few expressions in other organs (Figure 2A, Figure S1). Specifically, CsSUP has the highest expression in female flower buds (Figure 2A), while Csa001112 has the most abundant transcription in fruits 4 days before flower opening (fr-4) (Figure S1). Given that CsSUP shows the highest sequence similarity with SUP orthologs, and the expression of CsSUP appears to be enriched in younger female tissues than that of Csa001112, we focus on characterization of CsSUP thereafter. Cucumber fruit largely results from expansion of ovary, therefore, we next explored the transcript accumulation of CsSUP in different parts of cucumber fruits (Figure 2B–C). Epicarp, mesocarp, endocarp and ventricle were separated according to Figure 2B, and qRT-PCR was performed with these four types of tissues. Ventricle, where the ovules are located, showed the highest CsSUP expression (over 5 folds than that of epicarp), and mesocarp, which comprised of plentiful vascular system, displayed the second highest expression among the four parts of fruit (Figure 2C), suggesting that CsSUP may be highly expressed in ovules and vasculature of cucumber fruits.
Figure 2. Quantitative RT-PCR (qRT-PCR) analysis of CsSUP in different organs of cucumber (A) or different parts of cucumber fruit (B–C).
Three biological replicates were used for each sample, and 18S rRNA was used as internal control. Bars represent the standard error. (A) CsSUP is predominately expressed in the female flower bud and developing fruit. lf: leaves, te: tendrils, mb: male flower buds, fb: female flower buds, mf: male flowers, ff: female flowers, fr-4: fruit of 4 days before flower opening, fr: fruit on flower opening, fr+3: fruit of 3 days after flower opening. (B) Transverse sections of commercially mature cucumber fruit. ep: epicarp, me: mesocarp, en:endocarp, ve:ventricle. (C) CsSUP is highly expressed in the ventricle of cucumber fruit where the ovules are located.
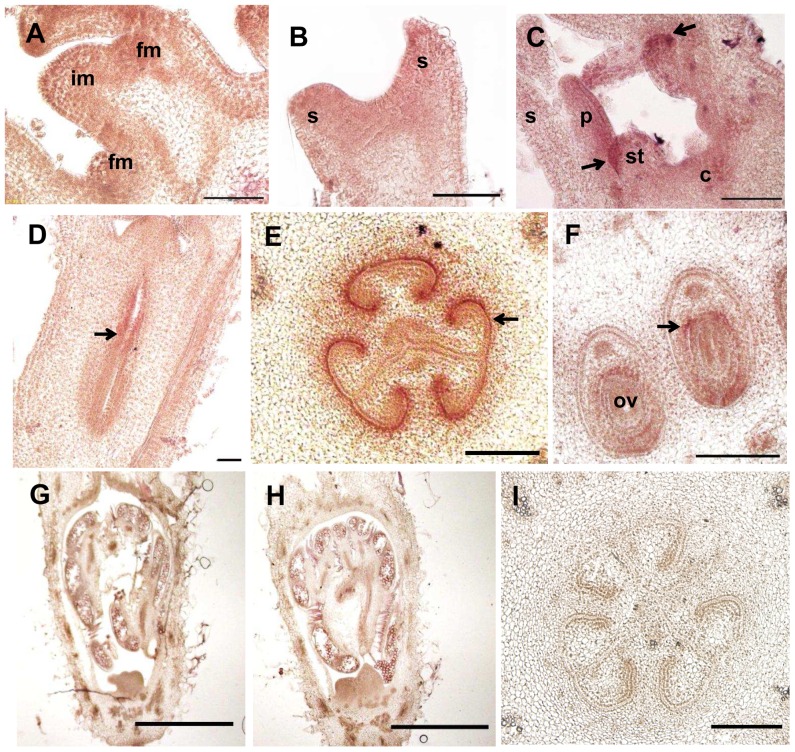
We further characterized the spatial and temporal expression pattern of CsSUP during flower and fruit development through in situ hybridization (Figure 3). Consistent with the results in qRT-PCR, CsSUP expression is detected throughout the inflorescence meristem (IM), floral meristem (FM) and young floral primordia during stage 1–2 [27]. By flowers develop into stage 4, CsSUP expression is limited to the boundary between petal and stamen (arrow in Figure 3C). CsSUP shows high expression in the developing ovary of female flower (arrow in Figure 3D), specifically, in the boundary of developing ovary (arrow in Figure 3E), and the developing ovules (arrow in Figure 3F). In male flower primordia (Figure 3G), CsSUP shows no detectable signal as compared to the sense control hybridizations (Figure 3H–I).
Figure 3. In situ hybridization of CsSUP transcripts in developing flowers and fruits of cucumber.
Longitudinal sections of the shoot apex (A), stage 2 flower (B), stage 4 flower (C) and stage 8 female flower (D) reveal that CsSUP is expressed throughout in the IM, FM and young floral primordia (stage 1–2), and then limited to the boundary between petal and stamen (arrow in C), and the developing ovary (arrow in D). Transverse sections reveal that CsSUP is specifically expressed in the boundary of developing ovary (arrow in E), and the developing ovules (arrow in F). CsSUP is undetectable in the male flower primordia (G). Control hybridizations with CsSUP sense probe in male (H) or female flower primordia (I) show no signal. Bar = 200 µm except for (I), in which bar = 100 µm.
Phenotypes of ectopic CsSUP expression in Arabidopsis
To understand the function of CsSUP, ectopic expression analysis was first performed in the sup-5 mutant Arabidopsis plants under AtSUP promoter (pAtSUP) or 35S promoter of Cauliflower mosaic virus (CaMV). As shown in Figure 4, ectopic CsSUP expression can partially rescue the sup-5 mutant phenotype. In the sup-5 mutant, number of stamens and carpels are increased, silique length is decreased and seed number is reduced (Figure 4A, 4D, 4G and 4H) [28], ectopic expression of CsSUP under native AtSUP promoter (pAtSUP::CsSUP;sup-5) (Figure 4B, 4E,4G and 4H) or constitutive 35S promoter (35S::CsSUP;sup-5) (Figure 4C, 4F and 4G) results in normal number of stamens and partially rescued silique length and morphology. For example, the ratio of normal silique is 23% in the sup-5 mutant, and it increases to 50% in the pAtSUP::CsSUP;sup-5, and to 83% in the 35S::CsSUP;sup-5 (Table 1). Further, 35S::CsSUP;sup-5 generally has better rescue effects than that of pAtSUP::CsSUP;sup-5 with regards to both flower and silique development (Figure 4). Consistently, expression of CsSUP is higher in the 35S::CsSUP;sup-5 lines as compared to those in the pAtSUP::CsSUP;sup-5 lines (Figure 4I). We further quantified the seed numbers in sup-5 and 35S::CsSUP;sup-5. As shown in Table 2, ectopic expression of CsSUP significantly increased the seed numbers per silique in the sup-5 mutant background, suggesting that the function of SUP is largely conserved between complete flower Arabidopsis and unisexual flower cucumber.
Figure 4. Ectopic CsSUP expression can partially rescue the phenotype of sup-5 mutant Arabidopsis.
Flowers of sup-5 (A, D), pAtSUP::CsSUP;sup-5 (B, E) and 35S::CsSUP;sup-5 (C, F) shows the complement of excess stamen upon ectopic CsSUP expression. (G) Representative siliques of sup-5 (top), pAtSUP::CsSUP;sup-5 (top middle), 35S::CsSUP;sup-5 (bottom middle) and Ler (bottom) indicate the partial rescue of the sup-5 silique development by ectopic expression of CsSUP in Arabidopsis. (H) Opened siliques of sup-5 (top) and pAtSUP::CsSUP;sup-5 (bottom) at similar developmental stages. (I) Expression of CsSUP in transgenic Arabidopsis. Lane 1–2: sup-5 plants, lane 3–4: 35S::CsSUP;sup-5 lines, lane 5–6: pAtSUP::CsSUP;sup-5 lines. Actin2 was used as internal control to normalize the expression data. Bars = 1 mm.
Table 1. Quantification of silique phenotype.
| Normal | Mild malformed | Severe malformed | Normal % | |
| sup-5 | 7 | 5 | 18 | 23.33% |
| pAtSUP::CsSUP;sup-5 | 15 | 4 | 11 | 50.00% |
| 35S::CsSUP;sup-5 | 25 | 2 | 3 | 83.33% |
The values shown are silique numbers. Total of 30 siliques were characterized for each genotype.
Table 2. Partial complement of sup-5 mutant upon ectopic expression of CsSUP.
| Control Plant | Seed Number | Transgenic Plants | Seed Number | |
| Ler | 46.10±0.78 | 35S::CsSUP/sup-5 | line1 | 23.67±1.20** |
| sup-5 | 14.80±0.64 | line2 | 26.00±0.87** | |
| line3 | 21.33±1.61** | |||
| line4 | 30.67±1.98** | |||
| Average | 25.41±4.81** | |||
The values shown are the means ± SE of 20 siliques from Arabidopsis Ler and sup-5 mutant, or 6 siliques from CsSUP transgenic lines (35S::CsSUP/sup-5). T-tests were used to determine whether differences between sup-5 and transgenic lines were statistically significant.
* and ** represent p<0.05 and p<0.01, respectively.
Next, we examined the effects upon overexpression of CsSUP in the Ler background (35S::CsSUP/Ler). As indicated in Figure 5, overexpression of CsSUP leads to disorganized petal patterning, suppressed stamen development, and reduced silique length (Figure 5). Compared to the flowers in wide-type plant (Figure 5A), 35S::CsSUP/Ler transgenic flowers display 10–20% petals with aberrant organization (Figure 5B), while petal number and petal size show no noticeable changes. Further, stamen development in the 35S::CsSUP/Ler transgenic flowers is largely suppressed. In contrast to the wild-type flower with six stamens, the 35S::CsSUP transgenic flowers developed an average of 4.39±0.06 stamens (Table 3), and about 10–15% stamens are greatly short (Figure 5C–D). Accordingly, silique length and seed numbers per silique are significantly reduced in the 35S::CsSUP plants (Figure 4E–F, Table 4 and Table 5). For example, the average silique length is 10 mm with 46 seeds per silique in the WT, while it decreases to 6.8 mm with 17 seeds per silique in the 35S::CsSUP transgenic plants (Table 4 and Table 5). At the whole plant level, transgenic plants are slightly dwarf as compared to wide-type plant (data not shown). Taken together with the expression data of CsSUP in Figure 2 and 3, CsSUP may function in the boundaries and ovules to modulate petal patterning, stamen and fruit development in Arabidopsis, probably through negative regulation of cell proliferation.
Figure 5. Phenotypes of overexpression of CsSUP in wild type Arabidopsis.
(A–B) Flowers of wild type (A) and 35S::CsSUP (B) show the disturbed petal organization. (C–D) Stamens of wild type (C) and 35S::CsSUP (D) indicate the suppressed stamen development in the transgenic lines. (E–F) Siliques (E) and opened siliques (F) of wild type (top) and 35S::CsSUP (bottom) show the reduced silique length and decreased seed numbers. Bars = 1 mm.
Table 3. Reduced stamen numbers in 35S::CsSUP transgenic plants.
| Control Plant | Stamen Number | Transgenic Plants | Stamen number | |
| Ler | 6.0±0.0 | 35S::CsSUP/Ler | line1 | 3.97±0.13** |
| line2 | 4.44±0.11** | |||
| line3 | 4.38±0.13** | |||
| line4 | 4.76±0.08** | |||
| Average | 4.39±0.06** | |||
The values shown are the means ± SE of 30 flowers from wild-type Arabidopsis, or 20 flowers from CsSUP transgenic plants (35S::CsSUP/Ler). T-tests were used to determine whether differences between Ler and transgenic lines were statistically significant.
* and ** indicate p<0.05 and p<0.01, respectively.
Table 4. Reduced silique length in 35S::CsSUP transgenic plants.
| Control Plant | Silique Length (mm) | Transgenic Plants | Silique Length (mm) | |
| Ler | 10.19±0.14 | 35S::CsSUP/Ler | line1 | 6.90±0.10** |
| line2 | 5.08±0.10** | |||
| line3 | 8.53±0.14** | |||
| Average | 6.84±0.16** | |||
The values shown are the means ± SE of 30 siliques from Arabidopsis Ler or CsSUP transgenic lines (35S::CsSUP/Ler). T-tests were used to determine whether differences between Ler and transgenic lines were statistically significant.
* and ** represent p<0.05 and p<0.01, respectively.
Table 5. Reduced seed numbers in 35S::CsSUP transgenic plants.
| Control Plant | Seed number/Silique | Transgenic Plants | Seed number/Silique | |
| Ler | 46.10±0.78 | 35S::CsSUP/Ler | line1 | 17.35±0.97** |
| line2 | 7.15±0.49** | |||
| line3 | 26.85±1.56** | |||
| Average | 17.12±1.22** | |||
The values shown are the means ± SE of 20 siliques from Arabidopsis Ler or CsSUP transgenic lines (35S::CsSUP/Ler). T-tests were used to determine whether differences between Ler and transgenic lines were statistically significant.
* and ** show p<0.05 and p<0.01, respectively.
Discussion
During early stages of flower development, all floral buds are bisexual, and unisexual flowers are formed by subsequent arrestment of either carpel or stamen development [29]. Phylogenetic analysis indicated that unisexual flowers evolved from bisexual flowers many times in the angiosperm lineage [20], [30], [31]. It is not surprising that unisexual species and bisexual species have distinct mechanism underlining flower development. So far, most studies about floral patterning are performed in bisexual flowers, while flower development in unisexual species is largely neglected. Here we cloned the ortholog of AtSUP in unisexual species cucumber (CsSUP) (Figure 1), and we characterized the expression pattern of CsSUP by tissue-specific qRT-PCR and in situ hybridization (Figures 2 and 3), and we explored the function of CsSUP by ectopic expression in Arabidopsis (Figures 4 and 5, Tables 1–5). Our data showed that CsSUP played shared as well as divergent roles during flower and fruit development.
CsSUP is expressed mostly in the female specific organs
Among the orthologs of SUP, expression patterns of PhSUP and SlSUP have been explored previously [13], [20], [22]. Transcripts of Arabidopsis SUP were first detected in late stage 3 flower primordia, and in boundaries between stamen and carpel primordia. Then, it was found in the adaxial side of the stamen primordia and later in stage 9, SUP RNA was detected in the ovary [13]. PhSUP, on the other hand, is expressed in the basal regions of developing petals and stamens, the interthecal regions of developing anthers, and the basal part of ovules [20]. While the SlSUP gene is exclusively expressed in female flowers in Silene latifolia, in developing petals, stamens and ovules [22]. In this study, we found that CsSUP is mostly expressed in the female specific organs: the female flower buds and fruits (Figure 2). Specifically, CsSUP is expressed throughout the IM, FM, and stage 1–2 floral primordia, then it is restricted to the boundary between petal and stamen at stage 4. By the time of unisexual flower is noticeable (stage 6 and on), CsSUP is expressed only in female flowers, in the boundary of developing ovary and in ovules (Figure 3). Therefore, the expression patterns of SUP orthologs are quite divergent, implying they may have distinct roles in bisexual flowers, dioecious species or monoecious species. Despite the specific domains are different, both CsSUP and SlSUP are expressed predominantly in female flowers, the reason of which may lie in the fact that both species produce unisexual flowers.
CsSUP has both conserved and divergent functions during flower and fruit development
Arabidopsis AtSUP has been shown to function through cell proliferation to regulate the balance between stamen fate and carpel fate, as well as to control the growth of outer integument of ovule [13], [16]. Loss-of-function of AtSUP results in increased number of stamen, defective carpel and infertile ovules [13], [16]. Ectopic expression of PhSUP or SlSUP in Arabidopsis can partially or fully rescue the excess stamen and infertile ovule phenotype in sup mutants [20], [22]. Similarly, our study showed that overexpression of CsSUP can partially complement the sup-5 mutant phenotype, particularly the number of stamens, silique length and number of seeds per silique (Figure 4, Tables 1–2), suggesting that CsSUP plays a conserved role during stamen and ovule development. However, constitutive expression of CsSUP in wild type Arabidopsis leads to disorganized patterning of petal, reduced number of stamen, decreased length of silique, reduced seed production and mild dwarfness of the whole plant (Figure 4). Dwarfness, suppressed petal and stamen development have been reported previously when ectopic expression of AtSUP or SUP orthologs under the 35S promoter [14], [19], [20], [22], while decreased silique length and seed production, and disorganized petal patterning appears to be specific to CsSUP, suggesting that monoecious CsSUP may evolved a distinct roles in petal patterning and fruit development as compared to SUP orthologs in bisexual or dioecious species. Considering the unique expression of CsSUP in the boundary between petal and stamen primordia (Figure 3C), the boundary of developing ovary (Figure 3E), and in ovules (Figure 3F), CsSUP may function in the boundaries and ovules to modulate petal patterning, stamen and fruit development in Arabidopsis, probably through negative regulation of cell proliferation. Previous studies showed that depending on the species, cell types and expression level, AtSUP and SUP orthologs stimulated or suppressed cell division, cell expansion or cell elongation so that enabling proper flower and ovule development [17], [18], [20], [21], [24], [32]. Recent studies showed that Arabidopsis and tobacco SUPs regulated cell proliferation through auxin and cytokinin signaling pathways in tobacco [24], and AtSUP suppresses cell division in floral meristem redundantly with a RNA helicase CARPEL FACTORY [33], and epigenetic modification-methylation is involved in the active or inactive state of AtSUP [34], [35] Therefore, tackling the underlining mechanism of CsSUP function in Arabidopsis, especially the potential relationship between CsSUP with hormones, RNA helicase and methylation modification during petal patterning and silique development, would be useful for gaining an entrance into understand how CsSUP might function in cucumber. Further studies through RNA interference and genetic transformation, to explore the precise roles of CsSUP in cucumber would shed light on the evolutional scenery of SUP function in hermaphrodite, dioecious and monoecious species of flowering plants.
Materials and Methods
Plant materials and growth
Cucumber (Cucumis sativus L.) inbred line 1461 was used in this study. Seeds were grown in pots in a regulated chamber at 28°C in a 16 h light/8 h dark cycle for about a month, and then they were moved to greenhouse for further development. The Arabidopsis Landsberg erecta (Ler) was used as wide-type control in the transgenic analysis. The sup-5 mutant was obtained from the Arabidopsis Biological Resource Center (ABRC). Arabidopsis seeds were sown in soil or Murashige-Skoog (MS) medium (0.2% agar and 1% sucrose) at 23°C on a 16 h light/8 h dark cycle.
Isolation of CsSUP
Total RNA was extracted from female flower buds using the Huayueyang RNA isolation kit (China), and 3 µg of samples were used to synthetize cDNA using M-MLV Reverse Transcriptase (Promega). The cDNA samples were amplified by PCR via the following system: 95°C for 5 min; 30 cycles of 95°C for 30 s, 53°C for 30 s, and 72°C for 30 s; then 72°C for 10 min.
Sequence comparison and phylogenetic analysis
Amino acid sequences of CsSUP and SUP-like genes were obtained by BLAST searches (http://www.ncbi.nlm.nih.gov/BLAST/) and aligned by ClustalW software (http://clustalw.ddbj.nig.ac.jp/top-j.html/). A phylogenetic tree based on their entire amino acid sequences was constructed by applying the neighbor joining (NJ) method using the bootstrap analysis with1000 replications [26]. The GeneBank accession numbers of the amino acid sequences are: AtSUP (U38946), PhSUP1 (AB117749), NtSUP (GQ227844), SlSUP (BAH59432), RBE (AB107371), Os05g0286100 (BAF17009). The sequence data for the three SUP-like genes in cucumber can be found in the Cucumber Genome Initiative databases (http://cucumber.genomics.org.cn) under the following accession numbers: CsSUP (Csa000134), Csa001112 and Cas010435.
Quantitative real-time RT-PCR
The leaves, tendrils, flower buds, flowers and fruits were frozen in liquid nitrogen and stored at −80°C prior to RNA extraction. Total RNA was extracted with Huayueyang RNA isolation kit (China), cDNA was synthetized using M-MLV Reverse Transcriptase (Promega). Quantitative RT-PCR was performed using an Applied Biosystems 7500 real-time PCR systems with SYBR Green as fluorescent dyes (TaKaRa). Three biological replicates were performed, upon which three technical replicates were used for the qRT-PCR analysis. 18S rRNA was used as reference control to normalize the expression data [36]. The gene specific primers are listed in Table S1.
Semi-quantitative RT-PCR
Inflorescence of sup-5, pAtSUP::CsSUP;sup-5, 35S::CsSUP;sup-5 were frozen in liquid nitrogen and stored at −80°C prior to RNA extraction. Total RNA was extracted with Huayueyang RNA isolation kit (China), cDNA was synthetized using M-MLV Reverse Transcriptase (Promega). Actin2 was used as internal control to normalize the expression data. The gene specific primers are listed in Table S1.
In situ hybridization
Cucumber flower buds and young fruits were fixed, embedded, sectioned, and hybridized with digoxigenin (DIG)-labeled sense and antisense RNA probes as described [37]. Through PCR amplification of cDNA, in situ probes were synthesized using gene specific primers including T7 and SP6 RNA polymerase-binding sites. T7 RNA polymerase was used for the synthesis of antisense probes and SP6 RNA polymerase was used for the generation of sense probes. The primer pairs are listed in Table S1.
Ectopic expression of CsSUP in Arabidopsis
The full-length CsSUP cDNA fragment was amplified by PCR using gene specific primers O-CsSUP-F and O-CsSUP-R containing Xba I and Sma I restriction enzyme sites respectively. The resulting fragment was digested and fused into the pBI121 vector. The resulting CsSUP-pBI121 construct driven by CaMV35S promoter was introduced to Agrobacterium tumefaciens by electric shock to transform wide-type (Ler) and sup-5 mutant of Arabidopsis using the floral-dip method [38]. Meantime, The 2.2 kb length of AtSUP promoter was amplified by PCR using gene specific primers pAtSUP-F and pAtSUP-F containing Cla I and Xba I restriction enzyme sites respectively. The resulting fragment was digested and fused into the CsSUP-pBI121 construct, and transformed into sup-5 mutant of as above. Transgenic plants were screened by MS medium with 40 mg/L kanamycin, and resistant plants were verified by CsSUP specific primers CsSUP-F and CsSUP-R.
Supporting Information
Quantitative RT-PCR (qRT-PCR) analysis of Cs001112 in different organs of cucumber. Csa001112 is predominately expressed in the fruit. lf: leaves, te: tendrils, mb: male flower buds, fb: female flower buds, mf: male flowers, ff: female flowers, fr-4: fruit of 4 days before flower opening, fr: fruit on flower opening, fr+3: fruit of 3 days after flower opening. Three biological replicates were used for each sample, and 18S rRNA was used as internal control. Bars represent the standard error.
(TIF)
Oligonucleotide primers used in this study.
(DOCX)
Acknowledgments
We thank members of the Zhang lab for discussion and technique help during the course of this study, the Arabidopsis Biological Resource Center for providing the sup-5 seeds, and Dr Jinsheng Lai for providing real time qRT-PCR machine and growth chambers.
Funding Statement
This work was supported by National basic research of China 973 program [2012CB113900], National Natural Science Foundation of China [31171399], and Chinese Universities Scientific Fund [2011JS067, 2013RC030] to XZ. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Yampolsky C, Yampolsky H (1922) Distribution of the sex forms in the phanerogamic flora. Bibl Genet 3: 1–62. [Google Scholar]
- 2. Renner SS, RicklefsI RE (1995) Dioecy and its correlates in the flowering plants. Am J Bot 82: 596–606. [Google Scholar]
- 3. Bowman JL, Smyth DR, Meyerowitz EM (1991) Genetic interactions among floral homeotic genes of Arabidopsis. Development 112: 1–20. [DOI] [PubMed] [Google Scholar]
- 4. Coen ES, Meyerowitz EM (1991) The war of the whorls: genetic interactions controlling flower development. Nature 353: 31–37. [DOI] [PubMed] [Google Scholar]
- 5. Weigel D, Meyerowitz EM (1994) The ABCs of floral homeotic genes. Cell 78: 203–209. [DOI] [PubMed] [Google Scholar]
- 6. Colombo L, Franken J, Koetje E, van Went J, Dons HJ, et al. (1995) The petunia MADS box gene FBP11 determines ovule identity. Plant Cell 7: 1859–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pelaz S, Ditta GS, Baumann E, Wisman E, Yanofsky MF (2000) B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405: 200–203. [DOI] [PubMed] [Google Scholar]
- 8. Favaro R, Pinyopich A, Battaglia R, Kooiker M, Borghi L, et al. (2003) MADS-box protein complexes control carpel and ovule development in Arabidopsis. Plant Cell 15: 2603–2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ditta G, Pinyopich A, Robles P, Pelaz S, Yanofsky MF (2004) The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr Biol 14: 1935–1940. [DOI] [PubMed] [Google Scholar]
- 10. Sridhar VV, Surendrarao A, Liu Z (2006) APETALA1 and SEPALLATA3 interact with SEUSS to mediate transcription repression during flower development. Development 133: 3159–3166. [DOI] [PubMed] [Google Scholar]
- 11. Schultz EA, Pickett FB, Haughn GW (1991) The FLO10 gene product regulates the expression domain of homeotic genes AP3 and PI in Arabidopsis flowers. Plant Cell 3: 1221–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bowman JL, Sakai H, Jack T, Weigel D, Mayer U, et al. (1992) SUPERMAN, a regulator of floral homeotic genes in Arabidopsis. Development 114: 599–615. [DOI] [PubMed] [Google Scholar]
- 13. Sakai H, Medrano LJ, Meyerowitz EM (1995) Role of SUPERMAN in maintaining Arabidopsis floral whorl boundaries. Nature 378: 199–203. [DOI] [PubMed] [Google Scholar]
- 14. Hiratsu K, Ohta M, Matsui K, Ohme-Takagi M (2002) The SUPERMAN protein is an active repressor whose carboxy-terminal repression domain is required for the development of normal flowers. FEBS Lett 514: 351–354. [DOI] [PubMed] [Google Scholar]
- 15. Hiratsu K, Mitsuda N, Matsui K, Ohme-Takagi M (2004) Identification of the minimal repression domain of SUPERMAN shows that the DLELRL hexapeptide is both necessary and sufficient for repression of transcription in Arabidopsis. Biochem Biophys Res Commun 321: 172–178. [DOI] [PubMed] [Google Scholar]
- 16. Gaiser JC, Robinson-Beers K, Gasser CS (1995) The Arabidopsis SUPERMAN Gene Mediates Asymmetric Growth of the Outer Integument of Ovules. Plant Cell 7: 333–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yun JY, Weigel D, Lee I (2002) Ectopic expression of SUPERMAN suppresses development of petals and stamens. Plant Cell Physiol 43: 52–57. [DOI] [PubMed] [Google Scholar]
- 18. Kater MM, Franken J, van Aelst A, Angenent GC (2000) Suppression of cell expansion by ectopic expression of the Arabidopsis SUPERMAN gene in transgenic petunia and tobacco. Plant J 23: 407–413. [DOI] [PubMed] [Google Scholar]
- 19. Bereterbide A, Hernould M, Castera S, Mouras A (2001) Inhibition of cell proliferation, cell expansion and differentiation by the Arabidopsis SUPERMAN gene in transgenic tobacco plants. Planta 214: 22–29. [DOI] [PubMed] [Google Scholar]
- 20. Nakagawa H, Ferrario S, Angenent GC, Kobayashi A, Takatsuji H (2004) The petunia ortholog of Arabidopsis SUPERMAN plays a distinct role in floral organ morphogenesis. Plant Cell 16: 920–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nandi AK, Kushalappa K, Prasad K, Vijayraghavan U (2000) A conserved function for Arabidopsis SUPERMAN in regulating floral-whorl cell proliferation in rice, a monocotyledonous plant. Curr Biol 10: 215–218. [DOI] [PubMed] [Google Scholar]
- 22. Kazama Y, Fujiwara MT, Koizumi A, Nishihara K, Nishiyama R, et al. (2009) A SUPERMAN-like gene is exclusively expressed in female flowers of the dioecious plant Silene latifolia. Plant Cell Physiol 50: 1127–1141. [DOI] [PubMed] [Google Scholar]
- 23. Huang S, Li R, Zhang Z, Li L, Gu X, et al. (2009) The genome of the cucumber, Cucumis sativus L. Nat Genet 41: 1275–1281. [DOI] [PubMed] [Google Scholar]
- 24. Nibau C, Di Stilio VS, Wu HM, Cheung AY (2011) Arabidopsis and Tobacco SUPERMAN regulate hormone signalling and mediate cell proliferation and differentiation. J Exp Bot 62: 949–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Takeda S, Matsumoto N, Okada K (2004) RABBIT EARS, encoding a SUPERMAN-like zinc finger protein, regulates petal development in Arabidopsis thaliana. Development 131: 425–434. [DOI] [PubMed] [Google Scholar]
- 26. Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425. [DOI] [PubMed] [Google Scholar]
- 27. Bai SL, Peng YB, Cui JX, Gu HT, Xu LY, et al. (2004) Developmental analyses reveal early arrests of the spore-bearing parts of reproductive organs in unisexual flowers of cucumber (Cucumis sativus L.). Planta 220: 230–240. [DOI] [PubMed] [Google Scholar]
- 28. Jacobsen SE, Meyerowitz EM (1997) Hypermethylated SUPERMAN epigenetic alleles in arabidopsis. Science 277: 1100–1103. [DOI] [PubMed] [Google Scholar]
- 29. Malepszy S, Niemirowicz-Szczytt K (1991) Sex determination in cucumber (Cucumis sativus) as a model system for molecular biology. Plant Science 80: 39–47. [Google Scholar]
- 30. LebelHardenack S, Grant SR (1997) Genetics of sex determination in flowering plants. Trends Plant Sci 2: 130–136. [Google Scholar]
- 31. Charlesworth D (2002) Plant sex determination and sex chromosomes. Heredity (Edinb) 88: 94–101. [DOI] [PubMed] [Google Scholar]
- 32. Bereterbide A, Hernould M, Farbos I, Glimelius K, Mouras A (2002) Restoration of stamen development and production of functional pollen in an alloplasmic CMS tobacco line by ectopic expression of the Arabidopsis thaliana SUPERMAN gene. Plant J 29: 607–615. [DOI] [PubMed] [Google Scholar]
- 33. Jacobsen SE, Running MP, Meyerowitz EM (1999) Disruption of an RNA helicase/RNAse III gene in Arabidopsis causes unregulated cell division in floral meristems. Development 126: 5231–5243. [DOI] [PubMed] [Google Scholar]
- 34. Cao X, Jacobsen SE (2002) Role of the arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr Biol 12: 1138–1144. [DOI] [PubMed] [Google Scholar]
- 35. Jackson JP, Lindroth AM, Cao X, Jacobsen SE (2002) Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416: 556–560. [DOI] [PubMed] [Google Scholar]
- 36. Wan H, Zhao Z, Qian C, Sui Y, Malik AA, et al. (2010) Selection of appropriate reference genes for gene expression studies by quantitative real-time polymerase chain reaction in cucumber. Anal Biochem 399: 257–261. [DOI] [PubMed] [Google Scholar]
- 37. Zhang X, Madi S, Borsuk L, Nettleton D, Elshire RJ, et al. (2007) Laser microdissection of narrow sheath mutant maize uncovers novel gene expression in the shoot apical meristem. PLoS Genet 3: e101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Quantitative RT-PCR (qRT-PCR) analysis of Cs001112 in different organs of cucumber. Csa001112 is predominately expressed in the fruit. lf: leaves, te: tendrils, mb: male flower buds, fb: female flower buds, mf: male flowers, ff: female flowers, fr-4: fruit of 4 days before flower opening, fr: fruit on flower opening, fr+3: fruit of 3 days after flower opening. Three biological replicates were used for each sample, and 18S rRNA was used as internal control. Bars represent the standard error.
(TIF)
Oligonucleotide primers used in this study.
(DOCX)