Abstract
Ku70 and Ku80 form a heterodimer called Ku that forms a holoenzyme with DNA dependent-protein kinase catalytic subunit (DNA-PKCS) to repair DNA double strand breaks (DSBs) through the nonhomologous end joining (NHEJ) pathway. As expected mutating these genes in mice caused a similar DSB repair-defective phenotype. However, ku70-/- cells and ku80-/- cells also appeared to have a defect in base excision repair (BER). BER corrects base lesions, apurinic/apyrimidinic (AP) sites and single stand breaks (SSBs) utilizing a variety of proteins including glycosylases, AP endonuclease 1 (APE1) and DNA Polymerase β (Pol β). In addition, deleting Ku70 was not equivalent to deleting Ku80 in cells and mice. Therefore, we hypothesized that free Ku70 (not bound to Ku80) and/or free Ku80 (not bound to Ku70) possessed activity that influenced BER. To further test this hypothesis we performed two general sets of experiments. The first set showed that deleting either Ku70 or Ku80 caused an NHEJ-independent defect. We found ku80-/- mice had a shorter life span than dna-pkcs-/- mice demonstrating a phenotype that was greater than deleting the holoenzyme. We also found Ku70-deletion induced a p53 response that reduced the level of small mutations in the brain suggesting defective BER. We further confirmed that Ku80-deletion impaired BER via a mechanism that was not epistatic to Pol β. The second set of experiments showed that free Ku70 and free Ku80 could influence BER. We observed that deletion of either Ku70 or Ku80, but not both, increased sensitivity of cells to CRT0044876 (CRT), an agent that interferes with APE1. In addition, free Ku70 and free Ku80 bound to AP sites and in the case of Ku70 inhibited APE1 activity. These observations support a novel role for free Ku70 and free Ku80 in altering BER.
Introduction
BER corrects a broad spectrum of DNA lesions [1] caused by reactive oxygen species (ROS) and alkylating agents [2] that would otherwise result in point mutations [3]. The damaged nucleotide is first recognized by one of many DNA damage specific glycosylases [4]. For example 8-oxoguanosine-glycosylase 1 (OGG1) is the primary glycosylase to excise the major ROS-induced base lesion, 8-oxoG. Glycosylases remove the damaged base to generate an apurinic/apyrimidinic (AP) site. AP endonuclease 1 (APE1) then makes a nick 5′ to the AP site, generating a dRP (deoxyribose phosphate) intermediate and a one base gap. DNA Polymerase β (Pol β) then fills in the missing nucleotide while its lyase activity generates a 5′ phosphorylated DNA strand by excising the 5′ terminal dRP residue so that DNA ligase can repair the nick. BER also repairs DNA single strand breaks (SSBs) that form spontaneously at AP sites, as a DNA repair intermediate or after exposure to ROS. XRCC1 is critical for repairing SSBs by interacting with a number of BER proteins including APE1 [5], [6]–[8], and PARP-1[6], [9]. Thus, deletion of BER components disables the repair of base lesions, AP sites and SSBs[10].
By contrast NHEJ repairs DNA DSBs. To initiate NHEJ, Ku70 and Ku80 form a heterodimer called Ku that forms a holoenzyme with DNA-PKCS [11]. Cells deleted for any of these proteins exhibited telomere end fusion [12], hypersensitivity to clastogenic agents, and premature replicative senescence [13]. Mice deleted for these proteins exhibited premature aging [14]–[18]. Thus, deletion of Ku70, Ku80 or DNA-PKCS resulted in a similar phenotype demonstrating a common defect in the holoenzyme. In addition, XRCC4 and DNA ligase IV form a heterodimer to join the broken ends. Cells deleted for either of these proteins also exhibited hypersensitivity to clastogens, premature replicative senescence [19] and early aging [20]. Thus, deletion of NHEJ proteins caused a similar phenotype.
However, our data also show that cells deleted for either Ku70 or Ku80 exhibited an NHEJ-independent phenotype. We found cells deleted for Ku70 or Ku80 were hypersensitive to ROS and alkylating agents implicating defective BER [21], [22]. However, cells deleted for Lig4 did not exhibit these hypersensitivities exonerating defective NHEJ. In addition, extracts from cells deleted for Ku80, but not Lig4, exhibited reduced BER capacity (correction of a U/G mismatch). Furthermore, ectopic expression of OGG1 or PARP-1 in Ku80-deleted cells rescued hypersensitivity to ROS [22] suggesting Ku80 deletion disabled BER. These data suggested Ku80-deletion caused a BER defect that was unrelated to NHEJ.
We hypothesized that free Ku70 (not bound to Ku80) and free Ku80 (not bound to Ku70) could influence BER. Cells deleted for either Ku70 or Ku80 did not exhibit the same sensitivity to ROS agents [21] and extracts from cells deleted for Ku80, but not Ku70, exhibited reduced BER capacity (correction of a U/G mismatch) [22]. We further hypothesized that free Ku70 could have activity in ku80-/- cells and free Ku80 could have activity in ku70-/- cells [21], [22]. This is possible since some Ku80 remains in the absence of Ku70 [23] and vice versa [24]. To support this hypothesis, we found deleting Ku70 resulted in a milder phenotype than deleting Ku80 in p53-mutant mice and this milder phenotype depended on Ku80 [21]. Thus, after deleting Ku70 in p53-mutant mice, there were two events that contributed to the phenotype: first, the loss of Ku70 and second, the activity of free Ku80. It is also possible that free Ku70 could contribute to the Ku80-mutant phenotype.
Here we test our model that proposes free Ku70 and/or free Ku80 alter BER. First, we provide more data that show deletion of either Ku70 or Ku80 caused a phenotype that was not due to defective NHEJ. We found that ku80-/- mice had a shorter life span than dna-pkcs-/- mice suggesting a phenotype greater than simply deleting NHEJ. Next we show that ku70-/- mice exhibited an elevated level of point mutations in the brain but only in the absence of p53 suggesting a defect in BER. We also show that Ku80-deletion in cells inhibited BER through a novel mechanism that is not epistatic to Pol β. Second, we present data that show free Ku70 and free Ku80 influence BER. Deletion of either Ku70 or Ku80, but not both, disabled the repair of AP sites. Interestingly, free Ku70 and free Ku80, but not the Ku heterodimer, associated with AP sites and free Ku70 inhibited APE1 activity in vitro. Thus, Ku70 and Ku80 appear to have activity outside NHEJ and the Ku heterodimer that influences BER.
Results
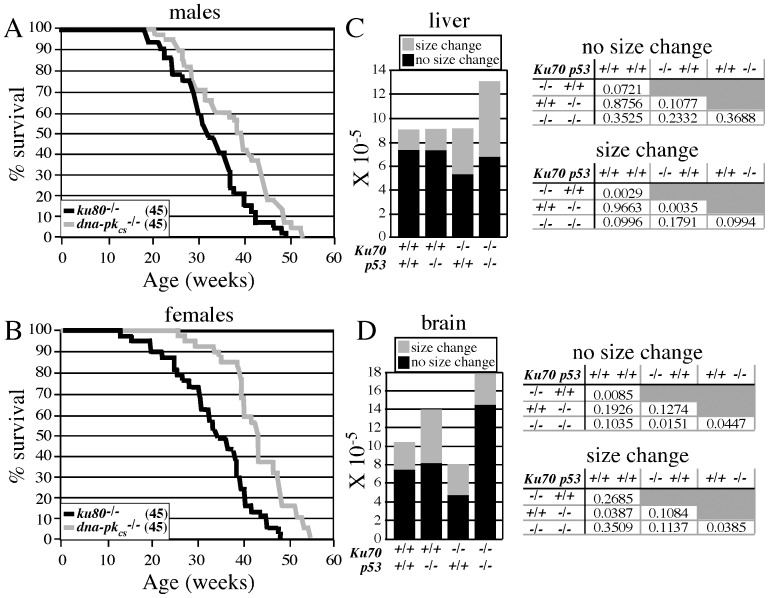
Ku80-deleted mice have a shorter life span than DNA-PKCS-deleted mice
Deletion of either Ku70 of Ku80 caused any early aging phenotype implicating a defect in NHEJ [16]. Yet in a p53-mutant background the ku70-/- mice lived significantly longer than the ku80-/- mice due to a lower incidence of pro-B cell lymphoma [21]. The presence of Ku80 was essential for this milder phenotype. Thus, we proposed that Ku80 functioned outside of the Ku heterodimer. To investigate the possibility that Ku80 functioned outside of the DNA-PK holoenzyme, we measured the life spans for mice deleted for either Ku80 or DNA-PKCS [11]. To guard against phenotypic variation due to differences in genetic background and environment, C57Bl6/J males were crossed to FVB females (both are Ku80+/- DNA-PKCS+/-) such that all mice were C57Bl6/J*FVB F1 hybrid brothers and sisters raised in the same cages. The ku80-/- mice exhibited a shorter life span than the dna-pkcs-/- mice for both males (Fig. 1A, p = 0.01) and females (Fig. 1B, p<0.0001). Thus, deleting Ku80 is more severe than deleting DNA-PKCS. Even though the mechanism for these different life spans is not known, this observation demonstrates that Ku80 has a function greater than the DNA-PK holoenzyme.
Figure 1. In vivo analysis. Life span for ku80-/- and dna-pkcs-/- (A) males and (B) females (45 mice in each cohort).
Mutation spectrum in ku70-/- mice with and without p53 in the (C) liver and (D) brain. Size changes are chromosomal rearrangements that include translocations and large insertions/deletions. No size changes are point mutations (base changes and small insertions/deletions). A student t test was performed for a statistical analysis and tables are presented showing all possible comparisons.
Ku70-mutant mice exhibited an elevated level of point mutations in the absence of p53
Previously we analyzed ku80-/- mice for the level of chromosomal rearrangements and point mutations in the liver and brain. We found Ku80-deletion increased levels of chromosomal rearrangements in a variety of tissues including the liver, as expected for defective NHEJ [25]. However, there was no significant increase in rearrangements in the brain, instead there was a decrease in point mutations. We surmised that p53-mediated DNA damage responses limited the number of recovered mutations. Here we tested if p53-dependent DNA damage responses influenced the level of mutations in ku70-/- livers (Fig. 1C) and brains (Fig. 1D) with and without p53 using a forward mutation detection system based on lacZ. We crossed the Ku70 mutation [26] and the p53 mutation [27] into mice that carry the pUR288-lacZ reporter [28]. These mice were sacrificed at 4 months and analyzed for the levels of rearrangements (size change) and point mutations (no size change) in their livers and brains.
First we looked at the level of chromosomal rearrangements. Similar to the results with ku80-/- mice [25], Ku70-deletion increased chromosomal rearrangements in liver, but not brain. In addition, we found that p53-deletion further increased the levels of chromosomal rearrangements in the liver, but not brain. However, this was not statistically significant because of one outlier among the replicate determinations in the double knockouts (refer to Fig. 2C, D for p values). Overall, however, it is possible to conclude that the liver is more prone to carry chromosomal rearrangements than the brain.
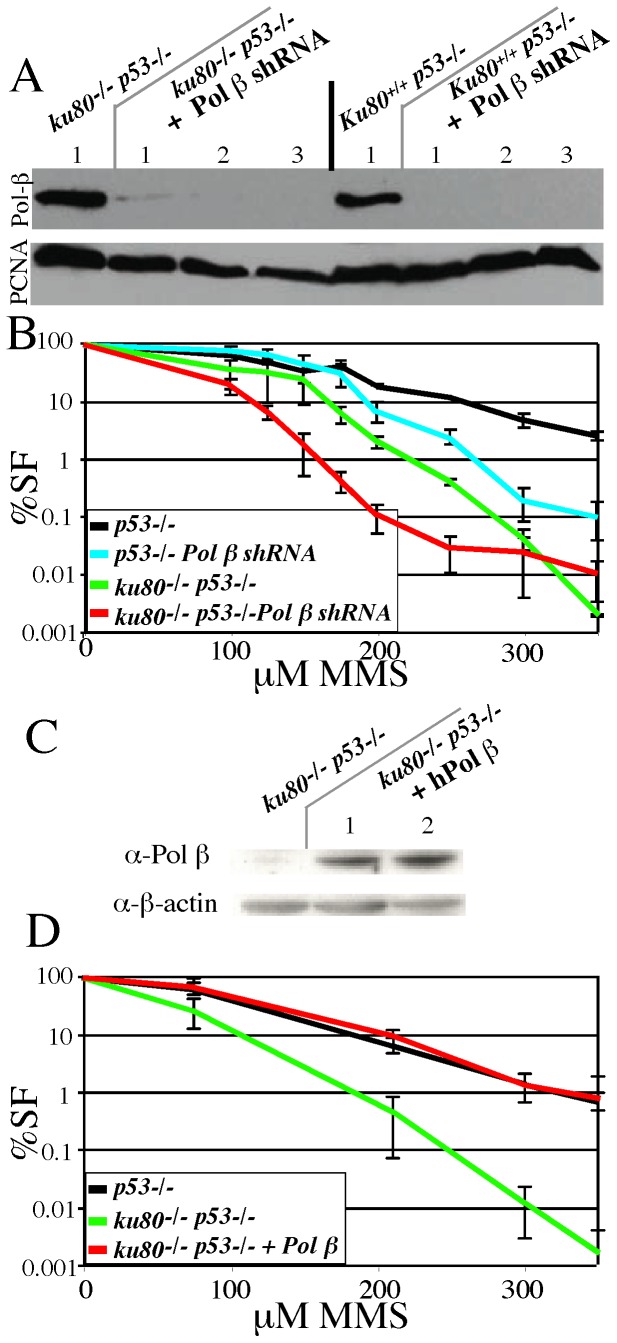
Figure 2. Epistatic analysis for Ku80 and Pol β.
All cells are deleted for p53 (even controls) to avoid early replicative senescence. Shown is the average of three experiments. (A) Western showing RNAi knockdown of Pol β in p53-/- control and ku80-/- p53-/- fibroblasts that stably express a shRNA plasmid specific for mouse Pol β (three clones). PCNA was loaded to normalize for nuclear protein levels. The expression of endogenous Pol β in the shRNA-transduced cell lines was undetectable by immunoblot, consistent with our earlier reports for this mouse Pol β -specific shRNA [30]. (B) Dose-response to MMS for p53-/- control and ku80-/- p53-/- fibroblasts with and without mouse Pol β shRNA expression. (C) Western showing increased Pol β levels for the ku80-/- p53-/- fibroblasts that stably express a Pol β expression plasmid (two clones). Beta-actin was loaded to normalize for cellular protein levels. (D) Pol β -overexpression rescues Ku80-mutant phenotype for MMS.
Next we looked at point mutations. Similar to ku80-/- brains [29], we found decreased levels of small mutations in ku70-/- p53+/+ brains compared to Ku70+/+ p53+/+ brains. The levels of small mutations were also lower as compared to Ku70+/+ p53-/- brains, but this was not significant. Yet, ku70-/- p53-/- brains exhibited increased point mutations as compared to ku70-/- p53+/+ and Ku70+/+ p53-/- brains. Thus, p53 reduced the level of small mutations in the ku70-/- brain. This observation supports the hypothesis that deletion of either Ku70 or Ku80 increased base lesions that are then limited through p53-mediated responses and corroborate our previous observations with tissue culture cells that either Ku70 and/or Ku80 influence the repair of base lesions [22].
The increased sensitivity to an alkylating agent in cells compromised for Ku80 and Pol β was additive
The above experiments support the possibility that deleting either Ku70 or Ku80 altered BER in mice. Therefore, we performed an epistatic analysis for Ku80 and Pol β to determine if they repaired methyl methanesulfonate (MMS)-induced lesions via the same sub-pathway [2]. We analyzed mouse embryonic fibroblasts deleted for p53 since NHEJ-deletion caused p53-mediated replicative senescence that prevents their proliferation; therefore, all cells were deleted for p53, including control cells [13], [21]. Pol β was depleted in p53-/- control and ku80-/- p53-/- MEFs by RNA interference using a mouse Pol β -specific shRNA-expressing lentivirus similar to one we previously reported [30]. We used this lentivirus to transduce p53-/- control and ku80-/- p53-/- MEF to generate corresponding cells with a deficiency in the expression of Pol β (Fig. 2A). Each group of cells was also transduced with a control, GFP-expressing lentivirus. These cells were then used to perform a dose-response analysis to MMS. We found Pol β depletion increased the sensitivity to MMS for control and ku80-/- p53-/- cells; yet the latter appear more sensitive than the former genotype (Fig. 2B). Thus, Pol β -mediated repair of MMS-induced lesions in both control and ku80-/- p53-/- MEFs. We also found Pol β over-expression (Fig. 2C) rescued sensitivity to MMS for ku80-/- p53-/- cells (Fig. 2D). Thus, these data suggest that Pol β and Ku80 are not epistatic and that Ku80 deletion negatively impacts the gap tailoring or DNA synthesis/ligation stages of BER without disabling Pol β [2].
Free Ku70 and free Ku80 sensitize cells to an APE1 inhibitor
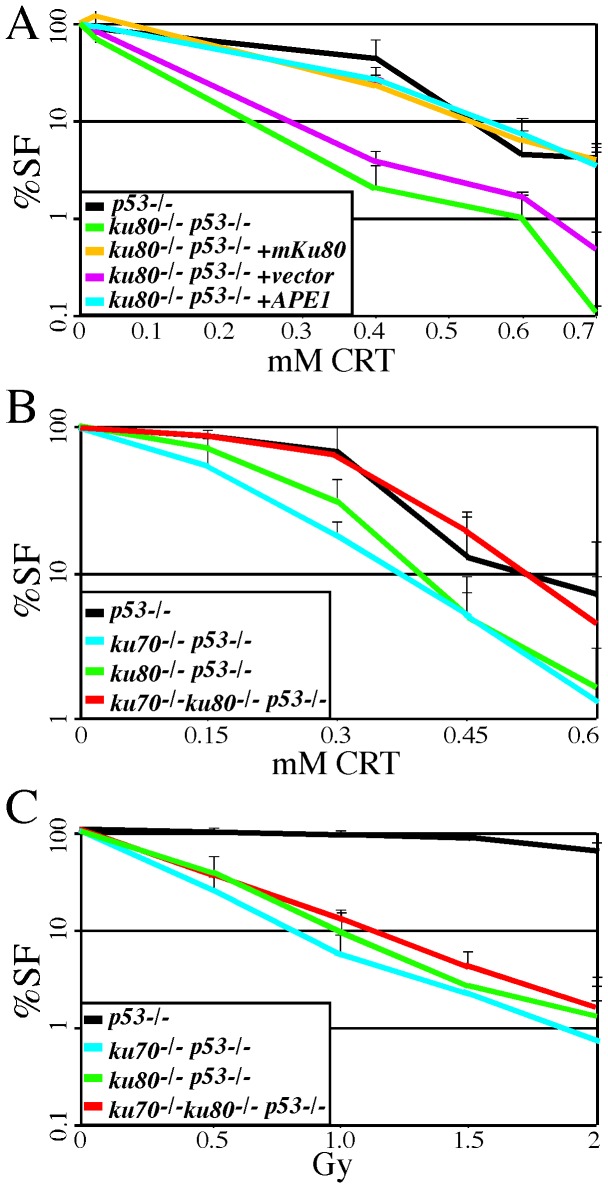
Our previously published results in mice [21] and tissue culture cells [22] suggest that either Ku70 or Ku80 function outside of the Ku heterodimer to influence BER. We also found that Ku80-deletion decreased the capacity to repair AP sites [22]. Therefore, we tested APE1 capacity in mouse fibroblasts deleted for Ku70 or Ku80 or both to provide a complementary biological analysis. We found that ku80-/- p53-/- mouse embryonic fibroblasts [13] were hypersensitive to CRT0044876 (CRT), an agent that specifically inhibits APE1 nicking [31] (Fig. 3A). Expression of mouse Ku80, but not vector alone, rescued hypersensitivity. Ectopic APE1 overexpression also rescued CRT hypersensitivity, supporting the possibility that free Ku70 competitively inhibited APE1 nicking. We then compared ku70-/- p53-/- and ku80-/- p53-/- dermal fibroblasts [21] to ku70-/- ku80-/- p53-/- dermal fibroblasts to determine if free Ku70 caused CRT hypersensitivity in ku80-/- p53-/- cells and if free Ku80 caused CRT hypersensitivity in ku70-/- p53-/- cells. We found ku70-/- p53-/- cells and ku80-/- p53-/- cells, but not ku70-/- ku80-/- p53-/- cells, were hypersensitive to CRT (Fig. 3B). Therefore, free Ku70 and free Ku80 were needed for CRT hypersensitivity. By comparison, all three genotypes were similarly hypersensitive to γ-radiation (Fig. 3C) showing the Ku heterodimer was needed to repair the γ-radiation-induced DSBs. Thus, free Ku70 and free Ku80 affected BER, but not NHEJ.
Figure 3. CRT0044876 (CRT) survival fraction (SF).
All cells are deleted for p53 (even controls) to avoid early replicative senescence. Shown is the average of three experiments. (A) Cells deleted for Ku80 are hypersensitive to CRT. Expression of APE1 or mouse Ku80 rescued CRT hypersensitivity for Ku80-mutant cells. (B) Cells deleted for either Ku70 or Ku80 but not both were hypersensitive to CRT demonstrating independent function for the individual Ku subunits as opposed to the Ku heterodimer. (C) Cells deleted for Ku70 or Ku80 or both were hypersensitive to γ-radiation demonstrating the Ku heterodimer repaired damage as opposed to independent function for the individual proteins.
The individual Ku70 and Ku80 subunits, but not the Ku heterodimer, preferentially bound to a substrate with an AP site
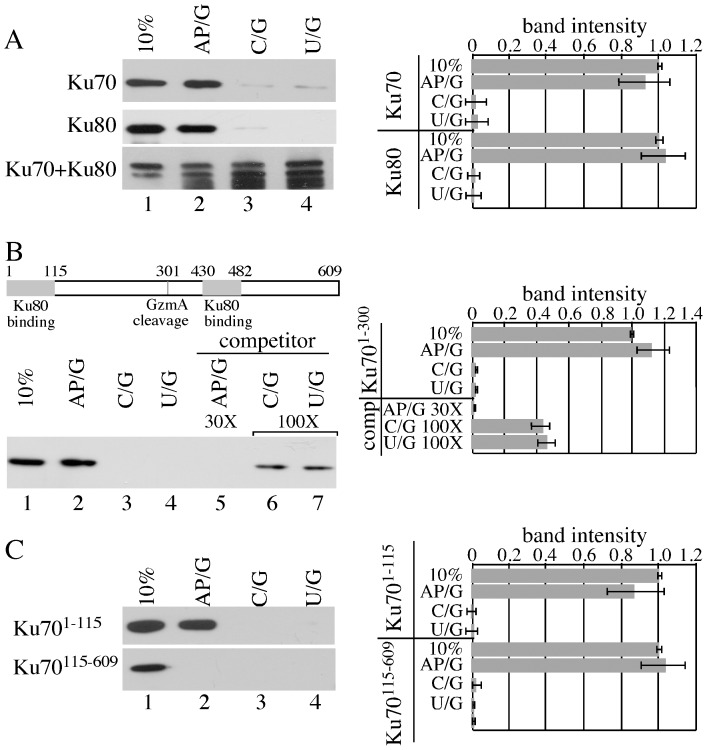
Previously Ku80 was shown to crosslink with AP sites in HeLa cell extracts and the Ku heterodimer along with DNA-PKCS was shown to inhibit AP site cleavage by APE1 [32]. For efficient end joining Ku was also shown to process AP sites at DNA ends with Ku70's 5′-dRP/AP lyase activity [33]. Here we analyzed the potential mechanism for the increased CRT sensitivity observed in cells deleted for Ku70 or Ku80, but not both. We hypothesized that free Ku70 and free Ku80 bound to AP sites to interfere with their repair. This is possible since some Ku80 remains in the absence of Ku70 [23] and vice versa [24]. For this purpose we used an oligonucleotide with an internal U/G mismatch and then converted the uracil to an AP site with uracil DNA glycosylase (UDG). Binding assays were performed with this AP/G substrate and the parental U/G substrate or the same substrate with a C instead of a U (C/G substrate). Biotinylated substrates were exposed to myc-tagged Ku70 (myc-Ku70) or myc-tagged Ku80 (myc-Ku80) after in vitro translation, diluted on ice with NaBH4 to trap proteins that form a Schiff base with the deoxyribose of the AP site [33], [34] and isolated with streptavidin-coated magnetic beads. Western blotting was performed with an anti-myc antibody. Myc-Ku70 exhibited preferential binding to the AP/G substrate as compared to the C/G (Fig. 4A, compare lanes 2 & 3, p = 0.000112, student t test) and U/G (Fig. 4A, compare lanes 2 & 4, p = 0.005) substrates. Myc-Ku80 also exhibited preferential binding to the AP/G substrate as compared to the C/G (Fig. 4A, compare lanes 2 & 3, p = 0.0022) and U/G (Fig. 4A, compare lanes 2 & 4, p = 0.0015) substrates. Thus, free Ku70 and free Ku80 bound to AP sites.
Figure 4. Ku70 and Ku80 bind to AP sites.
We show 10% input for in vitro translated product. The same concentration of biotinylated substrate were used for each reaction of the binding assays. Right panel shows the relative band intensity as measured with the Kodak document program and as compared to 10% input. Shown is the average of three experiments with error bars (standard deviation). Statistics are shown in the results. (A) Ku70 and Ku80, but not Ku70+Ku80, preferentially bind to the AP site. (B) GzmA cleaves Ku70 at Arg(301). Cleavage at this site separates the two Ku80 binding domains. The GzmA N-terminal Ku70 cleavage product (Ku701–300) binding and competition assays. (C) Ku701–115, but not Ku70115-609, bound to the AP/G substrate at a higher level than the C/G and U/G substrates.
The Ku heterodimer binds to DNA ends [35]; therefore, we tested if Ku would preferentially bind to an oligonucleotide substrate with an AP site by combining full length myc-Ku70 and myc-Ku80. However, unlike myc-Ku70 and myc-Ku80 alone, the combination did not preferentially bind to the AP/C substrate as compared to the C/G (Fig. 4A, compare lanes 2 & 3, p = 0.436) and U/G (Fig. 4A, compare lanes 2 & 4, p = 0.344) substrates. Instead, the Ku heterodimer bound to all substrates, implicating end binding. Thus, the Ku heterodimer did not preferentially bind to an AP site. This observation also suggests that free Ku70 and free Ku80 do not efficiently bind to DNA ends as compared to AP sites. Therefore, free Ku70 and free Ku80, but not the Ku heterodimer, bound to AP sites.
N-terminal Ku70, but not C-terminal Ku70, preferentially bound to a substrate with an AP site
Mechanistically, it is not clear how free Ku70 and free Ku80 could inhibit AP site repair in wild type cells or animals. One possibility is that one subunit is degraded while leaving the other intact. Granzyme A (GzmA) cleaves Ku70 at Arg301 to separate the two Ku80 binding domains (Fig. 4B) thereby preventing formation of the Ku heterodimer [36] as a part of the caspase-independent cell death pathway in killer cell cytotoxic granules [37]. Interestingly, GzmA also cleaves APE1 to induce cell death; thus, Ku70 cleavage could further diminish BER.
Since GzmA cleaves Ku70 at Arg301, we tested myc-Ku701–300 for its binding preference to these substrates. Similar to full length Ku70 (1–609), Myc-Ku701–300 exhibited preferential binding to the AP/G substrate as compared to the C/G (p = 0.0179) and U/G (p = 0.0131) substrates (Fig. 4B, lanes 2–4). In addition, a competition assay with myc-Ku701–300 was performed with hot (biotinylated) and cold (not biotinylated) substrate. We found 30X cold AP/G competed out hot AP/G when compared to no competitor (Fig. 4B compare lanes 2 & 5, p = 0.00037). However, 100X cold C/G (Fig. 4B compare lanes 2 & 6, p = 0.054) and 100X cold U/G (Fig. 4B compare lanes 2 & 7, p = 0.062) did not compete with the AP substrate to the same extent as 30X cold AP/G. Thus, Ku701–300 (the N-terminal GzmA cleavage product) preferentially bound to AP sites. We also found that preferential binding to the AP/G substrate was seen for myc-Ku701–115 as compared to the C/G (p = 0.031) and U/G (p = 0.036) substrates (Fig. 4C). However, myc-Ku70115–609 did not display preferential binding to the AP/G as compared to the C/G (p = 0.226) and U/G (p = 0.166) substrates (Fig. 4C). Thus, myc-Ku701–115 was necessary and sufficient to preferentially bind to the AP/G substrate, suggesting the N-terminal Ku70 cleavage product associates with AP sites to inhibit APE1.
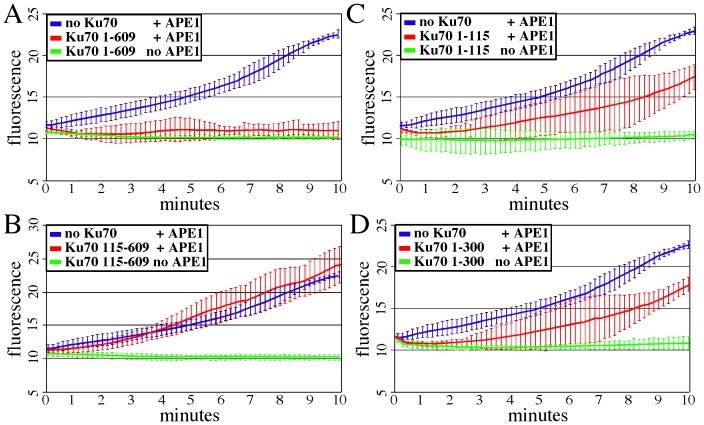
Full length and N-terminal Ku70, but not C-terminal Ku70 or Ku80, inhibited APE1 activity
Free Ku70 and free Ku80 binding to AP sites suggest a possible mechanism for interfering with APE1 activity. This activity was tested using a real-time molecular beacon assay [38] that measures fluorescence emitted after purified APE1 nicks 5′ to the AP site to release a FAM fluorophore from the dabsyl quench located on the complementary DNA strand. APE1 effectively released the FAM fluorophore from the DNA substrate without Ku70 (Fig. 5A, p<0.0001, student t test). Addition of myc-Ku701–609 (full length) prohibited APE1 nicking (Fig. 5A, p<0.0001). However, addition of myc-Ku70115–609 did not affect APE1 nicking (Fig. 5B, p = 0.7164). Furthermore, addition of myc-Ku701–115 (Fig. 5C, p<0.0001) myc-Ku701–300 (Fig. 5D, p<0.0001) impaired APE1 as compared to no myc-Ku70, though not as efficiently as full-length myc-Ku70. These data indicate that myc-Ku701–115, myc-Ku701–300 and myc-Ku701–609, but not myc-Ku70115–609, inhibited APE1 activity in keeping with the AP binding data. Thus, Ku70 binding to AP sites inhibits APE1 nicking. Interestingly, Ku80 did not inhibit APE1 activity by this assay (not shown), though a previous report showed that Ku and DNA-PKCS inhibited AP site cleavage by APE1 [32].
Figure 5. Molecular beacon assay to measure APE1 activity.
No Ku70 was compared to Ku70 added to substrate with or without APE1. Fluorescence: the excitation wavelength is 485 nm and the emission wavelength is 538 nm. Shown is the average of three experiments with error bars (standard deviation). (A) Ku701–609 (full-length Ku70) (B) Ku70115–609. (C) Ku701–115. (D) Ku701–300.
Disscussion
Mice and cells deleted for either Ku70 or Ku80 exhibit a complex phenotype that is not solely due to disruption of NHEJ; it is possible some of this complexity is due to altered BER. In mice Ku80-deletion had a more severe impact on life span than DNA-PKCS-deletion suggesting Ku80 does more than NHEJ. The level of small mutations also changed in mouse brains deleted for either Ku70 or Ku80 [25] implicating a change in BER. To further implicate altered BER, cells deleted for either Ku70 or Ku80, but not Lig4, exhibited increased sensitivity to ROS and alkylating agents and cells deleted for Ku80, but not Lig4, were deficient in repairing AP sites and cell extracts were deficient in correcting a U/G mismatch in an oligonucleotide substrate [21], [22]. Furthermore, ectopic expression of either OGG1 or PARP-1 recused hypersensitivity of ku80-/- cells to ROS while ectopic expression of Pol β or APE1 rescued hypersensitivity to an alkylating agent and an APE1 inhibitor, respectively. These rescue experiments suggest that deleting either Ku70 or Ku80 decreased BER capacity but still left the BER pathway intact.
Previously published data support the possibility that Ku70 and Ku80 have activity outside the heterodimer. For example, Ku70 and Ku80 passed through the nucleus using different nuclear localization signals [39], [40] and γ-radiation increased levels of Ku70 but not Ku80 [41]. Ku70 also associated with DNA [42] and with a variety of proteins independent of Ku80. These protein-protein interactions could influence cell death [43], [44], chromatin metabolism[45]–[47] and DNA repair [48]. Thus, Ku70 and Ku80 might function outside the Ku heterodimer to influence a variety of biological outcomes.
Our data also suggest that free Ku70 and free Ku80 could influence the phenotype of cells and mice deleted for their partner in the Ku heterodimer. In a p53-mutant background, we found free Ku80 ameliorated the severity of Ku70-deletion in mice [21]. We also show that deletion of either Ku70 or Ku80, but not both, increased sensitivity to an APE1 inhibitor. Based on these biological observations we observed extracts from cells deleted for Ku80, but not Ku70 or Lig4, were deficient in repairing an oligonucleotide with a U/G mismatch [22]. Now we show that both free Ku70 and free Ku80, but not the Ku heterodimer, preferentially bound to an oligonucleotide with an AP site and that free Ku70 inhibited APE1 activity. Thus, Ku70 and Ku80 have activity independent of the Ku heterodimer that can influence the phenotype of cells and mice deleted for their partner.
The impact free Ku70 and free Ku80 could have on BER might be physiologically relevant to GzmA-mediated cell death. GzmA is a serine protease that rapidly migrates from the cytosol to the mitochondria to cripple electron transport and increase ROS [2]. To further enhance cell death, GzmA cleaves APE1 to disable BER, the pathway mostly responsible for correcting ROS-induced lesions [37]. GzmA also cleaves Ku70 in cytotoxic T lymphocytes and natural killer cells [36] to generate an N-terminal Ku701–300 that binds to AP sites and impair APE1 activity. Our data suggests that Ku70 cleavage will further diminish BER to enhance cell death.
From a larger perspective our data suggests that nonequimolar ratios of Ku70 and Ku80 could impair BER in a wide range of cells, even those that do not express GzmA (or in cells that inappropriately express GzmA). For example, Ku70, but not Ku80, levels decline in lymphocytes [49]. This could lead to inadequate BER that would ultimately result in elevated DNA damage responses or, in the absence of these responses, to elevated point mutations. Thus, non-equimolar ratios of Ku70 and Ku80 could diminish BER and enhance p53-mediated cell clearance. This could contribute to generalized aging since Ku70 and Ku80 levels vary with age [50].
Materials and Methods
Ethics statement
All animal work was approved by the ethics committee of the National Institutes for Public Health and the Environment (RIVM), Antonie van Leeuwenhoeklaan, Bilthoven, The Netherlands, IACUC protocol #:99047x.
Life span analysis
F1 hybrid animals were generated with a C57Bl6/J*FVB background. Ku80+/- [51] and DNA-PKCS-/- mice [52] were re-derived and back crossed to C57Bl6/JIco (Charles River, France) and FVB/NHanTMHsd (Harlan, Germany) using a speed congenics approach [53]. ku80-/- and dna-pkcs-/- cohorts were generated using double heterozygous knock out breeders (Ku80+/- DNA-PKCS-/-). All male breeders were on C57Bl6/J-pUR288 and all female breeders on FVB background. Thus, the ku80-/- and dna-pkcs-/- cohorts are F1 brothers and sisters raised in the same cages to eliminate phenotypic variances due to genetic background and environment. They were kept on a 12-hour light/12-hour dark cycle at a standard temperature of 20°C. CRM pelleted maintenance diet (Special Diet Services, UK) and water supplied ad libitum. Survival (life span) was analyzed so there was no intervention. For the end of life analysis, moribund mice were sacrificed by exsanguination after sedation with an intramuscular injection of a Ketamine-Rompun mixture. The Criteria for euthanizing a moribund mouse was >15% weight loss within 2 weeks, not responsive to touch, prominent appearance of ribs, spine and hips, hunch body position, matted fur, prolapse of the rectum or uterus, or a visible tumor.
This study was carried out in strict accordance with institutional guidelines and regulations. All animal work was approved by the ethics committee of the National Institutes for Public Health and the Environment (RIVM), Antonie van Leeuwenhoeklaan, Bilthoven, The Netherlands, IACUC protocol #:99047x. These were survival studies; therefore, mice were monitored every day without intervention. Moribund mice were sacrificed with ketamine/xylazine anesthesia followed by cervical dislocation and all efforts were made to minimize suffering and discomfort. Criteria for moribund were >15% weight loss within 2 weeks, not responsive to touch, prominent appearance of ribs, spine and hips, hunch body position, matted fur, or a visible tumor.
Mutation analysis
The mutation spectrum was performed by crossing mice with a mutation in Ku70 [26] and p53 [27] into mice with the pUR288-lacZ reporter (line 60, integration sites on chromosomes 3 and 4) [28], [54]. The pUR288-lacZ reporter was bred to homozygosity. The animals were maintained in the UTHSCSA's animal facility. They were kept on a 12-hour light/12-hour dark cycle at a standard temperature of 23°C. Standard lab chow (Harlan Teklad, Madison WI) and water were supplied ad libitum. Mice were sacrificed by CO2 inhalation followed by cervical dislocation at 4 months of age. DNA preparation, plasmid rescue and mutation analysis was performed as described [25], [55].
CRT0044876 survival fraction
Dose response assays were performed as previously described for HeLa cells [56].
In vitro translation
For In vitro translation we followed the TNT® Quick Coupled Transcription/Translation System (Promega, WI) in 50 µl. For full length Ku70, Ku70 deletions and Ku80 PCR products were cloned into the pCS2/myc expression vector (Invitrogen, Carlsbad, CA) using Xho1 or EcoR1 and Asc1 sites (underlined) that adds an NH2-terminal c-Myc tag.
Ku70 1–609:
Ku70-XhoI-Top (5′-ACCGCTCGAGTCAGGGTGGAGTCATATTACAAA-3′)
Ku70-AscI-Bottom (5′-AAGGCGCGCCTCAGTCCTGGAAGTGCTTGGTGA-3′)
Ku70 1–115:
Ku70-XhoI-Top: 5′-ACCGCTCGAGTCAGGGTGGAGTCATATTACAAA-3′
Ku70N 1-115 Asc1-Bottom: 5′-AAGGCGCGCCTCGTTTTGCACCTGG-3′
Ku70 1–300:
Ku70-XhoI-Top: 5′-ACCGCTCGAGTCAGGGTGGAGTCATATTACAAA-3′
Ku70N 1–300 Asc1-Bottom: 5′-AAGGCGCGCCCTGTACTTGTATTAAAGGTCC-3′
Ku70-115-609:
Ku70-115-609 Xho1-Top: 5′-AACTCGAGCTCGACCAGTTTAAGGGACAAC-3′
Ku70-AscI-Bottom: 5′-AAGGCGCGCCTCAGTCCTGGAAGTGCTTGGTGA-3′
Ku80:
Ku80-EcoRI-Top (5′-TTGAATTCAGTGCGGTCGGGGAATAAGGCAG-3′)
Ku80-AscI-Bottom (5′-AAGGCGCGCCCTATATCATGTCCAATAAATCGTCCA-3′).
In vitro binding assay
To make the double strand substrate, single strand oligonucleotides were mixed in equimolar amounts (250 nM) as measured by UV spectrometry, incubated at 90°C (5 min) and slowly cooled to 24°C. The AP site was generated in the U/G substrate after treatment with E. coli UDG (5 U/pmol DNA, 50 mM Tris-HCl pH 8.0, 50 mM NaCl, 50 mM KCl, and 5 mM MgCl2). The substrate was biotinylated.
Sequences:
U strand: 5′-biotin-GCCCTGCAGGTCGAUTCTAGAGGATCCCCGGGTAC-3′
C strand: 5′-biotin-GCCCTGCAGGTCGACTCTAGAGGATCCCCGGGTAC-3′
Template strand: 5′-GTACCCGGGGATCCTCTAGA GTCGACCTGCAGGGC-3′.
For NaBH4 trapping [34] we used 10 µl reaction mixtures (50 mM Tris-HCl pH 8.0, 50 mM NaCl, 15 mM EDTA, 250 nM biotinylated-AP/G substrate) and in vitro translation (40 µg) prepared on ice and incubated at 37°C (15 min). Then reaction mixtures were diluted on ice with NaBH4 (final concentration 20 mM) and incubated at 0°C (30 min). 10 µg of streptavidin-coated magnetic beads (Dynabeads M280, Dynal) was added to the mixture on a rotator at 4°C (15 min). Beads (diameter ∼2.8 µm, 300pmol biotin-binding sites/mg and washed twice in 50 mM Tris-HCl pH 7.4 and 0.1% BSA to remove NaN3) were applied as a suspension (6-7×108 beads/ml, 2 µg of beads in 20 µl) in phosphate-buffered saline (PBS, 0.1% BSA and 0.02% NaN3). After incubation with mixture, beads were washed three times (1 ml PBS), resuspended in 20 µl of 1X SDS (50 mM Tris-HCl pH 6.8, 2% SDS, 10% glycerol, 1% β-mercaptoethanol, 12.5 mM EDTA and 0.02% bromophenol blue) and heated (5 min). Soluble fraction was separated by 10% SDS PAGE. Proteins were immunoblotted with mouse anti-human c-Myc antibody (BD Biosciences, MD) according to manufacturer's instructions.
Competition assay
AP/G competitor has same sequences to AP/G substrate but without 5′ end biotin labeling. The AP site was generated in U/G competitor after treatment with E. coli uracil DNA glycosylase (5 U/pmol DNA, 50 mM Tris-HCl pH 8.0, 50 mM NaCl, 50 mM KCl, and 5 mM MgCl2). C/G competitor has same sequences to C/G substrate without 5′ end biotin labeling. U/G competitor has same sequences to U/G substrate without 5′ end biotin labeling. For competition assay, each tube contained AP/G substrate (50 nM) with AP/G competitor (1.5 µM), C/G competitor (5 µM) and U/G competitor (5 µM) and did same process the incubation with in vitro translation samples, after that pulled down with streptoavidin beads and trapped by treating NaBH4. After washing, the beads were resuspended in 20 µl of 1X SDS sample buffer and heated for 5 min and soluble fraction were separated by 10% SDS PAGE. Proteins were immunoblotted with mouse anti-human c-Myc antibody (BD Biosciences, MD) according to manufacturer's instructions.
Molecular beacon assay for APE1 activity
APE1 activity was measured using a modification of our recently described BER molecular assay [57]–[59]. All oligonucleotides were purchased from Sigma-Aldrich (Saint Louis, MO): Top-FAM-5′-GAGAAΦATAGTCGCC-3′ Bottom-DAB-3′-CTCTTGTATCAGCGC-5′ [Φ; Tetrahydrofuran (THF)]. Oligonucleotides were mixed in equimolar amount (250 nM), heated at 90°C for 5 min, and then slowly cooled down to room temperature to form a double stranded DNA substrate. An oligonucleotide containing a THF AP site mimic was used in the APE1 molecular beacon substrate. The top strand was labeled on the 5′ end with a FAM fluorophore, the bottom on the 3′end with a dabsyl (DAB) quench. Upon APE1 cleavage, the fluorophore dissociated from the dabsyl quench, causing an increase in fluorescence, essentially as described [57]–[59]. All APE1 (New England Biolabs, Ipswich, MA) cleavage reactions were carried out in a buffer containing 20 mM Tris-acetate, pH 7.9, 10 mM Magnesium acetate, 50 mM Potassium acetate, 1 mM dithiothreitol. All reactions were performed at 37°C. Excitation and emission wavelengths for time curves were 485 and 538 mM, respectively. Standard reaction mixtures (100 µl) for the molecular beacon assay contained 20 mM Tris-acetate, pH 7.9, 10 mM Magnesium acetate, 50 mM Potassium acetate, 1 mM dithiothreitol, 250 nM AP DNA duplex and in vitro translation samples (40 µg) and APE1 (10 u) were prepared on ice and then measured at 37°C, every 10 sec. for 10 min.
Acknowledgments
We thank Dr. G.E. Taccioli for providing DNA-PKCS-mutant mice.
Funding Statement
This work was supported by NIH - UO1 ES11044, R01 CA76317-05A1, P01 AG17242, CA054174 to PH and by a Research Scholar grant (RSG-05-246-01-GMC) from the American Cancer Society, grants from the Susan G. Komen Breast Cancer Foundation (Grant # BCTR0403276), NIH (GM087798: CA148629: ES019498) and the Brain Tumor Foundation to RWS. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Holmquist GP (1998) Endogenous lesions, S-phase-independent spontaneous mutations, and evolutionary strategies for base excision repair. Mutat Res 400: 59–68. [DOI] [PubMed] [Google Scholar]
- 2. Almeida KH, Sobol RW (2007) A unified view of base excision repair: lesion-dependent protein complexes regulated by post-translational modification. DNA Repair (Amst) 6: 695–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barnes DE, Lindahl T (2004) Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu Rev Genet 38: 445–476. [DOI] [PubMed] [Google Scholar]
- 4. Parsons JL, Elder RH (2003) DNA N-glycosylase deficient mice: a tale of redundancy. Mutat Res 531: 165–175. [DOI] [PubMed] [Google Scholar]
- 5. Vidal AE, Boiteux S, Hickson ID, Radicella JP (2001) XRCC1 coordinates the initial and late stages of DNA abasic site repair through protein-protein interactions. Embo J 20: 6530–6539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Caldecott KW, Aoufouchi S, Johnson P, Shall S (1996) XRCC1 polypeptide interacts with DNA polymerase beta and possibly poly (ADP-ribose) polymerase, and DNA ligase III is a novel molecular ‘nick-sensor’ in vitro. Nucleic Acids Res 24: 4387–4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kubota Y, Nash RA, Klungland A, Schar P, Barnes DE, et al. (1996) Reconstitution of DNA base excision-repair with purified human proteins: interaction between DNA polymerase beta and the XRCC1 protein. Embo J 15: 6662–6670. [PMC free article] [PubMed] [Google Scholar]
- 8. Dantzer F, de La Rubia G, Menissier-De Murcia J, Hostomsky Z, de Murcia G, et al. (2000) Base excision repair is impaired in mammalian cells lacking Poly(ADP-ribose) polymerase-1. Biochemistry 39: 7559–7569. [DOI] [PubMed] [Google Scholar]
- 9. Masson M, Niedergang C, Schreiber V, Muller S, Menissier-de Murcia J, et al. (1998) XRCC1 is specifically associated with poly (ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol Cell Biol 18: 3563–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kuzminov A (2001) Single-strand interruptions in replicating chromosomes cause double-strand breaks. Proc Natl Acad Sci U S A 98: 8241–8246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Burma S, Chen BP, Chen DJ (2006) Role of non-homologous end joining (NHEJ) in maintaining genomic integrity. DNA Repair (Amst) 5: 1042–1048. [DOI] [PubMed] [Google Scholar]
- 12. Bailey SM, Meyne J, Chen DJ, Kurimasa A, Li GC, et al. (1999) DNA double-strand break repair proteins are required to cap the ends of mammalian chromosomes. Proc Natl Acad Sci U S A 96: 14899–14904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lim DS, Vogel H, Willerford DM, Sands AT, Platt KA, et al. (2000) Analysis of ku80-mutant mice and cells with deficient levels of p53. Mol Cell Biol 20: 3772–3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Espejel S, Klatt P, Menissier-de Murcia J, Martin-Caballero J, Flores JM, et al. (2004) Impact of telomerase ablation on organismal viability, aging, and tumorigenesis in mice lacking the DNA repair proteins PARP-1, Ku86, or DNA-PKcs. J Cell Biol 167: 627–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Espejel S, Martin M, Klatt P, Martin-Caballero J, Flores JM, et al. (2004) Shorter telomeres, accelerated ageing and increased lymphoma in DNA-PKcs-deficient mice. EMBO Rep 5: 503–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li H, Vogel H, Holcomb VB, Gu Y, Hasty P (2007) Deletion of Ku70, Ku80, or both causes early aging without substantially increased cancer. Mol Cell Biol 27: 8205–8214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vogel H, Lim DS, Karsenty G, Finegold M, Hasty P (1999) Deletion of Ku86 causes early onset of senescence in mice. Proc Natl Acad Sci U S A 96: 10770–10775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Holcomb VB, Vogel H, Hasty P (2007) Deletion of Ku80 causes early aging independent of chronic inflammation and Rag-1-induced DSBs. Mech Ageing Dev 128: 601–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Frank KM, Sekiguchi JM, Seidl KJ, Swat W, Rathbun GA, et al. (1998) Late embryonic lethality and impaired V(D)J recombination in mice lacking DNA ligase IV. Nature 396: 173–177. [DOI] [PubMed] [Google Scholar]
- 20. Chao C, Herr D, Chun J, Xu Y (2006) Ser18 and 23 phosphorylation is required for p53-dependent apoptosis and tumor suppression. Embo J 25: 2615–2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li H, Choi YJ, Hanes MA, Marple T, Vogel H, et al. (2009) Deleting Ku70 is milder than deleting Ku80 in p53-mutant mice and cells. Oncogene 28: 1875–1878. [DOI] [PubMed] [Google Scholar]
- 22.Li H, Marple T, Hasty P (2013) Ku80-deleted cells are defective at base excision repair. Mutat Res. [DOI] [PMC free article] [PubMed]
- 23. Gu Y, Jin S, Gao Y, Weaver DT, Alt FW (1997) Ku70-deficient embryonic stem cells have increased ionizing radiosensitivity, defective DNA end-binding activity, and inability to support V(D)J recombination. Proc Natl Acad Sci U S A 94: 8076–8081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nussenzweig A, Chen C, da Costa Soares V, Sanchez M, Sokol K, et al. (1996) Requirement for Ku80 in growth and immunoglobulin V(D)J recombination. Nature 382: 551–555. [DOI] [PubMed] [Google Scholar]
- 25. Busuttil RA, Munoz DP, Garcia AM, Rodier F, Kim WH, et al. (2008) Effect of Ku80 deficiency on mutation frequencies and spectra at a LacZ reporter locus in mouse tissues and cells. PLoS ONE 3: e3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gu Y, Seidl KJ, Rathbun GA, Zhu C, Manis JP, et al. (1997) Growth retardation and leaky SCID phenotype of Ku70-deficient mice. Immunity 7: 653–665. [DOI] [PubMed] [Google Scholar]
- 27. Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA Jr, et al. (1992) Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 356: 215–221. [DOI] [PubMed] [Google Scholar]
- 28. Dolle ME, Snyder WK, Gossen JA, Lohman PH, Vijg J (2000) Distinct spectra of somatic mutations accumulated with age in mouse heart and small intestine Proc Natl Acad Sci U S A. 97: 8403–8408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Busuttil RA, Garcia AM, Cabrera C, Rodriguez A, Suh Y, et al. (2005) Organ-specific increase in mutation accumulation and apoptosis rate in CuZn-superoxide dismutase-deficient mice. Cancer Res 65: 11271–11275. [DOI] [PubMed] [Google Scholar]
- 30. Trivedi RN, Almeida KH, Fornsaglio JL, Schamus S, Sobol RW (2005) The role of base excision repair in the sensitivity and resistance to temozolomide-mediated cell death. Cancer Res 65: 6394–6400. [DOI] [PubMed] [Google Scholar]
- 31. Madhusudan S, Smart F, Shrimpton P, Parsons JL, Gardiner L, et al. (2005) Isolation of a small molecule inhibitor of DNA base excision repair. Nucleic Acids Res 33: 4711–4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ilina ES, Lavrik OI, Khodyreva SN (2008) Ku antigen interacts with abasic sites. Biochim Biophys Acta 1784: 1777–1785. [DOI] [PubMed] [Google Scholar]
- 33. Roberts SA, Strande N, Burkhalter MD, Strom C, Havener JM, et al. (2010) Ku is a 5′-dRP/AP lyase that excises nucleotide damage near broken ends. Nature 464: 1214–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Haracska L, Prakash L, Prakash S (2003) A mechanism for the exclusion of low-fidelity human Y-family DNA polymerases from base excision repair. Genes Dev 17: 2777–2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Walker JR, Corpina RA, Goldberg J (2001) Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature 412: 607–614. [DOI] [PubMed] [Google Scholar]
- 36. Zhu P, Zhang D, Chowdhury D, Martinvalet D, Keefe D, et al. (2006) Granzyme A, which causes single-stranded DNA damage, targets the double-strand break repair protein Ku70. EMBO Rep 7: 431–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lieberman J (2011) Granzyme A activates another way to die. Immunol Rev 235: 93–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wilson DM 3rd, Simeonov A (2010) Small molecule inhibitors of DNA repair nuclease activities of APE1. Cell Mol Life Sci 67: 3621–3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Koike M, Shiomi T, Koike A (2000) Ku70 can translocate to the nucleus independent of Ku80 translocation and DNA-PK autophosphorylation. Biochem Biophys Res Commun 276: 1105–1111. [DOI] [PubMed] [Google Scholar]
- 40. Lim JW, Kim KH, Kim H (2008) NF-kappaB p65 regulates nuclear translocation of Ku70 via degradation of heat shock cognate protein 70 in pancreatic acinar AR42J cells. Int J Biochem Cell Biol 40: 2065–2077. [DOI] [PubMed] [Google Scholar]
- 41. Brown KD, Lataxes TA, Shangary S, Mannino JL, Giardina JF, et al. (2000) Ionizing radiation exposure results in up-regulation of Ku70 via a p53/ataxia-telangiectasia-mutated protein-dependent mechanism. J Biol Chem 275: 6651–6656. [DOI] [PubMed] [Google Scholar]
- 42. Wang J, Dong X, Myung K, Hendrickson EA, Reeves WH (1998) Identification of two domains of the p70 Ku protein mediating dimerization with p80 and DNA binding. J Biol Chem 273: 842–848. [DOI] [PubMed] [Google Scholar]
- 43. Gomez JA, Gama V, Yoshida T, Sun W, Hayes P, et al. (2007) Bax-inhibiting peptides derived from Ku70 and cell-penetrating pentapeptides. Biochem Soc Trans 35: 797–801. [DOI] [PubMed] [Google Scholar]
- 44.Mazumder S, Plesca D, Kinter M, Almasan A (2007) Interaction of a Cyclin E Fragment with Ku70 Regulates Bax-mediated Apoptosis. Mol Cell Biol. [DOI] [PMC free article] [PubMed]
- 45. Hsu HL, Gilley D, Galande SA, Hande MP, Allen B, et al. (2000) Ku acts in a unique way at the mammalian telomere to prevent end joining. Genes Dev 14: 2807–2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Song K, Jung D, Jung Y, Lee SG, Lee I (2000) Interaction of human Ku70 with TRF2. FEBS Lett 481: 81–85. [DOI] [PubMed] [Google Scholar]
- 47. Song K, Jung Y, Jung D, Lee I (2001) Human ku70 interacts with heterochromatin protein 1alpha. J Biol Chem 276: 8321–8327. [DOI] [PubMed] [Google Scholar]
- 48. Goedecke W, Eijpe M, Offenberg HH, van Aalderen M, Heyting C (1999) Mre11 and Ku70 interact in somatic cells, but are differentially expressed in early meiosis. Nat Genet 23: 194–198. [DOI] [PubMed] [Google Scholar]
- 49. Ju YJ, Lee KH, Park JE, Yi YS, Yun MY, et al. (2006) Decreased expression of DNA repair proteins Ku70 and Mre11 is associated with aging and may contribute to the cellular senescence. Exp Mol Med 38: 686–693. [DOI] [PubMed] [Google Scholar]
- 50. Li H, Mitchell JR, Hasty P (2008) DNA double-strand breaks: A potential causative factor for mammalian aging? Mech Ageing Dev 129: 416–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhu C, Bogue MA, Lim DS, Hasty P, Roth DB (1996) Ku86-deficient mice exhibit severe combined immunodeficiency and defective processing of V(D)J recombination intermediates. Cell 86: 379–389. [DOI] [PubMed] [Google Scholar]
- 52. Priestley A, Beamish HJ, Gell D, Amatucci AG, Muhlmann-Diaz MC, et al. (1998) Molecular and biochemical characterisation of DNA-dependent protein kinase-defective rodent mutant irs-20. Nucleic Acids Res 26: 1965–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wakeland E, Morel L, Achey K, Yui M, Longmate J (1997) Speed congenics: a classic technique in the fast lane (relatively speaking). Immunol Today 18: 472–477. [DOI] [PubMed] [Google Scholar]
- 54. Garcia AM, Busuttil RA, Rodriguez A, Cabrera C, Lundell M, et al. (2007) Detection and analysis of somatic mutations at a lacZ reporter locus in higher organisms: application to Mus musculus and Drosophila melanogaster. Methods Mol Biol 371: 267–287. [DOI] [PubMed] [Google Scholar]
- 55. Holcomb VB, Rodier F, Choi Y, Busuttil RA, Vogel H, et al. (2008) Ku80 deletion suppresses spontaneous tumors and induces a p53-mediated DNA damage response. Cancer Res 68: 9497–9502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Marple T, Li H, Hasty P (2004) A genotoxic screen: rapid analysis of cellular dose-response to a wide range of agents that either damage DNA or alter genome maintenance pathways. Mutat Res 554: 253–266. [DOI] [PubMed] [Google Scholar]
- 57. Tang JB, Svilar D, Trivedi RN, Wang XH, Goellner EM, et al. (2011) N-methylpurine DNA glycosylase and DNA polymerase beta modulate BER inhibitor potentiation of glioma cells to temozolomide. Neuro Oncol 13: 471–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mutamba JT, Svilar D, Prasongtanakij S, Wang XH, Lin YC, et al. (2011) XRCC1 and base excision repair balance in response to nitric oxide. DNA Repair (Amst) 10: 1282–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Svilar D, Conchita V, Sobol RW (2012) Quantitative, real-time analysis of base excision repair activity in cell lysates utilizing lesion-specific molecular beacons. J Vis Exp: (in press). [DOI] [PMC free article] [PubMed]