Abstract
Nonobese diabetic (NOD) mice spontaneously develop type 1 diabetes (T1D), progression of which is similar to that in humans, and therefore are widely used as a model for understanding the immunological basis of this disease. The incidence of T1D in NOD mice is influenced by the degree of cleanliness of the mouse colony and the gut microflora. In this report, we show that the T1D incidence and rate of disease progression are profoundly influenced by the pH of drinking water, which also affects the composition and diversity of commensal bacteria in the gut. Female NOD mice that were maintained on acidic pH water (AW) developed insulitis and hyperglycemia rapidly compared with those on neutral pH water (NW). Interestingly, forced dysbiosis by segmented filamentous bacteria (SFB)-positive fecal transfer significantly suppressed the insulitis and T1D incidence in mice that were on AW but not in those on NW. The 16S rDNA–targeted pyrosequencing revealed a significant change in the composition and diversity of gut flora when the pH of drinking water was altered. Importantly, autoantigen-specific T-cell frequencies in the periphery and proinflammatory cytokine response in the intestinal mucosa are significantly higher in AW-recipient mice compared with their NW counterparts. These observations suggest that pH of drinking water affects the composition of gut microflora, leading to an altered autoimmune response and T1D incidence in NOD mice.
Introduction
Based on the studies conducted under germ-free conditions, the incidence of type 1 diabetes (T1D) in nonobese diabetic (NOD) mice is thought to be influenced by environmental factors such as microbes (1–4). Importantly, the degree of cleanliness of the facility has been considered a primary reason for the difference in T1D incidence in NOD mice that are maintained around the world (5). Recently, the composition of gut microbiome has been implicated in the regulation of autoimmune fate in NOD mice (6–10). While germ-free NOD mice show no sexual dimorphism, female NOD mice from specific pathogen–free (SPF) facilities show higher disease incidence compared with their male counterparts (6,9,11). This sexual dimorphism of T1D in NOD mice has recently been linked to specific microbial communities that can alter testosterone levels in an androgen receptor–dependent manner and promote disease-protective commensal bacteria in the gut (6,12).
The Jackson Laboratory (JAX) is the primary vendor of NOD mice in the U.S. We have often observed that NOD mice purchased from JAX at varying ages develop hyperglycemia at different rates in our SPF facility and also relatively rapidly as reported by JAX (http://type1diabetes.jax.org/images/fine-mapping/1976%20cumulative%20inc.jpg) compared with their in-house counterparts (C.V., N.P., S.K.-M., unpublished observations). While the cleanliness of our SPF facility was assumed to be the reason for this difference, our inquiries have revealed a major difference in the dietary conditions. While JAX provides their mice with acidified drinking water (AW), other facilities, including ours, maintain them on neutral pH water (NW). Since dietary changes can reshape the gut microbial communities (13–16), we hypothesized that the difference in diabetes incidences observed with NOD mice of our SPF colony compared with those from JAX may be associated with pH of the drinking water. Hence, in this study we have examined the influence of drinking water pH on T1D incidence and associated changes in the gut microbiota (dysbiosis) of NOD mice.
Our results show that SPF NOD mice that received AW developed T1D rapidly and showed higher diabetes incidence compared with the NW recipients, and this effect was directly associated with difference in the composition and diversity of gut microbiome. Further, our results show that forced dysbiosis results in profound modulation of disease progression in AW recipients. This indicates that environmental factors such as drinking water pH can influence T1D incidence and the rate of disease progression, primarily by affecting the composition of gut microflora, which can modulate the overall peripheral immune response.
Research Design and Methods
NOD/ShiLtJ mice were purchased from JAX or used from our in-house SPF colony of NOD/ShiLtJ origin. In-house mice used for this study were 2–3 generations old. C57BL/6 mice were purchased from Taconic Farms, Inc. (TAC). Female mice were used throughout the study. All animal studies were approved by the animal care and use committee of the Medical University of South Carolina or University of Illinois at Chicago (UIC). These mice, including the NOD/ShiLtj breeding colony, were maintained on autoclaved neutral pH (7.0–7.2) water (NW) or acidic pH (3.0–3.2) water (AW) in SPF facilities. Water was acidified by adding HCl as described on the JAX website (http://jaxmice.jax.org/genetichealth/health_program.html; http://craniofacial.jax.org/new_standard_protocols.html).
Fecal Transplant
In some experiments, fecal pellets and bedding materials from cages of segmented filamentous bacteria (SFB+) 4-week-old C57BL/6 mice of TAC (on NW) were transferred (fecal transplant [FT]) into NOD/ShiLtj mouse cages that were on AW or NW. Whole bedding and fecal materials from one cage of 5 mice were transferred to two recipient cages (5 mice/cage), and this was repeated three times at 3-day intervals. Three days post–final transfer, these mice were maintained on fresh bedding materials.
Carboxyfluorescein Succinimidyl Ester Dilution Assay
Immunodominant β-cell antigen peptides [viz., 1) insulin B(9-23), 2) GAD65(206–220), 3) GAD65(524–543), 4) IA-2beta(755–777), and 5) IGRP(123–145)] were described in our earlier studies (17,18). These peptides were pooled at an equal molar ratio and used as β-cell antigen (Ag) for challenging T cells ex vivo. Single-cell suspension of lymphoid tissues was labeled with carboxyfluorescein succinimidyl ester (CFSE), cultured in the presence of β-cell Ag (5 μg/mL) for 4 days, stained for CD4 and CD8 cells, and analyzed for CFSE dilution by fluorescence-activated cell sorter (FACS).
Examination of T-Cell Phenotypes
Fluorochrome-conjugated anti-mouse CD4, CD8, CD62L, CD44, FoxP3, and isotype control antibodies (eBiosciences) were used for FACS analysis. Surface and intracellular staining were done using freshly isolated cells from indicated tissues as previously described (17,19). Stained cells were acquired using the FACS Calibur. Specific regions were marked and the gates and quadrants were set while analyzing the data based on isotype control antibody staining.
Qualitative and Quantitative PCR for Cytokines and Transcription Factors
Small intestine pieces (distal end of ileum) were weighed and snap frozen, ground using a mortar and pestle on dry ice, and total RNA was extracted using TRIZOL reagent (Invitrogen Life Technologies, Carlsbad, CA) according to the manufacturer’s recommendations. cDNA synthesis was done using the Superscript first-strand cDNA synthesis system (Invitrogen). Cytokine and transcription factor specific primer sets (Supplementary Table 1) were used for qualitative PCRs or real-time quantitative PCR by the SYBR Green method.
PCR for Detecting Bacterial 16S rDNA
Total DNA samples were prepared from small intestine (distal ileum) and fecal samples using TRIZOL reagent. Briefly, ground intestinal and fecal samples were processed using TRIZOL reagent as recommended by the manufacturer, centrifuged at 13,000 RPM, and DNA was precipitated from the interphase and organic phase using 0.1 mol/L trisodium citrate in 10% ethanol. DNA was washed, dried, and suspended in 8 mmol/L NaOH, and the insoluble material was removed by centrifuging at 13,000 RPM. DNA samples were diluted to 1:20 using water and amplified by PCR using universal or SFB-specific 16S rDNA–targeted primer sets.
Examination of Gut Microbial Communities
Pyrosequencing for bacterial community analysis was carried out by the microbiome core at the University of North Carolina, Chapel Hill. In brief, amplicons were prepared from small intestinal and fecal DNA samples by using a primer set that spans the V2–V3 regions of 16S rDNA, following which adaptors and 10 bp bar codes were added, and sequencing was done using a Roche 454 life Science Genome sequencer. Raw sequences were identified by their unique barcodes and analyzed using the bacterial community analysis application QIIME. For some samples, operational taxonomic units (OTUs) that were defined after removal of singleton sequences were taxonomically classified using BLAST against a curated GreenGenes database and compiled into each taxonomic level (20,21). The OTUs were compiled to the species level based upon the percentage identity to reference sequences (i.e., >97% identity), and the percentage values of sequences within each sample that mapped to specific species were calculated.
Histochemical Analysis of Pancreatic Tissues
Pancreata were fixed in 10% formaldehyde, and 5-µm paraffin sections were made and stained with hematoxylin-eosin (H-E). Stained sections were analyzed in a blinded fashion, using a grading system as described in our earlier studies: 0, no evidence of infiltration; 1, peri-islet infiltration; 2, <25%; 3, 25–50%; and 4, >50% infiltration of each islet (17,19,22). At least 150 islets were examined for each group.
Statistical Analyses
Various statistical approaches were used to calculate the mean, SD, and significance (P value). Because similar numbers of test and control values (data points) were compared, a two-tailed t test was used unless specified. Log-rank survival-curve analysis was performed to compare T1D incidence (hyperglycemia) of two different groups. Fisher exact test was used for comparing the total number of infiltrated islets in different groups. A P value of ≤0.05 was considered significant.
RESULTS
pH of Drinking Water Affects T1D Incidence in NOD Mice
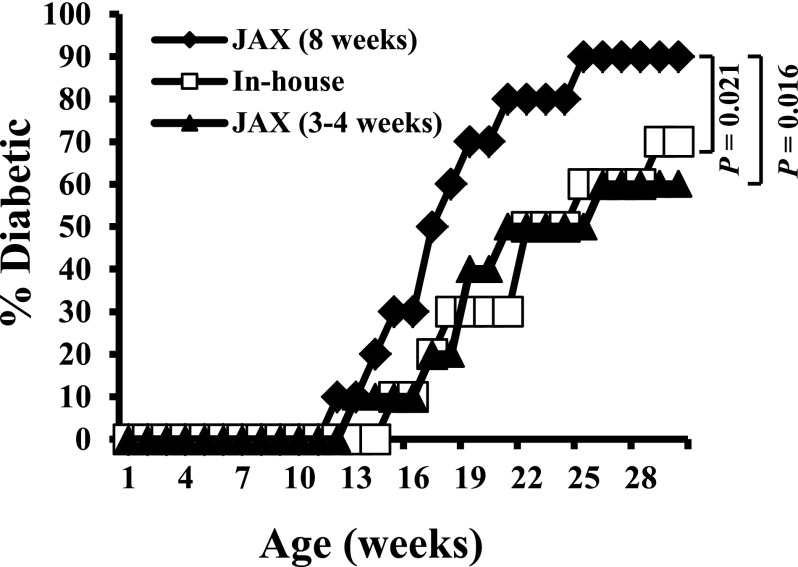
Our experiments using NOD mice in various projects showed that prediabetic (8 week old), but not young (3–4 week old), mice purchased from JAX develop hyperglycemia relatively rapidly compared with those from our SPF breeding colony at UIC. (An example of the results is shown in Fig. 1.) In addition, the diabetes incidence in prediabetic NOD mice obtained from JAX is higher compared with their in-house counterparts. For identification of the potential environmental factors involved in modulating insulitis in the initial stages of disease progression in NOD mice, prominent vendors and university animal cores were contacted. These personal communications indicated that while most housing conditions are similar, JAX, but not ours or other SPF facilities, maintains their NOD mice on AW. The JAX NOD mice demonstrate relatively rapid disease progression and higher T1D incidence compared with their counterparts of other facilities including ours.
Figure 1.
JAX NOD mice develop disease rapidly compared with their in-house counterparts. Female NOD/ShiLtJ mice were purchased from JAX at 3–4 weeks or prediabetic age (8 weeks) and maintained at our SPF facility (on NW as done traditionally; n = 10/group). These mice and female NOD mice from in-house SPF colony (NOD/ShiLtJ origin; 2nd generation) that were on NW water were examined for blood glucose levels every week starting at the age of week 9. Mice with glucose levels >250 mg/dL for two consecutive weeks were considered diabetic. An example of observations made in at least three different experiments using 10 mice/group is shown.
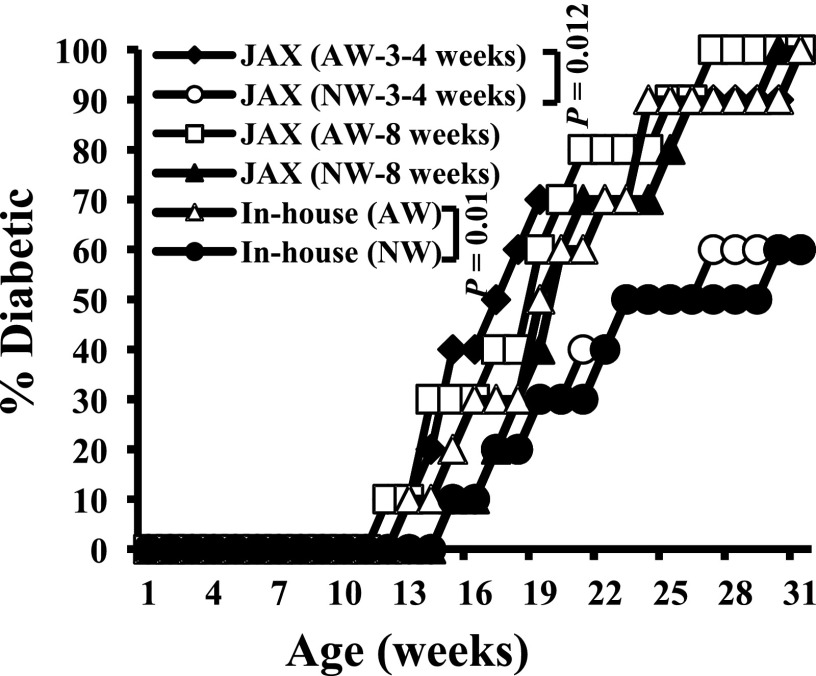
For validation of the effect of drinking water pH on T1D incidence, NOD mice obtained from JAX at 3–4 or 8 weeks of age were maintained on AW or NW and compared with the in-house SPF NOD mice. Figure 2 shows that JAX NOD mice that were purchased at 3–4 weeks of age and continued on AW developed hyperglycemia rapidly compared with those that were switched to NW. However, when 8-week-old prediabetic mice from JAX were switched to NW, no significant delay in hyperglycemia or overall diabetes incidence was observed compared with those continued on AW. In-house NOD mice that were on AW and NW showed trends of disease incidence and rate of disease progression that were comparable with those of young JAX NOD mice that were on AW and NW. These results show that the pH of drinking water has an influence on the disease initiation and progression in NOD mice, especially at early stages of insulitis.
Figure 2.
pH of drinking water affects the T1D incidence in NOD mice. Female NOD mice from in-house SPF colony (2nd- or 3rd-generation NOD/ShiLtJ mice) and 3- to 4- and 8-week-old mice purchased from JAX were maintained on either AW or NW and monitored for hyperglycemia (n = 10/group) as described for Fig. 1. While half the JAX mice were switched to NW or continued on AW upon arrival, in-house mice were on AW or NW starting at gestation stage.
Difference in T1D Incidence Observed in NOD Mice That Were on AW and NW Is Not Associated With SFB
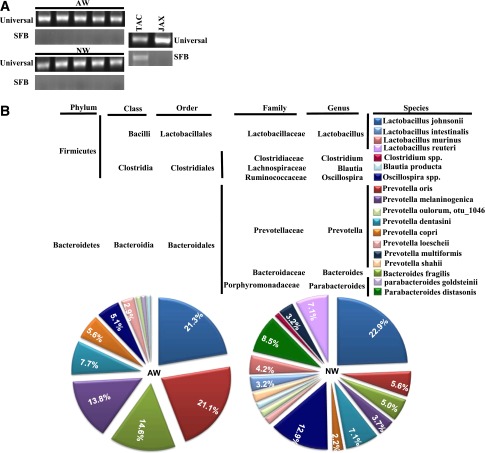
Recent studies have shown that SFB, a common mouse pathogenic bacteria that is often detected in the animals of some vendors (e.g., TAC) can skew the gut immune response and affect the autoimmune outcome, including in NOD mice (10,23). This prompted us to examine our in-house NOD mice that were on AW and NW for the presence of SFB. As observed in Fig. 3A, NOD mice of our SPF facility that were on AW and NW demonstrated no signs of infiltration by SFB. However, a fecal sample of C57BL/6 mice purchased from TAC showed the presence of SFB-specific 16S rDNA. This suggested the noninvolvement of SFB in drinking water pH–associated differences in T1D incidence in NOD mice.
Figure 3.
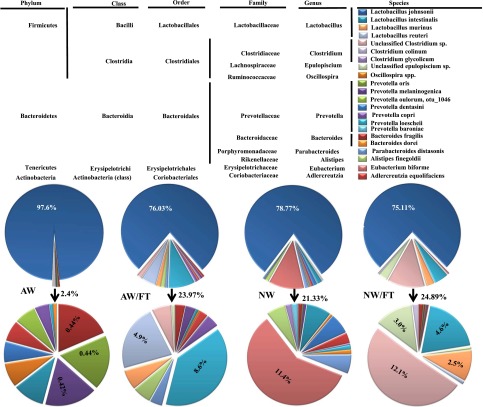
Drinking-water pH–dependent effect in NOD mice is not due to SFB but may be due to the difference in gut microflora. A: DNA prepared from the fecal samples of 8-week-old female NOD mice from our SPF facility (5 representative cages each) was tested for 16S rDNA using universal or SFB-specific primers by qualitative PCR. Fecal samples from JAX and B6 mice cages of TAC were used as negative and positive control, respectively. B: DNA prepared from the fecal samples of female NOD mice that were on AW or switched to NW at 4 weeks of age, for 30 days (at 8 weeks of age), were subjected to 16S rDNA targeted pyrosequencing as described in research design and methods. The OTUs were taxonomically classified using BLAST against a curated Green Genes database. The most relevant taxonomic levels of specific communities, based upon the percentage identity to reference sequences, were determined. The mean percentage values of bacterial sequences that were identified to species or phylum levels are shown. Communities that showed >1% of total flora are shown. Percentage values of communities that showed >2% of total flora are indicated on the chart.
For examination of whether the microflora communities are different in mice that were maintained on different pH drinking water, 3- to 4-week-old NOD mice were continued on AW or switched to NW for 30 days. Fecal samples from these mice were used for 16S rDNA–targeted pyrosequencing to identify changes in the microbial communities. As observed in Fig. 3B, mice that were switched to NW showed a significant change, not only with respect to the abundance of existing bacterial communities, but also in terms of the bacterial diversity, which was enhanced. While the Lactobacilli abundance remained about the same when the mice were switched to neutral pH drinking water, significantly reduced loads of Bacteroides and some of the Prevotella genus members were observed. On the other hand, mice switched to neutral pH drinking water acquired Parabacteroides as well as additional members of Prevotella genus. This indicates that the increase in diversity of the gut microbial communities may have a role in the enhanced immune regulation in NOD mice that were switched to neutral pH drinking water.
SFB Colonization of the Gut Delays T1D in NOD Mice That Were on AW but Not on NW
Previous studies have shown that TAC mice, but not JAX mice, carry SFB (10,23–25). Since TAC maintains their mice on NW, the effect of AW on gut colonization by SFB was examined. Further, a previous study has shown that colonization of the mouse gut by SFB leads to skewing of the immune response to Th17 and protects NOD mice from T1D (10). Therefore, the effect of gut colonization by SFB on T1D, under acidic and neutral pH drinking water conditions, was examined. Three- to four-week-old female NOD mice were purchased from JAX and continued on either AW or NW. Cages of one set of mice from each category were transplanted with fecal pellets (FT) from SFB+ C57BL/6 mice from TAC. Figure 4A shows that transfer of SFB+ fecal pellets resulted in effective colonization of NOD mouse gut by these bacteria, indicating that drinking water pH has no effect on SFB acquisition or gut colonization. While the AW group, which is SFB−, developed hyperglycemia rapidly compared with NW group (Fig. 1), the AW group that received SFB+ FT (AW/FT) showed a profound delay in hyperglycemia (Fig. 4B). Interestingly, T1D incidence in the NW group of mice that received SFB+ FT (NW/FT) was comparable with that of their non–FT recipient counterparts (Fig. 4B). These results suggest that protection of the AW group of mice that received SFB+ FT (AW/FT) from T1D compared with the AW group could be the result of difference in the overall composition of the gut microbiome in them but not due to SFB alone.
Figure 4.
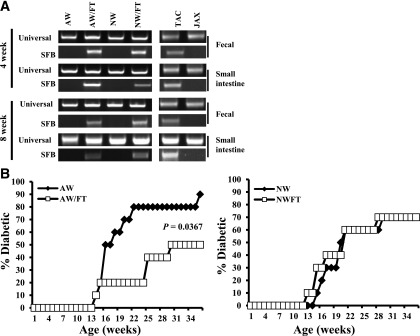
Dysbiosis induced by FT delays T1D in NOD mice that were on AW but not on NW. Female NOD mice were purchased from JAX at 3–4 weeks of age and continued on AW or switched to NW. Cages of one set of each group of mice were transplanted with fecal pellets (FT groups) of SFB+ C57BL/6 mice from TAC as described in research design and methods. A: Sets of three mice from each group were killed at 4 and 8 weeks post–drinking-water switch and FT (8 and 12 weeks of age), DNA was prepared from the distal ileum, and fecal pellets were tested for 16S rDNA using universal or SFB-specific primers by qualitative PCR (results obtained using representative samples are shown). Fecal samples from JAX and B6 mouse cages of TAC were used as negative and positive control, respectively. B: Sets of 10 mice/group were monitored for hyperglycemia as described for Fig. 1. Results of AW and NW groups are plotted against the results of their FT recipients and shown separately.
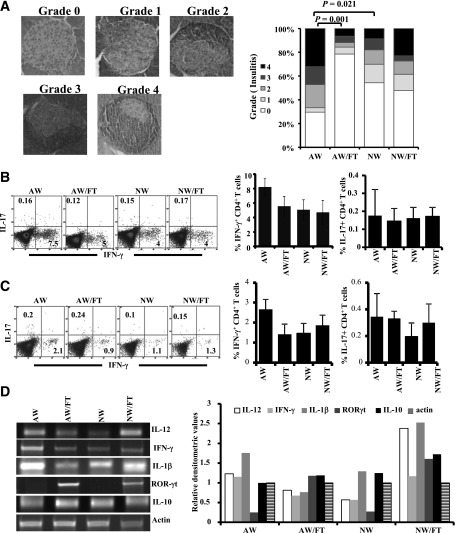
NOD Mice on AW Show Severe Insulitis and Difference in Gut Immune Response Compared With SFB+ FT and NW Recipients
Since NOD mice that were on AW showed rapid progression to hyperglycemia, insulitis severity and T-cell phenotype in these mice and SFB+ FT recipients were compared at the prediabetic stage. Approximately 70% of islets from mice of the AW group showed peri-insulitis to complete destruction of islets (grades 1–4), while ~80% of the islets of the NW group of mice and >50% of the islets of SFB+ FT recipients were found to be insulitis free (Fig. 5A). These results affirm the observations of Figs. 1 and 3 and suggest that SPF NOD mice that are on AW tend to develop autoimmunity rapidly and this can be reversed by introducing additional diversity in the gut microflora.
Figure 5.
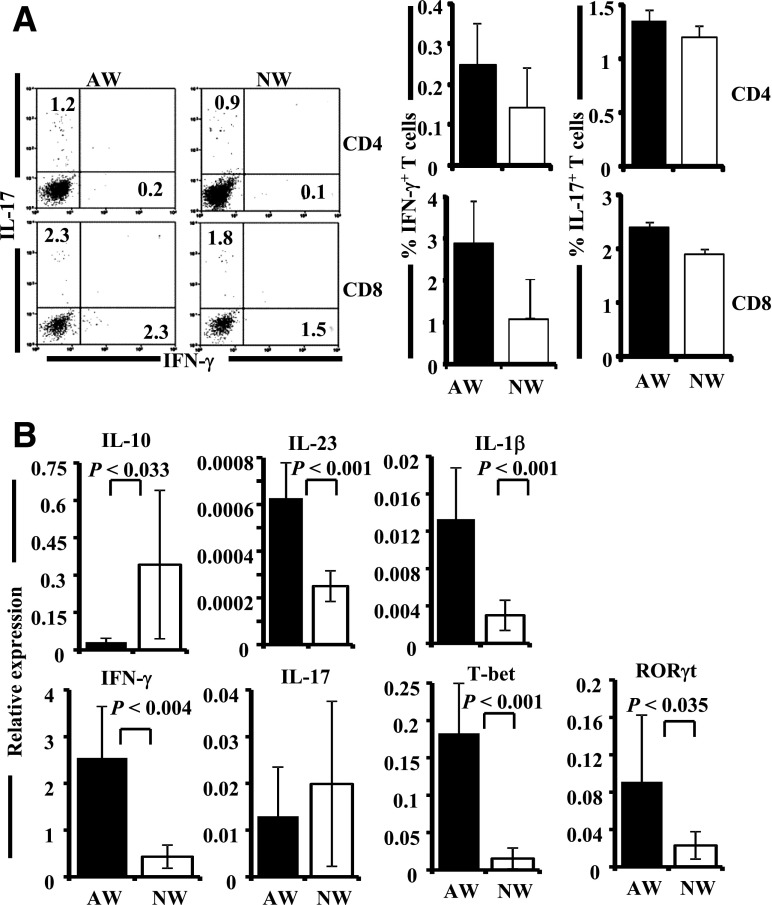
NOD mice on AW show severe insulitis and difference in the gut immune response compared with SFB+ FT and NW recipients. Female NOD mice were purchased from JAX at 3–4 weeks of age and continued on AW or switched to NW and subjected to FT as described for Fig. 4. Sets of three mice from each group were killed at 4 weeks post–drinking-water switch and FT (8 weeks of age) for histopathological and immunological analyses. A: Pancreatic tissues were sectioned and stained using H-E and examined for immune cell infiltration in the islets, and insulitis was scored based on the extent of infiltration. Representative images showing different insulitis grades (left panel) and bar diagrams on percentage of islets in each group with different grades of insulitis (right panel) are shown. At least 150 islets were examined for each group. Single-cell suspensions of spleen (B) and PP (C) were stimulated with phorbol myristic acid/ionomycin for 4 h and examined for intracellular cytokines in CD4+ T cells by FACS. Representative FACS analysis graphs with percentage values (left panels) and mean ± SD of percentage values of IFN-γ– and IL-17–positive cells (right panels) of three/group are shown in a bar diagram. This experiment was repeated once using a similar number of mice. D: cDNA were synthesized and used in a qualitative PCR assay to detect the expression levels of cytokines and transcription factors. This experiment was done twice using three mice/group, and gel profiles of representative samples (left panel) and mean densitometry values (right panel) are shown.
Since alteration of gut microflora by FT resulted in modulated insulitis and T1D incidence, Th1 and Th17 cell frequencies in spleen and Peyers patch (PP) tissues and the overall immune response in the intestine of these mice was examined. As observed in Fig. 5B and C, γ-interferon (IFN-γ)+, but not interleukin (IL)-17+, CD4 T-cell frequencies were relatively higher in the spleen and PP tissues of AW-recipient mice compared with NW-recipient or FT-recipient mice. Qualitative RT-PCR assay using cDNA prepared from distal ileum showed relatively higher IFN-γ expression in the AW group of mice compared with the NW and FT groups (Fig. 5D). Although higher expression of IL-12 was detected in the AW as well as NW/FT groups, expression levels of these factors did not correlate well with IFN-γ levels in different groups of mice. Importantly, retinoic acid receptor (RAR)-related orphan receptor gamma (RORγt), a transcription factor associated with IL-17–producing cells, was detectable primarily in SFB+ FT-recipient mice (AW/FT and NW/FT groups) but not in their control counterparts. These observations indicate that while the immune response in the AW group is predominantly Th1 type, FT resulted in suppressing/skewing this inflammatory response leading to protection from T1D. On the other hand, while mice in the NW group expressed lower Th1-related factors (IFN-γ and IL-12) compared with AW group, the NW/FT group showed somewhat balanced Th1- and Th17-associated factors, both of which appear to have caused a moderate protection from the disease.
AW- and NW-Recipient NOD Mice Show Comparable Number of Immune Regulatory T Cells
Since NOD mice that were on AW and NW as well as their FT-recipient counterparts showed a difference in T1D incidence, we have examined the frequencies of Foxp3+ T cells in these mice. Mice that were on AW and NW as well as their FT-recipient counterparts showed comparable Foxp3+ T-cell frequencies in all lymphoid organs including pancreatic lymph node (PnLN) and PP (Supplementary Fig. 1A). Previous studies have shown that CD62Lhigh T cells have a protective role in T1D (19,26,27). Therefore, we examined CD62L expression on T cells from AW and NW groups of mice and their FT-recipient counterparts. As observed in Supplementary Fig. 1B, frequency of both CD4+ and CD8+ T cells with higher CD62L expression in the PnLN was found to be relatively lower, albeit not statistically significant, in the AW group compared with the NW group of mice. On the other hand, CD62Lhigh T-cell frequencies were relatively higher in the PP of AW recipients compared with NW recipients. SFB+ FT recipient (AW/FT and NW/FT) mice showed CD62Lhigh T-cell frequencies similar to those of the NW group.
Drinking Water pH Affects the Acquisition of Microflora and Influences the Overall Composition of Gut Microbiome
Difference in the composition of fecal microflora (Fig. 3) and immune response in the intestine (Fig. 5) of AW and NW mice groups prompted us to examine the composition of microbiome in the small intestine of AW and NW groups of mice. 16S rDNA–targeted pyrosequencing was performed using DNA samples prepared from the distal ileum of AW, NW, AW/FT, and NW/FT groups of mice 30 days after FT and switching to NW. As observed in Fig. 6, ~98% of ileum commensals in JAX NOD mice that were continued on AW in our SPF facility were identified as Lactobacilli compared with ~79% in the ileum of NOD mice that were switched to NW for a brief period of 30 days. Importantly, ~60% (11.4% of total) of the newly acquired flora under the NW condition during this period was identified as Eubacterium bioforme. The remaining commensals (~10% of total flora) belonged to all major phyla (Firmicutes, Bacteroidetes, Tenericutes, and Actinobacteria). Interestingly, AW and NW groups of mice that received SFB+ FT showed an approximately similar level of Lactobacilli compared with the NW group. These two (AW and NW) groups, however, acquired different gut microbial profiles upon FT, implicating the influence of drinking water pH on the acquisition of gut flora. While the predominant non-Lactobacilli flora was Prevotella loescheii in the AW/FT group of mice (~40%; 8.6% of total flora), it was primarily Clostridium (~50%; 12.1% of total flora) species in the NW/FT group of mice. Our observation that NOD mice, maintained on AW, show relatively lower gut microflora diversity was also confirmed by results obtained in B6 mice that were initially on NW (TAC origin; mice that were used as SFB+ fecal source) but were switched to AW for 2 months (Supplementary Fig. 3). While the B6 mice that were continued on NW showed significantly diverse bacterial communities, >85% of the gut microflora in mice upon switching to AW was composed of Lactobacilli.
Figure 6.
Drinking water pH affects the acquisition of microflora and leads to dysbiosis upon switching from AW to NW. Three- to four-week-old female NOD mice were purchased from JAX and continued on AW or switched to NW and subjected to FT as described for Fig. 5. These mice were killed at 8 weeks of age, and the DNA prepared from distal ileum samples was subjected to 16S rDNA–targeted pyrosequencing. The data were analyzed as described for Fig. 4, and the mean percentage values of bacterial sequences that were identified to species or phylum levels are shown. Upper pie charts represent all communities, and lower pie charts show communities other than Lactobacillus johnsonii. Percentage values of major non–L. johnsonii flora are shown.
AW- and NW-Recipient Mice Show Comparable Overall Frequencies of Memory T Cells
Since our above observations suggested that drinking water pH does not affect the SFB colonization of mouse gut, and SFB may not have a dominant role in the drinking-water pH–dependent difference in the disease incidence in NOD mice, AW- and NW-recipient mice were further characterized for their immune cell properties. We have examined whether memory and autoreactive and/or inflammatory T-cell frequencies are different in AW and NW groups of mice. First, the frequencies of T cells with memory (CD62Llow/CD44high) and naïve (CD62Lhigh/CD44low) phenotypes were examined in mice that were switched to NW for 30 days compared with mice that were continued in the AW group. As observed in Supplementary Fig. 2, no profound differences in the frequencies of CD4+ and CD8+ T cells with memory or naïve phenotype, based on CD62L, CD44, CD127, and CCR7 expression, were observed in different lymphoid organs of NW and AW groups of mice.
AW Group of NOD Mice Shows Higher Frequency of Inflammatory T Cells Compared With NW Recipients
Although there was no significant difference in the frequencies of memory T cells detected in AW- and NW-recipient mice, whether T cells from these groups of mice have different abilities to respond to challenge with self-antigen was examined. As observed in Fig. 7A, a significantly higher number of PnLN and spleen T cells from AW-recipient mice showed proliferative response upon ex vivo challenge with immunodominant β-cell antigenic peptide compared with T cells from NW recipients. In addition, considerably higher frequencies of PnLN T cells were IFN-γ positive in AW recipients compared with mice that were switched to NW (Fig. 7B). These observations suggest that relatively higher numbers of pathogenic T cells are generated in AW compared with NW-recipient mice.
Figure 7.
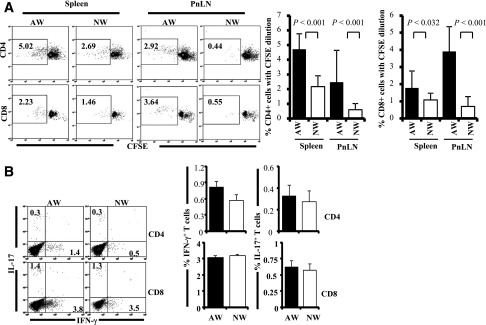
AW-recipient mice show higher autoreactive/proinflammatory T cells in the pancreatic microenvironment compared with NW recipients. Four-week-old NOD mice were continued on AW or switched to NW for 4 weeks and killed at 8 weeks of age, and single suspensions from spleen and PnLN were used for immunological analysis. A: PnLN and spleen cells were labeled with CFSE, incubated with immunodominant β-cell antigen for 4 days, and examined for CFSE dilution among CD4+ and CD8+ cells by FACS. Representative FACS analysis graphs with percentage values (left panel) and mean ± SD of percentage of CD4 and CD8 cells with CFSE dilution (4 mice/group tested independently) (right panel) are shown. B: PnLN cells were also activated ex vivo with phorbol myristic acid and ionomycin in the presence of brefeldin A for 4 h and stained for surface CD4 and CD8, followed by intracellular IFN-γ and IL-17. Representative FACS analysis graphs with percentage values (left panel) and mean ± SD of percentage values of IFN-γ– and IL-17–positive cells among CD4 and CD8 populations from 4 mice/group tested independently (right panel) are shown.
AW-Recipient Mice Show Relatively Higher Expression of Inflammatory Cytokines and Transcription Factors in the Intestinal Mucosa
Since the composition of gut microbiome was significantly different in AW and NW groups of mice and the immune response in the gut appear to be different (Figs. 5 and 6), gut immune function was studied further by FACS and real-time quantitative PCR. The CD4+ and CD8+ T cells from PP of AW- and NW-recipient mice were examined for IFN-γ– and IL-17–producing T cells. As observed in Fig. 8A, although not statistically significant, a relatively higher number of both CD4 and CD8 T cells from the PP of AW-recipient mice expressed IFN-γ compared with PP cells from NW-recipient mice. Other cytokines such as IL-10 and IL-4 were not detectable in the PP cells from these mice by FACS (not shown). Real-time quantitative PCR assay using cDNA prepared from the intestinal tissues shows significantly higher expression levels of a majority of the proinflammatory cytokines and transcription factors that were tested (IFN-γ, T-bet, IL-1β, IL-23, IL-17, and RORγt) in the intestine of the AW group of mice compared with NW recipients. However, NW-recipient mice showed significantly higher expression of IL-10 in the gut (Fig. 8B). These results indicate that mice that were given NW maintained better immune regulation, lesser inflammatory response, and slower breakdown to self-tolerance compared with the mice that were maintained on AW. Results from the microbiome analysis strongly indicate that the better immune regulation in NW groups of mice may directly be linked to higher diversity of gut microflora. Further, we believe that autoimmune progression and incidence of T1D disease are influenced by minor changes in the immune cell function that is, perhaps, initiated in the intestine by microflora. Overall, our observations indicate that in an SPF facility, drinking water of acidic pH restricts the microflora diversity compared with neutral pH drinking water and that higher diversity, in general, favors better immune regulation.
Figure 8.
AW-recipient mice show higher proinflammatory response in the gut mucosa. Four-week-old NOD mice were continued on AW or switched to NW for 4 weeks and killed at 8 weeks of age, and PP and intestinal tissues were used for immunological analysis. A: PP cells were activated ex vivo with phorbol myristic acid and ionomycin in the presence of brefeldin A for 4 h and stained for surface CD4 and CD8, followed by intracellular IFN-γ and IL-17 for FACS analysis. Representative FACS analysis graphs with percentage values (left panel) and mean ± SD of percentage values of IFN-γ– and IL-17–positive cells among CD4 and CD8 populations from 4 mice/group tested independently (right panel) are shown. B: cDNA prepared from the small intestine (distal ileum) was used in real-time quantitative PCR to examine the expression profiles of cytokines and transcription factors. Relative expression levels of these factors in each tissue were calculated against the expression level of a housekeeping gene (β-actin) of the same sample. Mean ± SD of values from 3 mice/group tested independently in triplicate is shown.
Importantly, how acidic pH of drinking water affects the gut microflora diversity is not clear, especially considering the fact that pH values of most parts of the alimentary tract of AW- and NW-recipient mice are comparable (Supplementary Table 2). Although more accurate real-time pH measurement in live animals during water consumption is needed, it appears that the pH is rapidly neutralized even in the oral cavity of the AW group of mice, indicating that the duration of exposure of oral cavity and esophagus to acidic pH may be limited to the consumption stage. It is possible that the oral and esophageal pH fluctuates frequently in AW group of mice leading to the creation of a less favorable condition, for at least some of the orally acquired microflora, resulting in reduced gut flora diversity and overall immune regulation.
Discussion
T1D in humans is believed to be caused by a combination of genetic and environmental factors (1–5,28,29). NOD mice are the primary animal model used for understanding the T1D disease etiology and for preclinical testing of therapeutics. In these mice, genetic background is the primary factor contributing to the development of T1D (6–8,30). T1D in this mouse model is shown to be influenced also by the degree of cleanliness of the mouse facility and exposure to microbial and dietary factors (5,31–37).
In this regard, T1D incidence in the NOD mouse colony housed in our SPF facilities was relatively slower than that in NOD mice from JAX. JAX provides their mice with AW (HCl added; pH 2.8–3.2); however, our SPF facilities, like many other research facilities, maintain them on autoclaved neutral pH water (pH 7.0–7.4). This prompted us to examine whether drinking water pH has an effect on T1D incidence in NOD mice. In fact, NOD mice from our in-house colony and JAX that were on AW showed comparable rates of disease progression and T1D incidence. However, relatively slower progression/lower T1D incidence was observed when in-house NOD mice were given NW or when JAX mice were switched to NW at an early age of 3–4 weeks. These observations confirmed that drinking-water pH does have an influence on T1D incidence in NOD mice.
It is not known whether the pH of drinking water has an influence on the prevalence of T1D or other autoimmune diseases in humans. Although the direct effect of pH on alimentary tract tissues is not well studied, it is assumed that pH of drinking water may affect only the tissues of the upper alimentary tract. The intestinal tissues, because of the naturally acidic stomach and the neutralization of stomach content upon exit to the small intestine, may not be affected directly by the pH of drinking water. Nevertheless, the chemicals that contribute to acidity of drinking water have the potential to affect the cellular function of tissues that are exposed to them. In fact, in vitro and ex vivo studies have demonstrated that HCl can induce direct inflammatory response in the esophageal mucosa involving IL-8, platelet activating factor, mucosa-released substance P, and H2O2 (38–41). The ability of IL-8 and platelet activating factor to promote T-helper cell pathways, including Th1 response, by induction of other proinflammatory cytokines such as IL-6 and TNF-α (39–41) has been recognized. Therefore, it is possible that drinking-water pH may have a direct effect in promoting Th1 response, leading to rapid progression of autoimmunity in T1D. Although our observations show that the pH of alimentary tract content of AW and NW groups of mice is comparable at the resting stage (while water is not being consumed), difference in the oral and esophageal pH during water consumption may be partially responsible for the inflammatory response.
While the overall gut microflora density was comparable in AW- and NW-recipient mice (not shown), results from the analysis of microbial communities in fecal samples of these mice suggest that brief exposures of the oral cavity and esophagus to AW upon water consumption or frequent changes in the pH of these parts of alimentary tract leads to restricted gut microbial diversity, perhaps by inhibiting the acquisition of certain communities. Our observations suggest that the direct effect of drinking-water pH on the microflora of upper alimentary tract needs to be investigated in the future. In addition, the existing microflora communities could determine which newly acquired bacteria have a place in the gut. Community profiling of small-intestinal microflora of FT-recipient mice that were on AW or those switched to NW indirectly supports this possibility. While switching the AW group of mice to NW resulted in an increase in the diversity of intestinal microbial communities, switching NW groups of mice to AW (as shown in Fig. 7) resulted in the enrichment of a small number of specific communities in the gut. However, FT from mice of a different facility (TAC) resulted in the acquisition of distinct communities of commensals under acidic and neutral water conditions.
Recent studies have shown the disease-modulatory effect of colonization of mouse gut by SFB in various autoimmune models in a Th17-dependent manner (10,23–25). Most importantly, natural colonization of NOD mouse gut by SFB resulted in a Th17 response in the intestine and protection of these mice from T1D (10). Our analyses showed that difference in the T1D disease incidence, observed in NOD mice that were maintained on AW and NW, is not linked to SFB infiltration. In addition, FT studies demonstrated that colonization of NOD mouse intestine by SFB was not affected by the pH of drinking water. Interestingly, although FT from SFB+ mice resulted in the protection of NOD mice that were on AW, SFB appeared to have no influence on the disease incidence in mice that were on NW. AW and NW groups that received SFB+ FT (AW/FT and NW/FT) showed a similar degree of gut microbial diversity for at least a short period, which is comparable with that of the NW group. These observations suggest that while SFB may have a disease-protective effect in NOD mice under specific conditions, disease progression and incidence are affected by overall composition of the gut microbiome, including the degree of diversity and, perhaps, the gut microflora–influenced immune response. In fact, our results show that maintaining NOD mice on AW, in an SPF facility, minimizes microbial diversity and promotes Th1 response in the gut, which is associated with rapid disease progression. However, altering and/or enhancing microflora diversity, through changes in drinking water pH or by FT, resulted in the expression of factors that are associated with a mixed T-helper cell response (involving both Th1 and Th17), leading to better immune regulation and slow disease progression and reduced T1D incidence. This notion has been substantiated by our results that show higher frequencies of pancreatic β-cell antigen–specific as well as IFN-γ–producing T cells and an overall higher inflammatory cytokine response in the intestine of the AW group of mice compared with NW recipients.
Overall, our observations show that NOD mice of an SPF facility that received AW developed T1D rapidly compared with NW recipients, and this effect could be due to difference in the composition and diversity of gut microbiome and the associated immune response. In this regard, recent epidemiological studies and experiments using animal models have shown that gut microbiome plays a key role affecting the disease outcome in T1D (7,42–44). These studies show a link between high levels of Bacteroides in the gut and autoimmunity in T1D diabetes (42–44). Since the majority of gut flora of the mice used in our study was composed of Lactobacillus (Firmicutes), it is not completely clear what effect drinking water pH has on members of the Bacteriodetes phylum. It appears that AW consumption substantially reduces the diversity of gut microflora including Bacteriodes but increases the proportion of Lactobacillus members resulting in rapid disease progression.
While future studies on what direct effect AW and the inflammatory response induced in the upper alimentary tract by this pH can have on autoimmunity are needed, our results show that drinking-water pH has a profound effect on the diversity of gut microflora and T1D incidence. In addition, the observation that forced dysbiosis can result in profound modulation of disease progression in AW recipients suggests that the direct effect of acidic pH on disease progression may not be as significant as the indirect effect produced through affecting the gut microbiome. In conclusion, environmental factors such as drinking-water pH can have a profound influence on autoimmunity primarily by affecting the composition of gut microflora.
Our observations indicate that a series of studies is needed in the future to understand 1) what effect the pH of drinking water has on the microflora of upper–alimentary tract and orally acquired flora, 2) what the long-term stability is of drinking-water pH–influenced gut microflora and the immune response of the gut, 3) which specific microbial communities are responsible for promoting or suppressing inflammatory and autoimmune response, and 4) what is the mechanism of immune modulation by these specific microbial communities.
Supplementary Material
Article Information
Acknowledgments. The authors thank Dr. M. Andrea Azcarate-Peril, Microbiome Core Facility, University of North Carolina, Chapel Hill, NC, for providing service with generating microbiome sequences. The authors also thank Dr. Scot E. Dowd, Molecular Research LP DNA (Molecular Research Center, Shallowater, TX), for help with analyzing the microbiome data and Dr. Joe Jones, EnGenCore, University of South Carolina, Columbia, SC, for help with using the QIIME application.
Funding. This work was supported by the internal funds from the Medical University of South Carolina and UIC, National Institutes of Health (NIH) grant R01-AI-073858, American Diabetes Association grant ADA-1-13-IN-57, and JDRF regular grant JDRF-32-2008-343 to C.V.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. M.H.S. designed experiments; researched data; and wrote, reviewed, and edited the manuscript. R.G. researched data and reviewed and edited the manuscript. S.K.-M., N.P., and B.M.J. researched data. C.V. designed experiments; analyzed data; and wrote, reviewed, and edited the manuscript. C.V. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db13-0981/-/DC1.
References
- 1.Rook GA. Hygiene hypothesis and autoimmune diseases. Clin Rev Allergy Immunol 2012;42:5–15 [DOI] [PubMed] [Google Scholar]
- 2.Bach JF. Protective role of infections and vaccinations on autoimmune diseases. J Autoimmun 2001;16:347–353 [DOI] [PubMed] [Google Scholar]
- 3.Germolec D, Kono DH, Pfau JC, Pollard KM. Animal models used to examine the role of the environment in the development of autoimmune disease: findings from an NIEHS Expert Panel Workshop. J Autoimmun 2012;39:285–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christen U, Bender C, von Herrath MG. Infection as a cause of type 1 diabetes? Curr Opin Rheumatol 2012;24:417–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pozzilli P, Signore A, Williams AJ, Beales PE. NOD mouse colonies around the world—recent facts and figures. Immunol Today 1993;14:193–196 [DOI] [PubMed] [Google Scholar]
- 6.Markle JG, Frank DN, Mortin-Toth S, et al. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science 2013;339:1084–1088 [DOI] [PubMed] [Google Scholar]
- 7.Hara N, Alkanani AK, Ir D, et al. The role of the intestinal microbiota in type 1 diabetes. Clin Immunol 2013;146:112–119 [DOI] [PubMed] [Google Scholar]
- 8.King C, Sarvetnick N. The incidence of type-1 diabetes in NOD mice is modulated by restricted flora not germ-free conditions. PLoS ONE 2011;6:e17049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wen L, Ley RE, Volchkov PY, et al. Innate immunity and intestinal microbiota in the development of type 1 diabetes. Nature 2008;455:1109–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kriegel MA, Sefik E, Hill JA, Wu HJ, Benoist C, Mathis D. Naturally transmitted segmented filamentous bacteria segregate with diabetes protection in nonobese diabetic mice. Proc Natl Acad Sci USA 2011;108:11548–11553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alam C, Bittoun E, Bhagwat D, et al. Effects of a germ-free environment on gut immune regulation and diabetes progression in non-obese diabetic (NOD) mice. Diabetologia 2011;54:1398–1406 [DOI] [PubMed] [Google Scholar]
- 12.Yurkovetskiy L, Burrows M, Khan AA, et al. Gender bias in autoimmunity is influenced by microbiota. Immunity 2013;39:400–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu GD, Chen J, Hoffmann C, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011;334:105–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faith JJ, McNulty NP, Rey FE, Gordon JI. Predicting a human gut microbiota’s response to diet in gnotobiotic mice. Science 2011;333:101–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muegge BD, Kuczynski J, Knights D, et al. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science 2011;332:970–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown K, DeCoffe D, Molcan E, Gibson DL. Diet-induced dysbiosis of the intestinal microbiota and the effects on immunity and disease. Nutrients 2012;4:1095–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karumuthil-Melethil S, Perez N, Li R, Prabhakar BS, Holterman MJ, Vasu C. Dendritic cell-directed CTLA-4 engagement during pancreatic beta cell antigen presentation delays type 1 diabetes. J Immunol 2010;184:6695–6708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perez N, Karumuthil-Melethil S, Li R, Prabhakar BS, Holterman MJ, Vasu C. Preferential costimulation by CD80 results in IL-10-dependent TGF-beta1(+) -adaptive regulatory T cell generation. J Immunol 2008;180:6566–6576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karumuthil-Melethil S, Perez N, Li R, Vasu C. Induction of innate immune response through TLR2 and dectin 1 prevents type 1 diabetes. J Immunol 2008;181:8323–8334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeSantis TZ, Hugenholtz P, Larsen N, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 2006;72:5069–5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McDonald D, Price MN, Goodrich J, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 2012;6:610–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li R, Perez N, Karumuthil-Melethil S, Vasu C. Bone marrow is a preferential homing site for autoreactive T-cells in type 1 diabetes. Diabetes 2007;56:2251–2259 [DOI] [PubMed] [Google Scholar]
- 23.Wu HJ, Ivanov II, Darce J, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity 2010;32:815–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc Natl Acad Sci USA 2011;108(Suppl. 1):4615–4622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ivanov II, Atarashi K, Manel N, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 2009;139:485–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szanya V, Ermann J, Taylor C, Holness C, Fathman CG. The subpopulation of CD4+CD25+ splenocytes that delays adoptive transfer of diabetes expresses L-selectin and high levels of CCR7. J Immunol 2002;169:2461–2465 [DOI] [PubMed] [Google Scholar]
- 27.Lepault F, Gagnerault MC. Characterization of peripheral regulatory CD4+ T cells that prevent diabetes onset in nonobese diabetic mice. J Immunol 2000;164:240–247 [DOI] [PubMed] [Google Scholar]
- 28.Eringsmark Regnéll S, Lernmark A. The environment and the origins of islet autoimmunity and type 1 diabetes. Diabet Med 2013;30:155–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beyan H, Wen L, Leslie RD. Guts, germs, and meals: the origin of type 1 diabetes. Curr Diab Rep 2012;12:456–462 [DOI] [PubMed] [Google Scholar]
- 30.Boerner BP, Sarvetnick NE. Type 1 diabetes: role of intestinal microbiome in humans and mice. Ann N Y Acad Sci 2011;1243:103–118 [DOI] [PubMed] [Google Scholar]
- 31.Cooke A, Zaccone P, Raine T, Phillips JM, Dunne DW. Infection and autoimmunity: are we winning the war, only to lose the peace? Trends Parasitol 2004;20:316–321 [DOI] [PubMed] [Google Scholar]
- 32.Saunders KA, Raine T, Cooke A, Lawrence CE. Inhibition of autoimmune type 1 diabetes by gastrointestinal helminth infection. Infect Immun 2007;75:397–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zaccone P, Burton OT, Gibbs SE, et al. The S. mansoni glycoprotein ω-1 induces Foxp3 expression in NOD mouse CD4⁺ T cells. Eur J Immunol 2011;41:2709–2718 [DOI] [PubMed] [Google Scholar]
- 34.Nikoopour E, Schwartz JA, Huszarik K, et al. Th17 polarized cells from nonobese diabetic mice following mycobacterial adjuvant immunotherapy delay type 1 diabetes. J Immunol 2010;184:4779–4788 [DOI] [PubMed] [Google Scholar]
- 35.Alam C, Valkonen S, Palagani V, Jalava J, Eerola E, Hänninen A. Inflammatory tendencies and overproduction of IL-17 in the colon of young NOD mice are counteracted with diet change. Diabetes 2010;59:2237–2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chamson-Reig A, Arany EJ, Summers K, Hill DJ. A low protein diet in early life delays the onset of diabetes in the non-obese diabetic mouse. J Endocrinol 2009;201:231–239 [DOI] [PubMed] [Google Scholar]
- 37.Funda DP, Kaas A, Tlaskalová-Hogenová H, Buschard K. Gluten-free but also gluten-enriched (gluten+) diet prevent diabetes in NOD mice; the gluten enigma in type 1 diabetes. Diabetes Metab Res Rev 2008;24:59–63 [DOI] [PubMed] [Google Scholar]
- 38.Ma J, Altomare A, de la Monte S, et al. HCl-induced inflammatory mediators in esophageal mucosa increase migration and production of H2O2 by peripheral blood leukocytes. Am J Physiol Gastrointest Liver Physiol 2010;299:G791–G798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheng L, Cao W, Fiocchi C, Behar J, Biancani P, Harnett KM. HCl-induced inflammatory mediators in cat esophageal mucosa and inflammatory mediators in esophageal circular muscle in an in vitro model of esophagitis. Am J Physiol Gastrointest Liver Physiol 2006;290:G1307–G1317 [DOI] [PubMed] [Google Scholar]
- 40.Lacasse C, Turcotte S, Gingras D, Stankova J, Rola-Pleszczynski M. Platelet-activating factor stimulates interleukin-6 production by human endothelial cells and synergizes with tumor necrosis factor for enhanced production of granulocyte-macrophage colony stimulating factor. Inflammation 1997;21:145–158 [DOI] [PubMed] [Google Scholar]
- 41.Kulcharyk PA, Heinecke JW. Hypochlorous acid produced by the myeloperoxidase system of human phagocytes induces covalent cross-links between DNA and protein. Biochemistry 2001;40:3648–3656 [DOI] [PubMed] [Google Scholar]
- 42.de Goffau MC, Luopajärvi K, Knip M, et al. Fecal microbiota composition differs between children with β-cell autoimmunity and those without. Diabetes 2013;62:1238–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Giongo A, Gano KA, Crabb DB, et al. Toward defining the autoimmune microbiome for type 1 diabetes. ISME J 2011;5:82–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brugman S, Klatter FA, Visser JT, et al. Antibiotic treatment partially protects against type 1 diabetes in the Bio-Breeding diabetes-prone rat. Is the gut flora involved in the development of type 1 diabetes? Diabetologia 2006;49:2105–2108 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.