Abstract
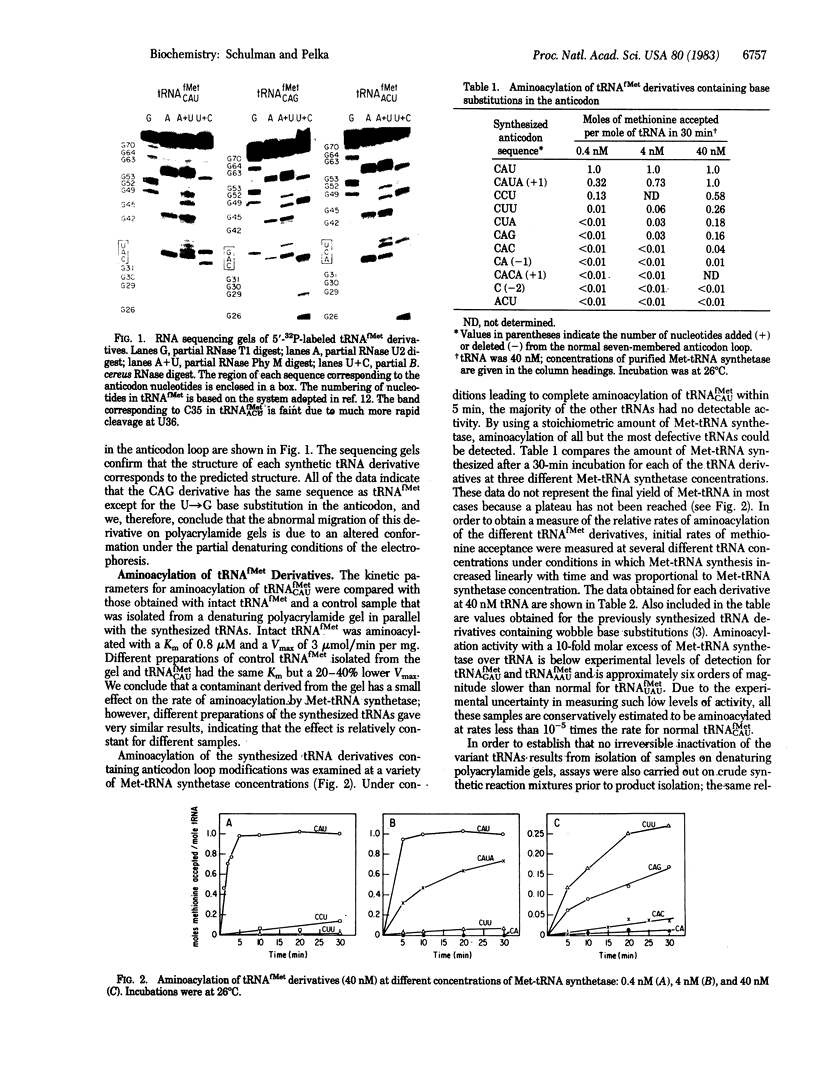
Previous work from our laboratory identified several specific sites in Escherichia coli tRNAfMet that are essential for recognition of this tRNA by E. coli methionyl-tRNA synthetase (EC 6.1.1.10). Particularly strong evidence indicated a role for the nucleotide base at the wobble position of the anticodon in the discrimination process. To further investigate the structural requirements for recognition in this region, we have synthesized a series of tRNAfMet derivatives containing single base changes in each position of the anticodon. In addition, derivatives containing permuted sequences and larger and smaller anticodon loops have been prepared. The variant tRNAs have been enzymatically synthesized in vitro. The procedure involves excision of the normal anticodon, CAU, by limited digestion of intact tRNAfMet with pancreatic RNase. This step also removes two nucleotides from the 3' CpCpA end. T4 RNA ligase is used to join oligonucleotides of defined length and sequence to the 5' half-molecule and subsequently to link the 3' and modified 5' fragment to regenerate the anticodon loop. The final step of the synthesis involves repair of the 3' terminus with tRNA nucleotidyltransferase. The synthetic derivative containing the anticodon CAU is aminoacylated with the same kinetics as intact tRNAfMet. Base substitutions in the wobble position reduce aminoacylation rates by at least five orders of magnitude. The rates of aminoacylation of derivatives having base substitutions in the other two positions of the anticodon are 1/55 to 1/18,500 times normal. Nucleotides that have specific functional groups in common with the normal anticodon bases are better tolerated at each of these positions than those that do not. A tRNAfMet variant having a six-membered loop containing only the CA sequence of the anticodon is aminoacylated still more slowly, and a derivative containing a five-membered loop is not measurably active. The normal loop size can be increased by one nucleotide with a relatively small effect on the rate of aminoacylation, indicating that the spatial arrangement of the nucleotides is less critical than their chemical nature. We conclude from these data that recognition of tRNAfMet requires highly specific interactions of methionyl-tRNA synthetase with functional groups on the nucleotide bases of the anticodon sequence.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beresten' S. F., Sheinker V. Sh, Biolotina I. A., Hurbekov M. K., Mashkova T. D. Rol' antikodonovoi petli v induktsii konformatsionnykh izmenenii triptofanil-tRNK-sintetazy pod desitviem tRNKtrp. Mol Biol (Mosk) 1981 Jul-Aug;15(4):805–815. [PubMed] [Google Scholar]
- Bonnet J., Ebel J. P. Interpretation of incomplete reactions in tRNA aminoacylation. Aminoacylation of yeast tRNA Val II with yeast valyl-tRNA synthetase. Eur J Biochem. 1972 Dec 4;31(2):335–344. doi: 10.1111/j.1432-1033.1972.tb02538.x. [DOI] [PubMed] [Google Scholar]
- Bruce A. G., Uhlenbeck O. C. Enzymatic replacement of the anticodon of yeast phenylalanine transfer ribonucleic acid. Biochemistry. 1982 Mar 2;21(5):855–861. doi: 10.1021/bi00534a007. [DOI] [PubMed] [Google Scholar]
- Bruce A. G., Uhlenbeck O. C. Specific interaction of anticodon loop residues with yeast phenylalanyl-tRNA synthetase. Biochemistry. 1982 Aug 17;21(17):3921–3926. doi: 10.1021/bi00260a003. [DOI] [PubMed] [Google Scholar]
- Cameron V., Soltis D., Uhlenbeck O. C. Polynucleotide kinase from a T4 mutant which lacks the 3' phosphatase activity. Nucleic Acids Res. 1978 Mar;5(3):825–833. doi: 10.1093/nar/5.3.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraburtty K. Effect of sodium bisulfite modification on the arginine acceptance of E. coli tRNA Arg. Nucleic Acids Res. 1975 Oct;2(10):1793–1804. doi: 10.1093/nar/2.10.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers R. W., Aoyagi S., Furukawa Y., Zawadzka H., Bhanot O. S. Inactivation of valine acceptor ativity by a C-U missense change in the anticodon of yeast valine transfer ribonucleic acid. J Biol Chem. 1973 Aug 10;248(15):5549–5551. [PubMed] [Google Scholar]
- Donis-Keller H., Maxam A. M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977 Aug;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- England T. E., Uhlenbeck O. C. Enzymatic oligoribonucleotide synthesis with T4 RNA ligase. Biochemistry. 1978 May 30;17(11):2069–2076. doi: 10.1021/bi00604a008. [DOI] [PubMed] [Google Scholar]
- Higgins N. P., Geballe A. P., Snopek T. J., Sugino A., Cozzarelli N. R. Bacteriophage T4 RNA ligase: preparation of a physically homogeneous, nuclease-free enzyme from hyperproducing infected cells. Nucleic Acids Res. 1977 Sep;4(9):3175–3186. doi: 10.1093/nar/4.9.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krug M., Uhlenbeck O. C. Reversal of T4 RNA ligase. Biochemistry. 1982 Apr 13;21(8):1858–1864. doi: 10.1021/bi00537a024. [DOI] [PubMed] [Google Scholar]
- Lefevre J. F., Bacha H., Renaud M., Ehrlich R., Gangloff J., Von der Haar F., Remy P. Fluorimetric study of yeast tRNAPheCCF in the complex with phenylalanyl-tRNA synthetase. Evidence for a correlation between the structural adaptation of both macromolecules and the appearance of the acylation activity. Eur J Biochem. 1981 Jul;117(3):439–447. [PubMed] [Google Scholar]
- Scheinker V. S., Beresten S. F., Mashkova T. D., Mazo A. M., Kisselev L. L. Role of exposed cytosine residues in aminoacylation activity of tRNATrp. FEBS Lett. 1981 Sep 28;132(2):349–352. doi: 10.1016/0014-5793(81)81195-7. [DOI] [PubMed] [Google Scholar]
- Schulman L. H., Goddard J. P. Loss of methionine acceptor activity resulting from a base change in the anticodon of Escherichia coli formylmethionine transfer ribonucleic acid. J Biol Chem. 1973 Feb 25;248(4):1341–1345. [PubMed] [Google Scholar]
- Schulman L. H., Pelka H. Alteration of the kinetic parameters for aminoacylation of Escherichia coli formylmethionine transfer RNA by modification of an anticodon base. J Biol Chem. 1977 Feb 10;252(3):814–819. [PubMed] [Google Scholar]
- Schulman L. H., Pelka H. Structural requirements for aminoacylation of Escherichia coli formylmethionine transfer RNA. Biochemistry. 1977 Sep 20;16(19):4256–4265. doi: 10.1021/bi00638a020. [DOI] [PubMed] [Google Scholar]
- Schulman L. H., Pelka H., Susani M. Base substitutions in the wobble position of the anticodon inhibit aminoacylation of E. coli tRNAfMet by E. coli Met-tRNA synthetase. Nucleic Acids Res. 1983 Mar 11;11(5):1439–1455. doi: 10.1093/nar/11.5.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman L. H. Structure and function of Escherichia coli formylmethionine transfer RNA. II. Effect of modification of guanosine residues on aminoacyl synthetase recognition. J Mol Biol. 1971 May 28;58(1):117–131. doi: 10.1016/0022-2836(71)90236-1. [DOI] [PubMed] [Google Scholar]
- Schulman L. H. Structure and function of Escherichia coli formylmethionine transfer RNA: loss of methionine acceptor activity by modification of a specific guanosine residue in the acceptor stem of formylmethionine transfer RNA from Escherichia coli. Proc Natl Acad Sci U S A. 1972 Dec;69(12):3594–3597. doi: 10.1073/pnas.69.12.3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squires C., Carbon J. Normal and mutant glycine transfer RNAs. Nat New Biol. 1971 Oct 27;233(43):274–277. doi: 10.1038/newbio233274a0. [DOI] [PubMed] [Google Scholar]
- Stern L., Schulman L. H. Role of anticodon bases in aminoacylation of Escherichia coli methionine transfer RNAs. J Biol Chem. 1977 Sep 25;252(18):6403–6408. [PubMed] [Google Scholar]
- THACH R. E., DOTY P. SYNTHESIS OF BLOCK OLIGONUCLEOTIDES. Science. 1965 Mar 12;147(3663):1310–1311. doi: 10.1126/science.147.3663.1310. [DOI] [PubMed] [Google Scholar]
- Uemura H., Imai M., Ohtsuka E., Ikehara M., Söll D. E. coli initiator tRNA analogs with different nucleotides in the discriminator base position. Nucleic Acids Res. 1982 Oct 25;10(20):6531–6539. doi: 10.1093/nar/10.20.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]