Abstract
Staphylococcal cassette chromosome (SCC) elements contribute considerably to virulence and resistance to antibiotic agents in staphylococci. SCC elements in coagulase-negative staphylococci (CoNS) are highly diverse and there is evidence suggesting that they serve as a reservoir for antibiotic resistance genes in methicillin-resistant Staphylococcus aureus (MRSA). However, only a small number of SCC elements have been characterized in CoNS and their exact roles in the emergence and evolution of MRSA remain to be demonstrated. Here, we determined the structure of an SCC composite island (CISH32) found in the clinical Staphylococcus haemolyticus isolate SH32 by whole-genome DNA sequencing. CISH32 was 48 kb in length and mainly composed of two imperfect SCC elements, namely (i) a ΨSCCmec(SH32) part containing a class C1 mec gene complex but lacking ccr genes and (ii) a SCCSH32 part with a ccrA5B3 gene complex but lacking mec genes. In addition, CISH32 contained a type III restriction-modification system and several resistance loci, for example genes conferring resistance to cadmium and arsenic. ΨSCCmec(SH32) is almost entirely identical to a pseudo SCCmec element found in S. haemolyticus WCH1 and shares pronounced sequence similarity to a ΨSCCmec element of S. haemolyticus JCSC1435. However, staphylococci other than S. haemolyticus, including S. aureus and S. epidermidis, contain homologs of SCCSH32 that are more similar to SCCSH32 than those elements found in S. haemolyticus, suggesting that CISH32 of S. haemolyticus SH32 was assembled in recent evolutionary events. Moreover, the composite structure of CISH32 indicates that the detection of class C1 mec and ccrA5B3 gene complexes in S. haemolyticus does not always indicate the existence of a UT9-type SCCmec element, which has remained questionable.
Introduction
The emergence and spread of methicillin-resistant Staphylococcus aureus (MRSA) has become a worldwide problem, which is in part due to the extensive repertoire of virulence factors present in many MRSA strains and the fact that some isolates also contain genes providing resistance to a wide array of other antibiotic agents [1]. Virulence and antibiotic resistance genes are mostly encoded by mobile genetic elements (MGEs), including plasmids, transposons and staphylococcal cassette chromosome (SCC) elements [2]. SCCmec elements represent a particular concern, because they harbor mec genes (mecA/mecC) providing resistance to methicillin and almost all other beta-lactam antibiotics.
In recent years, an extensive genetic diversity of SCCmec elements has been revealed in S. aureus and other staphylococci, by PCR assays, DNA microarrays, or genome sequencing [3]–[7]. These studies also showed that coagulase-negative staphylococci (CoNS) harbor more diverse SCCmec elements than S. aureus and are a potential reservoir for the transfer of SCCmec elements to S. aureus [8]. However, the exact role of SCCmec elements of CoNS in the emergence and evolution of MRSA is largely unknown and requires characterization of more SCC/SCCmec elements. In the present work, we identified an SCC composite island (CI) in the clinical S. haemolyticus isolate SH32 and analyzed its structure in comparison to related SCC elements.
Materials and Methods
Ethics statement, strain and DNA preparation
S. haemolyticus SH32 was isolated in 2003 from the blood of an inpatient at First Affiliated Hospital, College of Medicine, Zhejiang University. We obtained an exempt status from the Institutional Review Board of the First Affiliated Hospital, College of Medicine, Zhejiang University to use this strain to perform all experiments in this study. Antibiotic susceptibility of the strain was described previously [9]. Genomic DNA was prepared with a QIAamp DNA mini kit (Qiagen) according to the manufacturer's instructions.
Sequencing and gap closure
Both pair-end and mate-pair (3-kb insertion) libraries were constructed and sequenced using a HiSeq 2000 platform (Illumina, USA) in the Chinese National Human Genome Center, Shanghai. The acquired 2×100 bp reads were assembled by velvet software [10]. Identification of contigs representing parts of SCC elements was performed using BLAST tools [11], using mapping marker genes such as orfX, mecA and the ccr gene complex. Gap closure was then performed using standard PCR and PCR product sequencing using a 3730XL instrument (Applied Biosystems, USA).
Annotation and comparative analysis
Gene prediction in S. haemolyticus SH32 was performed using GeneMark-hmm (V2.08) [12]. The nucleotide acid sequence of predicted genes and their deduced amino acid sequences were subsequently compared against the non-redundant protein database provided by NCBI (www.nlm.nih.gov) using BLAST tools [11].
The comparison of different SCC elements was performed using BLASTN and displayed by in-house developed perl scripts. The sequences used in the comparative analysis were retrieved from the NCBI website.
Detection of mecA and ccrA5B3
A specific primer pair (forward: GAACCGCAGGTCTCTTCAGATCTAC; reverse: CACCTTGTCCGTAACCTGAATC) was used to determine the class C1 mec gene complex using standard PCR. Long-distance PCR was performed to detect ccrA5B3 (forward: CGCGCTATTATCACGAATCC; reverse: GCGTGATTAAGTGCGTTAGC). The PCR products were subsequently sequenced using an ABI 3730 sequence analyzer (Applied Biosystems).
Results and Discussion
Assembly and annotation
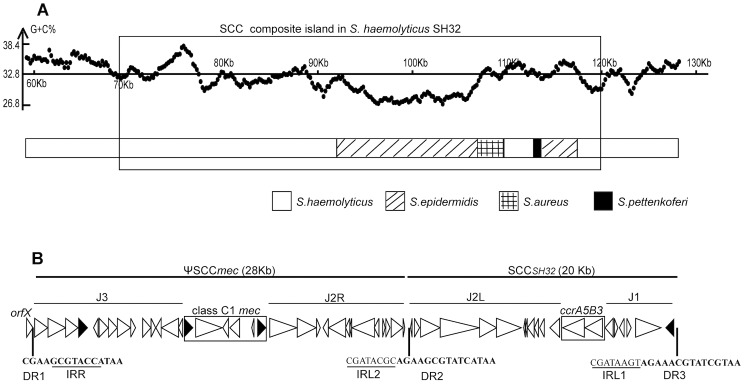
Genome sequencing of S. haemolyticus SH32 generated 339 contigs. Five of them harbored orfX, mecA and ccr genes or fragments, indicating that they contain fragments of an SCCmec element. The gaps between these contigs were then closed by standard PCR and PCR product sequencing, which yielded one contig with a length of 197,413 bp and an average G+C content of 33.3% (accession number: KF006347). Among a total of 201 genes that were identified in this contig, most had their best-hit homologs in S. haemolyticus JCSC1435. Almost half of the genes in the identified SCC composite island (hereafter referred to as CISH32) were more similar to their counterparts in S. epidermidis, S. aureus and other staphylococci than those reported from S. haemolyticus (Figure 1A). Partially, this may be due to the limited information available about SCC/SCCmec elements in S. haemolyticus, but it may also suggest events of horizontal transfer of those SCCmec elements between the respective staphylococcal species.
Figure 1. General features of the SCC composite island in Staphylococcus haemolyticus SH32.
(A) Distribution of the best-hit homologs of genes contained in the SH32 SCC composite island in staphylococci. The G+C content of the SCC region (from 70 to 120 kb) was calculated with a window size of 200 bp. 32.8% is the average G+C content of the whole chromosome. (B) Gene structure of the SH32 SCC composite island. Predicted genes in SCC elements are represented by triangles, while their direction indicates the encoding strands. Black triangles denote IS431 genes. Abbreviations: DR, direct repeat sequence; IR, inverted repeat sequence.
General features of the SCC composite island in SH32
The CISH32 region, which is characterized by the bracketing 15-bp sequences found in integration site sequences containing directed (DR) and inverted repeats (IR) (Figure 1B), is about 48 kb in length and comprises 54 predicted genes (SHP0065-118) (Table 1). The G+C content of CISH32 is 31.6%, slightly lower than that of the whole chromosome of S. haemolyticus (∼32.8%), which is possibly due to an enrichment of genes originating from horizontal gene transfer in SCC elements. In contrast to the composite islands found in S. epidermidis ATCC 12228 and S. haemolyticus JCSC1435, for which eight and six copies, respectively, of integration site sequences (ISS) were found and which are composed of multiple SCC elements (or remnants) [13], [14], only two copies of ISS were found in CISH32 in addition to that present in orfX. This suggests that less complex integration events occurred in the SCC element in S. haemolyticus SH32. According to the nomenclature of IWG-SCC [15], the structure of CISH32 was described as orfX-ΨSCCmec(SH32)-SCCSH32, i.e. a pseudo SCCmec element that contains a mec gene complex but lacks ccr recombinases [hereafter referred to as ΨSCCmec(SH32)], and an SCC element that harbors ccr but lacks mec genes (hereafter referred to as SCCSH32).
Table 1. Gene content of the SCCmec element in Staphylococcus haemolyticus SH32.
| Locus | Position1 | Regions | Characteristic genes | Description |
| SHP0065 | 70295–70774 | orfX | rRNA large subunit methyltransferase | |
| SHP0066 | 70903–71889 | J3 | ADP-ribosylglycohydrolase | |
| SHP0067 | 71908–73242 | Permease | ||
| SHP0068 | 73239–74177 | truncated ribokinase | ||
| SHP0069 | 74215–74889 | IS431 | transposase for IS431mec | |
| SHP0070 | 75693–75346 | transcriptional regulator | ||
| SHP0071 | 75771–76454 | ThiJ/PfpI family protein | ||
| SHP0072 | 76476–77141 | NAD dependent epimerase/dehydratase | ||
| SHP0073 | 77145–78149 | Oxidoreductase | ||
| SHP0074 | 78158–78523 | hypothetical protein | ||
| SHP0075 | 79059–79676 | cadD | cadmium binding protein CadD | |
| SHP0076 | 79695–80036 | cadX | cadmium resistant accessory protein | |
| SHP0077 | 80457–80056 | arsC | arsenate reductase | |
| SHP0078 | 81764–80475 | arsB | arsenical pump membrane protein | |
| SHP0079 | 82081–81764 | arsR | arsenical resistance operon repressor | |
| SHP0080 | 82211–82885 | mec gene complex | IS431 | transposase for IS-like element |
| SHP0081 | 83061–85067 | mecA | penicillin-binding protein 2′ | |
| SHP0082 | 85541–85113 | uncharacterized protein ydem | ||
| SHP0083 | 86381–85638 | glycerophosphoryl diester phosphodiesterase | ||
| SHP0084 | 87465–87298 | 3-hydroxy-3-methylglutaryl CoA synthase | ||
| SHP0085 | 87723–88397 | IS431 | transposase for IS-like element | |
| SHP0086 | 88660–90720 | J2R | copA | P-type ATPase copper (Cu2+) transporter |
| SHP0087 | 90735–92168 | multicopper oxidase mco | ||
| SHP0088 | 92188–92478 | putative lipoprotein | ||
| SHP0089 | 93079–92684 | arsC | arsenate reductase | |
| SHP0090 | 94390–93098 | arsB | arsenite-antimonite efflux pump | |
| SHP0091 | 94704–94390 | arsR | arsenic resistance operon repressor | |
| SHP0092 | 94844–94701 | hypothetical protein | ||
| SHP0093 | 96574–94844 | arsA | arsenite-activated ATPase | |
| SHP0094 | 96902–96555 | arsD | arsenical resistance operon trans-acting | |
| repressor | ||||
| SHP0095 | 97185–97379 | hypothetical protein | ||
| SHP0096 | 97425–97745 | ArsR family transcriptional regulator | ||
| SHP0097 | 97833–98717 | putative permease | ||
| SHP0098 | 98731–98859 | hypothetical protein | ||
| SHP0099 | 99411–99295 | J2L | hypothetical protein | |
| SHP0100 | 99585–99977 | type III R-M system enzyme, M subunit | ||
| SHP0101 | 100048–101532 | type III R-M system protein | ||
| SHP0102 | 101534–104503 | type III R-M, res subunit | ||
| SHP0103 | 104510–105802 | hypothetical protein | ||
| SHP0104 | 105786–107660 | type III R-M, res subunit | ||
| SHP0105 | 107961–107842 | hypothetical protein | ||
| SHP0106 | 108112–107981 | hypothetical protein | ||
| SHP0107 | 108630–108127 | hypothetical protein | ||
| SHP0108 | 108959–108648 | hypothetical protein | ||
| SHP0109 | 109396–109046 | hypothetical protein | ||
| SHP0110 | 110695–109961 | cyclopentanol dehydrogenase | ||
| SHP0111 | 112423–110777 | ccr gene complex | ccrB | cassette chromosome recombinase B |
| SHP0112 | 113793–112459 | ccrA | cassette chromosome recombinase A | |
| SHP0113 | 114595–113981 | J1 | cch | cassette chromosome helicase |
| SHP0114 | 114682–114957 | ΨccrA | cassette chromosome recombinase A | |
| SHP0115 | 115428–115069 | zinc/iron permease | ||
| SHP0116 | 115635–115961 | HTH-type transcriptional repressor CzrA | ||
| SHP0117 | 116291–118216 | cadmium-translocating P-type ATPase | ||
| SHP0118 | 119212–118538 | IS431 | transposase for IS431mec |
gene position in the contig (accession number: KF006347).
ΨSCCmec(SH32) is conserved in S. haemolyticus
ΨSCCmec(SH32) is about 28 kb in length (as determined by the size of the DNA region between DR1 and DR2), and contains three regions, a mec gene complex, a J3 region and a J2R region. The mec gene complex belongs to the C1 class with the common structure IS431-mecA-ΔmecR1-IS431, with two copies of IS431 arranged in the same direction. J2R, the region between the mec gene complex and DR2, is about 10 kb in length and encodes two heavy metal resistance genes/gene clusters, copA and arsCBRAD, conferring resistance to copper and arsenic, respectively. Interestingly, the J3 region (extending from IRR to the mec gene complex) harbors another cluster of arsenic resistance genes (arsCBR). The corresponding copies of the arsC, arsB and arsR genes in J2R and J3 share 84%, 86% and 63% amino acid identity, respectively. However, homologs of arsCBR and arsCBRAD with higher similarity were found in S. epidermidis and S. haemolyticus isolates, indicating that these two ars clusters in S. haemolyticus SH32 were not generated by duplication and divergence, but are derived from different ancestors. Finally, the J3 region contains additional cadmium resistance genes, cadD and cadX.
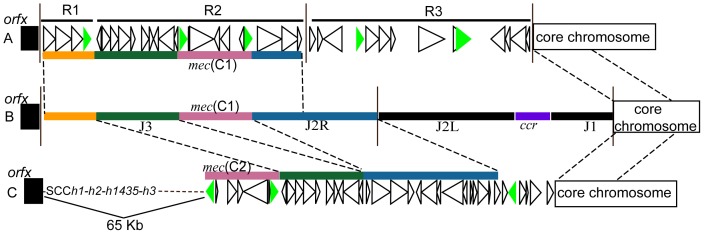
ΨSCCmec(SH32) is largely identical to ΨSCCWCH1 (Figure 2). However, ΨSCCWCH1 harbors an accessory region downstream of DR2 (ca. 17 kb, designated R3), which contains two IS elements, IS431 and ISSha1, as well as several genes encoding putative bacterial virulence factors, such as the proline permease PutP and the ion transporter FeoB [16], [17]. In contrast, ΨSCCmec(SH32) contains an accessory 6-kb insertion between the lip gene (SHP0088, encoding a lipoprotein) and DR1-2 (designated J2R-6k), which mainly encodes the ars gene cluster and is conserved in S. haemolyticus JCSC1435 (SH0089-98). Given the different positions of the R3 regions in ΨSCCWCH1 (lip-ISS-insertion) and the J2R-6k in ΨSCCmec(SH32) (lip-insertion-ISS), it is apparent that they were acquired by independent events. Zong et al. proposed that the R3 region resulted from homologous recombination, as no DR could be identified in the junction region [18]. However, considering that the J2R-6k region is well conserved in the isolates SH32 and JCSC 1435, this is most probably a common feature of a hypothetical ancestral S. haemolyticus clone that was lost in the isolate WCH1.
Figure 2. Comparison of SCCmec elements in three Staphylococcus haemolyticus isolates.
A: WCH1; B: SH32; C: JCSC 1435. mec(C1/C2): class C1/C2 mec complex. Rectangles with the same colors denote homologous regions. Dashed lines indicate their ends. Vertical solid lines indicate the site of integration site sequences. Green triangles denote insertion sequences.
Comparative analysis of SCCSH32
SCCSH32, bracketed by DR2 and DR3, is about 20 kb in length and divided into three regions, namely a ccr gene complex and the J2L and J1 regions. The recombinase genes in SCCSH32 are of the types ccrA5 and ccrB3 and are 1,335 bp and 1,647 bp in length, respectively. They are identical to those of S. haemolyticus H9, which were originally reported as ccrB SHP and ccrA SHP [9]. A ccrA5B3 complex has also been identified in another CoNS, S. cohnii WC28 (accession number GU370073), and the coagulase-positive S. pseudintermedius KM241 (accession number AM904731, originally reported as ccrA5B5) [19], [20]. The ccrA5 gene of SCCSH32 shares 89.7% and 91.5% nucleotide identity, and the ccrB3 gene 87.7% and 86.4%, with the corresponding genes of the SCCmec elements of S. cohnii WC28 and S. pseudintermedius KM241, respectively. Using a similarity search in the non-redundant NCBI nucleotide database, we also found highly similar (>99% identity) ccr gene complexes in four S. aureus isolates (accession numbers: HF569102, HF569097, HF569093 and GU066221).
J1, the region extending from the ccr gene complex to IRL1, is about 5.5 kb in length and contains six genes (SHP0113-118), including a truncated ccrA gene and genes encoding a cassette chromosome helicase and two ion permeases. Comparative analysis revealed that the J1 region is almost identical to those found in SCCpbp4 of S. epidermidis ATCC 12228 (accession number BK001539) and SCC elements in S. aureus isolates M1 (accession number HM030720, type IV) and BK20781 (accession number FJ670542, type VIII). The J2L region in SCCSH32, i.e. the region between the ccr gene complex and DR2, is about 10 kb in length. In contrast to type I restriction-modification (R-M) systems, such as hsdR, hsdS and hsdM, identified in most known SCCmec elements, a type III R-M system was found in SCCSH32 J2L. Interestingly, this R-M system is almost identical to an R-M system found in S. epidermidis (accession number NZ_AKGM01000027). In addition, we identified several hypothetical proteins in J2L, for example SHP0106-109, homologs of which are found in S. haemolyticus JCSC 1435 and isolates from other staphylococcal species like S. epidermidis and S. aureus. SHP0107 and SHP0108 are conserved in both species with an identity value of >92%, but SHP0106 and SHP0109 show higher similarity (98% and 97% identity values, respectively) to their counterparts in S. aureus or S. epidermidis than S. haemolyticus JCSC1435 (86% and 55%, respectively).
The ccrA5B3 gene complex has been identified in several staphylococcal species, both coagulase-negative and coagulase-positive staphylococci, with varied identity, revealing that it was exchanged between staphylococcal species during evolution. Remarkably, although ccr genes encoded by SCCSH32 are not found in S. epidermidis, several S. epidermidis isolates contain regions that are highly homologous to the two junction regions in SCCSH32, J1 and J2L, which include the type III R-M system. R-M systems are responsible for limiting the uptake of foreign DNA in bacteria; and both types I and III R-M systems have been identified as major barriers to lateral gene transfer in S. aureus [21]–[23]. Therefore, the apparent frequent gene transfer between S. haemolyticus and S. epidermidis in SCC regions may be ascribed to the fact that they have similar R-M systems.
Detection of ccrA5B3 in methicillin-resistant S. haemolyticus
We collected a total of 88 methicillin-resistant S. haemolyticus (MRSH) isolates in previous work [9], including 42 isolates that are positive for the arcA gene of the arginine catabolic mobile element (ACME) [24]_ENREF_10. By using PCR amplification and dot blotting, class C1 mec gene complexes and the ccrA5B3 gene were detected in eight of these 42 isolates, including the isolate SH32 [9]. In this work, we investigated the other 46 ACME-arcA negative isolates and found that another two isolates harbored ccrA5B3 gene complexes. Interestingly, both of them also contained class C1 mec gene complexes, suggesting that these isolates diverged after the assembly of class C1 mec and ccrA5B3 gene complexes.
Concluding remarks
More detailed knowledge of CoNS SCCmec elements is of special importance, as they are believed to serve as reservoirs for SCCmec elements transferred to S. aureus [25]. In this study we characterized the entire structure of a SCC composite island in S. haemolyticus, which we found is composed of two SCC remnants. Results from a previous study suggested the presence of a UT9-type SCCmec element in S. haemolyticus that encodes both a class C1 mec and a ccrA5B3 gene complex [6]. Notably, we here found that these two gene complexes are present in two separate SCC remnants, and are not assembled in one SCCmec element. Although the class C1 mec complex and ccrA5B3 genes have been identified in several other S. haemolyticus isolates, our results indicate that confirming the existence of a UT9 SCCmec element solely by PCR assays is problematic, necessitating the analysis of further genome sequences.
Funding Statement
This work was supported by the National Natural Science Foundation of China (no. NSFC81201327), the Ministry of Health of the People's Republic of China (no. 201002021), the Science Technology Department of Zhejiang Province (no. 2008C13029-1), and the Intramural Research Program of the National Institute of Allergy and Infectious Diseases (NIAID), U.S. National Institutes of Health (NIH). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Chambers HF, Deleo FR (2009) Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat Rev Microbiol 7: 629–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Malachowa N, DeLeo FR (2010) Mobile genetic elements of Staphylococcus aureus . Cell Mol Life Sci 67: 3057–3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shore AC, Coleman DC (2013) Staphylococcal cassette chromosome mec: Recent advances and new insights. Int J Med Microbiol: in press. [DOI] [PubMed]
- 4. Shore AC, Brennan OM, Deasy EC, Rossney AS, Kinnevey PM, et al. (2012) DNA microarray profiling of a diverse collection of nosocomial methicillin-resistant staphylococcus aureus isolates assigns the majority to the correct sequence type and staphylococcal cassette chromosome mec (SCCmec) type and results in the subsequent identification and characterization of novel SCCmec-SCCM1 composite islands. Antimicrob Agents Chemother 56: 5340–5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Garza-Gonzalez E, Morfin-Otero R, Llaca-Diaz JM, Rodriguez-Noriega E (2010) Staphylococcal cassette chromosome mec (SCCmec) in methicillin-resistant coagulase-negative staphylococci. A review and the experience in a tertiary-care setting. Epidemiol Infect 138: 645–654. [DOI] [PubMed] [Google Scholar]
- 6. Zong Z, Peng C, Lu X (2011) Diversity of SCCmec elements in methicillin-resistant coagulase-negative staphylococci clinical isolates. PLoS One 6: e20191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ruppe E, Barbier F, Mesli Y, Maiga A, Cojocaru R, et al. (2009) Diversity of staphylococcal cassette chromosome mec structures in methicillin-resistant Staphylococcus epidermidis and Staphylococcus haemolyticus strains among outpatients from four countries. Antimicrob Agents Chemother 53: 442–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Otto M (2013) Coagulase-negative staphylococci as reservoirs of genes facilitating MRSA infection: Staphylococcal commensal species such as Staphylococcus epidermidis are being recognized as important sources of genes promoting MRSA colonization and virulence. Bioessays 35: 4–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pi B, Yu M, Chen Y, Yu Y, Li L (2009) Distribution of the ACME-arcA gene among meticillin-resistant Staphylococcus haemolyticus and identification of a novel ccr allotype in ACME-arcA-positive isolates. J Med Microbiol 58: 731–736. [DOI] [PubMed] [Google Scholar]
- 10. Zerbino DR, Birney E (2008) Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18: 821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lukashin AV, Borodovsky M (1998) GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res 26: 1107–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Takeuchi F, Watanabe S, Baba T, Yuzawa H, Ito T, et al. (2005) Whole-genome sequencing of Staphylococcus haemolyticus uncovers the extreme plasticity of its genome and the evolution of human-colonizing staphylococcal species. J Bacteriol 187: 7292–7308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mongkolrattanothai K, Boyle S, Murphy TV, Daum RS (2004) Novel non-mecA-containing staphylococcal chromosomal cassette composite island containing pbp4 and tagF genes in a commensal staphylococcal species: a possible reservoir for antibiotic resistance islands in Staphylococcus aureus . Antimicrob Agents Chemother 48: 1823–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. International Working Group on the Classification of Staphylococcal Cassette Chromosome E (2009) Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob Agents Chemother 53: 4961–4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bayer AS, Coulter SN, Stover CK, Schwan WR (1999) Impact of the high-affinity proline permease gene (putP) on the virulence of Staphylococcus aureus in experimental endocarditis. Infect Immun 67: 740–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Aranda J, Cortes P, Garrido ME, Fittipaldi N, Llagostera M, et al. (2009) Contribution of the FeoB transporter to Streptococcus suis virulence. Int Microbiol 12: 137–143. [PubMed] [Google Scholar]
- 18. Zong Z (2013) Characterization of a complex context containing mecA but lacking genes encoding cassette chromosome recombinases in Staphylococcus haemolyticus . BMC Microbiol 13: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Descloux S, Rossano A, Perreten V (2008) Characterization of new staphylococcal cassette chromosome mec (SCCmec) and topoisomerase genes in fluoroquinolone- and methicillin-resistant Staphylococcus pseudintermedius . J Clin Microbiol 46: 1818–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zong Z, Lu X (2010) Characterization of a new SCCmec element in Staphylococcus cohnii . PLoS One 5: e14016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vasu K, Nagaraja V (2013) Diverse functions of restriction-modification systems in addition to cellular defense. Microbiol Mol Biol Rev 77: 53–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Waldron DE, Lindsay JA (2006) Sau1: a novel lineage-specific type I restriction-modification system that blocks horizontal gene transfer into Staphylococcus aureus and between S. aureus isolates of different lineages. J Bacteriol 188: 5578–5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Corvaglia AR, Francois P, Hernandez D, Perron K, Linder P, et al. (2010) A type III-like restriction endonuclease functions as a major barrier to horizontal gene transfer in clinical Staphylococcus aureus strains. Proc Natl Acad Sci U S A 107: 11954–11958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Diep BA, Gill SR, Chang RF, Phan TH, Chen JH, et al. (2006) Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus . Lancet 367: 731–739. [DOI] [PubMed] [Google Scholar]
- 25. Berglund C, Soderquist B (2008) The origin of a methicillin-resistant Staphylococcus aureus isolate at a neonatal ward in Sweden-possible horizontal transfer of a staphylococcal cassette chromosome mec between methicillin-resistant Staphylococcus haemolyticus and Staphylococcus aureus . Clin Microbiol Infect 14: 1048–1056. [DOI] [PubMed] [Google Scholar]