Abstract
Deregulated cell proliferation and apoptosis play a major role in hepatocellular carcinoma (HCC). MicroRNAs participate in the modulation of key molecules linked to hepatocarcinogenesis.
Purpose
This study aims to investigate the role of miR-221 in the modulation of Bmf, a proapoptotic BH3-only protein, and to characterize miR-221 contribution to hepatocarcinogenesis through modulation of apoptosis.
Experimental Design
Transfection of miR-221 and anti-miR-221 in HCC-derived cell lines and luciferase reporter assay were used to assess Bmf as a target of miR-221. Modulation of miR-221 and Bmf expression contributed to characterize their role in anoikis. Primary HCC tissues were analyzed to assess the clinical relevance of in vitro findings.
Results
Enforced miR-221 expression caused Bmf down-regulation, whereas anti-miR-221 induced its up-regulation. A luciferase reporter assay confirmed Bmf as a target of miR-221. Following matrix detachment, miR-221 silencing led to increased apoptotic cell death. The analysis of HCC tissues revealed an inverse correlation between miR-221 and Bmf expression and a direct correlation between Bmf and activated caspase-3, as a marker of apoptosis. High miR-221 levels were associated with tumor multifocality and reduced time to recurrence after surgery.
Conclusions
Our results indicate that miR-221, by targeting Bmf, inhibits apoptosis. Moreover, in HCC, miR-221 overexpression is associated with a more aggressive phenotype. These findings, together with the previously reported modulation of CDKN1B/ p27 and CDKN1C/p57, show that miR-221 simultaneously affects multiple pro-oncogenic pathways and suggest miR-221 as a potential target for nonconventional treatment against HCC.
Hepatocellular carcinoma (HCC) is one of the most common cancers worldwide, with an increasing trend in incidence (1). HCC results from the deregulation of multiple signaling pathways. Initial steps involve the disruption of a set of interdependent pathways controlling cell growth and apoptosis. At later stages, cells may acquire angiogenic, invasive, and metastatic properties in a process that involves the interactions of neoplastic cells with the surrounding microenvironment.
Among oncogenic factors in HCC, microRNAs (miRNA) participate in several carcinogenic mechanisms (2). We and other groups have reported previously the altered expression of miR-NAs in human HCC (3–14). miRNAs are short (19–25 nucleotides) RNA sequences able to modulate the expression of a wide range of target genes by pairing homologous sequences within 3′-untranslated region (3′-UTR) of mRNAs, thus preventing or impairing their translation or promoting RNA degradation. Among miRNAs deregulated in HCC, miR-221 is of particular interest, because it was reported to be up-regulated also in other tumor types, including glioblastoma, urinary bladder cancer, papillary tumors of the thyroid, pancreatic cancer, and prostate carcinoma cell lines (3, 4, 8, 9, 14–21). In addition, overexpression of miR-221 was shown to promote cancer cell proliferation by its ability to inhibit the expression of the cyclin-dependent kinase inhibitors CDKN1B/p27 (3, 8, 14, 21, 22) and CDKN1C/p57 (9, 14), which are important controllers of cell cycle progression, the down-regulation of which has been associated with a poor prognosis in HCC patients (23–25).
Molecular classification of HCC is still not defined; notwithstanding genes driving unregulated cell proliferation play a major role in the process of hepatocarcinogenesis (26). The balance between proliferating and proapoptotic signals has been extensively studied in liver diseases, with apoptosis triggered by Fas(CD95)/Fas ligand and Bcl-2 protein family playing a major role (27–29). Concerning the Bcl-2 family proteins, Bcl-2 expression was not found to affect prognosis following surgical resection of HCC (30); conversely, overexpression of the antiapoptotic gene Bcl-xL (31) independently predicts a decreased overall and disease-free survival (32). Furthermore, down-regulation of the proapoptotic genes bax, bcl-xS, and bid were observed in definite subgroups of HCCs (33, 34). Nevertheless, concerning HCC development, little is known on the transcriptional regulation of the >20 proapoptotic and antiapoptotic members of the Bcl-2 family. Very recently, an up-regulation of Bmf and Bim, two BH3-only members of the Bcl-2 family, have been reported during transforming growth factor-β (TGF-β)–induced apoptosis (35), a pathway directly involved in the progression of chronic liver disease and in the development of HCC (36, 37). Bmf belongs to the Bcl-2 family, which is composed of prosurvival members (Bcl-2, Bcl-xL, Bcl-w, Mcl-1, and A1) and proapoptotic members, including the Bax group bearing three Bcl-2 homology domains and the BH3-only proteins sharing only the BH3 interaction domain (Bmf, Bim, Bad, Bid, Bik, Puma, Noxa, and Hrk). BH3-only proteins monitor cellular well-being and, when activated by stress signals, engage prosurvival Bcl-2-like proteins and inactivate their function, thus promoting apoptosis. BH3-only members play key roles in development, tissue homeostasis, immunity, and tumor suppression, and compounds mimicking them are promising anti-cancer agents (38). Bmf protein is normally sequestered by the myosin V motor complex with dynein light chain 2, which may be important for sensing intracellular damage and triggering apoptosis. Following stress challenges, such as anoikis or exposure to UV irradiation, Bmf is released, together with dynein light chain 2, from the myosin V motor complex, thus allowing its binding to prosurvival Bcl-2 proteins (39). Bmf protein has been detected in many mouse organs, with abundant levels in pancreas, liver, kidney, and hematopoietic tissues. No data are available concerning Bmf expression in human HCC.
This study was undertaken to assay Bmf as a possible target of miR-221 and to further characterize miR-221 contribution to hepatocarcinogenesis.
Materials and Methods
Patients and HCC samples
Matched HCC and cirrhotic tissue were obtained from 51 consecutive patients (40 males and 11 females; median age 68 years, range 49–82) undergoing liver resection for HCC at the Department of Surgery of the University of Bologna from January 2002 to December 2006, after obtaining their informed consent. Tissue samples were collected at surgery and divided in two parts: the first one was immediately frozen in liquid nitrogen and stored at −80°C until RNA and protein extraction and the second one was fixed in 10% formalin and paraffin-embedded for histopathology examination. Histopathologic grading was scored according to Edmondson and Steiner’s criteria (40). Exclusion criteria were a previous history of local or systemic treatments for HCC and the presence of noncirrhotic tissue surrounding the HCC nodule(s). The characteristics of HCC patients included in this study are described in Table 1.
Table 1.
Characteristics of HCC patients analyzed in this study
| Serial no. | Gender | Age (y) | Cause of liver disease* | Focality† | Size‡ | AFP (ng/mL)§ | Grading|| | Bmf¶ |
|---|---|---|---|---|---|---|---|---|
| 1 | M | 65 | HBV + ethanol | Uni | 3 | 5 | G2 | X |
| 2 | M | 52 | None | Uni | 5 | 5 | G2 | |
| 3 | M | 70 | HCV | Uni | 4 | 35 | G3 | X |
| 4 | F | 65 | HCV | Uni | 3 | 3 | G3 | X |
| 5 | F | 62 | HCV | Uni | 3 | 76 | G3 | X |
| 6 | M | 67 | HCV | Uni | 6 | 3,352 | G3 | |
| 7 | M | 77 | HBV-Ab | Uni | 5 | 4 | G3 | X |
| 8 | M | 70 | Ethanol | Uni | 5 | 537 | G3 | X |
| 9 | M | 65 | HCV + ethanol | Uni | 3 | 20 | G3 | X |
| 10 | F | 80 | HCV | Uni | 5 | 76 | G3 | X |
| 11 | F | 59 | HCV | Uni | 6 | 7 | G4 | |
| 12 | F | 66 | HCV | Uni | 8 | 9,572 | G3 | |
| 13 | F | 68 | None | Uni | 5.5 | 2 | G3 | X |
| 14 | M | 59 | HCV | Uni | 5 | 86 | G3 | X |
| 15 | F | 82 | HCV | Uni | 7.5 | 964 | G3 | X |
| 16 | M | 60 | Ethanol | Uni | 2 | 156 | G2 | |
| 17 | M | 78 | HCV | Uni | 3 | 9 | G2 | X |
| 18 | M | 54 | HBV | Uni | 4 | 162 | G2 | X |
| 19 | M | 69 | HCV | Uni | 10 | 390 | G3 | |
| 20 | M | 74 | None | Uni | 11 | 78 | G3 | X |
| 21 | M | 73 | HCV | Uni | 4.5 | 2 | G3 | X |
| 22 | M | 77 | HCV | Uni | 2.2 | 48 | G2 | |
| 23 | F | 71 | HCV | Uni | 2 | 276 | G3 | X |
| 24 | M | 68 | HCV | Multi | 2.5 | 5 | G2 | X |
| 25 | M | 81 | HCV | Multi | 3 | 8 | G3 | |
| 26 | M | 74 | HCV | Multi | 3.5 | 2,198 | G3 | X |
| 27 | M | 74 | HCV + HBV | Multi | 3 | 75 | G4 | X |
| 28 | M | 82 | None | Multi | 5 | 4 | G3 | |
| 29 | M | 51 | HBV + HCV | Multi | 7 | 1,924 | G3 | X |
| 30 | M | 79 | HCV | Multi | 10 | 7 | G3 | X |
| 31 | M | 70 | HCV | Multi | 2.3 | 46 | G3 | |
| 32 | M | 72 | HCV | Multi | 3.4 | 18 | G3 | |
| 33 | M | 70 | None | Multi | 10 | 11 | G2 | |
| 34 | M | 75 | HCV | Multi | 7 | 9 | G3 | |
| 35 | M | 59 | HCV | Multi | 3 | 76 | G3 | X |
| 36 | M | 67 | None | Multi | 6 | 18,766 | G3 | |
| 37 | M | 65 | HCV | Multi | 6.5 | 167 | G3 | X |
| 38 | M | 76 | HBV-Ab | Multi | 5 | 10,000 | G4 | |
| 39 | M | 49 | HBV | Multi | 4 | 2,708 | G4 | |
| 40 | F | 53 | HCV | Multi | 4 | 60 | G3 | |
| 41 | F | 62 | HCV | Multi | 3 | 468 | G2 | |
| 42 | M | 79 | HBV | Multi | 7 | 540 | G4 | |
| 43 | M | 68 | HCV | Multi | 8 | 86 | G3 | |
| 44 | M | 65 | HCV | Multi | 7 | 139,313 | G4 | |
| 45 | M | 64 | HCV + ethanol | Multi | 6.5 | 8 | G3 | |
| 46 | M | 60 | HCV | Multi | 3.2 | 15 | G2 | |
| 47 | M | 60 | HCV | Multi | 1.3 | 257 | G4 | X |
| 48 | M | 58 | HCV | Multi | 3 | 132 | G3 | |
| 49 | M | 66 | HCV | Multi | 3 | 58 | G2 | X |
| 50 | M | 75 | HCV | Multi | 3 | 63 | G3 | X |
| 51 | M | 68 | HCV | Multi | 6.5 | 131 | G3 | X |
Cause of underlying liver disease: HBV, hepatitis B virus; HCV, hepatitis C virus; ethanol, history of ethanol abuse; HBV-Ab, presence of the antibodies against HBV; none, negative history for hepatitis virus infection and ethanol abuse.
Focality: Unifocality (Uni) or Multifocality (Multi) was assessed based on imaging techniques previous to surgery and by means of intraoperative ultrasound.
Size of the HCC nodule (in cm) used for RNA and protein extraction.
AFP: α-fetoprotein determination was made before surgery.
Grading of the HCC was assessed according to Edmondson and Steiner’s criteria.
X: Cases analyzed for Bmf expression.
Real-time reverse transcription-PCR analysis of miR-221
The expression of mature hsa-mir-221 and of U6 RNA, as housekeeping gene, was assayed using the TaqMan miRNA Assays (Applied Biosystems) as described previously (4). Reverse transcription reaction was done starting from 10 ng total RNA and using the looped primers. Real-time PCR was done in triplicate for each case using the standard TaqMan miRNA assays protocol on the iCycler iQ Real-time PCR Detection System as described by the manufacturer (Bio-Rad). miRNA expression was measured using Ct (threshold cycle). The ΔΔCt method for relative quantitation of gene expression was used to determine miRNA expression levels. The ΔCt was calculated by subtracting the Ct of U6 RNA from the Ct of the miRNA of interest. The ΔΔCt was calculated by subtracting the ΔCt of the reference sample (normal liver) from the ΔCt of each sample. Fold change was generated using the equation 2−ΔΔCt. A pool of three normal livers was used for the standard curve calculation and as reference sample for the ΔΔCt.
miRNA target prediction
The analysis of miR-221 predicted targets was determined using the algorithms TargetScan,6 PicTar,7 and miRanda.8 To identify the genes commonly predicted by the three different algorithms, miRGen9 was used.
Cell cultures
HCC-derived cell lines SNU449 (ATCC CRL-2234) and SNU398 (ATCC CRL-2233) were cultured with RPMI 1640, whereas Hep3B (ATCC HB-8064) and HepG2 (ATCC HB-8065) cell lines were cultured with MEM; both media were supplemented with 10% fetal bovine serum and antibiotics. Cells were transfected in 6-well plates with 100 pmol of stability-enhanced miR-221 precursor, anti-miR-221, or negative control #1. Stability-enhanced miR-221 precursor, anti-miR-221, and negative control #1 ribo-oligonucleotides were from Ambion. The day before transfection, cells were seeded in antibiotic-free medium; transfection of miRNAs was carried out using Lipofectamine 2000 in accordance with manufacturer’s procedure (Invitrogen). After transfection, cells were cultured for 72 h and intermediate samples at 24 and 48 h were collected and analyzed by Western blot. Each transfection experiment was done in triplicate. Because Bmf was reported to be up-regulated by anoikis (41), plates were coated with polyhema (Sigma), a synthetic polymer that prevents cell attachment, for 16 h at 37°C. SNU449 and Hep3B cells were transfected with anti-miR-221 and negative control #1 inhibitor miRNAs for 24 h as described above and then transferred into polyhema-coated plates to evaluate miR-221 effect. Cells were collected at 12, 24, and 48 h after cell suspension. Dead cells were detected by trypan blue staining and protein extracts were used to assay Bmf expression and caspase-3 activation.
The silencing of Bmf gene was obtained by using 20 nmol/L of a pool of three siRNAs against Bmf (TriFECTa Dicer-Substrate RNAi Kit, HSC.RNAI.N001003940.3; IDT).
Luciferase activity assay
A 301-bp fragment of the human BMF 3′-UTR (spanning from nucleotides 4,372 to 3,773) was amplified by PCR using primers BMF-3′UTR-F (5′-ATACTAGTTGGTGGGGACTT-TTGAGTCT-3′) and BMF-3′ UTR-R (5′-ATAAGCTTGCCCC-TTTCTTCTTCCTCTC-3′) and was cloned downstream of the firefly luciferase gene present in the pMIR-REPORT vector (Ambion) to develop the pMIR-BMF vector. For luciferase assay, Hep3B cells were cultured in 24-well plates and each transfected with 0.1 μg of either pMIR-BMF or pMIR-REPORT together with 0.01 μg pRL-TK vector (Promega), which contains the Renilla luciferase gene, and 30 pmol miR-221, anti-miR-221, or negative control #2. Transfection was done using Lipofectamine 2000 and Opti-MEM I reduced serum medium (Life Technologies) in a final volume of 0.6 mL. At 24 h after transfection, firefly and Renilla luciferase activity was measured using the Dual-Luciferase Reporter Assay (Promega). Each transfection was repeated twice in triplicate. As a further control, pMIR-BMF was mutated in the first three nucleotides of the seed-match sequence for miR-221 (mutagenesis and complete sequencing were done by Genescript) and cotransfected with miR-221 and anti-miR-221 as described above.
Western blot analysis
Twenty-seven matched HCC and cirrhotic tissues, selected based on tissue availability, were subjected to mechanical pulverization while in dry ice and then dissolved by repeated syringing in lysis buffer [10 mmol/L Tris-HCl (pH 7.4), 2.5 mmol/L MgCl2, 0.5% Triton X-100, 1 mmol/L DTT, and protease inhibitors]. Homogenates were then centrifuged at 14,000 × g and supernatants were collected. Frozen pellets from miRNA-transfected cells were resuspended in lysis buffer and protein extraction was carried out as described previously (4). A rabbit polyclonal antibody against Bmf (Novus Biologicals) was diluted 1:200 and incubated for 16 h at 4°C. A horseradish-conjugated secondary antibody (labeled polymer-horseradish peroxidase anti-rabbit, Envision system; DAKO Cytomation) diluted 1:200 was incubated for 45 min at room temperature and the corresponding band was revealed using the enhanced chemiluminescence method (Amersham). Digital images of autoradiography were acquired with Fluor-S Multi-Imager (Bio-Rad) and band signals were acquired in the linear range of the scanner using a specific densitometric software (Quantity-One; Bio-Rad). After autoradiography acquisition, the membranes were stripped and reprobed for 2 h at room temperature with anti-β-actin antibody (clone AC-40; Sigma). Ratio between intensities of Bmf and β-actin bands was used to normalize Bmf expression in each sample.
To evaluate apoptosis in HCC-derived cell lines and in HCC tissues, the cleaved fraction of caspase-3 was assessed on protein extracts by Western blot. Membranes were incubated at 4°C for 16 h with the polyclonal antibody anti-cleaved caspase-3 (Asp175; Cell Signaling) diluted 1:200. Normalization and assessment of intensities of protein signals was carried out as described above.
Semiquantitative reverse transcription-PCR
Semiquantitative reverse transcription-PCR was done to assess Bmf mRNA expression by using the following sense and antisense primers: BMF-F (540 bp) 5′-ATGGAGCCATCTCAGTGTGTG-3′ and BMF-R 5′-CCCCGTTCCT-GTTCTCTTCT-3′. β-Actin mRNA expression was chosen as an housekeeping gene and a 275-bp sequence was amplified by using the following primers: 5′-CAAGAGATGGCCACGGCTGCT-3′ and 5′-TCCTTCTGCATCCTGTCGGCA-3′. PCR was carried out in the exponential range of amplification with the following conditions: BMF 30 cycles of 30 s at 94°C, 30 s at 56.5°C, and 45 s at 72°C and β-actin 25 cycles of 30 s at 94°C, 30 s at 55°C, and 30 s at 72°C.
Statistical analysis
Differences between groups were analyzed using a double-sided Student’s t test, when two groups were present. Experimental data are expressed as mean ± SE of three separate experiments done in triplicate. Pearson’s correlation was used to explore the relationships between miR-221 and Bmf and between Bmf and caspase-3 expression in HCC protein extracts. Pearson’s correlation was also used to assay any relationship between miR-221 or Bmf expression and α-fetoprotein serum levels and tumor size. t test was used to explore any significant difference in miR-221 expression between low-grade (Edmondson and Steiner’s grades 1 and 2) and high-grade (Edmondson and Steiner’s grades 2 and 3) HCCs. ANOVA was used to explore any difference in miR-221 expression among different etiologic groups. Kaplan-Meier survival analysis was used to compare patient survival and time to recurrence based on different miR-221 and Bmf expression levels (cutoff values were chosen based on the median values) and statistical P value was generated by the Cox-Mantel log-rank test. Survival analysis was done considering only cancer-related deaths, whereas events related with other causes, including liver failure, were excluded. P values < 0.05 were considered statistically significant. Statistical analysis were done using SPSS version 8.0.
Results
Bmf is a target of miR-221
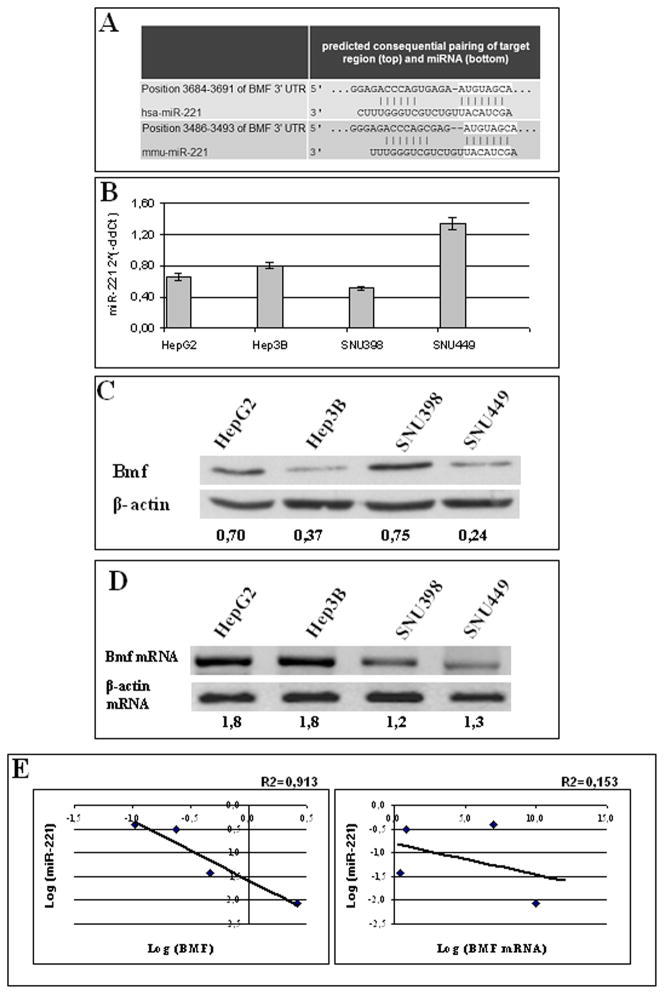
All the three prediction algorithms (miRanda, TargetScan, and PicTar) used in the bioinformatic analysis of miR-221 target genes identified Bmf, a proapoptotic BH3-only member of the Bcl-2 family, among the hypothetical targets of miR-221 (Fig. 1A). The pairing site of miR-221 with Bmf is highly conserved and can also be found in the 3′-UTR of the bmf gene of mouse, rat, dog, and chicken. A further hint of the potential role of miR-221 in the regulation of Bmf came from the analysis of four HCC-derived cell lines (HepG2, Hep3B, SNU398, and SNU449), which exhibited an inverse correlation between Bmf protein and miR-221 levels (Fig. 1; Supplementary Fig. S1), whereas Bmf mRNA did not show any correlation neither with Bmf protein (Fig. 1) nor with miR-221 levels (Supplementary Fig. S1).
Fig. 1.
Pairing site between miR-221 and BMF 3′-UTR. Analysis of HCC-derived cell lines. A, TargetScan predicted pairing of miR-221 to the 3′-UTR of the human BMF gene. The pairing site is highly conserved and can be found in the 3′-UTR of the BMF gene of mouse (shown in the figure), rat, dog, and chicken. miR-222 (not shown in the figure) shares with miR-221 the same predicted target pairing. B, real-time reverse transcription-PCR analysis of miR-221 expression in four HCC-derived cell lines (HepG2, Hep3B, SNU398, and SNU449). Each determination was done in triplicate. Mean ± SE. C, Western blot assay of Bmf protein expression in HCC-derived cell lines normalized based on β-actin expression. D, Bmf mRNA expression in HCC-derived cell lines assayed by reverse transcription-PCR.
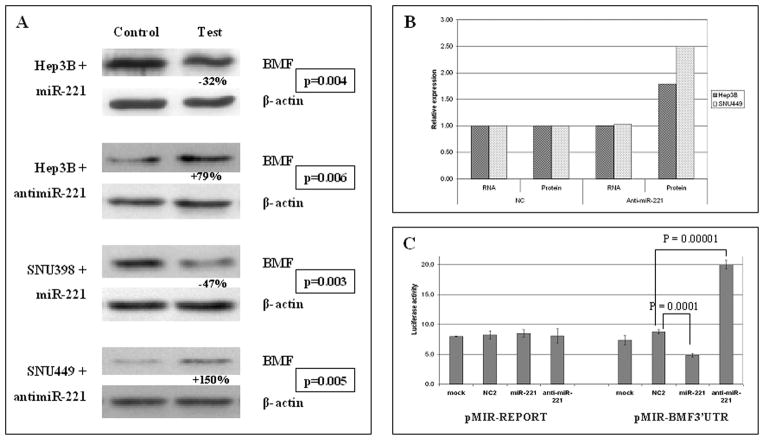
To directly verify if miR-221 is involved in Bmf regulation, we analyzed the expression of Bmf protein in response to enforced expression of miR-221 or anti-miR-221 in HCC-derived cell lines. SNU398 cells were chosen for transfection with miR-221 because they display the lowest miR-221 expression and the highest Bmf levels among the cell lines (Fig. 1B and C). Transfection of miR-221 in SNU398 cells resulted in a 47% decrease of Bmf levels when compared with cells transfected with negative control (P = 0.003, t test; Fig. 2A). SNU449 cell line, which expresses the highest levels of miR-221 and the lowest of Bmf protein, was chosen for transfection of anti-miR-221, which caused an increase of Bmf protein levels of 150% with respect to negative controls (P = 0.005, t test; Fig. 2A). As a final confirmation, Hep3B cells were transfected with either miR-221 or anti-miR-221 because they exhibit intermediate levels of miR-221 associated with low Bmf protein expression. After miR-221 transfection, Hep3B cells exhibited a 32% down-regulation of Bmf protein when compared with cells transfected with negative control (P = 0.004, t test), whereas anti-miR-221 transfection caused a 79% increase in Bmf expression levels when compared with negative control (P = 0.006, t test; Fig. 2A).
Fig. 2.
Bmf is a target of miR-221. A, transfection experiments in Hep3B, SNU398, and SNU449 HCC-derived cell lines. miR-221 and anti-miR-221 were transfected based on the basal expression levels of the different cell lines. Controls refer to negative control from Ambion. Enforced miR-221 expression caused a decrease of Bmf protein levels in Hep3B and SNU398 cells, whereas enforced anti-miR-221 caused an increased Bmf expression in Hep3B and SNU449 cells. Data are expressed as a mean of three replicates and P values resulting from t test are reported. B, analysis of Bmf mRNA and protein levels in Hep3B and SNU449 cells treated with anti-miR-221 and negative control. Following anti-miR-221 transfection, both cell lines do not display any variation of Bmf mRNA levels, whereas Bmf protein levels increase. This findings confirm that miR-221 controls Bmf expression at the translational level. C, luciferase assay in Hep3B cells. pMIR-BMF 3′-UTR vector was cotransfected with miR-221, anti-miR-221, or negative control. Luciferase activity displayed a significant decrease following miR-221 enforced expression and a significant increase following anti-miR-221 enforced expression when compared with cells cotransfected with negative control.
Because the level of Bmf mRNA expression did not change in response to miR-221 or anti-miR-221, our results show that the control of Bmf expression is at the translational level (Fig. 2B).
To prove a direct interaction between miR-221 and Bmf mRNA, Bmf 3′-UTR, which includes the potential target site for miR-221, was cloned downstream of the luciferase reporter gene of the pMIR-REPORT vector to generate the pMIR-BMF-3′ UTR vector. This vector was cotransfected into Hep3B cells together with miR-221, anti-miR-221, or scrambled miRNAs negative control. A Renilla luciferase vector (pRL-TK) was used to normalize differences in transfection efficiency. Luciferase activity in Hep3B cells cotransfected with pMIR-BMF-3′UTR vector showed a significant decrease with miR-221 (P = 0.0001, t test) and a significant increase with anti-miR-221 (P = 0.00001, t test) when compared with negative control (Fig. 2C). Conversely, no significant modulation of luciferase activity was observed following cotransfection of miR-221 and anti-miR-221 with a pMIR-BMF-3′UTR-M vector mutated in the first three nucleotides of the seed-match sequence.
Anti-miR-221 up-regulates Bmf and triggers apoptosis
Previous evidence reported that Bmf is involved in “anoikis,” a specific form of cell death induced by extracellular matrix detachment (41).
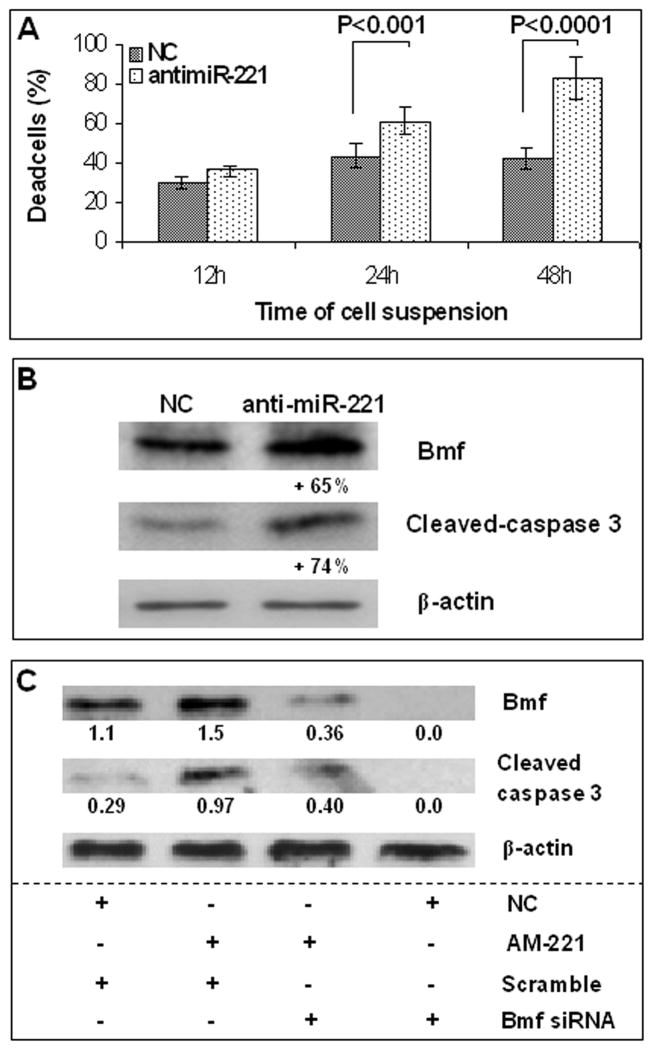
To test the role of miR-221, SNU449 cells, which exhibit the highest miR-221 and the lowest Bmf constitutive expression levels, were treated with anti-miR-221 and were grown in nonadherent conditions. Cell counting with trypan blue at 24 and 48 h after cell detachment resulted in a 1.4- and 2.0-fold increase of dead cells, respectively, in anti-miR-221 versus negative control transfected cells (P < 0.001 at 24 h and P < 0.0001 at 48 h, t test; Fig. 3A). At 24 h of nonadherent growth, anti-miR-221-transfected cells showed an increase of both Bmf (65%) and cleaved caspase-3 (74%) protein expression when compared with cells treated with negative control (Fig. 3B), in line with the activation of apoptosis. These findings suggest that Bmf promotes apoptosis of SNU449 cells grown in nonadherence, and by suppressing miR-221 activity, anti-miR-221 increases cell death and susceptibility to apoptosis.
Fig. 3.
Anti-miR-221 increases the susceptibility to apoptosis in SNU449 cells grown in nonadherent conditions. Bmf is released during nonadherent growth and triggers apoptosis. Following cell plate detachment, SNU449 cells progressively dye. Anti-miR-221 transfection in this experimental setting is associated with an increased number of dead cells (A), which is likely related with apoptosis as shown by increased Bmf and caspase-3 levels (B) at 24 h. NC, negative control. In nonadherent conditions (C), Bmf silencing (by Bmf siRNA) following anti-miR-221 (AM-221) transfection is associated with a decrease of caspase-3 activation (AM-221 + and Bmf siRNA +) when compared with anti-miR-221-transfected cells (AM-221 + and scramble +). Ratio between Bmf or cleaved caspase-3 (17 kDa isoform) and β-actin corresponding bands (absorbance).
To show that apoptotic cell death following anti-miR-221 transfection in SNU449 cells, grown in nonadherent conditions, was specifically triggered by Bmf protein, we performed Bmf silencing with a pool of siRNAs against Bmf. At 24 h following anti-miR-221 transfection, Bmf expression was silenced by siRNAs for further 24 h. Subsequently, SNU449 cells were transferred into a polyhema-coated plate for further 24 h. Bmf silencing following its up-regulation induced by anti-miR-221 in nonadherent SNU449 cells determined a decreased apoptotic cell death (as revealed by lower cleaved caspase-3 levels) when compared with negative control and anti-miR-221-treated cells. Moreover Bmf silencing prevented the cleavage of the 19 kDa caspase-3 isoform (Fig. 3C). Taken together, these data confirm the involvement of Bmf protein in apoptotic cell death in nonadherent conditions following miR-221 silencing.
miR-221 is up-regulated in HCC and inversely correlates with Bmf
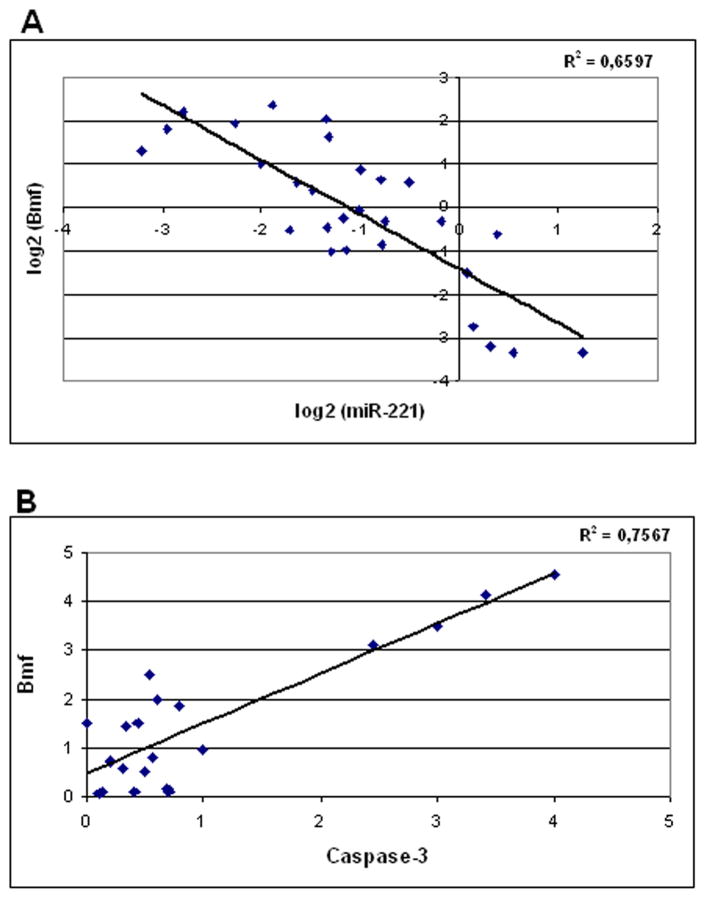
To assess the significance of miR-221/Bmf correlation in primary tumors, miR-221 expression was analyzed by real-time reverse transcription-PCR in 51 matched HCC and cirrhotic tissues and resulted to be up-regulated in 70% of HCCs versus matched cirrhosis. Twenty-seven matched HCC and cirrhotic tissues, selected based on tissue availability, were subjected to Western blot for the analysis of Bmf protein expression. miR-221 and Bmf levels of each HCC were referred to that of its matched cirrhosis. In line with a role of miR-221 in the regulation of Bmf expression, when Bmf protein levels were plotted against miR-221 expression in HCCs, an inverse correlation was found (P = 0.003, Pearson’s correlation; Fig. 4A).
Fig. 4.
Analysis of miR-221, Bmf, and activated caspase-3 expression in human HCCs. A, miR-221 (analyzed by real-time PCR) correlates with Bmf expression (analyzed by Western blot) in human HCCs (P = 0.003, Pearson’s correlation). Data are expressed in log2 form. B, Bmf protein expression directly correlates with activated caspase-3 expression in human HCC. Both molecules were assayed by Western blot (P < 0.0001, Pearson’s correlation).
To get an insight into Bmf function in HCC, we analyzed the cleaved fraction of caspase-3, as a marker of apoptosis, in the same HCC tissues assayed for Bmf protein expression. A significant correlation was found between Bmf protein expression and the cleaved fraction of caspase-3 (P < 0.0001, Pearson’s correlation; Fig. 4B), thus supporting a proapoptotic function of Bmf in vivo. Conversely, no correlation was found between miR-221 and the cleaved fraction of caspase-3.
miR-221 and Bmf correlates with tumor multifocality and time to recurrence after surgical resection of HCC
Because the deregulation of apoptosis is thought to play a crucial role in the development and progression of HCC, possible correlations between miR-221 and Bmf expression with different clinicopathologic features of HCCs were investigated. No correlation was found between miR-221 or Bmf expression and histopathologic grading, size of the tumor, viral infection, and α-fetoprotein serum levels. Conversely and significantly, higher miR-221 levels were observed in multifocal HCCs versus unifocal tumors (P = 0.009, t test). Moreover, in line with its inverse correlation with miR-221, Bmf expression was significantly lower in multifocal tumors than in unifocal HCCs (P = 0.01, t test).
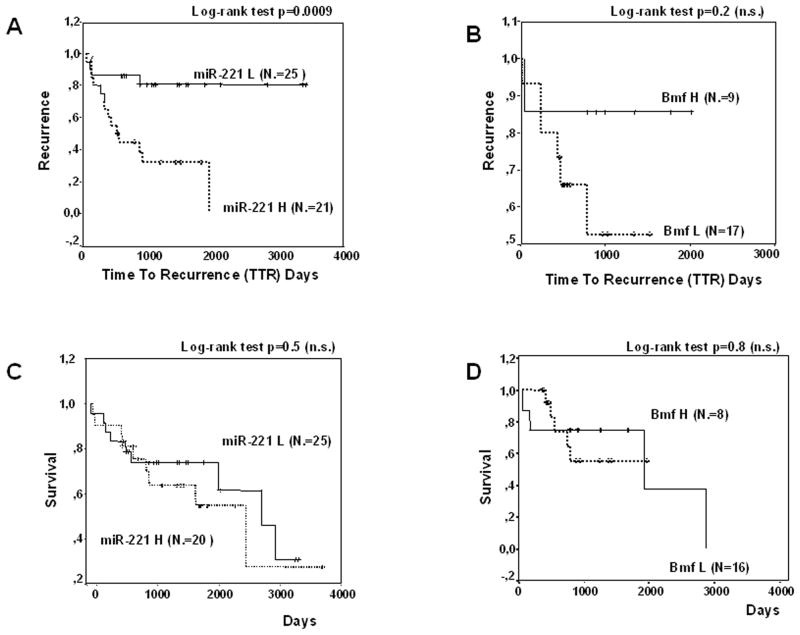
HCCs recurring within 2 years exhibited a higher miR-221 expression (P = 0.04, t test); conversely, Bmf levels were not significantly different in the two groups. When miR-221 and Bmf levels were plotted against time to recurrence after surgical resection for HCC, higher miR-221 levels (46 patients) turned out to be associated with a shorter time to recurrence (P = 0.0009, log-rank test), whereas low Bmf levels (26 HCCs) were associated with a trend toward shorter time to recurrence but did not achieve statistical significance (Fig. 5A and B) most likely due to the low number of patients. No statistically significant associations were instead found between miR-221 or Bmf levels and cancer-related death after surgery (Fig. 5C and D).
Fig. 5.
Time to recurrence and survival analysis. Kaplan-Meier survival curves exhibit higher recurrence rate in HCCs with higher miR-221 levels (A) and a trend toward a higher recurrence rate in HCC with lower Bmf expression levels (B). No correlation was instead found between miR-221 or Bmf expression and survival rate (C and D). miR-221 L, HCC with low expression of miR-221; miR-221 H, HCC with high expression of miR-221; Bmf L, HCC with low expression of Bmf; Bmf H, HCC with high expression of Bmf. Cutoff values of miR-221 and Bmf were chosen based on the respective median values.
Discussion
In this study, we have confirmed the up-regulation of miR-221 in 70% of HCCs and we have proven that Bmf is a target of miR-221. The direct molecular interaction between miR-221 and 3′-UTR of Bmf mRNA was ascertained by dual luciferase assay, whereas the modulation of Bmf protein by miR-221 and anti-miR-221 was confirmed in three HCC-derived cell lines. In agreement with these in vitro findings, an inverse correlation between miR-221 and Bmf protein levels was also recognized in HCC primary tumors. To get an insight into the mechanisms underlying Bmf function in HCC, we assessed the cleaved fraction of caspase-3 as a marker of apoptosis both in cell lines and in HCCs. The direct correlation between the cleaved fraction of caspase-3 and Bmf levels suggested that Bmf influences apoptosis in HCCs as well as in HCC-derived cell lines undergoing “anoikis.”
miR-221 was previously found to be up-regulated in several human malignancies and its oncogenic role was further supported by the identification of CDKN1B/p27/Kip1 and CDKN1C/p57/Kip2 among its targets (3, 8, 9, 14, 42), identifying cell cycle modulation as a mechanism involved in miR-221-induced hepatocarcinogenesis. Beside the down-regulation of cell cycle controlling molecules, here we describe the down-regulation of a proapoptotic molecule, Bmf, suggesting that miR-221 can simultaneously affect proliferation and apoptosis, thus acting at multiple levels in tumorigenesis.
Bmf is a well-known factor involved in the balance between proapoptotic and antiapoptotic stimuli in Bcl-2/Bcl-xL-induced apoptosis. Previous studies revealed that Bcl-2 is not generally expressed in HCC (43), whereas Bcl-xL, which exhibits an anti-apoptotic function similar to that of Bcl-2, is highly expressed in HCC and inhibits apoptosis initiated by various cellular stresses (31). Bmf can sequester Bcl-xL and other prosurvival proteins and the final outcome, in terms of apoptosis induction, relies on the balance between a complex network of proapoptotic and antiapoptotic factors (35).
In our study, the direct correlation between Bmf and the cleaved fraction of caspase-3 both in HCC tissues and in SNU449 cell line following proapoptotic stimuli supports a role of Bmf in modulating susceptibility to apoptotic stimuli of HCC cells. However, Bmf is not the only proapoptotic factor acting through caspases pathway, and miR-221 is not the only modulator of Bmf expression; thus, further investigations are warranted to clarify their functions in the network of events controlling apoptosis in HCC. Bmf can also be modulated at the transcriptional level. Indeed, Bmf and BCL2L11/Bim were shown to be up-regulated following TGF-β challenge in primary rat hepatocytes, rat hepatoma cells, lung carcinoma cell lines, B-cell lymphomas, and myeloid cells (35–37). Among pathways involved in HCC, the TGF-β signaling is known to play an important role. Moreover, it was recently reported that TGF-β response of cancer cells may be modulated by miRNAs. The miR-23a_27a_24 miRNA cluster, which is transcriptionally induced by TGF-β, acts as an antiapoptotic and growth-promoting effector in Huh-7 HCC cells (44), and gastric cancer cells become resistant to TGF-β-induced apoptosis by the aberrant up-regulation of the miR-106b-25 miRNA cluster, which can down-modulate the expression of BCL2L11/Bim (45). We can speculate that the up-regulation of miR-221 might potentially affect the TGF-β proapoptotic signals by post-transcriptionally down-regulating the expression of Bmf. However, this hypothesis deserves further investigations.
Aiming to uncover possible relationships between miR-221/ Bmf expression and clinicopathologic features, miR-221 and Bmf levels were correlated with tumor size, histopathologic grading, focality, α-fetoprotein serum levels, recurrence, and cancer-related death in HCC patients. Our results indicate that miR-221 expression is significantly higher in multifocal tumors and in HCCs from patients with a neoplastic recurrence within 2 years. The cutoff of 2 years was chosen according to Imamura et al. (46), who identified this time frame to differentiate true recurrence from new HCCs arisen on cirrhosis. In addition, higher miR-221 levels were associated with a shorter time to recurrence. Conversely, a significant difference in Bmf expression in patients with and without HCC recurrence could not be found, although lower Bmf levels were associated with a trend toward a shorter time to recurrence. Nevertheless, we cannot assess whether the different behavior of HCC patients with different miR-221 levels relies on Bmf, on other unidentified miR-221 target genes, or, more likely, on the entire set of miR-221 targets. The higher Bmf expression observed in unifocal versus multifocal HCCs, together with the correlation between Bmf and the activated fraction of caspase-3, suggests that an increased apoptotic rate triggered by Bmf might account, at least in part, for the lower trend toward multifocality and neoplastic dissemination. We might speculate that, by inhibiting Bmf expression, high miR-221 levels might contribute to a lower apoptotic rate, thus favoring the selection of more aggressive cancer cells, which thus generate multifocal tumors. No correlation was found between miR-221 or Bmf expression and survival. This can be explained by the fact that a percentage of patients curatively treated for HCC die for complication of cirrhosis or as a consequence of new HCCs.
Taken together, our data along with those of the literature show that miR-221 is able to modulate both cell proliferation, by regulating c-Kit, p27/CDKN1B, and p57/CDKN1C, and apoptosis, by regulating Bmf. Notably, these events occur in the same “oncogenic” direction, outlining the importance of the simultaneous action of one miRNA on multiple targets involved in distinct crucial tumorigenic processes. miR-221 represents therefore an interesting target for a therapeutic approach because its silencing would affect more than one oncogenic pathway. Recent advances in the knowledge of miRNA potentiality in cancer treatment have led to the development of antagomirs and locked nucleic acid–modified anti-miRNAs, chemically stabilized oligonucleotides able to inhibit miRNA functions (47). In addition, the liver represents an ideal organ for oligonucleotide-based treatments due to its high avidity in oligonucleotide uptake in both rodents and nonhuman primates (48–50). In this context, our findings on miR-221 are very encouraging and suggest that this miRNA could be a potentially useful target for the nonconventional treatment for HCC.
Supplementary Material
Translational Relevance.
MicroRNAs (miRNA) are emerging as important regulators of gene expression and their deregulation has been extensively reported in human malignancies. A restricted number of miRNAs was found up-regulated in several different human cancers. Among these “oncomiRs,” miR-221 was reported to be up-regulated in glioblastoma, urinary bladder cancer, papillary tumors of the thyroid, pancreatic cancer, hepatocellular carcinoma, and prostate carcinoma. Advances in the knowledge of miRNA potentiality in cancer treatment have led to the development of antagomirs and locked nucleic acid–modified anti-miRNAs, chemically stabilized oligonucleotides able to inhibit miRNA functions. Besides p27 and p57, we report Bmf as a target of miR-221, outlining the role of this miRNA in the control of both proliferation and apoptosis and contributing to characterize its role in liver cancerogenesis. Moreover, miR-221 overexpression was found to be associated with a more aggressive HCC phenotype, confirming miR-221 as a possible target for nonconventional molecular-targeted treatment against HCC.
Acknowledgments
Grant support: Associazione Italiana per la Ricerca sul Cancro (Regional), Italian Ministero dell’Università e della Ricerca Scientifica, and Italian Ministero della Salute (M. Negrini); Associazione Italiana per la Ricerca sul Cancro (Regional) and Fondazione CARISBO (L. Bolondi); National Cancer Institute Program Project Grants (C.M. Croce); Kimmel Foundation scholar award (G.A. Calin); Associazione Italiana per la Ricerca sul Cancro fellowship (F. Fornari); and Fondazione Italiana per la Ricerca sul Cancro fellowship (M. Ferracin).
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Bosch FX, Ribes J, Diaz M, Cleries R. Primary liver cancer: worldwide incidence and trends. Gastroenterology. 2004;127:S5–16. doi: 10.1053/j.gastro.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 2.Negrini M, Ferracin M, Sabbioni S, Croce CM. MicroRNAs in human cancer: from research to therapy. J Cell Sci. 2007;120:1833–40. doi: 10.1242/jcs.03450. [DOI] [PubMed] [Google Scholar]
- 3.Galardi S, Mercatelli N, Giorda E, et al. miR-221 and miR-222 expression affects the proliferation potential of human prostate carcinoma cell lines by targeting p27Kip1. J Biol Chem. 2007;282:23716–24. doi: 10.1074/jbc.M701805200. [DOI] [PubMed] [Google Scholar]
- 4.Gramantieri L, Ferracin M, Fornari F, et al. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res. 2007;67:6092–7. doi: 10.1158/0008-5472.CAN-06-4607. [DOI] [PubMed] [Google Scholar]
- 5.Huang S, He X, Ding J, et al. Upregulation of miR-23a approximately 27a approximately 24 decreases transforming growth factor-β-induced tumor-suppressive activities in human hepatocellular carcinoma cells. Int J Cancer. 2008;123:972–8. doi: 10.1002/ijc.23580. [DOI] [PubMed] [Google Scholar]
- 6.Jiang J, Gusev Y, Aderca I, et al. Association of microRNA expression in hepatocellular carcinomas with hepatitis infection, cirrhosis, and patient survival. Clin Cancer Res. 2008;14:419–27. doi: 10.1158/1078-0432.CCR-07-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ladeiro Y, Couchy G, Balabaud C, et al. Micro-RNA profiling in hepatocellular tumors is associated with clinical features and oncogene/tumor suppressor gene mutations. Hepatology. 2008;47:1955–63. doi: 10.1002/hep.22256. [DOI] [PubMed] [Google Scholar]
- 8.le Sage C, Nagel R, Egan DA, et al. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. 2007;26:3699–708. doi: 10.1038/sj.emboj.7601790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Medina R, Zaidi SK, Liu CG, et al. MicroRNAs 221 and 222 bypass quiescence and compromise cell survival. Cancer Res. 2008;68:2773–80. doi: 10.1158/0008-5472.CAN-07-6754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–58. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murakami Y, Saigo K, et al. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25:2537–45. doi: 10.1038/sj.onc.1209283. [DOI] [PubMed] [Google Scholar]
- 12.Wang Y, Lee AT, Ma JZ, et al. Profiling micro-RNA expression in hepatocellular carcinoma reveals microRNA-224 up-regulation and apoptosis inhibitor-5 as a microRNA-224-specific target. J Biol Chem. 2008;283:13205–15. doi: 10.1074/jbc.M707629200. [DOI] [PubMed] [Google Scholar]
- 13.Budhu A, Jia HL, Forgues M, et al. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology. 2008;47:897–907. doi: 10.1002/hep.22160. [DOI] [PubMed] [Google Scholar]
- 14.Fornari F, Gramantieri L, Ferracin M, et al. miR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene. 2008;27:5651–61. doi: 10.1038/onc.2008.178. [DOI] [PubMed] [Google Scholar]
- 15.Ciafre SA, Galardi S, Mangiola A, et al. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun. 2005;334:1351–8. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 16.Felicetti F, Errico MC, Bottero L, et al. The promyelocytic leukemia zinc finger-microRNA-221/-222 pathway controls melanoma progression through multiple oncogenic mechanisms. Cancer Res. 2008;68:2745–54. doi: 10.1158/0008-5472.CAN-07-2538. [DOI] [PubMed] [Google Scholar]
- 17.Gottardo F, Liu CG, Ferracin M, et al. Micro-RNA profiling in kidney and bladder cancers. Urol Oncol. 2007;25:387–92. doi: 10.1016/j.urolonc.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 18.He H, Jazdzewski K, Li W, et al. The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci U S A. 2005;102:19075–80. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee EJ, Gusev Y, Jiang J, et al. Expression profiling identifies microRNA signature in pancreatic cancer. Int J Cancer. 2007;120:1046–54. doi: 10.1002/ijc.22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pallante P, Visone R, Ferracin M, et al. MicroRNA deregulation in human thyroid papillary carcinomas. Endocr Relat Cancer. 2006;13:497–508. doi: 10.1677/erc.1.01209. [DOI] [PubMed] [Google Scholar]
- 21.Visone R, Russo L, Pallante P, et al. MicroRNAs (miR)-221 and miR-222, both overexpressed in human thyroid papillary carcinomas, regulate p27Kip1 protein levels and cell cycle. Endocr Relat Cancer. 2007;14:791–8. doi: 10.1677/ERC-07-0129. [DOI] [PubMed] [Google Scholar]
- 22.Gillies JK, Lorimer IA. Regulation of p27Kip1 by miRNA 221/222 in glioblastoma. Cell Cycle. 2007;6:2005–9. doi: 10.4161/cc.6.16.4526. [DOI] [PubMed] [Google Scholar]
- 23.Ito Y, Matsuura N, Sakon M, et al. Expression and prognostic roles of the G1-S modulators in hepatocellular carcinoma: p27 independently predicts the recurrence. Hepatology. 1999;30:90–9. doi: 10.1002/hep.510300114. [DOI] [PubMed] [Google Scholar]
- 24.Tannapfel A, Grund D, Katalinic A, et al. Decreased expression of p27 protein is associated with advanced tumor stage in hepatocellular carcinoma. Int J Cancer. 2000;89:350–5. doi: 10.1002/1097-0215(20000720)89:4<350::aid-ijc6>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 25.Nakai S, Masaki T, Shiratori Y, et al. Expression of p57(Kip2) in hepatocellular carcinoma: relationship between tumor differentiation and patient survival. Int J Oncol. 2002;20:769–75. [PubMed] [Google Scholar]
- 26.Roberts LR, Gores GJ. Hepatocellular carcinoma: molecular pathways and new therapeutic targets. Semin Liver Dis. 2005;25:212–25. doi: 10.1055/s-2005-871200. [DOI] [PubMed] [Google Scholar]
- 27.Fabregat I, Roncero C, Fernandez M. Survival and apoptosis: a dysregulated balance in liver cancer. Liver Int. 2007;27:155–62. doi: 10.1111/j.1478-3231.2006.01409.x. [DOI] [PubMed] [Google Scholar]
- 28.Guo XZ, Shao XD, Liu MP, et al. Effect of bax, bcl-2 and bcl-xL on regulating apoptosis in tissues of normal liver and hepatocellular carcinoma. World J Gastroenterol. 2002;8:1059–62. doi: 10.3748/wjg.v8.i6.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schattenberg JM, Galle PR, Schuchmann M. Apoptosis in liver disease. Liver Int. 2006;26:904–11. doi: 10.1111/j.1478-3231.2006.01324.x. [DOI] [PubMed] [Google Scholar]
- 30.Garcia EJ, Lawson D, Cotsonis G, Cohen C. Hepatocellular carcinoma and markers of apoptosis (bcl-2, bax, bcl-x): prognostic significance. Appl Immunohistochem Mol Morphol. 2002;10:210–7. doi: 10.1097/00129039-200209000-00004. [DOI] [PubMed] [Google Scholar]
- 31.Takehara T, Liu X, Fujimoto J, Friedman SL, Takahashi H. Expression and role of Bcl-xL in human hepatocellular carcinomas. Hepatology. 2001;34:55–61. doi: 10.1053/jhep.2001.25387. [DOI] [PubMed] [Google Scholar]
- 32.Watanabe J, Kushihata F, Honda K, et al. Prognostic significance of Bcl-xL in human hepatocellular carcinoma. Surgery. 2004;135:604–12. doi: 10.1016/j.surg.2003.11.015. [DOI] [PubMed] [Google Scholar]
- 33.Beerheide W, Tan YJ, Teng E, Ting AE, Jedpiyawongse A, Srivatanakul P. Downregulation of proapoptotic proteins Bax and Bcl-X (S) in p53 overexpressing hepatocellular carcinomas. Biochem Biophys Res Commun. 2000;273:54–61. doi: 10.1006/bbrc.2000.2891. [DOI] [PubMed] [Google Scholar]
- 34.Chen GG, Lai PB, Chan PK, et al. Decreased expression of Bid in human hepatocellular carcinoma is related to hepatitis B virus X protein. Eur J Cancer. 2001;37:1695–702. doi: 10.1016/s0959-8049(01)00182-4. [DOI] [PubMed] [Google Scholar]
- 35.Ramjaun AR, Tomlinson S, Eddaoudi A, Downward J. Upregulation of two BH3-only proteins, Bmf and Bim, during TGF β-induced apoptosis. Oncogene. 2007;26:970–81. doi: 10.1038/sj.onc.1209852. [DOI] [PubMed] [Google Scholar]
- 36.Bissell DM, Roulot D, George J. Transforming growth factor β and the liver. Hepatology. 2001;34:859–67. doi: 10.1053/jhep.2001.28457. [DOI] [PubMed] [Google Scholar]
- 37.Breuhahn K, Longerich T, Schirmacher P. Dysregulation of growth factor signaling in human hepatocellular carcinoma. Oncogene. 2006;25:3787–800. doi: 10.1038/sj.onc.1209556. [DOI] [PubMed] [Google Scholar]
- 38.Willis SN, Adams JM. Life in the balance: how BH3-only proteins induce apoptosis. Curr Opin Cell Biol. 2005;17:617–25. doi: 10.1016/j.ceb.2005.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Puthalakath H, Villunger A, O’Reilly LA, et al. Bmf: a proapoptotic BH3-only protein regulated by interaction with the myosin V actin motor complex, activated by anoikis. Science. 2001;293:1829–32. doi: 10.1126/science.1062257. [DOI] [PubMed] [Google Scholar]
- 40.Edmondson HA, Steiner P. Primary carcinoma of the liver: a study of 100 cases among 48,900 necropsies. Cancer. 1954;7:462–503. doi: 10.1002/1097-0142(195405)7:3<462::aid-cncr2820070308>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 41.Schmelzle T, Mailleux AA, Overholtzer M, et al. Functional role and oncogene-regulated expression of the BH3-only factor Bmf in mammary epithelial anoikis and morphogenesis. Proc Natl Acad Sci U S A. 2007;104:3787–92. doi: 10.1073/pnas.0700115104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Felli N, Fontana L, Pelosi E, et al. MicroRNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation. Proc Natl Acad Sci U S A. 2005;102:18081–6. doi: 10.1073/pnas.0506216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Charlotte F, L’Hermine A, Martin N, et al. Immunohistochemical detection of bcl-2 protein in normal and pathological human liver. Am J Pathol. 1994;144:460–5. [PMC free article] [PubMed] [Google Scholar]
- 44.Huang S, He X, Ding J, et al. Upregulation of miR-23a_27a_24 decreases transforming growth factor-β-induced tumor-suppressive activities in human hepatocellular carcinoma cells. Int J Cancer. 2008;132:972–8. doi: 10.1002/ijc.23580. [DOI] [PubMed] [Google Scholar]
- 45.Petrocca F, Visone R, Onelli MR, et al. E2F1-regulated microRNAs impair TGFβ-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13:272–86. doi: 10.1016/j.ccr.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 46.Imamura H, Matsuyama Y, Tanaka E, et al. Risk factors contributing to early and late phase intra-hepatic recurrence of hepatocellular carcinoma after hepatectomy. J Hepatol. 2003;38:200–7. doi: 10.1016/s0168-8278(02)00360-4. [DOI] [PubMed] [Google Scholar]
- 47.Krutzfeldt J, Rajewsky N, Braich R, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–9. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 48.Song E, Wang J, et al. RNA interference targeting Fas protects mice from fulminant hepatitis. Nat Med. 2003;9:347–51. doi: 10.1038/nm828. [DOI] [PubMed] [Google Scholar]
- 49.Morrissey DV, Lockridge JA, Shaw L, et al. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat Biotechnol. 2005;23:1002–7. doi: 10.1038/nbt1122. [DOI] [PubMed] [Google Scholar]
- 50.Elmen J, Lindow M, Schutz S, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–9. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.