Abstract
Papillomavirus (PV) E6 oncoproteins bind and often provoke the degradation of many cellular proteins important for the control of cell proliferation and/or cell death. Structural studies on E6 proteins have long been hindered by the difficulties of obtaining highly concentrated samples of recombinant E6. Here we show that recombinant E6 proteins from eight human and one bovine PV strains exist as oligomeric as well as multimeric species. These species were characterized using a variety of biochemical and biophysical techniques including analytical gel filtration, activity assays, SPR, EM and FTIR. The characterization of E6 oligomers is facilitated by the fusion to the maltose binding protein (MBP), which slows down the formation of higher-order multimeric species. The proportion of each oligomeric form vary depending on the viral strain considered. Oligomers appear to consist of folded units, which, in the case of high-risk mucosal HPV E6, retain binding to the ubiquitin ligase E6AP and the capacity to degrade the pro-apoptotic protein p53. In addition to the small-size oligomers, E6 proteins spontaneously assemble into large organized multimeric structures, a process which is accompanied by a significant increase in the β-sheet secondary structure content. Finally, co-localisation experiments using E6 equipped with different tags further demonstrate the occurrence of E6 self-association in eukaryotic cells. The ensemble of these data suggest that self-association is a general property of E6 proteins which occurs both in vitro and in vivo and might therefore be functionally relevant.
Keywords: HPV, E6, cervical cancer, self-association, oligomerization, ordered aggregation
INTRODUCTION
Papillomaviruses (PV) are small DNA viruses that induce squamous epithelial neoplasia. They are classified into phylogenetic groups with distinct tropisms for different body sites (skin, mouth and genitalia) and are divided into “low-risk” and “high-risk” types according to their carcinogenic potential. There are nearly 100 different Human papillomaviruses (HPV) so far identified. Cervical carcinomas are induced by high-risk genital HPVs and represent one of the main causes of cancer-related death for women 1. HPV16 is the most common oncogenic strain associated with 50% of the cervical cancers, followed by HPV18 covering 14% of the cancers 2. HPV types 5 and 8 are associated with squamous cutaneous carcinomas 3. Among the PV types infecting other animals, bovine papillomaviruses (BPV) have also been extensively studied due to their strong transforming properties 4.
In anogenital cancers caused by high-risk HPVs, two oncoproteins, E6 and E7, are expressed. E6 proteins from “high-risk” genital HPVs are thought to promote tumorogenesis by binding and often provoking the degradation of a number of proteins involved in cell proliferation and cell death control 5. In particular, E6 forms a complex with the ubiquitin ligase E6-associated protein (E6AP) 6, which in turn is able to target the tumor suppressor p53 7, leading to ubiquitin–mediated degradation of p53. Another group of E6 targets include the tumour suppressor human discs large hDlg 8 and the MAGI family of proteins 9; 10. However E6 proteins from other HPV species may carry out their functions via alternative mechanisms. For instance, high-risk cutaneous HPV5 and 8 E6 proteins do not appear to degrade p53, but promote the degradation of the Bak pro-apoptotic protein, which is induced after UV irradiation of the skin 11. BPV1 E6 interacts with the focal adhesion protein paxillin and the AP-1 adaptin complex and these interactions correlate with cellular transformation 12; 13.
Papillomavirus E6 proteins are rather small in size (about 150 amino acids). Most of them share a common architecture consisting of two zinc-binding domains (E6-N and E6-C) 14. While the structure of the E6-C domain has recently been solved in our laboratory 15, structures of full-length E6 proteins have not been reported yet, due to the difficulties encountered in the preparation of highly concentrated and monodisperse samples of these proteins. Most attempts to produce the recombinant full-length protein in E. coli have focussed on the main oncogenic HPV16 variant and mutants of this protein 16; 17; 18. Recently, we have also explored E6 proteins issued from other HPV types, such as high-risk genital HPV18, 33, 45, 52, 58, low-risk HPV11 and high-risk cutaneous HPV5. We found that all these E6 proteins form large soluble aggregates upon fusion to the carrier protein MBP in vivo during biosynthesis in bacteria 16; 19. Other authors have also observed multimeric structures in HPV16 and HPV18 E6 preparations obtained using refolding techniques 17; 20.
In the present work we focus on E6 self-association and show that this property is shared among E6 proteins issued from different phylogenetic viral groups. We identify and characterize both oligomeric and multimeric E6 species that exist in vitro. In addition, we demonstrate that heterologously expressed E6 proteins self-associate in eukaryotic cells. The conservation of self-association within the E6 family suggests that such mechanisms may have a functional role.
RESULTS
E6 proteins self-associate into oligomeric species
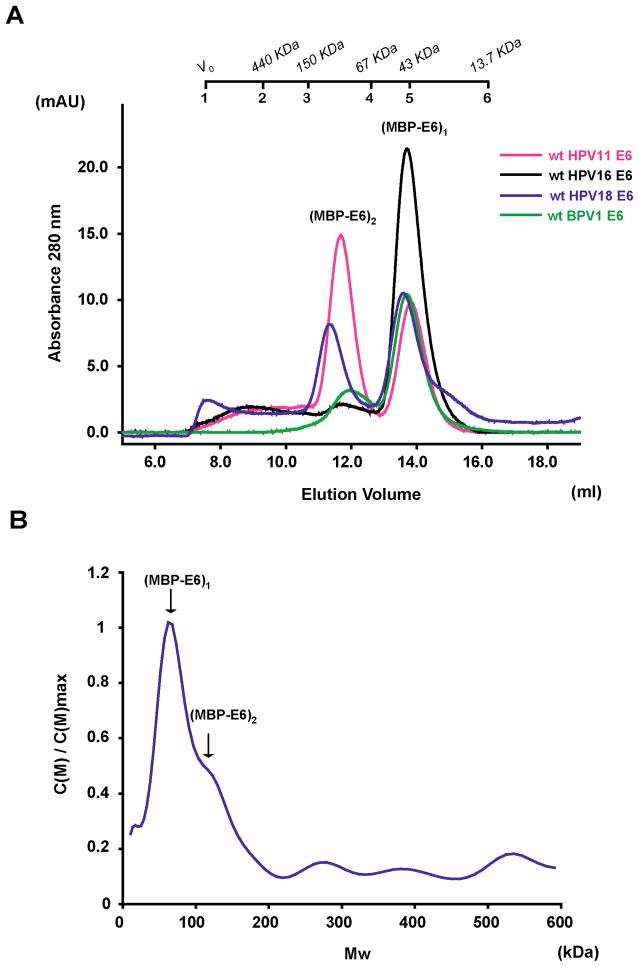
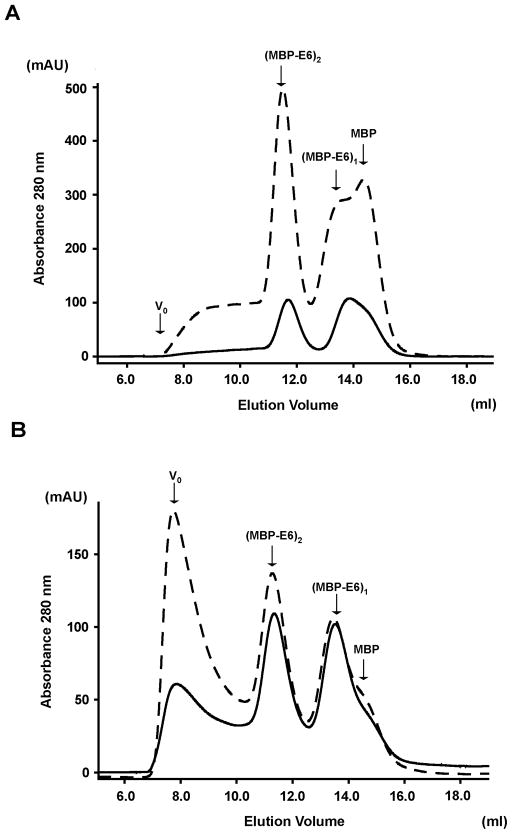
We examined the oligomeric state of a panel of E6 proteins issued from various papillomaviruses. The wild-type E6 proteins from HPV 11, 16 and 18 and from Bovine Papillomavirus (BPV1)21 were expressed as MBP-fusions, purified by amylose affinity and cleared from the high molecular weight MBP-E6 aggregates by ultracentrifugation as previously described 19. Sample concentrations of all MBP-E6 proteins were adjusted to 2.5 μM and analysed by size exclusion chromatography. The elution profiles indicate multiple oligomeric species in most of the preparations (Figure 1A). For MBP-fused HPV11 and HPV18 E6, two peaks of similar intensity corresponding to monomeric and dimeric forms are detected. On the other hand, the monomer is the most abundant form for MBP-fused E6 proteins from HPV16 and BPV1.
Figure 1.
Detection of soluble MBP-E6 oligomeric species. (A) Analytical gel filtration chromatography analysis. Affinity-purified MBP-E6 samples were cleared from soluble aggregates by ultracentrifugation. Concentrations were adjusted to approximately 2.5 μM and 100 μl volumes injected on a Superdex200 10/30 column. Elution peaks corresponding to monomer (MBP-E61, 60 kDa), dimer (MBP-E62, 120 kDa) and trimer (MBP-E63, 180 kDa) forms of MBP-E6 proteins are indicated. The shoulder at 15 ml corresponds to monomeric MBP arising from residual proteolytic activity in the protein preparations. Molecular size markers are reported on top of the figure. 1: V0; 2: ferritin (440 kDa); 3: mouse Igg (150 kDa); 4: BSA (67 kDa); 5: ovalbumin (43 kDa); 6: RNase (13.7 kDa). The elution profiles correspond to wt E6 proteins from the following viral stains: HPV11 (magenta line), HPV16 (black line), HPV18 (blue line) and BPV1 (green line). (B) Molecular weight distribution of wt HPV18 MBP-E6 oligomers derived from sedimentation velocity experiments. C(M) indicates arbitrary units.
To further confirm the gel filtration data we performed analytical ultracentrifugation sedimentation velocity experiments on wild-type HPV18 MBP-E6 preparations. Fitting of the sedimentation profiles points to the existence of two main species with molecular sizes consistent with those of monomeric and dimeric MBP-E6 (Figure 1B). Although the polydispersity of the system does not allow for a precise extrapolation of molecular weights, these results support the findings of analytical size exclusion chromatography described above.
In addition to these wild-type E6 proteins, we have also performed similar analysis on MBP-fused mutant E6 proteins from types 5, 11, 16, 33, 52 and 58. These E6 constructs have non-conserved cysteines mutated into serines in order to reduce intermolecular disulfide bridge formation as previously described 19. Gel-filtration profiles of all these mutant MBP-E6 constructs also reveal the presence of oligomeric forms (Supplementary Material, Figure 1). Therefore, cysteine mutagenesis of these constructs did not alter the general propensity of MBP-E6 proteins to form oligomers.
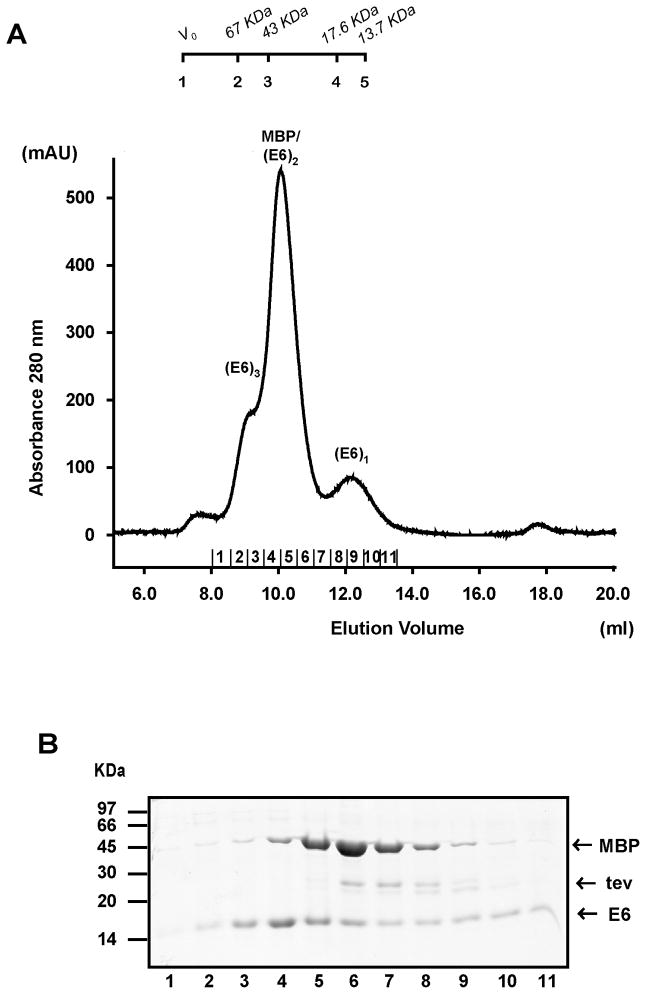
In order to rule out the possibility that MBP would drive the oligomerisation phenomenon, we proceeded to separate E6 from the MBP tag by TEV proteolysis and analyzed the digestion products by gel filtration chromatography (Figure 2). This experiment was performed on the cystein-mutated HPV11 MBP-E6 4C/4S. The rationale of using this mutant is based on the fact that all E6 proteins analysed in this study were prone to precipitation once separated from their MBP carrier. E6 precipitation becomes particularly severe if non conserved cysteines are kept in the sequence. Cysteine to serine mutagenesis limited disulfide cross-bridging effects, enabling us to obtained samples of pure E6 in absence of MBP.
Figure 2.
Gel filtration profiles of HPV11 MBP-E6 4C/4S preparations after separation from the MBP tag. Affinity-purified MBP-E6 preparations were cleared from soluble aggregates by ultracentrifugation and digested using TEV protease. Subsequently, 500 μl were injected on a Superdex75 10/30 column. (A) Peaks corresponding to monomeric (E61, 18 kDa), dimeric (E62, 36 kDa) and trimeric (E63, 54 kDa) E6 forms and MBP (MBP, 44 kDa) are indicated. Molecular size markers are reported on top of the figure. 1: V0; 2: BSA (67 kDa); 3: ovalbumin (43 kDa); 4: Myoglobin (17.6 kDa); 5: RNase (13.7 kDa). Numbers 1–11 reported on the x-axis indicate fractions collected for SDS-PAGE analysis. (B) Lanes 1–11 correspond to fractions 1–11 indicated on the elution profile. Fractions were precipitated with 20% trichloroacetic acid. Pellets were re-suspended in 50 μl of loading buffer and applied to a 12% SDS-polyacrylamide gel. Bands corresponding to MBP, TEV and E6 are indicated.
HPV11 MBP-E6 4C/4S displays monomeric, dimeric and trimeric forms (Supplementary Material, Figure 1A). After TEV digestion, the gel-filtration profile of this sample contains three separate peaks at 9.1, 10 and 12.0 ml. SDS-PAGE analysis shows that elution peaks at 9.1 and 12 ml contain predominantly E6, while the peak at 10 ml contains both E6 and the MBP tag (Figure 2A and 2B). Hence E6 elutes at three distinct volumes, which are expected for trimeric, dimeric and monomeric E6 according to the column’s calibration, showing that self-association is mediated by residues in the E6 protein rather than by the MBP carrier protein.
Stability of E6 oligomers upon dilution and concentration
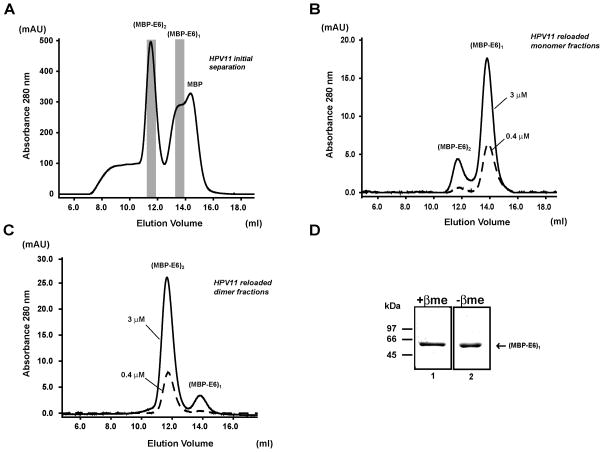
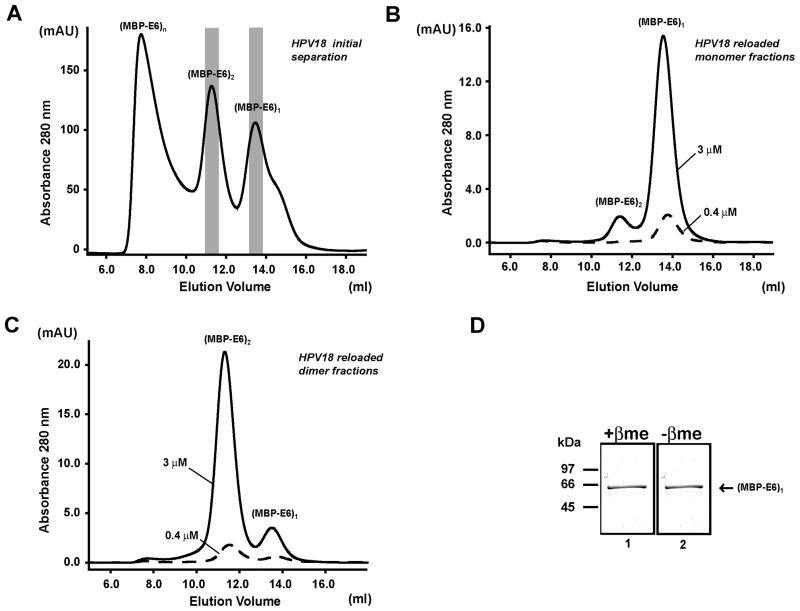
Next we investigated the influence of dilution on the stability of the E6 oligomers. Wild-type MBP-fused E6 proteins from HPV11 and 18 were affinity-purified, cleared from high-molecular weight aggregates by ultracentrifugation, applied onto a Superdex 200 10/30 gel filtration column and fractionated to the different oligomeric species. Protein concentrations in fractions corresponding to the isolated monomeric and dimeric species were adjusted by dilution to either 3 μM or 0.4 μM. After about two hours of incubation at room temperature, aliquots of these diluted samples were re-loaded individually for a second time on the same column. The resulting elution profiles are reported in Figures 3 and 4 for HPV11 and 18, respectively. Reloading of monomer fractions generates elution profiles with main monomer peaks (Figures 3B and 4B). When reloading the dimer fractions, we observe that dimers strongly persists in the second gel-filtration column, generating only small monomer peaks (accounting for about 10% of the total protein according to peak area integration values) (Figures 3C and 4C). This latter result highlights stability of the HPV11 and HPV18 dimeric species. Dimer fractions of HPV11 and HPV18 MBP-E6 were also analyzed by SDS-PAGE in the presence and in the absence of the reducing agent β-mercaptoethanol. We could not detect any band migrating at molecular weights higher than that corresponding to an MBP-E6 monomer (i.e. 60 kDa) ruling out the possibility that E6 dimer stability arises from disulfide cross-bridging (Figures 3D and 4D).
Figure 3.
Stability of wt HPV11 MBP-E6 oligomers upon dilution. (A) Affinity purified HPV11 MBP-E6 was cleared from soluble aggregates and applied onto a Superdex200 10/30 column. (B–C) Aliquots of fractions corresponding to the monomeric (B), dimeric (C) forms of MBP-E6 (corresponding to the indicated grey areas in (A)) were re-loaded individually onto the same gel filtration column at concentrations of 3.0 (continuous lines) and 0.4 (broken lines) μM. See also legend of Figure 1. (D) Dimer fractions of HPV11 MBP-E6 analyzed on a 12% SDS-PAGE in the presence (+βme) (Lane 1, L1) and in the absence (−βme) (L2) of the reducing agent β-mercaptoethanol. Samples were migrated on the same gel but were separated by two empty wells in order to avoid cross-contamination by β-mercaptoethanol. The migration of the molecular weight markers (in kDa) is reported on the left hand side.
Figure 4.
Stability of wt HPV18 MBP-E6 oligomers upon dilution. (A) wt HPV18 MBP-E6 preparations were loaded on a Superdex200 10/30 column as described in Figure 3. Monomer (B) and dimer (C) fractions (indicated by the grey areas) were re-loaded a second time on the same column at concentrations of 3.0 (continuous lines) and 0.4 μM (broken lines). (MBP-E6)n refer to larger oligomers eluting at the column’s void volume. See also legend of Figure 1. (D) Dimer fractions of HPV18 MBP-E6 analyzed on SDS-PAGE in the presence (+βme) (Lane 1, L1) and in the absence (−βme) (L2) of β-mercaptoethanol.
We also investigated E6 oligomerization upon concentration. HPV11 and 18 MBP-fused wild-type E6 preparations were affinity purified, cleared from soluble aggregates and analyzed by gel filtration before and after concentration. In the case of HPV11 MBP-E6, 5-fold concentration of the sample increases mainly the proportion of the dimeric form (Figure 5A). Instead, in the case of HPV18 MBP-E6, a 2-fold concentration leads to the assembly of higher-order oligomers, eluting at the column’s void volume (V0) (Figure 5B). It therefore appears that HPV18 E6 dimers could represent intermediates in the build-up of larger oligomers. These larger species are instead less readily formed upon concentration of HPV11 MBP-E6 samples.
Figure 5.
MBP-E6 oligomeric forms upon concentration. Affinity purified HPV11 (A) and HPV18 (B) MBP-fused wt E6 protein samples were ultracentrifugated to remove soluble aggregates. Aliquots of the supernatants were injected on a Superdex200 10/30 column (continous lines). The remaining samples were concentrated either 5-fold (A) or 2-fold (B) and injected on the same column (broken lines). See also legend of Figure 1.
Oligomers possess native-like E6 in vitro activities
Next, we characterized the zinc coordination properties of the different E6 forms. Zinc concentrations were measured in gel filtration fractions of monomeric and oligomeric forms of most wild-type and mutated MBP-E6 constructs investigated in this study (Table 1). These experiments were performed using the selective fluorophore TSQ 17. For all samples we observed a 2:1 Zn2+:E6 stoichiometry that is expected from the common architecture of E6 proteins consisting of two zinc-binding domains. These data indicate that the E6 self-association into small oligomers is not related to loss of metal ion coordination.
Table 1.
Experimentally derived Zn2+:protein stoichiometries for wt and mutant MBP-E6 samples from HPV strains 11, 16 and 18. Monomer, dimer and trimer species are indicated as (MBP-E6)1, (MBP-E6)2 and (MBP-E6)3 respectively. Zinc concentrations were measured by the TSQ assay and calculated relative to ZnSO4 standards.
| Sample | Zn2+: protein stoichiometry |
|---|---|
|
| |
| wt HPV 11 (MBP-E6)1 | 2.08 ± 0.17 |
| wt HPV 11 (MBP-E6)2 | 2.01 ± 0.12 |
| HPV 11 4C/4S (MBP-E6)1 | 2.30 ± 0.33 |
| HPV 11 4C/4S (MBP-E6)2 | 1.90 ± 0.03 |
| HPV 11 4C/4S (MBP-E6)3 | 2.10 ± 0.11 |
| wt HPV 16 (MBP-E6)1 | 2.06 ± 0.29 |
| HPV 16 6C/6S (MBP-E6)1 | 1.97 ± 0.24 |
| wt HPV 18 (MBP-E6)1 | 1.70 ± 0.09 |
| wt HPV 18 (MBP-E6)2 | 2.00 ± 0.04 |
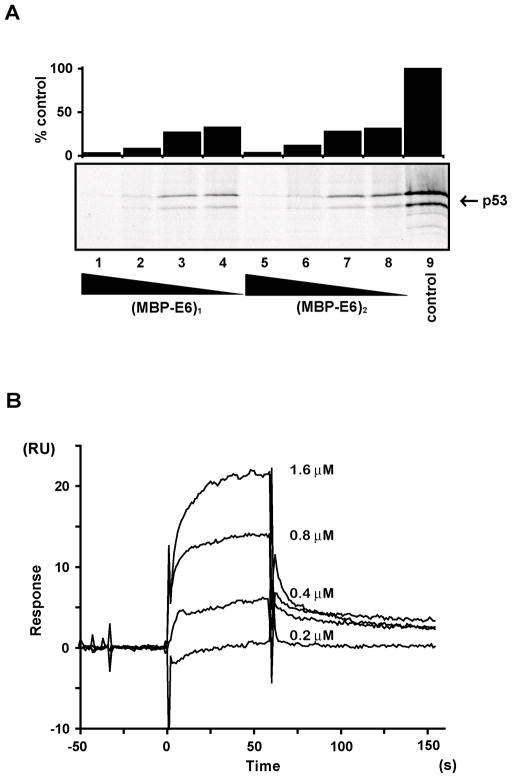
Monomeric and dimeric species of the MBP fused wild-type HPV18 E6 were tested for their p53 degradation activities immediately after fractionation on the gel filtration column. Radiolabelled 35S p53 was incubated with varying concentrations of monomeric and dimeric MBP-E6, corresponding to 400, 200, 100 and 50 nM for 1 hour at 29 °C. Given the observations on wild-type HPV18 E6 dimer stability reported above, it is likely that the dimer persists at least for the highest concentrations (i.e. 400 and 200 nM). The results clearly show very similar degradation profiles for monomeric and dimeric E6 at all concentrations tested (Figure 6A).
Figure 6.
Characterization of the activities of HPV18 wt E6 oligomers. Oligomeric MBP-E6 forms were fractionated on a Superdex 200 10/60 column. Protein concentrations were determined based on the calculated extinction coefficients of the monomeric forms. (A) Degradation of in vitro translated p53 by wt HPV18 MBP-E6 monomer (lanes (L): 1–4) and dimer (L: 5–8) species. p53 degradation reactions contained the following concentrations of HPV18 MBP-E6: 400 nM (L1 and L5); 200 nM (L2 and L6); 100 nM (L3 and L7); 50 nM (L4 and L8). The histogram indicates the residual p53 signal expressed as percentage relative to the intensity of the input control band (L9). (B) Binding of wt HPV18 MBP-E6 dimer to a peptide containing the LxxφLsh motif of E6AP as monitored by SPR. The figure illustrates the concentration dependent binding of MBP-E6 to approximately 600 RU of affinity-captured GST-E6AP peptide ligand. The MBP-E6 analyte concentrations for each sensorgram are indicated on the figure.
Subsequently, we investigated the binding activity of dimeric wild-type HPV18 MBP-E6 to a 15-mer peptide containing the LxxφLsh motif of E6AP using Biacore technology. For such purpose we followed the protocols described in a previous report 22, where we carried out an extensive analysis of the interactions of different monomeric E6 proteins to the same peptide. Dimer fractions of HPV18 MBP-E6 were injected at concentrations ranging from 1.6 to ~0.2 μM on a surface capturing GST-E6AP peptide fusions. The resulting sensorgrams show concentration dependent binding (Figure 6B). Unfortunately, like in the case of monomeric HPV18 E6, extrapolation of precise kinetic and/or affinity parameters for this interaction is difficult. Kinetic analysis of these sensorgrams is hampered by the fast dissociation rates and the slight biphasic profiles, while equilibrium analysis requires sampling of high analyte concentrations that, in the case of E6 proteins, promote aggregation processes (as previously discussed in Zanier et al. 22). However the magnitudes of binding responses are comparable for the dimeric HPV18 MBP-E6 analyzed herein and the monomeric HPV18 MBP-E6 form analyzed in our previous BIAcore study 22. This suggests that monomeric and dimeric HPV18 MBP-E6 have similar affinities for the cognate peptide corresponding to a KD of approximately 30 μM as estimated previously for monomeric HPV18 MBP-E6 22.
Overall these observations indicate that the HPV 18 E6 dimer is an active species with respect to p53 degradation and E6AP binding. Together with the finding of a 2:1 Zn2+: protein stoichiometry our results suggest that the E6-N and E6-C domains possess a native-like fold within the oligomeric E6 species.
Monomeric E6 proteins spontaneously assemble into large organized multimeric structures
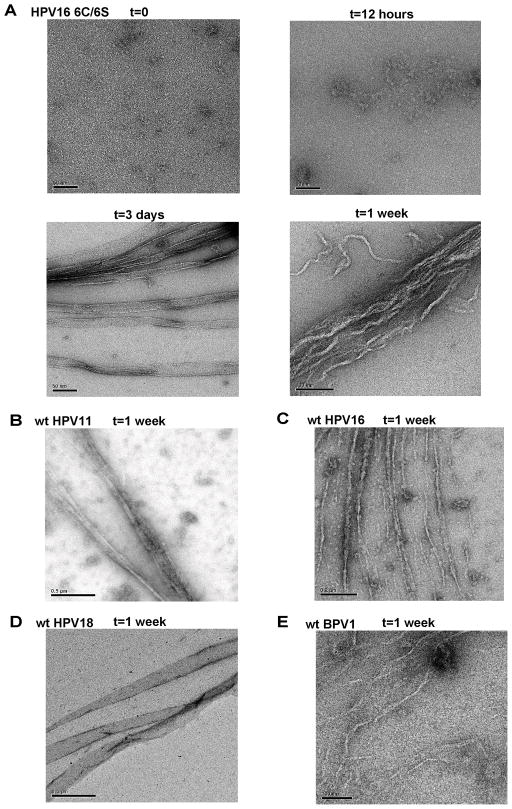
Recombinant E6 proteins, once separated from the MBP tag, progressively loose activity upon storage (data not shown). To investigate the physical processes involved in activity loss, we selected the HPV16 E6 6C/6S construct, which, in our hands displays the highest concentration treshold of aggregation once separated from the MBP tag (50 μM, data not shown) and thus is the E6 protein most amenable to purification. HPV16 E6 6C/6S devoid of MBP tag was isolated by gel-filtration as a monomer as described previously 22, adjusted to 40 μM and incubated at 20 °C for one week in sustained reducing conditions. Aliquots were taken at time intervals of 0, 1/2, 3 and 7 days, and analysed by electron microscopy (Figure 7A). At t = 0, we observe low molecular weight particles (most likely monomeric protein) together with a few small spherical deposits (approximate diameter 4 nm). At t = 12 hours, larger structures resembling a collection of spherical particles are visible in some parts of the grid. At t = 3 days, we observe remarkable ribbon-like structures with widths varying between 20 and 30 nm and measuring several hundreds of nanometers in length. At t = 1 week, bundles of fibrous filaments are still visible, with an overall less regular morphology than the structures observed at day 3. Structures similar to those reported in Figure 7A were also detected in preparations of monomeric E6 6C/6S incubated at either low temperature (5 °C) or at lower salt concentration (150 mM NaCl instead of 400 mM) (data not shown). Similar results were also obtained with monomeric MBP-E6 6C/6S fusions, indicating that the presence of the MBP tag does not hinder the fiber assembly process. This latter finding allowed us to investigate other E6 proteins, which require fusion to MBP for the isolation of their monomeric form. We purified monomeric wild-type MBP-E6 proteins issued from HPV11, 16, 18 and BPV1 and incubated them as indicated above. EM images taken immediately after purification show that all MBP-E6 proteins are homogeneous preparations of low molecular weight particles (Supplementary Material, Figures 2A–D). As the incubation time increases we detect formation of similar structures as those formed by E6 6C/6S. Images in Figures 7B–7E show that all MBP-E6 proteins analyzed develop fiber bundles and ribbon-like structures after one week of incubation. Hence, the ensemble of these data show that spontaneous self-association into large multimeric structures is a property shared by distantly related E6 proteins and that this is independent of mutagenesis of non-conserved cysteins.
Figure 7.
Spontaneous assembly of monomeric E6 and MBP-E6 proteins into ordered multimeric structures visualized by EM. (A) Partial field images of monomeric HPV16 E6 6C/6S taken at 0, 1/2, 3 and 7 days of incubation. Images of monomeric wt MBP-E6 proteins from (B) HPV11, (C) HPV16, (D) HPV18 and (D) BPV-1 viruses after one week incubation. Bar = 50 nm for images in (A) t=0, t=12 hours, t=3 days. Bar = 100 nm for images in (A) t=1 week and in (E). Bar=200 nm for image in (C). Bar = 500 nm for image in (B) and (D).
Next, we purified HPV16 MBP-E6 6C/6S samples on amylose resin but purposedly omitted the subsequent steps of ultracentrifugation and gel-filtration. Such preparations are dominated by the well-defined spherical structures previously described 16; 19. After few days of incubation, these samples also developed fiber bundles although with an overall less regular morphology as the structures shown in Figure 7 (Supplementary Material, Figure 2E). Remarkably, when incubation was performed at low temperature (5 °C) to slow down the kinetics of self-assembly, we clearly observed small E6 filaments growing from the spherical aggregates (indicated by an arrow in Supplementary Material, Figure 2F), suggesting that soluble MBP-E6 spherical structures can also act as nuclei for fiber formation.
Secondary structure changes implicated in E6 self-assembly
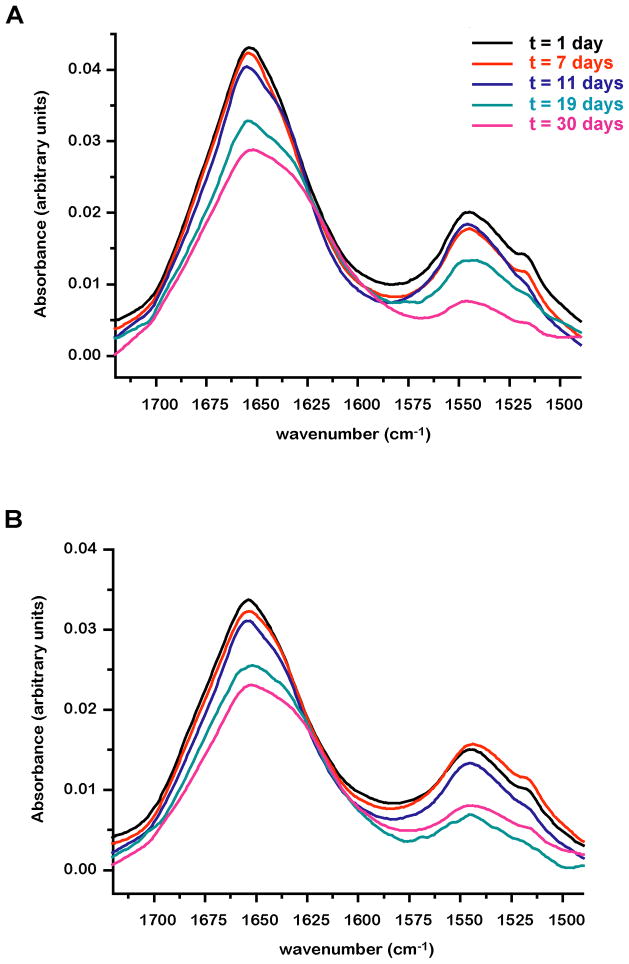
In an attempt to further characterize the structure of E6 multimers, we monitored the changes in Thioflavin-T fluorescence in preparations of non-fused monomeric HPV16 E6 6C/6S. We detected no significant differences in the intensity of the Thioflavin-T emission signal at the different time points of E6 incubation (data not shown), suggesting that E6 multimers do not contain the cross-beta structures typical of amyloid aggregation 23. In addition, far UV Circular Dichroism (CD) experiments failed to provide conclusive results, mainly due to the difficulties imposed by sample precipitation upon incubation and light scattering during the measurements. We therefore opted for FTIR spectroscopy, which enables to overcome these problems. Experiments were performed on 40 μM monomeric HPV16 E6 6C/6S preparations incubated at 20 °C in buffer containing either 150 or 400 mM NaCl (see Materials and Methods). E6 samples did not show any evolution in the amide I region (spectral range 1700/1600 cm−1) during the first week of measurement. However a significant change in the bandwidth of the amide I band is observed starting from the second week of incubation which terminates after one month (Figure 8A and B), Similar changes were also observed for HPV16 E6 6C/6S samples which have been dialyzed against buffer containing 98% D2O immediately after purification (data not shown). Overall it appears that salt concentration has no significant effect on the time-evolution of E6 samples.
Figure 8.
FTIR characterization of aggregates in samples of monomeric HPV16 E6 6C/6S. Superposition of IR spectra of E6 in buffer containing either 150 mM (A) or 400 mM NaCl (B) recorded at different time points of the incubation.
Secondary structure analysis was carried out on the Amide I band of spectra of the E6 protein in 150 mM NaCl (Table 2 and Supplementary Material, Figure 3). For monomeric E6 at t=0 we obtained the following estimate: 30% β-sheet, 28% α-helix, 24% β-turn and 13% random coil. Error is typically about ±3%, however it is lower for β-sheet and higher for α-helical elements, since the latter are hardly distinguishable from random contributions. Another source of error is the fitting procedure which requires a baseline correction for the spectrum. These results are in agreement with what has been previously reported by Lipari and coworkers on the HPV16 E6(Δ143–151) construct, which lacks the last 8 unfolded C-terminal residues (27% β-sheet, 26% α-helix, 24% β-turn) 24.
Table 2.
Secondary structure content (expressed as % secondary structure (ss)) of monomeric HPV16 E6 immediately after purification (t=0) and after 30 days of incubation (t = 30 days). Secondary structure analysis was carried out by deconvolution of spectra of E6 in 20 mM sodium phosphate (pH 6.8), 150 mM NaCl, 2 mM DTT (Figure 8B). The frequency assignments are based on previousy published criteria 38.
| HPV 16 E6 (t=0) | HPV 16 E6 (t=30 days) | |||
|---|---|---|---|---|
|
|
||||
| Position (cm−1) | % ss | Position (cm−1) | % ss | |
| β-sheet | 1623, 1631, 1639, 1690 | 30 | 1625, 1637, 1685, 1695 | 38 |
| α-helix | 1657 | 28 | 1653 | 17 |
| β-turn | 1669, 1679 | 24 | 1663, 1674 | 27 |
| random coil | 1647 | 13 | 1654 | 10 |
On the other hand, for the E6 sample incubated for 30 days we obtain a significantly different profile with 38% β-sheet, 17% α-helix, 27% β-turn and 10% random coil. It therefore seems that there is a significant increase in β-sheet structure (from 30 to 38%), which is complemented by a decrease in α-helix (from 28 to 17%).
Detection of E6 self-association in vivo
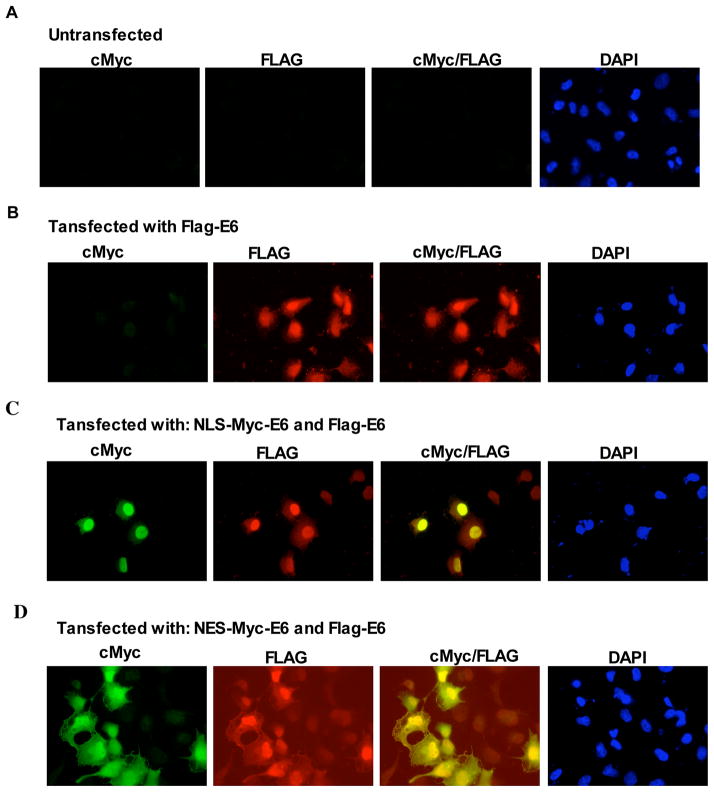
To investigate E6 self-association in vivo, we used nucleo-cytoplasmic delocalization techniques which allow detection of protein-protein interactions in transiently co-transfected eukaryotic cells 25. This assay requires one of the proteins to be fused to either a nuclear localisation or a nuclear export sequence (NLS or NES). If the other protein is an interaction partner, it will be co-targeted to either the nucleus or the cytoplasm. We cloned fusion constructs containing the wild-type HPV18 E6 sequence N-terminally fused to either a Flag or a cMyc tag to allow for immunodetection. The cMyc-E6 constructs were additionally appended with either the SV40 T-antigen based-NLS sequence or a leucine-rich NES sequence at the N-terminus (NLS-cMyc-E6 and NES-cMyc-E6).
Results are shown in Figure 9. When transfected in HPV-negative H1299 cells, Flag-E6 localises preferentially, but not completely, to the nucleus (panel B). When an excess of NLS-Myc-E6 is co-transfected with Flag-E6 (panel C), the NLS-Myc-E6 exhibites a strong nuclear signal and co-transfected Flag-E6 also adopts a comparably strong nuclear signal. On the other hand, when an excess of NES-cMyc-E6 is co-transfected with Flag-E6 (panel D), NES-Myc-E6 displays a more even distribution between nucleus and cytoplasm and a similarly even distribution is then observed for the co-transfected Flag-E6. Therefore the distribution of Flag-E6 is modified by co-transfection with an excess of cMyc-E6 equipped with localisation signals (compare Flag detection views in A, B and C).
Figure 9.
Detection of wt HPV18 E6 self-association in vivo using nucleo-cytoplasmic delocalization techniques. (A) Images of untransfected H1299 cells. (B–D) Localisation of Flag-E6, NLS-Myc-E6 and NES-Myc-E6 in H1299 cells transiently transfected: with: 0.7 μg of Flag-E6 and 2.3 μg of empty pXj vector (B); 2.3 μg of NLS-Myc-E6 and 0.7 μg of Flag-E6 (C) and 2.3 μg of NES-Myc-E6 and 0.7 μg of Flag-E6 (D).
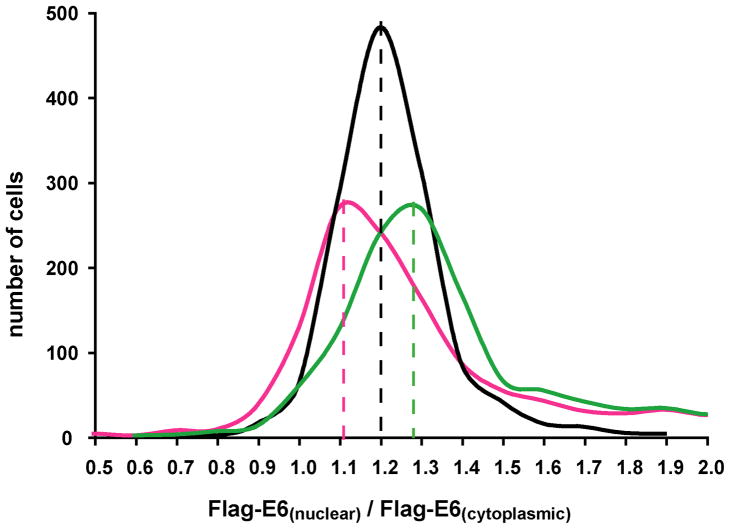
To avoid any bias due to the visual inspection of only a few transfected cells, we performed high throughput image analysis of a large number of cells. In each condition, 1320 transfected cells were automatically selected by the analysis program, and the Flag signal intensities in cytoplasm and in nucleus were quantified. For each individual cell, the ratio between the nuclear and cytoplasmic Flag signal intensities was calculated. The distribution of the values of this nuclear/cytoplasmic ratio for the population of transfected cells was then plotted for each experiment (Figure 10). Whereas the distribution of this ratio centers at a mean value of 1.2 for the cells transfected with Flag-E6 only, it shifts up to 1.3 (increased relative nuclear signal) for cells co-transfected with Flag-E6 and NLS-Myc-E6, and down to 1.1 (decreased relative nuclear signal) for cells co-transfected with Flag-E6 and NES-Myc-E6.
Figure 10.
Distribution of the ratios of nuclear/cytoplasmic Flag-E6 expression in the populations of H1299 transfected cells transfected with Flag-E6 alone (black line), NLS-Myc-E6 and Flag-E6 (green line) and NES-Myc-E6 and Flag-E6 (magenta line). Positions of the mean values for the three distribution curves are indicated by dashed lines. See also legend of Figure 9.
Altogether, these data indicate that differentially tagged E6 monomers interact with each other inside the cells, reinforcing the notion that E6 proteins self-associate in their natural in vivo environment.
DISCUSSION
E6 proteins share a common domain architecture but have variable surface properties 15. We have previously described soluble aggregates possessing miscellar structure that are formed upon biosynthesis by PV E6 proteins upon fusion to MBP 16; 19. Besides us, other authors also reported on the existence of oligomeric E6 forms of HPV16 and 18 E6 17; 18; 20. In this work we find that distantly related E6 proteins such as HPV16 E6 and BPV1 E6 form both soluble oligomers and multimeric ordered ribbon-like structures in vitro, which clearly highlights a conservation of self-association in the E6 protein family.
We found that E6 oligomers have in vitro properties very similar to the monomeric counterparts. E6 monomers, dimers and trimers all possess a native 2:1 Zn2+:protein stoichiometry expected from the E6 domain architecture, indicating that E6 oligomerization is not caused by loss of metal ion coordination. In addition, we found no detectable difference in the p53 degradation and E6AP binding activities of monomeric and dimeric HPV18 E6. We also observed that the MBP-E6 oligomers can slowly re-dissociate into the monomeric form upon dilution. The ensemble of these observations suggest that the small-size oligomers are formed by natively folded monomers engaging in reversible non-covalent interactions which do not alter the structural features required for their activities.
In this study we also observe that monomeric E6 proteins from different PV strains purified in native conditions can spontaneously assemble into large organized ribbon structures upon incubation at 20 °C for several days. FTIR experiments have shown that E6 self-association into these large multimers is accompanied by secondary structure changes involving a significant increase in the β-sheet content. However, the absence of thioflavin-T binding, which is in agreement with the findings of Garcia-Alai et al. 20 on spheroid multimers of HPV18 and HPV16 E6 refolded in acidic conditions, suggests that E6 multimers do not possess amyloid structure. At present we do not know whether the conformational change related to the increase in β-sheet takes place at the level of E6 oligomerization or upon formation of larger multimers. Unfortunately we cannot measure FTIR spectra on the different oligomers of HPV11 and 18 E6, since these species can only be purified as MBP-E6 fusions and cannot be separated from the MBP tag that would dominate the spectra. These E6 oligomeric species might act as intermediates in the processes leading to formation of larger multimers, as suggested by the concentration experiments described in this work. We have also observed that MBP-E6 soluble aggregates purified from bacterial extracts might serve as nuclei for the formation of small protofilaments. In a previous publication 19 we have proposed that E6 moieties in MBP-E6 soluble aggregates possess a native-like structure based on the observation of an expected 2:1 zinc:protein stoichiometry. It is therefore possible that MBP-E6 soluble aggregates and the small oligomers described here share the same zinc-binding domain structure and pattern of intermolecular interactions. However, at present we cannot exclude that the mechanisms of E6 assembly into larger multimeric structures are distinct from those leading to small size oligomers, involving, for example, covalent interactions such as disulfide bridging and/or inter-molecular cross-linking via zinc ions. Interestingly, we also observed fibrous structures in preparations of the isolated HPV16 E6-N and E6-C domains (data not shown), albeit at higher protein concentrations (300 μM), suggesting that the sequence and/or structure requirements for self-association are located on both of the E6 domains. However, our present data do not indicate whether it is a particular domain or the combination of both which drive the self-association of full-length E6 protein. Indeed, in a recent biophysical study of HPV 16 E6 and its isolated E6N and E6C domains, Liu et al. 26 have obtained data indicating that E6 aggregation is mediated more through the N-terminal domain than the C-terminal domain. Further structural work will be needed to identify more precisely the particular regions involved in self-association.
In the last part of our work we demonstrated E6 self-association in vivo in transiently transfected Hela cells. The assay used relies on the indirect nucleo-cytoplasmic delocalization of E6 as a result of association with another E6 molecule equipped with a signal peptide encoding for either nuclear localization or nuclear export. The finding, that E6 self-associates in vivo is partly in agreement with the previous work of Garcia-Alai et al. 20. Their data indicated that antibodies raised against preparations of oligomers of HPV16 and 18 E6 can detect large amounts of oligomeric endogenous E6 in immunofluorescence studies of HPV-positive Hela and CaSki cells. In the course of this work, we thoroughly investigated the detection of endogeneously expressed E6 in extracts of Hela and Caski cells performed in various conditions (Supplementary Material, Figure 4). These experiments showed that the amounts of extracted endogeneous E6 protein did not substantially change when the extraction was performed under native or denaturing conditions. Therefore, Hela and Caski cells do not seem to possess significant deposits of insoluble E6 multimers. However, this does not preclude that these cells contain soluble E6 oligomers such as those identified in the present in vitro studies.
What could be the functional role of E6 self-association? Here we show that p53 degradation and E6AP binding activity profiles are similar for monomeric and oligomeric E6 forms. On the other hand we have repeatedly observed a loss of both E6AP peptide binding and p53 degradation activity in recombinant E6 samples that were stored for several weeks (data not shown), which is likely to be related to the formation of larger E6 multimers. It would be interesting to understand what are the physical events leading to activity loss, whether it is simple steric hindrance in the larger complexes or real conformational changes which occur at more advanced stages of self-association. It is possible that binding to certain ligands may promote E6 self-association. Results by Medcaf and coworkers have shown that complexes of HPV16 E6 and p53 consist of p53 tetramers and E6 dimers 27. One intriguing hypothesis for the functional role of active E6 oligomers could be the simultaneous targeting of multiple cellular proteins, resulting in the coordination of various virus-host interaction pathways. For example, it is well known that E6 binds to several host proteins and targets them to degradation 28; 29. The best characterized E6-mediated degradation pathway is the one occurring via the 26S proteasome, which requires ubiquitination of the substrate by the E6AP ligase. A single oligomer could bind at the same time to different cellular proteins which target either the same or different E6 sites (such as cellular proteins containing the LxxφLsh motif 30 or PDZ domains 31). Consequently a single ubiquitination event on any of the substrates would be sufficient to target all these proteins simultaneously to the 26S proteasome. Such mechanism would increase the efficiency of the E6 proteins and therefore enhance their oncogenic potential.
MATERIALS AND METHODS
Expression and purification of HPV E6 proteins
The wild-type sequences of the E6 proteins issued from HPV11, HPV16, HPV18 and BPV1 viral strains were cloned in the NcoI and KpnI sites of the pETM-41 vector (kindly provided by G. Stier, EMBL, Heidelberg, Germany). The HPV5 5C/5S, HPV11 E6 4C/4S, HPV16 E6 6C/6S, HPV33 2C/2S, HPV52 1C/1S and HPV58 1C/1S constructs have nonconserved cysteines mutated into serinesand have been previously described 14; 19; 32. These latter constructs have also been cloned in the pETM-41 vector.
MBP-E6 and E6 protein samples used in this work were over-expressed in E. Coli BL21 DE3 by low temperature induction at 15°C for 17 hours. MBP-E6 fusion proteins were purified using amylose affinity chromatography and separated from soluble aggregates by extensive ultracentrifugation as described previously 19. Buffer A (50 mM Tris pH 6.8, 400 mM NaCl and 2 mM DTT) was used throughout the purification. Preparations of monomeric MPB-E6 and E6 samples were obtained by further fractionation on a Superdex 75 16/60 gel filtration column equilibrated in buffer A. The individual oligomeric forms of MBP-E6 were instead obtained from fractionation on a Superdex 200 16/60 column equilibrated in buffer A as described in the text.
Concentrations were determined by absorbance at 280 nm based on the calculated extinction coefficient of the monomeric form of the protein.
Analytical gel filtration chromatography
Sample volumes of 100 or 500 μl were injected on Superdex200 10/30 and/or Superdex75 10/30 columns equilibrated in buffer A. The experiments were performed using an AKTA Purifier System (GE Healthcare) maintained at 10 °C.
Analytical ultracentrifugation
Sedimentation velocity experiments were done at 4 °C using 2-channel charcoal centerpices and speed 46 000 rpm in a Beckman Optima XL-A centrifuge fitted with a four-hole AN-60 rotor. Sedimentation velocity profiles were collected by monitoring the absorbance signal at 280 nm in buffer A. Sedimentation coefficient and molecular weight distributions were analysed by the C(s) method implemented in the Sedfit software package 33. Buffer density and viscosity corrections were made according to published data 34.
Determination of Zn2+ content (TSQ assay)
Zn2+ concentrations in MBP-E6 samples were determined monitoring changes in fluorescence of the zinc selective fluorophore TSQ ((N-6-methoxy-8-quinolyl)-p-toluenesulfonamide) (Molecular Probes, Eugene, OR) according to published protocols 17 with minor modifications described in detail elsewhere 19.
p53 degradation assays
p53 degradation assays were carried out as described previously 35. Briefly, p53 proteins were synthesized in vitro using the TNT T7/SP6 reticulocyte lysate transcription/translation system (Promega). The p53 translation product was incubated with wild-type HPV18 MBP-E6 concentrations between 400 and 25 nM in 25 mM Tris-HCl pH 7.5, 100 mM NaCl and 2 mM DTT, at 29 °C for 1 hour. Control reactions were made by replacing the MBP-E6 with equivalent volumes of Buffer A. Reactions were stopped by addition of SDS-loading buffer, subjected to SDS-PAGE and analysed by autoradiography.
BIAcore experiments
Experiments were performed at 25 °C on a Biacore 2000 instrument (Biacore). All experiments were carried out using the protocols described previously 22. In brief, the minimal E6 binding site of E6AP (i.e. ESSELTLQELLGEER) was expressed fused to GST and captured onto the Biacore’s surface via a polyclonal anti-GST antibody. Subsequently, the analyte wild-type HPV18 E6 protein was injected and the interaction with the E6AP peptide monitored. A negative control GST-fusion peptide (peptide sequence: GNSGNSGNS) was used to correct the binding sensorgrams for buffer effects and non-specific binding.
Electron microscopy
Monomeric HPV16 E6 6C/6S was purified as described previously 22. Monomeric MBP-E6 fusions were isolated by gel-filtration as described above. MBP-E6 preparations containing spherical aggregates (“S.A.”) were prepared by a single step of amylose resin as previously described 19. All samples were incubated at 20 °C for one week. During this period, freshly prepared DTT was added to the samples every 24 hours to maintain reducing conditions and avoid disulfide cross-bridging. Aliquots were taken at time intervals of 0, 1/2, 3 and 7 days, and diluted to a final concentration of 30 ng/μl in buffer A. 5ul were deposited on an air glow-discharged carbon-coated grid. After 2 min adsorption samples were negatively stained with a 2% (w/v) uranyl acetate solution. Images were acquired on a Philips CM120 transmission electron microscope operating at 100 kV with a LaB6 filament. Areas covered with individual molecules were recorded under low dose condition, at a magnification of x 45.000 on a Pelletier cooled CCD camera (Model 794, Gatan, Pleasanton, CA).
FT-IR
40 μM preparations of monomeric HPV16 E6 6C/6S in 20 mM phosphate buffer (pH 6.8), 400 NaCl (unless otherwise stated), 2 mM DTT were incubated at 20 °C for a period of up to 30 days. Middle infrared measurements were carried out at room temperature on a Vertex 70 (Bruker) Fourier transform spectrometer equipped with a globar source, a LN-MCT Photovoltaic detector, a KBr beam splitter and an ATR cell (Harrick) fitted with a diamond crystal (6 mm2) as the reflecting element. The spectrometer compartments were purged with dry air to reduce interference with H2O and CO2. At the different time points, 2–3 μl of sample solution were deposited on the ATR crystal and allowed to dry at room temperature for 10 minutes. For each spectrum, a resolution of 2 cm−1 was chosen and 256 scans were accumulated. Residual water in the gas phase was subtracted and all spectra were smoothed with 9 points. The data are presented in terms of arbitrary units. We noticed that sample concentration decreases during incubation due to adherence to the eppendorf’s walls. However the relative ratio of each structural element is unaffected and secondary structure analysis was performed on the Amide I band (spectral range 1700/1600 cm−1) using PeakFit software for deconvolution. The fitting procedure performed has an error of approximately 3% percent as discussed in the results section.
Cell transfections and immunofluorescence assays
Plasmid constructions. DNA fragments were inserted into the pXj vector 36 allowing the expression of proteins under hCMV promoter. The E6 ORF of HPV type 18 was modified at its splicing donor site to prevent the formation of the truncated form of E6 namely E6* 37. Three following wild-type HPV 18 E6 fusion constructs were generated: Flag-E6, NLS-cMyc-E6 and NES-cMyc-E6. The amino acid sequences for NLS and NES sequences used are as described by 25.
Exponentially growing H1299 cells were transfected lipofectamine (Invitrogen) with the different plasmids. The following transfections were performed: i) 0.7 μg of Flag-18 E6 and 2.3 μg of empty pXj vector; ii) 2.3 μg of NLS-Myc-18 E6 and 0.7 μg of Flag-18 E6; iii) 2.3 μg of NES-Myc-18 E6 and 0.7 μg of Flag-18 E6. Transfected cells were let grow for 24 hours. Hence cells were washed with PBS, fixed in PFA 3% for 15 min and permeabilised by PBS supplemented by 0.1% TritonX100. Upon fixation, the cells were incubated with the anti-FLAG rabbit polyclonal antibody F7425 (Sigma) diluted in PBS for 1 hour at room temperature, then with the anti-Myc mouse monoclonal antibody 9E10 (kindly provided by Dr. M. Oulad, IGBMC, Strasbourg) subsequently with Alexa Fluor 568-conjugated anti-rabbit goat immunoglobulin and Alexa Fluor 488-conjugated anti-mouse goat immunoglobulin (Invitrogen) and finally with DAPI (Sigma). After extensive washing with PBS, cells were dried and mounted with Fluoromount (Southern Biotech) on a slide. The cells were then examined with a Zeiss Axioplan Fluorescence microscope equipped with an Olympus DP50 camera. Images were collected with Zeiss 40X plan-neofluar objectives.
High throughput image analysis of nucleo-cytoplasmic translocation assay
Image acquisition of nuclei (DAPI staining), Myc (A488 labelling) and Flag epitopes (Cy3 labelling) were performed using the automated cell imaging device IN Cell Analyzer 1000 from GE Healthcare Life Sciences. An average of approximately 1600 cells per experimental condition were analyzed and individual cellular parameters covering object labelling, object intensities, local background were determined for each cell using the cellular segmentation tool “Multi Target Analysis module” of the IN Cell Investigator high content analysis software v1.3 (GE Healthcare Life Sciences). Filters were then applied to extract the subpopulation of cells harbouring either the Flag labelling in the control experiment (Flag-E6) or the dual Flag and Myc labelling in the nucleo-cytoplasmic translocation assays (Flag-E6 and NLS-Myc-E6; Flag-E6 and NES-Myc-E6). From these three population sets, 1320 cells were blindly selected and their respective Nucleo-Cytoplasmic ratio of Flag labelling were determined and their distributions compared side by side. Plots of the distribution of the values of this ratio through the entire cell population allow to visualise the Nucleo-Cytoplasmic distribution adopted by the largest amount of transfected cells in each condition (Figure 10). Noteworthy, in the doubly transfected Flag-E6 plus cMyc-E6 experiments, 12 to 14 % of the cells had abnormally high nuclear/cytoplasmic Flag-E6 ratio (between 2 and 7). Visual inspection suggested that such cells have a condensed nuclear appearance most probably due to apoptotic processes, which are likely to be provoked by the high levels of expression of the E6 protein.
Supplementary Material
Acknowledgments
We thank Prof. G. Orth (Institut Pasteur, Paris) for providing the plasmids containing the HPV genomes, Dr. Johannes Schweizer (Arbor Vita Corporation, Sunnyvale, CA) for providing the anti-HPV18 E6 antibodies, and Sadek Fournane (ESBS, Strasbourg), Dr. Dymalla Susanne (DKFZ, Heidelberg) and Dr. Raimon Sabate (Universitat Autonoma de Barcelona, Barcelona) for useful comments and suggestions. We grateful to Anne Chappelle for technical support. K.Z. was supported by a fellowship from the Fondation pour la Recherche Medicale (FRM). A.O.M was supported by a fellowship from ARC. This work was funded by the following organizations: Arbor Vita Corporation (Sunnyvale, CA), Association pour la Recherche sur le Cancer (ARC), Institut National de la Santé et de la Recherche Médicale, Centre National pour la Recherche Scientifique (CNRS) and Université Louis Pasteur (Strasbourg).
Abbreviations
- MBP
maltose binding protein
- HPV
human papillomavirus
- BPV
bovine papillomavirus
- TEV
tobacco etch virus
- SPR
surface plasmon resonance
- RU
resonance units
- EM
electron microscopy
- FTIR
Fourier Transform Infrared Spectroscopy
References
- 1.zur Hausen H. Papillomaviruses in human cancers. Proc Assoc Am Physicians. 1999;111:581–587. doi: 10.1046/j.1525-1381.1999.99723.x. [DOI] [PubMed] [Google Scholar]
- 2.Bosch FX, Manos MM, Munoz N, Sherman M, Jansen AM, Peto J, Schiffman MH, Moreno V, Kurman R, Shah KV. Prevalence of human papillomavirus in cervical cancer: a worldwide perspective. International biological study on cervical cancer (IBSCC) Study Group. J Natl Cancer Inst. 1995;87:796–802. doi: 10.1093/jnci/87.11.796. [DOI] [PubMed] [Google Scholar]
- 3.Pfister H. Chapter 8: Human papillomavirus and skin cancer. J Natl Cancer Inst Monogr. 2003;31:52–56. doi: 10.1093/oxfordjournals.jncimonographs.a003483. [DOI] [PubMed] [Google Scholar]
- 4.Lambert PF, Baker CC, Howley PM. The genetics of bovine papillomavirus type 1. Annu Rev Genet. 1988;22:235–58. doi: 10.1146/annurev.ge.22.120188.001315. [DOI] [PubMed] [Google Scholar]
- 5.Narisawa-Saito M, Kiyono T. Basic mechanisms of high-risk human papillomavirus-induced carcinogenesis: roles of E6 and E7 proteins. Cancer Sci. 2007;98:1505–11. doi: 10.1111/j.1349-7006.2007.00546.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scheffner M, Huibregtse JM, Vierstra RD, Howley PM. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- 7.Werness BA, Levine AJ, Howley PM. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science. 1990;248:76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- 8.Kiyono T, Hiraiwa A, Fujita M, Hayashi Y, Akiyama T, Ishibashi M. Binding of high-risk human papillomavirus E6 oncoproteins to the human homologue of the Drosophila discs large tumor suppressor protein. Proc Natl Acad Sci U S A. 1997;94:11612–11616. doi: 10.1073/pnas.94.21.11612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glaunsinger B, Lee S, Thomas M, Banks L, Javier R. Interactions of the PDZ-protein MAGI-1 with adenovirus E4-ORF1 and high-risk papillomavirus E6 oncoproteins. Oncogene. 2000;19:5270–5280. doi: 10.1038/sj.onc.1203906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomas M, Laura R, Hepner K, Guccione E, Sawyers C, Lasky L, Banks L. Oncogenic human papillomavirus E6 proteins target the MAGI-2 and MAGI-3 proteins for degradation. Oncogene. 2002;21:5088–5096. doi: 10.1038/sj.onc.1205668. [DOI] [PubMed] [Google Scholar]
- 11.Jackson S, Harwood C, Thomas M, Banks L, Storey A. Role of Bak in UV-induced apoptosis in skin cancer and abrogation by HPV E6 proteins. Genes Dev. 2000;14:3065–73. doi: 10.1101/gad.182100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tong X, Howley PM. The bovine papillomavirus E6 oncoprotein interacts with paxillin and disrupts the actin cytoskeleton. Proc Natl Acad Sci U S A. 1997;94:4412–4417. doi: 10.1073/pnas.94.9.4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tong X, Boll W, Kirchhausen T, Howley PM. Interaction of the bovine papillomavirus E6 protein with the clathrin adaptor complex AP-1. J Virol. 1998;72:476–482. doi: 10.1128/jvi.72.1.476-482.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nominé Y, Charbonnier S, Ristriani T, Stier G, Masson M, Cavusoglu N, Van Dorsselaer A, Weiss E, Kieffer B, Travé G. Domain substructure of HPV E6 protein: biophysical characterization of E6 C-terminal DNA-binding domain. Biochemistry. 2003;42:4909–4917. doi: 10.1021/bi026980c. [DOI] [PubMed] [Google Scholar]
- 15.Nomine Y, Masson M, Charbonnier S, Zanier K, Ristriani T, Deryckere F, Sibler AP, Desplancq D, Atkinson RA, Weiss E, Orfanoudakis G, Kieffer B, Trave G. Structural and functional analysis of E6 oncoprotein: insights in the molecular pathways of human papillomavirus-mediated pathogenesis. Mol Cell. 2006;21:665–78. doi: 10.1016/j.molcel.2006.01.024. [DOI] [PubMed] [Google Scholar]
- 16.Nominé Y, Ristriani T, Laurent C, Lefèvre JF, Weiss E, Travé G. Formation of soluble inclusion bodies by HPV E6 oncoprotein fused to Maltose-binding protein. Protein Expr Purif. 2001;23:22–32. doi: 10.1006/prep.2001.1451. [DOI] [PubMed] [Google Scholar]
- 17.Degenkolbe R, Gilligan P, Gupta S, Bernard HU. Chelating agents stabilize the monomeric state of the zinc binding huma, papillomavirus 16 E6 oncoprotein. Biochemistry. 2003;42:3868–3873. doi: 10.1021/bi027390h. [DOI] [PubMed] [Google Scholar]
- 18.Daniels PR, Sanders CM, Coulson P, Maitland NJ. Molecular analysis of the interaction between HPV type 16 E6 and human E6-associated protein. FEBS Letters. 1997;416:6–10. doi: 10.1016/s0014-5793(97)01160-5. [DOI] [PubMed] [Google Scholar]
- 19.Zanier K, Nomine Y, Charbonnier S, Ruhlmann C, Schultz P, Schweizer J, Trave G. Formation of well-defined soluble aggregates upon fusion to MBP is a generic property of E6 proteins from various human papillomavirus species. Protein Expr Purif. 2007;51:59–70. doi: 10.1016/j.pep.2006.07.029. [DOI] [PubMed] [Google Scholar]
- 20.Garcia-Alai MM, Dantur KI, Smal C, Pietrasanta L, de Prat-Gay G. High-risk HPV E6 oncoproteins assemble into large oligomers that allow localization of endogenous species in prototypic HPV-transformed cell lines. Biochemistry. 2007;46:341–9. doi: 10.1021/bi602457q. [DOI] [PubMed] [Google Scholar]
- 21.Bohl J, Das K, Dasgupta B, Vande Pol SB. Competitive binding to a charged leucine motif represses transformation by a papillomavirus E6 oncoprotein. Virology. 2000;271:163–170. doi: 10.1006/viro.2000.0316. [DOI] [PubMed] [Google Scholar]
- 22.Zanier K, Charbonnier S, Baltzinger M, Nomine Y, Altschuh D, Trave G. Kinetic analysis of the interactions of Human Papillomavirus E6 oncoproteins with the ubiquitin ligase E6AP using Surface Plasmon Resonance. J Mol Biol. 2005;349:401–412. doi: 10.1016/j.jmb.2005.03.071. [DOI] [PubMed] [Google Scholar]
- 23.Nilsson MR. Techniques to study amyloid fibril formation in vitro. Methods. 2004;34:151–60. doi: 10.1016/j.ymeth.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 24.Lipari F, McGibbon GA, Wardrop E, Cordingley MG. Purification and biophysical characterization of a minimal functional domain and of an N-terminal Zn2+-binding fragment from the human papillomavirus type 16 E6 protein. Biochemistry. 2001;40:1196–204. doi: 10.1021/bi001837+. [DOI] [PubMed] [Google Scholar]
- 25.Sibler AP, Nordhammer A, Masson M, Martineau P, Trave G, Weiss E. Nucleocytoplasmic shuttling of antigen in mammalian cells conferred by a soluble versus insoluble single-chain antibody fragment equipped with import/export signals. Exp Cell Res. 2003;286:276–87. doi: 10.1016/s0014-4827(03)00093-4. [DOI] [PubMed] [Google Scholar]
- 26.Liu Y, Cherry JJ, Dineen JV, Androphy EJ, Baleja JD. Determinants of Stability for the E6 Protein of Papillomavirus Type 16. J Mol Biol. 2009;386:1123–1137. doi: 10.1016/j.jmb.2009.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Medcalf EA, Milner J. Targeting and degradation of p53 by E6 of human papillomavirus type 16 is preferential for the 1620+ p53 conformation. Oncogene. 1993;8:2847–51. [PubMed] [Google Scholar]
- 28.Liu Y, Baleja JD. Structure and function of the papillomavirus E6 protein and its interacting proteins. Front Biosci. 2008;13:121–34. doi: 10.2741/2664. [DOI] [PubMed] [Google Scholar]
- 29.Tungteakkhun SS, Duerksen-Hughes PJ. Cellular binding partners of the human papillomavirus E6 protein. Arch Virol. 2008;153:397–408. doi: 10.1007/s00705-007-0022-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen JJ, Hong Y, Rustamzadeh E, Baleja JD, Androphy EJ. Identification of an alpha helical motif sufficient for association with papillomavirus E6. J Biol Chem. 1998;273:13537–13544. doi: 10.1074/jbc.273.22.13537. [DOI] [PubMed] [Google Scholar]
- 31.Thomas M, Dasgupta J, Zhang Y, Chen X, Banks L. Analysis of specificity determinants in the interactions of different HPV E6 proteins with their PDZ domain-containing substrates. Virology. 2008;376:371–8. doi: 10.1016/j.virol.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 32.Nominé Y, Ristriani T, Laurent C, Lefèvre JF, Weiss E, Travé G. A strategy for optimizing the monodispersity of fusion proteins: application to purification of recombinant HPV E6 oncoprotein. Protein Eng. 2001;14:297–305. doi: 10.1093/protein/14.4.297. [DOI] [PubMed] [Google Scholar]
- 33.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J. 2000;78:1606–19. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laue T, Shah B, Ridgeway T, Pelletier S. Analytical Ultracentrifugation in Biochemistry and Polymer Science. The Royal Society of Chemistry; Cambridge: 1992. pp. 90–125. [Google Scholar]
- 35.Ristriani T, Nomine YCL, Weiss E, Trave G. Protein mutagenesis with monodispersity-based quality probing: selective inactivation of p53 degradation and DNA-binding properties of HPV E6 oncoprotein. Protein Expr Purif. 2002;26:357–367. doi: 10.1016/s1046-5928(02)00570-3. [DOI] [PubMed] [Google Scholar]
- 36.Xiao JH, Davidson I, Matthes H, Garnier JM, Chambon P. Cloning, expression, and transcriptional properties of the human enhancer factor TEF-1. Cell. 1991;65:551–68. doi: 10.1016/0092-8674(91)90088-g. [DOI] [PubMed] [Google Scholar]
- 37.Pim D, Massimi P, Banks L. Alternatively spliced HPV-18 E6* protein inhibits E6 mediated degradation of p53 and suppresses transformed cell growth. Oncogene. 1997;15:257–64. doi: 10.1038/sj.onc.1201202. [DOI] [PubMed] [Google Scholar]
- 38.Barth A. Infrared spectroscopy of proteins. Biochim Biophys Acta. 2007;1767:1073–101. doi: 10.1016/j.bbabio.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 39.Lagrange M, Charbonnier S, Orfanoudakis G, Robinson P, Zanier K, Masson M, Lutz Y, Travé G, Weiss E, Deryckere F. Binding of HPV16 E6 to p53 and E6AP is impaired by monoclonal antibodies directed against the second zinc binding domain of E6. J Gen Virol. 2005;86:1001–7. doi: 10.1099/vir.0.80607-0. [DOI] [PubMed] [Google Scholar]
- 40.Giovane C, Trave G, Briones A, Lutz Y, Wasylyk B, Weiss E. Targetting of the N-terminal domain of the human papillomavirus type 16 E6 oncoprotein with monomeric ScFvs blockes the E6-mediated degradation of cellular p53. Journal of Molecular Recognition. 1999;12:141–152. doi: 10.1002/(SICI)1099-1352(199903/04)12:2<141::AID-JMR453>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.