Abstract
In the presence of O2 and 5 mM dithiothreitol, penta-N-methylpyrrolecarboxamide-EDTA X Fe(II) [P5E X Fe(II)] at 0.5 microM cleaves pBR322 plasmid DNA (50 microM in base pairs) on opposite strands to afford discrete DNA fragments as analyzed by agarose gel electrophoresis. High-resolution denaturing gel electrophoresis of a 32P-end-labeled 517-base-pair restriction fragment containing a major cleavage site reveals that P5E X Fe(II) cleaves 3-5 base pairs contiguous to a 6-base-pair sequence, 5'-T-T-T-T-T-A-3' (4,323-4,328 base pairs). The major binding orientation of the pentapeptide occurs with the amino terminus at the adenine side of this sequence. In the presence of 5 mM dithiothreitol, 0.01 microM P5E X Fe(II) converts form I pBR322 DNA at 0.22 microM plasmid (1.0 mM in base pairs) to 40% form II, indicating the cleavage reaction is catalytic, turning over a minimum of nine times. This synthetic molecule achieves double-strand cleavage of DNA (pH 7.9, 25 degrees C) at the 6-base-pair recognition level and may provide an approach to the design of "artificial restriction enzymes".
Full text
PDF
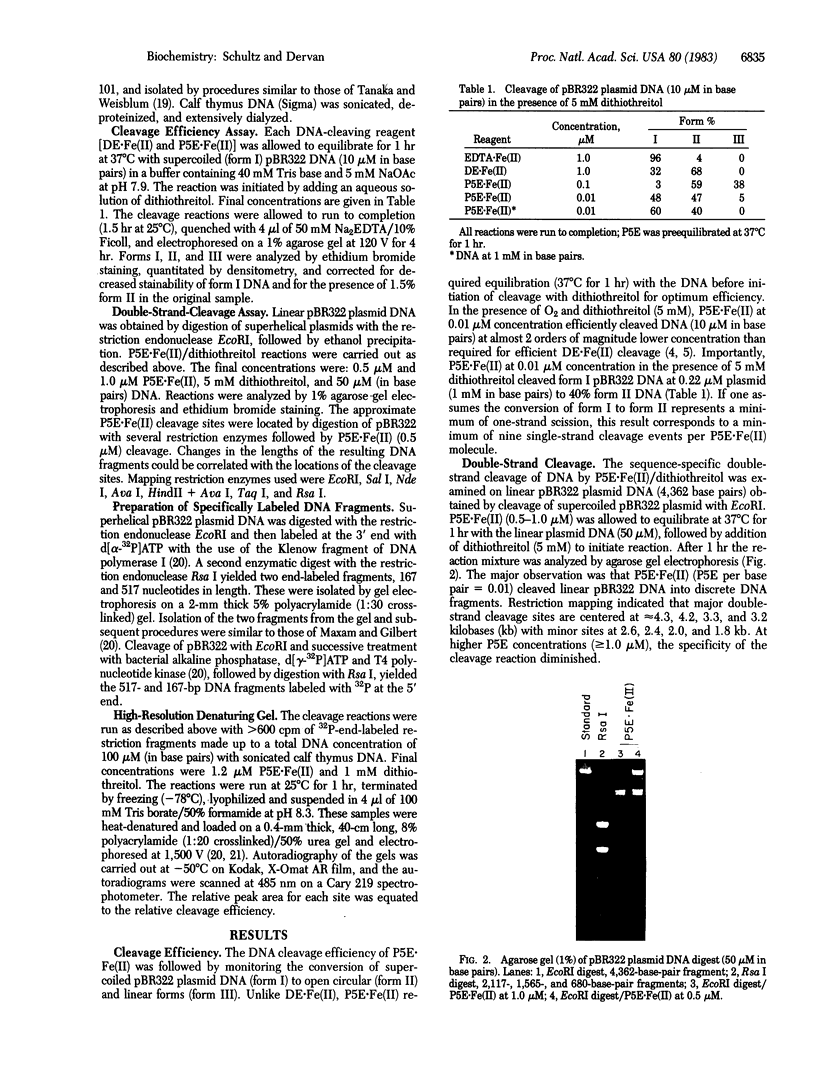


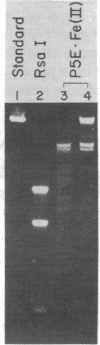
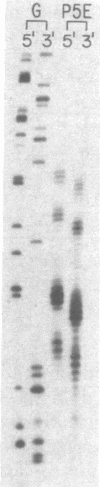
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berman H. M., Neidle S., Zimmer C., Thrum H. Netropsin, a DNA-binding oligopeptide structural and binding studies. Biochim Biophys Acta. 1979 Jan 26;561(1):124–131. doi: 10.1016/0005-2787(79)90496-9. [DOI] [PubMed] [Google Scholar]
- Giloni L., Takeshita M., Johnson F., Iden C., Grollman A. P. Bleomycin-induced strand-scission of DNA. Mechanism of deoxyribose cleavage. J Biol Chem. 1981 Aug 25;256(16):8608–8615. [PubMed] [Google Scholar]
- Krylov A. S., Grokhovsky S. L., Zasedatelev A. S., Zhuze A. L., Gursky G. V., Gottikh B. P. Quantitative estimation of the contribution of pyrrolcarboxamide groups of the antibiotic distamycin A into specificity of its binding to DNA AT pairs. Nucleic Acids Res. 1979 Jan;6(1):289–304. doi: 10.1093/nar/6.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck G., Zimmer C., Reinert K. E., Arcamone F. Specific interactions of distamycin A and its analogs with (A-T) rich and (G-C) rich duplex regions of DNA and deoxypolynucleotides. Nucleic Acids Res. 1977 Aug;4(8):2655–2670. doi: 10.1093/nar/4.8.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marky L. A., Blumenfeld K. S., Breslauer K. J. Calorimetric and spectroscopic investigation of drug-DNA interactions. I. The binding of netropsin to poly d(AT). Nucleic Acids Res. 1983 May 11;11(9):2857–2870. doi: 10.1093/nar/11.9.2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Modrich P. Studies on sequence recognition by type II restriction and modification enzymes. CRC Crit Rev Biochem. 1982;13(3):287–323. doi: 10.3109/10409238209114231. [DOI] [PubMed] [Google Scholar]
- Nosikov V. V., Sain B. Protection of particular endonuclease R. Hind III cleavage sites by distamycin A, propyl-distamycin and netropsin. Nucleic Acids Res. 1977 Jul;4(7):2263–2273. doi: 10.1093/nar/4.7.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinert K. E. Adenosine-thymidine cluster-specific elongation and stiffening of DNA induced by the oligopeptide antibiotic netropsin. J Mol Biol. 1972 Dec 30;72(3):593–607. doi: 10.1016/0022-2836(72)90178-7. [DOI] [PubMed] [Google Scholar]
- Roberts R. J. Restriction and modification enzymes and their recognition sequences. Nucleic Acids Res. 1983 Jan 11;11(1):r135–r167. [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol Biol. 1975 May 25;94(3):441–448. doi: 10.1016/0022-2836(75)90213-2. [DOI] [PubMed] [Google Scholar]
- Smith H. O. Nucleotide sequence specificity of restriction endonucleases. Science. 1979 Aug 3;205(4405):455–462. doi: 10.1126/science.377492. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Weisblum B. Construction of a colicin E1-R factor composite plasmid in vitro: means for amplification of deoxyribonucleic acid. J Bacteriol. 1975 Jan;121(1):354–362. doi: 10.1128/jb.121.1.354-362.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyke M. W., Dervan P. B. Footprinting with MPE.Fe(II). Complementary-strand analyses of distamycin- and actinomycin-binding sites on heterogeneous DNA. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):347–353. doi: 10.1101/sqb.1983.047.01.040. [DOI] [PubMed] [Google Scholar]
- Van Dyke M. W., Hertzberg R. P., Dervan P. B. Map of distamycin, netropsin, and actinomycin binding sites on heterogeneous DNA: DNA cleavage-inhibition patterns with methidiumpropyl-EDTA.Fe(II). Proc Natl Acad Sci U S A. 1982 Sep;79(18):5470–5474. doi: 10.1073/pnas.79.18.5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wartell R. M., Larson J. E., Wells R. D. Netropsin. A specific probe for A-T regions of duplex deoxyribonucleic acid. J Biol Chem. 1974 Nov 10;249(21):6719–6731. [PubMed] [Google Scholar]
- Zimmer C. Effects of the antibiotics netropsin and distamycin A on the structure and function of nucleic acids. Prog Nucleic Acid Res Mol Biol. 1975;15(0):285–318. doi: 10.1016/s0079-6603(08)60122-1. [DOI] [PubMed] [Google Scholar]
- Zimmer C., Luck G., Thrum H., Pitra C. Binding of analogues of the antibiotics distamycin A and netropsin to native DNA. Effect of chomophore systems and basic residues of the oligopeptides on thermal stability, conformation and template activity of the DNA complexes. Eur J Biochem. 1972 Mar 15;26(1):81–89. doi: 10.1111/j.1432-1033.1972.tb01742.x. [DOI] [PubMed] [Google Scholar]