Abstract
Genome-wide association studies (GWAS) have uncovered many genetic associations for cardiovascular disease (CVD). However, data are limited regarding causal genetic variants within implicated loci. We sought to identify regulatory variants (cis- and trans-eQTLs) affecting expression levels of 93 genes selected by their proximity to SNPs with significant associations in prior GWAS for CVD traits. Expression levels were measured by qRT–PCR in leukocytes from 1846 Framingham Heart Study participants. An additive genetic model was applied to 2.5 million imputed SNPs for each gene. Approximately 45% of genes (N = 38) harbored at least one cis-eSNP after a regional multiple-test adjustment. Applying a more rigorous significance threshold (P < 5 × 10−8), we found the expression level of 10 genes was significantly associated with more than one cis-eSNP. The top cis-eSNPs for 7 of these 10 genes exhibited moderate-to-strong association with ≥1 CVD clinical phenotypes. Several eSNPs or proxy SNPs (r2 = 1) were replicated by other eQTL studies. After adjusting for the lead GWAS SNPs for the 10 genes, expression variances explained by top cis-eSNPs were attenuated markedly for LPL, FADS2 and C6orf184, suggesting a shared genetic basis for the GWAS and expression trait. A significant association between cis-eSNPs, gene expression and lipid levels was discovered for LPL and C6orf184. In conclusion, strong cis-acting variants are localized within nearly half of the GWAS loci studied, with particularly strong evidence for a regulatory role of the top GWAS SNP for expression of LPL, FADS2 and C6orf184.
INTRODUCTION
Cardiovascular disease (CVD), particularly coronary artery disease (CAD) and stroke, is the leading cause of death in the United States (1) and other industrialized countries. Atherothrombosis is the main pathophysiology underlying CVD, and the burden of subclinical atherosclerosis in major arteries such as the coronary and carotid arteries predicts future risk of CVD (2,3). Beyond well-established traditional atherosclerosis risk factors, such as elevated levels of LDL cholesterol and blood pressure, there is evidence that biomarkers of hemostasis and thrombosis as well as blood cell counts may be important risk markers. For example, elevated levels of coagulation factors including fibrinogen, factor VII (FVII), factor VIII (FVIII) and von Willebrand Factor (vWF) are associated with increased risk of thrombosis and of CVD (4–6). Variation in the blood-group antigen ABO is a known risk factor for venous thrombosis and may have a role in clinically apparent CAD (7). Erythrocyte disorders such as anemia and erythrocytosis are associated with hypertension and other CVDs (8), and variation in erythrocyte measures even within normal ranges has been related to mortality and prognosis in clinically apparent CAD (9). Genome-wide association studies (GWAS) have uncovered many genetic loci that are strongly associated with CVD traits (10), including clinically apparent CAD (11–13), subclinical atherosclerosis, traditional risk factors such as lipids, and blood biomarkers of hemostasis/thrombosis and hematology (8–10,14–20).
However, despite the abundance of SNP association findings, data remain sparse regarding causal genes and genetic variants within implicated loci, limiting the rapid translation of genetic findings into clinical applications. Variants discovered in GWAS, or nearby variants in linkage disequilibrium (LD), may act via various regulatory mechanisms that act transcriptionally or post-transcriptionally (21). Recently, expression quantitative trait locus (eQTL) studies in a variety of cells and tissues have identified genetic variants that strongly affect gene regulation and are often reproduced across tissues and datasets (22–24). However, aside from eQTL studies in monocytes (25) and whole blood (26), there are limited additional studies of eQTLs for CVD and its risk factors, particularly within samples of single large population.
Our study objective was to identify genetic regulatory mechanisms that may contribute to biological pathways underlying GWAS associations with CVD phenotypes. For this objective, we selected 93 genes for gene expression profiling based upon their proximity to SNPs with strong and significant associations in prior GWAS for CAD (11), subclinical atherosclerosis (20), blood lipids (27) and other blood risk factors (9,18). We used quantitative reverse-transcriptase polymerase chain reaction (qRT–PCR) to measure expression levels in leukocytes collected from 1846 Framingham Heart Study (FHS) Offspring participants. After excluding 10 genes with low levels of expression, we further conducted GWAS analysis for each of 83 genes to assess cis and trans regulatory variants affecting gene expression. Additionally, we assessed the overlap of significant associations with prior GWAS signals and known eQTLs and examined the association of gene expression with clinical lipid traits.
RESULTS
Gene expression measurements of genes at loci for clinically apparent CAD, subclinical atherosclerosis and blood risk factors
We identified a total of 93 genes from top loci in published GWAS studies associated with CAD and blood risk factor traits including lipid levels, hemostatic factors and red blood cell (RBC) indices. In addition, three housekeeping genes were measured (B2M, GAPDH and ACTB) for normalization purposes, as previously described (28). The mean gene expression levels (cycle thresholds) for all measured genes are shown in Supplementary Material, Table S1 along with a description of the rationale for gene selection. The majority of genes targeted (83/93 = 90%) showed moderate-to-high expression levels in large numbers of leukocyte samples, with a few exceptions (N = 10), namely ADH4, LPA, APOA4, APOA5, APOC4, APOB, EDNRA, FGB, HNF1A and PHACTR1. After excluding these 10 genes with low levels of expression, a total of 83 non-housekeeping genes were available for subsequent analysis.
cis-eQTL associations
Applying a regionally adjusted P-value based on the effective number of independent tests at each locus (29), we found that the expression levels of 38 genes are associated with at least one cis-eSNP as shown in Table 1. Greater than 45% of genes (38/83) harbor at least one cis-eSNP that survives a regional multiple-test correction. The top cis-eQTL results for other 45 genes not passing this threshold are shown in Supplementary Material, Table S2.
Table 1.
cis-eSNPs results for 38 genes with evidence for a significant association
| Gene | Clinical phenotype | #SNP < 100 kb | #Eff SNPs | Top 1 cis-eSNP | C hr | Physical location | MAF | Major > minor | Beta (s.e.) | P, cis-eSNP | Distance from gene (bp) | Variance explained by top cis-eSNP (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ABOa | Hemostatic, CAD, others | 190 | 40 | rs8176731 | 9 | 135122171 | 0.36 | T>C | −1.43 (0.07) | 1.12 × 10−91 | intron | 17.73 |
| FADS2a | Lipid, CAD | 144 | 44 | rs968567 | 11 | 61352140 | 0.15 | C>T | −1.53 (0.08) | 5.72 × 10−73 | −149 | 6.85 |
| LPLa | Lipid, CAD | 281 | 41 | rs6993414 | 8 | 19947198 | 0.10 | A>G | −1.83 (0.12) | 1.21 × 10−52 | +78148 | 5.63 |
| APOC2a | Lipid, CAD | 85 | 31 | rs2288912 | 19 | 50141039 | 0.45 | C>G | 0.82 (0.07) | 1.98 × 10−36 | −44 | 7.33 |
| FNTBa | RBC traits | 218 | 35 | rs7148144 | 14 | 64569203 | 0.07 | G>A | −1.00 (0.10) | 1.90 × 10−22 | intron | 1.49 |
| CDKN2Ba | CAD, CAC | 151 | 30 | rs598664 | 9 | 22017551 | 0.11 | T>C | 0.50 (0.07) | 1.86 × 10−13 | +18239 | 1.09 |
| FAM117Ba | CAD | 93 | 22 | rs1971739 | 2 | 203192869 | 0.30 | C>G | 0.32 (0.05) | 2.93 × 10−11 | −15277 | 0.35 |
| C6orf184a | CAD | 250 | 30 | rs7773213 | 6 | 109708578 | 0.53 | C>T | −0.36 (0.06) | 7.07 × 10−11 | −13621 | 0.88 |
| CDKN2BAS_S/ANRILa | CAD, CAC | 254 | 49 | rs1360590 | 9 | 22031443 | 0.48 | C>T | 0.36 (0.06) | 4.58 × 10−10 | intron | 2.63 |
| SCARA5a | Hemostatic (FVII) | 447 | 80 | rs7003622 | 8 | 27792549 | 0.08 | T>C | −0.67 (0.12) | 3.70 × 10−8 | intron | 0.90 |
| PRDX2a | RBC traits | 58 | 15 | rs897804 | 19 | 12737964 | 0.42 | G>C | −0.49 (0.09) | 2.26 × 10−7 | −30670 | 0.92 |
| GCKRa | Hemostatic (FVII), others | 90 | 16 | rs2010087 | 2 | 27490739 | 0.34 | C>T | 0.32 (0.06) | 2.57 × 10−7 | −82471 | 1.21 |
| COL4A2a | CAD, CAC | 594 | 151 | rs9559792 | 13 | 109857094 | 0.09 | C>T | 0.60 (0.12) | 5.93 × 10−7 | intron | 0.87 |
| PCCBa | Hemostatic (fibrinogen) | 98 | 18 | rs2290131 | 3 | 137463363 | 0.19 | C>T | −0.27 (0.05) | 6.51 × 10−7 | intron | 0.26 |
| CITED2a | RBC traits | 160 | 43 | rs1131431 | 6 | 139735527 | 0.18 | G>A | −0.31 (0.07) | 2.20 × 10−6 | 3′ UTR | 0.34 |
| CLEC4Ma | Hemostatic (vWF) | 133 | 48 | rs4804800 | 19 | 7711128 | 0.12 | A>G | 0.65 (0.14) | 2.35 × 10−6 | −22907 | 3.04 |
| MYBPHLa | CAD | 147 | 33 | rs10858092 | 1 | 109745416 | 0.26 | T>C | 0.29 (0.06) | 4.78 × 10−6 | +94230 | 0.74 |
| VWFa | Hemostatic | 354 | 76 | rs4764482 | 12 | 6039994 | 0.43 | C>T | 0.31 (0.07) | 7.72 × 10−6 | intron | 0.75 |
| HALa | CAC | 276 | 60 | rs17024981 | 12 | 94912183 | 0.05 | G>A | 0.70 (0.16) | 8.23 × 10−6 | intron | 0.42 |
| ICA1La | CAD | 88 | 18 | rs2036927 | 2 | 203348958 | 0.26 | C>T | −0.35 (0.08) | 1.26 × 10−5 | 3′ UTR | 0.49 |
| APOA1 | Lipid, CAD | 179 | 30 | rs583219 | 11 | 116228381 | 0.05 | T>C | 0.55 (0.14) | 8.30 × 10−5 | +14833 | 0.75 |
| CDC7 | CAC | 271 | 51 | rs17501937 | 1 | 91794293 | 0.46 | G>A | −0.32 (0.08) | 9.17 × 10−5 | +30385 | 0.50 |
| PRKCE | RBC traits | 1017 | 208 | rs12986554 | 2 | 46006618 | 0.54 | G>A | −0.31 (0.08) | 1.03 × 10−4 | intron | 0.36 |
| CELSR2 | Lipid, CAD | 179 | 40 | rs4246519 | 1 | 109585078 | 0.49 | G>A | 0.35 (0.09) | 1.18 × 10−4 | −9086 | 1.09 |
| CDKN2A | CAD, CAC | 148 | 30 | rs12335941 | 9 | 21945669 | 0.39 | A>G | −0.14 (0.04) | 1.36 × 10−4 | −12083 | 0.20 |
| PRKAG2 | RBC traits | 478 | 115 | rs1029945 | 7 | 150906805 | 0.26 | C>T | 0.28 (0.08) | 1.69 × 10−4 | intron | 0.23 |
| EPO | RBC traits | 71 | 21 | rs314298 | 7 | 100209050 | 0.39 | T>C | 0.31 (0.08) | 1.94 × 10−4 | +49791 | 1.82 |
| CHURC1 | CAC, RBC traits | 232 | 33 | rs1951487 | 14 | 64442101 | 0.23 | T>C | −0.22 (0.06) | 2.84 × 10−4 | −8792 | 0.14 |
| FADS1 | Lipid, CAD | 131 | 36 | rs174548 | 11 | 61327924 | 0.29 | C>G | −0.21 (0.06) | 3.25 × 10−4 | intron | 0.18 |
| COL4A1 | CAD, CAC | 430 | 117 | rs9559792 | 13 | 109857094 | 0.09 | C>T | 0.38 (0.11) | 3.83 × 10−4 | +99597 | 0.55 |
| CBS | Lipid, CAD | 208 | 37 | rs234702 | 21 | 43350612 | 0.05 | C>G | 0.65 (0.19) | 5.13 × 10−4 | intron | 0.75 |
| PSRC1 | CAD, others | 157 | 36 | rs12036884 | 1 | 109529052 | 0.03 | G>A | −0.87 (0.25) | 5.24 × 10−4 | −94648 | 0.49 |
| MAX | RBC traits | 239 | 45 | rs11622366a | 14 | 64569509 | 0.18 | C>T | 0.27 (0.08) | 6.01 × 10−4 | intron | 0.26 |
| SARS | CAD | 165 | 39 | rs12036884 | 1 | 109529052 | 0.03 | G>A | −0.96 (0.29) | 8.78 × 10−4 | −28986 | 0.46 |
| PCSK9 | CAD | 201 | 48 | rs12117661 | 1 | 55259934 | 0.20 | C>G | 0.50 (0.15) | 9.08 × 10−4 | −17803 | 5.46 |
| KCNE2 | CAD | 186 | 44 | rs7283334 | 21 | 34647521 | 0.27 | G>A | 0.48 (0.15) | 9.14 × 10−4 | −10672 | 1.31 |
| APOC1 | Lipid, CAD | 90 | 33 | rs204907 | 19 | 50153836 | 0.02 | A>G | −0.64 (0.20) | 1.41 × 10−3 | +39390 | 0.52 |
| APOC3 | Lipid, CAD | 183 | 30 | rs17092646 | 11 | 116117174 | 0.02 | G>C | −0.96 (0.30) | 1.46 × 10−3 | −88660 | 1.92 |
aSignificant after correcting for the total effective number of independent SNPs tested (N = 3692). Twenty genes pass a global multiple-test adjusted P-value cut off (P < 1.35 × 10−5), which was calculated as dividing 0.05 by the total effective number of independent SNPs tested across 83 genes.
3 eQTL positive control genes (PEX6, OAS1 and MTRR) are excluded even though they pass multiple-test corrections. Distances from genes are from the maximal boundaries of all transcript isoforms; negative values indicate upstream (5′) from genes and positive values reflect positions downstream (3′) from genes.
SNP, single-nucleotide polymorphism; #Eff, effective number of; Chr, chromosome; MAF, minor allele frequency; Beta, β-coefficient; bp, base pair; CAD, clinically apparent coronary artery disease; RBC, red blood cell; CAC, coronary artery calcium; FVII, coagulation factor 7; vWF, von Willebrand factor.
Ten genes have more than one cis-eSNP strongly associated (P < 5 × 10−8) including ABO (P < 1.12 × 10−91), FADS2 (P < 5.72 × 10−73), LPL (P < 1.21 × 10−52), APOC2 (P < 1.98 × 10−36), FNTB (P < 1.90 × 10−22), CDKN2B (P < 1.86 × 10−13), FAM117B (P < 2.93 × 10−11), C6orf184 (P < 7.07 × 10−11), CDKN2BAS-S/ ANRIL (P < 4.58 × 10−10) and SCARA5 (P < 3.70 × 10−8) (Table 1). Regional plots and expression by genotype plots are shown in Figure 1A and C for LPL, Figure 1B and D for FADS2 and Figure 2A and C for ABO as examples. Similar plots for the other seven top genes with P < 5 × 10−8 are presented in Supplementary Material, Figs S1 and S2. Among these 10 genes, 6 show down-regulation of expression with the minor allele (negative beta coefficient), suggesting potential reduced function (Table 1). The magnitude of effect sizes ranged from 0.32 to 1.83 ΔCT units per allele, with the largest (3.6-fold) change per allele being observed for the SNP rs6993414, associated with LPL expression. The distance of the top eSNP from the index cis gene, among the 38 genes surviving multiple-test correction, ranged from 0 bases (15 eSNPs within the regulated gene region) to 94 kb (MYBPHL), with a clear trend toward gene-centricity of significant findings.
Figure 1.
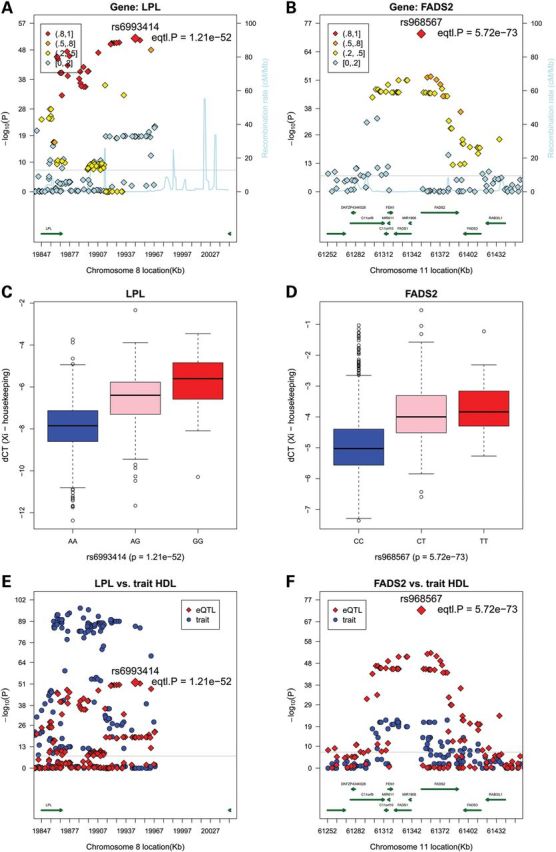
Regional plots and box plots for gene LPL and FADS2 top cis-SNPs whose signals were attenuated markedly after adjusting for the lead GWAS SNPs. Top panel: observed –log10(P) and recombination rates by chromosomal position for all associated SNPs near rs6993414 near LPL on chromosome 8 (A) and rs968567 near FADS2 on chromosome 11 (B). Association plots were conducted with the use of R. Top SNP of interest and P-values in each region are indicated (red diamonds). Color coding indicates the strength of LD of each SNP with the top SNP in each region (measured by r2). The light blue line represents the estimated recombination rates. Gene annotations are shown as dark green lines. Middle panel: linear regression analysis of normalized log transformed ΔCT expression values (ΔCT =CTgene − mean CT3 housekeeping genes) for gene LPL with genotypes of SNP rs6993414, giving a P-value of 1.21 × 10−52 and beta-coefficient of −1.83 (C), and for gene FADS2 with genotypes of SNP rs968567, giving a P-value of 5.72 × 10−73 and β-coefficient of −1.53 (D). Bottom panel: regional plots for eQTL associations (red triangles) and GWAS clinical traits associations (blue circles). Colors indicate LD with the peak eSNPs (red diamonds). LPL cis-associations versus. GWAS HDL results (E), and FADS2 cis-associations versus. GWAS HDL results (F).
Figure 2.
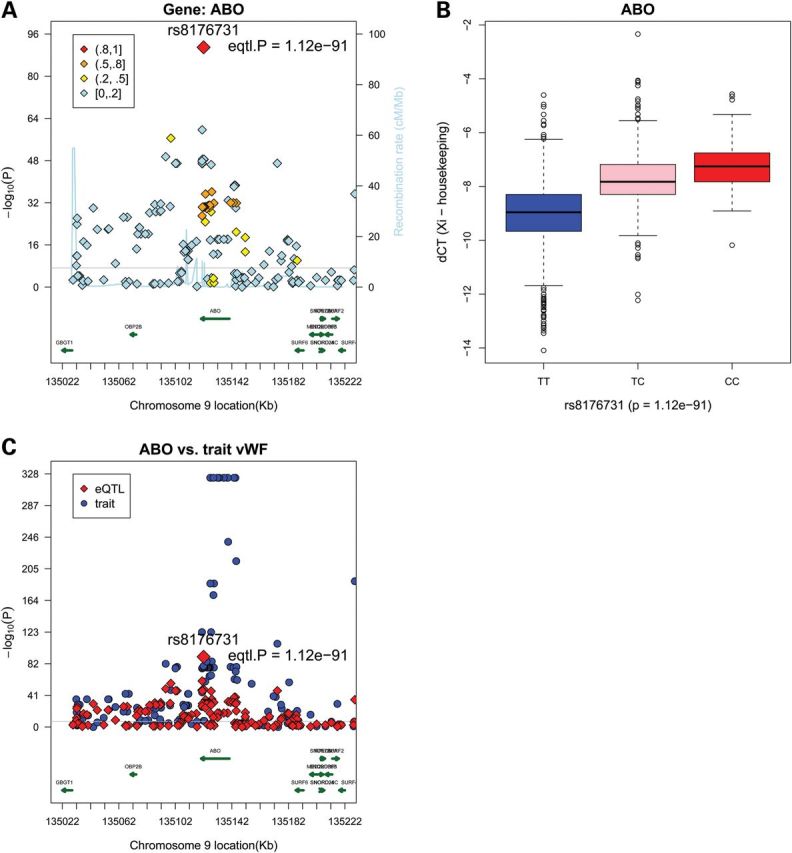
Regional plots and box plots for gene ABO top cis-SNPs whose signal was not attenuated after adjusting for the lead GWAS SNPs. (A) Observed −log10(P) and recombination rates by chromosomal position for all associated SNPs near rs8176731 near ABO on chromosome 9. Top SNP of interest and P-values in each region are indicated (red diamonds). Color coding indicates the strength of LD of each SNP with the top SNP in each region (measured by r2). The light blue line represents the estimated recombination rates. Gene annotations are shown as dark green lines. (B) Linear regression analysis of normalized log transformed ΔCT expression values (ΔCT = CTgene − mean CT3 housekeeping genes) for gene ABO with genotypes of SNP rs8176731, giving a P-value of 1.12 × 10−91 and β-coefficient of −1.43. (C) ABO cis-associations versus. GWAS vWF results. Regional plots for eQTL associations (red triangles) and GWAS clinical traits associations (blue circles). Colors indicate LD with the peak eSNPs (red diamonds).
Among the top 10 cis-eQTL associations, the strongest cis-eSNPs explained between ∼1 and 17% of the expression variance (Table 1), which is generally larger than the trait variance explained by GWAS SNPs (30). For example, the cis-eSNP rs8176731 accounts for 17.7% of ABO expression variance. In order to distinguish between the contribution of the clinical/GWAS SNP and the eSNP to the gene expression variance, we performed conditional analyses for each of the top loci (Table 2). An eQTL association that disappears/attenuates markedly after conditioning upon the GWAS SNP implies that the two traits might share the same genetic basis. After adjusting for the lead GWAS SNP, the expression variance explained by top cis-eSNPs generally was not markedly attenuated, with the exception of LPL and FADS2 [associated with lipid levels (27)] and C6orf184 [associated with CAD (11)] (Table 2). Furthermore, in order to fine-map the top 10 eQTL regions, we conducted the same cis-eQTL analysis using 1000G imputation SNP data for the top 10 genes in Table 2. This strategy identified four times more cis-eSNPs at a threshold of P < 5 × 10−8 (Supplementary Material, Table S3a), which is not surprising because 1000G imputation data contain many more SNPs than 2.5 million HapMap data. As shown in Supplementary Material, Table S3b, the top cis-eSNP result using HapMap 2.5 million SNP imputation data is different from that of using 1000 Genomes Phase 1 imputation data except for FADS2. Regional plots for LPL, FADS2 and C6orf184 are shown in Supplementary Material, Fig. S3.
Table 2.
Top cis-eSNP associations after adjustment for previously reported GWAS SNPs
| Gene | Clinical phenotype | C hr | Top 1, cis-eSNP | P, cis-eSNP | #cis- eSNP P < 5 × 10−8 | Clinical SNP used for adjustment (clinSNP) | R2 (eSNP versus. clinSNP) | GWAS clinical traits | P, clinSNP in GWAS | P, cis-eSNP after adjust for clinSNP |
|---|---|---|---|---|---|---|---|---|---|---|
| ABO | Hemostatic, CAD, others | 9 | Rs8176731 | 1.12 × 10−91 | 120 | rs687621 | 0.04 | vWF | <5.00 × 10−324 | 6.61 × 10−87 |
| rs687289 | 0.04 | FVIII | <5.00 × 10−324 | 6.35 × 10−87 | ||||||
| rs579459 | 0.10 | CAD | 4.00 × 10−14 | 8.84 × 10−81 | ||||||
| FADS2 | Lipid, CAD | 11 | Rs968567a | 5.72 × 10−73 | 64 | rs174546 | 0.47 | TG, HDL, TC,LDL | 5.40 × 10−24 | 6.71 × 10−22 |
| rs174626b | 0.32 | CAD | 5.30 × 10−3 | 1.33 × 10−54 | ||||||
| LPL | Lipid, CAD | 8 | rs6993414 | 1.21 × 10−52 | 127 | rs12678919c | 1.00 | TG,HDL | 1.50 × 10−115 | 3.00 × 10−3 |
| rs3779788 | 0.34 | CAD | 2.40 × 10−7 | 1.74 × 10−27 | ||||||
| APOC2 | Lipid, CAD | 19 | rs2288912 | 1.98 × 10−36 | 32 | rs4420638 | 0.00 | LDL,TC | 8.72 × 10−147 | 3.20 × 10−36 |
| rs4420638d | 0.00 | CAD | 2.14 × 10−4 | 3.20 × 10−36 | ||||||
| FNTB | RBC traits | 14 | rs7148144 | 1.90 × 10−22 | 7 | rs4466998 | 0.06 | MCV | 4.90 × 10−8 | 1.14 × 10−20 |
| CDKN2B | CAD, CAC | 9 | rs598664 | 1.86 × 10−13 | 18 | rs4977574 | 0.08 | CAD, CAC | 1.35 × 10−22 | 9.49 × 10−14 |
| FAM117B | CAD | 2 | rs1971739 | 2.93 × 10−11 | 9 | rs6705330 | 0.27 | CAD | 8.98 × 10−11 | 4.17 × 10−7 |
| C6orf184e | CAD | 6 | rs7773213 | 7.07 × 10−11 | 17 | rs9374080 | 0.68 | MCV | 3.70 × 10−8 | 3.20 × 10−2 |
| CDKN2BAS-S* | CAD, CAC | 9 | rs1360590 | 4.58 × 10−10 | 18 | rs4977574 | 0.43 | CAD, CAC | 1.35 × 10−22 | 1.85 × 10−6 |
| SCARA5 | Hemostatic | 8 | rs7003622 | 3.70 × 10−8 | 1 | rs2726953 | 0.01 | vWF | 1.30 × 10−16 | 2.80 × 10−8 |
| rs9644133 | 0.00 | FVIII | 4.40 × 10−15 | 2.71 × 10−8 |
ars968567 was also reported to be significantly associated with the expression level of FADS2 in other blood-related eQTL studies (Supplementary Material, Table S3a)
brs174626 is not found in GWAS catalog, and FADS2 is not found to be associated with CAD in GWAS catalog.
crs12678919, a lead SNP in TG and HDL GWAS, is also reported to be significantly associated with the expression level of LPL in lymphocytes (Supplementary Material, Table S3a)
drs4420638 was associated with C-reactive protein (P = 9 × 10−139) and LDL cholesterol (P = 2 × 10−40) in GWAS catalog (APOC2 is not found to be associated with CAD in GWAS catalog).
eC6orf184 is an alias of gene CCDC162P. CDKN2BAS-S is an alias for a short transcript isoform of ANRIL.
All previously reported GWAS SNPs reach genome-wide significant (P < 5 × 10−8) except 3 SNPs associated with CAD (12): rs174626 (P = 5.30 × 10−3), rs3779788 (P = 2.40 × 10−7), and rs4420638 (P = 2.14 × 10−4).
Chr, chromosome; SNP, single-nucleotide polymorphism; clinSNP, SNP from clinical genome-wide association study used for adjustment in conditional analyses; GWAS, genome-wide association study; CAD, clinically apparent coronary artery disease; RBC, red blood cell; CAC, coronary artery calcium.
Co-occurrence of cis-eQTLs with GWAS results for CVD and related risk factors
We assessed the co-occurrence of eSNPs and GWAS associations with CAD and related phenotypes, by checking the associations of eSNPs within respective GWAS scans for coronary artery calcium (CAC), lipids, RBC traits, hemostatic factors and blood pressure (Table 3). Several eSNPs exhibit strong association with gene expression levels and moderate-to-strong association with CAD or one or more related traits. These eSNPs include SNPs associated with ABO (for ABO, eQTL P < 1.12 × 10−91, vWF P < 1.94 × 10−8, LDL P < 3.45 × 10−10), FADS2 (eQTL P < 5.72 × 10−73, LDL P < 3.87 × 10−7), LPL (eQTL P < 1.21 × 10−52, TG P < 2.91 × 10−104), APOC2 (eQTL P < 1.98 × 10−36, TG P < 1.38 × 10−6), CDKN2B (eQTL P < 1.86 × 10−13, LDL P < 5.3 × 10−3), FAM117B (eQTL P < 2.93 × 10−11, TC P < 4.5 × 10−4), C6orf184 (eQTL P < 7.07 × 10−11, HDL P < 1.7 × 10−3, MCV P < 2.9 × 10−6) and CDKN2BAS-S (eQTL P < 4.58 × 10−10, CAC P < 1.78 × 10−5). SNPs located in 9p21 are strongly cis associated with the expression of two genes within the same locus (CDKN2B and CDKN2BAS-S). These associations suggest functional genetic regulation of the genes at these loci by the same SNPs that are associated with related biomarkers.
Table 3.
Comparison of GWAS results for clinical CVD traits with eQTL results
| Gene | Clinical phenotype | Top1 cis-eSNP | P, cis-eSNP | Hemostatic factors (18) | Blood lipids (27) | Red blood cell traits (9) | Subclinical CAD (20) | Blood pressure (35) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| vWF | FVII | FVIII | TC | HDL | LDL | TG | HB | HCT | MCHC | MCH | MCV | RBC | CAC | SBP | DBP | ||||
| Below are the P-values from previously reported GWAS results of relevant clinical traits | |||||||||||||||||||
| ABO | Hemostatic, CAD, others | rs8176731 | 1.12 × 10−91 | 1.94 × 10−8 | 0.91 | 2.83 × 10−6 | 6.51 × 10−8 | 0.21 | 3.45 × 10−10 | 0.69 | 0.02 | 0.02 | 0.26 | 0.52 | 0.80 | 0.89 | 0.79 | 0.98 | 0.85 |
| FADS2 | Lipid, CAD | rs968567 | 5.72 × 10−73 | 0.45 | 0.76 | 0.40 | 5.39 × 10−7 | 8.32 × 10−5 | 3.87 × 10−7 | 4.96 × 10−7 | 0.04 | 0.08 | 0.05 | 0.91 | 0.89 | 0.40 | 0.74 | 0.05 | 0.21 |
| LPL | Lipid, CAD | rs6993414 | 1.21 × 10−52 | 0.68 | 0.36 | 0.56 | 0.93 | 3.22 × 10−87 | 0.77 | 2.91 × 10−104 | 0.17 | 0.46 | 0.83 | 0.15 | 0.53 | 0.13 | 0.23 | 0.39 | 0.03 |
| APOC2 | Lipid, CAD | rs2288912 | 1.98 × 10−36 | 0.85 | 0.35 | 0.55 | 0.83 | 1.40 × 10−4 | 6.10 × 10−3 | 1.38 × 10−6 | 0.39 | 0.76 | 0.24 | 0.40 | 0.38 | 0.84 | 0.70 | 0.23 | 0.57 |
| FNTB | RBC traits | rs7148144 | 1.90 × 10−22 | 0.56 | 0.58 | 0.91 | 0.86 | 0.50 | 0.48 | 0.41 | 0.44 | 0.38 | 0.25 | 0.55 | 0.16 | 0.27 | 0.21 | 0.68 | 0.63 |
| CDKN2B | CAD, CAC | rs598664 | 1.86 × 10−13 | 0.27 | 0.56 | 0.96 | 0.04 | 0.46 | 5.30 × 10−3 | 0.74 | 0.30 | 0.38 | 0.23 | 0.71 | 0.58 | 0.75 | 0.05 | 0.98 | 0.79 |
| FAM117B | CAD | rs1971739 | 2.93 × 10−11 | 0.16 | 0.05 | 0.38 | 4.50 × 10−4 | 4.00 × 10−3 | 0.14 | 0.05 | 0.13 | 0.33 | 0.33 | 0.82 | 0.55 | 0.81 | 0.94 | 0.03 | 0.16 |
| C6orf184 | CAD | rs7773213 | 7.07 × 10−11 | 0.95 | 0.47 | 0.43 | 8.00 × 10−3 | 1.70 × 10−3 | 0.52 | 0.17 | 0.63 | 0.08 | 0.43 | 5.74 × 10−5 | 2.90 × 10−6 | 0.05 | 0.43 | 0.94 | 0.96 |
| CDKN2BAS_S | CAD, CAC | rs1360590 | 4.58 × 10−10 | 0.58 | 0.45 | 0.59 | 0.40 | 0.86 | 0.78 | 0.38 | 0.42 | 0.76 | 0.51 | 0.62 | 0.12 | 0.42 | 1.78 × 10−5 | 0.29 | 0.03 |
| SCARA5 | Hemostatic (FVII) | rs7003622 | 3.70 × 10−8 | 0.22 | 0.68 | 0.29 | 0.37 | 0.51 | 0.49 | 0.94 | 0.93 | 0.51 | 0.42 | 0.60 | 0.27 | 0.69 | 0.93 | 0.03 | 0.67 |
P-value of <0.05 is given in bold.
FVII, coagulation factor VII; FVIII, coagulation factor VIII; vWF, von Willebrand Factor; TC, total cholesterol; HDL, high-density lipoprotein cholesterol; LDL, low-density lipoprotein cholesterol; TG, triglycerides; HB, hemoglobin concentration; HCT, hematocrit; MCHC, mean corpuscular hemoglobin concentration; MCH, mean corpuscular hemoglobin; MCV, mean corpuscular volume; RBC, red blood cell count; CAD, clinically apparent coronary artery disease; CAC, coronary artery calcium; SBP, systolic blood pressure; DBP, diastolic blood pressure.
While these concordant associations may reflect a true causal component, they could also reflect partial LD with the actual causal variants at these loci. To address this, we plotted regional eQTL associations on the same plot with regional GWAS (clinical or biomarker traits) to visualize the concordance of signals (Figs 1 and 2). LD was also assessed between top cis-eQTLs and top GWAS SNPs, conditional analysis of eQTL associations was conducted adjusting for the top GWAS SNP, and eQTL analysis was repeated for evidence of attenuation of the top signal. As shown in Table 2, the top cis-eSNP associated with ABO expression is completely independent from the other three GWAS SNPs known to be strongly associated with CAD, vWF and FVIII, respectively, indicating that SNPs associated with expression and clinical phenotypes are mutually independent (Fig. 2C). Similar results are also observed for several other top genes (APOC2, FNTB, CDKN2B and SCARA5). However, after controlling for the TG/HDL GWAS SNP (rs174546), the signal of the top cis-eSNP for FADS2 (rs968567) is dramatically attenuated (from 5.72 × 10−73 to 6.71 × 10−22). rs968567 is moderately associated with LDL (P < 3.87 × 10−7) and TG (P < 4.96 × 10−7) (Table 3 and Fig. 1F). These two SNPs are partially correlated (r2 = 0.47), suggesting that this cis-eSNP, rs968567, could play a regulatory role for clinical lipid traits, although future research is warranted to confirm the functional molecular mechanism of this eSNP. Similar evidence for attenuation after controlling for the clinical GWAS SNPs was noted for LPL, FAM117B, C6orf184 and CDKN2BAS-S (Table 2).
Association between gene expression and clinical lipid traits
We examined the association of the expression signal of the top 10 genes in Table 2 (cis-eSNP P < 5 × 10−8) with four clinical lipid traits measured at the same blood draw as the RNA sample collection (Examination 8): HDL, triglycerides (TG), LDL and total cholesterol (TC). The transcript abundance for LPL, a gene known to be associated with HDL, is moderately associated with HDL level (P = 0.02). Another gene, C6Orf184, is more significantly associated with HDL (P = 0.002). As the dosage of the minor allele increases, gene expression increases and HDL level decreases, as observed in the Global Lipid HDL GWAS results (27). From this targeted examination of LPL and C6orf184, we identify a possible causal molecular role of these cis-eSNPs in the regulation of lipid metabolism.
Comparison of eQTL results with publicly available GWAS results
Beyond comparing our results to available full results of GWAS scans of CAD and blood risk factor traits, we also examined the top GWAS results reported in the NHGRI GWAS catalog to assess whether our most significant cis-eSNPs are the same as SNPs reported in prior GWAS to be associated with other clinical traits. Among 377 cis-eSNPs with P < 5 × 10−8 in the current study (120 associated with ABO, 64 with FADS2, 127 with LPL), 17 cis-eSNPs were significantly associated in prior GWAS (Supplementary Material, Table S4). For example, seven eSNPs associated with ABO gene expression in our study were also associated in prior GWAS with multiple traits including hematological and biochemical traits, plasma E-selectin levels, pancreatic cancer and CAD. Extensive pleiotropy for the ABO gene region has previously been described (31). One eSNP, rs1535 associated with FADS2 expression level (P = 4.02 × 10−46) is associated with metabolic syndrome (P = 4.0 × 10−7) in the GWAS catalog. Out of 127 SNPs associated with LPL expression level (P < 5 × 10−8), nine have been reported to be associated with hypertriglyceridemia, HDL cholesterol, TG or metabolic syndrome at a genome-side significant P-value (Supplementary Material, Table S4). We employed a conservative threshold (P < 5 × 10−8) for selecting cis-eSNPs; however, as shown in Supplementary Material, Table S5, a greater number of cis-eSNPs are identified as the significance threshold is loosened, but proportion of SNPs that overlap between cis-eSNPs and GWAS SNPs decreases as the significance threshold becomes less restrictive (Chi-squared test result for difference across P-value categories P < 1.1 × 10−11).
trans-eQTL associations
In analyses to detect important trans-eQTL associations, we found SNPs strongly associated with the expression level of target genes on different chromosomes (P < 5 × 10−8) (Table 4). Expression levels of VWF, transcribed from chromosome 12, are associated with the SNP rs1354034 on chromosome 3 (P = 5.43 × 10−20) proximal to ARHGEF3. The ARHGEF3 locus (rs1354034) is also associated in trans with expression levels of COL4A2 on chromosome 13 (P = 7.01 × 10−11), a target locus that was reported in a recent large GWAS analysis for CAD (11), and also within a large analysis of CAC levels (20). Two loci on chromosome 8 (rs4909812 and rs727582) are associated with the expression of CITED2 and CCND3, respectively. Both of these target genes were found within top loci in RBC traits GWAS.
Table 4.
Top trans-eSNP results from GWAS for 83 genes
| Gene | Clinical phenotype | Chr_gene | Top 1 trans-eSNP | Chr_trans-eSNP | Physical location | MAF | P, trans-eSNP | Nearest genesa | Additional trans-eQTL Support? | References |
|---|---|---|---|---|---|---|---|---|---|---|
| VWF | Hemostatic | 12 | rs1354034 | 3 | 56824789 | 0.38 | 5.43 × 10−20 | ARHGEF3 | Y | (25) |
| COL4A2 | CAD, CAC | 13 | rs1354034 | 3 | 56824789 | 0.38 | 7.01 × 10−11 | ARHGEF3 | – | – |
| CITED2 | RBC traits | 6 | rs4909812 | 8 | 139714828 | 0.04 | 9.30 × 10−9 | COL22A1 | Y | (32) |
| CCND3 | RBC traits | 6 | rs727582 | 8 | 116719643 | 0.33 | 3.13 × 10−8 | TRPS1 | – | – |
| LPL | Lipid, CAD | 8 | rs7085130 | 10 | 63159783 | 0.45 | 3.59 × 10−8 | C10orf107 | – | – |
| EPO | RBC traits | 7 | rs723580 | 6 | 46155599 | 0.05 | 4.68 × 10−8 | ENPP4 | – | – |
| PCCB | Hemostatic (fibrinogen) | 3 | rs10802704 | 1 | 236766283 | 0.03 | 4.85 × 10−8 | ZP4 | – | – |
aThe nearest gene to the trans-eSNP is given in bold.
Chr, chromosome; SNP, single-nucleotide polymorphism; MAF, minor allele frequency; CAD, clinically apparent coronary artery disease; RBC, red blood cell; CAC, coronary artery calcium.
The expression level of LPL is trans-associated with rs7085130 on chromosome 10 (P = 3.59 × 10−8) located within C10orf107 region. Notably, the C10orf107 locus was associated with blood pressure levels in recent GWAS (33–35). Another interesting trans-eSNP is rs2815067, located in a locus on chromosome 6 containing the KIF6 gene and associated with expression levels of CDKN2A (P = 5.06 × 10−8). CDKN2A is located at the 9p21.3 CAD locus, and the KIF6 region was previously postulated to be a CAD gene, although replication attempts have failed (11,36).
Among seven genes in Table 4, the expression levels of six genes are also associated with a cis-SNP that survives a regional multiple-test correction (VWF, COL4A2, CITED2, LPL, EPO and PCCB). For LPL, COL4A2 and PCCB, it is notable that the associated cis-eSNP nearly reaches genome-wide significance (P < 7 × 10−7) (Table 1 and Supplementary Material, Fig. S4).
In silico validation of top cis- and trans-eQTLs
Each top cis-eSNP at P < 5 × 10−8 (Table 1) was mapped to previously published eQTL results. As shown in Supplementary Material, Table S6a, 4 of 10 genes have expression levels with evidence of significant cis-effects (<100 kb) by top eSNPs or their proxy SNPs (r2 = 1) in at least one other blood-related eQTL study (FADS2, LPL, FNTB and CDKN2B). The lead eSNP rs968567 (P = 5.72 × 10−73), found to be cis-associated with the expression of FADS2 in our study, was also previously reported as a cis-eSNP for FADS2 expression in primary peripheral-blood CD4+ lymphocytes(37), lymphocytes (38), peripheral-blood mononuclear cells (PBMC) (39), leukocytes (40) and blood (26), indicating a robust blood-related eQTL for FADS2. For LPL, the lead eSNP rs6993414 identified in our study was not reported by other blood-related eQTL studies. However, five proxy SNPs (r2 = 1) are strongly associated with expression levels of LPL in monocytes (P < 1.73 × 10−37) (25). Another proxy SNP rs12678919, the lead SNP in HDL and TG GWAS, is also associated with LPL in lymphocytes (38) (P = 2.96 × 10−15). Similarly, for CDKN2B, the lead eSNP rs598664 was not found to be associated with CDKN2B in published eQTL results. However, four proxy SNPs are significantly cis associated with CDKN2B expression level in lymphocytes (38), blood (41) and monocytes (25).
The top trans-eSNPs are reported in Table 4. We first mapped them to trans-eQTLs that are publicly available. The lead trans-eSNP rs1354034 on chromosome 3 was found to be associated with the expression of VWF (on chromosome 12) (P = 5.43 × 10−20) and is in close LD (r2 = 0.6) with a lead SNP rs12485738 associated with mean platelet volume (MPV) (P = 6.00 × 10−31). rs12485738 was previously shown to have trans-association with VWF expression (eQTL P = 7.70 × 10−13) (42). Interestingly, the most proximal gene for rs12485738 is ARHGEF3 on chromosome 3 too, consistent with our trans-eQTL finding for VWF (Table 4). Furthermore, several trans-eSNPs within a tight LD block located on chromosome 3 affect VWF expression level in our data (P < 5 × 10−8) and were also found in prior eQTL studies in monocytes (25) and blood (26) (Supplementary Material, Table S6b). For example, a trans-eSNP rs1344142 on chromosome 3 was found in monocytes (P = 4.78 × 10−21) and in blood (P = 5.20 × 10−10). In our data, we confirmed that rs1344142 is associated with VWF expression (P = 2.19 × 10−15). This SNP is located 21.5 kb from ARHGEF3 and is in a close LD (r2 = 0.64) with the top trans-eSNP, rs1354034, with VWF in our data.
DISCUSSION
With many complex trait GWAS results published and additional genomic data available, including data on expression, transcription factor binding and protein–protein interactions, there is growing interest in identifying genes and gene-regulatory networks that may predict the function of disease-associated loci by integrating various sources of data (43–46). In our cis-eQTL analysis of 83 cardiovascular trait-related genes within a large population sample, >45% of genes harbor eQTLs that surpass a regional multiple-test correction, suggesting that a high proportion of disease-associated loci harbor cis-acting regulatory variants. These results are consistent with recent publications (47,48) and broad surveys of allele-specific SNP expression in biologically relevant human tissues that indicate 30–50% of genes are subject to cis-acting variation (49). Notably, at least one cis-eSNP (P < 5 × 10−8) is significantly associated with the expression level of 10 genes from the total of 83 GWAS loci tested, indicating the advantage of combining prior GWAS knowledge with eQTL study data to efficiently distinguish potentially functional associations. This approach was successfully adopted in recent reports applying eQTL analysis to GWAS studies followed by the use of in vivo knock-out mice models to help identify the functional regulatory mechanism of 9p21 SNPs for CAD (50) and a potential drug target (SORT1) for LDL lowering (51). In our cis-eQTL results, ABO and VWF could be candidates for follow-up studies of von Willebrand factor, LPL and FADS2 for dysregulated levels of lipids and CAD, and C6orf184 for abnormal RBC-related traits and CAD.
Up to 17% of expression variance is explained by top cis-eSNPs, an upper bound of variance that is typically larger than the variance explained by SNPs in GWAS. We performed conditional analysis to distinguish the contribution to the gene expression variance of GWAS clinical SNPs versus cis-eSNPs. After adjusting for top SNPs selected from clinical biomarker GWAS results, we found that the expression variance explained by top cis-eSNPs is generally not dramatically attenuated except for two genes associated with lipid levels (FADS2 and LPL) and gene C6orf184 associated with CAD, suggesting that cis-eSNPs are often independent from clinical GWAS SNPs or may contribute to secondary association signals at these loci.
On the other hand, several genes including C6orf184, APOC2, FADS2 and LPL showed strong eQTL associations for SNPs that are themselves among the strongest for a GWAS trait of interest, suggesting coinciding signals. rs7773213 is associated with the expression of C6orf184 (also known as CCDC162, a pseudo gene) and is also strongly associated with RBC traits. Polymorphisms associated with expression levels of APOC2, FADS2 and LPL are also strongly associated with lipid levels in the largest lipid GWAS study to date (27) and thus well powered to detect concordance between eSNPs and associations with lipid levels.
In our data, top associated SNPs in RBC GWAS results are not strongly cis-associated with their proximal gene expression. One possible reason is that the gene expression may be tissue/cell type specific, and we studied leukocyte gene expression rather than proerythroblast or erythroid cell lines. Similar observations have been reported by other studies. Folkersen et al. (52) identified a variant rs6725887 affecting the expression level of a well-known myocardial infarction gene NBEAL1 in aorta media (P = 4.23 × 10−5), but in our data, the signal is only moderate after adjusting for multiple testing (P = 0.08). This provides support that using tissues/cells appropriate to the disease/trait of interest may be necessary to detect eQTL association. Data from the Genotype-Tissue Expression (GTEx) project may be helpful for such future studies (available at: http://www.broadinstitute.org/gtex/).
When we examined the NHGRI GWAS catalog (available at: www.genome.gov/gwastudies), we found the most significant cis-eSNPs (P < 5 × 10−8) were significantly associated with CAD risk factors and other disease traits. For example, eSNPs associated with ABO gene expression in our study are also associated with multiple traits including hematological and biochemical traits, plasma E-selectin levels, pancreatic cancer and CAD. Further, significant ABO eSNPs are also reported by other genome-wide cis-eQTL studies in monocyte (25) and CD4+ cells (37). These strong ABO associations in both GWAS and eQTL studies indicate multiple phenotypic influences of the ABO locus (pleiotropy) that may be because of one or a few similar underlying molecular mechanisms.
trans-eQTLs have moderate effect sizes and may be difficult to detect and replicate (53). In our trans-eQTL analyses, we found many SNPs located on different chromosomes and strongly associated with the expression level of our target genes (P < 5 × 10−8). Several of these trans-associations have also been reported by other studies. Using blood cells, Fehrmann et al. (26) reported a MPV lead SNP rs12485738 on 3p26 (42), which independently affects some blood coagulation genes in trans, including VWF. This trans-association is also found in our data (P = 7.67 × 10−13), and this SNP is also in close LD with the top trans-eSNP for VWF in our data rs1354034 (r2 = 0.61). Furthermore, ARHGEF3 is a gene nearby these two trans-eSNPs, whose expression is significantly cis affected (data not shown).
There are several genes whose expression tends to be regulated by both cis- and trans-eQTLs, including LPL and VWF both in our data (Supplementary Material, Fig. S4) and other studies. For example, we identified a trans-eSNP affecting VWF expression, and Fehrmann et al. (26) reported a cis-SNP rs4764482 affecting VWF expression in blood (P = 2.80 × 10−40), consistent with the result in our data (P = 7.72 × 10−6) for the same SNP rs4764482. However, when we conducted an interaction analysis between all the significant cis-SNPs and trans-SNPs (P < 5.0 × 10−8) for seven genes, there is no significant cis–trans interaction effect observed (data not shown). The absence of an interaction might occur because our current trans-eQTL analyses are not comprehensive, as we only measured 93 target genes rather than all genes in the genome.
Our study has several strengths and limitations. Previous eQTL analyses have been conducted using microarray gene expression data and genotyped/selected SNPs. In this study, we applied qRT–PCR, a gold standard for measuring mRNA expression levels, to measure expression levels in a single well-characterized large population of 1846 FHS participants.
We have selected only a subset of genes to assay at key cardiovascular risk loci; thus, we may have missed important functional genes and have limited data to address full trans-eQTL networks. Furthermore, for all but one gene (CDKN2BAS), we have selected an assay that detects an exon that is common in all transcripts for each gene; thus we cannot address issues of alternative transcript isoforms and their regulation in most cases. Although we observed concordant associations among SNPs, gene expression and blood lipid traits, there was a temporal difference in sample collection for the majority of the CVD-related biomarkers and the collection of RNA samples for gene expression measurements. Therefore, we cannot conduct systematic correlation studies between gene expression and many of the clinically relevant CVD biomarkers. Thus, we are limited in our ability to definitively establish genetic regulatory variation as the causal molecular mechanism of outcomes and biomarkers. More experimental validation and mechanistic studies are necessary to pinpoint specific regulatory variants. However, this study may provide incremental functional information for several key GWAS loci where regulatory variation is strongly suspected to play a role in disease etiology.
In the future, with improvements in data quantity and quality for key regulatory pathways from ENCODE (54) and other large projects, integrating genomic, expression, epigenomic and protein interaction data may allow a more mature systems biology approach for insights into the networks and key functions that underlie CVD. This may in turn lead to novel molecular approaches to prevention, diagnosis and treatment.
MATERIALS AND METHODS
Study population and sample collection
Participants of the FHS Offspring cohort who attended examination 8 and had blood samples available for gene expression were included (n = 1846). The FHS started in 1948 with 5209 randomly ascertained participants from Framingham, MA, USA, who have undergone biannual examinations to investigate CVD and its risk factors (55). In 1971, the Offspring cohort (56,57) (comprised of 5124 children of the original cohort and the children's spouses) and in 2002, the third generation (consisting of 4095 children of the offspring cohort), were recruited (58). FHS participants in this study are mainly of European ancestry. The FHS was reviewed by the Boston University Medical Center Institutional Review Board, and all participants gave written informed consent. Among the FHS Offspring cohort participants included in this study, 723 are male and 890 are female. The mean age at time of blood sampling was 66.6 years with a standard deviation of 8.9 years.
Samples were processed and assayed within 2 months of completion of Offspring cohort participant visits. The PBMCs were isolated on site at the FHS center. After isolation, PBMCs were transferred into a clean tube, washed with phosphate-buffered saline, pelleted by centrifugation and lysed with RNA lysis solution (RLT; Qiagen, Germantown, MD, USA). PBMC lysates were kept at –80°C for further RNA isolations.
Expression profiling
At the Boston University School of Medicine, lysed cells were thawed and total RNA was isolated in both platelets and PBMCs using RNeasy Mini Kits (Qiagen). RNA concentration and quality was assessed using a ND-1000 NanoDrop Spectrophotometer (NanoDrop, Wilmington, DE, USA). RNA samples were stored at –80°C until qRT–PCR.
After cDNA conversion and preamplification, quantitative reverse-transcriptase polymerase chain reaction (qRT–PCR) was performed with a high-throughput RT–PCR instrument (BioMark; Fluidigm, San Francisco, CA, USA). Preamplified cDNA samples were loaded into the DynamicArray 48.48 chips (Fluidigm). TaqMan Gene Expression Assays (Applied Biosystems, Foster City, CA, USA) were diluted and pipetted out into the DynamicArray chips. Then, the DynamicArray was placed into the NanoFlex controller. All qRT–PCR reactions were performed in the BioMark Real-Time PCR system. Protocol details were described previously (28). To minimize the batch effect in this high-throughput experiment, three repeated housekeeping genes were targeted in each plate/run to serve as the internal controls. As Life Technologies does not disclose the exact probe and primer sequences, for each gene, we provided the context sequence to which the TaqMan probe binds, and the Life Technologies Assay ID as shown in Supplementary Material, Table S7.
A total of 93 genes were chosen from top loci in published GWAS studies associated with CAD(11), CAC(20) and blood risk factor traits including hemostatic factors (17,18), lipid levels (27) and RBC traits (9) (Supplementary Material, Table S1 for the gene list). In general, a single candidate gene was selected for most loci based upon proximity to the top associated SNP, though more than one gene were targeted for a few loci [e.g., at 9p21.3: CDKN2A, CDKN2B, CDKN2BAS-S (short isoform), CDKN2BAS-L (long isoform)]. After excluding 10 genes with low levels of expression, a total of 83 non-housekeeping genes were available for subsequent analysis.
Genotyping
Genotyping was carried out as a part of the SNP Health Association Resource project (SHARe) using the Affymetrix 500K mapping array (250K Nsp and 250K Sty arrays) and the Affymetrix 50K supplemental gene focused array on 9274 individuals. Genotyping resulted in 503 551 SNPs with successful call rate >95% and HWE P > 10−6. Finally, out of a total 9274 participants, 8481 individuals with call rate >97% were remained. Imputation of ∼2.5 million autosomal SNPs in HapMap with reference to release 22 CEU samples and imputation of ∼36 million SNPs (autosomal + chromosome X) using 1000 Genomes Phase 1 data were conducted using the algorithm implemented in MACH (59). There were 1846 Offspring cohort participants with genotype imputation and expression measurements available for analysis. The deposition of single-nucleotide polymorphisms imputed SNPs in dbGaP for the FHS have been are described on the dbGaP website (http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?Study_id=phs000342.v4.p6).
Statistical analysis
For each gene, we excluded samples that were >5 SD away from the mean of the transformed ΔCT expression values (ΔCT = CTgene − mean CT3 housekeeping genes). This left up to 1594 samples out of a total 1846 included in the downstream GWAS analysis.
A GWAS analysis for each of the 83 genes was conducted on 2.5 million imputed SNPs assuming an additive genetic model and adjusting for age, sex and the mean expression level of three housekeeping genes (B2M, GAPDH, and ACTB). SNPs positively associated and within 100 kb of each gene, respectively, were defined as cis-eQTLs, otherwise they were labelled as potential trans-eQTLs. The gene boundary for each gene is based on NCBI human reference genome builds 36/hg18, and was calculated as the maximum union of overlapping transcripts isoforms based on RefSeq genes. Between 6 (MIA3) and 1017 (PRKCE) SNPs were genotyped or imputed and within 100 kb of the genes measured (median of 183 SNPs) (Supplementary Material, Fig. S1).
Missing expression measurements for a given gene and sample were defined by measurements that were beyond detection limits (30 CT). Genes with detectable expression levels for at least 20% of the samples (n = 83) were analyzed as continuous outcomes using an efficient score statistic. Only four genes (PCSK9, APOC3, EPO, SCO2) had missing values of >50%. The efficient score statistic employed here uses a robust measure of the score variance that makes the statistic robust to departure from the normality assumption on the continuous outcome. Family structure is controlled for in a random effects term by including the matrix of the kinship coefficients in the genetic covariance matrix. Specific details are described further in Dupuis et al. (60). All models were run on expression measured from leukocyte cells. A regional multiple-test correction method by computing the effective number of independent SNPs was applied to account for multiple testing (29).
Analysis for testing the possible batch effect of gene expression data
To eliminate a possible impact on our qRT–PCR expression data from batch effects, we have first calculated the Principle Components (PCs) using our rtPCR expression data, as shown in Supplementary Material, Table S8. For each of the top 10 genes in Table 2, we recomputed the P-value for the top eQTL after adjusting for seven top PCs. As shown in Supplementary Material, Table S8, the adjusted P-value is in general slightly attenuated from the unadjusted P-value, but all of the P-values remain highly statistically significant (P < 5.0 × 10−8) except for SCARA5. These results provide further reassurance that batch effects are not playing a major role for our highly significant results.
Analysis for testing the possible impact of population stratification effects
Although all FHS participants in this study are of European ancestry, we still tested a possible impact of population stratification effects using the 10 top PCs derived from 503 551 genotyping SNPs, as shown in Supplementary Material, Table S9. For each of these top 10 PCs, we first checked for evidence of association with the expression level of each of the top 10 genes in Table 2, We found no strong evidence for population stratification effects for any of the genes (no evidence for population stratification at P < 0.01). For six genes, expression levels are nominally associated with one or two PCs (0.01<P < 0.05). For these six genes, we recomputed the P-value for their top eQTLs after adjusting for those PCs nominally associated with the respective genes. As shown in Supplementary Material, Table S9, the adjusted P-value for each of these genes is not materially different from the unadjusted P-value.
Comparison of expression findings to GWAS results for disease and clinical biomarkers
GWAS results were reviewed from the CARDIoGRAM (Coronary ARtery DIsease Genome-wide Replication And Meta-Analysis) Consortium of CAD results including 22 233 cases and 64 762 controls of European descent (11); the Global Lipids Genetics Consortium of >100 000 individuals of European ancestry (27); the ICBP (International Consortium for Blood Pressure) GWAS comprising ∼200 000 individuals of European descent (35); and GWAS for CAC (n = 15 993) (20), RBC traits (n = 24 167) (9), hemostatic factors including FVII, FVIII and vWF (n = 23 608) (18), and circulating fibrinogen levels (n = 22 096) (17) from the CHARGE (Cohorts for Heart and Aging Research in Genome Epidemiology) Consortium. Results in the NHGRI GWAS catalog (Jan-16-2011) were downloaded from http://www.genome.gov/gwastudies/. Expression-associated SNPs were mapped to the GWAS catalog directly by matching SNP rsids.
Association analysis of gene expression and clinical lipid traits
Regression analyses were performed to look for the association between gene expression measured by qRT–PCR, as the outcome, and lipid traits including HDL, TG, LDL collected at Examination 8, as predictors. To be consistent with the eQTL analysis, a linear mixed effects model was used to control for family structure, and other covariates such like age, sex and the mean of the three housekeeping genes were also included in the model. Subjects with lipid treatments at Examination 8 were excluded from the analysis.
In silico validation of cis- and trans-eQTLs
Twelve genome-wide eQTL datasets from distinct tissues including whole blood (26,41), monocytes (25), CD4+lymphocytes cells (37), lymphocytes (38), PBMCs (39), leukocytes (40), brain (61), liver, stomach and subcutaneous adipose tissue (48) were collected from nine published articles. The sample size, definition of cis- and trans-eQTLs, and the number of SNPs and genes tested in the original publications are shown in Supplementary Material, Table S10. This catalog is limited by the prior methods and platforms of the source eQTL studies and the availability of partial, top results. The selected boundaries for defining cis-eQTLs, and coverage of SNPs, genes and trans-eQTLs varied across studies (Supplementary Material, Table S10). cis- and trans-eSNPs from the current study were cross-referenced with significant eSNPs reported in the twelve eQTL datasets directly by matching SNP rsids.
Accession numbers
The expression levels for these 93 genes and three housekeeping genes were deposited in the NCBI database of genotypes and phenotypes (dbGaP) under the Dataset Name (l_rnatrans_ex08_1_0552 s) and Dataset Accession (pht002077.v2.p6).
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the National Heart, Lung and Blood Institute, Division of Intramural Research.
Supplementary Material
ACKNOWLEDGEMENTS
This research was conducted in part using data and resources from the Framingham Heart Study of the National Heart Lung and Blood Institute of the National Institutes of Health and Boston University School of Medicine. The analyses reflect intellectual input and resource development from the Framingham Heart Study investigators participating in the SNP Health Association Resource (SHARe) project. The authors acknowledge the essential role of the Cohorts for Heart and Aging Research in Genome Epidemiology (CHARGE) Consortium in development and support of this manuscript, especially acknowledging members from the CHARGE Consortium Hematological, Hemostasis, and Subclinical CAC Working Groups for making their GWAS data fully available (Supplementary Material, Acknowledgement).
Conflict of Interest statement. None declared.
REFERENCES
- 1.Roger V.L., Go A.S., Lloyd-Jones D.M., Benjamin E.J., Berry J.D., Borden W.B., Bravata D.M., Dai S., Ford E.S., Fox C.S., et al. Executive summary: heart disease and stroke statistics – 2012 update: a report from the American heart association. Circulation. 2012;125:188–197. doi: 10.1161/CIR.0b013e3182456d46. [DOI] [PubMed] [Google Scholar]
- 2.Detrano R., Guerci A.D., Carr J.J., Bild D.E., Burke G., Folsom A.R., Liu K., Shea S., Szklo M., Bluemke D.A., et al. Coronary calcium as a predictor of coronary events in four racial or ethnic groups. N. Engl. J. Med. 2008;358:1336–1345. doi: 10.1056/NEJMoa072100. doi:10.1056/NEJMoa072100. [DOI] [PubMed] [Google Scholar]
- 3.Polak J.F., Pencina M.J., Pencina K.M., O'Donnell C.J., Wolf P.A., D'Agostino R.B., Sr Carotid-wall intima–media thickness and cardiovascular events. N. Engl. J. Med. 2011;365:213–221. doi: 10.1056/NEJMoa1012592. doi:10.1056/NEJMoa1012592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Danesh J., Lewington S., Thompson S.G., Lowe G.D., Collins R., Kostis J.B., Wilson A.C., Folsom A.R., Wu K., Benderly M., et al. Plasma fibrinogen level and the risk of major cardiovascular diseases and nonvascular mortality: an individual participant meta-analysis. JAMA. 2005;294:1799–1809. doi: 10.1001/jama.294.14.1799. doi:10.1001/jama.294.14.1799. [DOI] [PubMed] [Google Scholar]
- 5.Smith A., Patterson C., Yarnell J., Rumley A., Ben-Shlomo Y., Lowe G. Which hemostatic markers add to the predictive value of conventional risk factors for coronary heart disease and ischemic stroke? The caerphilly study. Circulation. 2005;112:3080–3087. doi: 10.1161/CIRCULATIONAHA.105.557132. doi:10.1161/CIRCULATIONAHA.105.557132. [DOI] [PubMed] [Google Scholar]
- 6.Folsom A.R., Cushman M., Heckbert S.R., Ohira T., Rasmussen-Torvik L., Tsai M.Y. Factor VII coagulant activity, factor VII -670A/C and -402G/A polymorphisms, and risk of venous thromboembolism. J. Thromb. Haemost. 2007;5:1674–1678. doi: 10.1111/j.1538-7836.2007.02620.x. doi:10.1111/j.1538-7836.2007.02620.x. [DOI] [PubMed] [Google Scholar]
- 7.Carpeggiani C., Coceani M., Landi P., Michelassi C., L'Abbate A. ABO Blood group alleles: a risk factor for coronary artery disease. An angiographic study. Atherosclerosis. 2010;211:461–466. doi: 10.1016/j.atherosclerosis.2010.03.012. doi:10.1016/j.atherosclerosis.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 8.Zakai N.A., Katz R., Hirsch C., Shlipak M.G., Chaves P.H., Newman A.B., Cushman M. A prospective study of anemia status, hemoglobin concentration, and mortality in an elderly cohort: the cardiovascular health study. Arch. Intern. Med. 2005;165:2214–2220. doi: 10.1001/archinte.165.19.2214. doi:10.1001/archinte.165.19.2214. [DOI] [PubMed] [Google Scholar]
- 9.Ganesh S.K., Zakai N.A., van Rooij F.J., Soranzo N., Smith A.V., Nalls M.A., Chen M.H., Kottgen A., Glazer N.L., Dehghan A., et al. Multiple loci influence erythrocyte phenotypes in the CHARGE consortium. Nat. Genet. 2009;41:1191–1198. doi: 10.1038/ng.466. doi:10.1038/ng.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Donnell C.J., Nabel E.G. Genomics of cardiovascular disease. N. Engl. J. Med. 2011;365:2098–2109. doi: 10.1056/NEJMra1105239. [DOI] [PubMed] [Google Scholar]
- 11.Schunkert H., Konig I.R., Kathiresan S., Reilly M.P., Assimes T.L., Holm H., Preuss M., Stewart A.F., Barbalic M., Gieger C., et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat. Genet. 2011;43:333–338. doi: 10.1038/ng.784. doi:10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang F., Xu C.Q., He Q., Cai J.P., Li X.C., Wang D., Xiong X., Liao Y.H., Zeng Q.T., Yang Y.Z., et al. Genome-wide association identifies a susceptibility locus for coronary artery disease in the Chinese Han population. Nat. Genet. 2011;43:345–349. doi: 10.1038/ng.783. doi:10.1038/ng.783. [DOI] [PubMed] [Google Scholar]
- 13.The Coronary Artery Disease (C4D) Genetics Consortium. A genome-wide association study in Europeans and south Asians identifies five new loci for coronary artery disease. Nat. Genet. 2011;43:339–344. doi: 10.1038/ng.782. doi:10.1038/ng.782. [DOI] [PubMed] [Google Scholar]
- 14.Reiner A.P., Lettre G., Nalls M.A., Ganesh S.K., Mathias R., Austin M.A., Dean E., Arepalli S., Britton A., Chen Z., et al. Genome-wide association study of white blood cell count in 16,388 African Americans: the continental origins and genetic epidemiology network (COGENT) PLoS Genet. 2011;7:e1002108. doi: 10.1371/journal.pgen.1002108. doi:10.1371/journal.pgen.1002108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson A.D., Yanek L.R., Chen M.H., Faraday N., Larson M.G., Tofler G., Lin S.J., Kraja A.T., Province M.A., Yang Q., et al. Genome-wide meta-analyses identifies seven loci associated with platelet aggregation in response to agonists. Nat. Genet. 2010;42:608–613. doi: 10.1038/ng.604. doi:10.1038/ng.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lovely R.S., Yang Q., Massaro J.M., Wang J., D'Agostino R.B., Sr, O'Donnell C.J., Shannon J., Farrell D.H. Assessment of genetic determinants of the association of {gamma}’ fibrinogen in relation to cardiovascular disease. Arterioscler Thromb. Vasc. Biol. 2011;31:2345–2352. doi: 10.1161/ATVBAHA.111.232710. doi:10.1161/ATVBAHA.111.232710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dehghan A., Yang Q., Peters A., Basu S., Bis J.C., Rudnicka A.R., Kavousi M., Chen M.H., Baumert J., Lowe G.D., et al. Association of novel genetic loci with circulating fibrinogen levels: a genome-wide association study in 6 population-based cohorts. Circ. Cardiovasc. Genet. 2009;2:125–133. doi: 10.1161/CIRCGENETICS.108.825224. doi:10.1161/CIRCGENETICS.108.825224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith N.L., Chen M.H., Dehghan A., Strachan D.P., Basu S., Soranzo N., Hayward C., Rudan I., Sabater-Lleal M., Bis J.C., et al. Novel associations of multiple genetic loci with plasma levels of factor VII, factor VIII, and von willebrand factor: the CHARGE (cohorts for heart and aging research in genome epidemiology) consortium. Circulation. 2010;121:1382–1392. doi: 10.1161/CIRCULATIONAHA.109.869156. doi:10.1161/CIRCULATIONAHA.109.869156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bis J.C., Kavousi M., Franceschini N., Isaacs A., Abecasis G.R., Schminke U., Post W.S., Smith A.V., Cupples L.A., Markus H.S., et al. Meta-analysis of genome-wide association studies from the CHARGE consortium identifies common variants associated with carotid intima media thickness and plaque. Nat. Genet. 2011;43:940–947. doi: 10.1038/ng.920. doi:10.1038/ng.920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Donnell C.J., Kavousi M., Smith A.V., Kardia S.L., Feitosa M.F., Hwang S.J., Sun Y.V., Province M.A., Aspelund T., Dehghan A., et al. Genome-wide association study for coronary artery calcification With follow-up in myocardial infarction. Circulation. 2011;124:2855–2864. doi: 10.1161/CIRCULATIONAHA.110.974899. doi:10.1161/CIRCULATIONAHA.110.974899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sadee W., Wang D., Papp A.C., Pinsonneault J.K., Smith R.M., Moyer R.A., Johnson A.D. Pharmacogenomics of the RNA world: structural RNA polymorphisms in drug therapy. Clin. Pharmacol. Ther. 2011;89:355–365. doi: 10.1038/clpt.2010.314. doi:10.1038/clpt.2010.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ioannidis J.P., Thomas G., Daly M.J. Validating, augmenting and refining genome-wide association signals. Nat. Rev. Genet. 2009;10:318–329. doi: 10.1038/nrg2544. doi:10.1038/nrg2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheung V.G., Spielman R.S. Genetics of human gene expression: mapping DNA variants that influence gene expression. Nat. Rev. Genet. 2009;10:595–604. doi: 10.1038/nrg2630. doi:10.1038/nrg2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cookson W., Liang L., Abecasis G., Moffatt M., Lathrop M. Mapping complex disease traits with global gene expression. Nat. Rev. Genet. 2009;10:184–194. doi: 10.1038/nrg2537. doi:10.1038/nrg2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zeller T., Wild P., Szymczak S., Rotival M., Schillert A., Castagne R., Maouche S., Germain M., Lackner K., Rossmann H., et al. Genetics and beyond – the transcriptome of human monocytes and disease susceptibility. PLoS ONE. 2010;5:e10693. doi: 10.1371/journal.pone.0010693. doi:10.1371/journal.pone.0010693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fehrmann R.S., Jansen R.C., Veldink J.H., Westra H.J., Arends D., Bonder M.J., Fu J., Deelen P., Groen H.J., Smolonska A., et al. Trans-eQTLs reveal that independent genetic variants associated with a complex phenotype converge on intermediate genes, with a major role for the HLA. PLoS Genet. 2011;7:e1002197. doi: 10.1371/journal.pgen.1002197. doi:10.1371/journal.pgen.1002197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teslovich T.M., Musunuru K., Smith A.V., Edmondson A.C., Stylianou I.M., Koseki M., Pirruccello J.P., Ripatti S., Chasman D.I., Willer C.J., et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. doi:10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Freedman J.E., Larson M.G., Tanriverdi K., O'Donnell C.J., Morin K., Hakanson A.S., Vasan R.S., Johnson A.D., Iafrati M.D., Benjamin E.J. Relation of platelet and leukocyte inflammatory transcripts to body mass index in the Framingham heart study. Circulation. 2010;122:119–129. doi: 10.1161/CIRCULATIONAHA.109.928192. doi:10.1161/CIRCULATIONAHA.109.928192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li J., Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity (Edinburgh) 2005;95:221–227. doi: 10.1038/sj.hdy.6800717. doi:10.1038/sj.hdy.6800717. [DOI] [PubMed] [Google Scholar]
- 30.Manolio T.A., Collins F.S., Cox N.J., Goldstein D.B., Hindorff L.A., Hunter D.J., McCarthy M.I., Ramos E.M., Cardon L.R., Chakravarti A., et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. doi:10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang J., Johnson A.D., O'Donnell C.J. PRIMe: a method for characterization and evaluation of pleiotropic regions from multiple genome-wide association studies. Bioinformatics. 2011;27:1201–1206. doi: 10.1093/bioinformatics/btr116. doi:10.1093/bioinformatics/btr116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dixon A.L., Liang L., Moffatt M.F., Chen W., Heath S., Wong K.C., Taylor J., Burnett E., Gut I., Farrall M., et al. A genome-wide association study of global gene expression. Nat. Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. doi:10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 33.Newton-Cheh C., Johnson T., Gateva V., Tobin M.D., Bochud M., Coin L., Najjar S.S., Zhao J.H., Heath S.C., Eyheramendy S., et al. Genome-wide association study identifies eight loci associated with blood pressure. Nat. Genet. 2009;41:666–676. doi: 10.1038/ng.361. doi:10.1038/ng.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wain L.V., Verwoert G.C., O'Reilly P.F., Shi G., Johnson T., Johnson A.D., Bochud M., Rice K.M., Henneman P., Smith A.V., et al. Genome-wide association study identifies six new loci influencing pulse pressure and mean arterial pressure. Nat. Genet. 2011;43:1005–1011. doi: 10.1038/ng.922. doi:10.1038/ng.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ehret G.B., Munroe P.B., Rice K.M., Bochud M., Johnson A.D., Chasman D.I., Smith A.V., Tobin M.D., Verwoert G.C., Hwang S.J., et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. doi: 10.1038/nature10405. doi:10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kathiresan S., Voight B.F., Purcell S., Musunuru K., Ardissino D., Mannucci P.M., Anand S., Engert J.C., Samani N.J., Schunkert H., et al. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat. Genet. 2009;41:334–341. doi: 10.1038/ng.327. doi:10.1038/ng.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murphy A., Chu J.H., Xu M., Carey V.J., Lazarus R., Liu A., Szefler S.J., Strunk R., Demuth K., Castro M., et al. Mapping of numerous disease-associated expression polymorphisms in primary peripheral blood CD4+ lymphocytes. Hum. Mol. Genet. 2010;19:4745–4757. doi: 10.1093/hmg/ddq392. doi:10.1093/hmg/ddq392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goring H.H., Curran J.E., Johnson M.P., Dyer T.D., Charlesworth J., Cole S.A., Jowett J.B., Abraham L.J., Rainwater D.L., Comuzzie A.G., et al. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat. Genet. 2007;39:1208–1216. doi: 10.1038/ng2119. doi:10.1038/ng2119. [DOI] [PubMed] [Google Scholar]
- 39.Heinzen E.L., Ge D., Cronin K.D., Maia J.M., Shianna K.V., Gabriel W.N., Welsh-Bohmer K.A., Hulette C.M., Denny T.N., Goldstein D.B. Tissue-specific genetic control of splicing: implications for the study of complex traits. PLoS Biol. 2008;6:e1. doi: 10.1371/journal.pbio.1000001. doi:10.1371/journal.pbio.1000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Idaghdour Y., Czika W., Shianna K.V., Lee S.H., Visscher P.M., Martin H.C., Miclaus K., Jadallah S.J., Goldstein D.B., Wolfinger R.D., et al. Geographical genomics of human leukocyte gene expression variation in southern morocco. Nat. Genet. 2010;42:62–67. doi: 10.1038/ng.495. doi:10.1038/ng.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Emilsson V., Thorleifsson G., Zhang B., Leonardson A.S., Zink F., Zhu J., Carlson S., Helgason A., Walters G.B., Gunnarsdottir S., et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–428. doi: 10.1038/nature06758. doi:10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- 42.Soranzo N., Spector T.D., Mangino M., Kuhnel B., Rendon A., Teumer A., Willenborg C., Wright B., Chen L., Li M., et al. A genome-wide meta-analysis identifies 22 loci associated with eight hematological parameters in the haemGen consortium. Nat. Genet. 2009;41:1182–1190. doi: 10.1038/ng.467. doi:10.1038/ng.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu J., Zhang B., Smith E.N., Drees B., Brem R.B., Kruglyak L., Bumgarner R.E., Schadt E.E. Integrating large-scale functional genomic data to dissect the complexity of yeast regulatory networks. Nat. Genet. 2008;40:854–861. doi: 10.1038/ng.167. doi:10.1038/ng.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang X., Deignan J.L., Qi H., Zhu J., Qian S., Zhong J., Torosyan G., Majid S., Falkard B., Kleinhanz R.R., et al. Validation of candidate causal genes for obesity that affect shared metabolic pathways and networks. Nat. Genet. 2009;41:415–423. doi: 10.1038/ng.325. doi:10.1038/ng.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen Y., Zhu J., Lum P.Y., Yang X., Pinto S., Macneil D.J., Zhang C., Lamb J., Edwards S., Sieberts S.K., et al. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008;452:429–435. doi: 10.1038/nature06757. doi:10.1038/nature06757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heinig M., Petretto E., Wallace C., Bottolo L., Rotival M., Lu H., Li Y., Sarwar R., Langley S.R., Bauerfeind A., et al. A trans-acting locus regulates an anti-viral expression network and type 1 diabetes risk. Nature. 2010;467:460–464. doi: 10.1038/nature09386. doi:10.1038/nature09386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nicolae D.L., Gamazon E., Zhang W., Duan S., Dolan M.E., Cox N.J. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. doi:10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Greenawalt D.M., Dobrin R., Chudin E., Hatoum I.J., Suver C., Beaulaurier J., Zhang B., Castro V., Zhu J., Sieberts S.K., et al. A survey of the genetics of stomach, liver, and adipose gene expression from a morbidly obese cohort. Genome Res. 2011;21:1008–1016. doi: 10.1101/gr.112821.110. doi:10.1101/gr.112821.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson A.D., Zhang Y., Papp A.C., Pinsonneault J.K., Lim J.E., Saffen D., Dai Z., Wang D., Sadee W. Polymorphisms affecting gene transcription and mRNA processing in pharmacogenetic candidate genes: detection through allelic expression imbalance in human target tissues. Pharmacogenet. Genomics. 2008;18:781–791. doi: 10.1097/FPC.0b013e3283050107. doi:10.1097/FPC.0b013e3283050107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harismendy O., Notani D., Song X., Rahim N.G., Tanasa B., Heintzman N., Ren B., Fu X.D., Topol E.J., Rosenfeld M.G., et al. 9p21 DNA variants associated with coronary artery disease impair interferon-gamma signalling response. Nature. 2011;470:264–268. doi: 10.1038/nature09753. doi:10.1038/nature09753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Musunuru K., Strong A., Frank-Kamenetsky M., Lee N.E., Ahfeldt T., Sachs K.V., Li X., Li H., Kuperwasser N., Ruda V.M., et al. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature. 2010;466:714–719. doi: 10.1038/nature09266. doi:10.1038/nature09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Folkersen L., van't Hooft F., Chernogubova E., Agardh H.E., Hansson G.K., Hedin U., Liska J., Syvanen A.C., Paulsson-Berne G., Franco-Cereceda A., et al. Association of genetic risk variants with expression of proximal genes identifies novel susceptibility genes for cardiovascular disease. Circ. Cardiovasc. Genet. 2010;3:365–373. doi: 10.1161/CIRCGENETICS.110.948935. doi:10.1161/CIRCGENETICS.110.948935. [DOI] [PubMed] [Google Scholar]
- 53.Montgomery S.B., Dermitzakis E.T. From expression QTLs to personalized transcriptomics. Nat. Rev. Genet. 2011;12:277–282. doi: 10.1038/nrg2969. doi:10.1038/nrg2969. [DOI] [PubMed] [Google Scholar]
- 54.Birney E., Stamatoyannopoulos J.A., Dutta A., Guigo R., Gingeras T.R., Margulies E.H., Weng Z., Snyder M., Dermitzakis E.T., Thurman R.E., et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. doi:10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dawber T.R., Kannel W.B., Lyell L.P. An approach to longitudinal studies in a community: the Framingham study. Ann. N. Y. Acad. Sci. 1963;107:539–556. doi: 10.1111/j.1749-6632.1963.tb13299.x. doi:10.1111/j.1749-6632.1963.tb13299.x. [DOI] [PubMed] [Google Scholar]
- 56.Kannel W.B., Feinleib M., McNamara P.M., Garrison R.J., Castelli W.P. An investigation of coronary heart disease in families. The Framingham offspring study. Am. J. Epidemiol. 1979;110:281–290. doi: 10.1093/oxfordjournals.aje.a112813. [DOI] [PubMed] [Google Scholar]
- 57.Feinleib M., Kannel W.B., Garrison R.J., McNamara P.M., Castelli W.P. The Framingham offspring study. Design and preliminary data. Prev. Med. 1975;4:518–525. doi: 10.1016/0091-7435(75)90037-7. doi:10.1016/0091-7435(75)90037-7. [DOI] [PubMed] [Google Scholar]
- 58.Splansky G.L., Corey D., Yang Q., Atwood L.D., Cupples L.A., Benjamin E.J., D'Agostino R.B., Sr, Fox C.S., Larson M.G., Murabito J.M., et al. The third generation cohort of the national heart, lung, and blood institute's Framingham heart study: design, recruitment, and initial examination. Am. J. Epidemiol. 2007;165:1328–1335. doi: 10.1093/aje/kwm021. doi:10.1093/aje/kwm021. [DOI] [PubMed] [Google Scholar]
- 59.Li Y., Willer C.J., Ding J., Scheet P., Abecasis G.R. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. doi:10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dupuis J., Siegmund D.O., Yakir B. A unified framework for linkage and association analysis of quantitative traits. Proc. Natl. Acad. Sci. USA. 2007;104:20210–20215. doi: 10.1073/pnas.0707138105. doi:10.1073/pnas.0707138105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Myers A.J., Gibbs J.R., Webster J.A., Rohrer K., Zhao A., Marlowe L., Kaleem M., Leung D., Bryden L., Nath P., et al. A survey of genetic human cortical gene expression. Nat. Genet. 2007;39:1494–1499. doi: 10.1038/ng.2007.16. doi:10.1038/ng.2007.16. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


