Abstract
Surface plasmon resonance (SPR) is a medical diagnosis technique with high sensitivity and specificity. In this research, a new method based on SPR is proposed for rapid, 10-minute detection of the anti-dengue virus in human serum samples. This novel technique, known as rapid immunoglobulin M (IgM)-based dengue diagnostic test, can be utilized quickly and easily at the point of care. Four dengue virus serotypes were used as ligands on a biochip. According to the results, a serum volume of only 1 μl from a dengue patient (as a minimized volume) is required to indicate SPR angle variation to determine the ratio of each dengue serotype in samples with 83–93% sensitivity and 100% specificity.
Dengue is an acute febrile illness, globally recognized as one of the most significant vector-borne human diseases. It is caused by the dengue virus, which is carried by, and introduced into a human host by a female Aedes aegypti mosquito. In the past, the disease was geographically restricted to tropical and subtropical zones, but more recently with factors such increasing human migration and unplanned urbanization, the spread of disease has expanded1. Dengue fever (DF) and its more serious forms, dengue hemorrhagic fever (DHF) and dengue shock syndrome (DSS), have become a major global health problem. These were formally included within the disease portfolio of the World Health Organization's (WHO) special program for research and training in tropical diseases by the Joint Coordination Board in June 1999. The global prevalence of dengue has grown dramatically in recent decades. According to WHO, around 3.6 billion people, or more than half of the world's population, are now at risk from dengue2,3,4,5. Currently, the disease is endemic in over 100 tropical and sub-tropical countries and roughly 390 million cases of dengue infections are estimated worldwide every year6,7.
The treatment of this disease, however, can be simple, inexpensive and effective provided that correct and early diagnosis is performed. This is only feasible if the clinical problems and disease phases are known, especially when patients are first seen and examined in triage. For proper disease management, a full blood count should be done during the first visit. A hematocrit (HCT) test establishes the patient's individual baseline, from which a subsequent decrease in white blood cell count indicates a high probability of dengue. A rapid decrease in platelets with rising HCT suggests advancement towards a critical disease phase. Current biomedical diagnostics procedures include the enzyme-linked immune sorbent assay (ELISA) technique and Rapid Diagnostic Tests (RDTs)8,9, which are commonly used to detect Non Structural protein 1 (NS1)10,11,12, Immunoglobulin M (IgM)13,14, and Immunoglobulin G (IgG)8. ELISA is limited by slow processing due to the required incubation period (from a few hours to 2 days) and does not provide sensitive detection in non-laboratory settings typical of point of care (POC)15. Additionally, the automated ELISA system requires high-level expertise to operate the expensive, bulky equipment and consumes considerable amounts of chemicals, for which reasons it is not available in many hospitals. In rapid immune chromatography characterized by ease of use and rapid detection rate, only one drop of blood is necessary for diagnosis16,17. However, this method is only suitable for screening since it is cannot deliver high sensitivity and specificity results.
There are several commercial antibody detection kits for dengue virus identification. The most popular screening methods are the immunoassay method (ELISA), dipstick and rapid testing with the immune-chromatographic dot blot. In conventional methods18,19,20 the diagnostic procedure is time-consuming, requiring a lengthy process conducted by well-trained staff. The ELISA method requires several sequential, time-controlled steps that may take more than 6 hours to complete. Since the method relies on manual intervention, it can render expensive and provide inaccurate results.
Surface plasmon resonance (SPR) is an optical technique with prospective application in probing for refractive index changes that generally occur within the immediate vicinity of a sensor surface. It additionally forms the basis of many sensing tools for measuring material adsorption on planar metal surfaces (typically gold and silver) or on the surfaces of metal nanoparticles, such as several color-based biosensors and lab-on-a-chip sensors21,22,23,24,25. Initially, SPR was used to investigate the inherent optical properties of thin metal films. Subsequent usage has been extended to a variety of other applications26,27,28,29,30. In these sensors, a surface plasmon mode (wave) is excited at the interface between a metal film and a dielectric medium using a light wave. A change in the dielectric medium's refractive index produces a modification in surface plasmon mode propagation. Consequently, the coupling condition between the light wave and surface plasmon wave is altered, which becomes evident as a change in one of the characteristics of the optical wave interacting with the surface plasmon mode31,32,33.
The aim of this study is to propose a technique for the early detection of the dengue virus using the surface plasmon resonance method. The technique assumes the immobilized antigen of all four dengue serotypes is a ligand as opposed to an antibody commonly assumed in conventional methods.
Results
Surface plasmon resonance is proposed for the rapid detection of anti-dengue virus in human serum samples within 10 minutes. All four dengue virus serotypes were immobilized onto the biochip surface (Fig. 1). Following the immobilization stage, the patient sera were categorized as high positive (HP), mid positive (MP), and low positive (LP) samples. These were optimized such that only 1 μl was required, rendering the method extremely suitable for POC environments. Some samples without the dengue virus were provided in order to taking specificity into account as well. The experiments were carried out using samples supplied by the Department of Microbiology, University of Malaya. The samples included high positive (antibody titre 1280–10240 or more), mid positive (antibody titre 160–640), and low positive (antibody titre 10–160), and were classified via hemagglutination inhibition (HI) antibody and ELISA tests (Table 1)5,34. Furthermore, some samples with tick-borne encephalitis (TBE) and hepatitis C (HC) viruses were provided in conjunction with the specificity investigation as shown in Table 2. The samples were tested with the SPR method to ensure high sensitivity and specificity.
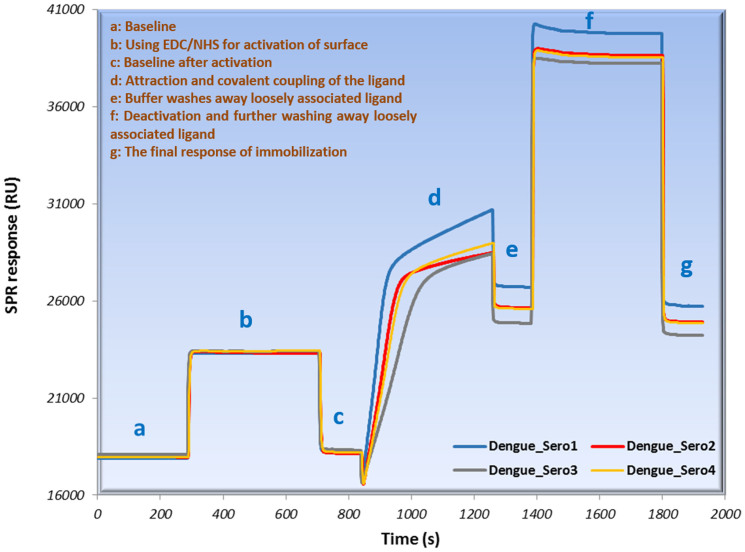
Figure 1. Immobilization sensorgram of four serotypes of dengue antigen on sensor surface.
Table 1. Comparative data base of ELISA and proposed SPR biosensor in low, mid and high positive patient samples of dengue virus.
| ELISA results | Biosensor results (ΔθSPR) | |||||||
|---|---|---|---|---|---|---|---|---|
| Type of sample (HI) | Patient serum | P/N ratio | NS1 | IgM | S.T. 1 | S.T. 2 | S.T. 3 | S.T. 4 |
| Low positive (Antibody titre 10–160) | AH01 | 2.48 | − | + | 1.0110 | 0.7454 | 0.5808 | 0.5923 |
| AH02 | 2.67 | − | + | 1.0570 | 0.7186 | 0.5845 | 0.6344 | |
| AH03 | 2.59 | + | + | 1.0688 | 0.7211 | 0.5785 | 0.6369 | |
| AH04 | 2.47 | + | + | 1.0013 | 0.6498 | 0.5499 | 0.5957 | |
| AH05 | 2.81 | + | + | 1.0102 | 0.6694 | 0.5162 | 0.5572 | |
| AH06 | 2.47 | + | + | 1.0060 | 0.6560 | 0.5080 | 0.5467 | |
| AH07 | 2.79 | + | + | 0.9665 | 0.6910 | 0.5548 | 0.6105 | |
| AH08 | 2.14 | + | + | 1.0019 | 0.7015 | 0.5002 | 0.5844 | |
| AH09 | 2.93 | − | + | 1.1015 | 0.7528 | 0.5775 | 0.6512 | |
| AH10 | 2.58 | − | + | 1.0104 | 0.7501 | 0.5771 | 0.6081 | |
| Mid positive (Antibody titre 160–640) | AH11 | 3.01 | + | + | 0.9983 | 0.8863 | 0.6866 | 0.7051 |
| AH12 | 3.93 | + | + | 1.0574 | 0.9095 | 0.5849 | 0.8077 | |
| AH13 | 3.45 | − | + | 1.1691 | 0.7965 | 0.5987 | 0.7498 | |
| AH14 | 3.75 | + | + | 0.9862 | 0.9380 | 0.6720 | 0.7899 | |
| AH15 | 3.34 | − | + | 1.1001 | 0.8754 | 0.6972 | 0.7386 | |
| AH16 | 4.59 | − | + | 1.1124 | 0.9566 | 0.7399 | 0.8211 | |
| AH17 | 4.5 | + | + | 1.1092 | 0.9487 | 0.7410 | 0.8222 | |
| AH18 | 4.96 | + | + | 1.1978 | 0.9637 | 0.7405 | 0.8230 | |
| AH19 | 4.34 | − | + | 1.0543 | 0.7688 | 0.7337 | 0.6968 | |
| AH20 | 4.63 | + | + | 1.1733 | 0.9598 | 0.7234 | 0.8176 | |
| High positive (Antibody titre 1280–10240) | AH21 | 5.25 | + | + | 1.1255 | 1.0114 | 0.7154 | 0.9045 |
| AH22 | 5.69 | + | + | 1.1469 | 0.9778 | 0.7740 | 0.8304 | |
| AH23 | 5.12 | + | + | 1.1344 | 0.9463 | 0.7595 | 0.7983 | |
| AH24 | 5.6 | + | + | 1.1226 | 0.9462 | 0.7519 | 0.8084 | |
| AH25 | 6.53 | − | + | 1.1215 | 0.9084 | 0.7585 | 0.7845 | |
| AH26 | 7.13 | − | + | 1.1987 | 1.0695 | 0.7021 | 0.8793 | |
| AH27 | 7.32 | + | + | 1.2731 | 1.1128 | 0.8676 | 0.9519 | |
| AH28 | 6.06 | + | + | 1.1996 | 0.9954 | 0.7567 | 0.8922 | |
| AH29 | 7.02 | − | + | 1.2453 | 1.0532 | 0.8063 | 0.9476 | |
| AH30 | 7.38 | − | + | 1.2782 | 0.9869 | 0.7631 | 0.8541 | |
Table 2. The negative controls and the number of the serum samples were examined for the specificity evaluation in this study.
| ELISA results | Biosensor results (|ΔθSPR|) | ||||||
|---|---|---|---|---|---|---|---|
| Type of sample | Patient serum | NS1 | IgM | S.T. 1 | S.T. 2 | S.T. 3 | S.T. 4 |
| Tick-Borne Encephalitis | SAM01 | − | − | 0.31852 | 0.3760 | 0.19469 | 0.32879 |
| SAM02 | − | − | 0.15085 | 0.2557 | 0.21550 | 0.19386 | |
| SAM 03 | − | − | 0.37746 | 0.2860 | 0.00040 | 0.19946 | |
| SAM 04 | − | − | 0.16855 | 0.2940 | 0.04890 | 0.16955 | |
| SAM 05 | − | − | 0.14738 | 0.1254 | 0.02943 | 0.13294 | |
| SAM 06 | − | − | 0.02807 | 0.1651 | 0.25771 | 0.08001 | |
| SAM 07 | − | − | 0.25098 | 0.0770 | 0.18170 | 0.10140 | |
| SAM 08 | − | − | 0.03523 | 0.0973 | 0.02780 | 0.07478 | |
| SAM 09 | − | − | 0.04094 | 0.0646 | 0.07810 | 0.04010 | |
| SAM 10 | − | − | 0.05042 | 0.0050 | 0.01400 | 0.01260 | |
| SAM 11 | − | − | 0.24308 | 0.0911 | 0.25786 | 0.04538 | |
| SAM 12 | − | − | 0.11004 | 0.0011 | 0.25924 | 0.09303 | |
| SAM 13 | − | − | 0.25980 | 0.1937 | 0.21978 | 0.09314 | |
| SAM 14 | − | − | 0.35171 | 0.2944 | 0.31243 | 0.20877 | |
| SAM 15 | − | − | 0.2852 | 0.1905 | 0.23039 | 0.10882 | |
| SAM 16 | − | − | 0.21199 | 0.1115 | 0.14507 | 0.01650 | |
| SAM 17 | − | − | 0.22769 | 0.1312 | 0.17126 | 0.03061 | |
| SAM 18 | − | − | 0.28431 | 0.1407 | 0.15434 | 0.00509 | |
| SAM 19 | − | − | 0.25883 | 0.1396 | 0.17210 | 0.04012 | |
| Hepatitis C | SAM 20 | − | − | 0.21092 | 0.1737 | 0.23510 | 0.11329 |
| SAM 21 | − | − | 0.23587 | 0.1497 | 0.24237 | 0.09972 | |
| SAM 22 | − | − | 0.30437 | 0.1798 | 0.19558 | 0.06360 | |
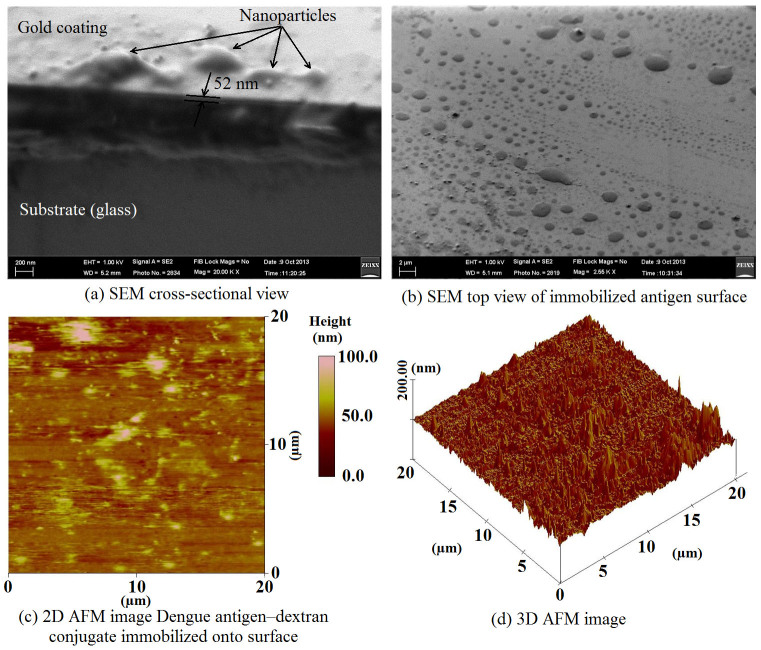
The surface on which the antigens (Ag) were immobilized was characterized using scanning electron microscopy (FE/SEM Quanta FEG250) and atomic force microscopy (VEECO DIMENSION 3000 AFM). AFM imaging was performed in contact mode using 0.01–0.025 Ohm-cm antimony (n)-doped silicon probes. Figure 2 (a) exemplifies a typical cross-sectional view of gold coating on glass accompanied by nanoparticles such as amine groups and immobilized antigens. According to the SEM, the gold coating was around 52 nm thick. Figure 2 (b) displays the top view of the gold surface with immobilized antigens on it. For further investigation 2D and 3D images of AFM were employed to prove the existence of immobilized antigens (Figs. 2 (c) and (d)). The 3D AFM image shows the surface of the gold-coated glass with two distinctive types of hills: first, homogenous, dense and low gold hills resulting from amine groups and second, sporadic higher hills created by immobilized antigens. The amine groups acted as a binding protein to the sensing antigen, which anchored very well to the sensor surface.
Figure 2. Characterization of the immobilization process on the gold surface using SEM and AFM equipments.
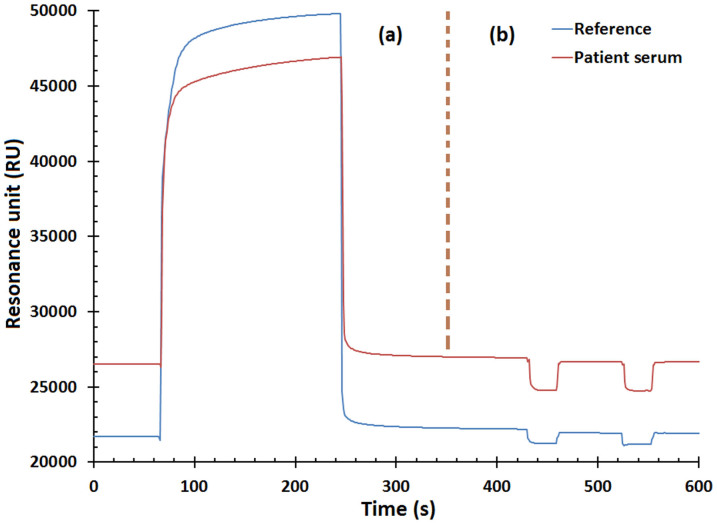
The binding response sensorgram resulting from dengue high-positive serum (as an example) along with the control experiment (as reference) shows the amount of binding interaction on the biosensor's gold surface (Fig. 3). Upon injecting the patient's sample, the response increased exponentially representing the amount of dengue antibody bound with its antigens during injection. Binding will not occur if the patient sample does not carry dengue antibody or if it carries non-dengue antibodies.
Figure 3. The binding response curve termed by sensorgram, (a) the binding process and (b) the regeneration of biosensor surface.
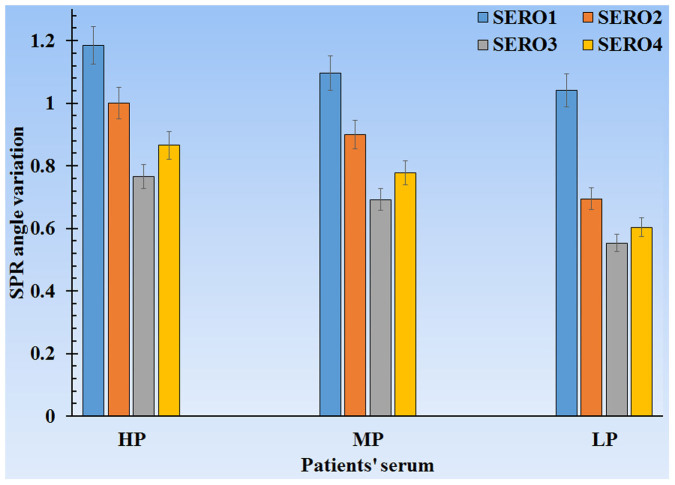
As previously mentioned, the samples are categorized into three groups: HP, MP, and LP. Each category has four bars that represent the four dengue virus types (Figure 4).
Figure 4. SPR angle variation via patient's serum- dengue virus diagnosis graph.
Discussion
In the current study, a means of early detection of all four dengue virus serotypes via surface plasmon resonance was introduced. The technique considers the immobilized antigen to be a ligand as an alternative to the conventionally used antibody. Although Sunita Kumbhat et al.35 reported that with the SPR technique it is possible to detect the dengue virus they did not categorize all four dengue serotypes. Moreover, we have demonstrated that only 1 μl of dengue-patient serum could indicate the SPR angle variation that determines the ratio of each dengue serotype in the samples. Then all four virus serotypes were categorized with high sensitivity and specificity.
Figure 1 ((a) to (g)) illustrates the immobilization process of four dengue virus serotypes in seven steps. The SEM and AFM images confirm that virus immobilization took place correctly (Fig. 2). Figure 3 (b) shows two dips at the end of the sensorgram caused by SPR setup adjustment. The dips indicate that the chip surface regenerated twice and all Ag-Ab binding was removed by the glycine buffer11. The gold surface of the sensor should ideally be cleaned with minimal influence from previous binding, something substantiated by the control experiment. It is also worth noting that the entire assay took only ten minutes.
Figure 4 manifests the SPR angle variations in terms of patient serum. Clearly, the maximum SPR angle variations occurred in HP, MP, and LP. Each HP, MP, and LP is the average of ten samples of its type, and every bar may also serve as reference for diagnosing every dengue virus type. This method can determine the ratio of four dengue virus serotypes, and with this ratio, specialists can effortlessly and correctly decide whether to keep patients for observation (hospitalize) or send them home. The SPR angle variations in each dengue virus serotype tend to have a linear slope. Such linearity disparities can generate boundaries distinguished by HP, MP, and LP.
A comparative study was performed between the proposed SPR technique and the conventional ELISA method to validate the results (Table 1). The NS1, positive/negative (P/N) ratio and IgM results of each patient serum are displayed based on the ELISA method. The first test run was NS1, whose positive result indicates the presence of dengue virus in blood; however, to achieve further validity, an IgM test needed to be conducted as well. It is particularly vital to perform an IgM test for final confirmation in samples with negative NS1.
Table 1 lists the positive IgM results proving the presence of the dengue virus in all samples, along with IgM antibody quantity (P/N ratio). The proposed method outcome shows the binding ratio between IgM antibodies in samples with all four serotypes of the immobilized dengue virus on the chip surface. The changes in the quantity of surface-bound antibodies in the four serotypes were measured by monitoring each sample's surface plasmon resonance angle.
To identify the sensor's detection sensitivity, the change in angle matching the minimum reflection coefficient, or the SPR angle (ΔθSPR), was calculated for every experiment (Table 1). The sensitivity of the sensor then related to the test's ability to identify positive results in contrast to conventional method results, which is 83–93% in this research.
The samples with TBE and HC viruses (negative dengue NS1 and subsequently negative dengue IgM results) are presented in Table 2. The final outcome of the proposed method indicates there is an insignificant change in the SPR angle of all four serotypes. Such minute change implies there is no binding between TBE/HC antibody present in samples and the dengue virus serotypes. According to Table 2, there are no false positive results and the specificity of 100% was obtained.
Methods
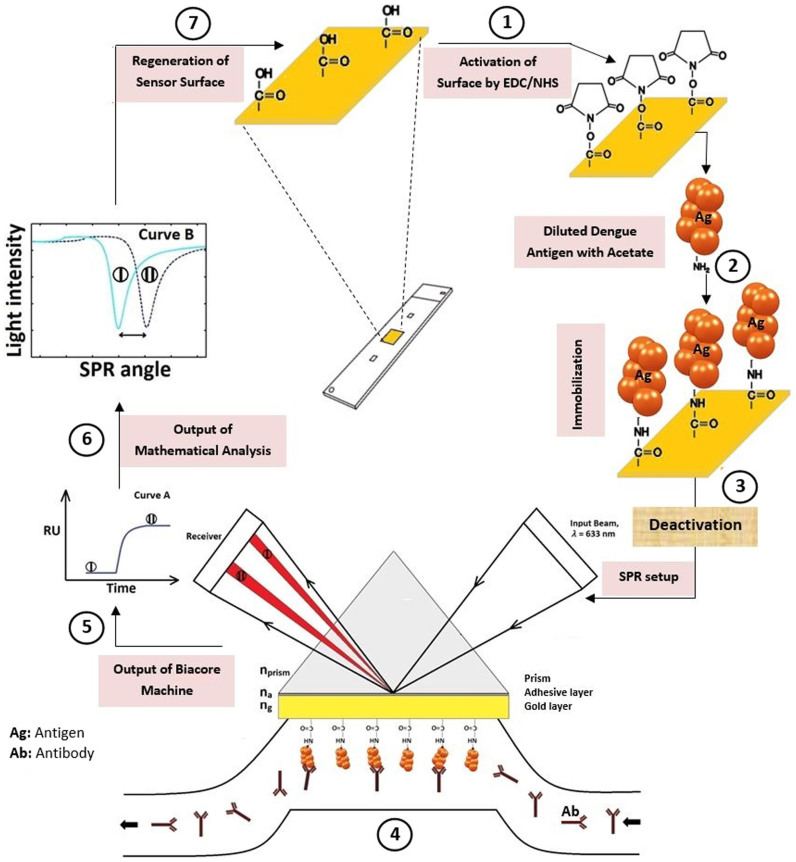
In the current work, a CM5 sensor chip was used for dengue virus detection. Figure 5 shows the schematic of the dengue virus diagnosis process. A BIAcore 3000 (GE Healthcare) system36,37 was utilized for real-time biomolecular interaction analyses based on surface plasmon resonance (SPR) technique. This method monitors the forming and dissociation of biomolecular complexes on the chip surface. By covalently attaching a molecule (as a ligand) to the chip surface, the binding of another molecule in solution (as analyte) with the immobilized molecule is achievable38. Unlike conventional methods, the SPR technique does not require the labeling of interacting components.
Figure 5. Schematic of the dengue virus diagnosis process.
In the serological approach, immunoglobulins (IgM, IgG, and IgA) are produced from the immune system's reaction to dengue infection. These are distinct to virus (E) protein. Depending on the patient's condition of whether or not they have a primary or secondary infection, the sharpness of the response changes. Usually, for a primary infection the IgM response has higher titre than for secondary34. Due to this distinctive characteristic of the IgM antibody and owing to the importance of presenting a rapid diagnostic method, the authors decided to utilize IgM.
The serum samples containing IgM were provided by the University of Malaya (UM) Medical Center. N-hydroxysuccinimide (NHS) and N-ethyl-N-(dimethylaminopropyl) carbodiimide (EDC) were used to activate the biosensor's sensor prior to injecting the ligand. To wash and remove bounded material from the sensor surface and to complete the immobilization procedure 80 μl ethanolamine and 500 μl 10 mM glycine-HCI buffer with pH 2.0 were used. 10 mM sodium acetate with pH 4.5 diluted the sample to obtain adequate concentration for the assay process.
There are different ways to immobilize substances on a sensor surface. Immobilization method selection depends on the substance properties. The immobilization approaches may be directed towards amine, carboxyl, thiol or hydroxyl groups on the ligand, or using specific tags attached to the ligand. An amine coupling chemistry was chosen, as it is the most widely applicable method to covalently attach biomolecules to the sensor chip surface and is suitable for the ligand. With this method, the dextran matrix on the sensor chip surface was initially activated with a mixture of 120 μl EDC and 120 μl NHS to produce reactive succinimide esters. The ligand was subsequently passed over the surface and the esters reacted spontaneously with amine groups to covalently link the ligand to the dextran.
The 6 μl ligand (each serotype of dengue antigen) was diluted to 194 μl acetate buffer (concentration of 3:100). After injecting the ligand, ethanolamine was passed over the sensor surface to deactivate remaining active esters. The chip was then inserted into the SPR apparatus to measure the SPR angle variations for each sample. The SPR device generated two sensorgrams (RU via time) for every experiment (sample result and its control experiment), which measured the response in terms of resonance units (RU) or in other words, is proportional to the molecular mass on the surface. For an interactant of a given mass, the response is proportional to the number of molecules at the surface. A sensorgram provides a plot of response against time showing the interaction progress that can be monitored in real-time throughout the analysis. The BIAcore machine results were then converted to display a graph of intensity via incident angle, where 1000 RU is equivalent to 0.1 angle variations.
The optimal level of immobilized ligand depends on the objective of the analysis. In this case, the ligand was immobilized on a gold surface to act as a probe on the chip surface. EDC/NHS helped activate the sensor surface (Fig. 5, step 1). After surface activation, attraction and covalent coupling of the ligand occurred, after which a buffer washed away loosely-associated ligand (Fig. 5, step 2). The response level at this point provided the first indication of the immobilized amount. Deactivation and further washing away of loosely-associated ligand were done using ethanolamine (Fig. 5, step 3). In addition, a moderate flow rate (10 μl/min) was employed for immobilization.
As shown in Fig. 5 (step 4), the dengue antibodies bound to immobilized antigens on the gold surface. The binding interaction between Ags and Abs was monitored by changing the SPR angle in real-time.
Changes in SPR angle were investigated with BIAcore 3000 to identify the existence of the anti-dengue virus IgM in samples. Data from BIAcore (Fig. 5, step 5) was converted to intensity via incident angle (Fig. 5, step 6) using Matlab programming to monitor SPR angle variations. The sensor surface was regenerated at the end of each experiment to remove the bound analyte from the immobilized ligands on the surface (Fig. 5, step 7).
Author Contributions
According to theoretical surface plasmon resonance method which is proposed by P.J., he has done all experiments through SPR setup in Microbiology Department, University of Malaya. He has also collected and analyzed the experimental data. All figures including SEM and AFM surface characterization and graphs are provided by Dr. E.Z. along with P.J. and Prof. S.D.S. who is our advisor in the Microbiology field from Medicine Faculty. Prof. F.R.M.A. supervised the project and commented on the manuscript. All authors have investigated and discussed the results and then approved the manuscript contents.
Acknowledgments
This work has been supported by the University of Malaya High Impact Research Grant (MOHE-HIRG A000007-50001). The authors would like to thank Ms. Adeline Yeo Kin Lian for assisting in laboratorial works. We also thank Dr Keivan Zandi for the TBE samples and Dr Shankar Esaki Muthu for the Hepatitis C serum samples.
References
- Hunsperger E. a. et al. Evaluation of Commercially Available Anti–Dengue Virus Immunoglobulin M Tests. Emerging Infect. Dis. 15, 436–440 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dussart P. et al. Evaluation of two new commercial tests for the diagnosis of acute dengue virus infection using NS1 antigen detection in human serum. PLoS Negl. Trop. Dis. 2, e280 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman M. G. et al. Multi-country evaluation of the sensitivity and specificity of two commercially-available NS1 ELISA assays for dengue diagnosis. PLoS Negl. Trop. Dis. 4, e811 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman O., Fong M. Y. & Devi S. A preliminary study of dengue infection in Brunei. Jpn. J. Infect. Dis. 60, 205 (2007). [PubMed] [Google Scholar]
- Organization, W. H. & others. Comprehensive guidelines for prevention and control of dengue and dengue haemorrhagic fever. WHO Reg. Publ. SEARO (2011). [Google Scholar]
- Murray N. E. A., Quam M. B. & Wilder-Smith A. Epidemiology of dengue: past, present and future prospects. Clin. Epidemiol. 5, 299 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt S. et al. The global distribution and burden of dengue. Nature (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. M. & Sekaran S. D. Early diagnosis of Dengue infection using a commercial Dengue Duo rapid test kit for the detection of NS1, IGM, and IGG. Am. J. Trop. Med. Hyg. 83, 690–5 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. M. & Sekaran S. D. Evaluation of a commercial SD dengue virus NS1 antigen capture enzyme-linked immunosorbent assay kit for early diagnosis of dengue virus infection. J. Clin. Microbiol. 48, 2793–7 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumarasamy V. et al. Evaluation of a commercial dengue NS1 antigen-capture ELISA for laboratory diagnosis of acute dengue virus infection. J. Virol. Methods 140, 75–9 (2007). [DOI] [PubMed] [Google Scholar]
- Gopinath S. C. B. Regeneration of commercial Biacore chips to analyze biomolecular interactions. Opt. Eng. 50, 034402 (2011). [Google Scholar]
- Sekaran S. D., Ew C. L., Subramaniam G. & Kanthesh B. M. Sensitivity of dengue virus NS-1 detection in primary and secondary infections. African J. Microbiol. Res. 2, 105–110 (2009). [Google Scholar]
- Nunes M. R. T. et al. Evaluation of an immunoglobulin M-specific capture enzyme-linked immunosorbent assay for rapid diagnosis of dengue infection. J. Virol. Methods 171, 13–20 (2011). [DOI] [PubMed] [Google Scholar]
- Shu P.-Y. et al. Comparison of capture immunoglobulin M (IgM) and IgG enzyme-linked immunosorbent assay (ELISA) and nonstructural protein NS1 serotype-specific IgG ELISA for differentiation of primary and secondary dengue virus infections. Clin. Diagn. Lab. Immunol. 10, 622–630 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yager P., Domingo G. J. & Gerdes J. Point-of-care diagnostics for global health. Annu. Rev. Biomed. Eng. 10, 107–44 (2008). [DOI] [PubMed] [Google Scholar]
- Rich R. L. & Myszka D. G. Survey of the 2009 commercial optical biosensor literature. J. Mol. Recognit. 24, 892–914 (2011). [DOI] [PubMed] [Google Scholar]
- Xu J. et al. A surface plasmon resonance biosensor for direct detection of the rabies virus. Acta Vet. Brno 81, 107–111 (2012). [Google Scholar]
- Lazcka O., Del Campo F. J. & Muñoz F. X. Pathogen detection: a perspective of traditional methods and biosensors. Biosens. Bioelectron. 22, 1205–17 (2007). [DOI] [PubMed] [Google Scholar]
- Nawa M., Takasaki T., Ito M., Kurane I. & Akatsuka T. Detection of Dengue Virus Serotype-specific IgM by IgM Capture ELISA in the Presence of Sodium thiocyanate (NaSCN). Dengue Bull. 28, 119 (2004). [Google Scholar]
- Kuruvilla J. G., Troyer R. M., Devi S. & Akkina R. Dengue virus infection and immune response in humanized RAG2−/− γc−/− (RAG-hu) mice. Virology 369, 143–152 (2007). [DOI] [PubMed] [Google Scholar]
- Liu C., Cui D. & Li H. Biosensors and Bioelectronics A hard – soft microfluidic-based biosensor flow cell for SPR imaging application. Biosens. Bioelectron. 26, 255–261 (2010). [DOI] [PubMed] [Google Scholar]
- Mandal S., Goddard J. M. & Erickson D. A multiplexed optofluidic biomolecular sensor for low mass detection. Lab Chip 9, 2924–32 (2009). [DOI] [PubMed] [Google Scholar]
- Nilsson C. E. et al. A novel assay for influenza virus quantification using surface plasmon resonance. Vaccine 28, 759–66 (2010). [DOI] [PubMed] [Google Scholar]
- Huy T. Q. et al. A novel biosensor based on serum antibody immobilization for rapid detection of viral antigens. Talanta 86, 271–7 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutse S. W. & Yusof N. A. Microfluidics-based lab-on-chip systems in DNA-based biosensing: An overview. Sensors 11, 5754–5768 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan R. J. & Zare R. N. Surface plasmon resonance detection for capillary electrophoresis separations. Anal. Chem. 75, 1542–7 (2003). [DOI] [PubMed] [Google Scholar]
- Watanabe K. et al. High resolution imaging of patterned model biological membranes by localized surface plasmon microscopy. Appl. Opt. 49, 887–891 (2010). [DOI] [PubMed] [Google Scholar]
- Eltzov E. & Marks R. Parameters to consider in the construction of fiber-optic biosensors as alternative bioanalytical tools. IEEE Instrum. Meas. Mag. 12, 10–16 (2009). [Google Scholar]
- Kumbhat S., Sharma K., Gehlot R., Solanki A. & Joshi V. Surface plasmon resonance based immunosensor for serological diagnosis of dengue virus infection. J. Pharm. Biomed. Anal. 52, 255–9 (2010). [DOI] [PubMed] [Google Scholar]
- Fontana E. Thickness optimization of metal films for the development of surface-plasmon-based sensors for nonabsorbing media. Appl. Opt. 45, 7632–7642 (2006). [DOI] [PubMed] [Google Scholar]
- Lin S., Lee A. S., Lin C. & Lee C. Determination of Binding Constant and Stoichiometry for Antibody-Antigen Interaction with Surface Plasmon Resonance. Curr. Proteomics 3, 271–282 (2006). [Google Scholar]
- Pitarke J. M., Silkin V. M., Chulkov E. V. & Echenique P. M. Theory of surface plasmons and surface-plasmon polaritons. Reports Prog. Phys. 70, 1 (2007). [Google Scholar]
- De Leon I. & Berini P. Theory of surface plasmon-polariton amplification in planar structures incorporating dipolar gain media. Phys. Rev. B 78, 1–4 (2008). [Google Scholar]
- Peeling R. W. et al. Evaluation of diagnostic tests: dengue. Nat. Rev. Microbiol. 8, S30–S37 (2010). [DOI] [PubMed] [Google Scholar]
- Kumbhat S., Sharma K., Gehlot R., Solanki A. & Joshi V. Surface plasmon resonance based immunosensor for serological diagnosis of dengue virus infection. J. Pharm. Biomed. Anal. 52, 255–9 (2010). [DOI] [PubMed] [Google Scholar]
- BIACORE. [Application Wizards] BIAevaluation Version 3 Software Handbook. [Chapter 5, 5.1–5.6] (2008). [Google Scholar]
- Dennis G., Neeliyath A. & M. J. D. Surface Plasmon Resonance (SPR). Analysis of Binding Interactions of Proteins in Inner-Ear Sensory Epithelia. Methods Mol Biol. 493, 323–343 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijaya E. et al. Surface plasmon resonance-based biosensors: from the development of different SPR structures to novel surface functionalization strategies. Curr. Opin. Solid State Mater. Sci. 15, 208–224 (2011). [Google Scholar]