Background: Processing of pre-miRNAs by Dicer is essential for miRNA biogenesis.
Results: The nuclear-cytoplasmic dynamics of TAZ and YAP control Dicer levels and activity through regulation of the LIN28/Let-7 axis.
Conclusion: The Hippo pathway effectors TAZ and YAP control miRNA biogenesis.
Significance: Our work provides crucial insight into the poorly understood signaling mechanisms controlling miRNA biogenesis.
Keywords: Cell adhesion, Dicer, MicroRNA, RNA Processing, Signal Transduction, Hippo Pathway, Let-7, TAZ/YAP
Abstract
MicroRNAs (miRNAs) are genome-encoded small double-stranded RNAs that have emerged as key regulators of gene expression and are implicated in most aspects of human development and disease. Canonical miRNA biogenesis involves processing of ∼70-nucleotide pre-miRNA hairpins by Dicer to generate mature ∼22-nucleotide miRNAs, which target complementary RNA sequences. Despite the importance of miRNA biogenesis, signaling mechanisms controlling this process are poorly defined. Here we demonstrate that the post-transcriptional regulation of Dicer is controlled by the cell density-mediated localization of the Hippo pathway effectors TAZ (transcriptional co-activator with PDZ-binding motif) and YAP (Yes-associated protein) (TAZ/YAP). We show that nuclear TAZ/YAP, which are abundant at low cell density, are required for efficient pre-miRNA processing. Knockdown of TAZ/YAP in low density cells, or density-mediated sequestration of TAZ/YAP into the cytoplasm, results in the defective processing of pre-miRNAs. Strikingly, one exception is Let-7, which accumulates upon loss of nuclear TAZ/YAP, leading to Let-7-dependent reduction in Dicer levels. Accordingly, inhibition of Let-7 rescues the miRNA biogenesis defects observed following TAZ/YAP knockdown. Thus, density-regulated TAZ/YAP localization defines a critical and previously unrecognized mechanism by which cells relay cell contact-induced cues to control miRNA biogenesis.
Introduction
A large body of work has described that when untransformed cells adhere to one another, cells cease to proliferate, grow, and migrate (1). This process is known as “contact inhibition” and serves as a powerful tumor suppressive mechanism (2). A major signaling pathway influenced by cell contacts is the Hippo tumor suppressor pathway (1, 3). The localization of the Hippo pathway effectors TAZ and YAP (TAZ/YAP)3 intimately responds to cytoskeletal changes that occur upon cell-cell contact. When nuclear, TAZ/YAP foster the activity of various transcription factors to promote proliferation, control apoptosis, and direct stem/progenitor cell fate (4). Upon cell contact-mediated adhesion/polarity, TAZ/YAP accumulate in the cytoplasm, which in large part is mediated by the activity of the LATS1 and LATS2 (LATS1/2) kinases (3). LATS1/2 phosphorylate TAZ and YAP on conserved serine residues (5), which promotes TAZ/YAP binding to 14-3-3 proteins and consequent cytoplasmic sequestration (6, 7). Cytoplasmic TAZ/YAP localization has important tumor suppressive functions by inhibiting cell proliferation and growth factor-induced cues (8), and thus, TAZ/YAP have emerged as primary mediators of contact inhibition (1).
Evidence has indicated that cell contact-mediated cues affect the global levels of miRNAs (9). In the canonical pathway, the primary miRNA transcript (pri-miRNA) is processed by Drosha generating an ∼70-nucleotide pre-miRNA hairpin (10). The pre-miRNA hairpin is then exported out of the nucleus where it is further processed by Dicer to generate the mature ∼22-bp miRNA. It is clear that miRNA-mediated gene targeting is important in development (11) and is deregulated in a wide range of diseases (12). However, despite detailed insight gained into the miRNA processing steps, little is known regarding the signaling networks controlling miRNA biogenesis, particularly those that relate to disease.
Here we report that the nuclear-cytoplasmic dynamics of TAZ/YAP control miRNA processing. Specifically, we show that nuclear TAZ/YAP is required to support Dicer-mediated pre-miRNA processing. Accordingly, cell contact-induced localization of TAZ/YAP to the cytoplasm, or siRNA-mediated knockdown of TAZ/YAP, decreases Dicer levels and leads to aberrant maturation of miRNAs. We further demonstrate that TAZ/YAP mediate Dicer activity through the LIN28/Let-7 axis and that inhibition of Let-7 can rescue the miRNA processing defects observed upon TAZ/YAP knockdown. Thus, our data detail novel cell contact-mediated cues that control miRNA biogenesis and outline new and unappreciated roles for Hippo pathway signaling.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfections
MCF10A cells were cultured using DMEM/F12 medium (1:1) supplemented with 5% horse serum, 20 ng/ml epithelial growth factor (EGF; PeproTech), 0.5 μg/ml hydrocortisone (Sigma), 100 ng/ml cholera toxin (Sigma), 10 μg/ml insulin (Sigma). Nearly confluent cultures of MCF10A cells were trypsinized, counted, and plated. For low density cultures, 1 × 105 cells were plated in 10-cm dishes and then grown for 48 h. For high density cultures, 5 × 106 cells were plated on 10-cm dishes and then grown for 72 h. For siRNA-mediated knockdown, cells were reverse transfected as the cells were plated using Lipofectamine RNAiMAX (Life Technologies) according to manufacturer's protocol. The following siRNAs were used (Thermo Scientific): control siRNA, GGGCAAGACGAGCGGGAAG; TAZ, siGENOME WWTR1 siRNA#1,2,4 (MQ-016083-00-0002); YAP, siGENOME YAP1 siRNA siRNA#1–4 (MQ-012200-00-0002); TAZ/YAP, UGUGGAUGAGAUGGAUACA; Dicer, CAUUGAUCCUGUCAUGGAU.
Let-7b inhibition was performed by co-transfecting siRNA together with the Let-7b miRIDIAN microRNA inhibitor (Thermo Scientific, catalog number: IH-300476-07-0005) using Lipofectamine RNAiMAX (Life Technologies). The miRIDIAN microRNA Hairpin Inhibitor Negative Control #1 (Thermo Scientific, catalog number: IN-001005-01-05) was used as a control.
Northern Blot miRNA Analysis and Quantitative PCR
Total RNA was prepared with TRIzol (Invitrogen). 25–35 μg of total RNA was resolved by 15% (19:1) denaturing PAGE and transferred to GeneScreen Plus membrane (PerkinElmer). The membranes were probed at 42 °C with 5′-32P-end-labeled DNA oligonucleotides (in ULTRAhyb-Oligo buffer (Ambion)) with exact complementarity to the mature miRNA sequences. U6 was probed as a loading control. The expression of miRNAs was quantified with ImageQuant (5.2) software. Bar graphs were generated with Excel software and represent the average of three independent experiments ± S.E. To collect cellular nuclear fractions, cells (∼10 million) were lysed in cell lysis buffer (25 mm Tris, pH 8.0, 25 mm Hepes 8.0, 50 mm NaCl, 2 mm EDTA, 1 mm phenylmethylsulfonyl fluoride), incubated on ice for 15 min, and then centrifuged (6000 × g, 1 min), and supernatant was removed. For quantitative PCR analysis, cDNA was synthesized from RNA using iScript cDNA synthesis kit (Bio-Rad). qPCR was performed using Fast SYBR Green enzyme (Applied Biosystems) and measured on a ViiA 7 real time PCR system (Applied Biosystems). Transcript levels were analyzed using the ΔΔCT method and normalized to GAPDH. Primer sequences used for qPCR are as follows: TAZ, CCATCACTAATAATAGCTCAGATC (forward) and GTGATTACAGCCAGGTTAGAAAG (reverse); YAP, CTCGAACCCCAGATGACTTC (forward) and CCAGGAATGGCTTCAAGGTA (reverse); Dicer, GCGCATGAGGGCCGCCTTTC (forward) and GACACCACCATGCGGCTGGG (reverse); KSRP, GCTGCCACGACAGTGAATAA (forward) and TCTTGCTCTCCGGTTGATCT (reverse); TRBP, TCAGCAGTCTGAGTGCAACC (forward) and CCAGACTCCTGGGTCACTGT (reverse); PACT, GCAGAGGCTGCCATAAACAT (forward) and TGGAAGGGTCAGGCATTAAG (reverse); LIN28B, ATCCCAGCCATGCACTTCAA (forward) and AGGTAGACTTTGCAACCGGG (reverse); Pri-Let-7, GCGCGCGGTACCATGGAAGGGATGCAGTGTGG (forward) and AACCACACAACCTACTACCTCA (reverse).
Transcriptional Reporter Assays
HEK293 cells were transfected with a transcriptional reporter that drives firefly luciferase expression from either a 1-kb or a 1.5-kb fragment of the Let-7 promoter (13). A pCMV-β-galactosidase plasmid was co-transfected in these cells, and the levels of luciferase activity were normalized against the levels of β-galactosidase activity, as we have previously done for other reporters (14). The Let-7 promoter luciferase reporters were a kind gift from Dr. Hillary Coller (Princeton University).
Immunoblotting
Cells were lysed in radioimmune precipitation buffer (50 mm Tris/HCl, pH 8.0, 1 mm EDTA, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS) containing protease inhibitor mixture. Lysates were fractionated on SDS-PAGE gels, transferred to nitrocellulose membrane, and probed with anti-TAZ/YAP antibody (Cell Signaling Technology, catalog number: 8418), anti-Dicer antibody (Sigma, catalog number: SAB4200087), anti-LIN28B antibody (Cell Signaling Technology, catalog number: 4196), or anti-GAPDH antibody (Sigma, catalog number: G9545). The mouse anti-rabbit IgG (Conformation Specific) HRP-conjugated antibody (Cell Signaling Technology, catalog number: 5127) was used to visualize our immunoblots on a Bio-Rad ChemiDoc imager.
Immunofluorescence Microscopy
Cells were fixed with 4% paraformaldehyde and permeabilized in 0.1% Triton X-100 in PBS. Cells were then blocked with 2% BSA in PBS for 1 h and probed with mouse anti-TAZ antibody (BD Bioscience, catalog number: 560235) and rabbit anti-YAP (Cell Signaling Technology, catalog number: 4912). Nuclei were visualized by DAPI staining. Cells were visualized using confocal microscopy (LSM 700), and images were processed using Volocity software (PerkinElmer).
RESULTS
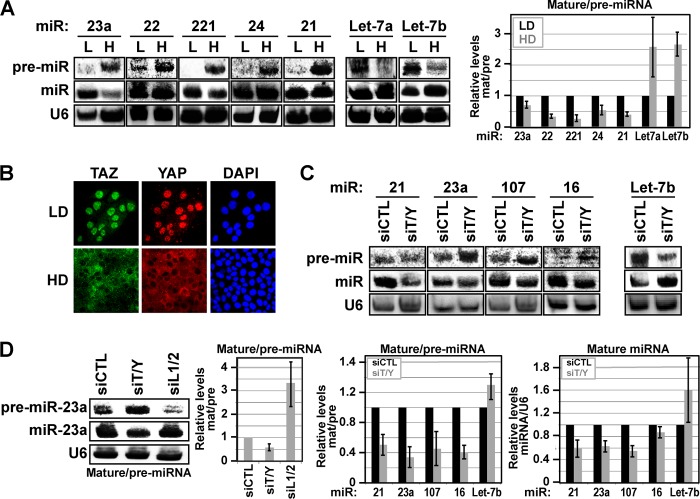
Recent work has revealed that cell-cell contact-mediated cues affect miRNA levels (9). Given that aberrant cell contacts are a hallmark of many diseases, including several aggressive cancers (2), we set out to understand the mechanisms of how cell contact-regulated cues intersect with miRNA processing. We started by examining miRNA processing in low density and high density human MCF10A mammary epithelial cells, an immortalized, but non-transformed, cell model for which expressed miRNAs are well documented (15). Northern blot analysis, which allows direct visualization of both pre-miRNAs and mature miRNAs, revealed cell density-dependent differences in miRNA processing that were surprisingly different from that previously described in transformed cells (9). At low cell density, miRNA expression was robust with no visible pri-miRNA and near complete processing of pre-miRNA to mature miRNA for the unrelated miRNAs miR-23a, miR-22, miR-221, miR-24, and miR-21 (Fig. 1A). However, at high cell density, we observed a striking accumulation of pre-miRNA with very apparent differences in the relative ratios of mature to pre-miRNA levels (Fig. 1A). The exceptions were Let-7a and Let-7b, two miRNAs expressed from the same pri-Let-7a-3∼Let-7b transcript, which accumulated at high levels in their mature form in high density, as compared with low density, MCF10A cells (Fig. 1A).
FIGURE 1.
Cell density-mediated TAZ/YAP localization controls miRNA processing. A, the levels of indicated pre-miRNAs and mature miRNAs were examined by Northern blotting in lysates isolated from MCF10A cells grown at low (L) and high (H) density. The relative levels of mature and pre-miRNAs were quantitated, and the ratios of mature/pre-miRNAs are shown as the average of three independent experiments ± S.D. (p < 0.05). U6 was probed as a loading control. LD, low density; HD, high density. B, MCF10A cells were grown at low and high cell density, and TAZ and YAP localization was examined by immunofluorescence confocal microscopy. Nuclei were stained with DAPI. C and D, MCF10A cells transfected with control siRNA, siRNA targeting TAZ/YAP (siT/Y), or siRNA targeting LATS1/2 (siL1/2 in D) were lysed and examined by Northern blotting for levels of the indicated pre-miRNA and mature miRNA. U6 was probed as a loading control. The relative levels of mature and pre-miRNAs from three independent experiments were quantitated, and the ratios of mature/pre-miRNAs are shown as the average ± S.E. (p < 0.05). The ratios of mature miRNAs/U6 from three independent experiments are also shown as the average ± S.E.
To understand how cell density-mediated miRNA differences arise, we examined signaling factors that govern cell density-induced events. The localization of the Hippo pathway effectors TAZ and YAP is regulated by mechanical and cell contact-induced cues, which critically impact cell proliferation and cell fate specification (16–18). MCF10A cells grown at low cell density exhibit predominantly nuclear TAZ/YAP, but upon density-mediated cell compaction TAZ/YAP markedly shift localization to accumulate in the cytoplasm (Fig. 1B) (18). Transfection of low density MCF10A cells with siRNA targeting both TAZ and YAP led to decreased conversion of endogenous miR-23a, miR-21, miR-107, and miR-16 pre-miRNAs to their mature state (Fig. 1C). Mature Let-7b levels, however, were increased upon TAZ/YAP knockdown (Fig. 1C). Thus, depletion of TAZ/YAP led to similar effects on miRNA processing as that observed at high cell density, a condition where TAZ/YAP are cytoplasmic.
TAZ/YAP localization is in large part defined by the activity of the Hippo pathway-regulated LATS1 and LATS2 kinases (5). To directly test whether the nuclear-cytoplasmic dynamics of TAZ/YAP affect miRNA biogenesis, we used siRNA to knock down the LATS1 and LATS2 kinases in MCF10A cells grown at high cell density. We observed that the knockdown of LATS1/2 resulted in the increased conversion of pre-miR-23a to mature miR-23a (Fig. 1D), suggesting that nuclear TAZ/YAP stimulate miRNA processing.
Nuclear TAZ/YAP function as strong transcriptional regulators (5). Therefore, to test whether the observed differences in miRNA biogenesis were related to the ability of TAZ/YAP to regulate the expression of pri-miRNAs, we examined the effect of TAZ/YAP depletion in HEK-293 cells expressing either the miR-23a∼24 or the miR-17∼92 miRNA clusters from a constitutive CMV promoter. As we detected with endogenous miRNA, the processing of overexpressed pre-miRNA to mature miRNA was defective following TAZ/YAP knockdown (Fig. 2A). Consequently, a striking deficiency in the amount of mature miRNAs was observed (Fig. 2A). Interestingly, knockdown of either TAZ or YAP alone had minimal effects on the maturation of miRNAs, indicating that TAZ and YAP function redundantly to regulate miRNA biogenesis (Fig. 2A). Such important redundancy in TAZ/YAP function is supported by genetic observations in animal development (19).
FIGURE 2.
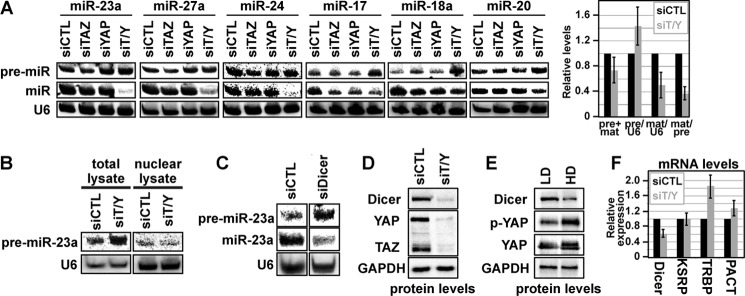
TAZ/YAP control miRNA biogenesis by mediating Dicer stability. A, HEK-293 cells expressing the indicated pri-miRNA clusters were co-transfected with control siRNA (siCTL), siRNA targeting TAZ (siTAZ) or YAP (siYAP), or siRNA targeting both TAZ and YAP (siT/Y). Cell lysates were then examined for miRNA processing efficiency by Northern blotting for the levels of pre-miRNA and mature miRNA. The data from all the miRNAs from each cluster was quantitated, and the combined average ± S.E. of all experiments is shown in the right panel. U6 was probed as a loading control. B, MCF10A cells were transfected with siCTL or siT/Y, and then either total cell lysates or lysates from nuclear fractions were examined for pre-miRNAs. C, MCF10A cells were transfected with siCTL or siRNA targeting Dicer (siDicer), and the lysates were examined by Northern blotting for the levels of pre-miR-23a and miR-23a. U6 was probed as a loading control. D, MCF10A cells were transfected with siCTL or siT/Y, and the levels of Dicer protein were then examined by immunoblotting. TAZ and YAP, as well as GAPDH levels, were analyzed as controls. E, MCF10A cells were grown at low (LD) and high (HD) density, and the levels of Dicer, phospho-Ser-127-YAP (p-YAP), total YAP, and GAPDH were examined by immunoblotting. F, MCF10A cells were transfected with siCTL or siT/Y, and mRNA levels of the indicated genes were quantitated by qPCR. The data represents the average of three experiments ± S.E.
The processing of pre-miRNAs requires exportin-5-mediated nuclear export of pre-miRNAs, which facilitates cytoplasmic processing by the Dicer enzyme. We hypothesized that the general accumulation of pre-miRNAs was a result of either impaired nuclear export or reduction of Dicer activity. A comparison of total and nuclear extracts from low density MCF10A cells transfected with siRNAs targeting TAZ/YAP revealed that pre-miRNAs did not accumulate in the nucleus of TAZ/YAP-depleted cells (Fig. 2B), indicating that the defects arising in these cells are not a result of defective pre-miRNA transport. Our data therefore suggested that TAZ/YAP impact miRNA biogenesis at the level of Dicer activity. Confirming this possibility, we observed that siRNA-mediated knockdown of Dicer in low density MCF10A cells resulted in accumulation of pre-miRNAs and defective processing of mature miRNAs (Fig. 2C), as expected based on prior work (20). Given that TAZ/YAP knockdown emulates Dicer knockdown, we examined whether TAZ/YAP regulate Dicer. We did not observe any detectable binding between Dicer and TAZ or YAP (data not shown). We did, however, detect striking decreases in Dicer protein levels following TAZ/YAP knockdown (Fig. 2D), indicating that TAZ/YAP are required to maintain Dicer levels. Similar decreases in Dicer protein levels were revealed in high density cell cultures (Fig. 2E), suggesting that the reduced maturation of miRNAs under these conditions results from the depletion of Dicer. We also observed moderate decreases in Dicer mRNA (Fig. 2F) following TAZ/YAP knockdown, with no negative effect on mRNA levels of other pre-miRNA processing factors, such as KSRP, TRBP, and PACT (10, 21). Given that Dicer protein levels were affected more intensely than mRNA levels following TAZ/YAP knockdown, we hypothesized that TAZ/YAP control Dicer post-transcriptionally, potentially via an miRNA-mediated process.
When screening the pre-miRNA processing defects in TAZ/YAP-depleted cells, all miRNAs probed exhibited similar processing defects, with the exception of mature Let-7, which was strongly elevated following TAZ/YAP knockdown (Fig. 1C). Although Let-7 has been reported to regulate Dicer levels (20) and high Let-7 leads to the increased expression of thousands of proteins (22), to date a mechanism of general miRNA regulation via Let-7 has not been proposed. We hypothesized that the decreased pre-miRNA processing observed following the depletion of TAZ/YAP is due to increased Let-7 expression. Consistent with this premise, inhibition of Let-7b activity with a targeted miRNA inhibitor completely reversed the miRNA processing defects observed following TAZ/YAP knockdown in low density MCF10A cells (Fig. 3A).
FIGURE 3.
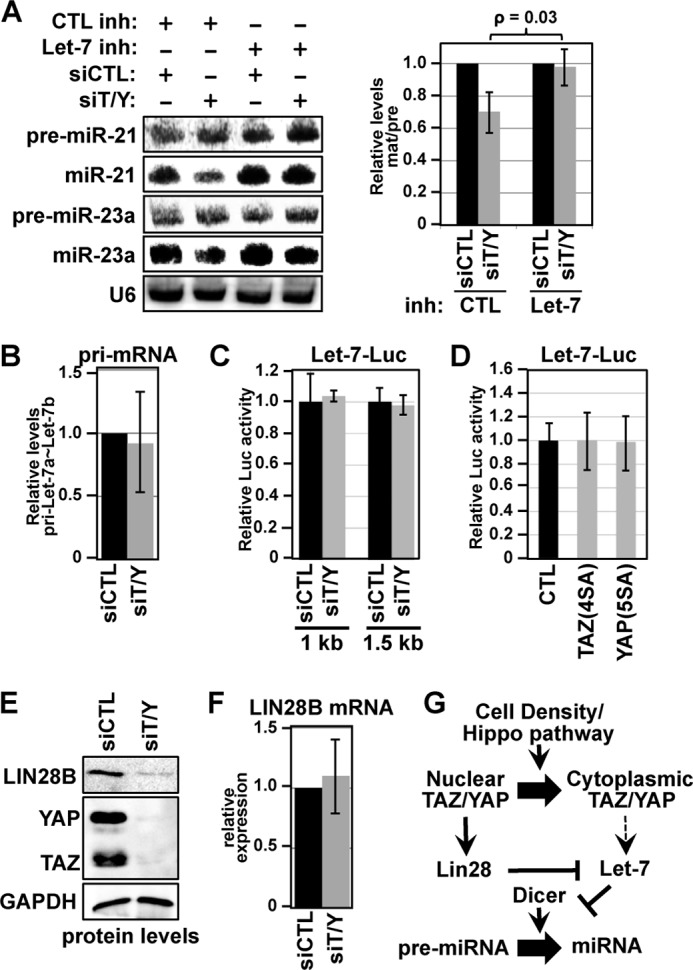
TAZ/YAP govern the LIN28/Let-7 axis. A, MCF10A cells were co-transfected with the indicated siRNA and a control inhibitor (CTL inh) or Let-7b microRNA inhibitor (Let-7 inh), and cell lysates were subsequently examined by Northern blotting for the indicated pre-miRNAs and mature miRNAs. The relative expression of mature miRNA to pre-miRNA was quantified, comparing control siRNA (siCTL) with siRNA targeting TAZ/YAP (siT/Y) in the absence or presence of a Let-7 inhibitor. Experiments were performed three times independently, and the average ± S.E. is shown. B, MCF10A cells were transfected with siCTL or siT/Y, and the levels of pri-Let7a∼Let-7b were quantitated by real time qPCR. C, HEK293 cells were transfected with siCTL or siT/Y along with either a 1-kb or 1.5-kb Let-7 promoter-luciferase reporter, previously used to assess Let-7 transcriptional regulation (13). Relative normalized luciferase activity from the indicated cell lysates is shown. D, nuclear TAZ-4SA or YAP-5SA was co-expressed in HEK293 cells with the 1.5-kb Let-7 promoter-luciferase reporter, and the relative luciferase activity was examined. E, MCF10A cells transfected with siCTL or siT/Y were lysed and examined for the indicated proteins by immunoblotting. F, MCF10A cells were transfected with siCTL or siT/Y, and the mRNA levels of LIN28B were quantitated by real time qPCR. G, model for how cell density-mediated TAZ/YAP localization controls miRNA biogenesis.
Given the observed differences between Let-7 levels versus the other miRNAs, we examined whether Let-7 pri-miRNA transcripts are regulated by TAZ/YAP-mediated transcriptional activity. Knockdown of TAZ/YAP in MCF10A cells did not significantly affect the levels of pri-Let-7a∼Let-7b transcripts (Fig. 3B). Additionally, knockdown of TAZ/YAP, or ectopic expression of mutants of TAZ (TAZ-4SA (23)) or YAP (YAP-5SA (18)) that localize to the nucleus, did not affect the activity of Let-7 promoter luciferase reporters (Fig. 3C). These data therefore indicate that TAZ/YAP do not regulate Let-7 pri-miRNA expression and that the observed differences in Let-7 levels are a consequence of post-transcriptional events.
LIN28 is a well established regulator of Let-7 biogenesis through mechanisms that depend on LIN28 recognition of the Let-7 pre-miRNA hairpin (25, 26). We therefore speculated that deregulation of LIN28 expression might contribute to the deregulated Let-7 levels in TAZ/YAP knockdown cells. Indeed, LIN28B protein levels were markedly reduced in low density MCF10A cells depleted of TAZ/YAP (Fig. 3E), indicating that nuclear TAZ/YAP foster increased LIN28 levels. LIN28B mRNA levels, however, were unaffected by TAZ/YAP knockdown (Fig. 3F), indicating that, like pri-Let-7, LIN28B is not transcriptionally regulated by TAZ/YAP. Thus, TAZ/YAP control post-transcriptional events that regulate the Let-7/LIN28 axis. Taken together, our data indicate that cell density-mediated control of TAZ/YAP activity is a critical mechanism by which cells direct Let-7 biogenesis, which ultimately directs Dicer activity and global miRNA processing (see model in Fig. 3G).
DISCUSSION
The Hippo signaling pathway has emerged as a major regulator of tissue and organ growth, and deregulated Hippo signaling is associated with developmental defects and a range of diseases, including many cancers (5). Here we show that the Hippo pathway effectors TAZ/YAP govern miRNA processing, revealing novel and unexpected molecular insight into the roles of TAZ/YAP. Strikingly, we observed that depletion of nuclear TAZ/YAP, with siRNAs or through activation of the Hippo pathway in response to increased cell density, results in decreased maturation of nine unique miRNAs. The miRNAs we examined were selected based on their abundance as they are included in the top 15 miRNAs expressed in MCF10A cells (15). Since these 15 miRNAs account for ∼80% of the total miRNAs in MCF10A cells (15), our analysis spanned more than 40% of the total miRNAs, strongly suggesting that TAZ/YAP control mechanisms that direct global miRNA biogenesis. Our data indicate that mechanistically TAZ/YAP control miRNA biogenesis by mediating the levels and activity of Dicer. Signaling mechanisms controlling Dicer are poorly defined, and thus, our observations that nuclear TAZ/YAP are required to maintain high Dicer levels provide key insight into how miRNA processing might be dynamically controlled in development and disease.
Interestingly, we found that Let-7 is regulated by TAZ/YAP conversely to other miRNAs and that inhibition of Let-7 mitigates the deleterious effect of TAZ/YAP depletion on pre-miRNA processing. Our data therefore implicate Let-7 as a central factor downstream of TAZ/YAP that controls miRNA biogenesis. Dicer is a validated target of Let-7 (20), and thus, our data indicate that Let-7 targeting of Dicer is an underappreciated mechanism controlling miRNA biogenesis. Such a role for Let-7 provides a mechanistic explanation for the broad increases in protein levels previously seen following Let-7 overexpression (22).
LIN28 has emerged as a major regulator of Let-7 (27). The concomitant reduction of LIN28B with TAZ/YAP depletion suggests that LIN28B stability is associated with the mechanisms by which TAZ/YAP are functioning to control Let-7 levels. Interestingly, however, nuclear TAZ/YAP do not control LIN28B transcription, suggesting that TAZ/YAP impact LIN28B via post-transcriptional events. Such regulation may include direct stabilization of LIN28B by cytoplasmic TAZ/YAP as protein-stabilizing functions for TAZ and YAP have been shown in other contexts (28, 29). Alternatively, TAZ/YAP localization may regulate Let-7 levels or activity via LIN28-independent mechanisms, thereby initiating the Let-7/LIN28B feedback loop and consequent decreases in LIN28B translation. Regardless of whether TAZ/YAP control these events in the nucleus, cytoplasm, or both, it is clear that TAZ/YAP localization is critical for defining miRNA processing. Thus, given that changes in localization define TAZ/YAP activity in a variety of contexts, it is likely that miRNA biogenesis is a major aspect of TAZ/YAP-mediated biology.
Let-7 is a well established tumor suppressor that is typically reduced in tumors (30). Our work shows that nuclear TAZ/YAP suppress mature Let-7 levels, and thus, complements the known roles of TAZ/YAP as tumorigenic factors. Moreover, the LIN28/Let-7 axis controls cell fate dynamics (27) in a manner consistent with TAZ/YAP. For example, nuclear TAZ/YAP are required to maintain embryonic stem cell pluripotency (17, 31), a state that exhibits high expression of LIN28 and decreased levels of Let-7 (24, 26). Thus, cell density-mediated TAZ/YAP localization changes provide a mechanistic explanation for how miRNAs are controlled during cell fate specification. Taken together our data demonstrate the first association between the Hippo pathway effectors TAZ/YAP and miRNA biogenesis, providing novel insight into their roles in development and disease.
Footnotes
- TAZ
- Transcriptional co-activator with PDZ-binding motif
- YAP
- Yes-associated protein
- miRNA
- microRNA
- miR
- microRNA
- pri-miRNA
- primary miRNA transcript
- pre-miRNA
- precursor miRNA
- qPCR
- quantitative PCR.
REFERENCES
- 1. Zeng Q., Hong W. (2008) The emerging role of the Hippo pathway in cell contact inhibition, organ size control, and cancer development in mammals. Cancer Cell 13, 188–192 [DOI] [PubMed] [Google Scholar]
- 2. Hanahan D., Weinberg R. A. (2011) Hallmarks of cancer: the next generation. Cell 144, 646–674 [DOI] [PubMed] [Google Scholar]
- 3. Yu F. X., Guan K. L. (2013) The Hippo pathway: regulators and regulations. Genes Dev. 27, 355–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hiemer S. E., Varelas X. (2013) Stem cell regulation by the Hippo pathway. Biochim. Biophys. Acta 1830, 2323–2334 [DOI] [PubMed] [Google Scholar]
- 5. Pan D. (2010) The Hippo signaling pathway in development and cancer. Dev. Cell 19, 491–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kanai F., Marignani P. A., Sarbassova D., Yagi R., Hall R. A., Donowitz M., Hisaminato A., Fujiwara T., Ito Y., Cantley L. C., Yaffe M. B. (2000) TAZ: a novel transcriptional co-activator regulated by interactions with 14-3-3 and PDZ domain proteins. EMBO J. 19, 6778–6791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Basu S., Totty N. F., Irwin M. S., Sudol M., Downward J. (2003) Akt phosphorylates the Yes-associated protein, YAP, to induce interaction with 14-3-3 and attenuation of p73-mediated apoptosis. Mol. Cell 11, 11–23 [DOI] [PubMed] [Google Scholar]
- 8. Varelas X., Wrana J. L. (2012) Coordinating developmental signaling: novel roles for the Hippo pathway. Trends Cell Biol. 22, 88–96 [DOI] [PubMed] [Google Scholar]
- 9. Hwang H. W., Wentzel E. A., Mendell J. T. (2009) Cell-cell contact globally activates microRNA biogenesis. Proc. Natl. Acad. Sci. U.S.A. 106, 7016–7021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yates L. A., Norbury C. J., Gilbert R. J. (2013) The long and short of microRNA. Cell 153, 516–519 [DOI] [PubMed] [Google Scholar]
- 11. Stefani G., Slack F. J. (2008) Small non-coding RNAs in animal development. Nat. Rev. Mol. Cell Biol. 9, 219–230 [DOI] [PubMed] [Google Scholar]
- 12. Olive V., Li Q., He L. (2013) mir-17–92: a polycistronic oncomir with pleiotropic functions. Immunol. Rev. 253, 158–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang D. J., Legesse-Miller A., Johnson E. L., Coller H. A. (2012) Regulation of the let-7a-3 promoter by NF-κB. PLoS One 7, e31240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Varelas X., Miller B. W., Sopko R., Song S., Gregorieff A., Fellouse F. A., Sakuma R., Pawson T., Hunziker W., McNeill H., Wrana J. L., Attisano L. (2010) The Hippo pathway regulates Wnt/β-catenin signaling. Dev. Cell 18, 579–591 [DOI] [PubMed] [Google Scholar]
- 15. Landgraf P., Rusu M., Sheridan R., Sewer A., Iovino N., Aravin A., Pfeffer S., Rice A., Kamphorst A. O., Landthaler M., Lin C., Socci N. D., Hermida L., Fulci V., Chiaretti S., Foà R., Schliwka J., Fuchs U., Novosel A., Müller R. U., Schermer B., Bissels U., Inman J., Phan Q., Chien M., Weir D. B., Choksi R., De Vita G., Frezzetti D., Trompeter H. I., Hornung V., Teng G., Hartmann G., Palkovits M., Di Lauro R., Wernet P., Macino G., Rogler C. E., Nagle J. W., Ju J., Papavasiliou F. N., Benzing T., Lichter P., Tam W., Brownstein M. J., Bosio A., Borkhardt A., Russo J. J., Sander C., Zavolan M., Tuschl T. (2007) A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129, 1401–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dupont S., Morsut L., Aragona M., Enzo E., Giulitti S., Cordenonsi M., Zanconato F., Le Digabel J., Forcato M., Bicciato S., Elvassore N., Piccolo S. (2011) Role of YAP/TAZ in mechanotransduction. Nature 474, 179–183 [DOI] [PubMed] [Google Scholar]
- 17. Varelas X., Samavarchi-Tehrani P., Narimatsu M., Weiss A., Cockburn K., Larsen B. G., Rossant J., Wrana J. L. (2010) The Crumbs complex couples cell density sensing to Hippo-dependent control of the TGF-β-SMAD pathway. Dev. Cell 19, 831–844 [DOI] [PubMed] [Google Scholar]
- 18. Zhao B., Wei X., Li W., Udan R. S., Yang Q., Kim J., Xie J., Ikenoue T., Yu J., Li L., Zheng P., Ye K., Chinnaiyan A., Halder G., Lai Z. C., Guan K. L. (2007) Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 21, 2747–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nishioka N., Inoue K., Adachi K., Kiyonari H., Ota M., Ralston A., Yabuta N., Hirahara S., Stephenson R. O., Ogonuki N., Makita R., Kurihara H., Morin-Kensicki E. M., Nojima H., Rossant J., Nakao K., Niwa H., Sasaki H. (2009) The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev. Cell 16, 398–410 [DOI] [PubMed] [Google Scholar]
- 20. Tokumaru S., Suzuki M., Yamada H., Nagino M., Takahashi T. (2008) let-7 regulates Dicer expression and constitutes a negative feedback loop. Carcinogenesis 29, 2073–2077 [DOI] [PubMed] [Google Scholar]
- 21. Bartel D. P. (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 [DOI] [PubMed] [Google Scholar]
- 22. Selbach M., Schwanhäusser B., Thierfelder N., Fang Z., Khanin R., Rajewsky N. (2008) Widespread changes in protein synthesis induced by microRNAs. Nature 455, 58–63 [DOI] [PubMed] [Google Scholar]
- 23. Lei Q. Y., Zhang H., Zhao B., Zha Z. Y., Bai F., Pei X. H., Zhao S., Xiong Y., Guan K. L. (2008) TAZ promotes cell proliferation and epithelial-mesenchymal transition and is inhibited by the Hippo pathway. Mol. Cell Biol. 28, 2426–2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Richards M., Tan S. P., Tan J. H., Chan W. K., Bongso A. (2004) The transcriptome profile of human embryonic stem cells as defined by SAGE. Stem Cells 22, 51–64 [DOI] [PubMed] [Google Scholar]
- 25. Heo I., Joo C., Cho J., Ha M., Han J., Kim V. N. (2008) Lin28 mediates the terminal uridylation of let-7 precursor microRNA. Mol. Cell 32, 276–284 [DOI] [PubMed] [Google Scholar]
- 26. Viswanathan S. R., Daley G. Q., Gregory R. I. (2008) Selective blockade of microRNA processing by Lin28. Science 320, 97–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shyh-Chang N., Daley G. Q. (2013) Lin28: primal regulator of growth and metabolism in stem cells. Cell Stem Cell 12, 395–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Levy D., Adamovich Y., Reuven N., Shaul Y. (2007) The Yes-associated protein 1 stabilizes p73 by preventing Itch-mediated ubiquitination of p73. Cell Death Differ. 14, 743–751 [DOI] [PubMed] [Google Scholar]
- 29. Zhao D., Zhi X., Zhou Z., Chen C. (2012) TAZ antagonizes the WWP1-mediated KLF5 degradation and promotes breast cell proliferation and tumorigenesis. Carcinogenesis 33, 59–67 [DOI] [PubMed] [Google Scholar]
- 30. Büssing I., Slack F. J., Grosshans H. (2008) let-7 microRNAs in development, stem cells and cancer. Trends Mol. Med. 14, 400–409 [DOI] [PubMed] [Google Scholar]
- 31. Lian I., Kim J., Okazawa H., Zhao J., Zhao B., Yu J., Chinnaiyan A., Israel M. A., Goldstein L. S., Abujarour R., Ding S., Guan K. L. (2010) The role of YAP transcription coactivator in regulating stem cell self-renewal and differentiation. Genes Dev. 24, 1106–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]