Background: Mechanisms underlying carbon catabolite repression (CCR) control of the anaerobic degradation of aromatic compounds have previously remained elusive.
Results: Phosphorylated AccR was identified as a transcriptional repressor of aromatic degradation operons expressed under anaerobic conditions.
Conclusion: The response regulator AccR controls the succinate-dependent CCR in Azoarcus sp. CIB.
Significance: AccR is a master regulator that controls anaerobic CCR in bacteria.
Keywords: Bacterial Transcription, Gene Regulation, Microbiology, Protein-DNA Interaction, Signal Transduction, Transcription Regulation, Anaerobic Degradation, Azoarcus, Carbon Catabolite Repression
Abstract
Here we characterized the first known transcriptional regulator that accounts for carbon catabolite repression (CCR) control of the anaerobic catabolism of aromatic compounds in bacteria. The AccR response regulator of Azoarcus sp. CIB controls succinate-responsive CCR of the central pathways for the anaerobic catabolism of aromatics by this strain. Phosphorylation of AccR to AccR-P triggers a monomer-to-dimer transition as well as the ability to bind to the target promoter and causes repression both in vivo and in vitro. Substitution of the Asp60 phosphorylation target residue of the N-terminal receiver motif of AccR to a phosphomimic Glu residue generates a constitutively active derivative that behaves as a superrepressor of the target genes. AccR-P binds in vitro to a conserved inverted repeat (ATGCA-N6-TGCAT) present at two different locations within the PN promoter of the bzd genes for anaerobic benzoate degradation. Because the DNA binding-proficient C-terminal domain of AccR is monomeric, we propose an activation mechanism in which phosphorylation of Asp60 of AccR alleviates interdomain repression mediated by the N-terminal domain. The presence of AccR-like proteins encoded in the genomes of other β-proteobacteria of the Azoarcus/Thauera group further suggests that AccR constitutes a master regulator that controls anaerobic CCR in these bacteria.
Introduction
Carbon catabolite repression (CCR)3 is widespread in bacteria and imparts a competitive advantage and improved fitness by establishing priorities in carbon utilization. This enables bacteria to optimize their growth rates in natural environments that provide complex mixtures of nutrients. The regulatory mechanisms of CCR operate through subverting expression of genes required for the uptake and/or metabolism of secondary (less preferred) carbon sources in the presence of a preferred substrate (1). Such secondary carbon sources include aromatic compounds, which, after carbohydrates, are the most widely distributed class of organic compounds in nature.
Where studied, CCR mechanisms impact expression of pathways for the degradation of aromatic compounds. In contrast to enterics, such as Escherichia coli, which utilize glucose as the preferred carbon source, soil bacteria (e.g. Pseudomonas) metabolize many organic acids or amino acids in preference to sugars (2). However, not all organic acids mediate CCR; nor do all organic acids provoke the same effect in different microorganisms (3, 4).
The regulatory elements involved in CCR of aerobic catabolism of aromatic compounds in different bacteria are diverse (2, 4–8). In E. coli, the cAMP-responsive CRP transcriptional regulator imparts CCR control over catabolism of aromatic compounds (8). In Pseudomonas strains, the Crc protein exerts CCR on the assimilation of certain aromatic compounds (9–11) as well as some sugars and amino acids (12–14). In contrast to CRP, Crc acts post-transcriptionally (3, 4, 15). Other proteins described to be involved in CCR control of aerobic catabolism of aromatics in Pseudomonas include PtsN and PtsO (nitrogen phosphotransferase system) (6, 16) and the redox-responsive o-type terminal oxidase CyoB (17, 18), whereas in Acidovorax sp. KKS102 CCR of biphenyl degradation in response to some organic acids is mediated by the response regulator BphQ (7).
Far less is known about CCR control of anaerobic catabolism of aromatic compounds, although cases of this phenomenon have been reported. For example, some organic acids cause CCR of genes involved in the anaerobic degradation of aromatic compounds in Thauera aromatica (19) and Azoarcus sp. CIB (20), whereas in closely related strains, such as Aromatoleum aromaticum EbN1, aromatic compounds are preferred carbon sources over aliphatic organic acids (21). However, the mechanism(s) underlying CCR in these cases are still unknown.
Elucidating the molecular mechanism(s) underlying CCR is important for both basic understanding of how metabolism is regulated in the environment and for potential biotechnological applications, such as optimizing bioremediation strategies and the design of tailor-made biocatalysts/biosensors (4). This is particularly relevant for aromatic compounds that are difficult to degrade and tend to accumulate in the environment under both aerobic and anaerobic conditions. In this work, we sought to use the denitrifying β-proteobacterium Azoarcus sp. CIB to analyze the mechanism(s) underlying CCR of anaerobic pathways for the dissimilation of aromatic compounds. Azoarcus sp. CIB is able to degrade aromatic compounds both aerobically and anaerobically (20, 22), and it has been used as a model organism to study the specific regulation of the benzoyl-CoA central pathways for the aerobic (box genes) and anaerobic (bzd genes) degradation of benzoate (20, 23–27).
The Azoarcus sp. CIB bzd genes responsible for the anaerobic degradation of benzoate are clustered and consist of the PN promoter-driven bzdNOPQMSTUVWXYZA catabolic operon and the bzdR regulatory gene (20). BzdR-mediated repression of PN is alleviated by the inducer molecule benzoyl-CoA, the first intermediate of the catabolic pathway (23, 24). However, activity of the PN promoter also strictly requires the AcpR transcriptional activator, an ortholog of the E. coli Fnr global regulator, which specifically drives expression of the bzd catabolic operon in an oxygen-dependent manner (25). In addition to this dual repressor/activator control, the PN promoter is also subject to control by the benzoyl-CoA dependent BoxR repressor, a BzdR paralog that regulates the expression of the box genes, indicating the existence of cross-regulation between the aerobic and anaerobic benzoate degradation pathways of Azoarcus sp. CIB (27).
In our previous work (20), we found that anaerobic expression of the bzd genes from the PN promoter is additionally subject to CCR control in response to organic acids, such as succinate, malate, and acetate, although the mechanism(s) underlying this level of control was not determined. In this work, we report the characterization of AccR as a response regulator that mediates anaerobic CCR control on the PN promoter. Based on our finding that AccR exerts anaerobic CCR control on central pathways for catabolism of other aromatics in Azoarcus sp. CIB, AccR constitutes the first master regulator described that controls CCR of anaerobic catabolism of aromatic compounds in bacteria. Because of the existence of AccR orthologs within the genomes of closely related Azoarcus/Thauera strains, we suggest that AccR is probably the global mediator of CCR control for anaerobic assimilation of aromatic compounds in this group of β-proteobacteria.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Plasmids, and Growth Conditions
Bacterial strains and plasmids used are listed in Table 1. Plasmids were constructed by standard molecular techniques (35) with the fidelity of all PCR-derived DNA (Table 2) verified by sequencing. E. coli cells were routinely grown at 37 °C in lysogeny broth (LB) medium (35). When required, E. coli AFMCPN was grown aerobically or anaerobically (using 10 mm nitrate as terminal electron acceptor) at 30 °C in M63 minimal medium (36), supplemented with 0.1 mg/ml thiamine and 20 mm glycerol as the carbon source. Azoarcus strains were similarly grown aerobically or anaerobically at 30 °C in MC medium containing the indicated carbon source(s) as described previously (20). Where appropriate, antibiotics were added at the following concentrations: ampicillin (100 μg/ml), kanamycin (50 μg/ml), gentamicin (7.5 μg/ml), and streptomycin (50 μg/ml). An AccR null derivative of Azoarcus sp. CIB was generated by homologous recombination using the R6K-based suicide plasmid pKNG101ΔaccR (Table 1), which allows positive selections of double site recombinants using the sacB gene of Bacillus subtilis (31). Conjugation of pKNG101ΔaccR from replication-permissive E. coli SM10 λpir was used to introduce the plasmid into Azoarcus sp. CIB. Derivatives containing first site recombinants were selected on streptomycin-containing MC medium with 10 mm glutarate as the sole carbon source. Second site recombination was selected by growth on the same medium supplemented with 5% sucrose. Correct allelic exchange in sucrose-resistant and streptomycin-sensitive derivatives was verified using primers 5′AccRmut (BamHI) and 3′AccRmut (SpeI) (Table 2).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or phenotypea | Reference or source |
|---|---|---|
| E. coli strains | ||
| DH10B | F′, mcrA Δ(mrr hsdRMS-mcrBC) φ80dlacΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (Smr) endA1 nupG | Invitrogen |
| M15 | Strain for regulated high level expression with pQE vectors | Qiagen |
| MC4100 | araD139 Δ(argF-lac)U169 rpsL150 (Smr) relA1 flbB5301 deoC1 ptsF25 rbsR | 28 |
| AFMCPN | Kmr Rfr, MC4100 spontaneous rifampicin-resistant mutant harboring a chromosomal insertion of the PN::lacZ translational fusion | 23 |
| SM10 λpir | Kmr thi-1 thr leu tonA lacY supE recA::RP4–2-Tc::Mu λpir lysogen | 29 |
| S17-1λpir | Tpr Smr recA thi hsdRM+ RP4::2-Tc::Mu::Km Tn7 λpir lysogen | 30 |
| XLI-Bue MRA (P2) | Δ(mcrA)183, Δ(mcrCB-hsdSMR-mrr)173, endA1, gyrA96, supE44, relA1, thi-1, lac, P2 lysogen | Stratagene |
| Azoarcus sp. CIB strains | ||
| CIB | Wild-type strain | 20 |
| CIBΔaccR | Azoarcus sp. strain CIB with a deletion of the accR gene | This work |
| Plasmids | ||
| pQE32 | Apr, oriColE1, T5 promoter lac operator, λ to/E. coli rrnB T1 terminators, N-terminal His6 | Qiagen |
| pQE32-His6-AccR | Apr, pQE32 derivative for expression of His6-accR; carries a 636-bp BamHI to PstI fragment generated using primers 5′HisaccR and 3′HisaccR (Table 2) | This work |
| pQE32-His6-CAccR | Apr, pQE32 derivative for expression of His6-CaccR; carries a 232-bp BamHI to PstI fragment generated using 5′HisCaccR and 3′HisaccR (Table 2) | This work |
| pQE32-His6-AccRD60E | Apr, pQE32 derivative for expression of His6-accRD60E; carries a 636-bp BamHI to PstI fragment generated using flanking primers 5′HisaccR and 3′HisaccR and overlapping PCR mutagenesis primers 5′AccRD60E and 3′AccRD60E | This work |
| pREP4 | Kmr, plasmid that expresses the lacI repressor | Qiagen |
| pECOR7 | Apr, pUC19 harboring a 7.1-kb EcoRI fragment containing the bzdRNO genes | 20 |
| pKNG101 | Smr, oriR6K, Mob+. Suicide vector with a sacB selection marker for gene replacement by double site homologous recombination | 31 |
| pKNG101ΔaccR | Smr, pKNG101 derivative carrying the ΔaccR allele as a 1.4-kb BamHI to SpeI fragment assembled using the four AccRmut primers listed in Table 2 | This work |
| pIZ1016 | Gmr, oripBBR1, Mob+, lacZα, Ptac/lacIq, broad-host-range cloning and expression vector | 32 |
| pIZAccSR | Gmr, pIZ1016 derivative expressing accR from the Ptac promoter; carries a 3.3-kb SalI to SpeI fragment amplified using primers 5′AccSR and 3′AccSR (Table 2) | This work |
| pIZAccRD60E | Gmr, pIZ1016 derivative expressing the His6-accRD60E from the Ptac promoter; carries a 0.6-kb BamHI to PstI fragment derived from pQE32-His6-AccRD60E | This work |
| pBBR1MCS-5 | Gmr, oripBBR1, Mob+, lacZα, Plac, broad-host-range cloning and expression vector | 33 |
| pBBR5AccR | Gmr, pBBR1MCS-5 derivative expressing the accR gene from the Plac promoter; carries a 697-bp BamHI to XbaI fragment generated using primers 5′AccRExt and 3′AccRExt (Table 2) | This work |
| pBBR5PN | Gmr, pBBR1MCS-5 derivative harboring a PN::lacZ translational fusion | 23 |
| pBBR5PNI | Gmr, pBBR1MCS-5 derivative harboring a 4.2-kb BamHI to XbaI fragment that expresses the PNI::lacZ translational fusion. The truncated PNI promoter spans from position −61 to +79 with respect to the transcription start site | This work |
| pIZPB1 | Gmr, pIZ1016 derivative harboring a PB1::lacZ translational fusion | 34 |
| pJCD-PN | Apr, pJCD01 derivative harboring a 585-bp EcoRI fragment that includes the PN promoter | 25 |
a Apr, ampicillin-resistant; Gmr, gentamicin-resistant; Kmr, kanamycin-resistant; Rfr, rifampicin-resistant; Smr, streptomycin-resistant.
TABLE 2.
Oligonucleotides used in this study
| Primers | Sequencea | Use |
|---|---|---|
| 5′HisaccR | 5′-CGGGATCCTTACCGCACCGTCCGCCGTA (BamHI) | Amplification of the 5′-end of accR; used to construct His-tagged accR and accRD60E genes |
| 3′HisaccR | 5′-AACTGCAGTCAGCACGTTCTGCACGAGTTCG (PstI) | Amplification of the 3′-end of accR gene; used to construct the accR, CaccR, and accRD60E genes |
| 5′6HisCAccR | 5′-CGGGATCCTTCGGCAGCTCGAGGCGGAGA (BamHI) | Amplification of the 5′-end of CaccR; used to construct His-tagged CaccR |
| 5′AccRD60E | 5′-CTCGTGCTCGAAGTGCGCATG | Internal accR primers used to introduce the accRD60E substitution |
| 3′AccRD60E | 5′-CATGCGCACTTCGAGCACGAG | |
| 5′AccSR | 5′-ACGCGTCGACTGACCTAAGGAGGTAAATAATGTCCGACTCTGCCGAATCCG (SalI) | Used to amplify and clone the accSR genes into plasmid pIZAccSR |
| 3′AccSR | 5′-GGACTAGTTCAGCGGCCGCCACCTCC (SpeI) | |
| 5′AccRext | 5′-CGGGATCCTAGTTAACTAGGAAGGGGGTACCATCTTCTC (BamHI) | Amplification of the accR gene into plasmid pBBR5AccR |
| 3′AccRext | 5′-GCTCTAGACAGGCCGGCACATCCGTCAGCG (XbaI) | |
| 5′AccRmut(BamHI) | 5′-CGGGATCCGCGGAGGTCTCGCGCCAGC (BamHI) | Amplification of a 762-bp BamHI/XbaI fragment, spanning the upstream region of accR, for constructing the ΔaccR allele in pKNG101ΔaccR |
| 3′AccRmut(XbaI) | 5′-GCTCTAGACATTGTTCAGGTACGGCGGACG (XbaI) | |
| 5′AccRmut(XbaI) | 5′-GCTCTAGAGGCGGCCGCTGACGGATG (XbaI) | Amplification of a 642-bp XbaI/SpeI fragment, spanning the downstream region of accR, for constructing the ΔaccR allele in pKNG101ΔaccR |
| 3′AccRmut(SpeI) | 5′-CCACTAGTCCATTACGCTGAGGAACCTTGCG (SpeI) | |
| 5′pboxdQ | 5′-CAACTCCAGTGCCTGTGCG | RT-PCR amplification of cDNA, a 153-bp product that reports PD promoter activity |
| 3′pboxdQ | 5′-GAGAGGAGACAATCAGGTGAAGC | |
| 5′RTpN1 | 5′-GCAACACATCAGAGGAGATAG | RT-PCR amplification of cDNA, a 141-bp product that reports PN promoter activity |
| 3′RTpN2 | 5′-GTGTAGGTACACATCGTTGC | |
| 5′POLIIIHK | 5′-CGAAACGTCGGATGCACGC | RT-PCR amplification of cDNA, a 166-bp product of dnaE used as internal control in RT-PCR assays |
| 3′POLIIIHK | 5′-GCGCAGGCCTAGGAAGTCGAAC | |
| FPAdh 5′ | CGGAATTCGCACCTTCGATCCATTGCCC (EcoRI) | Amplification of a 234-bp boxDR intergenic fragment, PD probe |
| FPAdh 3′ bis | AAAAGTACTCGGTTGCTGTGATGCTGTGTC (ScaI) | |
| 5IVTPN | CGGAATTCCGTGCATCAATGATCCGGCAAG(EcoRI) | 5′-end of PN promoter, used with 3IVTPN to amplify the 376-bp PN probe |
| 3IVTPN | CGGAATTCCATCGAACTATCTCCTCTGATG(EcoRI) | 3′-end of PN promoter, used to amplify the PN, PN2, and PN2mut probes |
| 5′PN3 | TTTCCGCTCGCGTTCGCTCTC | Amplification of the 93-bp PN3 probe |
| 3′PN3 | AATCTTTCTTGCCGCAACGC | |
| 5′PN2 | AATGCAATCAAGTGCATGCAAACG | Used with 3IVTPN to amplify the 162-bp PN2 probe |
| 5′PN mut3 | AAACCCCTGATCAAGTGCATGCA | Used with 3IVTPN to amplify the 165-bp PN2mut probe |
| 5′ScaI PB1 | AAAAGTACTGGTATTACGGTAAGTGCTCCA (ScaI) | Amplification of the 251-bp PB1 probe |
| 3′EcoRI PB1 | CCGGAATTCCCTGCGCGCGGCACTATG (EcoRI) |
a Engineered restriction sites are underlined, and the corresponding restriction enzyme is shown in parentheses.
Sequence Analyses
The nucleotide sequence of the accR gene of Azoarcus sp. CIB has been submitted to GenBankTM with accession number KF233996. Pairwise and multiple sequence alignments of AccR-like proteins were made with the ClustalW (37) program at the EMBL-EBI server. Phylogenetic analysis, carried out according to Kimura's two-parameter method (38), was reconstructed using the neighbor-joining method (39) PHYLIP program (40).
β-Galactosidase Assays
The β-galactosidase activities from promoter-lacZ reporter fusions were measured with permeabilized cells, when cultures reached mid-exponential phase, as described by Miller (36).
Purification of AccR Derivatives
His-tagged AccR derivatives, all with a 13-amino acid (MRGSHHHHHHGIL) N-terminal fusion, were expressed from the isopropyl-1-thio-β-d-galactopyranoside-inducible T5 promoter of the pQE32 expression vector (His6-AccR from pQE32-His6-AccR, His6-AccRD60E from pQE32-His6-AccRD60E, and His6-CAccR from pQE32-His6-CAccR; Table 1). Proteins were purified from E. coli M15 that additionally harbored pREP4, a LacI expression plasmid that aids isopropyl-1-thio-β-d-galactopyranoside-dependent expression. Cultures were grown at 37 °C until the mid-exponential growth phase in LB containing appropriate antibiotics, and then expression was induced by the addition of isopropyl-1-thio-β-d-galactopyranoside (0.1 mm). After a further 5-h incubation, cells were harvested from 1 liter of culture at 4 °C, resuspended in 25 ml of lysis buffer (50 mm NaH2PO4, pH 8.0, 300 mm KCl, 20 mm imidazole), and disrupted by passage through a French press (Aminco Corp.) operated at a pressure of 20,000 p.s.i. Cell lysates were clarified by centrifugation (26,000 × g for 25 min at 4 °C) prior to loading on nickel-nitrilotriacetic acid-agarose columns (Qiagen) equilibrated with lysis buffer. Columns were washed at 4 °C with 50 volumes of lysis buffer, and the His-tagged proteins were subsequently eluted with elution buffer (50 mm NaH2PO4, pH 8.0, 300 mm KCl, 75 mm imidazole). Peak fractions were pooled, dialyzed at 4 °C into modified FP buffer (20 mm Tris-HCl, pH 7.5, 5% glycerol, and 50 mm KCl), and stored as independent aliquots at −20 °C.
In Vitro Phosphorylation of AccR Derivatives
AccR and its derivatives were phosphorylated by treatment with 20 mm carbamoyl phosphate in phosphorylation buffer (20 mm Pipes, 10 mm magnesium acetate, 10 mm KCl, 170 mm NaCl, and 5% (v/v) glycerol, pH 7.5) for 90 min at 28 °C.
Mass Spectrometry Analyses
Mass spectrometry experiments with the AccR and AccR-P proteins (20 μm) were performed on an Autoflex III MALDI-TOF-TOF instrument (Bruker Daltonics, Bremen, Germany) with a smartbeam laser. Spectra were acquired using a laser power just above the ionization threshold. Samples were analyzed in the positive ion detection and delayed extraction linear mode. Typically, 1000 laser shots were summed into a single mass spectrum. External calibration was performed using bovine albumin (Sigma), covering the range from 30,000 to 70,000 Da. A saturated solution of sinapinic acid in 3:1 water/acetonitrile and 0.1% trifluoroacetic acid was used as the matrix. For sample preparation, the sample solution and the matrix were premixed (1:1, v/v), and then 1 μl of the mixture was spotted on the stainless steel target and allowed to dry at room temperature.
Analytical Ultracentrifugation Methods
Analyses were performed using several protein concentrations (from 5 to 40 μm) equilibrated with 20 mm Tris-HCl, pH 7.5, 50 mm KCl, 5% glycerol buffer. Sedimentation velocity runs (48,000 rpm at 20 °C) were carried out in an XL-I analytical ultracentrifuge (Beckman-Coulter Inc.) equipped with UV-visible optic detection system, using an An50Ti rotor and 12-mm double sector centerpiece. Sedimentation profiles were registered every 1–5 min at 280 nm in the experiments with His6-AccR and His6-AccR-P, and 235 nm in the experiments with His6-CAccR. Sedimentation coefficient distributions were calculated by least squares boundary modeling of sedimentation velocity data using the c(s) method (41), as implemented in the SEDFIT program. These s values were corrected to standard conditions (water at 20 °C and infinite dilution) using the SEDNTERP program (42) to obtain the corresponding standard s values (s20,w). Sedimentation equilibrium assays were carried out using the same experimental conditions and instrument as in the sedimentation velocity experiments, with speeds ranging from 5,000 to 15,000 rpm (depending upon the sample analyzed) and at several wavelengths (235, 260, 280, and 290 nm) with short columns (85–95 μl). After each equilibrium scan, a high speed centrifugation run (40,000 rpm) was performed to estimate the corresponding base-line offset. The measured low speed equilibrium concentration (signal) gradients of His6-AccR, His6-AccR-P, and His6-CAccR were fitted using an equation that characterizes the equilibrium gradient of an ideally sedimenting solute (using a MATLAB program, kindly provided by Dr. Allen Minton, National Institutes of Health) to obtain the corresponding buoyant signal average molecular weight.
Gel Retardation Assays
Gel retardation reaction mixtures (9 μl) contained FP buffer (20 mm Tris-HCl, pH 7.5, 10% glycerol, 2 mm β-mercaptoethanol, 50 mm KCl), 0.05 nm 32P-labeled DNA probe, 250 μg/ml bovine serum albumin, 50 μg/ml herring sperm (competitor) DNA, and the indicated concentration of purified protein. After incubation for 20 min at 30 °C, samples were analyzed by electrophoresis using 5% polyacrylamide gels buffered with 0.5× TBE (45 mm Tris borate, 1 mm EDTA). Dried gels were subsequently imaged using Hyperfilm MP (Amersham Biosciences). Radiolabeled DNA probes (PN (376 bp), PN2 (162 bp), PN3 (93 bp), PN2mut (165 bp), PB1 (251 bp), and PD (234 bp)) were generated by PCR amplification (Table 2). For all but the PB1 probe, 32P was incorporated by the use of a primer 5′-end-labeled with phage T4 polynucleotide kinase and [γ-32P]ATP (3,000 Ci/mmol; PerkinElmer Life Sciences). For the PB1 probe, 32P was incorporated by digestion of the PCR-derived DNA with EcoRI and subsequent filling in of the resulting overhang end with [α-32P]dATP (6,000 Ci/mmol; PerkinElmer Life Sciences) and the Klenow fragment of E. coli DNA polymerase I.
DNase I Footprinting Assays
DNase I footprinting reactions (15 μl) in FP buffer contained 2 nm radiolabeled PN probe (see above), 1 mg/ml bovine serum albumin, and the indicated concentration of in vitro phosphorylated His6-AccR (AccR-P). Reactions were incubated for 20 min at 25 °C prior to the addition of 3 μl (0.05 units) of DNase I solution (prepared in 10 mm CaCl2, 10 mm MgCl2, 125 mm KCl, and 10 mm Tris-HCl, pH 7.5). After a 20-s incubation at 37 °C, reactions were quenched by the addition of 180 μl of a solution containing 0.4 m sodium acetate, 2.5 mm EDTA, 50 μg/ml calf thymus DNA, and 0.3 μg/ml glycogen. DNA fragments were phenol-extracted, precipitated, and analyzed as previously described (23). An A+G reaction (43) was carried out with the same fragment. Images were obtained as described for gel retardation assays.
In Vitro Transcription Experiments
Multiple-round in vitro transcription reactions (50 μl) were performed essentially as described previously (25) in a buffer consisting of 40 mm Tris-HCl, pH 7.5, 100 mm KCl, 10 mm MgCl2, 0.1 mm bovine serum albumin, and 10 mm dithiothreitol. Reactions contained 5 nm supercoiled plasmid pJCD-PN as the DNA template, 50 nm E. coli σ70-RNA polymerase (1 unit/μl; U.S. Biochemical Corp.), 200 nm His6-Fnr* activator (a constitutively active Fnr derivative that is able to promote transcription in the presence of oxygen) (25, 44), and different concentrations of purified AccR or AccR-P proteins. Reactions were equilibrated for 10 min at 37 °C prior to initiation of transcription by the addition of nucleotides (final concentrations: 500 μm of ATP, CTP, and GTP; 50 μm UTP; and 1 μCi of [α-32P]UTP (3,000 Ci/mmol; PerkinElmer Life Sciences)). After incubation for 20 min at 37 °C, reactions were terminated by the addition of an equal volume of a solution containing 50 mm EDTA, 350 mm NaCl, and 0.5 mg/ml of carrier tRNA. Ethanol-precipitated transcripts were dissolved in loading buffer (7 m urea, 1 mm EDTA, 0.6 m glycerol, 0.9 mm bromphenol blue, and 1.1 mm xylene cyanol); resolved by electrophoresis on a denaturing 7 m urea, 4% polyacrylamide gel; and visualized by autoradiography.
Real-time RT-PCR Assays
Total RNA was extracted and DNase I-treated using reagents provided with the RNeasy minikit (Qiagen) according to the manufacturer's instructions. The concentration and purity of the RNA samples were assessed using an ND1000 spectrophotometer (Nanodrop Technologies) according to the manufacturer's protocols. Synthesis of total cDNA was performed in 20-μl reactions containing 1 μg of RNA, a 0.5 mm concentration of each dNTP, 200 units of SuperScript II reverse transcriptase (Invitrogen), and 5 μm random hexamers in the buffer provided by the manufacturer. Samples were initially heated at 65 °C for 5 min and then incubated at 42 °C for 2 h. Reactions were terminated by incubation at 70 °C for 15 min, and the cDNA was further purified using Geneclean Turbo kit (MP Biomedicals). An IQ5 multicolor real-time PCR detection system (Bio-Rad) was used for real-time PCR of triplicate samples. Reactions (25 μl) contained 5 ng of cDNA, 0.2 μm two target-specific primers, and 12.5 μl of SYBR Green Mix (Applied Biosystems). Primer pairs used to amplify transcripts from the PD and PN promoters and the transcript of the dnaE gene (encoding the α-subunit of DNA polymerase III) as an internal control to normalize the sample data are given in Table 2. Amplifications were carried with one denaturation cycle (95 °C for 4 min), followed by 30 cycles of amplification (95 °C for 1 min, 60 °C for 1 min, and 72 °C for 30 s). After amplification, melting curves were generated to confirm amplification of a single product. For relative quantification, a calibration curve was constructed for each amplicon by using 5-fold serial dilutions of Azoarcus sp. CIB genomic DNA ranging from 2.5 ng to 1.6 × 10−6 ng. These standard curves were used to extrapolate the relative abundance of the cDNA targets within the linear range of the curve. Results were normalized relative to those obtained for the dnaE internal control.
RESULTS
The accR Gene Is Required for CCR Control of the bzd Genes in Azoarcus sp. CIB
In silico analysis of the unpublished draft Azoarcus sp. CIB genome sequence identified a gene, hereafter referred to as accR (aromatic catabolic control regulator; accession number KF233996), whose product shows 47% amino acid sequence identity with the response regulator BphQ involved in CCR control of the biphenyl degradation pathway in Acidovorax sp. KKS102 (7). Orthologs of accR are also found in the genomes of other β-proteobacteria Azoarcus and Thauera strains (supplemental Fig. S1). These observations prompted us to test AccR as a potential candidate for CCR control of the assimilation of aromatic compounds in Azoarcus sp. CIB. To this end, we generated an AccR null derivative (Azoarcus sp. CIBΔaccR; Table 1), and measured the activity of the PN promoter, which drives the anaerobic expression of the bzd genes, in comparison with PN activity in the wild-type strain. Transcripts from PN were monitored by real-time RT-PCR using RNA extracted from cultures grown anaerobically at the expense of benzoate alone (control condition) or benzoate plus succinate (CCR condition). As shown in Fig. 1, both strains exhibited similar levels of PN activity when grown on benzoate, but the AccR null mutant was unable to exert CCR control in response to succinate, a defect that could be complemented by introduction of plasmid pBBR5AccR (Table 1) carrying accR under control of the Plac promoter. Hence, these results support the idea that AccR acts to repress the PN promoter under CCR conditions.
FIGURE 1.
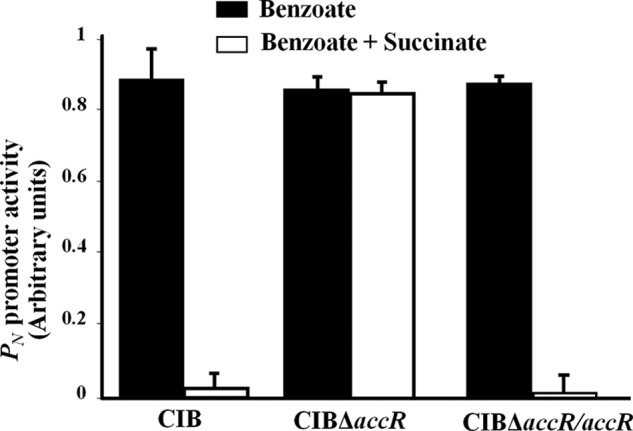
The accR gene is required for CCR control of the PN promoter. Azoarcus sp. CIB (CIB), its AccR null counterpart (CIBΔaccR), and CIBΔaccR harboring plasmid pBBR5AccR, which expresses AccR from the Plac promoter (CIBΔaccR/accR), were grown anaerobically in minimal medium supplemented with 3 mm benzoate (black bars) or a mixture of 3 mm benzoate and 0.4% succinate (white bars). Total RNA was isolated from cells grown until mid-exponential phase, and the activity of the PN promoter (in arbitrary units) was measured by real-time RT-PCR. Graphed values are the average of three independent experiments ± S.D. (error bars).
AccR Possess in Vitro Properties of Response Regulators
Sequence comparisons indicated that AccR belongs to the FixJ/NarL family of response regulators (45–47). AccR exhibits the typical two-domain architecture of this class of proteins, consisting of an N-terminal domain (NAccR; residues 1–122) and a C-terminal region (CAccR; residues 145–212) connected by a 23-residue-long linker (supplemental Fig. S1A). The NAccR domain contains the highly conserved receiver motif (REC motif) (residues 20–122) (supplemental Fig. S1A) that is involved in phosphorylation and dephosphorylation reactions of response regulators (48). The CAccR domain contains a NarL/FixJ helix-turn-helix DNA-binding motif (residues 162–189) responsible for interaction with the DNA (supplemental Fig. S1A).
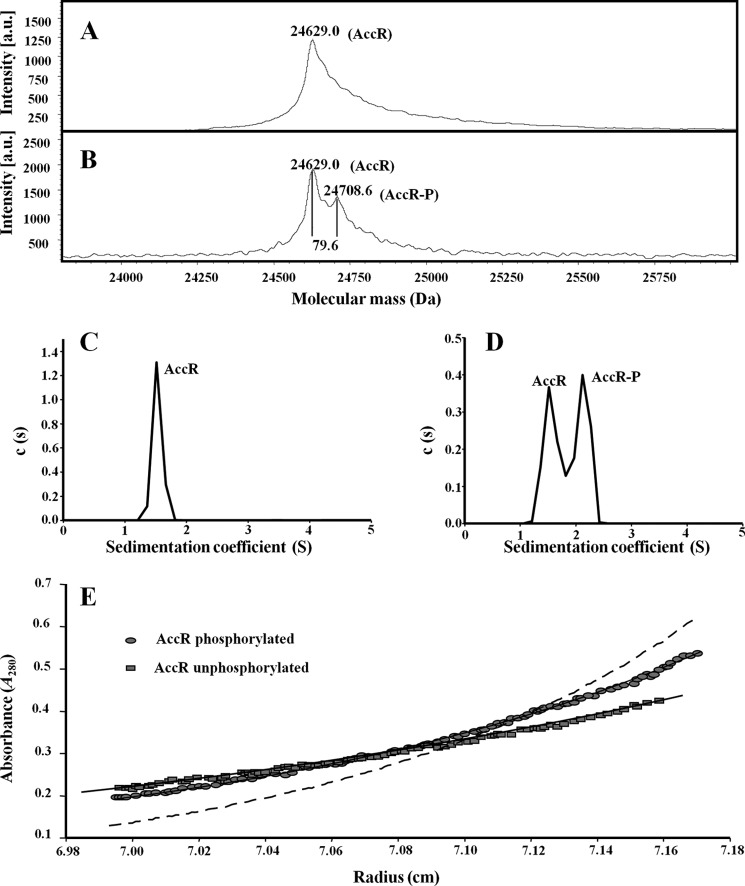
To determine whether AccR behaves as a typical response regulator in vitro, we expressed and purified a N-terminally His-tagged version of the AccR protein (His6-AccR). Response regulators usually receive a phosphate group from a cognate histidine kinase in intact cells; however, REC motifs can also catalyze autophosphorylation using small phosphodonor molecules, such as carbamoyl phosphate or acetyl phosphate (48). Hence, we analyzed purified AccR before and after treatment with carbamoyl phosphate. Mass spectrometry analysis of untreated AccR revealed a single peak that corresponds to the theoretical mass of monomeric AccR (24,629 Da; Fig. 2A). When carbamoyl phosphate-treated AccR was similarly analyzed, two peaks were resolved: one corresponding to the unadorned monomer and the other compatible with the addition of a phosphate group (24,708.6 Da; Fig. 2B). We conclude from these results that AccR becomes phosphorylated (AccR-P) in the presence of carbamoyl phosphate.
FIGURE 2.
In vitro phosphorylation of AccR provokes a monomer-to-dimer transition. A and B, mass spectrometry (MALDI-TOF) of untreated His6-AccR (A) and His6-AccR preincubated with 20 mm carbamoyl phosphate (B). Molecular masses of the peaks corresponding to unphosphorylated (AccR) and phosphorylated (AccR-P) proteins are indicated. C and D, sedimentation coefficient distribution c(s) corresponding to 10 μm untreated His6-AccR (C) and carbamoyl phosphate-treated His6-AccR (D) are represented in relation to the sedimentation coefficient (S). The peaks corresponding to the AccR and AccR-P species are indicated. The s value of the proteins did not change significantly with protein concentration over the range examined (5–40 μm). E, sedimentation equilibrium data of untreated (squares) and carbamoyl phosphate-treated His6-AccR (circles). Best fit analysis assuming a theoretical protein monomer (solid line) or dimer (dashed line) is also shown.
Analysis of the REC motif suggested that autophosphorylation of the AccR protein would occur at the highly conserved Asp60 residue that is located at the C-terminal end of β3-strand (supplemental Fig. S1A). To experimentally verify this supposition, we purified a His-tagged mutant variant with Asp60 substituted to Glu (AccRD60E), a substitution that serves as a phosphomimic in some response regulators (49, 50). In contrast to the wild-type protein, AccRD60E exhibited no change in mass upon treatment with carbamoyl phosphate, supporting the idea that Asp60 is the target residue for AccR autophosphorylation.
AccR Undergoes a Monomer-to-dimer Shift upon Phosphorylation
Phosphorylation of some response regulators results in a change in their oligomeric state (51). To determine if such a change occurs with AccR, we performed analytical ultracentrifugation analyses with different concentrations (5–40 μm) of untreated and carbamoyl phosphate-treated AccR. Sedimentation velocity analysis of 10 μm untreated AccR revealed a single species with an s value of 1.5 S (Fig. 2C). Carbamoyl phosphate treatment resulted in two species of similar abundance, one with an s value of 1.5 S and another with an s value of 2.1 S (Fig. 2D), suggesting that ∼50% of AccR becomes phosphorylated under the experimental conditions used.
The molecular mass of the 1.5 S species, as measured by sedimentation equilibrium (24,000 ± 500 Da), is compatible with the mass of the AccR monomer (Fig. 2E, squares). However, because the frictional ratio f/f0 of untreated AccR was 1.7, the shape deviates from that expected for a globular protein and suggests an elongated monomer. Sedimentation equilibrium experiments of the carbamoyl phosphate-treated AccR sample indicated a molecular mass (38,500 ± 400 Da) between that of a monomer and a dimer of the protein (Fig. 2E, circles), which is in agreement with the co-existence of approximately equal amounts of the 1.5 S (monomer) and 2.1 S (dimer) species within this preparation (Fig. 2D). Together, these results demonstrate that unmodified AccR exists primarily as an elongated monomer, whereas phosphorylation induces a monomer-to-dimer transition but does not significantly change the global shape of the protein that retains a frictional ratio f/f0 of 1.7.
DNA Binding by AccR Is Phosphorylation-dependent
Having established that AccR undergoes a phosphorylation-dependent conformational change (Fig. 2) and influences expression of the bzd genes through the PN promoter (Fig. 1), we next determined the ability of AccR to bind to DNA. For this, we used a 376-bp DNA fragment spanning from −293 to +83 with respect to the transcription start site of the PN promoter (PN probe) in gel retardation assays. The AccR protein was unable to bind to the PN probe unless phosphorylated by prior carbamoyl phosphate treatment (Fig. 3, A and B). In contrast, the Asp60 to Glu phosphomimic substitution in AccRD60E rendered the protein DNA binding-competent in its non-phosphorylated state (Fig. 3C). These results show that the DNA binding capacity of AccR is dependent on its phosphorylated state, which can be mimicked by an Asp60 to Glu substitution. Because phosphorylation induces a monomer-to-dimer transition, it appears likely that AccR binds DNA primarily as a dimer.
FIGURE 3.
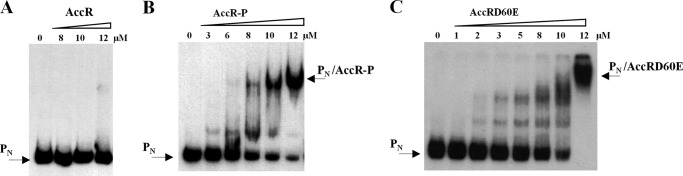
DNA binding by AccR is phosphorylation-dependent. Shown are gel retardation analyses of the PN probe by AccR derivatives (0–12 μm) as described under “Experimental Procedures.” A, assays with increasing concentrations of untreated His6-AccR protein (AccR). Free PN probe is indicated by an arrow. B, assays performed with increasing concentrations of carbamoyl phosphate-treated His6-AccR (AccR-P). Free PN probe and the major PN·AccR-P complex are indicated by arrows. C, assays performed with increasing concentrations of His6-AccRD60E (AccRD60E). Free PN probe and the major PN·AccRD60E complex are indicated by arrows.
AccR Binds to Palindrome Operator Sites within PN to Confer CCR Control
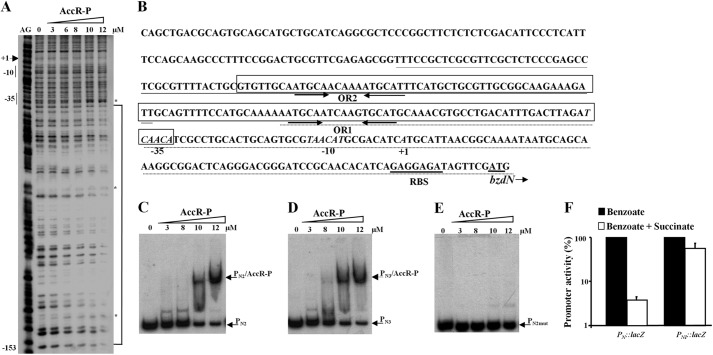
To further characterize the DNA binding sites of AccR-P within the PN promoter, we performed DNase I footprinting assays. As shown in Fig. 4A, AccR-P protected a DNA region spanning from position −34 to −153 with respect to the transcription start site of the PN promoter. This protected region contains two repetitions of the palindromic sequence (ATGCA-N6-TGCAT), denoted OR1 (from −67 to −82) and OR2 (from −130 to −145) (see Fig. 4B). To determine if AccR-P was able to recognize the two palindromic regions independently, we used two probes (PN2, which spans OR1, and PN3, which spans OR2) (Fig. 4B) in gel retardation assays. AccR-P bound to both probes with an affinity similar to that observed with the complete PN probe (compare Fig. 4, C and D, with Fig. 3B), indicating that AccR-P is able to recognize and bind to two different operator regions within the PN promoter. To verify the role of the ATGCA-N6-TGCAT palindrome in AccR binding to PN, we constructed a mutant derivative of the PN2 probe (PN2mut) in which the left-half site of OR1 (ATGCA) was replaced by the sequence CCCTG. As shown in Fig. 4E, AccR-P was unable to bind to the PN2mut probe. Similarly, AccR-P was unable to bind a truncated PN2 probe lacking the left-half site of OR1 (data not shown).
FIGURE 4.
AccR-P binds to two palindromic operator sequences within the PN promoter. A, DNase I footprinting analyses of the PN probe with 0–12 μm carbamoyl phosphate-treated His6-AccR (AccR-P) as described under “Experimental Procedures.” Lane AG shows an A + G Maxam and Gilbert sequencing reaction with the location of the −10 and −35 boxes and the transcription initiation site (+1) indicated to the left. The AccR-P-protected region between positions −153 and −34 is highlighted by a bracket, with phosphodiester bonds mildly hypersensitive to DNase I cleavage indicated by asterisks. B, expanded view of the PN promoter sequence. Inferred −10 and −35 promoter elements and the transcription start site (+1) are indicated in italic type. The ribosome-binding site (RBS) and the bzdN ATG initiation codon are underlined. The region protected by AccR-P (as in A) is boxed with the location of the palindrome operator sequences (OR1 and OR2) indicated by convergent arrows. Nucleotides corresponding to the PN2 and PN3 probes used in C and D are underlined with discontinuous and continuous lines, respectively. C–E, gel retardation analyses of the indicated probes as in Fig. 3B. Gel retardation assays were performed using increasing concentrations (0–12 μm) of carbamoyl phosphate-treated His6-AccR (AccR-P). The free probes and the major DNA·AccR-P complex are indicated by arrows. F, effect of succinate on the activity of the native PN promoter and the truncated PNI promoter that lacks the OR1 and OR2 operators. Azoarcus sp. CIB cells harboring the PN::lacZ translational reporter plasmid pBBR5PN or the PNI::lacZ translational reporter plasmid pBBR5PNI were grown anaerobically in minimal medium containing 3 mm benzoate (black bars) or a mixture of 3 mm benzoate and 0.2% succinate (white bars) until the mid-exponential phase. Values are the average of three independent experiments ± S.D. (error bars) and are graphed setting the β-galactosidase activities obtained for each promoter in the absence of succinate (22,000 and 2,500 Miller units for PN and PNI, respectively) as 100%.
The results above demonstrate that AccR-P independently binds to two palindromic operator regions within the PN promoter. To confirm that the AccR operator regions are the ones responsible for carbon catabolite control of PN, we constructed a truncated PN promoter that lacks the AccR binding regions (PN1; −61 to +79) in a PN1::lacZ fusion reporter plasmid and compared its activity with that of an otherwise identical wild-type PN::lacZ reporter plasmid in Azoarcus sp. CIB. As anticipated, the PN1::lacZ fusion was essentially immune to CCR control in response to succinate (Fig. 4F).
AccR Acts as a Repressor of the PN Promoter in Vitro
Our in vivo experiments (Figs. 1 and 4F) suggested a repressor role for AccR on transcription from the PN promoter, which is consistent with the location of the AccR-P binding region spanning −35 to −153 (Fig. 4). To confirm this hypothesis, we performed in vitro transcription assays using a supercoiled DNA template bearing the PN promoter. Within these assays, activity from PN was elicited by constant concentrations of RNA polymerase and the Fnr* activator, an E. coli homolog of the AcpR activator of PN that retains activity in the presence of oxygen (25). The effect of different concentrations of AccR or AccR-P on PN output was then assessed. Whereas the formation of the expected 184-nucleotide transcript due to the activity of the PN promoter was unperturbed by the presence of AccR (Fig. 5, lanes 5–7), the levels of this transcript decreased in the presence of increasing amounts of AccR-P (Fig. 5, lanes 2–4). These data confirm the repressor role of AccR and demonstrate that its inhibitory action on PN activity is strictly dependent on the phosphorylated status of AccR.
FIGURE 5.
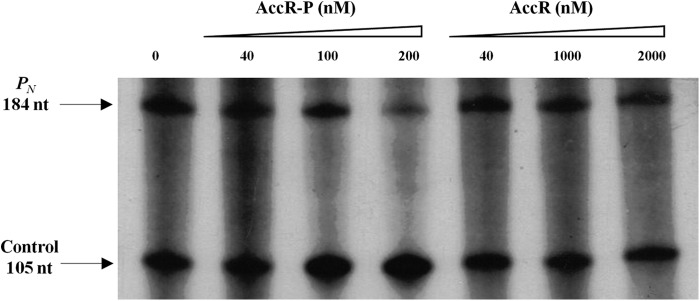
AccR-P represses transcription from the PN promoter in vitro. Multiple-round in vitro transcription reactions, as detailed under “Experimental Procedures,” contained 5 nm supercoiled pJCD-PN plasmid DNA (which produces a 184-nucleotide mRNA from PN and a 105-nucleotide control mRNA), 40 nm E. coli RNA polymerase, and 200 nm His6-Fnr* activator. Reactions were carried out in the absence of AccR or in the presence of increasing concentrations of phosphorylated His6-AccR (AccR-P) or unphosphorylated His6-AccR (AccR).
CAccR Functions as an Independent Monomeric DNA Binding Domain
As outlined above, Asp60 within the REC motif of the NAccR domain is involved in autophosphorylation, and CAccR contains the NarL/FixJ helix-turn-helix DNA-binding motif predicted to mediate DNA binding. To determine if CAccR could function independently in this capacity, we purified a His-tagged version of CAccR (His6-CAccR) and monitored its DNA binding capacity in gel retardation assays using the PN probe (as in Fig. 3). The data in Fig. 6A demonstrate that CAccR binds to the PN probe in a concentration-dependent manner, experimentally verifying that this domain functions as an independent DNA binding domain.
FIGURE 6.
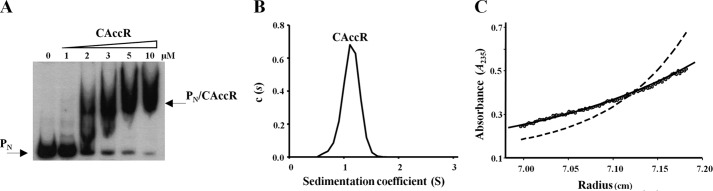
CAccR functions as an independent monomeric DNA binding domain. A, gel retardation analysis of the PN probe, as under Fig. 3A, using increasing concentrations (0–10 μm) of His6-CAccR (CAccR). The free PN probe and the major PN·CAccR complex are indicated by arrows. B, sedimentation coefficient distribution c(s) corresponding to 10 μm His6-CAccR is represented in relation to the sedimentation coefficient (S). The peak corresponding to the single His6-CAccR species detected is indicated. C, sedimentation equilibrium data of His6-CAccR (circles) and best fit analysis assuming a theoretical protein monomer (solid line) or dimer (dashed line).
Because phosphorylation of AccR provokes a monomer-to-dimer transition, we also determined the oligomeric status of CAccR as described previously for full-length AccR. Sedimentation velocity experiments revealed a single species of CAccR with an s value of 1.1 S (Fig. 6B). The molecular weight of the 1.1 S species, as measured by sedimentation equilibrium of CAccR at different concentrations, is compatible with the mass of a monomer (Fig. 6C). We conclude from this data that, under our experimental conditions, DNA binding-proficient CAccR is a monomer in solution. As expanded upon under “Discussion,” this suggests that dimerization of AccR after phosphorylation is carried out through the N-terminal domain and that dimerization is not a prerequisite for DNA binding per se.
AccRD60E Acts as a Superrepressor
The Asp60 → Glu substitution renders AccR constitutively active (i.e. phosphorylation-independent) in terms of its DNA binding capacity (Fig. 3C). Therefore, we were interested to test if it also exhibited constitutive repressor activity in vivo. To this end, we used the heterologous E. coli AFMCPN strain, which carries a PN::lacZ translational fusion integrated into its chromosome (Table 1) and would be expected to lack a histidine kinase capable of phosphorylating AccR. Similar activities from the PN promoter were found in E. coli AFMCPN harboring either a vector control or a plasmid expressing AccR, whereas PN activity was reduced ∼4-fold in the strain harboring a plasmid expressing AccRD60E (Fig. 7A).
FIGURE 7.
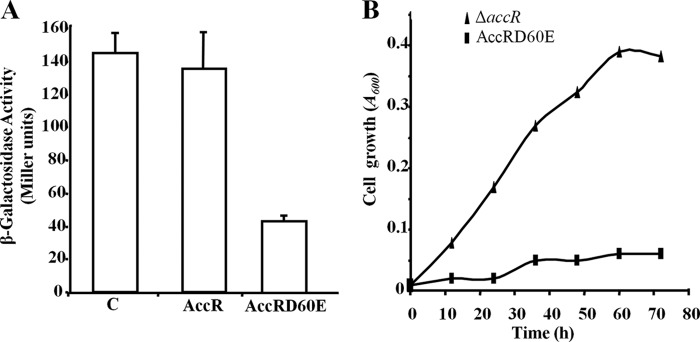
The AccRD60E protein acts as a constitutive repressor of the PN promoter. A, effect of the AccRD60E protein on the activity of the PN promoter in E. coli AFMCPN cells that contain a chromosomal PN::lacZ fusion. Cells harboring a plasmid expressing the AccRD60E (pIZAccRD60E), AccR (pIZAccSR), or a cognate vector control (pIZ1016) (C), were grown anaerobically in glycerol-containing minimal medium until mid-exponential phase. Graphed values of β-galactosidase activities (in Miller units) are the average from three independent experiments ± S.D. (error bars). B, anaerobic growth of Azoarcus sp. CIBΔaccR (triangles) and Azoarcus sp. CIBΔaccR harboring pIZAccRD60E (squares) in minimal medium containing 3 mm benzoate as the sole carbon source.
The results outlined above demonstrate that the unphosphorylated AccRD60E protein is able to effectively bind and repress PN promoter activity in E. coli. To test if this was also the case in Azoarcus sp. CIB, we monitored the effect of expressing AccRD60E on the ability of the AccR null strain to grow anaerobically at the expense of benzoate. Whereas Azoarcus sp. CIBΔaccR was able to grow on benzoate (Fig. 7B, triangles), Azoarcus sp. CIBΔaccR harboring a plasmid expressing AccRD60E protein could not (Fig. 7B, squares). These results strongly support the idea that AccRD60E is constitutively active in Azoarcus and behaves as a superrepressor of the bzd cluster through inhibiting the activity of the PN promoter.
AccR Is a Master Regulator Controlling All Central Pathways for the Anaerobic Catabolism of Aromatics in Azoarcus sp. CIB
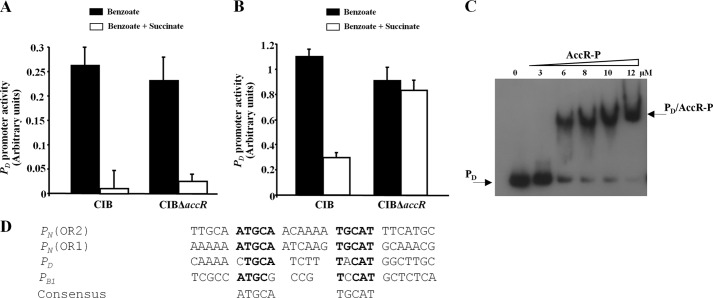
In addition to the bzd-pathway, the Azoarcus sp. CIB mbd pathway for anaerobic assimilation of 3-methylbenzoate/m-xylene is also subject to CCR control in response to some organic acids (34). To explore whether AccR also controls expression of the mbd genes, we monitored the activity of the PB1 promoter that drives expression of the mbd genes using a PB1::lacZ translational fusion reporter plasmid. Although succinate caused inhibition of PB1 promoter activity in the wild-type CIB strain, no inhibition of PB1 activity was observed in the AccR null strain (Fig. 8A). A direct interaction between AccR-P with the PB1 promoter was confirmed by gel retardation (Fig. 8B). Taken together, these results demonstrated that AccR is also involved in succinate-mediated anaerobic CCR control of the mbd genes in Azoarcus sp. CIB.
FIGURE 8.
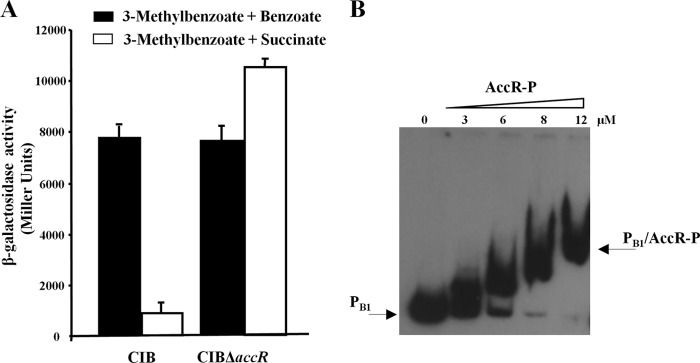
AccR controls the activity of the PB1 promoter. A, Azoarcus sp. CIB (CIB) and Azoarcus sp. CIBΔaccR (CIBΔaccR) cells harboring the PB1::lacZ reporter plasmid pIZPB1 were grown anaerobically in minimal medium containing a mixture of 3 mm 3-methylbenzoate and 1 mm benzoate as a co-carbon source that does not cause CCR of the PB1 promoter (black bars) or in minimal medium containing a mixture of 3 mm 3-methylbenzoate and 0.2% succinate as a co-carbon source that causes CCR of the PB1 promoter (white bars). Values are the average of three independent experiments ± S.D. (error bars) obtained for cells grown to the mid-exponential phase. B, gel retardation analyses of PB1 probe as in Fig. 3B. Assays were performed using increasing concentrations (0–12 μm) of carbamoyl phosphate-treated His6-AccR (AccR-P). The free PB1 probe and the major PB1·AccR-P complex are indicated by arrows.
A third distinct central pathway for anaerobic degradation of aromatics in Azoarcus sp. CIB is the hbd pathway devoted to 3-hydroxybenzoate catabolism (22). To determine whether AccR also controls the expression of the genes for 3-hydroxybenzoate catabolism, Azoarcus sp. CIBΔaccR harboring a plasmid expressing wild-type AccR or AccRD60E was grown anaerobically in minimal medium containing 3-hydroxybenzoate as the sole carbon source. As was the case for growth at the expense of benzoate (Fig. 7B), cells expressing AccR were able to grow on 3-hydroxybenzoate, whereas those expressing the constitutively active AccRD60E superrepressor could not. Hence, these results strongly suggest that the genes responsible for the anaerobic catabolism of 3-hydroxybenzoate are also negatively controlled by AccR. Together with the impact of AccR on the bzd and mbd pathways, the results provide additional support for the idea that AccR is a master regulator of the anaerobic catabolism of aromatic compounds in Azoarcus sp. CIB.
The AccR-mediated CCR Control Is Only Operational under Anaerobic Growth Conditions
To explore whether AccR also mediates CCR control of aerobic degradation of aromatic compounds, we used real-time RT-PCR to monitor the activity of the PD promoter, which drives the expression of the box genes for the aerobic degradation of benzoate in Azoarcus sp. CIB (27). When the cells were grown aerobically in benzoate, succinate significantly inhibited the activity of PD promoter; however, this inhibition was not attributable to AccR because it was also observed in the AccR null strain (Fig. 9A). Because we have previously shown that the box genes can be also expressed under anaerobic growth conditions (27), we tested whether such anaerobic expression was controlled by AccR. In contrast to the results obtained under aerobic growth conditions, when the cells were grown anaerobically in the presence of benzoate, succinate-mediated inhibition was found in the wild-type but not in the AccR null strain (Fig. 9B). As is the case for the PN and PB1P promoters, the PD promoter was bound by AccR-P in gel retardation assays (Fig. 9C) and also contains a sequence that bears homology to the ATGCA-N6-TGCAT motif identified on the basis of AccR-P binding to PN (Fig. 9D). Taken together, these results indicate that the AccR regulatory circuit operates through binding to common DNA motifs and is only operational under anaerobic growth conditions.
FIGURE 9.
AccR controls the activity of the PD promoter under anaerobic conditions. A and B, Azoarcus sp. CIB (CIB) and Azoarcus sp. CIBΔaccR (CIBΔaccR) cells were grown aerobically (A) or anaerobically (B) in minimal medium containing 3 mm benzoate (black bars) or a mixture of 3 mm benzoate and 0.2% succinate (white bars) until mid-exponential phase. Transcripts from the PD promoter were measured by real-time RT-PCR. Graphed values (in arbitrary units) are the average from three independent experiments ± S.D. (error bars). C, gel retardation analyses of PD probe as in Fig. 3B. Assays were performed with increasing concentrations (0–12 μm) of carbamoyl phosphate-treated His6-AccR (AccR-P). The free PD probe and the major PD·AccR-P complex are indicated by arrows. D, sequence alignment of AccR binding regions in different catabolic promoters of Azoarcus sp. CIB. PN (OR2), operator region 2 of the PN promoter; PN (OR1), operator region 1 of the PN promoter; PD, PD promoter of the box cluster; PB1, PB1 promoter of the mbd cluster. A consensus sequence of the AccR operator is shown at the bottom. Nucleotides identical to the consensus are shown in boldface type.
DISCUSSION
When faced with a mixture of alternative carbon sources, bacteria usually strategically utilize the substrate that yields the highest energy return. The utilization of preferred carbon sources is controlled by global regulatory mechanisms, termed CCR, which depend on the energy status of the cell. CCR control systems are also responsive to other environmental conditions, such as growth temperature and carbon source concentrations, and thus confer a competitive advantage in natural environments (4, 52, 53). Aromatic compounds usually constitute a secondary (non-preferred) carbon source for many bacteria. Diverse mechanisms underlie CCR control in aerobic metabolism of aromatic compounds (6–11, 16–18). Here we report the first known mechanism that accounts for CCR control of anaerobic catabolism of aromatic compounds in bacteria, namely the transcriptional repressor AccR of Azoarcus sp. CIB. Our evidence that AccR can be considered as a master regulator of anaerobic catabolism of aromatics is based on the finding that this protein controls expression of all known central anaerobic aromatic catabolic pathways of this strain. These include the bdz pathway for dissimilation of benzoate, the 3-methybenzoate mbd pathway, the 3-hydroxybenzoate hbd pathway, and the benzoate box pathway as expressed under anaerobic growth conditions (Figs. 1, 4F, 8, and 9). Because expression of the box pathway is only affected by the lack of AccR under anaerobic but not aerobic conditions (Fig. 9), we suggest that the AccR circuitry is only operational under anaerobic growth conditions.
In silico analysis assigns AccR to the FixJ/NarL family of response regulators (supplemental Fig. S1), whose members participate in the control of a wide variety of biological functions (45–47). We found that, like some previously characterized members of this family (e.g. FixJ, OmpR, PhoB, and StyR (54–56)), phosphorylation of AccR to AccR-P with high energy small molecule phosphodonors (e.g. carbamoyl phosphate) (Fig. 2B) triggers a monomer-to-dimer transition (Fig. 2, D and E); however, this reconfiguration does not change the apparent overall elongated shape of the AccR protomers. Under our in vitro conditions, only ∼50% phosphorylation of AccR was achieved (Fig. 2, B and D). This could be attributable to partial phosphorylation or spontaneous dephosphorylation; alternatively, it may be due to an innate phosphatase activity of AccR. In this respect, it is well established that despite extensive structural and chemical similarities, the half-life of the phosphorylated state of response regulators varies extensively (from seconds to hours), depending on both their innate autophosphorylation and autodephosphorylation kinetics (42).
In addition to subunit configuration, phosphorylation controls the ability of AccR to bind DNA (Fig. 3) and its capacity to cause repression of promoter output in in vitro transcription assays (Fig. 5). Conversely, substitution of the highly conserved Asp60 residue of the N-terminal REC motif of AccR to a phosphomimic Glu residue (AccRD60E) renders the protein constitutively active (phosphorylation-independent) both in terms of its DNA binding ability (Fig. 3C) and its capacity to repress promoter output in intact cells (Fig. 7), suggesting that Asp60 is the target residue of phosphorylation. Thus, AccR behaves like a typical response regulator that upon activation represses promoter output both in vivo and in vitro. The lack of anaerobic growth in benzoate of an AccR null derivative of Azoarcus sp. CIB expressing the AccRD60E protein (Fig. 7B) is consistent with the role of this mutant regulator as a superrepressor of the bzd central pathway.
The phosphorylation-triggered monomer-to-dimer transition of AccR suggests that the AccR-P would bind DNA as a dimer, a configuration consistent with our identification of the inverted repeat ATGCA-N6-TGCAT as the AccR DNA binding site within the PN promoter. The PN promoter harbors two of these sites (denoted OR1 and OR2), which can each be bound independently by AccR with approximately equal affinity (Fig. 4), whereas the PB1 and PD promoters, which are also targeted by AccR (Figs. 8 and 9), possess one copy of this motif (Fig. 9D). A truncated PN promoter that lacks the AccR binding regions (PN1) did not show succinate-dependent CCR control (Fig. 4F), which illustrates how a rational design of a target promoter can be an interesting strategy to circumvent CCR in bacteria. The location of the AccR binding region within PN (spanning positions −35 to −153 relative to the transcriptional start site) is consistent with the observed repressor role of AccR. We can envisage two plausible and non-mutually exclusive means by which DNA binding by AccR-P could inhibit promoter output: (i) inhibition of closed complex formation by occluding RNA polymerase and/or (ii) inhibition of open complex formation by preventing binding of the AcpR activator, whose operator region spans from −42.5 to −39.5 (25).
In contrast to the apparent dimerization-dependent DNA binding of intact AccR-P, the DNA binding-proficient C-terminal domain of AccR (CAccR) is monomeric in solution (Fig. 6). These findings suggest that (i) dimerization per se is not a prerequisite for DNA binding, and (ii) dimerization is carried out through the N-terminal domain, as exemplified by NarL (46). Further, the results suggest a regulatory scenario in which phosphorylation of Asp60 of AccR alleviates interdomain repression mediated by the N-terminal domain to allow reconfiguration to a DNA binding-proficient configuration. Because linker regions can be crucial for correct interdomain communication (57), it is plausible that the long linker region of AccR may play a role in transmitting the phosphorylation-dependent conformational changes (from the NAccR domain to the CAccR domain) to result in derepression and thus activation of its capacity to bind to target promoters. However, like a potential autophosphatase activity of AccR, these mechanistic possibilities require experimental verification.
As described in the Introduction, among the diverse proteins that have been found to be involved in CCR control of aerobic catabolism of aromatic compounds, only one, BphQ, is a response regulator. A search for accR homologs in bacterial genomes revealed the presence of AccR-like proteins (amino acid sequence identity higher than 70%) encoded in the genomes of other Azoarcus and Thauera strains (supplemental Fig. S1A). These AccR-like regulators cluster in a branch of the phylogenetic tree different from that of the BphQ-like regulators (supplemental Fig. S1B), which suggests that both types of proteins, although evolutionarily related, constitute different subgroups of response regulators. In this respect, it is worth noting that AccR from Azoarcus sp. CIB and BphQ from Acidovorax sp. KKS102 have completely different regulatory features; whereas BphQ is a transcriptional activator that binds its target DNA in the absence of phosphorylation (7), AccR is a repressor that requires phosphorylation in order to bind to a target operator that is completely different from that described for BphQ.
Many β-proteobacteria Azoarcus and Thauera strains are able to anaerobically metabolize aromatic compounds. The presence of accR orthologs within their genomes raises the possibility that AccR constitutes a new global regulator that controls anaerobic carbon catabolite repression in this group of organisms. Our observation that the accR genes are always located adjacent to a gene that encodes a putative sensor histidine kinase strongly suggests that the latter would be responsible for maintaining AccR in its active phosphorylated state in the presence of preferred carbon sources, such as succinate. The work presented here should greatly facilitate our future efforts aimed at characterizing this predicted two-component regulatory system and deciphering the environmental signals that trigger the activation/inactivation status of AccR.
Acknowledgments
The technical work of A. Valencia is greatly appreciated. The assistance of Secugen S. L. with DNA sequencing, of C. A. Botello and J. R. Luque with ultracentrifugation experiments, and of V. de los Ríos with the MALDI-TOF analyses is gratefully acknowledged. We are grateful to G. Durante-Rodríguez for providing plasmid pBBR5PNI.
This work was supported by Ministry of Economy and Competitiveness of Spain Grants BIO2009-10438, CSD2007-00005, and BIO2012-39501), European Union FP7 Grant 311815, and Swedish Research Council Grant 621-2011-4791.

This article contains supplemental Fig. S1.
- CCR
- carbon catabolite repression
- REC motif
- receiver motif.
REFERENCES
- 1. Görke B., Stülke J. (2008) Carbon catabolite repression in bacteria. Many ways to make the most out of nutrients. Nat. Rev. Microbiol. 6, 613–624 [DOI] [PubMed] [Google Scholar]
- 2. Collier D. N., Hager P. W., Phibbs P. V., Jr. (1996) Catabolite repression control in the Pseudomonads. Res. Microbiol. 147, 551–561 [DOI] [PubMed] [Google Scholar]
- 3. Moreno R., Fonseca P., Rojo F. (2012) Two small RNAs, CrcY and CrcZ, act in concert to sequester the Crc global regulator in Pseudomonas putida, modulating catabolite repression. Mol. Microbiol. 83, 24–40 [DOI] [PubMed] [Google Scholar]
- 4. Rojo F. (2010) Carbon catabolite repression in Pseudomonas. Optimizing metabolic versatility and interactions with the environment. FEMS Microbiol. Rev. 34, 658–684 [DOI] [PubMed] [Google Scholar]
- 5. Shingler V. (2003) Integrated regulation in response to aromatic compounds. From signal sensing to attractive behaviour. Environ. Microbiol. 5, 1226–1241 [DOI] [PubMed] [Google Scholar]
- 6. Marqués S., Aranda-Olmedo I., Ramos J. L. (2006) Controlling bacterial physiology for optimal expression of gene reporter constructs. Curr. Opin. Biotechnol. 17, 50–56 [DOI] [PubMed] [Google Scholar]
- 7. Ohtsubo Y., Goto H., Nagata Y., Kudo T., Tsuda M. (2006) Identification of a response regulator gene for catabolite control from a PCB-degrading β-proteobacteria, Acidovorax sp. KKS102. Mol. Microbiol. 60, 1563–1575 [DOI] [PubMed] [Google Scholar]
- 8. Prieto M. A., Galán B., Torres B., Ferrández A., Fernández C., Miñambres B., García J. L., Díaz E. (2004) Aromatic metabolism versus carbon availability. The regulatory network that controls catabolism of less-preferred carbon sources in Escherichia coli. FEMS Microbiol. Rev. 28, 503–518 [DOI] [PubMed] [Google Scholar]
- 9. Morales G., Linares J. F., Beloso A., Albar J. P., Martínez J. L., Rojo F. (2004) The Pseudomonas putida Crc global regulator controls the expression of genes from several chromosomal catabolic pathways for aromatic compounds. J. Bacteriol. 186, 1337–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Moreno R., Fonseca P., Rojo F. (2010) The Crc global regulator inhibits the Pseudomonas putida pWW0 toluene/xylene assimilation pathway by repressing the translation of regulatory and structural genes. J. Biol. Chem. 285, 24412–24419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hernández-Arranz S., Moreno R., Rojo F. (2013) The translational repressor Crc controls the Pseudomonas putida benzoate and alkane catabolic pathways using a multi-tier regulation strategy. Environ. Microbiol. 15, 227–241 [DOI] [PubMed] [Google Scholar]
- 12. MacGregor C. H., Wolff J. A., Arora S. K., Phibbs P. V. (1991) Cloning of a catabolite repression control (crc) gene from Pseudomonas aeruginosa, expression of the gene in Escherichia coli, and identification of the gene product in Pseudomonas aeruginosa. J. Bacteriol. 173, 7204–7212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Moreno R., Martínez-Gomariz M., Yuste L., Gil C., Rojo F. (2009) The Pseudomonas putida Crc global regulator controls the hierarchical assimilation of amino acids in a complete medium. Evidence from proteomic and genomic analyses. Proteomics 9, 2910–2928 [DOI] [PubMed] [Google Scholar]
- 14. Hester K. L., Madhusudhan K. T., Sokatch J. R. (2000) Catabolite repression control by crc in 2xYT medium is mediated by posttranscriptional regulation of bkdR expression in Pseudomonas putida. J. Bacteriol. 182, 1150–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Milojevic T., Grishkovskaya I., Sonnleitner E., Djinovic-Carugo K., Bläsi U. (2013) The Pseudomonas aeruginosa catabolite repression control protein Crc is devoid of RNA binding activity. PloS One 8, e64609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pflüger K., de Lorenzo V. (2008) Evidence of in vivo cross talk between the nitrogen-related and fructose-related branches of the carbohydrate phosphotransferase system of Pseudomonas putida. J. Bacteriol. 190, 3374–3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Morales G., Ugidos A., Rojo F. (2006) Inactivation of the Pseudomonas putida cytochrome o ubiquinol oxidase leads to a significant change in the transcriptome and to increased expression of the CIO and cbb3-1 terminal oxidases. Environ. Microbiol. 8, 1764–1774 [DOI] [PubMed] [Google Scholar]
- 18. Petruschka L., Burchhardt G., Müller C., Weihe C., Herrmann H. (2001) The cyo operon of Pseudomonas putida is involved in carbon catabolite repression of phenol degradation. Mol. Genet. Genomics 266, 199–206 [DOI] [PubMed] [Google Scholar]
- 19. Heider J., Boll M., Breese K., Breinig S., Ebenau-Jehle C., Feil U., Gad'on N., Laempe D., Leuthner B., Mohamed M. E., Schneider S., Burchhardt G., Fuchs G. (1998) Differential induction of enzymes involved in anaerobic metabolism of aromatic compounds in the denitrifying bacterium Thauera aromatica. Arch. Microbiol. 170, 120–131 [DOI] [PubMed] [Google Scholar]
- 20. López Barragán M. J., Carmona M., Zamarro M. T., Thiele B., Boll M., Fuchs G., García J. L., Díaz E. (2004) The bzd gene cluster, coding for anaerobic benzoate catabolism, in Azoarcus sp. strain CIB. J. Bacteriol. 186, 5762–5774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Trautwein K., Grundmann O., Wöhlbrand L., Eberlein C., Boll M., Rabus R. (2012) Benzoate mediates repression of C(4)-dicarboxylate utilization in “Aromatoleum aromaticum” EbN1. J. Bacteriol. 194, 518–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Carmona M., Zamarro M. T., Blázquez B., Durante-Rodríguez G., Juárez J. F., Valderrama J. A., Barragán M. J., García J. L., Díaz E. (2009) Anaerobic catabolism of aromatic compounds. A genetic and genomic view. Microbiol. Mol. Biol. Rev. 73, 71–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Barragán M. J., Blázquez B., Zamarro M. T., Mancheño J. M., García J. L., Díaz E., Carmona M. (2005) BzdR, a repressor that controls the anaerobic catabolism of benzoate in Azoarcus sp. CIB, is the first member of a new subfamily of transcriptional regulators. J. Biol. Chem. 280, 10683–10694 [DOI] [PubMed] [Google Scholar]
- 24. Durante-Rodríguez G., Valderrama J. A., Mancheño J. M., Rivas G., Alfonso C., Arias-Palomo E., Llorca O., García J. L., Díaz E., Carmona M. (2010) Biochemical characterization of the transcriptional regulator BzdR from Azoarcus sp. CIB. J. Biol. Chem. 285, 35694–35705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Durante-Rodríguez G., Zamarro M. T., García J. L., Díaz E., Carmona M. (2006) Oxygen-dependent regulation of the central pathway for the anaerobic catabolism of aromatic compounds in Azoarcus sp. strain CIB. J. Bacteriol. 188, 2343–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Durante-Rodríguez G., Zamarro M. T., García J. L., Díaz E., Carmona M. (2008) New insights into the BzdR-mediated transcriptional regulation of the anaerobic catabolism of benzoate in Azoarcus sp. CIB. Microbiology 154, 306–316 [DOI] [PubMed] [Google Scholar]
- 27. Valderrama J. A., Durante-Rodríguez G., Blázquez B., García J. L., Carmona M., Díaz E. (2012) Bacterial degradation of benzoate. Cross-regulation between aerobic and anaerobic pathways. J. Biol. Chem. 287, 10494–10508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Casadaban M. J. (1976) Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage λ and μ. J. Mol. Biol. 104, 541–555 [DOI] [PubMed] [Google Scholar]
- 29. Miller V. L., Mekalanos J. J. (1988) A novel suicide vector and its use in construction of insertion mutations. Osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170, 2575–2583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. de Lorenzo V., Timmis K. N. (1994) Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 235, 386–405 [DOI] [PubMed] [Google Scholar]
- 31. Kaniga K., Delor I., Cornelis G. R. (1991) A wide-host-range suicide vector for improving reverse genetics in gram-negative bacteria. Inactivation of the blaA gene of Yersinia enterocolitica. Gene 109, 137–141 [DOI] [PubMed] [Google Scholar]
- 32. Moreno-Ruiz E., Hernáez M. J., Martínez-Pérez O., Santero E. (2003) Identification and functional characterization of Sphingomonas macrogolitabida strain TFA genes involved in the first two steps of the tetralin catabolic pathway. J. Bacteriol. 185, 2026–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kovach M. E., Elzer P. H., Hill D. S., Robertson G. T., Farris M. A., Roop R. M., 2nd, Peterson K. M. (1995) Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166, 175–176 [DOI] [PubMed] [Google Scholar]
- 34. Juárez J. F., Zamarro M. T., Eberlein C., Boll M., Carmona M., Díaz E. (2013) Characterization of the mbd cluster encoding the anaerobic 3-methylbenzoyl-CoA central pathway. Environ. Microbiol. 15, 148–166 [DOI] [PubMed] [Google Scholar]
- 35. Sambrook J., Rusell D. W. (2001) Molecular Cloning. A Laboratory Manual, 3rd Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 36. Miller J. H. (1972) Experiments in Molecular Genetics, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 37. Thompson J. D., Higgins D. G., Gibson T. J. (1994) CLUSTAL W. improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kimura M. (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120 [DOI] [PubMed] [Google Scholar]
- 39. Saitou N., Nei M. (1987) The neighbor-joining method. A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 [DOI] [PubMed] [Google Scholar]
- 40. Felsenstein J. (1993) PHYLYP (Phylogenetic Inference Package), version 3.5.1, University of Washington, Seattle, WA [Google Scholar]
- 41. Schuck P. (2000) Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 78, 1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Laue T. M., Shah B. D., Ridgeway T. M., Pelletier S. L. (1992) Analytical Ultracentrifugation in Biochemistry, Royal Society of Chemistry, Cambridge, UK [Google Scholar]
- 43. Maxam A. M., Gilbert W. (1980) Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 65, 499–560 [DOI] [PubMed] [Google Scholar]
- 44. Kiley P. J., Reznikoff W. S. (1991) Fnr mutants that activate gene expression in the presence of oxygen. J. Bacteriol. 173, 16–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kurashima-Ito K., Kasai Y., Hosono K., Tamura K., Oue S., Isogai M., Ito Y., Nakamura H., Shiro Y. (2005) Solution structure of the C-terminal transcriptional activator domain of FixJ from Sinorhizobium meliloti and its recognition of the fixK promoter. Biochemistry 44, 14835–14844 [DOI] [PubMed] [Google Scholar]
- 46. Maris A. E., Sawaya M. R., Kaczor-Grzeskowiak M., Jarvis M. R., Bearson S. M., Kopka M. L., Schröder I., Gunsalus R. P., Dickerson R. E. (2002) Dimerization allows DNA target site recognition by the NarL response regulator. Nat. Struct. Biol. 9, 771–778 [DOI] [PubMed] [Google Scholar]
- 47. Milani M., Leoni L., Rampioni G., Zennaro E., Ascenzi P., Bolognesi M. (2005) An active-like structure in the unphosphorylated StyR response regulator suggests a phosphorylation-dependent allosteric activation mechanism. Structure 13, 1289–1297 [DOI] [PubMed] [Google Scholar]
- 48. Bourret R. B. (2010) Receiver domain structure and function in response regulator proteins. Curr. Opin. Microbiol. 13, 142–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Klose K. E., Weiss D. S., Kustu S. (1993) Glutamate at the site of phosphorylation of nitrogen-regulatory protein NTRC mimics aspartyl-phosphate and activates the protein. J. Mol. Biol. 232, 67–78 [DOI] [PubMed] [Google Scholar]
- 50. Lan C. Y., Igo M. M. (1998) Differential expression of the OmpF and OmpC porin proteins in Escherichia coli K-12 depends upon the level of active OmpR. J. Bacteriol. 180, 171–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gao R., Stock A. M. (2010) Molecular strategies for phosphorylation-mediated regulation of response regulator activity. Curr. Opin. Microbiol. 13, 160–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fonseca P., Moreno R., Rojo F. (2013) Pseudomonas putida growing at low temperature shows increased levels of CrcZ and CrcY sRNAs, leading to reduced Crc-dependent catabolite repression. Environ. Microbiol. 15, 24–35 [DOI] [PubMed] [Google Scholar]
- 53. Valentini M., Lapouge K. (2013) Catabolite repression in Pseudomonas aeruginosa PAO1 regulates the uptake of C4-dicarboxylates depending on succinate concentration. Environ. Microbiol. 15, 1707–1716 [DOI] [PubMed] [Google Scholar]
- 54. Da Re S., Schumacher J., Rousseau P., Fourment J., Ebel C., Kahn D. (1999) Phosphorylation-induced dimerization of the FixJ receiver domain. Mol. Microbiol. 34, 504–511 [DOI] [PubMed] [Google Scholar]
- 55. Leoni L., Ascenzi P., Bocedi A., Rampioni G., Castellini L., Zennaro E. (2003) Styrene-catabolism regulation in Pseudomonas fluorescens ST. Phosphorylation of StyR induces dimerization and cooperative DNA-binding. Biochem. Biophys. Res. Commun. 303, 926–931 [DOI] [PubMed] [Google Scholar]
- 56. Bachhawat P., Swapna G. V., Montelione G. T., Stock A. M. (2005) Mechanism of activation for transcription factor PhoB suggested by different modes of dimerization in the inactive and active states. Structure 13, 1353–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mattison K., Oropeza R., Kenney L. J. (2002) The linker region plays an important role in the interdomain communication of the response regulator OmpR. J. Biol. Chem. 277, 32714–32721 [DOI] [PubMed] [Google Scholar]