Background: Sld2 is required for the initiation of DNA replication.
Results: An Sld2-Mcm2-7 binding mutant exhibits premature helicase assembly, and an Sld2-DNA binding mutant displays no helicase assembly.
Conclusion: Sld2-Mcm interaction is critical to prevent the premature assembly of the replicative helicase, and Sld2-DNA interaction is required for proper helicase assembly.
Significance: Sld2 regulates assembly of the replicative helicase.
Keywords: CDK (Cyclin-dependent Kinase), Cell Cycle, DNA-binding Protein, DNA Helicase, DNA Replication
Abstract
Assembly of the Cdc45-Mcm2-7-GINS (CMG) replicative helicase complex must be regulated to ensure that DNA unwinding is coupled with DNA synthesis. Sld2 is required for the initiation of DNA replication in budding yeast. We identified a mutant of Sld2, Sld2-m1,4, that is specifically defective in Mcm2-7 binding. When this sld2-m1,4 mutant is expressed, cells exhibit severe inhibition of DNA replication. Furthermore, the CMG complex assembles prematurely in G1 in mutant cells, but not wild-type cells. These data suggest that Sld2 binding to Mcm2-7 is essential to block the inappropriate formation of a CMG helicase complex in G1. We also study a mutant of Sld2 that is defective in binding DNA, sld2-DNA, and find that sld2-DNA cells exhibit no GINS-Mcm2-7 interaction. These data suggest that Sld2 association with DNA is required for CMG assembly in S phase.
Introduction
DNA unwinding at a replication fork is a highly regulated event to ensure that the unwinding of genomic DNA is coordinated with DNA synthesis (1). Uncoupled DNA unwinding from DNA synthesis may result in genome instability and chromosome damage (2, 3). The Cdc45-Mcm2-7-GINS (CMG)2 complex unwinds parental DNA at a replication fork in eukaryotes during S phase, and the CMG assembles during S phase (4–7). To ensure the proper regulation of DNA unwinding, the Mcm2-7 heterohexameric ring assembly is loaded to encircle double-stranded DNA during G1 (8, 9). The biochemical details of this loading event have recently been elucidated (10, 11). Electron microscopy and biochemical data demonstrate that the Mcm2-7 complex, on its own, exists in equilibrium between an open-ring and closed-ring state (12, 13). The intrinsic ability of the Mcm2-7 ring-shaped complex to open is required for the ring to encircle double-stranded DNA in G1 (12, 13). The Cdc45 protein associates with Mcm2-7 proteins in G1 in a manner that depends upon the Dbf4-dependent kinase (DDK) (14–16).
During S phase, the Mcm2-7 transitions from encircling two strands of DNA to encircling one strand of DNA (4, 17). Thus, a single-strand of DNA is extruded from the central channel of the Mcm2-7 ring during S phase (17). The open-ring conformation of the Mcm2-7 complex is required to allow a single-strand of DNA to exit from the central channel of the Mcm2-7 complex (12, 13). Once the Mcm2-7 complex encircles a single-strand of DNA, GINS binds to the Cdc45-Mcm2-7 complex to form the CMG complex (12). GINS association with Cdc45-Mcm2-7 depends upon activity of the cyclin-dependent kinase (CDK), and it may also require the activities of three essential initiation proteins: Sld2, Sld3, and Dpb11 (16, 18, 19). The CMG complex, unlike the Mcm2-7 complex, is a sealed ring-shaped assembly that cannot open (12). The sealed CMG ring allows for processive translocation of the helicase while surrounding single-stranded DNA (17). The CMG does not dissociate from DNA during replication of the genome because of its closed-ringed structure (6).
Sld2 is required for the initiation of DNA replication in budding yeast, but Sld2 does not travel with the replication fork (20). Sld2 contains a domain that is conserved in human is RecQL4 (21). Sld2, along with Sld3 and Dpb11, assist in the recruitment of GINS to replication origins during S phase in a manner that depends upon CDK activity (18, 19). A phosphomimetic mutant of Sld2, Sld2T84D, functionally substitutes for CDK-activated Sld2 in vivo (22). CDK-phosphorylated Sld2 and CDK-phosphorylated Sld3 bind to Dpb11 (18, 19). Sld2 also forms part of the pre-loading complex, which is a CDK-dependent assembly composed of Sld2, Pol ϵ, Dpb11, and GINS (23).
It has previously been reported that in vitro, GINS and Sld2 compete with one another for Mcm2-7 binding (24). However, the biological significance of this in vitro observation has not been elucidated. It has also been reported that single-stranded origin DNA binds to CDK-activated Sld2; furthermore, CDK-activated-Sld2 binding to origin ssDNA dissociates Sld2 from the Mcm2-7 complex in vitro (24). A mutant of Sld2 that does not bind DNA, Sld2-K44E,K50E,K438E (Sld2-DNA), is lethal when expressed in budding yeast cells, and DNA replication is substantially inhibited when this sld2-DNA mutant is expressed in budding yeast (24). Further investigation of the Sld2-DNA-binding mutant has not yet been accomplished.
In this report, we identify a mutation of Sld2 that is specifically defective in binding Mcm2-7. This mutant, Sld2-m1,4 (Sld2-K416E,R435E,R436E), in contrast to wild-type Sld2, does not compete with GINS for Mcm2-7 binding in vitro. When sld2-m1,4 is expressed in budding yeast cells, there is a severe growth defect and DNA replication is substantially inhibited. Co-immunoprecipitation analyses reveal that in sld2-m1,4 mutant cells, GINS associates with Cdc45-Mcm2-7 prematurely in G1, while wild-type cells reveal no GINS association with Cdc45-Mcm2-7 until S phase. Furthermore, RPA-chromatin immunoprecipitation assays of cells exposed to hydroxyurea demonstrate that RPA is not recruited to origins in S phase in sld2-m1,4 mutant cells, unlike wild-type cells. These data suggest that Sld2-Mcm2-7 interaction is required to restrict the association of GINS with Mcm2-7 to S phase. We also find that for cells expressing sld2-DNA, GINS does not associate with Mcm2-7. These data suggest that Sld2-ssDNA interaction is essential to allow GINS to bind Mcm2-7 during S phase.
EXPERIMENTAL PROCEDURES
Purified Proteins
Full-length GST-Sld2, PKA-Sld2, and Sld2 (wild-type and mutants) were purified as described (24, 25). Sld2 fragments were purified as described (24). GINS and PKA-GINS were purified as described (26). GST-Dpb11 and PKA-Dpb11 were purified as described (25). PKA-Mcm2-7 (PKA tag at the N terminus of Mcm3), GST-Mcm2-7 (GST-tag at the N terminus of Mcm2), and Mcm2-7 were purified as described (26). GST-Pol ϵ was purified as described (27). Protein kinase A was a generous gift from Susan Taylor. α-Factor peptide pheromone was obtained from Zymo research (catalogue number Y1001).
DNA
The sequence of ssARS1-1 is TTACATCTTGTTATT TTACAGATTTTATGTTTAGATCTTTTATGCTTGCTTTTCAAAAGGCCTGCAGGCAAGTGCACAAA.
Kinase Labeling
PKA kinase labeling was performed as described (26). Proteins containing a PKA tag at the N terminus (Sld2, Mcm2-7 (Mcm3), Dpb11, GINS (Psf1)) were radiolabeled in a reaction volume 100 μl that contained 20 μm PKA-tagged protein in kinase reaction buffer (5 mm Tris-HCl, pH 8.5, 10 mm MgCl2, 1 mm DTT, 500 μm ATP, 500 μCi [γ-32P]ATP) containing 5 μg of PKA. Reactions were incubated for 1 h at 30 °C.
GST Pulldown
The GST pulldown reactions were performed as described (26). GST-pulldown reactions were in a volume of 100 μl and contained GST-tagged protein in GST-binding buffer (40 mm Tris-HCl, pH 7.5, 100 mm NaCl, 0.1 mm EDTA, 10% glycerol, 0.1% Triton X-100, 1 mm DTT, 0.7 μg/ml pepstatin, 0.1 mm PMSF, and 0.1 mg/ml BSA) and varying amounts of radiolabeled DNA or protein as described in each figure. Reactions were incubated at room temperature for 1 h. Following incubation, reactions were added to 40 μl of prepared glutathione-Sepharose and gently mixed. Binding of GST-tagged protein to the beads we performed for 20 min with gentle mixing every few minutes. When the binding was complete, the beads were allowed to settle, the supernatant was removed, and the glutathione beads were washed two times with 0.5 ml of GST-binding buffer. After the last wash, 30 μl of 5× SDS sample buffer was added to each reaction, and the samples were boiled for 10 min. Samples (20 μl) were then analyzed by SDS-PAGE.
Biotinylated-DNA Pulldown
Biotinylated-DNA pulldown assays were performed as described (28). Biotinylated DNA conjugated to streptavidin-agarose magnetic beads (Dynal) was incubated 5 min at 30 °C with increasing concentrations of radiolabeled protein in a solution containing 0.1 mm EDTA, 0.2 mm DTT, 10 mm magnesium acetate, 10% glycerol, 40 μg/ml BSA, and 20 mm Tris-HCl pH 7.5 in a final volume of 25 μl. Following the 5-min incubation the beads were collected at room temperature using a magnet (Dynal). The supernatant was removed, and the beads were washed twice with a solution containing 0.1 mm EDTA, 0.2 mm DTT, 10 mm magnesium acetate, 10% glycerol, 40 μg/ml BSA, and 20 mm Tris-HCl pH 7.5. The beads were collected with a magnet, the supernatant was removed, and the beads were heated at 95 °C for 10 min in a solution containing 2% SDS, 2 mm DTT, 4% glycerol, 4 mm Tris-HCl, and 0.01% bromphenol blue. The reactions were analyzed by SDS/PAGE. The gel was dried for 1 h at 80 °C and exposed to a phosphorimaging screen for 1 h.
Size-exclusion Chromatography
Size-exclusion chromatography was performed as described (26). Unlabeled protein was mixed with radiolabeled protein as described in the figure legend in a final volume of 200 μl, and incubated at 30 °C for 1 h in column buffer (50 mm Tris pH 7.5, 100 mm NaCl, 1 mm EDTA, 5% glycerol). The protein mixture was then subjected to 24 ml Superose 6 (GE Life Sciences) size exclusion chromatography in the same column buffer. Each 250 μl fraction was then subjected to SDS-PAGE analysis and quantified using phosphorimager analysis.
Yeast Strains and Plasmids
The sld2-td degron strain was generated as described (24). The sld2-td degron strain was transformed with a PRS415 vector containing either an empty vector, SLD2-wild-type, sld2-m1,4, or sld2-DNA under the control of the native Sld2 promoter. Positive transformants were selected on CSM-leu plates. PSF2-5FLAG and MCM4-9Myc were generated using reagents from Yeast Genetic Resource Center (Strain BY 25927) and Karim Labib (Strain YAG-355-2).
Antibodies
Antibodies directed against RPA, Mcm2, and Pol2 were purchased (RPA-Pierce MA1-25889, Mcm2-Santa Cruz SC-28551, Pol2-Santa Cruz SC-6752). Antibodies against Psf2 and Cdc45 were generated and purified as described (27). Open Biosystems produced polyclonal antibodies directed against Sld2 or Dpb11 (we supplied the antigens). Crude serum was purified against immobilized antigen to remove nonspecific antibodies. The specificity of each antibody was analyzed by Western analysis of purified protein and wild-type yeast extract (not shown). Antibodies directed against the flag or myc epitopes were commercially purchased.
Serial Dilution Analysis
Serial dilution was performed as described (24). 10-fold serial dilutions were performed on the indicated media and incubated at the indicated temperatures.
Fluorescence Activated Cell Sorting (FACS)
FACS was performed as described (24). For G1 arrest and release experiments 6 × 106 cells/ml were treated with α-factor (Zymo Research) for 3 h. Following extensive washes and addition of 50 μg/ml Pronase (Calbiochem), cells were further incubated for the indicated time. Cell cycle progression was followed by flow cytometry (FACS) stained with propidium iodide using FACSAria.
Co-immunoprecipitation
For G1 arrest and release experiments 6 × 106 cells/ml were treated with α-factor (Zymo Research) for 3 h. Cells were then subjected to extensive washes, followed by the addition of 50 μg/ml Pronase (Calbiochem). Cells were further incubated in 0.2 m hydroxyurea for the indicated time. Cells (4 × 108) were collected and lysed at 4 °C with glass beads (BeadBeater) in IP buffer (100 mm Hepes-KOH pH 7.9, 100 mm potassium acetate, 10 mm magnesium acetate, 2 mm NaF, 1 mm PMSF, 0.1 mm Na3VO4, 20 mm β-glycerophosphate, 1% Triton X-100, leupeptin, pepstatin, 1% protease inhibitor mixture (Sigma P8215), 1× Complete protease inhibitor mixture without EDTA (Roche 04693132001). Lysed material was treated with 200 units of Benzonase nuclease (Novagen 70746–3) on ice for 1 h. Clarified extract was then mixed with 2 μl of specified antibody and rotated for 2 h in the cold room, and then 5 μl of Dynabeads Protein A (Invitrogen 100.01D) beads equilibrated with IP buffer, were added and further incubated for 2 h. Fixed samples (formaldehyde) were prepared as described (23). Beads were washed with two times with 1 ml of IP buffer and finally resuspended in SDS-sample buffer. Western analysis was performed using the enhanced chemiluminescence system (GE Lifesciences).
Chromatin Immunoprecipitation (ChIP)
For G1 arrest and release experiments 6 × 106 cells/ml were treated with α-factor (Zymo Research) for 3 h. Following extensive washes and addition of 50 μg/ml Pronase (Calbiochem), cells were further incubated for the indicated time. Chromatin immunoprecipitation was performed as described (27). We performed PCR with [32P-α]dCTP as a component of the PCR reaction to quantify the amplified product. Formaldehyde cross-linked cells were lysed with glass beads in a Bead Beater. DNA was fragmented by sonication (Branson 450). Antibody and magnetic protein A beads (Dynabeads Protein A, Invitrogen 100.02D) were added to the cleared lysate to immunoprecipitate the DNA. Immunoprecipitates were washed extensively to remove nonspecific DNA. Eluted DNA was then subjected to PCR analysis using primers directed against ARS305, ARS306, or a region midway between ARS305 and ARS306 as described (29). The radioactive band in the agarose gel, representing specific PCR-amplified DNA product, was quantified by phosphorimaging and normalized by a reference standard run in the same gel. The reference standard was a PCR reaction accomplished with known quantity of template DNA replacing immunoprecipitate.
RESULTS
Identification of an Sld2 Mutant (Sld2-m1,4) That Is Defective in Mcm2-7 Binding
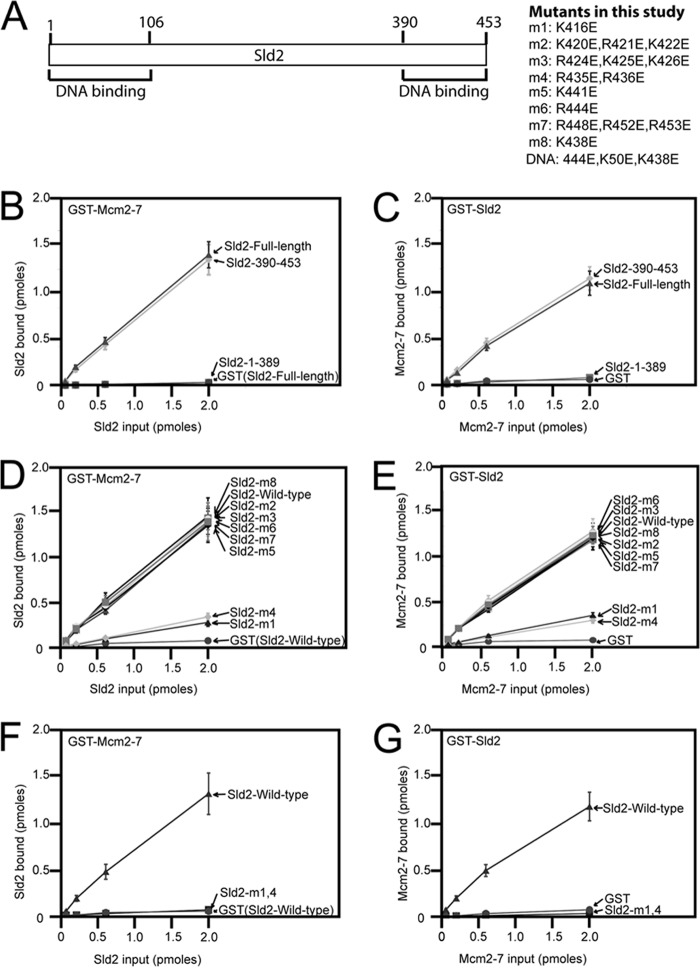
We previously reported that Sld2 binds directly to the Mcm2-7 complex by GST-pulldown assays of purified proteins and size-exclusion chromatography analysis (24). We sought to identify a mutant of Sld2 that is specifically defective in Mcm2-7 binding, to characterize the role of Sld2-Mcm2-7 interaction in vivo. We previously found that region 1–106 and region 390–453 of Sld2 bind to single-stranded DNA (Fig. 1A) (24). To identify the region of Sld2 that binds to Mcm2-7, we divided Sld2 into an N-terminal region (amino acids 1–389, Sld2–1-389), and a C-terminal region (amino acids 390–453, Sld2–390-453). We engineered a PKA tag at the N termini of the proteins to allow for protein radiolabeling with 32P and subsequent quantitation of the pulldown assay. We found that GST-Mcm2-7 pulls down [32P]Sld2-full-length protein like the C-terminal [32P]Sld2–390-453 (Fig. 1B). In contrast, GST-Mcm2-7 does not pull down the N-terminal [32P]Sld2–1-389 (Fig. 1B). Similarly, GST-Sld2–390-453 pulls down [32P]Mcm2-7 like wild-type GST-Sld2, while GST-Sld2–1-389 does not pull down [32P]Mcm2-7 (Fig. 1C).
FIGURE 1.
Identification of an Sld2 mutant (Sld2-m1,4) that is defective in Mcm2-7 binding. A, schematic of the region/function of Sld2. The N-terminal (1–106) and C-terminal (390–453) regions each bind to ssDNA (24). Eight mutations (labeled m1-m8) that reverse positive charge in the C-terminal region (amino acids 390–453) of Sld2. “DNA” is a mutation of Sld2 that does not bind to ssDNA (24). B, 2 pmol GST-Mcm2-7 or GST was used to pull-down various concentrations of radiolabeled Sld2-full-length, Sld2–390-453, or Sld2–1-389. GST-Mcm2-7 has a GST tag at the N terminus of Mcm2. Sld2 proteins have an N-terminal PKA tag that was phosphorylated with purified PKA and [γ-32P]ATP. Bound protein was analyzed by SDS-PAGE and quantified with phosphorimaging. For detailed methods, see “Experimental Procedures.” C, 2 pmol GST-Sld2-full-length, GST-Sld2–390-453, GST-Sld2–1-389, or GST was used to pull down radiolabeled Mcm2-7. To generate radiolabeled Mcm2-7, we generated Mcm3 with a N-terminal PKA tag that was phosphorylated with purified PKA and [γ-32P]ATP. D, 2 pmol GST-Mcm2-7 or GST was used to pull down various concentrations of radiolabeled Sld2-full-length protein. Sld2-full-length was wild-type, m1, m2, m3, m4, m5, m6, m7, or m8. E, 2 pmol GST-Sld2 (full-length or m1, m2, m3, m4, m5, m6, m7, or m8) of GST was used to pull down radiolabeled Mcm2-7. F, 2 pmol GST-Mcm2-7 or GST was used to pull down various concentrations of radiolabeled full-length Sld2-wild-type or Sld2-m1,4 (K416E,R435E,R436E). G, 2 pmol full-length GST-Sld2-wild-type or Sld2-m1,4 was used to pull down various concentrations of radiolabeled Mcm2-7.
The C-terminal region (390–453) of Sld2 is rich in positively charged amino acids, and we postulated that the interaction between Sld2 and Mcm2-7 is mediated at least in part by one or more of these positively charged amino acids. We reversed the charge of the lysines and arginines in the 390–453 region of Sld2, generating 8 mutations, designated Sld2-m1 to Sld2-m8 (Fig. 1A). We screened each of these 8 mutants with the GST-pulldown assays, and we found that two of these mutants, m1 and m4, were each partially defective in binding to Mcm2-7 (Fig. 1, D and E). We combined mutations m1 and m4 to create the combined mutation Sld2-m1,4 (Sld2-K416E,R435E,R436E). This Sld2-m1,4 mutation was completely defective in binding Mcm2-7 as determined by the GST-pulldown assays (Fig. 1, F and G).
Sld2-m1,4 Binds to Dpb11, ssDNA, and Polϵ-like Wild-type Sld2
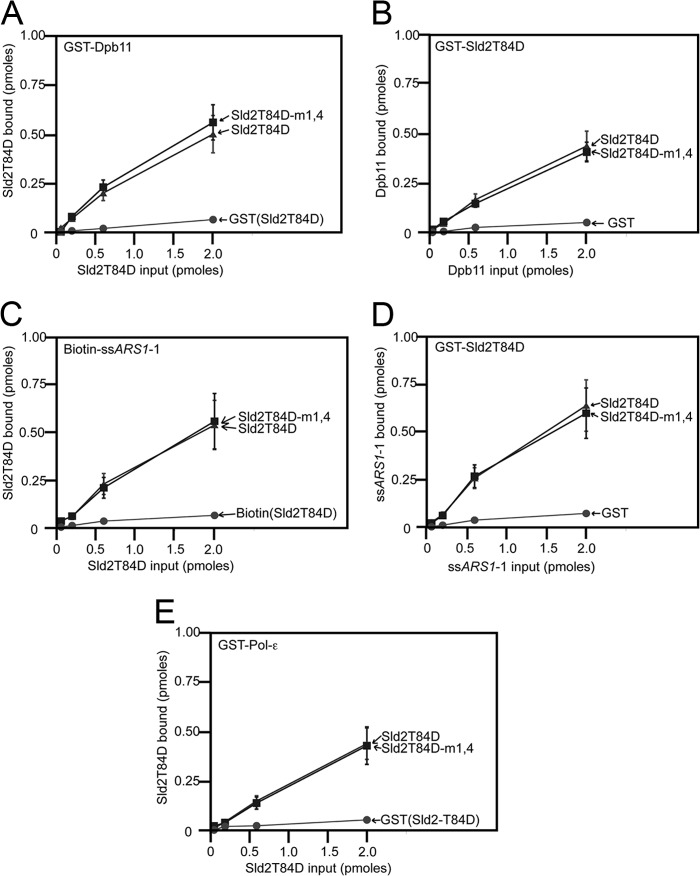
We next determined whether the Sld2-m1,4 mutant was defective in binding to its other known binding partners, Dpb11, ssDNA, or Polϵ (Fig. 2). These interactions are activated by CDK (23, 25); thus, we utilized the phosphomimetic mutant of Sld2, Sld2T84D, to assess whether the Sld2-m1,4 mutation inhibited binding to Dpb11 (Fig. 2, A and B), ssDNA (Fig. 2, C and D), or Polϵ (Fig. 2E). We found no significant difference in binding between Sld2-T84D and Sld2T84D-m1,4 in these pulldown assays. These data suggest that the Sld2-m1,4 mutation is specifically defective in Mcm2-7 binding.
FIGURE 2.
Sld2-m1,4 binds to Dpb11, ssDNA and Polϵ-like wild-type Sld2. A, 2 pmol GST-Dpb11 or GST was used to pull down various concentrations of radiolabeled full-length Sld2T84D or Sld2T84D-m1,4 (K416E,R435E,R436E). Sld2 proteins have an N-terminal PKA tag that was phosphorylated with purified PKA and [γ-32P]ATP. Bound protein was analyzed by SDS-PAGE and quantified with phosphorimaging. For detailed methods, see “Experimental Procedures.” B, 2 pmol full-length GST-Sld2T84D, GST-Sld2T84D-m1,4 or GST was used to pull down various concentrations of radiolabeled Dpb11. Dpb11 has an N-terminal PKA tag that was phosphorylated with purified PKA and [γ-32P]ATP. C, 2 pmol biotinylated ssARS1-1 (sequence under “Experimental Procedures”) or biotin was used to pull down various concentrations of radiolabeled full-length Sld2T84D or Sld2T84D-m1,4. D, 2 pmol full-length GST-Sld2T84D, GST-Sld2T84D-m1,4 or GST was used to pull down various concentrations of radiolabeled ssARS1-1. E, 2 pmol GST-Pol-ϵ or GST was used to pull down various concentrations of full-length radiolabeled Sld2T84D or Sld2T84D-m1,4.
Sld2-m1,4 Does Not Compete with GINS for Mcm2-7 Binding
We previously found that Sld2 competes with GINS for binding to Mcm2-7 (24). To determine whether Sld2-m1,4 is defective in competing with GINS for Mcm2-7, we performed the GST pulldown assay with GST-Mcm2-7, radiolabeled GINS, and various concentrations of unlabeled Sld2-wild-type or Sld2-m1,4 (Fig. 3). Wild-type Sld2 competes with GINS for binding to Mcm2-7 in a concentration-dependent manner, as expected (Fig. 3A). In contrast, Sld2-m1,4 does not compete with GINS for binding to Mcm2-7 (Fig. 3A), even as the molar ratio of Sld2-m1,4/Mcm2-7 was varied from 0 to 2, 5, and 10. These data show that Sld2-m1,4 does not interfere with GINS-Mcm2-7 interaction, unlike wild-type Sld2.
FIGURE 3.
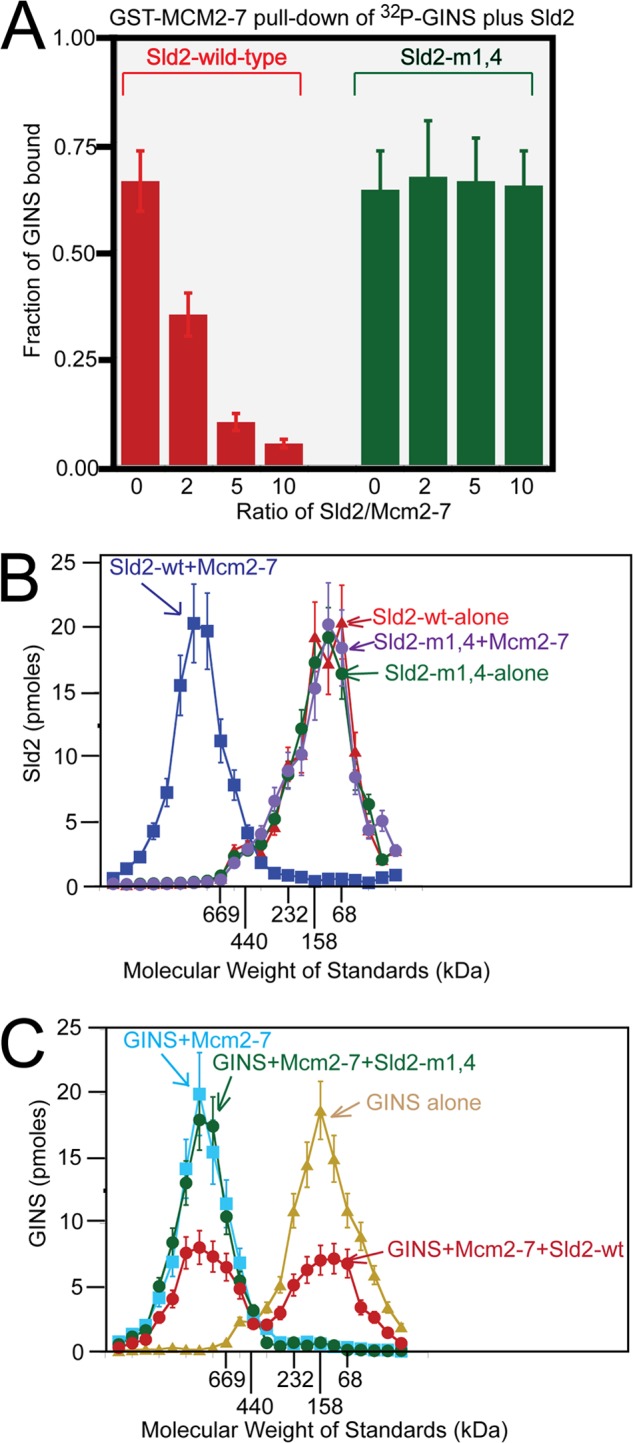
Sld2-m1,4 does not compete with GINS for Mcm2-7 binding. A, 2 pmol GST-Mcm2-7 was used to pull down 2 pmol of radiolabeled GINS in the presence of various concentrations of unlabeled full-length Sld2-wild-type or Sld2-m1,4. Radiolabeled GINS has a PKA tag at the N terminus of Psf1 that was phosphorylated with purified PKA and [γ-32P]ATP. Bound protein was analyzed by SDS-PAGE and quantified with phosphorimaging. For detailed methods, see “Experimental Procedures.” B, 100 pmol radiolabeled full-length Sld2-wild-type or Sld2-m1,4 was incubated with equimolar Mcm2-7 or no protein and subjected to size exclusion chromatography as described in “Experimental Procedures.” The radioactive counts in each fraction were used to calculate the pmol of Sld2 in each fraction. Sld2 (pmol) was then plotted versus the elution of molecular mass standards. C, 100 pmol of radiolabeled GINS was incubated with no protein, equimolar Mcm2-7, equimolar Mcm2-7, and Sld2-wild-type, or equimolar Mcm2-7 and Sld2-m1,4. The proteins were then subjected to size exclusion chromatography as described in “Experimental Procedures.” The radioactive counts in each fraction were used to calculate the pmol of GINS in each fraction. GINS (pmol) was then plotted versus the elution of molecular mass standards.
Sld2-wild-type interaction with Mcm2-7 is observable by size exclusion chromatography analysis (24). To determine if size exclusion chromatography detects an interaction between Sld2-m1,4 and Mcm2-7, we performed size exclusion chromatography with the relevant proteins (Fig. 3B). Whereas wild-type Sld2 interaction with Mcm2-7 is observable in this assay, Sld2-m1,4 does not exhibit an interaction with Mcm2-7 under the same conditions (Fig. 3B). These data further support that Sld2-m1,4 does not bind directly to Mcm2-7.
We next determined whether Sld2-m1,4 competes with GINS for Mcm2-7 by size exclusion chromatography (Fig. 3C). Wild-type Sld2 displaces roughly half of the GINS bound to Mcm2-7 in this assay, as expected, confirming that wild-type Sld2 competes with GINS for Mcm2-7 (Fig. 3C). In contrast, Sld2-m1,4 does not affect the interaction between GINS and Mcm2-7 as determined by the gel filtration assay (Fig. 3C). The in vitro data from Figs. 1, 2, and 3 suggest that Sld2-m1,4 is specifically defective in binding Mcm2-7; furthermore, Sld2-m1,4 unlike wild-type Sld2, does not compete with GINS for Mcm2-7 binding.
Expression of sld2-m1,4 Severely Inhibits Cell Growth and DNA Replication
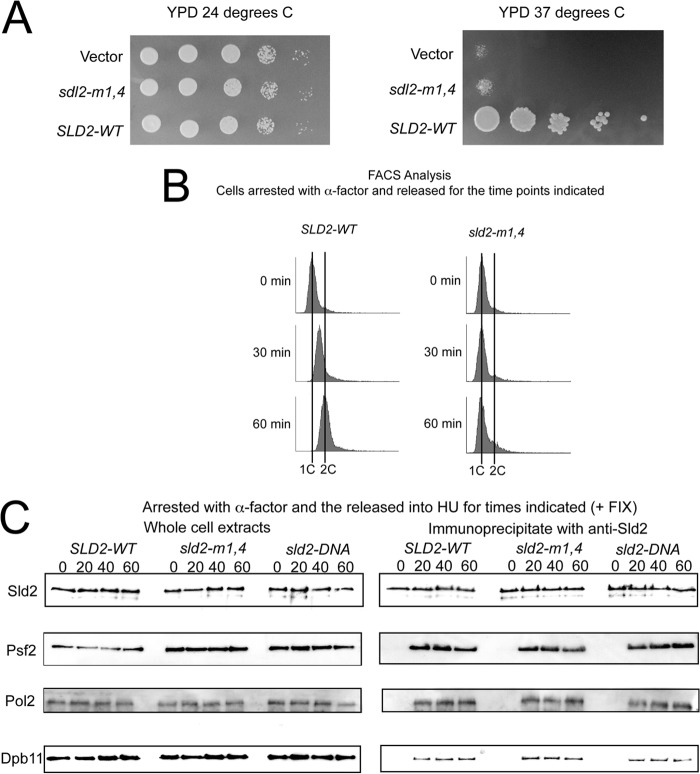
To elucidate the in vivo function of Sld2-Mcm2-7 interaction, we constructed wild-type and mutant SLD2 plasmids with expression driven by the endogenous promoter. We expressed wild-type-SLD2, sld2-m1,4 or vector alone in a yeast strain that bears a temperature-controlled degron for genomic SLD2. We plated the cells in 10-fold serial dilutions onto YPD agar plates at the permissive (24 °C) or restrictive (37 °C) temperature (Fig. 4A). At the permissive temperature, SLD2-WT, sld2-m1,4, and vector alone each grow to similar levels, suggesting that the genomic copy of SLD2 is supporting growth under permissive conditions (Fig. 4A, left panel). However, at the restrictive temperature, there is a severe growth defect in the sld2-m1,4 and vector plasmids compared with SLD2-Wild-type, suggesting that sld2-m1,4 cannot support yeast growth in the absence of wild-type SLD2 (Fig. 4A, right panel).
FIGURE 4.
Expression of sld2-m1,4 severely inhibits cell growth and DNA replication. A, 10-fold serial dilutions of sld2-td cells expressing SLD2, sld2-m1,4 (K416E,R435E,R436E), or vector alone at the permissive (24 °C, left panel) or restrictive temperature (37 °C, right panel). B, fluorescence-activated cell sorting was performed on cells expressing SLD2-wt (left) or sld2-m1,4 (right) at the restrictive temperature (37 °C). Cells were synchronized in G1 with α-factor and then released into medium lacking α-factor for the indicated time points. C, SLD2-Wild-type and mutant cells exhibit similar levels of Pre-LC Assembly. C, sld2-td cells expressing SLD2-wt, sld2-m1,4 (K416E,R435E,R436E), or sld2-DNA (K44E,K50E,K438E) at the restrictive temperature (37 °C) were arrested with α-factor and then released into HU for the indicated times. C, left panel, whole cell extracts were analyzed by Western for expression of Sld2, Psf2 (a component of GINS), Pol2 (a component of Polϵ), or Dpb11. C, right panel, cell extracts were fixed and immunoprecipitated with antibodies directed against Sld2, followed by Western analysis for Sld2, Psf2, Pol2, or Dpb11.
We next determined whether sld2-m1,4 cells were defective in DNA replication. Wild-type-SLD2 and mutant sld2-m1,4 cells were arrested in G1 with α-factor at the restrictive temperature, and then released into growth media at the restrictive temperature. DNA replication was measured by FACS analysis, with propidium iodide stain as a measure of double-stranded DNA content (Fig. 4B). Wild-type SLD2 cells progressed through S phase in 60 min (Fig. 4B, left panel). In contrast, mutant sld2-m1,4 cells exhibited little DNA replication after 60 min (Fig. 4B, right panel). These data show that sld2-m1,4 mutant cells are defective in DNA replication at the restrictive temperature.
SLD2-Wild-type and Mutant Cells Exhibit Similar Levels of Pre-LC Assembly
We next performed co-immunoprecipitation analysis to determine the assembly of proteins required for replication initiation in wild-type-SLD2 and mutant sld2-m1,4 at the restrictive temperature (Fig. 4C). We also analyzed sld2-DNA (sld2-K44E,K50E,K438E) cells in the same assay (Fig. 4C). sld2-DNA cells are defective in cell growth and DNA replication at the restrictive temperature (24), but co-immunoprecipitation studies have not been reported for this mutant. We first probed for components of the pre-loading complex, which is a CDK-dependent assembly of Sld2, Dpb11, GINS, and Polϵ (Fig. 4C) (23). We arrested cells in G1 with α-factor at the restrictive temperature, and then released the cells into HU for 0, 20, 40, or 60 min (Figs. 4C). Cells were fixed with formaldehyde prior to analysis because this treatment is required to observe Pre-LC assembly (23).
We probed for Sld2 alone to determine the protein levels for wild-type compared with mutant (Fig. 4C). Sld2 was expressed in whole cell extracts at roughly equal levels in wild-type and mutant cells (Fig. 4C). Immunoprecipitation of cells with antibodies directed against Sld2 followed by probing for Sld2 by Western analysis revealed similar levels of Sld2 in mutant and wild-type cells (Fig. 4C). These data suggest that the mutations do not alter the expression levels of the Sld2 protein. We next probed whole cell extracts for Psf2 (a component of GINS), Pol2 (a component of Polϵ), or Dpb11 and found similar levels of expression for each of these three proteins in mutant cells compared with wild-type cells (Fig. 4C). We then immunoprecipitated cells with antibody directed against Sld2, and probed for co-immunoprecipitation with Psf2, Pol2, or Dpb11 (Fig. 4C). At 0 min there is no signal for wild-type or mutant cells, consistent with the CDK-dependence of the assembly (23). At 20, 40, and 60 min time points, there is a similar interaction observed for wild-type and mutant cells, suggesting that the pre-LC assembles normally in mutant cells (Fig. 4C). These data suggest that the DNA replication defect of sld2-m1,4 and sld2-DNA is not the result of inhibition of pre-LC assembly.
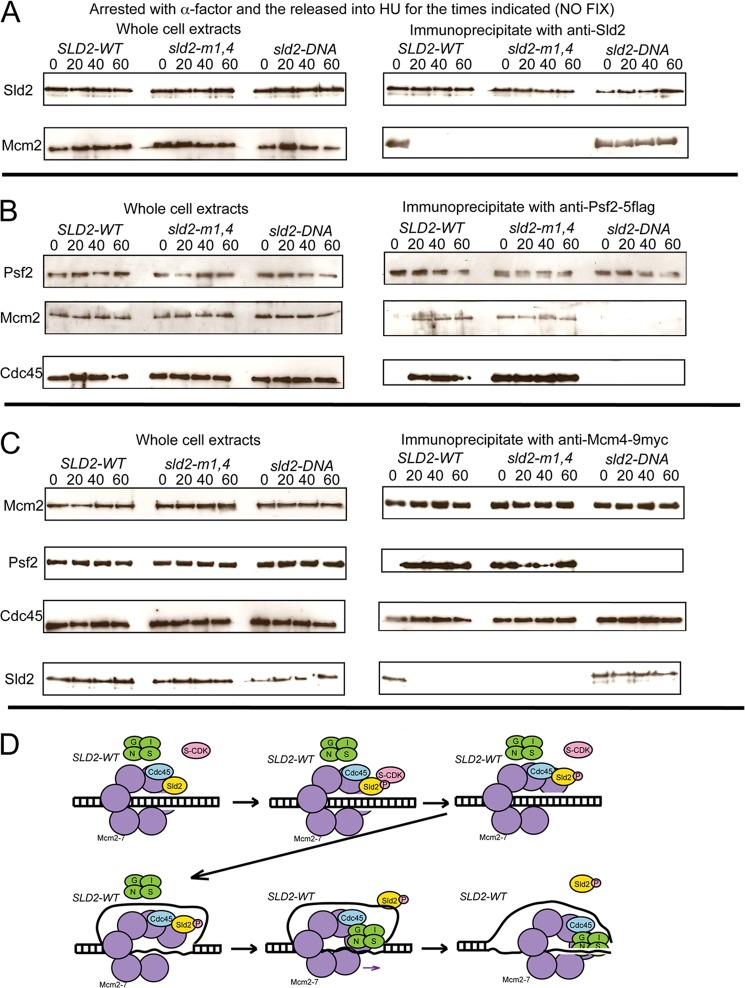
Cells Expressing sld2-m1,4 Exhibit Premature GINS-Mcm2-7 Interaction in G1, and Cells Expressing sld2-DNA Exhibit No GINS-Mcm2-7 Interaction
We next performed a similar experiment to determine if Sld2 is bound to Mcm2-7 in mutant compared with wild-type cells (Fig. 5A). In this experiment, cells were not fixed. Sld2 was found at similar levels in wild-type and mutant cells, and immunoprecipitation of Sld2 followed by probing with Sld2 revealed similar levels of Sld2 protein in mutant compared with wild-type cells (Fig. 5A). When whole cell extracts were probed with Mcm2, similar levels were present in mutant and wild-type cells (Fig. 5A). However, for cell extracts precipitated with antibodies against Sld2 and then probed for Mcm2 at 0 min release from α-factor arrest, there is a substantial decrease in sld2-m1,4 interaction compared with SLD2-wild-type cells or sld2-DNA cells (Fig. 5A). These data suggest that Sld2-Mcm2-7 interaction in G1 is defective in sld2-m1,4 cells. At 20, 40, and 60 min release from α-factor arrest, there is an increased Sld2-Mcm2 interaction observable in sld2-DNA cells compared with SLD2-wild-type cells or sld2-m1,4 cells (Fig. 5A). These data suggest that Sld2-ssDNA interaction is required to release Sld2 from Mcm2-7 after cells are released from G1 arrest.
FIGURE 5.
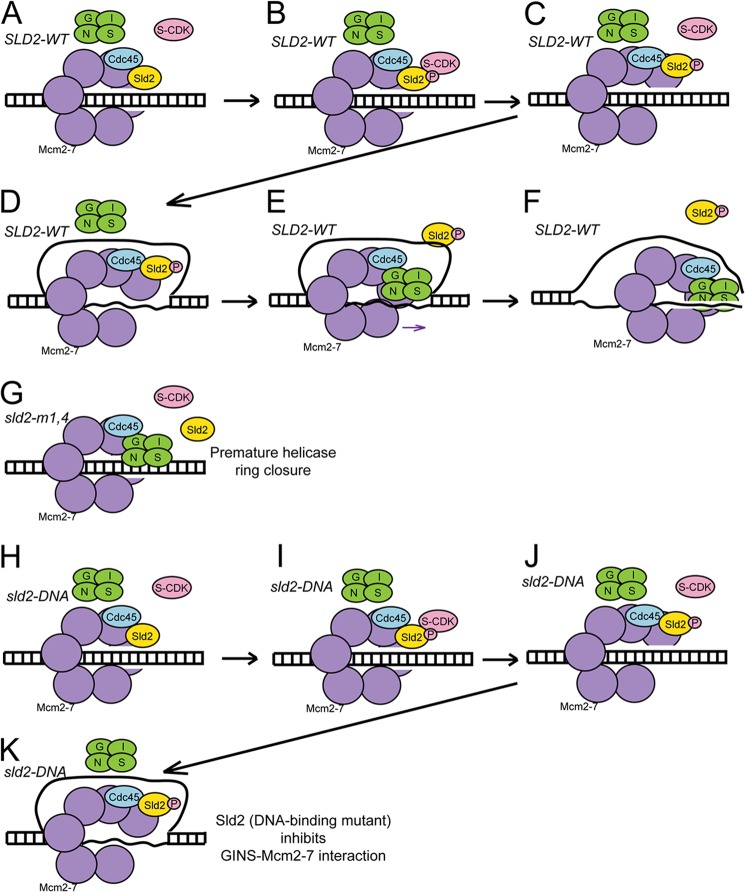
Cells expressing sld2-m1,4 exhibit premature GINS-Mcm2-7 interaction in G1, and cells expressing sld2-DNA exhibit no GINS-Mcm2-7 interaction. A, sld2-td cells expressing SLD2-wt, sld2-m1,4 (K416E,R435E,R436E), or sld2-DNA (K44E,K50E,K438E) at the restrictive temperature (37 °C) were arrested with α-factor and then released into HU for the indicated times. A–C, left panel) Whole cell extracts were analyzed by Western for expression indicated proteins. A, right panel, cell extracts were immunoprecipitated with antibodies directed against Sld2, followed by Western analysis for Sld2 or Mcm2. B, right panel, cell extracts were immunoprecipitated with antibodies directed against Psf2–5flag (a component of GINS), followed by Western analysis for Psf2, Mcm2, or Cdc45. C, right panel, cell extracts were prepared to isolate loaded Mcm2-7 complexes as described (29), using antibodies directed against Mcm4–9myc, followed by Western analysis for Mcm2, Psf2, Cdc45, or Sld2. D, model for assembly of CMG complex in wild-type yeast cells.
To assess the interaction between GINS and Mcm2-7, immunoprecipitation was performed using antibodies directed against Psf2–5flag, followed by probing for Psf2 or Mcm2 (Fig. 5B). Cells were not fixed in this assay. Psf2 was found at similar levels in whole cell extracts in mutant and wild-type cells, and immunoprecipitation with antibodies directed against Psf2–5flag followed by probing with Psf2 revealed similar levels of Psf2 in wild-type and mutant cells (Fig. 5B). Levels of Mcm2 were also similar in whole cell extracts in wild-type and mutant cells (Fig. 5B). However, when cells were immunoprecipitated with antibodies directed against Psf2–5flag followed by probing for Mcm2, differences were observed between mutant and wild-type cells (Fig. 5B). Wild-type-SLD2 cells show no Mcm2 signal at 0 min, but a visible Mcm2 signal at 20, 40, and 60-min time points. These data are consistent with published literature, since GINS associates with Mcm2-7 in S phase, but not G1 phase (30, 31). However, the sld2-m1,4 mutant exhibits a visible signal for Mcm2 at time 0, suggesting that GINS is associating with Mcm2-7 in G1 in sld2-m1,4 cells. In contrast, sld2-DNA cells exhibit no interaction between GINS and Mcm2-7 at any time point, suggesting that GINS does not associate with Mcm2-7 in sld2-DNA cells.
We next examined the interaction between GINS and Cdc45 with the co-immunoprecipitation assay (Fig. 5B). Whole cell extracts exhibited similar levels of Cdc45 in mutant compared with wild-type cells. Furthermore, immunoprecipitation with antibodies directed against anti-Psf2–5flag followed by probing with Cdc45 revealed enhanced interaction at 0 min in sld2-m1,4 cells compared with wild-type cells (Fig. 5B). These data suggest that the CMG complex is assembling prematurely in G1 phase in Sld2-m1,4 cells. At 20, 40, and 60-min time points, there is diminished interaction between Cdc45 and Psf2 in sld2-DNA cells. These data suggest that the CMG complex does not properly assemble in sld2-DNA cells.
A recent report demonstrates a method for isolating Mcm2-7 complexes that are loaded on chromosomal DNA from budding yeast cells (29). We used this approach to determine interactions between loaded Mcm2-7 and GINS, Cdc45, or Sld2 (Fig. 5C). Levels of Mcm2, Psf2, Cdc45, or Sld2 are equivalent in wild-type and mutant cells (Fig. 5C). Furthermore, the interaction between loaded Mcm4–9myc and Mcm2 is similar in wild-type compared with mutant cells (Fig. 5C). However, the interaction between loaded Mcm4–9myc and Psf2 at 0 min release from α-factor is enhanced in sld2-m1,4 cells compared with SLD2-wild-type cells (5C). These data suggest that GINS prematurely interacts with loaded Mcm2-7 in sld2-m1,4 cells in G1. There is diminished interaction between loaded Mcm4–9myc and Psf2 at 20, 40, and 60 min in sld2-DNA cells compared with wild-type cells (Fig. 5C). These data suggest that Sld2-ssDNA interaction is important for GINS to interact with loaded Mcm2-7 after cells are released from G1 arrest. The interaction between loaded Mcm4-9myc and Cdc45 is similar in wild-type and mutant cells (Fig. 5C), suggesting that Sld2 interaction with Mcm2-7 or ssDNA does not regulate Cdc45-Mcm2-7 interaction. The interaction between Sld2 and loaded Mcm4-9myc is diminished in sld2-m1,4 cells at 0 min compared with SLD2-wild-type cells (Fig. 5C), suggesting that interaction between Sld2 and loaded Mcm2-7 is defective in sld2-m1,4 cells. The interaction between Sld2 and loaded Mcm4–9myc is increased in sld2-DNA cells at 20, 40, and 60 min compared with SLD2-wild-type cells (Fig. 5C), suggesting that interaction between Sld2 and ssDNA is important to release Sld2 from loaded Mcm2-7. A model for assembly of the CMG complex in wild-type yeast cells is shown in Fig. 5D.
RPA Is Not Observed at Early Origins in sld2-m1,4 Cells Exposed to Hydroxyurea
The Mcm2-7 complex converts from encircling double-stranded DNA to single-stranded DNA during S phase, resulting in the extrusion of single-stranded DNA to the exterior of the Mcm2-7 ring (12, 17). When cells are arrested in G1 and then released into hydroxyurea, the extrusion of single-stranded DNA at an early origin can be revealed by chromatin immunoprecipitation with antibodies directed against RPA (RPA-ChIP) (29). Wild-type SLD2 cells exhibit an increase in RPA-CHIP signal at the early replication origins ARS305 and ARS306 as cells are released into hydroxyurea (Fig. 6), while no increase in signal is observed at a site between these origins, consistent with the published literature (29). However, sld2-m1,4 mutants exhibit little increase in RPA-CHIP signal at ARS305 and ARS306 as cells are released into hydroxyurea, suggesting that single-stranded DNA is not extruded from the central channel of Mcm2-7 in sld2-m1,4 mutant cells (Fig. 6). Intermediate results are seen in sld2-DNA mutants, suggesting that ssDNA is extruded from the central channel of Mcm2-7 in these sld2-DNA mutant cells, but origin firing is not initiated (Fig. 6).
FIGURE 6.
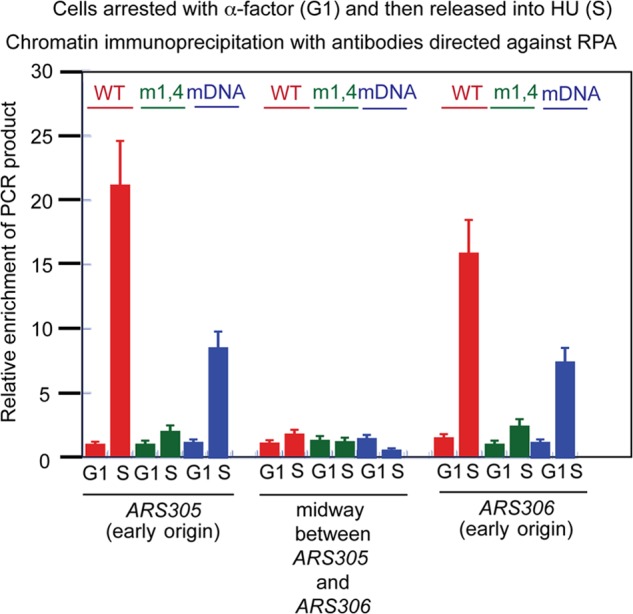
RPA is not observed at early origins in sld2-m1,4 cells exposed to hydroxyurea. sld2-td cells expressing SLD2-wt, sld2-m1,4 (K416E,R435E,R436E), or sld2-DNA (K44E,K50E,K438E) at the restrictive temperature (37 °C) were arrested with α-factor and then released into hydroxyurea for 30 min. Cells extracts were subjected to chromatin immunoprecipitation with antibodies directed against the RPA (yeast single-stranded binding protein). PCR primers were used that target the early yeast replication origins ARS305 or ARS306, or a region positioned midway between ARS305 and ARS306 (not an origin). [32P-α]dCTP was included in the PCR reaction for quantitation. Radioactive PCR bands were quantified and plotted.
DISCUSSION
Sld2 Regulates CMG Helicase Assembly
We identified a mutant of Sld2, Sld2-K416E,R435E,R436E (Sld2-m1,4), that is defective in binding Mcm2-7 in vitro (Fig. 1). Sld2-R436E is conserved from yeast to human (21). Sld2-m1,4 binds to Dpb11, ssDNA, and pol ϵ like wild-type Sld2 (Fig. 2), and Sld2-m1,4 does not block GINS interaction with Mcm2-7 in vitro (Fig. 3). Cells expressing sld2-m1,4 exhibit a severe defect in growth and DNA replication (Fig. 4). Co-immunoprecipitation analysis of wild-type SLD2 cells exhibit GINS-Mcm2-7 interaction in S phase but not G1 phase (Fig. 5). In contrast, sld2-m1,4 mutant cells exhibit premature GINS-Mcm2-7 interaction in G1 phase (Fig. 5). These data suggest that Sld2-Mcm2--7 interaction is important to prevent the premature interaction of GINS with Mcm2-7 in G1. We previously identified a mutant of Sld2, Sld2-DNA, which is specifically defective in Sld2-ssDNA interaction (24). Expression of the sld2-DNA mutant results in little GINS-Mcm2-7 interaction as cells are released from α-factor arrest (Fig. 5). These data suggest that Sld2-ssDNA interaction is important to allow GINS to bind Mcm2-7 in S phase. Furthermore, RPA is observed at high levels at early origins in wild-type SLD2 cells exposed to hydroxyurea, while RPA is at low levels under the same conditions in sld2-m1,4 cells (Fig. 6). These data suggest that single-stranded DNA is extruded from the central channel of Mcm2-7 in SLD2, but not in sld2-m1,4 cells. The premature association of GINS with Mcm2-7 in G1 in sld2-m1,4 cells may create a sealed helicase ring in G1, preventing the extrusion of ssDNA from the central channel of Mcm2-7 in these cells.
A Model for Sld2 Action at a Replication Origin in Budding Yeast Cells
We propose a model for Sld2 action at a replication origin based upon the data (Fig. 7). For wild-type SLD2 cells (Fig. 7, A–F), Sld2 binds directly to Mcm2-7 in G1 (Fig. 7A). Sld2, while bound to Mcm2-7, blocks the interaction between GINS and Mcm2-7 (Fig. 7A). During S phase, S-CDK phosphorylates Sld2 (Fig. 7, B and C), and a single-strand of DNA is extruded from the central channel of Mcm2-7 (Fig. 7D). CDK-phosphorylated Sld2 binds to ssDNA and dissociates from Mcm2-7 (Fig. 7E). Once Sld2 is bound to ssDNA, GINS can bind to Mcm2-7 (Fig. 7E). The CMG complex is now assembled, and it can unwind DNA to initiate DNA replication (Fig. 7F).
FIGURE 7.
Sld2 regulates CMG helicase assembly. A–F, model for cells expressing SLD2-wt. A, in G1, Sld2 binds directly to Mcm2-7 and Sld2 blocks the interaction between GINS and Mcm2-7. B–D, in S phase, S-CDK phosphorylates Sld2, and a single-strand of DNA is extruded from the central channel of Mcm2-7. E, CDK-phosphorylated-Sld2 dissociates from Mcm2-7, and CDK-phosphorylated-Sld2 binds to single-stranded DNA. GINS now binds directly to Mcm2-7. F, assembled CMG complex unwinds duplex DNA. G, model for cells expressing sld2-m1,4. Sld2-m1,4 does not bind to Mcm2-7 in G1, and GINS binds directly to Mcm2-7 in G1. The formation of a CMG complex in G1 in these mutant cells generates a sealed helicase ring that does not extrude single-stranded DNA from the central channel of Mcm2-7. H–K, model for cells expressing sld2-DNA. H, in G1, Sld2 binds directly to Mcm2-7, and Sld2 blocks the interaction between GINS and Mcm2-7. I–K, in S phase, S-CDK phosphorylates Sld2, and a single-strand of DNA is extruded from the central channel of Mcm2-7. However, since Sld2-DNA is defective in binding single-stranded DNA, Sld2-DNA remains bound to Mcm2-7, and GINS does not bind to Mcm2-7.
We also propose models to account for the mutant data. For sld2-m1,4 cells, Sld2 does not bind to Mcm2-7 in G1 (Fig. 7G), and GINS binds prematurely to Mcm2-7 in G1. The prematurely sealed CMG complex that is formed in G1 cannot open to allow extrusion of single-stranded DNA. Therefore, the prematurely closed CMG helicase ring cannot function to unwind DNA because it encircles two DNA strands (Fig. 7G). For sld2-DNA cells (Fig. 7, H–K), Sld2-DNA binds to Mcm2-7 in G1 as in wild-type cells (Fig. 7H). A single-strand of DNA is extruded from the central channel of Mcm2-7 in S phase (Fig. 7K), but CDK-phosphorylated Sld2-DNA cannot bind to DNA, and the Sld2-DNA remains bound to Mcm2-7 (Fig. 7K). The prolonged interaction between Sld2-DNA with Mcm2-7 in S phase blocks the interaction between GINS and Mcm2-7 in S phase, and the CMG complex does not form (Fig. 7K).
A Key Role for Sld2 Is to Regulate the Interaction between GINS and Mcm2-7
We show here that Sld2 plays a complex role in regulating the recruitment of GINS to the Mcm2-7 complex. During G1, Sld2 blocks the interaction between GINS and Mcm2-7. This inhibition is important to prevent in G1 the premature assembly of the CMG assembly, a sealed helicase ring that does not allow extrusion of ssDNA from the central channel (12). During S phase, the single-strand of DNA extruded from the central channel of Mcm2-7 dissociates CDK-phosphorylated Sld2 from Mcm2-7, allowing GINS to bind Mcm2-7. Thus, the role of Sld2 is to regulate the interaction between GINS and Mcm2-7. Sld2 ensures that GINS can only bind to Mcm2-7 when CDK phosphorylates Sld2, and single-stranded DNA is extruded from the central channel of Mcm2-7. Thus, Sld2 ensures that the helicase ring is sealed at the appropriate time.
An Additional Role for CDK in the Cell May be Revealed
The phosphomimetic mutant of Sld2, Sld2T84D, mimics the CDK-phosphorylated state of Sld2 (22). This mutant, when combined with a fusion of Sld3 and Dpb11, bypasses the requirement for CDK in the cell (19). Sld2T84D binds to Dpb11 (18, 19), and Sld2T84D also binds to single-stranded DNA (25). Thus, the Sld2T84D mutation may bypass the cellular requirement for CDK activity for two different reasons: (1) Sld2T84D binds to Dpb11, thereby recruiting GINS to replication origins, and (2) Sld2T84D binds to single-stranded DNA, thereby allowing GINS to bind directly to Mcm2-7. Thus, an additional role of CDK in the cell may be revealed in this study.
Will This Mechanism Apply to Human Cells?
Sld2 interaction with single-stranded DNA involves three lysines, K44, K50, and K438 (24). The N-terminal lysines, K44 and K50, are conserved in human RecQL4 as an arginine and a lysine, respectively (32). It was recently found that the N terminus of human RecQL4 binds to single-stranded DNA (32). Furthermore, the homologous residues of Sld2-K44 and Sld2-K50 of human RecQL4 are involved in the interaction with DNA (32). The alignment of C-terminal Sld2 with human RecQL4 was accomplished recently (21). This alignment reveals conservation of the K438 involved in Sld2-DNA binding, although the residue is an asparagine in human cells (21). This alignment also reveals now that one of the residues involved in Mcm2-7 binding, R438, is also conserved in human cells (21). It also has been reported that human RecQL4 may associate with Mcm2-7 in vivo (33, 34). Based upon this data, we propose that the model shown in Fig. 7 may apply to human cells, with RecQL4 substituting for Sld2, but clearly this is speculative until definitive experiments in human cells can be accomplished. RecQL4 likely participates in additional functions not related to Sld2, since RecQL4, but not Sld2, is a helicase.
A Key Direction for the Future Is to Reveal How Single-stranded DNA Is Extruded from the Central Channel of Mcm2-7 in S Phase
This study has revealed how the interaction between GINS and Mcm2-7 is regulated in the cell. In the future, it will be interesting to determine what regulates the extrusion of single-strand of DNA from the central channel of Mcm2-7. Mcm10 may play a role, as may Sld2, Sld3, and possibly other proteins (26, 28, 29, 35). This activity will likely be complex and highly regulated. One key feature of this process is that it involves opening of the Mcm2-7 ring. While it is clear that the Mcm2-7 complex is capable of spontaneously cracking at the Mcm2-Mcm5 subunit interface (12, 36), we speculate that one or more replication initiation proteins may favor the opening or closing of the Mcm2-7 ring to extrude a single-strand of DNA from the central channel. Thus, while critical aspects of helicase activation in eukaryotic cells have been revealed, additional discoveries in this field are likely to be forthcoming.
Acknowledgments
We thank Susan Taylor for supplying purified PKA. We thank Mike O'Donnell for providing expression constructs for GINS.
This work was supported by National Science Foundation Grant 1265431 (to D. L. K.).
- CMG
- Cdc45-Mcm2-7-GINS
- CDK
- cyclin-dependent kinase
- FACS
- fluorescence activated cell sorting
- PKA
- protein kinase A.
REFERENCES
- 1. Diffley J. F. X. (2011) Quality control in the initiation of eukaryotic DNA replication. Phil. Trans. Royal Society B. 366, 3545–3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cimprich K. A., Cortez D. (2008) ATR: an essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 9, 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Byun T., Pacek M., Yee M., Walter J., Cimprich K. (2005) Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 19, 1040–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yardimci H., Loveland A., Habuchi S., van Oijen A., Walter J. (2010) Uncoupling of Sister Replisomes during Eukaryotic DNA Replication. Mol. Cell 40, 834–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gambus A., Jones R. C., Sanchez-Diaz A., Kanemaki M., van Deursen F., Edmondson R. D., Labib K. (2006) GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nature Cell Biology 8, 358–366 [DOI] [PubMed] [Google Scholar]
- 6. Ilves I., Petojevic T., Pesavento J., Botchan M. (2010) Activation of the MCM2–7 Helicase by Association with Cdc45 and GINS Proteins. Mol. Cell 37, 247–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pacek M., Tutter A., Kubota Y., Takisawa H., Walter J. (2006) Localization of MCM2–7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol. Cell 21, 581–587 [DOI] [PubMed] [Google Scholar]
- 8. Evrin C., Clarke P., Zech J., Lurz R., Sun J., Uhle S., Li H., Stillman B., Speck C. (2009) A double-hexameric MCM2–7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc. Natl. Acad. Sci. U.S.A. 106, 20240–20245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Remus D., Beuron F., Tolun G., Griffith J., Morris E., Diffley J. (2009) Concerted loading of Mcm2–7 double hexamers around DNA during DNA replication origin licensing. Cell 139, 719–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Frigola J., Remus D., Mehanna A., Diffley J. (2013) ATPase-dependent quality control of DNA replication origin licensing. Nature 495, 339–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fernández-Cid A., Riera A., Tognetti S., Herrera M., Samel S., Evrin C., Winkler C., Gardenal E., Uhle S., Speck C. (2013) An ORC/Cdc6/MCM2–7 Complex Is Formed in a Multistep Reaction to Serve as a Platform for MCM Double-Hexamer Assembly. Mol. Cell 50, 577–588 [DOI] [PubMed] [Google Scholar]
- 12. Costa A., llves I., Tamberg N., Petojevic T., Nogales E., Botchan M., Berger J. (2011) The structural basis for MCM2–7 helicase activation by GINS and Cdc45. Nat. Struct. Mol. Biol. 18, 471–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bochman M., Schwacha A. (2010) The Saccharomyces cerevisiae Mcm6/2 and Mcm5/3 ATPase active sites contribute to the function of the putative Mcm2–7 'gate'. Nucleic Acids Res. 38, 6078–6088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sheu Y.-J., Stillman B. (2006) Cdc7-Dbf4 phosphorylates MCM proteins via a docing site-mediated mechanism to promote S phase progression. Mol. Cell 24, 101–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Masai H., Taniyama C., Ogino K., Matsui E., Kakusho N., Matsumoto S., Kim J., Ishii A., Tanaka T., Kobayashi T., Tamai K., Ohtani K., Arai K. (2006) Phosphorylation of MCM4 by Cdc7 kinase facilitates its interaction with Cdc45 on the chromatin. J. Biol. Chem. 281, 39249–39261 [DOI] [PubMed] [Google Scholar]
- 16. Heller R., Kang S., Lam W., Chen S., Chan C., Bell S. (2011) Eukaryotic origin-dependent DNA replication in vitro reveals sequential action of DDK and S-CDK kinases. Cell 146, 80–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fu Y., Yardimci H., Long D., Ho T., Guainazzi A., Bermudez V., Hurwitz J., van Oijen A., Schärer O., Walter J. (2011) Selective bypass of a lagging strand roadblock by the eukaryotic replicative DNA helicase. Cell 146, 931–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tanaka S., Umemori T., Hirai K., Muramatsu S., Kamimura Y., Araki H. (2007) CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature 445, 328–332 [DOI] [PubMed] [Google Scholar]
- 19. Zegerman P., Diffley J. F. (2007) Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature 445, 281–285 [DOI] [PubMed] [Google Scholar]
- 20. Kamimura Y., Masumoto H., Sugino A., Araki H. (1998) Sld2, which interacts with Dpb11 in Saccharomyces cerevisiae, is required for chromosomal DNA replication. Mol. Cell Biol. 18, 6102–6109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Marino F., Vindigni A., Onesti S. (2013) Bioinformatic analysis of RecQ4 helicases reveals the presence of a RQC domain and a Zn knuckle. Biophys. Chem. 177, 34–39 [DOI] [PubMed] [Google Scholar]
- 22. Tak Y., Tanaka Y., Endo S., Kamimura Y., Araki H. (2006) A CDK-catalysed regulatory phosphorylation for formation of the DNA replication complex Sld2-Dpb11. EMBO J. 25, 1987–1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Muramatsu S., Hirai K., Tak Y., Kamimura Y., Araki H. (2010) CDK-dependent complex formation between replication proteins Dpb11, Sld2, Pol (epsilon}, and GINS in budding yeast. Genes Dev. 24, 602–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bruck I., Kanter D. M., Kaplan D. L. (2011) Enabling association of the GINS tetramer with the Mcm2–7 complex by phosphorylated Sld2 protein and single-stranded origin DNA. J. Biol. Chem. 286, 36414–36426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kanter D., Kaplan D. (2011) Sld2 binds to origin single-stranded DNA and stimulates DNA annealing. Nucleic Acids Res. 39, 2580–2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bruck I., Kaplan D. (2011) GINS and Sld3 compete with one another for Mcm2–7 and Cdc45 binding. J. Biol. Chem. 286, 14157–14167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bruck I., Kaplan D. (2013) Cdc45 protein-single-stranded DNA interaction is important for stalling the helicase during replication stress. J. Biol. Chem. 288, 7550–7563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bruck I., Kaplan D. (2011) Origin Single-stranded DNA Releases Sld3 Protein from the Mcm2–7 Complex, Allowing the GINS Tetramer to Bind the Mcm2–7 Complex. J. Biol. Chem. 286, 18602–18613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. van Deursen F., Sengupta S., De Piccoli G., Sanchez-Diaz A., Labib K. (2012) Mcm10 associates with the loaded DNA helicase at replication origins and defines a novel step in its activation. EMBO J. 31, 2195–2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kanemaki M., Labib K. (2006) Distinct roles for Sld3 and GINS during establishment and progression of eukaryotic DNA replication forks. EMBO J. 25, 1753–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Araki H. (2010) Cyclin-dependent kinase-dependent initiation of chromosomal DNA replication. Curr. Opin. Cell Biol. 22, 766–771 [DOI] [PubMed] [Google Scholar]
- 32. Ohlenschläger O., Kuhnert A., Schneider A., Haumann S., Bellstedt P., Keller H., Saluz H.-P., Hortschansky P., Hänel F., Grosse F., Görlach M., Pospiech H. (2012) The N-terminus of the human RecQL4 helicase is a homeodomain-like DNA interaction motif. Nucleic Acids Res. 40, 8309–8324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Im J.-S., Ki S.-H., Farina A., Jung D.-S., Hurwitz J., Leea J.-K. (2009) Assembly of the Cdc45-Mcm2–7-GINS complex in human cells requires the Ctf4/And-1, RecQL4, and Mcm10 proteins. Proc. Natl. Acad. Sci. U.S.A. 106, 15628–15632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xu X., Rochette P. J., Feyissa E. A., Su T. V., Liu Y. (2009) MCM10 mediates RECQ4 association with MCM2–7 helicase complex during DNA replication. EMBO J. 28, 3005–3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kanke M., Kodama Y., Takahashi T., Nakagawa T., Masukata H. (2012) Mcm10 plays an essential role in origin DNA unwinding after loading of the CMG components. EMBO J. 31, 2182–2194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bochman M., Schwacha A. (2008) The Mcm2–7 complex has in vitro helicase activity. Mol. Cell 31, 287–293 [DOI] [PubMed] [Google Scholar]