Background: The biochemistry of noscapine metabolism in opium poppy has not been established.
Results: CYP82Y1 catalyzes the efficient 1-hydroxylation of N-methylcanadine. Silencing CYP82Y1 reduced noscapine accumulation in opium poppy plants.
Conclusion: CYP82Y1 is N-methylcanadine 1-hydroxylase and functions in noscapine biosynthesis.
Significance: The enzyme catalyzing the first step in the conversion of N-methylcanadine to noscapine has been identified and characterized.
Keywords: Biosynthesis, Cytochrome P-450, Enzyme Catalysis, Gene Silencing, Metabolism, Plant Biochemistry, Secondary Metabolism
Abstract
Noscapine is a phthalideisoquinoline alkaloid investigated for its potent pharmacological properties. Although structurally elucidated more than a century ago, the biosynthesis of noscapine has not been established. Radiotracer studies have shown that noscapine is derived from the protoberberine alkaloid (S)-scoulerine and has been proposed to proceed through (S)-N-methylcanadine. However, pathway intermediates involved in the conversion of N-methylcanadine to noscapine have not been identified. We report the isolation and characterization of the cytochrome P-450 CYP82Y1, which catalyzes the 1-hydroxylation of N-methylcanadine to 1-hydroxy-N-methylcanadine. Comparison of transcript and metabolite profiles of eight opium poppy chemotypes revealed four cytochrome P-450s, three from the CYP82 and one from the CYP719 families, that were tightly correlated with noscapine accumulation. Recombinant CYP82Y1 was the only enzyme that accepted (R,S)-N-methylcanadine as a substrate with strict specificity and high affinity. As expected, CYP82Y1 was abundantly expressed in opium poppy stems where noscapine accumulation is highest among plant organs. Suppression of CYP82Y1 using virus-induced gene silencing caused a significant reduction in the levels of noscapine, narcotoline, and a putative downstream secoberbine intermediate and also resulted in increased accumulation of the upstream pathway intermediates scoulerine, tetrahydrocolum-bamine, canadine, and N-methylcanadine. The combined biochemical and physiological data support the 1-hydroxylation of (S)-N-methylcanadine catalyzed by CYP82Y1 as the first committed step in the formation of noscapine in opium poppy.
Introduction
Noscapine, also known as narcotine, anarcotine, or gnoscopine, is a benzylisoquinoline alkaloid (BIA)3 belonging to the phthalideisoquinoline alkaloid subgroup and displaying potent pharmacological properties. Noscapine was first detected in 1803 by the French industrial chemist Charles Derosne and referred to as “sel narcotique de Derosne” (Derosne's narcotic salt) (1). Although more than two centuries have passed since the discovery of noscapine, the chemistry and molecular biochemistry of phthalideisoquinoline alkaloid biosynthesis have not been established. Extensive radiotracer experiments in the 1960s showed that the structural scaffold of noscapine shares the same biosynthetic origin as other BIAs. Labeled noscapine was isolated when opium poppy (Papaver somniferum) plants were fed radioactive tyrosine, norlaudanosoline, or laudanosoline (2, 3), suggesting that the noscapine carbon skeleton, apart from the lactonic carbonyl group, is formed by the condensation of two tyrosine derivatives. Later studies with labeled laudanosoline and reticuline showed that the lactone carbonyl carbon of phthalideisoquinoline alkaloids originates from the N-methyl moiety of the same pathway intermediate yielding the C8 position of protoberberine and protopine alkaloids. The biosynthetic relationship among these alkaloid subgroups was further demonstrated by the efficient incorporation of labeled scoulerine and canadine into noscapine (2, 4). Recently, the role of scoulerine and canadine as intermediates in noscapine biosynthesis has been corroborated by the isolation and characterization of the genes encoding SOMT1 (scoulerine 9-O-methyltransferase) and the cytochrome P-450 canadine synthase (CYP719A21) from opium poppy (see Fig. 1) (5, 6). A concomitant decrease in the levels of noscapine, tetrahydrocolumbamine, and canadine in plants with suppressed SOMT and CYP719A21 transcript abundance was demonstrated (5, 6). The detection of several naturally occurring phthalideisoquinoline alkaloids such as macrantaldehyde, papaveroxynoline, papaveroxine, and narcotinehemiacetal in certain Papaver species (7) suggested a biosynthetic process involving the N-methylation of a protoberberine alkaloid, most likely canadine, yielding N-methylcanadine (8), which is hydroxylated at C1 and subsequently methylated to form 1-methoxy-N-methylcanadine (9). Hypothetical C8 oxidation of 1-methoxy-N-methylcanadine yields macrantaldehyde. Hydroxylation of macrantaldehyde at C13 would produce narcotinehemiacetal, which could be dehydrogenated to noscapine.
FIGURE 1.
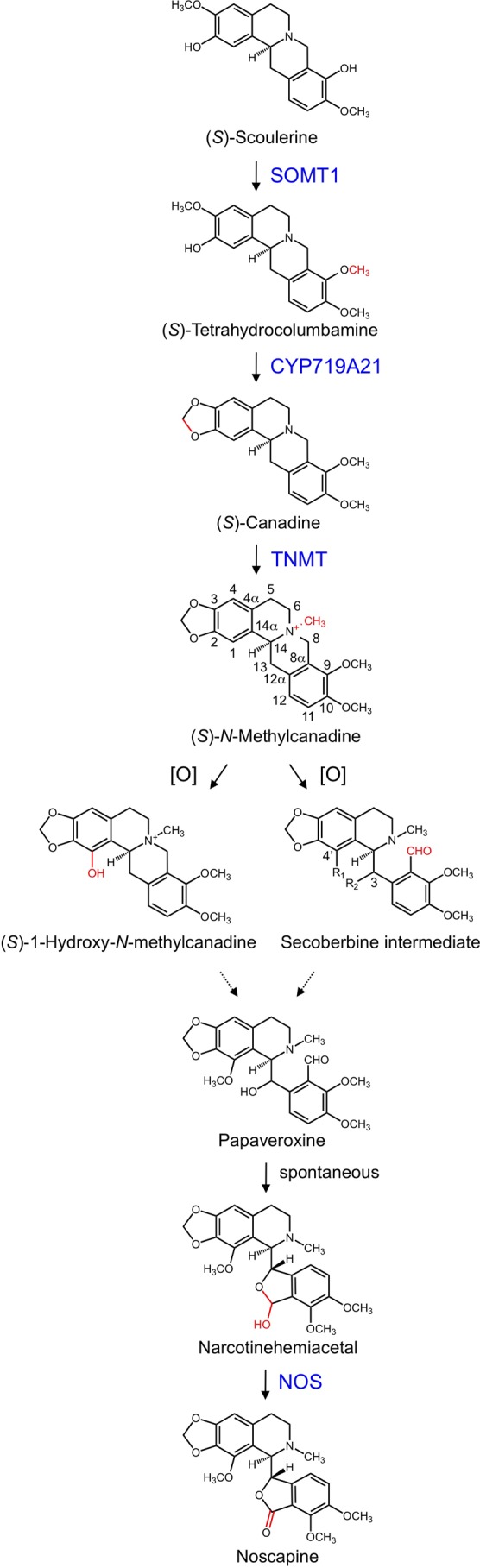
Proposed noscapine biosynthetic pathway. Enzymes shown in blue have been functionally characterized. Dashed arrows represent several uncharacterized reactions. R1 and R2 = H or OH. Partial carbon numbering is shown for N-methylcanadine and the putative secoberbine intermediate. [O], uncharacterized oxidative conversions.
The recent discovery of a 10-gene cluster in the opium poppy genome putatively encoding all noscapine biosynthetic enzymes, except TNMT (tetrahydroprotoberberine N-methyltransferase), operating downstream of (S)-scoulerine prompted a re-evaluation of the pathway model (10). Accordingly, N-methylcanadine was proposed to undergo ring opening via C8 oxidation catalyzed by a cytochrome P-450 (CYP) yielding a 3-hydroxysecoberbine intermediate potentially prior to C4′ hydroxylation, corresponding to C1 on N-methylcanadine (see Fig. 1). Subsequent hydroxylation followed by O-methylation at C4′ and O-acetylation of the 3-hydroxyl group on the secoberbine intermediate was suggested to yield 3-O-acetylpapaveroxine, which would be O-deacetylated by a carboxylesterase to form narcotinehemiacetal. The hemiacetal moiety of narcotinehemiacetal ultimately yields the lactone ring in noscapine. The final step was validated by the recent isolation and characterization of noscapine synthase (NOS), a short chain dehydrogenase/reductase that converts narcotinehemiacetal to noscapine in opium poppy (see Fig. 1) (11). O-Methylation of a secoberbine intermediate at the position equivalent to the C4′ hydroxyl group of noscapine was attributed to one of three O-methyltransferases in the gene cluster. Both proposed schemes are solely supported by either the discovery of potential pathway intermediates in various Papaver species (7, 9) or the accumulation of putative intermediates associated with the VIGS-mediated suppression of specific gene transcripts (10). The recent characterization of CYP719A21 as canadine synthase supports the subsequent oxidation of canadine or N-methylcanadine as the first committed step in noscapine biosynthesis (see Fig. 1) (6). Determination of the oxidative substrate as either canadine or N-methylcanadine and characterization of the enzyme responsible are central to establishing the sequence of reactions in noscapine metabolism. The reaction must involve either (i) breakage of the N7/C8 bridge or (ii) 1-hydroxylation of the isoquinoline moiety on canadine or N-methylcanadine. Either oxidative reaction could be catalyzed by a CYP.
Members of the CYP superfamily catalyze several key conversions in BIA metabolism. Three CYP families, CYP80, CYP82, and CYP719, have been implicated in BIA biosynthesis (12). Enzymes in the CYP80 family catalyze the aromatic ring 3′-hydroxylation of N-methylcoclaurine (13), C-O coupling between 1-benzylisoquinoline monomers to form the bisbenzylisoquinoline alkaloid berbamunine (14), and intramolecular C-C phenol coupling of (S)-reticuline yielding the aporphine alkaloid (S)-corytuberine (15). Members of the CYP719 family are involved in the formation of methylenedioxy bridges on protoberberine alkaloids (16–18), such as tetrahydrocolumbamine to yield canadine (Fig. 1) (6, 17), and the C-C phenol coupling of (R)-reticuline to form the promorphinan alkaloid salutaridine (19). Recently, enzymes of the CYP82 family have been shown to catalyze regiospecific aliphatic hydroxylations of (i) quaternary protoberberine alkaloids including N-methylcanadine, which causes tautomerization to the protopine backbone (20), and (ii) protopine alkaloids resulting in spontaneous intramolecular rearrangement to the benzo[c]phenanthridine scaffold (21). Although there are three CYP82s and one CYP719 in the proposed noscapine biosynthetic gene cluster (10), only CYP719A21 has been functionally characterized as canadine synthase (6). In this paper, we report the isolation and characterization of CYP82Y1 in opium poppy as N-methylcanadine 1-hydroxylase. The gene was isolated based on the strict association of CYP82Y1 gene transcripts with opium poppy chemotypes accumulating noscapine, compared with noscapine-free chemotypes. CYP82Y1 catalyzed the regiospecific 1-hydroxylation of the quaternary protoberberine alkaloids (R,S)-N-methylcanadine and (S)-N-methylstylopine. The physiological role of CYP82Y1 in opium poppy was partially supported using VIGS. Formation of 1-hydroxy-N-methylcanadine is the gateway to noscapine biosynthesis in opium poppy.
EXPERIMENTAL PROCEDURES
Plant Material
The opium poppy chemotypes Bea's Choice and Veronica (22) were cultivated at 20/18 °C (light/dark) in a growth chamber (Conviron, Winnipeg, Canada) with a photoperiod of 16 h and a combination of Cool White fluorescent (Sylvania, Mississauga, Canada) and incandescent lighting (5). Plant samples were collected as described previously (6).
Chemicals
(R,S)-Canadine was purchased from Latoxan (Valence, France). (R,S)-N-Methylcanadine was produced from (R,S)-canadine using purified recombinant Eschscholzia californica TNMT (8). Enzymatic conversion was performed for 4 h with 100 μm (R,S)-canadine, 200 μm S-adenosyl-l-methionine, and 75 μg of purified TNMT. Reactions were terminated by the addition of an equal volume of methanol, and precipitated protein was removed by centrifugation. The supernatant was concentrated under reduced pressure, resuspended in 0.1 n NaOH, and loaded on a StrataTM-X-CW 33-μm polymeric weak cation, 30 mg ml−1 column (Phenomenex, Torrance, CA). (S)-N-Methylcanadine was selectively eluted from the column using formic acid:methanol (5:95). The purity and identity of (S)-N-methylcanadine was confirmed by LC-MS/MS. (S)-N-Methylstylopine was similarly prepared from (S)-stylopine (8). Other alkaloids were obtained as described previously (5, 8, 23). S-Adenosyl-l-methionine was purchased from Sigma-Aldrich. All other chemicals were purchased from Bioshop (Ontario, Canada).
Phylogenetic Analysis
Amino acid alignments were performed using ClustalW (24), and a phylogenetic tree was built from bootstrap values generated using the Geneious (Biomatters, Newark, NJ) software package. GenBankTM accession numbers for sequences used to construct the phylogenetic tree are as follows: PsCYP82N4 (N-methylstylopine 14-hydroxylase), P. somniferum N-methylstylopine 14-hydroxylase (KC154003); PsCYP82Y1, P. somniferum CYP82Y1 (JQ659005); PsCYP82X1, P. somniferum CYP82X1 (JQ659002); PsCYP82X2, P. somniferum CYP82X2 (JQ659004); CjCYP82R1, Coptis japonica CYP82R1 (BAF98472), PsCYP82N3 (protopine 6-hydroxylase), P. somniferum protopine 6-hydroxylase (KC154002); EcCYP82N2v2 (protopine 6-hydroxylase), E. californica protopine 6-hydroxylase (AB598834); PsCYP719A21 (canadine synthase (CAS)), P. somniferum canadine synthase (JQ659003); PsCYP719B1 (SalSyn), P. somniferum salutaridine synthase (ABR14720); PsCYP719A25 (cheilanthifoline synthase), P. somniferum cheilanthifoline synthase (GU325749); PsCYP80B3 (N-methylcoclaurine 3′-hydroxylase), P. somniferum N-methylcoclaurine 3′-hydroxylase (AF191722); EcCYP80B1 (N-methylcoclaurine 3′-hydroxylase), E. californica N-methylcoclaurine 3′-hydroxylase (AF014801); CjCYP80G2 (corytuberine synthase), C. japonica corytuberine synthase (AB288053); AtCYP82C2, Arabidopsis thaliana CYP82C2 (O49394); NtCYP82E4v1, Nicotiana tabacum CYP82E4v1 (DQ131886); AtCYP82G1, A. thaliana CYP82G1; VvCYP82C4, Vitis vinifera CYP82C4 (XP_02284810); and GmCYP82G4, Glycine max CYP82G4 (XP_003523888).
Isolation of CYP82 Candidates
Total RNA and alkaloid extracts from the latex of eight opium poppy chemotypes were subjected to transcript and metabolite profiling, respectively, as described previously (5, 22). CYP82 candidates were identified among genes differentially expressed in noscapine-free (Deborah, Przemko, 40, and T) and noscapine-producing (Natasha, Marianne, Roxanne, and Veronica) chemotypes of opium poppy. The full-length coding regions of CYP82X1, CYP82X2, and CYP82Y1 were assembled in silico by searching each database using the tBLASTn algorithm. Relative transcript abundance was determined as the number of reads corresponding to each selected candidate compared with the total number of reads in each database (5).
Gene Expression and Noscapine Accumulation in Different Plant Organs
Total RNA from different opium poppy Veronica organs were collected by flash freezing tissues in liquid N2 prior to excision from the plant to preserve the latex content. Total RNA was extracted from each organ, and cDNA was synthesized as described previously (5). The relative abundance of CYP82Y1 transcripts in each organ was determined by quantitative real time PCR (qRT-PCR) analysis using the primer set: 5′-TGACACTTTCGCCTGTTATTA-3′ and 5′-CTATGACTACCGAATCTTAACAG-3′. Plant tissues were lyophilized, and alkaloids were extracted to measure noscapine content.
Cloning and Expression of CYP82 Candidates
Full-length coding regions of CYP82X1 and CYP82Y1 were amplified from cDNA derived from total stem RNA of the Bea's Choice chemotype using Takara Ex Taq DNA polymerase (Fisher Scientific, Ottawa, Canada) and the following primer sets: CYP82X1, 5′-GCGGCCGCGCCATGGAGTTATTCATAAAG and 5′-CATACCTAGTGCAACCCATGAATAAGAGCCGC; CYP82Y1, 5′-ATTAGCGGCCGCACCATGGCGTATTTGATGATCAA-3′ and 5′-CATAACTAGTGCATCTAGTGTGCGTGGGGTGA-3′. A synthetic CYP82X2 gene based on native nucleotide sequences was constructed (Genscript, Piscataway, NJ) by replacing the first 60 N-terminal amino acids of CYP82X2 with 43 N-terminal amino acids from lettuce germacrene A oxidase to improve heterologous expression in yeast (25). For heterologous production of FLAG-tagged CYP82 cDNAs, the cognate cDNAs were ligated into the NotI and SpeI restriction sites of the dual plasmid pESC-leu2d::CPR (20, 26) yielding pESC-Leu2d::CYP82X1/CPR, pESC-Leu2d::CYP82X2/CPR, and pESC-Leu2d::CYP82Y1/CPR. Yeast harboring pESC-leu2d::CPR was used as the negative control.
Yeast Culture, Preparation of Microsomes, and Immunoblot Analysis
The protease-deficient Saccharomyces cerevisiae strain YPL 154C:Pep4 was transformed with pESC-Leu2d::CYP82X1/CPR, pESC-Leu2d::CYP82X2/CPR, and pESC-Leu2d::CYP82Y1/CPR. Transformed yeast were used to inoculate 2 ml of synthetic complete (SC) medium lacking leucine (SC−Leu), but containing 2% (w/v) glucose, and cultured overnight on a gyratory shaker at 250 rpm and at 30 °C. Initial cultures were then diluted 100-fold in SC−Leu medium supplemented with 2% (w/v) glucose and cultured for 16 h. Yeast cultures were harvested and transferred to fresh SC−Leu containing 2% (w/v) galactose for 24 h to induce expression of recombinant genes. Microsomes were prepared as described previously (6, 27). Recombinant enzymes were detected by immunoblot analysis as described previously (6).
CYP82 Enzyme Assays
Standard enzyme assays were performed at 30 °C for 30 min in 200 μl of 100 mm HEPES-NaOH, pH 7.5, containing 5 mg of total microsomal proteins, 500 μm NADPH, and 50 μm (R,S)-canadine or (R,S)-N-methylcanadine on a gyratory shaker with gentle agitation (60 rpm). Reactions were stopped by the addition of 800 μl of methanol. Microsomal protein extracted from yeast harboring pESC-leu2d::CPR was used as a negative control.
In Vitro Biochemical Characterization of CYP82Y1
Standard in vitro enzyme assays were performed as described above. A variety of alkaloids belonging to several different BIA subgroups (1-benzylisoquinoline, morphinan, protoberberine, aporphine, benzo[c]phenanthridine, protopine, phthalideisoquinoline, and bisbenzylisoquinoline) were tested to determine the substrate range of CYP82Y1 (see Table 1). Steady-state enzyme kinetics for (R,S)-N-methylcanadine were determined by varying the alkaloid substrate concentration in the standard in vitro assay between 0 and 400 μm at a fixed concentration of 500 μm NADPH. Kinetic constants for CYP82Y1 were calculated by fitting initial velocity versus substrate concentration to the Michaelis-Menten equation using the GraphPad Prism 5.0 (GraphPad Software, San Diego, CA).
TABLE 1.
Benzylisoquinoline alkaloids tested as potential CYP82Y1 substrates
| Allocryptopine | Cryptopine | Papaverine |
|---|---|---|
| Berbamine | Dehydroreticuline | Pavine |
| Berberine | (S)-Glaucine | Protopine |
| (R,S)-Boldine | Hydrastine | (S)-Reticuline |
| (S)-Bulbocapnine | (R,S)-Isothebaine | Salutaridine |
| Bicuculline | (R,S)-N-Methylcanadine | Sanguinarine |
| Canadaline | (S)-N-Methylstylopine | Scoulerine |
| (R,S)-Canadine | Narcotinehemiacetal | (S)-Stylopine |
| Chelerythrine | Narcotoline | (R,S)-Tetrahydrocolumbamine |
| (S)-Coclaurine | (R,S)-Norlaudanosoline | (R,S)-Tetrahydropalmatine |
| Codeine | (S)-Norreticuline | (R,S)-Tetrahydropapaverine |
| Corytuberine | Noscapine | Thebaine |
Virus-induced Gene Silencing
The tobacco rattle virus (TRV) vector system (28) was used to suppress transcript levels of CYP82Y1 in opium poppy plants. A unique sequence encompassing part of the 3′-UTR and coding region of CYP82Y1 was amplified with the primer set: 5′-TAATCTAGACTCACCCCACGCACACTAGAT-3′ and 5′-AGGCTCGAGATCCACCAAAGGAATTATATTAAG-3′. The fragment was cloned into pTRV2, and vectors were mobilized in Agrobacterium tumefaciens as described previously (6). Apical meristems of 2–3-week-old seedlings were infiltrated with a 1:1 mixture of A. tumefaciens harboring pTRV1 and either pTRV2::CYP82Y1 or pTRV2. Infiltrated plants were grown in the greenhouse for 8–10 weeks. Visual confirmation of gene silencing was monitored using the pTRV2-PDS construct encoding phytoene desaturase (29). Latex and stems of infiltrated opium poppy were collected immediately prior to anthesis as described previously (5). Infiltration with A. tumefaciens was confirmed by detection of the RNA corresponding to the TRV2 transgene cassette in VIGS-treated plants using TRV-MCS primer sets specific to sequences (i) flanking the multiple cloning site of pTRV2 (5′-GGTCAAGGTACGTAGTAGAG-3′ and 5′-CGAGAATGTCAATCTCGTAGG-3′) (29) and (ii) encoding GAPDH as a positive control (5′-CTCATTTGAAGGGTGGAGC-3′ and 5′-GTCATTGCGTGGACAGTGG-3′). Latex samples from infiltrated plants were lyophilized, resuspended in methanol at a concentration of 0.1 μg μl−1, and extracted overnight at −80 °C. Transcript analysis of infiltrated plants was performed by qRT-PCR using a 7300 real time PCR system (Applied Biosystems, Burlington, Canada) for 40 cycles of template denaturation, primer annealing, and primer extension. Each 10-μl PCR contained 1 μl of cDNA, 300 nm forward and reverse primers, and a 1× KAPA SYBR FAST qPCR kit (Kapa Biosystems, Boston, MA). The opium poppy gene encoding ubiquitin was used as an endogenous reference and amplified with the primer set: 5′-CCATTTGGTGCTTCGTCTAC-3′ and 5′-CAAGCCATAGCTGAAACACC-3′. Primer sets used to perform qRT-PCR on other genes were: CYP82Y1, 5′-ACACTTTCGCCTGTTATTA-3′ and 5′-CTATGACTACCGAATCTTAACAG-3′; CYP82X1, 5′-GAGGAAGAAAGCACCAAATGC-3′ and 5′-CACTGTTTTACCATCTCCCAG-3′; CYP82X2, 5′-ACGAATGGTTGGCTGACATGAT-3′ and 5′-ACAAGACCTCTCAATCGGTCT-3′; CYP719A21 (CAS), 5′-CTACCAATCCATATCGTCATTACACTGCCG-3′ and 5′-GGTAACTTGCACATTCTTGGAGGCACTGC-3′; TNMT, 5′-AGTGGATGGTTGCGCAGCTG-3′ and 5′-CAAGCCAAACCCCTGTGATACA-3′; SOMT1, 5′-TATGGTCATAATCATCAATCA-3′ and 5′-TTGGAATGCATATTAACTTCC-3′; SOMT2, 5′-CGTCCAGGTGTTGAGTTCATC-3′ and 5′-TCCCAATTGTGCAGGACATAC-3′; SOMT3, 5′-GCTTCAGCATTGGTTAACGAG-3′ and 5′-GCTTTGGATATGGCTTTCACTG-3′; AT1, 5′-CGCTTTGATATGGAAGTCAGCAG-3′ and 5′-CATGGCATGGGTTACAGTGGAT-3′; CXE1 (carboxylesterase), 5′-CGTTTATTGTAATCCGAATATTGATCAT-3′ and 5′-CATAAAATCCTACCACAAAACATCTCTC-3′; and NOS, 5′-TTTTCAAATGAATAAACGGGCAG-3′ and 5′-AACAACATATAGCCAAAGGCTCTTC-3′; 6OMT, 5′-CAACAATGTCAAACCCATGTCTTT-3′ and 5′-CGGAACAGACGGTCTTCGTT-3′; 4′OMT2, 5′-CCCGGTGATATGTTCAAGTCTG-3′ and 5′-ATCACTTTCCCTGTCTCTTTCG-3′; BBE, 5′-GTGCAGAAGCTTAACATTAC-3′ and 5′-GACGAGAGGAGATTATCATT-3′. Plant lines showing the highest expression level served as the calibrator for each target gene. Dissociation curve analysis was used to validate PCR specificity. Relative gene expression data were statistically analyzed as described previously (5, 6).
LC-MS/MS Analysis
Enzyme assay and VIGS samples were diluted 1:100 and 1:1000, respectively, with solvent A (10 mm ammonium acetate:acetonitrile (95:5)) and analyzed using a 6410 Triple Quadruple LC-MS/MS (Agilent Technologies, Santa Clara, CA). Liquid chromatography was carried out using a Poroshell 120 SB C18 column (2.1 × 50 mm, 2.7-μm particle size; Agilent Technologies) at a flow rate of 0.7 ml min−1. The column was equilibrated in solvent A, and the following elution conditions were used: (i) for enzyme assays, 0–6 min 60% solvent B (acetonitrile), 6–7 min-ramp to 99% solvent B, 7–9 min isocratic at 99% solvent B, and 9–13 min ramp to 0% solvent B; (ii) for VIGS, 0–6 min 60% solvent B, 6–9-min ramp to 99% solvent B, 9–14 min isocratic at 99% solvent B, and 14–18-min ramp to 0% solvent B. Samples were injected into the mass analyzer via an electrospray ionization (ESI) probe inlet. Ions were generated and focused using the following parameters: (i) for enzyme assays, capillary voltage, 4000 kV; gas flow, 9 liter min−1, fragmentor voltage, 100 V; nebulizer pressure, 40 psi; gas temperature, 330 °C; (ii) for VIGS, capillary voltage, 4000 kV; gas flow, 10 liter min−1; fragmentor voltage, 100 V; nebulizer pressure, 50 psi; gas temperature, 350 °C. Mass spectrometry data were acquired in positive ion mode in the range of m/z 200–700. Positive mode electrospray ionization (ESI[+]), collision-induced dissociation (CID) spectra were recorded using collision energy of −25.0 eV applied in quadrupole 2 and an argon collision gas pressure of 1.8 × 10−3 torr. MS2 fragments were analyzed in quadrupole 3 by scanning from m/z 40 to m/z 2 greater than that of the precursor ion. Compounds were identified based on retention times and ESI[+]-CID spectra compared with authentic standards, and alkaloid content was calculated as ng alkaloid μg−1 dry weight of latex based on standard quantification curves. LC-MS/MS data for reported alkaloids have been described previously (10, 30).
LTQ-Orbitrap XL Analysis
High resolution MSn experiments were performed using an LTQ-Orbitrap XL equipped with a syringe pump and an Accela HPLC system (ThermoFisher Scientific). Alkaloids (1 μg ml−1) were introduced continuously with a syringe pump (5 μl min−1) into the HPLC flow at a rate of 500 ml min−1 using acetonitrile, and positive ions were generated by heated ESI with the following parameters: heater, 400 °C; sheath gas, 60 au; auxiliary gas, 20 au; spray voltage, 3 kV. Ion interface settings were 380 °C and 38 V (capillary) and 85 V (tube lens). MSn experiments were performed by conducting CID on target ions isolated in the linear ion trap followed by high resolution (60,000 full width at half-maximum; <2 ppm error) mass analysis of the resulting fragment ions in the Orbitrap XL. Full scan data were collected in centroid mode over mass ranges extending from the lowest permissible value up to 10 atomic mass units beyond the parent ion. Detection methods consisted of three scan events of ∼1 s each with a total run time of 10 min. External and internal instrument calibration ensured an error of <2 ppm.
NMR Analysis
1-Hydroxy-N-methylcanadine was produced from N-methylcanadine in scaled-up CYP82Y1 reactions terminated with an equal volume of methanol. Precipitated protein was removed by centrifugation, the supernatant was concentrated under reduced pressure, resuspended in 10 mm ammonium acetate:acetonitrile (95:5), and loaded on a StrataTM-X-33μ polymeric reversed phase, 200 mg ml−1 column (Phenomenex, Torrance, CA) to remove non-alkaloid components. N-Methylcanadine and 1-hydroxy-N-methylcanadine were eluted from the column using 10 mm ammonium acetate:acetonitrile (70:30), and the sample was concentrated under reduced pressure. Compounds were separated on a 250-μm silica-coated glass TLC plate using a mobile phase of isopropanol:ammonia hydroxide:water 9:2:2 (Moravek Biochemical, Brea, CA). Bands corresponding to N-methylcanadine and 1-hydroxyl-N-methylcanadine were excised, eluted with 50% (v/v) ethanol, dried under reduced pressure, and lyophilized with water. One milligram of each compound was dissolved in CD3OD, and 1H COSY NMR spectra were recorded in 5-mm Shigemi NMR tubes (Shigemi, Allison Park, PA) on a Varian VNMRS three-channel 600 MHz spectrometer. Data were interpreted based on 1H NMR spectra for (−)-cis-N-methylcanadine reported previously (31, 32).
RESULTS
Identification of CYP Candidates
Available opium poppy chemotypes display two distinct phenotypes with respect to the accumulation of noscapine (Fig. 2A) (5). Whereas the chemotypes Marianne, Natasha, Roxanne, and Veronica accumulate high levels of noscapine, the chemotypes Deborah, 40, T, and Przemko display only trace to undetectable noscapine content. Marianne contained the highest levels of noscapine, and Roxanne and Veronica accumulated papaverine in addition to noscapine. Compared with noscapine-free plants, the high noscapine chemotypes accumulated substantially less morphinan alkaloids.
FIGURE 2.

Heat maps showing the relative abundance of major latex alkaloids (A) and transcripts encoding selected CYPs (B) in eight opium poppy chemotypes. CYP719B1 is salutaridine synthase, CYP719A20 is stylopine synthase, and CYP80B3 is N-methylcoclaurine 3′-hydroxylase.
Comparing the Marianne and Deborah transcriptome databases for differentially expressed genes revealed a variety of CYPs, O-methyltransferases (5), a short chain dehydrogenase/reductase (11), an acetyltransferase, and a carboxyl esterase for which transcripts were detected only in noscapine-producing chemotypes. Focusing on CYPs that displayed (i) sequence similarity with previously reported BIA biosynthetic enzymes (i.e., members of the CYP80, CYP82, and CYP719 families) and (ii) gene expression profiles that strictly correlated with the accumulation of noscapine in different opium poppy chemotypes yielded four candidate genes. One candidate belongs to CYP719 family, whereas three others are members of the CYP82 family (Fig. 2B). Other characterized CYPs involved in BIA biosynthesis did not exhibit a transcript accumulation pattern that correlated with the accumulation of noscapine. Except for CYP719A21, which has been characterized as canadine synthase (6), biochemical functions have not been assigned for CYP82X1, CYP82X2, and CYP82Y1.
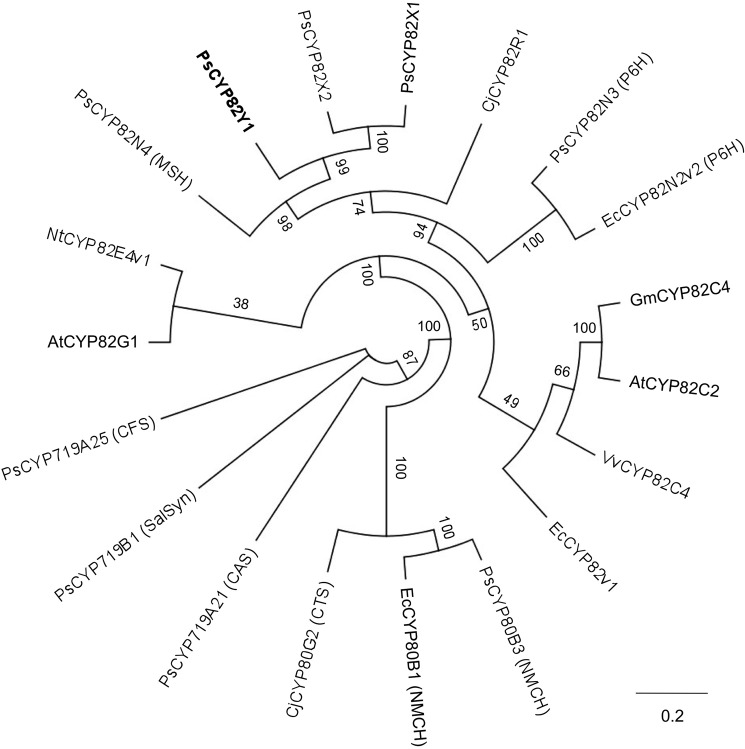
The CYP82X1, CYP82X2, and CYP82Y1 cDNAs contained open reading frames encoding 540, 554, and 556 amino acids, respectively. Phylogenetic analysis based on an alignment of CYP protein sequences involved in BIA biosynthesis highlighted the distinction between CYP82 members, and the CYP719 and CYP80 families (Fig. 3). CYP82Y1 shares the highest amino acid sequence identity with other opium poppy CYPs shown or purported to play a role in BIA metabolism: CYP82N4 (53%), CYP82X1 (50%), CYP82N3 (47%), putatively protopine 6-hydroxylase, and CYP82X2 (43%). Among functionally characterized CYPs from other plant species, CYP82Y1 displayed the highest amino acid sequence identity with EcCYP82N2v2 (44%), which is protopine 6-hydroxylase (21). Based on substantial amino acid sequence similarity with CYP82N4, which was functionally characterized as N-methylstylopine 14-hydroxylase and also acted on N-methylcanadine (20), CYP82Y1 might also accept quaternary protoberberine alkaloids as substrates.
FIGURE 3.
Unrooted neighbor-joining phylogenetic tree for selected CYP candidates and others shown to function in BIA metabolism. Bootstrap frequencies for each clade were based on 1,000 iterations. Abbreviations and GenBankTM accessions numbers for each protein are provided under “Experimental Procedures.”
Expression and Characterization of CYP82Y1
A galactose-inducible yeast dual expression vector pESC-Leu2d was used to simultaneously express opium poppy CYP82 and CPR cDNAs in S. cerevisiae. The CYP82 coding regions were inserted into the multiple cloning site of pESC-Leu2d under the control of the Gal-10 promoter and in-frame with the FLAG epitope. In the same pESC-Leu2d vector, CPR was inserted under the Gal-1 promoter and in-frame with the c-Myc epitope. Co-expression of CYP82 and CPR cDNAs was confirmed by immunoblot analysis of microsomal fractions prepared from S. cerevisiae cultures harboring the pESC-leu2d::CYP82X1/CPR, pESC-leu2d::CYP82X2/CPR, and pESC-leu2d::CYP82Y1/CPR vectors using FLAG and c-Myc antibodies to detect epitope-tagged recombinant proteins (Fig. 4A).
FIGURE 4.
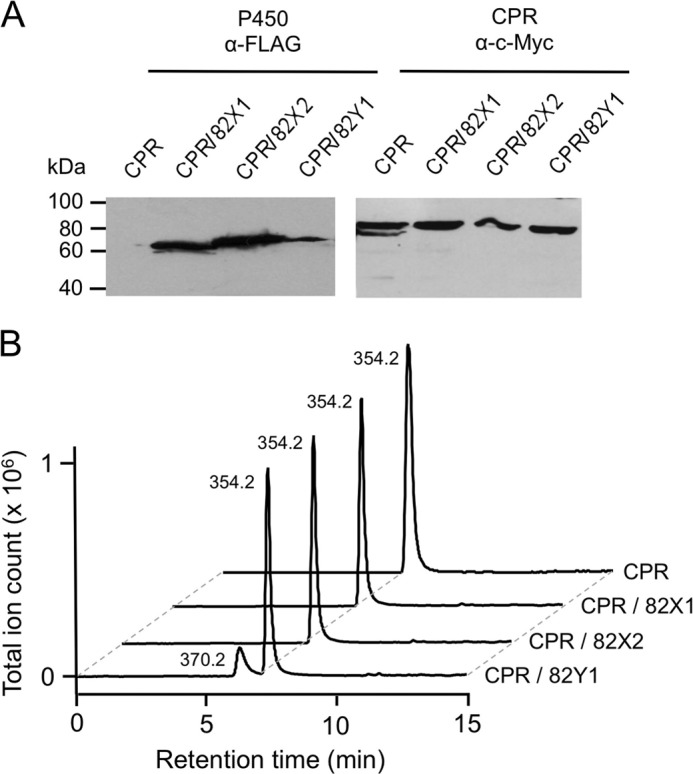
Heterologous expression of CYP82X1, CYP82X2, CYP82Y1, and catalytic function of the recombinant enzymes. A, S. cerevisiae harboring pESC-leu2d::CPR (CPR), pESC-Leu2d::CYP82X1/CPR (CPR/82X1), pESC-Leu2d::CYP82X2/CPR (CPR/82X2), or pESC-leu2d::CYP82Y1/CPR (CPR/82Y1) were induced on galactose, and CPR, CYP82X1, CYP82X2, or CYP82Y1 recombinant proteins were detected using α-FLAG (CYP) and α-c-Myc (CPR) antibodies. Each lane contained 2 μg of total microsomal proteins. B, extracted ion chromatograms showing the in vivo catalytic activity of CYP82X1 (CPR/82X1), CYP82X2 (CPR/82X2), or CYP82Y1 (CPR/82Y1) using (R,S)-N-methylcanadine as the enzymatic substrate, compared with the negative control (CPR). Only the incubation of microsomal preparations containing recombinant CYP82Y1 with N-methylcanadine (m/z 354) resulted in the formation of a reaction product at m/z 370.
(R,S)-Canadine and (R,S)-N-methylcanadine were tested as potential substrates for the three CYP82 candidates. In the presence of NADPH, only N-methylcanadine (m/z 354) was accepted by CYP82Y1, resulting in the formation of a reaction product with m/z 370 (Fig. 4B). CYP82X1 and CYP82X2 did not show activity with (R,S)-canadine or (R,S)-N-methylcanadine. The substrate specificity of CYP82Y1 was determined using 36 BIAs representing several different structural subgroups including 1-benzylisoquinoline, protoberberine, benzophenanthridine, aporphine, protopine, morphinan, pavine, and phthalideisoquinoline alkaloids (Table 1). In addition to (R,S)-N-methylcanadine (m/z 354), only (S)-N-methylstylopine was accepted as a substrate, although with considerably lower efficiency (38% of the substrate turnover rate compared with (R,S)-N-methylcanadine). Microsomal fractions of yeast harboring pESC-leu2d::CPR (i.e., lacking a CYP82 candidate) did not catalyze the formation of any products with either (R,S)-N-methylcanadine or (S)-N-methylstylopine. CYP82Y1 showed an absolute requirement for the quaternary N-methyl moiety because neither (R,S)-canadine nor (S)-stylopine were accepted as substrates. No compounds belonging to other BIA subgroups were accepted as substrates. Notably, no conversions to compounds with m/z reduced by 2 atomic mass units compared with the potential substrate were detected, ruling out the possible occurrence of C-C bond formation. The increased m/z by 16 atomic mass units of the reaction products compared with the m/z of N-methylcanadine and N-methylstylopine suggested the hydroxylation of these substrates.
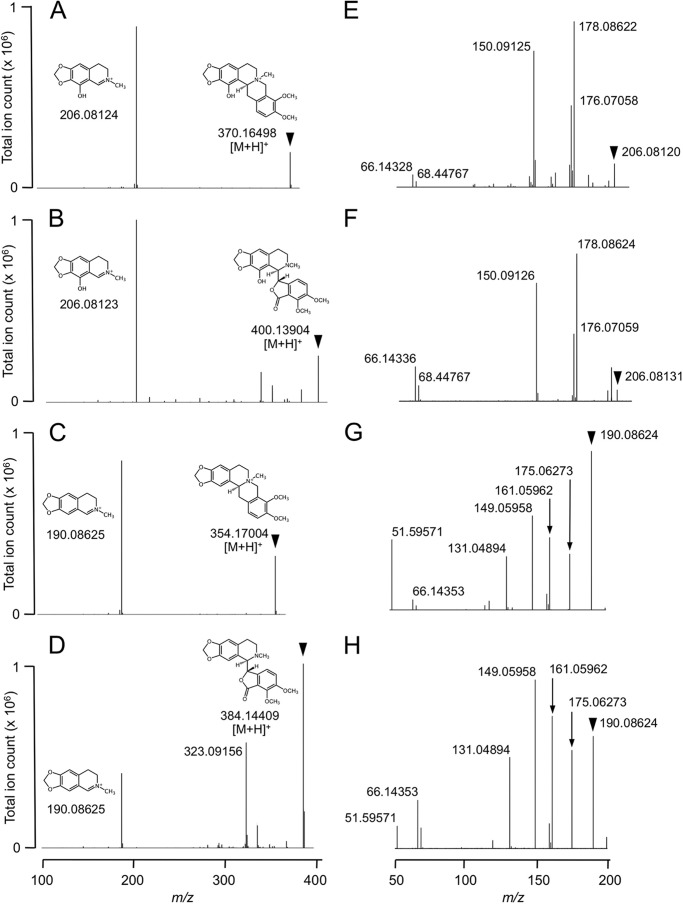
Structural Elucidation of the CYP82Y1 Reaction Product
High resolution MSn analysis suggested aromatic ring hydroxylation at positions C1 or C4 on the isoquinoline moiety of N-methylcanadine. The ion trap portion of a hybrid LTQ-Orbitrap-XL instrument isolated and fragmented parent ions and subsequent daughter ions, and the Orbitrap analyzer acquired high resolution mass data for all MS2 and MS3 fragment ions. Because authentic standards were not available, mass fragmentation data for the enzymatic reaction product (1-hydroxy-N-methylcanadine) and the substrate (N-methylcanadine) were compared with authentic standards of the phthalideisoquinoline alkaloids narcotoline and hydrastine. Narcotoline and 1-hydroxyl-N-methylcanadine both possess a 1-hydroxyl group, whereas hydrastine and N-methylcanadine are not hydroxylated at the C1 position. CID of both phthalideisoquinoline and protoberberine alkaloids generally yielded a principal isoquinoline fragment (33), which allowed MS3 fragmentation analysis of equivalent MS2 daughter ions. Narcotoline and the N-methylcanadine reaction product yielded isoquinoline fragments at 206.08124 and 206.08123 m/z, respectively, corresponding to an elemental formula of C11H12O3N and indicating that either C1 or C4 hydroxylation of N-methylcanadine had occurred (Fig. 5, A and B). In contrast, MS2 of hydrastine and the N-methylcanadine substrate both yielded fragments at 190.08625 m/z corresponding to an elemental formula of C11H12O2N, which is consistent with an isoquinoline moiety lacking a 1-hydroxyl moiety (Fig. 5, C and D). To support the 1-hydroxylation of N-methylcanadine, CID analysis of all isoquinoline ions was performed. MS3 revealed nearly identical spectra for both 1-hydroxylated isoquinoline moieties of 1-hydroxyl-N-methylcanadine and narcotoline (Fig. 5, E and F) and similarly identical spectra for both nonhydroxylated isoquinoline ions (Fig. 5, G and H) of N-methylcanadine and hydrastine. Because the product derived from N-methylstylopine produced a similar MS/MS fragmentation pattern, we conclude that a regiospecifically identical hydroxylation event occurred with N-methylstylopine (m/z 338.2) yielding 1-hydroxy-N-methylstylopine.
FIGURE 5.
Putative identification of the CYP82Y1 reaction product as 1-hydroxy-N-methylcanadine by LTQ-Orbitrap analysis. A–D, the reaction product putatively identified as 1-hydroxy-N-methylcanadine (A) and the authentic standards narcotoline (B), N-methylcanadine (C), and hydrastine (D) each produced a single major fragment ion in MS2 at m/z 206 for 1-hydroxy-N-methylcanadine and narcotoline or m/z 190 for N-methylcanadine and hydrastine. E and F, fragmentation in MS3 of m/z 206 daughter ions of 1-hydroxy-N-methylcanadine (E) and narcotoline (F) yielded identical spectra. G and H, similarly, fragmentation of m/z 190 daughter ions of N-methylcanadine (G) and hydrastine (H) produced equivalent spectra in MS3. Arrowheads indicate parent ions.
The CYP82Y1 reaction product derived from N-methylcanadine was confirmed as 1-hydroxyl-N-methylcandine by NMR spectroscopy. The 1H COSY NMR (600 MHz, CD3OD, δH) spectrum of N-methylcanadine showed 16 proton signals consisting of 7.07 (1H, d, J = 8.4, H12), 6.97 (1H, d, J = 8.4, H11), 6.83 (1H, s, H1), 6.80 (1H, s, H4), 5.98, 5.99 (2H, s, -O-CH2-O-), 4.85, 4.84 (2H, broad, H8), 4.72 (1H, m, H14), 3.89 (3H, s, 9-O-Me), 3.86 (3H, s, 10-O-Me), 3.80, 3.83 (1H, m, H6), 3.27 (3H, s, N-Me), 3.43, 3.19 (2H, m, H13), which is comparable to previously assigned spectra for N-methylcanadine from Zanthoxylum sprucei (31) and Corydalis turtschaninovii (32). The 1H COSY NMR (600 MHz, CD3OD, δH) spectrum of 1-hydroxy-N-methylcanadine showed only 15 proton signals consisting of 7.06 (1H, d, J = 8.4, H12), 6.95 (1H, d, J = 8.4, H11), 6.31 (1H, s, H4), 5.91, 5.92 (2H, s, -O-CH2-O-), 4.84, 4.85 (2H, broad, H8), 4.99 (1H, m, H14), 3.89 (3H, s, 9-O-Me), 3.87 (3H, s, 10-O-Me), 3.75, 3.77 (1H, m, H6), 3.24 (3H, s, N-Me), 3.42, 3.19 (2H, m, H13). Comparing the two spectra showed that the proton signal at C1 on the enzymatic substrate N-methylcanadine was absent from the reaction product. The chemical shift of the proton signal at C4 from 6.8 in N-methylcanadine to 6.3 in the reaction product is attributable to the effect of a hydroxyl group at C1. To further confirm the 1-hydroxylation regiospecificity, the correlation between protons at C1 and C14 in N-methylcanadine was absent in the reaction product, indicating a replacement of the proton at C1 by a hydroxyl moiety.
Biochemical Properties
CYP82Y1 was designated N-methylcanadine 1-hydroxylase because of the high substrate specificity and conversion rate with respect to N-methylcanadine. Optimal activity was detected in a HEPES-buffered reaction at pH 7.5 and 30 °C. When determined using microsomal fractions and varying concentrations of (R,S)-N-methylcanadine, CYP82Y1 followed Michaelis-Mention-type reaction kinetics with a Km of 19.5 μm and an estimated Vmax value of 98 pmol min−1 mg−1 total protein (Fig. 6). No significant substrate or product inhibition was detected. Limited availability of (S)-N-methylstylopine precluded the determination of kinetic data for this substrate.
FIGURE 6.
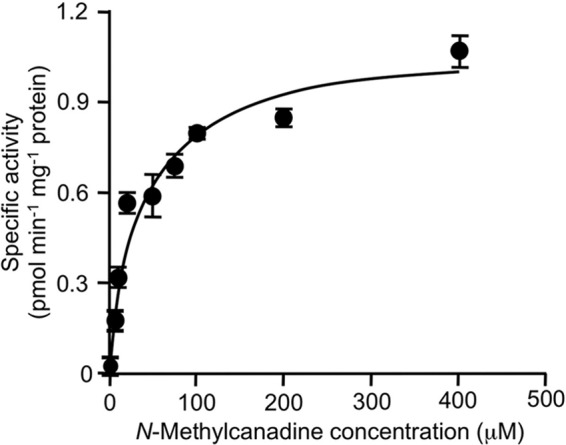
Steady-state enzyme kinetics of opium poppy CYP82Y1 in total microsomal protein extracts of S. cerevisiae. Various concentrations of (R,S)-N-methylcanadine were provided as the substrate.
Physiological Role of CYP82Y1 in Noscapine Biosynthesis
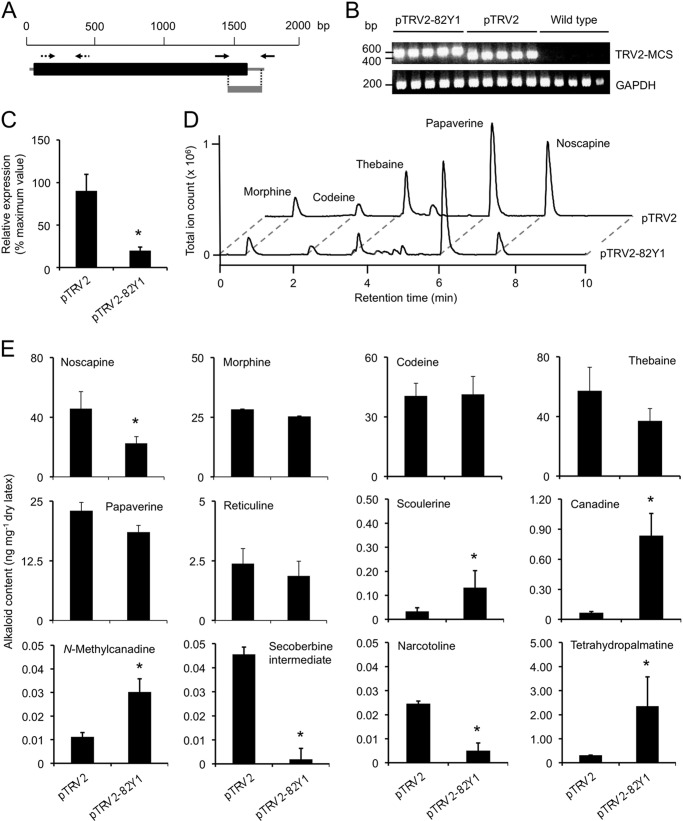
A unique region of the coding region and 3′-UTR was used to construct pTRV2–82Y1 to suppress CYP82Y1 transcript levels in opium poppy plants using VIGS (Fig. 7A). The Bea's Choice chemotype accumulates noscapine in addition to papaverine, morphine, and other BIAs. TRV infection of infiltrated plants was confirmed by reverse transcription-PCR amplification of TRV2 RNA (Fig. 7B). CYP82Y1 transcript levels were significantly reduced in plants infiltrated with A. tumefaciens harboring the pTRV2–82Y1 construct compared with the empty pTRV2 vector control (Fig. 7C). Total alkaloid content and the levels of major alkaloids including morphine, codeine, reticuline, thebaine, and papaverine were not altered in CYP82Y1-silenced plants compared with controls (Fig. 7, D and E). However, the suppression of CYP82Y1 transcript levels significantly reduced the accumulation of noscapine (Fig. 7D). CYP82Y1-silenced plants also accumulated significantly higher levels of scoulerine, canadine, N-methylcanadine, and tetrahydropalmatine. Suppression of CYP82Y1 transcript abundance also caused a significant reduction in the levels of narcotoline and an alkaloid displaying a secoberbine-type CID spectrum (Fig. 7E). Notably, CYP82Y1 silencing also significantly reduced the relative abundance of SOMT1, SOMT2, CAS, CYP82X1, and CYP82X2 transcripts, but did not affect transcript levels of other tested genes (Fig. 8).
FIGURE 7.
Virus-induced gene silencing confirms the involvement of CYP82Y1 in noscapine biosynthesis. A, region (gray line) of the CYP82Y1 cDNA inserted into the pTRV2 vector used to suppress CYP82Y1 transcript levels. The thick black line represents the coding region, whereas the flanking thin lines indicate noncoding 5′- and 3′-untranslated regions. Solid arrows show annealing sites of primers used to amplify cDNA fragments, whereas dotted arrows show annealing sites of primers used for qRT-PCR analysis. B, ethidium bromide-stained agarose gel showing the detection by reverse transcription-PCR of TRV coat protein transcripts in total RNA extracted from opium poppy plants infiltrated with A. tumefaciens cultures harboring pTRV1 and pTRV2 constructs. C, relative CYP82Y1 transcript abundance in control (pTRV2) and silenced (pTRV2–82Y1) plants. D, total ion chromatograms showing the major alkaloid profiles of control (pTRV2) and CYP82Y1-silenced (pTRV2–82Y1) plants. E, relative abundance of major alkaloids and other alkaloids affected by the suppression of CYP82Y1 transcript levels in control (pTRV2) and silenced (pTRV2–82Y1) plants. Asterisks represent significant differences determined using an unpaired, two-tailed Student t test (p < 0.05).
FIGURE 8.
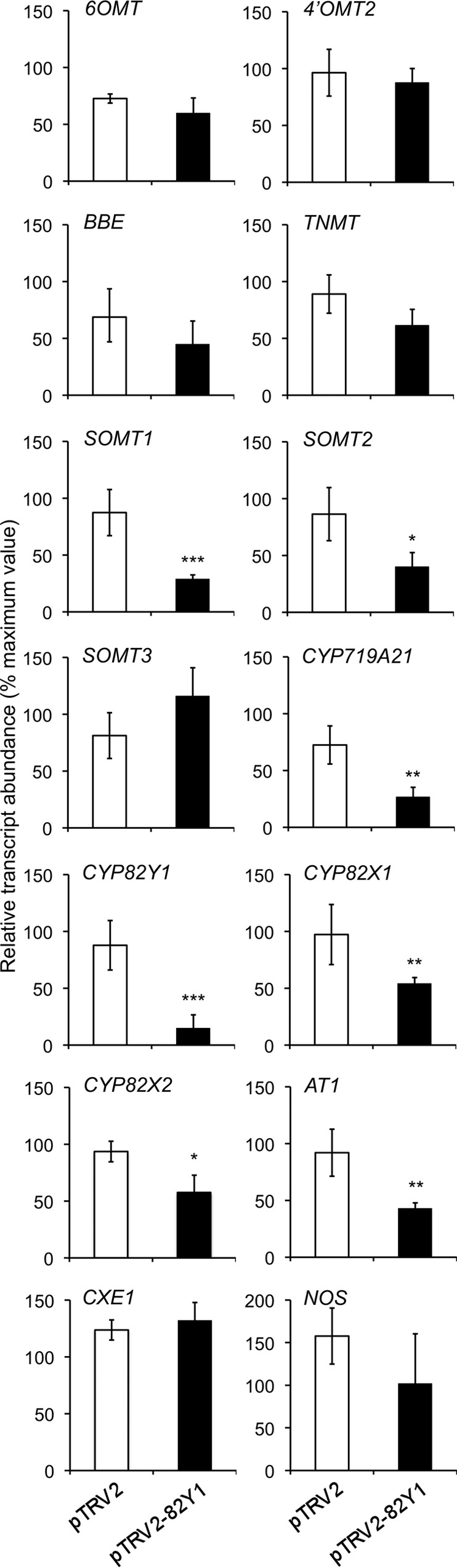
Effects of suppressing CYP82Y1 transcript levels by virus-induced gene silencing on the relative abundance of other putative noscapine biosynthetic genes in opium poppy. Differences in transcript abundance between control (white bars) and CYP82Y1-silenced (black bars) plants were analyzed by unpaired, two-tailed Student's t test. *, p < 0.05; **, p < 0.01; ***, p < 0.001. 6OMT, norcoclaurine 6-O-methyltransferase; 4′OMT2, 3′-hydroxy-N-methylcoclaurine 4′-O-methyltransferase; BBE, berberine bridge enzyme.
Expression of CYP82Y1 in Opium Poppy
Noscapine levels were highest in opium poppy stems, but the alkaloid was detected in all organs (Fig. 9A). Expression analysis by qRT-PCR also revealed the occurrence of CYP82Y1 transcripts in all plant organs, with the highest levels detected in stems (Fig. 9B).
FIGURE 9.
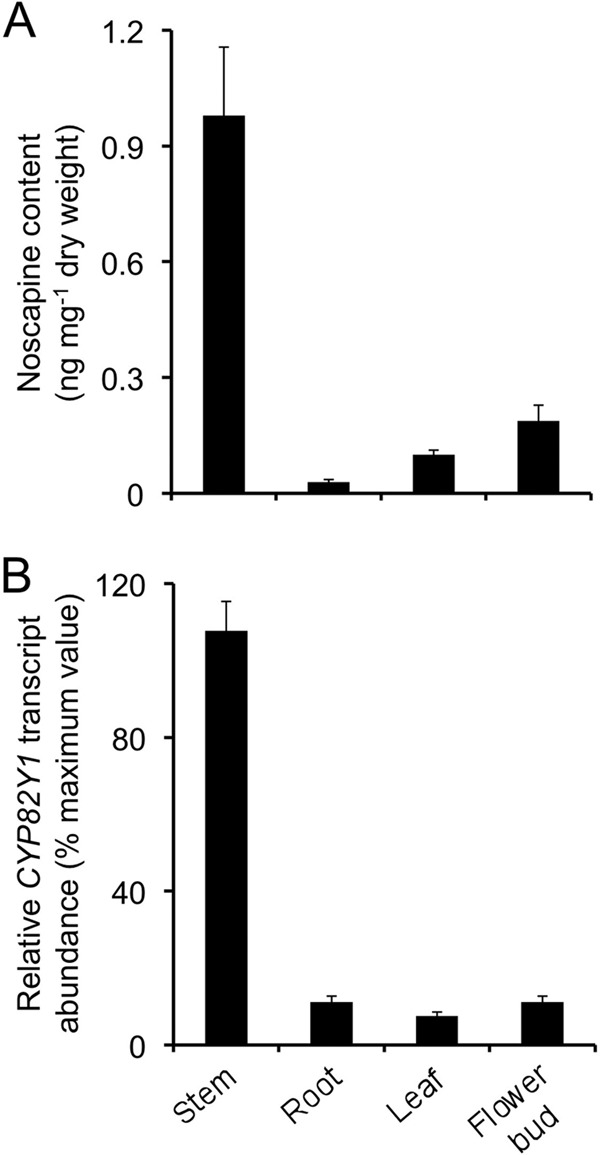
Relative noscapine accumulation (A) and CYP82Y1 transcript levels (B) in different opium poppy organs. cDNA synthesized using total RNA extracted from each organ was used to perform qRT-PCR. The same tissue was used for RNA and alkaloid extraction.
DISCUSSION
CYP82Y1 was shown to catalyze the conversion of (R,S)-N-methylcanadine to (R,S)-1-hydroxy-N-methylcanadine and also accepted (S)-N-methylstylopine as a substrate. Targeted metabolite and transcript profiling of eight opium poppy chemotypes that either accumulate or lack noscapine was used to identify CYP candidates potentially involved in phthalideisoquinoline alkaloid metabolism (Fig. 2). The same approach was used to isolate scoulerine 9-O-methyltransferase (SOMT1), canadine synthase (CYP719A21), and noscapine synthase (NOS) (Fig. 1), along with two other O-methyltransferases (SOMT2 and SOMT3) (5, 6, 11). CYP82Y1, CYP82X1, CYP82X2, and CYP719A21 transcripts were detected in 454 pyrosequencing databases of opium poppy chemotypes accumulating noscapine or narcotoline (Fig. 2). These CYPs were also reported as four of a ten-member genomic cluster consisting of biosynthetic genes absent in a noscapine-free opium poppy chemotype (10). The extended correlation between the occurrence of CYP transcripts corresponding to members of the ten-gene cluster and noscapine in several different opium poppy chemotypes suggests that the presence or absence of the gene cluster determines the ability to produce phthalideisoquinoline alkaloids.
Three CYP families have been implicated in BIA metabolism (12). Whereas several enzymes in the CYP80 and CYP719 families have been shown to catalyze aromatic hydroxylation and inter- and intramolecular C-O and C-C coupling reactions involved in 1-benzylisoquinoline, protoberberine, aporphine, bisbenzylisoquinoline, and morphinan alkaloid biosynthesis, only two members in the CYP82 family have been functionally characterized: CYP82N4 catalyzes aliphatic hydroxylation at C14 of N-methylcanadine and N-methylstylopine, resulting in tautomerization to allocryptopine and protopine, respectively (20), whereas CYP82N2v2 from E. californica catalyzes aliphatic hydroxylation at C6 of protopine and allocryptopine leading to intramolecular rearrangement to the benzo[c]phenenthridine scaffold (21). CYP82Y1 displays considerable sequence identity with CYP82N4 (53%) and CYP82N2v2 (44%) and, similar to CYP82N4, also accepted only N-methylcanadine and N-methylstylopine as substrates. In contrast, CYP82Y1 and CYP82N4 catalyze regiospecific hydroxylation at C1 and C14, respectively. Also, CYP82N4 converted both N-methylstylopine and N-methylcanadine at similar rates (20), whereas PsCYP82Y1 converted N-methylcanadine with greater efficiency than N-methylstylopine. Alterations in the residues responsible for substrate recognition and alkaloid binding might be attributed to differences in substrate preference and oxidation activity among CYP82 variants. In contrast with the KPIAPXXXPH substrate recognition site motif of enzymes in the CYP719 family, CYP82Y1, CYP82X1, and CYP82X2 contain a distinct YPA(G/S)XXX(E/D)R domain and a conserved Gly-370 residue. However, whereas the residues specific for alkaloid binding in N-methylstylopine 14-hydroxylase (CYP82N4) (20) are Ile and Leu, the corresponding residues in CYP82Y1 are Leu and Ser (6). CYP82N2v2 was relatively promiscuous with protopine alkaloid substrates, including protopine, allocryptopine, 13-oxoprotopine, and corycavine (21). Other substituted quaternary protoberberine alkaloids beyond N-methylcanadine and N-methylstylopine could also serve as substrates for CYP82Y1 and CYP82N4, although such reactions might not be physiologically relevant. A narrow substrate range is common among CYPs involved in the biosynthesis of BIAs (16–19) and other plant-specialized metabolites. For example, CYP82G1 from tobacco (N. tabacum), which catalyzes the final step in the biosynthesis of common homoterpene volatiles, displays substrate specificity for (E,E)-geranyllinalool and its C15 analog (E)-nerolidol (34), and CYP82C2 and CYP82C4 from mouse ear cress (A. thaliana) show substrate specificity for 8-methoxypsoralen (35).
A major unresolved aspect of noscapine biosynthesis concerns the transformation of the protoberberine to the secoberbine backbone as a component in the ultimate formation of the phthalideisoquinoline scaffold. On structural grounds, secoberbine alkaloids, such as macrantaline and macrantoridine, were considered as possible intermediates between a protoberberine precursor, such as scoulerine or 1-methoxyscoulerine, and noscapine (36). An alternative proposal involves the stereospecific N-methylation of scoulerine followed by multistage oxidation to egenine, which is further oxidized to phthalideisoquinoline alkaloids, such as bicuculline (7). A protoberberine, such as scoulerine, has also been hypothesized to potentially undergo N-methylation and C8 oxidation to the secoberbine macrantaldehyde, which could be reduced to macrantaline or oxidized to macrantoridine (7). Theoretically, macrantaldehyde could undergo oxidation to narcotinehemiacetal (37). As such, a hypothetical scheme for noscapine biosynthesis in opium poppy was proposed whereby N-methylcanadine is hydroxylated at C1 and possibly O-methylated to form 1-methoxy-N-methylcanadine (9). Oxidation of 1-methoxy-N-methylcanadine at C8 was seen as the entry point to the formation of a secoberbine intermediate, such as macrantaldehyde. The drawback of this hypothesis is the lack of empirical evidence for the occurrence of secoberbine alkaloids in opium poppy. Most known secoberbine alkaloids have been isolated from different Papaver species such as Papaver pseudo-oriental, Papaver armeiacum, and Papaver fugax (Sariyar 2002). However, the proposed pathway extrapolated from the partial physiological characterization of a putative noscapine biosynthetic gene cluster supports the natural occurrence of secoberbine alkaloids in opium poppy (10). The individual suppression of transcript levels for six of ten clustered genes using VIGS was interpreted to suggest a biosynthetic route involving an initial ring opening (i.e., cleavage of the berberine bridge) on N-methylcanadine catalyzed by either CYP82X1 or CYP82Y1 and yielding a secoberbine intermediate. CYP82X2 was proposed to then catalyze hydroxylation at the C3 position, yielding a derivative of this secoberbine intermediate. The two schemes agree on the involvement of an enzymatic product derived from N-methylcanadine as the entry point to unidentified secoberbine intermediates in noscapine biosynthesis.
High resolution MSn and 1H NMR analyses showed unequivocally that the CYP82Y1 reaction product derived from N-methylcanadine was 1-hydroxy-N-methylcanadine. Since none of the three CYP82 candidates accepted canadine as a substrate, and because neither CYP82X1 nor CYP82X2 acted on N-methylcanadine, the conversion of N-methylcanadine to 1-hydroxyl-N-methylcanadine by CYP82Y1 clearly represents the first committed step in noscapine biosynthesis and appears crucial to further elaboration of further pathway by other enzymes. CYP82X1 and CYP82X2 are likely responsible for cleavage of the N7/C8 bridge and hydroxylation of the C13 position. 1-Hydroxylation of N-methylcanadine distinguishes the biosynthesis of noscapine and narcotoline in opium poppy from the formation of other phthalideisoquinoline alkaloids, such as hydrastine and bicuculline which are found mostly in plants outside of the genus Papaver and lack a C4′ hydroxyl or methoxyl moiety. Should downstream enzymes in opium poppy depend on the C1 hydroxylation of N-methylcanadine in order to catalyze subsequent ring opening and lactone bridge conversions, the biosynthesis of compounds such as hydrastine and bicuculline might involve relatively unrelated genes. It will be interesting to determine whether homologs of CYP82Y1 and genomic clusters of phthalideisoquinoline alkaloid biosynthetic genes occur in plants such as goldenseal (Hydrastis canadensis) and Corydalis chaerophylla, which accumulate hydrastine and bicuculline, respectively. Interestingly, the C4′ methoxyl group contributes to the potent pharmacological properties of noscapine compared with other phthalideisoquinolines.
The VIGS-mediated suppression of CYP82Y1 transcript levels resulted in the increased accumulation of several protoberberine alkaloids including the noscapine biosynthetic intermediates scoulerine, canadine, tetrahydrocolumamine, and N-methylcanadine (Fig. 1). However, the silencing of CYP82Y1 was also associated with a significant reduction in the accumulation of transcripts encoding enzymes operating, putatively in some cases, immediately upstream (SOMT1 and CYP719A21) and downstream (CYP82X1, CYP82X2, SOMT2, and an O-acetyltransferase, AT1) of 1-hydroxy-N-methylcanadine (Fig. 8). Co-suppression of several pathway genes hinders the utility of VIGS as a functional genomics tool to validate the physiological role of CYP82Y1 in noscapine metabolism. Nevertheless, the co-silencing of only clustered genes suggests some form of coordinated regulation, although a mechanism whereby post-transcriptional suppression of CYP82Y1 would affect the expression of other genes, many with entirely unrelated nucleotide sequences, is not known. Frequent co-expression of biosynthetic genes involved in the same metabolic pathway has been proposed as a factor in co-suppression phenomena (38). A negative correlation between intergenic distance and co-expression has also been suggested (39).
The concomitant accumulation of the structurally related protoberberine tetrahydropalmatine, which is not a noscapine biosynthetic intermediate, supports a divergence in alkaloid pathway flux when a bottleneck is introduced at (S)-N-methylcanadine. Interestingly, the expression of biosynthetic genes outside of the gene cluster (6OMT, 4′OMT2, BBE, and TNMT) and two clustered genes encoding enzymes operating at the end of the noscapine pathway (CXE1 and NOS) were not affected by the silencing of CYP82Y1. 6OMT (norcoclaurine 6-O-methyltransferase) and 4′OMT2 (3′-hydroxy-N-methylcoclaurine 4′-O-methyltransferase) catalyze early conversions in BIA metabolism (12). In contrast, VIGS-mediated silencing of NOS did not affect the expression of other genes, which is perhaps related to the localization of CXE1 and NOS in a different cell type compared with other enzymes encoded by genes in the cluster (11).
Acknowledgments
We thank Dr. Jillian Hagel for performing the LTQ-Orbitrap analysis and Dr. Dae-Kyun Ro for providing the pESC-leu2d vector and the YPL 154C:Pep4 yeast strain.
This work was supported through funding from Genome Canada, Genome Alberta, the Government of Alberta, the Natural Sciences and Engineering Research Council of Canada, and the Canada Foundation for Innovation Leaders Opportunity Fund.
- BIA
- benzylisoquinoline alkaloid
- CID
- collision-induced dissociation
- CPR
- cytochrome P-450 reductase
- CYP
- cytochrome P-450
- ESI
- electrospray ionization
- NOS
- noscapine synthase
- TRV
- tobacco rattle virus
- VIGS
- virus-induced gene silencing
- qRT
- quantitative real time.
REFERENCES
- 1. Anderson T. (1862) On the chemistry of opium. J. Chem. Soc. Perkin Trans. I 15, 446–455 [Google Scholar]
- 2. Battersby A. R., Hirst M., McCaldin D. J., Southgate R., Staunton J. (1968) Alkaloid biosynthesis. XII. The biosynthesis of narcotine. J. Chem. Soc. Perkin Trans. I 17, 2163–2172 [DOI] [PubMed] [Google Scholar]
- 3. Kleinschmidt G., Mothes K. (1959) Physiology and biosynthesis of alkaloids of Papaver sominferum. Z. Naturforsch. 14B, 52–56 [PubMed] [Google Scholar]
- 4. Battersby A. R., Staunton J., Wiltshire H. R., Bircher B. J., Fuganti C. (1975) Studies of enzyme-mediated reactions. Part V. Synthesis of (13S)- and (13R)-[13-3H1]scoulerine from stereospecifically labelled (S)-[α-3Hl] benzyl alcohols. Stereochemistry of enzymic reactions at saturated benzylic carbon. J. Chem. Soc. Perkin Trans. 1, 1162–1171 [Google Scholar]
- 5. Dang T. T., Facchini P. J. (2012) Characterization of three O-methyltransferases involved in noscapine biosynthesis in opium poppy. Plant Physiol. 159, 618–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dang T. T., Facchini P. J. (2014) Cloning and characterization of canadine synthase involved in noscapine biosynthesis in opium. FEBS Lett. 588, 198–204 [DOI] [PubMed] [Google Scholar]
- 7. Sariyar G., Shamma M. (1986) Six alkaloids from Papaver species. Phytochemistry 25, 2403–2406 [Google Scholar]
- 8. Liscombe D. K., Facchini P. J. (2007) Molecular cloning and characterization of tetrahydroprotoberberine cis-N-methyltransferase, an enzyme involved in alkaloid biosynthesis in opium poppy. J. Biol. Chem. 282, 14741–14751 [DOI] [PubMed] [Google Scholar]
- 9. Facchini P. J., Hagel J. M., Liscombe D. K., Loukanina N., MacLeod B. P., Samanani N., Zulak K. G. (2007) Opium poppy. Blueprint for an alkaloid factory. Phytochem. Rev. 6, 97–124 [Google Scholar]
- 10. Winzer T., Gazda V., He Z., Kaminski F., Kern M., Larson T. R., Li Y., Meade F., Teodor R., Vaistij F. E., Walker C., Bowser T. A., Graham I. A. (2012) Papaver somniferum 10-gene cluster for synthesis of the anticancer alkaloid noscapine. Science 336, 1704–1708 [DOI] [PubMed] [Google Scholar]
- 11. Chen X., Facchini P. J. (2014) Short-chain dehydrogenase/reductase catalyzing the final step of noscapine biosynthesis is localized to laticifers in opium poppy. Plant J. 10.1111/tpj.12379 [DOI] [PubMed] [Google Scholar]
- 12. Hagel J. M., Facchini P. J. (2013) Benzylisoquinoline alkaloid metabolism. A century of discovery and a brave new world. Plant Cell Physiol. 54, 647–672 [DOI] [PubMed] [Google Scholar]
- 13. Nguyen D. T., Göpfert J. C., Ikezawa N., MacNevin G., Kathiresan M., Conrad J., Spring O., Ro D. K. (2010) Biochemical conservation and evolution of germacrene A oxidase in Asteraceae. J. Biol. Chem. 285, 16588–16598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kraus P. F., Kutchan T. M. (1995) Molecular cloning and heterologous expression of a cDNA encoding berbamunine synthase, a C-O phenol-coupling cytochrome P450 from the higher plant Berberis stolonifera. Proc. Natl. Acad. Sci. U.S.A. 92, 2071–2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ikezawa N., Iwasa K., Sato F. (2008) Molecular cloning and characterization of CYP80G2, a cytochrome P450 that catalyzes an intramolecular C-C phenol coupling of (S)-reticuline in magnoflorine biosynthesis, from cultured Coptis japonica cells. J. Biol. Chem. 283, 8810–8821 [DOI] [PubMed] [Google Scholar]
- 16. Díaz Chávez M. L., Rolf M., Gesell A., Kutchan T. M. (2011) Characterization of two methylenedioxy bridge-forming cytochrome P450-dependent enzymes of alkaloid formation in the Mexican prickly poppy Argemone mexicana. Arch. Biochem. Biophys. 507, 186–193 [DOI] [PubMed] [Google Scholar]
- 17. Ikezawa N., Tanaka M., Nagayoshi M., Shinkyo R., Sakaki T., Inouye K., Sato F. (2003) Molecular cloning and characterization of CYP719, a methylenedioxy bridge-forming enzyme that belongs to a novel P450 family, from cultured Coptis japonica cells. J. Biol. Chem. 278, 38557–38565 [DOI] [PubMed] [Google Scholar]
- 18. Ikezawa N., Iwasa K., Sato F. (2007) Molecular cloning and characterization of methylenedioxy bridge-forming enzymes involved in stylopine biosynthesis in Eschscholzia californica. FEBS J. 274, 1019–1035 [DOI] [PubMed] [Google Scholar]
- 19. Gesell A., Rolf M., Ziegler J., Díaz Chávez M. L., Huang F.-C., Kutchan T. M. (2009) CYP719B1 is salutaridine synthase, the C-C phenol-coupling enzyme of morphine biosynthesis in opium poppy. J. Biol. Chem. 284, 24432–24442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Beaudoin G. A., Facchini P. J. (2013) Isolation and characterization of a cDNA encoding (S)-cis-N-methylstylopine 14-hydroxylase from opium poppy, a key enzyme in sanguinarine biosynthesis. Biochem. Biophys. Res. Commun. 431, 597–603 [DOI] [PubMed] [Google Scholar]
- 21. Takemura T., Ikezawa N., Iwasa K., Sato F. (2013) Molecular cloning and characterization of a cytochrome P450 in sanguinarine biosynthesis from Eschscholzia californica cells. Phytochemistry 91, 100–108 [DOI] [PubMed] [Google Scholar]
- 22. Desgagné-Penix I., Khan M. F., Schriemer D. C., Cram D., Nowak J., Facchini P. J. (2010) Integration of deep transcriptome and proteome analyses reveals the components of alkaloid metabolism in opium poppy cell cultures. BMC Plant Biol. 10, 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hagel J. M., Facchini P. J. (2010) Dioxygenases catalyze the O-demethylation steps of morphine biosynthesis in opium poppy. Nat. Chem. Biol. 6, 273–275 [DOI] [PubMed] [Google Scholar]
- 24. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 25. Ro D.-K., Ouellet M., Paradise E. M., Burd H., Eng D., Paddon C. J., Newman J. D., Keasling J. D. (2008) Induction of multiple pleiotropic drug resistance genes in yeast engineered to produce an increased level of anti-malarial drug precursor, artemisinic acid. BMC Biotechnol. 8, 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pauli H. H., Kutchan T. M. (1998) Molecular cloning and functional heterologous expression of two alleles encoding (S)-N-methylcoclaurine 3′-hydroxylase (CYP80B1), a new methyl jasmonate-inducible cytochrome P450-dependent mono-oxygenase of benzylisoquinoline alkaloid biosynthesis. Plant J. 13, 793–801 [DOI] [PubMed] [Google Scholar]
- 27. Pompon D., Louerat B., Bronine A., Urban P. (1996) Yeast expression of animal and plant P450s in optimized redox environments. Methods Enzymol. 272, 51–64 [DOI] [PubMed] [Google Scholar]
- 28. Dinesh-Kumar S. P., Anandalakshmi R., Marathe R., Schiff M., Liu Y. (2003) Virus-induced gene silencing. Methods Mol. Biol. 236, 287–294 [DOI] [PubMed] [Google Scholar]
- 29. Hileman L. C., Drea S., Martino G., Litt A., Irish V. F. (2005) Virus-induced gene silencing is an effective tool for assaying gene function in the basal eudicot species Papaver somniferum (opium poppy). Plant J. 44, 334–341 [DOI] [PubMed] [Google Scholar]
- 30. Desgagné-Penix I., Farrow S. C., Cram D., Nowak J., Facchini P. J. (2012) Integration of deep transcript and targeted metabolite profiling of eight cultivars of opium poppy. Plant Mol. Biol. 79, 295–313 [DOI] [PubMed] [Google Scholar]
- 31. Binutu O. A., Cordell G. A. (2000) Constituents of Zanthoxylum sprucei. Pharm. Biol. 38, 210–213 [DOI] [PubMed] [Google Scholar]
- 32. Lee J. K., Cho J. G., Song M. C, Yoo J. S, Lee D. Y, Yang H. J., Han K. M, Kim D. H, Oh Y. J, Jeong T. S, Baek N. I. (2009) Isolation of isoquinoline alkaloids from tuber of Corydalis turtschaninovii and their inhibition activity on low density lipoprotein oxidation. J. Korean Soc. Appl. Biol. Chem. 52, 646–654 [Google Scholar]
- 33. Le P. M, McCooeye M., Windust A. (2013) Characterization of the alkaloids in goldenseal (Hydrastis canadensis) root by high resolution Orbitrap LC-MS(n). Anal. Bioanal. Chem. 405, 4487–4498 [DOI] [PubMed] [Google Scholar]
- 34. Lee S., Badieyan S., Bevan D. R., Herde M., Gatz C., Tholl D. (2010) Herbivore-induced and floral homoterpene volatiles are biosynthesized by a single P450 enzyme (CYP82G1) in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 107, 21205–21210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kruse T., Ho K., Yoo H.-D., Johnson T., Hippely M., Park J.-H., Flavell R., Bobzin S. (2008) In planta biocatalysis screen of P450s identifies 8-methoxypsoralen as a substrate for the CYP82C subfamily, yielding original chemical structures. Chem. Biol. 15, 149–156 [DOI] [PubMed] [Google Scholar]
- 36. Sariyar G., Phillipson D. (1977) Macrantaline and macrantoridine, new alkaloids from a Turkish sample of Papaver pseudo-orientale. Phytochemistry 16, 2009–2013 [Google Scholar]
- 37. Sariyar G. (2002) Biodiversity in the alkaloids of Turkish Papaver species. Pure Appl. Chem. 74, 557–574 [Google Scholar]
- 38. Williams E. J., Bowles D. J. (2004) Coexpression of neighboring genes in the genome of Arabidopsis thaliana. Genome Res. 14, 1060–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Michalak P. (2008) Coexpression, coregulation, and cofunctionality of neighboring genes in eukaryotic genomes. Genomics 91, 243–248 [DOI] [PubMed] [Google Scholar]