Background: Smads require transcriptional cofactors to regulate transcription of transforming growth factor (TGF)-β target genes.
Results: ZNF451 binds to both Smads and p300, and disrupts the Smad-p300 interaction required for TGF-β-induced transcription.
Conclusion: ZNF451 physically interacts with Smads and functionally represses TGF-β signaling.
Significance: Identification and characterization of ZNF451 add a new regulatory component in TGF-β signaling.
Keywords: Signal Transduction, SMAD Transcription Factor, Transcription Repressor, Transforming Growth Factor β (TGFβ), Zinc Finger
Abstract
ZNF451 is a transcriptional cofactor localized to promyelocytic leukemia bodies. Here, we present evidence demonstrating that ZNF451 physically interacts with Smad3/4 and functionally inhibits TGF-β signaling. Increased expression of ZNF451 attenuates TGF-β-induced growth inhibitory and gene transcriptional responses, whereas depletion of ZNF451 enhances TGF-β responses. Mechanistically, ZNF451 blocks the ability of Smad3/4 to recruit p300 in response to TGF-β, which causes reduction of histone H3K9 acetylation on the promoters of TGF-β target genes. Taken together, ZNF451 acts as a transcriptional corepressor for Smad3/4 and negatively regulates TGF-β signaling.
Introduction
The transforming growth factor β (TGF-β) superfamily comprises TGF-βs, bone morphogenetic proteins (BMPs),2 Activins, and related proteins (1–4). Members of the TGF-β superfamily control diverse developmental processes and the pathogenesis of many diseases through direct transcriptional regulation (5). TGF-β and related proteins signal through heteromeric serine-threonine kinase receptors at the cell surface and intracellular effectors called Smads (1–4). There are five receptor-associated Smads (R-Smads) that can be recruited and phosphorylated by type I receptor kinases (1–4). Among these, Smad2 and Smad3 transduce signals from TGF-β and Activin, whereas Smad1, Smad5, and Smad8 transduce signals from BMPs (1–4). The activated R-Smads form heterocomplexes with Smad4 (co-Smad) and are transported to the nucleus. As transcriptional factors, Smads orchestrate the transcription of specific target genes through interactions with specific DNA response elements, sequence-specific DNA-binding transcription factors, and non-DNA-binding transcription cofactors (coactivators or corepressors) (1, 2). Transcription cofactors act to modify local chromatin structure and/or engage the basal transcription machinery. Among all the known transcription co-factors involved in TGF-β signaling, cAMP-responsive element-binding protein (CREB)-binding protein (CBP) and the structurally closely related p300 have been defined as two of the most important transcriptional co-activators (6–8). CBP/p300 possesses intrinsic histone acetyltransferase activity, thereby generating a “loose” state of chromatin structure that is required for gene transcription (6–8). Most corepressors involved in TGF-β signaling often interfere with the integrity of the Smads complex, disrupt Smad-coactivator interactions, or directly modifies chromatin; examples of corepressors include c-Ski (9, 10), Bcl6 (11), c-Myc (12, 13), and Evi-1 (14, 15). Viral oncoprotein E1A also represses transcriptional activation dependent on its ability to bind and sequester CBP/p300 (16, 17).
In an effort to understand the molecular mechanisms underlying Smad transcriptional regulation of TGF-β target gene expression, we performed a yeast two-hybrid screen to search for Smad co-factors and identified ZNF451 as a novel Smad4-binding protein. ZNF451 has previously been reported to localize in promyelocytic leukemia bodies in a SUMO-modified form, and SENP1-mediated de-sumoylation can disrupt its subnuclear localization (18). In this study, we validated the physical interaction between Smad3/4 and ZNF451 in cells and in vitro, and demonstrated that ZNF451 down-regulates TGF-β signaling by blocking formation of the p300-Smad3/4 complex and the recruitment of p300 to the promoter of TGF-β target genes. As a result, the acetylation at lysine 9 residue on histone H3 was reduced, which leads to a chromatin conformation favoring transcription repression. Furthermore, mutations at the sumoylated site Lys-706 or SUMO-interacting motif (SIM) in ZNF451 did not affect the inhibitory function of ZNF451, suggesting that ZNF451 negatively regulates TGF-β signaling in a p300-dependent and sumoylation-independent manner.
EXPERIMENTAL PROCEDURES
Plasmids and Antibodies
HA- or FLAG-tagged Smads, FLAG-tagged constitutively active TβRI (T202D), HA-tagged p300, and reporter gene plasmids (Smad-binding element (SBE)-Luc, plasminogen activator inhibitor type 1 (PAI-1)-Luc, and pFR-Luc) were described previously (11, 19). Full-length ZNF451 cDNA was amplified by PCR from a HeLa cell cDNA library and subcloned into pXF6F vector. The deletion/point mutants were generated by PCR. The antibodies were purchased as follows: anti-ZNF451 (number SAB2102880), anti-FLAG (number F3165), anti-β-actin (number A5441), and anti-γ-tubulin (number T5326) from Sigma; anti-Myc (number sc-40), anti-Smad4 (number sc-7966), and anti-PAI-1 (number sc-5297) from Santa Cruz; anti-HA (number 3724), anti-p21 (number 2947), anti-phospho-Smad2 (number 3108), anti-phospho-Smad3 (number 9520), and anti-Smad2/3 (number 8685) from Cell Signaling Technology; anti-p15 (number C0287) antibody from Assaybiotech; anti-His (number 04 905 318 001) antibody was from Roche Applied Science; Alexa Fluor 488 donkey anti-mouse, rabbit IgG (numbers A21202 and A21206), and Alexa Fluor 546 donkey anti-mouse and rabbit IgG (numbers A10036 and A10040) from Invitrogen; peroxidase-conjugated goat anti-rabbit and rabbit anti-mouse IgG (number 315–035-048) from Jackson ImmunoResearch.
Cell Line and Transfection
Human HEK293T, A549, and HaCaT were grown as described previously (19). A549 and HaCaT cells were transfected with X-tremeGENE (Roche Applied Science) and HEK293T cells were transfected with PEI (Polyscience).
RNA Interference
Small interfering RNA (siRNA) targeting human ZNF451 was transfected into cells for 24 h using Lipofectamine® RNAiMAX Reagent (Invitrogen). siRNAs were made by the RiboBio Co. (number 1 target sequence: nucleotides 1143–1161 of coding region, GCAGAATTCAGGACACAAA; number 2 target sequence: nucleotides 1615–1633 of coding region, GAGTTAACAAGGAAAGATA).
Transcription Reporter Assay
Eighteen hours after transfection, cells were treated with TGF-β (2 ng/ml, 8 h) as described (19). Cells were then harvested and analyzed with the Dual Luciferase Reporter Assay system (Promega). All assays were done in triplicates and all values normalized for transfection efficiency against Renilla luciferase activities.
Immunoprecipitation and Western Blotting Analysis
Twenty-four hours after transfection, cell lysates were harvested by NET lysis buffer (150 mm NaCl, 20 mm Tris-HCl, pH 7.4, 5 mm EDTA, and 0.5% Nonidet P-40) and incubated with protein A-Sepharose CL-4B (GE Healthcare) and appropriate antibodies for 4 h. After extensive washes, immunoprecipitated proteins were eluted in SDS sample loading buffer, separated by SDS-PAGE, transferred onto PVDF membranes (Millipore), and detected by Western blotting analysis (19).
In Vitro Translation and GST Pull-down Assay
In vitro translation was performed with the TnT® Quick Coupled Transcription/Translation System (Promega). Proteins fused with GST in pGEX vector were expressed in Escherichia coli BL21(DE3) strain and purified according to the manufacturer's instructions. GST pull-down experiments were carried out as previously described (19).
DNA Pull-down Assay
Cells were lysated in DNAP-lysis buffer (50 mm NaCl, 25 mm Tris-HCl, pH 7.4, and 0.5% Triton X-100), then incubated with 1 μmol of biotinylated SBE oligonucleotides in DNAP binding buffer (50 mm KCl, 10 mm Tris-HCl, pH 7.4, and 5% glycerol) at 24 °C for 30 min. DNA-protein complexes were collected by streptavidin beads (GE Healthcare) for 15 min, washed extensively with the binding buffer, and identified by Western blotting analysis.
Real-time RT-PCR (qRT-PCR)
Total RNAs were obtained by TRIzol reagent (Invitrogen) (19). RNAs were reverse-transcribed to complementary DNA using the PrimeScript® RT reagent kit (TaKaRa). Quantitative reverse transcriptase (qRT)-PCR was performed on an ABI PRISM 7500 Sequence Detector System (Applied Biosystems) using gene-specific primers for p15, p21, PAI-1, and ZNF451, and SYBR Green Master Mix (Applied Biosystems). Primers used for each gene are listed as follows: 5′-AAGCTGAGCCCAGGTCTCCTA-3′ and 5′-CCACCGTTGGCCGTAAACT-3′ for human p15, 5′-ACCATGTGGACCTGTCACTGT-3′ and 5′-TTAGGGCTTCCTCTTGGAGAA-3′ for human p21, 5′-GTGTTTCAGCAGGTGGCGC-3′ and 5′-CCGGAACAGCCTGAAGAAGTG-3′ for human PAI-1, 5′-AATGCCAGGACTCAAAACAGGCA-3′ and 5′-GAGAGTGATCGAACCTTTTCAAA-3′ for human ZNF451, 5′-CAAAGTTCACAATGTGGCCGAGGA-3′ and 5′-GGGACTTCCTGTAACAACGCATCT-3′ for human β-actin.
Lentivirus Production and Stable Cell Line Generation
ZNF451 and mutant cDNAs were subcloned into pWPI-puro vector and transfected into HEK293T cells together with packaging plasmid psPAX2 and envelope plasmid pMD2.G. After 48 h, lentiviral supernatants were collected and infected onto host cells. Stable cells were selected in the presence of 1 ng/ml of puromycin (Sigma) (19).
Immunofluorescence
HaCaT cells were grown on coverslips and transfected with X-tremeGENE. After 18 h, cells were fixed with 4% paraformaldehyde for 15 min, followed by 0.5% Triton X-100 treatment for 10 min, and blocked in 5% BSA. Cells were then probed with appropriate primary antibodies, followed by Alexa Fluor 488 or Alexa Fluor 546 secondary antibodies (Invitrogen). Fluorescence images were captured by Zeiss Axiovert 200M microscope (Carl Zeiss).
Cell Proliferation Assays
Cell proliferation was measured using bromodeoxyuridine (BrdU) ELISA kit (Roche Applied Science) according to the manufacturer's instructions. Briefly, cells cultured in 96-well culture plates (5000 cells per well) were treated for 4 h with BrdU labeling buffer. After fixation and denaturation, cells were labeled by anti-BrdU antibody and incubated with a colorimetric substrate. All assays were done in triplicates and BrdU incorporation was determined with a luminometer (Mikrolumat CB 96P; Berthold, Germany) using MikroWinTM software (Mikrotek, Germany).
Chromatin Immunoprecipitation (ChIP) Assay
HaCaT cells were treated with TGF-β for 4 h. Cells were cross-linked with 1% formaldehyde at 37 °C for 10 min and then quenched with 0.125 m glycine for 5 min at room temperature. Cell pellets were suspended in 1 ml of SDS lysis buffer. After sonication, 2 μg of anti-acetyl-H3K9 (number 9649, Cell Signaling Technology) or anti-rabbit IgG was used for each ChIP sample. After extensive washes, the immunoprecipitated protein-DNA complex was amplified by qPCR, and the amplification product was expressed as a percentage of the input for each condition. ChIP primers for the human p15 promoter were: forward primer, 5′-CTGCCTGGGGATGAATTTAAC-3′ and reverse primer, 5′-GGTTTCACTGTGGAGACGTTG-3′; ChIP primers for the human p21 promoter were: forward primer, 5′-TGTGTCCTCCTGGAGAGTGC-3′ and reverse primer, 5′-CAGTCCCTCGCCTGCGTTG-3′.
RESULTS
ZNF451 Is a Smad4-binding Protein
To identify cofactors of Smad4, we performed a yeast two-hybrid screen for Smad4-interacting proteins with a human placenta cDNA library (data not shown). A multi-C2H2-type zinc finger protein ZNF451 (ZNF451) was identified as a potential Smad4-binding protein for further characterization. There are three isoforms of ZNF451 present in mammalian cells generated by alternative splicing (18). The longest isoform encodes a 121-kDa protein of 1061 amino acids (aa), which can be divided into a coiled-coil domain, 11 Krüppel-like C2H2-type zinc fingers, an SIM, and a ubiquitin-interacting motif (see Fig. 2A) (18).
FIGURE 2.
Mapping the interacting regions of ZNF451 and Smad4. A, diagram of various ZNF451 and Smad4 deletion mutants used for ZNF451-Smad4 interaction experiments. The start and end amino acid residues for each fragment are indicated. “−” marks a lack of detectable interaction, whereas “+” marks a detectable interaction. B, Smad4 binds to the N-terminal zinc figure region (aa 168–525) of ZNF451 in vivo. FLAG-ZNF451 mutants and HA-Smad4 were co-transfected into HEK293T cells. Smad4-bound ZNF451 mutant was immunoprecipitated by anti-HA antibody (α-HA) and detected by Western blotting (WB) with anti-FLAG antibody (α-FLAG). C, Smad4 interacts directly with ZNF451 (aa 168–525). Purified GST-Smad4 was prebound to GST beads and mixed with in vitro synthesized (IVT) FLAG-ZNF451 truncations. Smad4-bound ZNF451 were eluted by SDS loading buffer and detected by Western blotting. GST-Smad4 input was detected by Ponceau S staining. D, the MH2 domain of Smad4 mediates the Smad4-ZNF451 interaction in vivo. E, the MH2 domain of Smad4 mediates Smad4-ZNF451 interaction in vitro.
To confirm the interaction between ZNF451 and Smad4 in mammalian cells, we examined whether ZNF451 bound to Smad4 in protein interaction assays. Using cell lysates from 293T cells transfected with expression plasmids for HA-tagged Smad4 and FLAG-tagged ZNF451, we carried out anti-FLAG immunoprecipitation (IP). We detected HA-Smad4 from anti-FLAG-ZNF451 immunoprecipitates (Fig. 1A). To further investigate the specificity and physiological relevance of the ZNF451-Smad4 interaction, we sought to perform co-IP experiments with endogenous levels of interacting proteins. In the absence of good ZNF451 antibodies, we first established stable expression of FLAG-ZNF451 in lung epithelial A549 cells (data not shown). We then examined the association between stably expressed ZNF451 and endogenous Smad4 and found that FLAG-ZNF451 was detected in the immunoprecipitates of Smad4, but not in that of IgG (Fig. 1B, lane 2). We next determined if TGF-β regulates the ZNF451 and Smad4 interaction. As also shown in Fig. 1B (lane 3), TGF-β (2 ng/ml) had no effect on the ZNF451-Smad4 association. In addition, we found that ZNF451 interacted with Smad2 and Smad3, two R-Smads that transduce TGF-β signals, but not with BMP-activated Smad1 (data not shown).
FIGURE 1.

ZNF451 interacts with Smad4. A, ZNF451 interacts with Smad4 in vivo. FLAG-ZNF451 and HA-Smad4 were co-transfected into HEK293T cells. The cell lysates were harvested and IP with anti-FLAG antibody (α-FLAG). Smad4 and ZNF451 were detected from the immunoprecipitates by Western blotting (WB) with appropriate antibodies as indicated. B, the association between ZNF451 and Smad4 is not induced or enhanced by TGF-β treatment. ZNF451 overexpressed A549 stable cells were harvested after TGF-β (2 ng/ml) treatment (2 h), and then immunoprecipitated by anti-IgG antibody (α-IgG) or anti-Smad4 antibody (α-Smad4). ZNF451 and Smad4 were detected by Western blotting with the indicated antibodies. C, ZNF451 interacts with Smad4 in vitro. GST-Smad4 was expressed and purified from the E. coli BL21(DE3) strain and incubated with in vitro synthesized FLAG-ZNF451 (IVT). After an extensive wash, GST-Smad4-bound ZNF451 was resolved by SDS-PAGE and detected by Western blotting. GST-Smad4 input was detected by Ponceau S staining.
To evaluate whether the ZNF451-Smad4 interaction is direct, we conducted an in vitro interaction assay where only recombinant proteins were used. Smad4 was expressed and purified from bacteria as a glutathione S-transferase (GST) fusion protein, whereas ZNF451 was obtained from in vitro coupled transcription/translation in rabbit reticulocyte lysate. As seen in Fig. 1C, in vitro synthesized ZNF451 was pulled down by GST-fused Smad4, but not GST alone, indicating that ZNF451 directly interacts with Smad4. Taken together, the data in Fig. 1 suggest that ZNF451 directly interacts with Smad4 under physiological conditions.
The N-terminal Zinc Finger Region of ZNF451 Binds to the Smad4 MH2 Domain
ZNF451 contains 11 zinc finger motifs distributed between aa 168 and 813 flanked by SIM and a ubiquitin-interacting motif. Zinc fingers 6–9 are prerequisite for its self-oligomerization (18). To determine the structural features for ZNF451 binding to Smad4, we mapped the region in ZNF451 that mediates the ZNF451-Smad4 interaction using a series of ZNF451 truncation mutants (Fig. 2A). The truncated ZNF451 (aa 168–813) and ZNF451 (aa 168–525) mutants were pulled down by HA-Smad4 in vivo (Fig. 2B) and GST-Smad4 in vitro (Fig. 2C). The region of aa 526–813 (self-oligomerization domain) did not bind to Smad4 (Fig. 2, B, lane 5, and C, lane 5). These results provide strong evidence showing that the region of aa 168–525 in ZNF451, which contains five Krüppel-like C2H2-type zinc fingers in the N terminus, is the segment for Smad4 binding (Fig. 2A). This aa 168–525 region in ZNF451, but not other mutants, contained a NLS motif and was localized in the nucleus (data not shown), suggesting that the ZNF451-Smad4 interaction might occur in the nucleus.
We also mapped the regions of Smad4 (Fig. 2A) that could bind to ZNF451, and found that the MH2 domain in Smad4 mediated the ZNF451-Smad4 interaction in vivo (Fig. 2D) and in vitro (Fig. 2E). In comparison, Smad4 (aa 1–322) and Smad4 (aa 143–322) did not interact with ZNF451 (Fig. 2D). Taken together with results in Fig. 2, B and C, these data strongly indicate that the N-terminal zinc finger region of ZNF451 preferentially recognizes the MH2 domain of Smad4 (Fig. 2, A and B).
ZNF451 Inhibits TGF-β-induced Signaling
Having established that ZNF451 directly interacts with Smad4, we then examined the effects of stably expressed ZNF451 on endogenous TGF-β-induced, Smad-dependent transcriptional responses. HaCaT and A549 cells stably expressing ZNF451 were generated by the lentivirus system (Fig. 3A). The expression level of exogenous ZNF451 in HaCaT cells was comparable with that of endogenous ZNF451, as assessed by qRT-PCR (Fig. 3A). The effect of ZNF451 on TGF-β-induced transcriptional activity was first determined using the SBE-Luc reporter assay. As shown in Fig. 3B, ZNF451 apparently inhibited TGF-β-induced SBE-Luc expression. In addition to its potent growth-inhibitory response, TGF-β strongly stimulates extracellular matrix production by inducing expression of genes such as PAI-1 through a Smad-dependent mechanism (20–22). Consistent with the effect on SBE-Luc, ZNF451 showed similar inhibitory effects on the promoter activity of PAI-1 (Fig. 3C). Because TGF-β-induced growth arrest is partly mediated through up-regulation of the CDK inhibitors (CKI) p15Ink4b (23) and p21Cip1 (24), we examined the effect of ZNF451 on expression of these CDK inhibitors. In HaCaT cells, TGF-β treatment induced rapid accumulation of p15Ink4b (Fig. 3D), p21Cip1 (Fig. 3E), and PAI-1 (Fig. 3F) mRNAs. Overexpression of ZNF451 significantly attenuated these accumulations (Fig. 3, D–F). A similar result could be observed in PAI-1 mRNA accumulation in A549 cells (data not shown). Consistently, TGF-β-induced p21Cip1 and PAI-1 protein expression was inhibited in HaCaT cells stably expressing ZNF451 (Fig. 3G). A similar result could also be found in the PAI-1 protein level in A549 cells (data not shown). TGF-β potently inhibits cell cycle progression at the G1 phase. Thus, we investigated whether ZNF451 affected the ability of cells to undergo growth inhibition in response to TGF-β. We found ZNF451 could lead to HaCaT cell resistance to TGF-β-induced cell growth inhibition (Fig. 3H). These results collectively showed a specific inverse correlation between ZNF451 and TGF-β signaling.
FIGURE 3.
ZNF451 inhibits TGF-β-induced signaling responses. A, stable expression of FLAG-ZNF451 in HaCaT cells. The mRNA level of ZNF451 was detected by using qRT-PCR, and the protein level by Western blotting (inset). Note that the mRNA level of exogenous FLAG-ZNF451 is similar to that of endogenous ZNF451. B and C, ZNF451 inhibits TGF-β-induced reporter gene activity. ZNF451-expressing HaCaT stable cells were transfected with SBE-Luc (B) or PAI-1-Luc (C) reporter. Relative luciferase activity was measured after an 8-h TGF-β treatment (2 ng/ml). Values are the mean ± S.E. of three separate experiments performed in triplicates and normalized for transfection efficiency against Renilla luciferase activities. D–F, ZNF451 blocks TGF-β-induced target gene transcription. ZNF451-expressing HaCaT stable cells were stimulated with 2 ng/ml of TGF-β for 8 h. Total mRNAs were analyzed by qRT-PCR using primers specific to p15 (D), p21 (E), and PAI-1 (F). Values are the mean ± S.E. of three separate experiments performed in triplicates. G, ZNF451 abolishes TGF-β-induced protein expression. ZNF451-expressing HaCaT stable cells (or control cells expressing GFP) were stimulated with 2 ng/ml of TGF-β for 12 h and then harvested for cell lysates. The expression levels of various proteins were examined by immunoblotting with appropriate antibodies as indicated. H, ZNF451 renders cells resistant to TGF-β-induced growth arrest. HaCaT stable cells expressing ZNF451 or control cells expressing GFP were seeded in 96-well plates and treated with or without TGF-β. Cell proliferation was examined using BrdU staining at 2 days, according to the manufacturer's instructions. The number of BrdU-stained cells with TGF-β treatment was scored as the percentage over untreated cells. Values are the mean ± S.E. of three separate experiments performed in triplicates. *, p < 0.05; **, p < 0.01 (Student's t test).
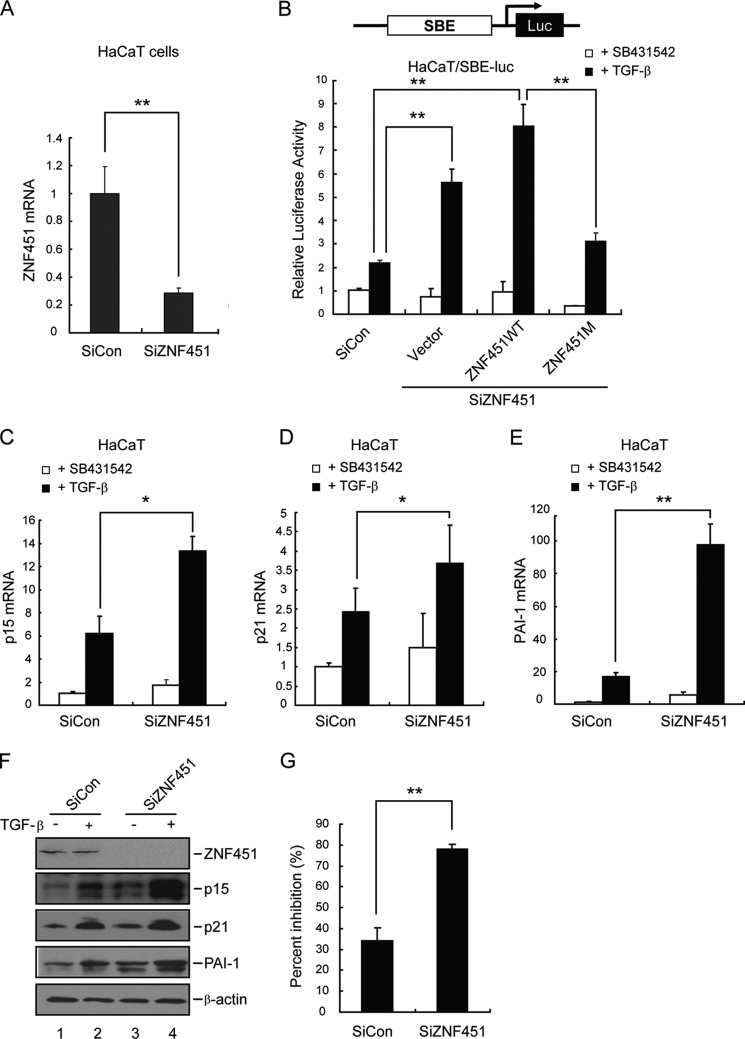
Knockdown of ZNF451 Enhances TGF-β-induced Transcriptional and Growth Inhibitory Responses
To further evaluate the significance of the ZNF451-Smad4 interaction in modulating TGF-β signaling, we sought to examine whether disrupting this interaction would affect TGF-β transcriptional responses. To confirm the physiological function of endogenous ZNF451 on regulating TGF-β signaling, we knocked down ZNF451 expression and investigated the effect of ZNF451 depletion on TGF-β-induced responses. We had made two ZNF451 sequence-specific siRNAs against ZNF451, which could effectively reduce ZNF451 mRNA and protein levels; the knockdown effect of one siRNA was shown in Fig. 4A. Then, we determined if ZNF451 knockdown specifically modulated TGF-β gene responses at the transcriptional level. We found that knockdown of ZNF451 expression resulted in an increase in TGF-β-dependent transcriptional activation in HaCaT (Fig. 4B) and A549 cells (data not shown). Moreover, co-expression of an siRNA-resistant ZNF451 mutant (ZNF451M) could reverse the siZNF451-mediated enhancing effect (Fig. 4B). To further characterize the effect of ZNF451 knockdown on expression of endogenous TGF-β target genes, we examined the levels of p15Ink4b and p21Cip1 mRNAs and proteins in siZNF451 cells. Although TGF-β induced levels of mRNAs of both p15Ink4b (Fig. 4C) and p21Cip1 (Fig. 4D), depletion of ZNF451 apparently increased even higher levels of both p15Ink4b (Fig. 4C) and p21Cip1 (Fig. 4D) mRNAs in HaCaT cells. The PAI-1 mRNA showed the same response to TGF-β and knockdown of ZNF451 (Fig. 4E). As expected, TGF-β induced p15Ink4b and p21Cip1 proteins levels in control HaCaT cells (Fig. 4F); notably, knockdown of ZNF451 profoundly enhanced TGF-β-induced p15Ink4b and p21Cip1 expression, as detected by Western blotting (Fig. 4F). Similarly, TGF-β-induced expression of PAI-1 was also enhanced (Fig. 4F). Furthermore, knockdown of ZNF451 expression rendered cells more sensitive to TGF-β-induced cell growth inhibition, assessed by BrdU staining (Fig. 4G). These data support the notion that depletion of ZNF451 expression enhances TGF-β transcriptional and growth inhibitory responses.
FIGURE 4.
Knockdown of ZNF451 enhances TGF-β signaling. A, ZNF451 expression is effectively reduced after siRNA-mediated knockdown. qRT-PCR analysis using primers specific to ZNF451 indicates about 70% knockdown of the ZNF451 mRNA level (with β-actin as an internal control). B, a siRNA-resistant mutant of ZNF451 (ZNF451M) reverses the knockdown phenotype. HaCaT cells were transfected with siZNF451, and 12 h later, ZNF451 were cotransfected with SBE-Luc, and ZNF451 plasmids (siRNA-resistant ZNF451 ZNF451M or ZNF451WT). Luciferase activity was measured after TGF-β treatment (2 ng/ml for 8 h). C–E, depletion of ZNF451 increases TGF-β-induced target gene transcription. HaCaT cells transfected with control or siZNF451 were stimulated with 2 ng/ml of TGF-β for 8 h. Total mRNAs were then isolated and analyzed by qRT-PCR using primers specific to p15, p21, and PAI-1. F, ZNF451 knockdown increases the TGF-β-targeted protein level. Control or ZNF451 knockdown HaCaT cells were stimulated with 2 ng/ml of TGF-β for 12 h. The expression levels of various proteins were examined by Western blotting with appropriate antibodies as indicated. G, depletion of ZNF451 enhances the sensitivity of cells to TGF-β-induced growth arrest. HaCaT cells transfected with the indicated siZNF451 were seeded in 96-well plates and treated with or without 2 ng/ml of TGF-β. Cell proliferation was examined and scored as described in the legend to Fig. 3H. Values are the mean ± S.E. of three separate experiments performed in triplicates. *, p < 0.05; **, p < 0.01 (Student's t test).
ZNF451 Inhibits the Transcriptional Action of Smad4
To elucidate the molecular mechanisms underlying ZNF451-mediated repression of TGF-β signaling, we tested whether ZNF451 could affect Smad complex formation, its nuclear localization, and presence on the target gene promoters.
We first tested the effect of ZNF451 on Smad complex formation. In HEK293T cells transfected with various Smads, we found that ZNF451 did not alter the ability of Smad4 to form a ligand-induced complex with either Smad2 (Fig. 5A) or Smad3 (Fig. 5B). Likewise, ZNF451 did not interfere with the complex formation between endogenous Smad4 and Smad2/3 (Fig. 5C). In addition, in A549 cells, stably expressed ZNF451 had no effect on TGF-β-induced phosphorylation of endogenous Smad2/3 (Fig. 5D) or Smad4 nuclear accumulation (Fig. 5E). Moreover, ZNF451 did not alter the DNA-binding activity of Smad4 (Fig. 5F).
FIGURE 5.
ZNF451 affects the transcriptional action of Smad4. A and B, ZNF451 has no effect on complex formation between exogenously expressed Smad4 and Smad2/3. HEK293T cells were cotransfected with expression plasmids for HA-Smad4, FLAG-ZNF451, together with Myc-Smad2 (A) or Myc-Smad3 (B) as indicated. Smad4-bound Smad2/3 was immunoprecipitated with anti-Myc antibody (α-Myc) and detected by anti-HA antibody (α-HA) Western blotting. C, ZNF451 has no effect on endogenous complex formation between Smad4 and Smad2/3. HEK293T cells were cotransfected with an expression plasmid for FLAG-ZNF451. The Smad2/3-Smad4 complex formation was detected by IP-coupled Western blotting (WB). D, ZNF451 exhibits no effect on TGF-β-induced Smad2/3 phosphorylation. ZNF451-expressing A549 stable cells (or Control cells expressing GFP) were stimulated with 2 ng/ml of TGF-β for 1 h. Endogenous levels of P-Smad2/3 were detected with appropriate antibodies as indicated. E, ZNF451 does not affect the nuclear accumulation of Smad4. HaCaT cells transfected with FLAG-ZNF451 were subjected to immunofluorescence analysis with appropriate antibodies. Nuclei were visualized by DNA staining with DAPI. F, ZNF451 does not affect the DNA binding ability of Smad4. Biotinylated SBE oligonucleotides were incubated with cell lysates from HEK293T. The DNA-Smad complex was affinity-purified using streptavidin beads, and then examined by Western blotting with anti-FLAG antibody (α-FLAG). G, knockdown of ZNF451 stimulates the transactivation activity of Gal4-Smad4C. HaCaT cells were transfected with siZNF451, Gal4-Smad4C, and pFR-Luc. Relative luciferase assay was performed after treatment with 2 ng/ml of TGF-β for 8 h. H, ZNF451 inhibits the transactivation activity of Gal4-Smad4C. ZNF451-expressing HaCaT stable cells were transfected with Gal4-Smad4C and pFR-Luc. Relative luciferase assay was performed after treatment with 2 ng/ml of TGF-β for 8 h. *, p < 0.05; **, p < 0.01 (Student's t test).
Because ZNF451 resided mainly in the nucleus (data not shown), we tested whether nuclear ZNF451 would repress Smad4-mediated transcriptional activation using the heterologous Gal4 system. As expected, TGF-β stimulation could induce transcription activity of the Gal4-fused Smad4C (without the DNA-binding MH1 domain), which was reflected by expression of the Gal4-driven luciferase reporter FR-Luc (Fig. 5G). Notably, knockdown of ZNF451 dramatically increased transcriptional activity of Gal4-Smad4C (Fig. 5G). In contrast, ectopic expression of ZNF451 suppressed the transcriptional activity of Smad4 (Fig. 5H). Our results identified ZNF451 as a critical corepressor that modulates the transcriptional activity of Smad4 in mammalian cells.
ZNF451 Blocks p300-Smad3/4 Complex Formation
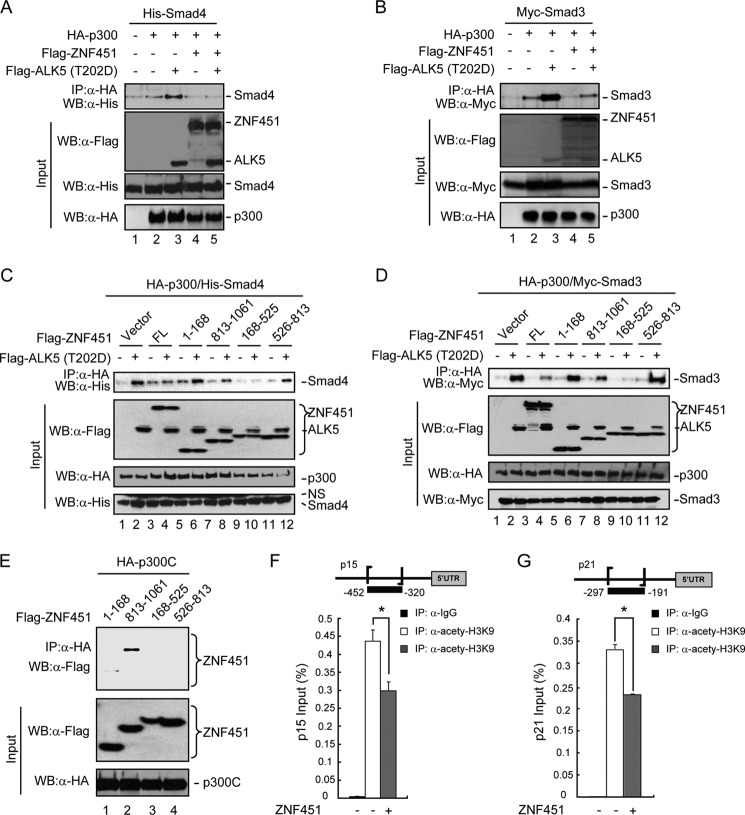
Transcription coactivators increase gene activation by bringing the sequence-specific transcription factors into physical proximity with the RNA polymerase II complex. Some of these coactivators, such as the closely related coactivators CBP and p300, possess histone acetyltransferase activity to modify the chromatin structure. It has previously been reported that CBP/p300 cooperates with the Smads complex to regulate TGF-β target gene transcription by facilitating the formation of multiple and/or larger Smad activator complex and stabilizing the transcriptional activity of the Smad complex. The interaction between Smads and p300 was mediated through the MH2 domain of Smads and C-terminal region of p300 (aa 1572–2378) (8). To further characterize the mechanism of the inhibitory effect of ZNF451 on TGF-β signaling, we sought to determine the effects that ZNF451 acted on Smad3/4. We first examined whether ZNF451 affected TGF-β-induced formation of the Smad4/p300 complex. The Smad4-p300 interaction was induced by the constitutively active TGF-β type I receptor, ALK5 (T202D) (Fig. 6A, lane 3). This increased Smad4-p300 interaction was profoundly suppressed by ectopic expression of ZNF451 (Fig. 6A, lane 5). Similarly, the interaction between Smad3 and p300 was attenuated by ZNF451 (Fig. 6B), consistent with the observation that Smad3 also interacts with ZNF451 (data not shown). These results prompted us to further examine which region of ZNF451 could affect the association of p300 with Smad3/4. We found that the near N-terminal region of ZNF451 (aa 168–525), which inhibited TGF-β signaling, exhibited a strong inhibitory effect on the Smad4-p300 interaction induced by ALK5 (T202D) (Fig. 6C). Consistently, ZNF451 (aa 168–525) also profoundly decreased the Smad3-p300 interaction (Fig. 6D). Interestingly, we noticed that the C-terminal region (aa 813–1061) of ZNF451 also dampened the Smad3/4-p300 association, although the efficiency was less than that of ZNF451 (aa 168–525) (Fig. 6, C and D). It has been reported that the C terminus of p300 (p300C) is the Smad3/4 docking site (8, 25). To map the region of ZNF451 binding to p300C, we performed co-IP experiments using the same set of ZNF451 deletion mutants (Fig. 2A). As shown in Fig. 6E, ZNF451 (aa 813–1061), but not the other mutants, could interact strongly with p300C. These results suggest that ZNF451 negatively regulated TGF-β signaling through its binding to either Smads or p300 to interfere with formation of the Smad-p300 complex.
FIGURE 6.
ZNF451 blocks p300-Smad3/4 complex formation. A and B, ZNF451 disrupts the p300-Smad3/4 interaction. HEK293T cells were cotransfected with expression plasmids for HA-p300, FLAG-ZNF451, and FLAG-ALK5 (T202D), together with His-Smad4 (A) or Myc-Smad3 (B) as indicated. Smad3/4-bound p300 was immunoprecipitated with the indicated antibodies and detected by Western blotting with anti-FLAG antibody (α-FLAG). C and D, the aa 168–525 and 813–1061 fragments of ZNF451 inhibit Smad interaction with p300. HEK293T cells were cotransfected with FLAG-ZNF451 full-length and mutants, HA-p300, and His-Smad4 (C) or Myc-Smad3 (D). p300-Smad3/4 complex formation was detected by IP-coupled Western blotting (WB). E, p300C (aa 1572–2378) interacts with ZNF451 (aa 813–1061). HEK293T cells were transfected with the indicated FLAG-ZNF451 mutants and HA-p300C. p300C-bound ZNF451 mutant was immunoprecipitated by anti-HA antibody (α-HA) and detected by Western blotting with anti-FLAG antibody (α-FLAG). F and G, ZNF451 down-regulates the acetylation level on K9 (lysine 9) of histone H3 at the p15 and p21 promoters. HaCaT cells stably expressing ZNF451 or control cells were treated with 2 ng/ml of TGF-β for 4 h. The H3K9 acetylation level at the promoters induced by TGF-β was analyzed by ChIP assay. Isolated chromatins were incubated with anti-IgG (α-IgG) control antibody or anti-acetyl-H3K9 (α-acetyl-H3K9) antibody and the 132-bp fragment at the p15 promoter (F) and the 106-bp fragment at the p21 promoter (G) were amplified and analyzed by qPCR. Values are the mean ± S.E. of three separate experiments performed in triplicates. *, p < 0.05; **, p < 0.01 (Student's t test).
As a histone acetyltransferase for histone H3 and H4, p300 plays a critical role in modifying the chromatin structure during transcriptional activation. Disruption of p300-Smad complex formation may severely compromise the local chromatin acetylation, resulting in gene transcription suppression (26). We next examined whether the ZNF451-p300 interplay acts on the promoter in the context of TGF-β signaling. We analyzed the acetylation level of histone H3 lysine 9 (AcH3K9) at the p15Ink4b and p21Cip1 promoters by chromatin immunoprecipitation (ChIP) assays. Antibody specific to AcH3K9 was used for immunoprecipitation in control cells or cells stably expressing ZNF451. The level of AcH3K9 on the p15Ink4b and p21Cip1 promoters was determined by qPCR with primers specifically recognizing promoter regions of p15Ink4b or p21Cip1. We found that stably expressed ZNF451 could attenuate the TGF-β-induced AcH3K9 level on both p15Ink4b (Fig. 6F) and p21Cip1 promoters (Fig. 6G).
ZNF451 Represses TGF-β Signaling Independently of the Sumoylation Machinery
Because the ZNF451 protein contains an SIM and resides in promyelocytic leukemia bodies in its sumoylated form (18), we were curious if SUMO modification would regulate ZNF451 activity. We generated two ZNF451 mutants in which either SIM (L48A/V49A) or the SUMO acceptor lysine residue (L706R) were mutated, and established stable cell lines expressing these individual mutants (Fig. 7A). The inhibitory effects of these mutants on TGF-β signaling were then assessed. We found that these mutants had similar activities as that of wild-type ZNF451 in blocking the TGF-β-induced SBE-Luc response (Fig. 7B) and transcription activity of Smad4C (Fig. 7C). Furthermore, these mutants attenuated the interaction between Smad3 and p300 to the same extent as wild-type ZNF451 (Fig. 7D). These data suggest that ZNF451 inhibits TGF-β signaling in a manner independent of SUMO modification.
FIGURE 7.
ZNF451 inhibits TGF-β signaling in a sumoylation-independent manner. A, similar expression levels of ZNF451 and variants in A549 stable cells. B, ZNF451 L48A/V49A mutant and the K706R mutant inhibit TGF-β-induced SBE-Luc response as potently as wild-type ZNF451. A549 stable cells expressing ZNF451 or mutants were transfected with the SBE-Luc reporter. Relative luciferase activity was measured after an 8-h TGF-β treatment (2 ng/ml). C, ZNF451 L48A/V49A and K706R mutants attenuate the TGF-β-induced transcription activity of Gal4-Smad4C as potently as wild-type ZNF451. A549 stable cells expressing ZNF451 or mutants were transfected with Gal4-Smad4C and pFR-Luc. Relative luciferase assay was performed after treatment with 2 ng/ml of TGF-β for 8 h. D, ZNF451 wild-type and mutants equally disrupt the p300-Smad3/4 interaction. HEK293T cells were cotransfected with expression plasmids for HA-p300, Myc-Smad3, FLAG-ZNF451, and FLAG-ALK5 (T202D) as indicated. p300-bound Smad3 was immunoprecipitated with anti-HA antibody (α-HA) and detected by Western blotting with anti-Myc antibody (α-Myc). Inputs are shown as indicated. *, p < 0.05; **, p < 0.01 (Student's t test).
DISCUSSION
Smad4 is a key signal transducer in TGF-β induced signaling pathways, and has been identified as a tumor suppressor that is inactivated in ∼50% of pancreatic carcinomas and 15% of colorectal cancers (27, 28). In an effort to further elucidate the functions of Smad4, we identified ZNF451 as a co-factor for Smad4. How ZNF451 functions in cells has not been characterized. We first validated the physical interactions of ZNF451 with Smad4. Our study not only adds ZNF451 as a new member to the Smad cofactor list, but also assigns one function to ZNF451 in cellular response (Fig. 8). ZNF451 also interacts with Smad2/3, but with low affinity. It is possible that ZNF451 binds to each component of the Smad2/3/4 complex. Alternatively, binding to Smad2/3 is indirectly through Smad4. In our functional assays (using both gain-of-function and loss-of-function approaches), ZNF451 apparently inhibits TGF-β-induced transcriptional and growth inhibitory responses. ZNF451 is down-regulated upon TGF-β stimulation (data not shown), suggesting that ZNF451 serves as a gatekeeper to tightly constrain TGF-β signaling under normal conditions. Its down-regulation provides a mechanism whereby the sensitivity to TGF-β signals is enhanced.
FIGURE 8.

A working model for the functions of ZNF451 in TGF-β signaling. A, schematic diagram of the domains of ZNF451 and their functions. Domains for newly assigned Smad-binding and p300-binding are shown. CC, coil-coil domain; ZF, zinc fingers; UIM, ubiquitin-interacting domain. B, a working model of ZNF451 as a Smad corepressor. ZNF451 binds to both Smads and p300, and disrupts the Smad-CBP/p300 interactions, thereby disrupting the coactivator function of CBP/p300 in Smad-mediated transcriptional activation. p300-mediated histone H3 acetylation is indicated.
Because Smad4 is the central player for both the BMP and TGF-β signaling pathways, ZNF451 should also regulate the BMP pathway, and thus may be involved in the control of many aspects of tissue differentiation and development controlled by BMP signaling. It should be pointed out that our finding differs from a previous report showing the positive role of ZNF451 in transcription. Karvenon et al. (18) found that ZNF451 acts as a transcriptional coactivator for the androgen receptor, which also requires SUMO modification of ZNF451. In our study, SUMO modification exhibited no effects on TGF-β signaling. The opposite effects of ZNF451 on TGF-β and androgen receptor signaling suggest a context-dependent role of ZNF451 during regulation of transcription. Therefore, more investigations on ZNF451 and its activities in transcription are needed for better understanding of this under-characterized transcription regulator.
A series of biochemical and cellular assays were carried out to analyze which steps ZNF451 acts on to repress TGF-β signaling. We reason that ZNF451 may affect Smad activation steps or Smad activity as transcriptional factors. Although ZNF451 binds to the MH2 domain of Smad4, which is responsible for its transcriptional activity and Smad oligomerization, ZNF451 did not reduce Smad3-Smad4 complex formation. As a nuclear protein with potential DNA binding C2H2 zinc finger motifs (29, 30), ZNF451 was unable to alter Smad4 DNA binding activity (Fig. 5F), yet it represses the transcriptional activity of Smad4 (Fig. 5, G and H). ZNF451 achieves this repressing effect by binding to either Smad4 or p300, thereby completely blocking TGF-β-induced formation of the p300-Smad4 complex. It binds to the MH2 domain of Smad4, where p300 also binds (8), and thus competes with p300 for Smad4 binding. Furthermore, ZNF451 also binds to the C-terminal domain of p300 (aa 1572–2378), the docking region for Smad4, further interfering with the Smad4-p300 interaction. These data suggest that ZNF451 blocks the p300-Smad4 interaction not only through its interaction with Smad4 but also through its interaction with p300 (Fig. 8). A similar mechanism to repress TGF-β signaling has been reported with viral oncoprotein E1A, in which E1A interacts with both Smad and p300, leading to a complete inhibition of the TGF-β signaling pathway (16, 17). As a consequence of disrupting the Smad-p300 interaction, ZNF451 leads to loss of histone H3K9 acetylation on the promoters of TGF-β target genes p15Ink4b and p21Cip1, which may explain, at least in part, down-regulation of TGF-β target gene expression.
The growth inhibitory effect of TGF-β in epithelial cells is mostly achieved through the induction of CDK inhibitors such as p15Ink4b and p21Cip1. We found that ZNF451 could inhibit TGF-β-induced expression of p15Ink4b and p21Cip1 and attenuate TGF-β-induced cell growth inhibition, whereas ZNF451 knockdown enhanced TGF-β-induced p15Ink4b and p21Cip1 expression and growth arrest. Because loss of the anti-proliferative response to TGF-β is often correlated with cancer progression at early stages (5) and many Smad corepressors are oncoproteins (9–15), it is conceivable that ZNF451 can possess oncogenic property. Indeed, preliminary analysis of the GEO database implicates higher expression levels of ZNF451 in breast tumor samples (data not shown). Nonetheless, further studies are required to characterize the physiological functions of ZNF451 in tumorigenesis.
Acknowledgments
We thank David Luskutoff for p800(PAI-1)-luc and Bert Vogelstein for SBE-luc. We are grateful to laboratory members for helpful discussion and technical assistance.
This work was supported, in whole or in part, by NSFC Grant 31090360, MOST Grants 2012CB966600 and 2011CB910501, NSFZ Grant Z2110591, Project 985, the Fundamental Research Funds for the Central Universities. National Institutes of Health Grants R01GM63773, R01AR053591, R01CA108454, and R01DK073932.
- BMP
- bone morphogenetic protein
- CBP
- CREB-binding protein
- SIM
- SUMO-interacting motif
- SBE
- Smad-binding element
- IP
- immunoprecipitation
- PAI-1
- plasminogen activator inhibitor type 1
- qPCR
- quantitative PCR
- aa
- amino acid(s)
- Luc
- luciferase.
REFERENCES
- 1. Feng X.-H., Derynck R. (2005) Specificity and versatility in TGF-β signaling through Smads. Annu. Rev. Cell Dev. Biol. 21, 659–693 [DOI] [PubMed] [Google Scholar]
- 2. Massagué J. (2012) TGF-β signalling in context. Nat. Rev. Mol. Cell Biol. 13, 616–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen Y. G., Meng A. M. (2004) Negative regulation of TGF-β signaling in development. Cell Res. 14, 441–449 [DOI] [PubMed] [Google Scholar]
- 4. Watabe T., Miyazono K. (2009) Roles of TGF-β family signaling in stem cell renewal and differentiation. Cell Res. 19, 103–115 [DOI] [PubMed] [Google Scholar]
- 5. Derynck R., Akhurst R. J., Balmain A. (2001) TGF-β signaling in tumor suppression and cancer progression. Nat. Genet. 29, 117–129 [DOI] [PubMed] [Google Scholar]
- 6. Janknecht R., Wells N. J., Hunter T. (1998) TGF-β-stimulated cooperation of Smad proteins with the coactivators CBP/p300. Genes Dev. 12, 2114–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chan H. M., La Thangue N. B. (2001) p300/CBP proteins. HATs for transcriptional bridges and scaffolds. J. Cell Sci. 114, 2363–2373 [DOI] [PubMed] [Google Scholar]
- 8. Feng X.-H., Zhang Y., Wu R. Y., Derynck R. (1998) The tumor suppressor Smad4/DPC4 and transcriptional adaptor CBP/p300 are coactivators for Smad3 in TGF-β-induced transcriptional activation. Genes Dev. 12, 2153–2163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Akiyoshi S., Inoue H., Hanai J., Kusanagi K., Nemoto N., Miyazono K., Kawabata M. (1999) c-Ski acts as a transcriptional co-repressor in transforming growth factor-β signaling through interaction with Smads. J. Biol. Chem. 274, 35269–35277 [DOI] [PubMed] [Google Scholar]
- 10. Luo K., Stroschein S. L., Wang W., Chen D., Martens E., Zhou S., Zhou Q. (1999) The Ski oncoprotein interacts with the Smad proteins to repress TGF-β signaling. Genes Dev. 13, 2196–2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang D., Long J., Dai F., Liang M., Feng X.-H., Lin X. (2008) BCL6 represses Smad signaling in transforming growth factor-β resistance. Cancer Res. 68, 783–789 [DOI] [PubMed] [Google Scholar]
- 12. Feng X.-H., Liang Y. Y., Liang M., Zhai W., Lin X. (2002) Direct interaction of c-Myc with Smad2 and Smad3 to inhibit TGF-β-mediated induction of the CDK inhibitor p15Ink4B. Mol. Cell 9, 133–143 [DOI] [PubMed] [Google Scholar]
- 13. Chen C. R., Kang Y., Massagué J. (2001) Defective repression of c-myc in breast cancer cells. A loss at the core of the transforming growth factor β growth arrest program. Proc. Natl. Acad. Sci. U.S.A. 98, 992–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kurokawa M., Mitani K., Irie K., Matsuyama T., Takahashi T., Chiba S., Yazaki Y., Matsumoto K., Hirai H. (1998) The oncoprotein Evi-1 represses TGF-β signalling by inhibiting Smad3. Nature 394, 92–96 [DOI] [PubMed] [Google Scholar]
- 15. Alliston T., Ko T. C., Cao Y., Liang Y. Y., Feng X.-H., Chang C., Derynck R. (2005) Repression of bone morphogenetic protein and activin-inducible transcription by Evi-1. J. Biol. Chem. 280, 24227–24237 [DOI] [PubMed] [Google Scholar]
- 16. Datto M. B., Hu P. P., Kowalik T. F., Yingling J., Wang X. F. (1997) The viral oncoprotein E1A blocks transforming growth factor β-mediated induction of p21WAF1/Cip1 and p15INK4B. Mol. Cell Biol. 17, 2030–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chakravarti D., Ogryzko V., Kao H. Y., Nash A., Chen H., Nakatani Y., Evans R. M. (1999) A viral mechanism for inhibition of p300 and PCAF acetyltransferase activity. Cell 96, 393–403 [DOI] [PubMed] [Google Scholar]
- 18. Karvonen U., Jääskelainen T., Rytinki M., Kaikkonen S., Palvimo J. J. (2008) ZNF451 is a novel PML body- and SUMO-associated transcriptional coregulator. J. Mol. Biol. 382, 585–600 [DOI] [PubMed] [Google Scholar]
- 19. Dai F., Lin X., Chang C., Feng X.-H. (2009) Nuclear export of Smad2 and Smad3 by RanBP3 facilitates termination of TGF-β signaling. Dev. Cell 16, 345–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Datta P. K., Blake M. C., Moses H. L. (2000) Regulation of plasminogen activator inhibitor-1 expression by transforming growth factor-β-induced physical and functional interactions between Smads and Sp1. J. Biol. Chem. 275, 40014–40019 [DOI] [PubMed] [Google Scholar]
- 21. Dennler S., Itoh S., Vivien D., ten Dijke P., Huet S., Gauthier J. M. (1998) Direct binding of Smad3 and Smad4 to critical TGF-β-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J. 17, 3091–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stroschein S. L., Wang W., Luo K. (1999) Cooperative binding of Smad proteins to two adjacent DNA elements in the plasminogen activator inhibitor-1 promoter mediates transforming growth factor β-induced Smad-dependent transcriptional activation. J. Biol. Chem. 274, 9431–9441 [DOI] [PubMed] [Google Scholar]
- 23. Hannon G. J., Beach D. (1994) p15INK4B is a potential effector of TGF-β-induced cell cycle arrest. Nature 371, 257–261 [DOI] [PubMed] [Google Scholar]
- 24. Datto M. B., Li Y., Panus J. F., Howe D. J., Xiong Y., Wang X. F. (1995) Transforming growth factor β induces the cyclin-dependent kinase inhibitor p21 through a p53-independent mechanism. Proc. Natl. Acad. Sci. U.S.A. 92, 5545–5549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shen X., Hu P. P., Liberati N. T., Datto M. B., Frederick J. P., Wang X. F. (1998) TGF-β-induced phosphorylation of Smad3 regulates its interaction with coactivator p300/CREB-binding protein. Mol. Biol. Cell 9, 3309–3319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kundu T. K., Palhan V. B., Wang Z., An W., Cole P. A., Roeder R. G. (2000) Activator-dependent transcription from chromatin in vitro involving targeted histone acetylation by p300. Mol. Cell 6, 551–561 [DOI] [PubMed] [Google Scholar]
- 27. Tascilar M., Skinner H. G., Rosty C., Sohn T., Wilentz R. E., Offerhaus G. J., Adsay V., Abrams R. A., Cameron J. L., Kern S. E., Yeo C. J., Hruban R. H., Goggins M. (2001) The SMAD4 protein and prognosis of pancreatic ductal adenocarcinoma. Clin. Cancer Res. 7, 4115–4121 [PubMed] [Google Scholar]
- 28. Salovaara R., Roth S., Loukola A., Launonen V., Sistonen P., Avizienyte E., Kristo P., Järvinen H., Souchelnytskyi S., Sarlomo-Rikala M., Aaltonen L. A. (2002) Frequent loss of SMAD4/DPC4 protein in colorectal cancers. Gut. 51, 56–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Laity J. H., Lee B. M., Wright P. E. (2001) Zinc finger proteins. New insights into structural and functional diversity. Curr. Opin. Struct. Biol. 11, 39–46 [DOI] [PubMed] [Google Scholar]
- 30. Beerli R. R., Barbas C. F., 3rd (2002) Engineering polydactyl zinc-finger transcription factors. Nat. Biotechnol. 20, 135–141 [DOI] [PubMed] [Google Scholar]