Background: Actinobacillus pleuropneumoniae N-glycosyltransferase is a cytoplasmic glycosyltransferase catalyzing N-glycosylation of polypeptides.
Results: In depth analysis of a reconstituted A. pleuropneumoniae glycosylation system in Escherichia coli showed a surprisingly relaxed peptide substrate specificity of N-glycosyltransferase.
Conclusion: N-Glycosyltransferase constitutes a general glycosylation system with a preference for autotransporters.
Significance: Our study could provide the basis for a novel route for the engineering of N-glycoproteins in bacteria.
Keywords: Bacteria, Glucose, Glycosylation, Glycosyltransferases, Post-translational Modification, Actinobacillus pleuropneumoniae
Abstract
N-Linked protein glycosylation is a frequent post-translational modification that can be found in all three domains of life. In a canonical, highly conserved pathway, an oligosaccharide is transferred by a membrane-bound oligosaccharyltransferase from a lipid donor to asparagines in the sequon NX(S/T) of secreted polypeptides. The δ-proteobacterium Actinobacillus pleuropneumoniae encodes an unusual pathway for N-linked protein glycosylation. This pathway takes place in the cytoplasm and is mediated by a soluble N-glycosyltransferase (NGT) that uses nucleotide-activated monosaccharides to glycosylate asparagine residues. To characterize the process of cytoplasmic N-glycosylation in more detail, we studied the glycosylation in A. pleuropneumoniae and functionally transferred the glycosylation system to Escherichia coli. N-Linked glucose specific human sera were used for the analysis of the glycosylation process. We identified autotransporter adhesins as the preferred protein substrate of NGT in vivo, and in depth analysis of the modified sites in E. coli revealed a surprisingly relaxed peptide substrate specificity. Although NX(S/T) is the preferred acceptor sequon, we detected glycosylation of alternative sequons, including modification of glutamine and serine residues. We also demonstrate the use of NGT to glycosylate heterologous proteins. Therefore, our study could provide the basis for a novel route for the engineering of N-glycoproteins in bacteria.
Introduction
N-Linked protein glycosylation is a frequent post-translational modification of secretory proteins in eukaryotes, but it occurs in all three domains of life (reviewed in Ref. 1). It entails the formation of an N-glycosidic bond between the amide of an asparagine side chain in polypeptides and a glycan moiety. The classical pathway of N-linked glycosylation follows the same basic scheme in all three domains of life: assembly of an oligosaccharide precursor on an isoprenoid lipid carrier starts in the cytoplasm, and the resulting lipid-linked oligosaccharide is then translocated into the extracytoplasmic space (the endoplasmic reticulum in eukaryotes, the periplasm in bacteria), further elaborated in most eukaryotes, and then transferred en bloc to the asparagine side chain in the consensus sequon NX(S/T) (where X ≠ Pro) by a membrane-bound oligosaccharyltransferase (2). In eukaryotes, N-linked glycans are involved in many important biological functions including but not limited to protein folding and quality control in the endoplasmic reticulum (3, 4), cell-cell recognition (5), immune modulation, and inflammation (6, 7). In total, over half of all eukaryotic proteins are predicted to be N-glycosylated (8).
Recently, a novel type of N-linked protein glycosylation pathway has been described in the Gram-negative bacterium Haemophilus influenzae. Asparagine residues of the adhesin protein HMW1A were found to be modified with hexose and dihexose residues (9, 10). The enzyme responsible for this modification, HMW1C, is a soluble, cytoplasmic protein that uses nucleotide-activated sugars to transfer single glycan moieties to the Asn side chain of proteins (11). Like for the eukaryotic oligosaccharyltransferases, NX(S/T) has been proposed to represent the acceptor sequon in this pathway. HMW1C belongs to the GT41 family of glycosyltransferases and is related to UDP-GlcNAc:peptide N-acetylglucosaminyltransferase (O-GlcNAc transferase) (CAZY, Carbohydrate Active enZYmes database (12)) but is completely unrelated to STT3 type oligosaccharyltransferase that performs the classical N-linked protein glycosylation. Therefore, it constitutes a novel class of N-glycosylation-catalyzing enzymes. Homologous enzymes can be found in various other Pasteurellaceae species as well as Yersinia spp. For the HMW1C homologs of Actinobacillus pleuropneumoniae and Yersinia enterocolitica, N-glycosylation activity has been demonstrated in vitro (13, 14). The A. pleuropneumoniae N-glycosyltransferase (ApNGT)6 is the best studied enzyme of this family. It catalyzes the formation of a β1-N-glycosidic bond between the δ-amino group of asparagine residues and glucose or galactose (14), and the crystal structure of this enzyme has been determined (15). In this study we first analyzed protein glycosylation in A. pleuropneumoniae. Because of the difficulty of genetic manipulation of this organism and the lack of an efficient enrichment procedure for A. pleuropneumoniae glycoproteins, we functionally transferred the glycosylation machinery into Escherichia coli for in-depth analysis of a simplified and readily accessible experimental system. We describe the use of N-linked hexose-specific human antisera for the detection of N-glycoproteins and demonstrate a relaxed peptide substrate specificity of ApNGT in vivo. We further show the glycosylation of heterologous proteins by ApNGT in the cytoplasm of E. coli, establishing a novel bacterial system for the expression of glycoproteins with interesting biotechnological applications.
EXPERIMENTAL PROCEDURES
Construction of Plasmids
All strains, plasmids and primers are shown in supplemental Table S1. For plasmids pMLBAD(His10-AtaC-1866–2428) and pMLBAD(HIS10-COK_1394-62-640), part of the genes encoding AtaC and COK_1394 were amplified by PCR from genomic DNA of A. pleuropneumoniae (strain AP76, locus APP7_0520, 582076–583761) and Mannheimia haemolytica (strain 984, locus ACZY01000043, 11352–13511), respectively, using primers ataC-fw and ataC-rev for AtaC and primers COK_1394-fw and COK_1394-rev for COK_1394. The resulting PCR fragments (1686 and 1979 bp in length) were digested with NcoI/XhoI and NcoI/XbaI, respectively, and ligated into the correspondingly digested pMLBAD vector. The gene encoding ApNGT (APP76_1697) was amplified from plasmid pFLA91 using primers NGTmyc-fw and NGTmyc-rev, thereby replacing the hexahistidine tag for the c-Myc epitope. Digestion with NdeI/EcoRI and ligation into the correspondingly cut vector pEC415 yielded plasmid pEC415-AP1697myc. The cholera toxin B (CtxB) coding sequence without the signal peptide was amplified by PCR from plasmid pMIK14 using primers CtxB-fw and CtxB-rev. Digestion with NdeI/XhoI and ligation into the pET22b vector yielded plasmid pKP04. The sequences of all plasmids were verified by Sanger sequencing (Microsynth AG)
Protein Identification by nanoLC-ESI-MS/MS
Unless stated otherwise, proteins were prepared for mass spectrometric analysis using the filter-aided sample preparation method (16), and the resulting peptides were cleaned-up by C18 ZipTip (Millipore). The analysis was performed on a calibrated LTQ-Orbitrap Velos mass spectrometer (Thermo Fischer Scientific, Bremen, Germany) coupled to an Eksigent-Nano-HPLC system (Eksigent Technologies, Dublin, CA). Peptides were resuspended in 2.5% acetonitrile and 0.1% formic acid, loaded on a self-made tip column (75 μm × 80 mm) packed with C18 material (AQ, 3 μm 200 Å; Bischoff GmbH, Leonberg, Germany), and eluted with a flow rate of 200 nl/min with a gradient from 3 to 30% acetonitrile, 0.1% formic acid in 22 min, 50% acetonitrile, 0.1% formic acid in 25 min, 97% acetonitrile, 0.1% formic acid in 27 min. One scan cycle comprised of a full scan MS survey spectrum, followed by up to 20 sequential CID MS/MS on the most intense signals above a threshold of 1500. Full scan MS spectra (300–2000 m/z) were acquired in the FT-Orbitrap at a resolution of 60,000 at 400 m/z, whereas CID MS/MS spectra were recorded in the linear ion trap (target value 1e4, collision energy 35 V, Q value 0.25, and activation time 30 ms). Auto gain control (ACG) target values were 5e5 for full FTMS scans and 1e4 for ion trap MSn scans. For all experiments, dynamic exclusion was used with one repeat count, 15-s repeat duration, and 60-s exclusion duration.
To confirm hexosylated peptides, further measurements using higher energy collisional dissociation (HCD) and electron transfer dissociation (ETD) were performed in targeted mode with an including list. The AGC target values were 1e5 for HCD and 1e4 for ETD, respectively. MS/MS spectra of higher energy collisional dissociation were acquired in the FT-Orbitrap at a resolution of 15,000 at 400 m/z, whereas ETD MS/MS spectra were recorded in the linear ion trap. The ETD anion target value was set at 1e6, and activation time was set at 100 ms. Supplementary activation was employed to enhance the fragmentation efficiency for 2+ precursors, and charge state-dependent ETD time was enabled. The ETD reaction time was 120 ms, and isolation width was 2 m/z.
Database Analysis and Identification of Modified Residues
MS and MS/MS data were searched against a database containing the A. pleuropneumoniae, M. hemolytica, H. influenzae, E. coli, and Saccharomyces cerevisiae sequences extracted from UniProtKB database version 201208 (217,946 sequences in total) through Mascot engine (version 2.2) with the consideration of carbamidomethylation at Cys, oxidation at Met, and N-hexosylation at Asn, Gln, Ser, or Thr. The monoisotopic masses of 2+ or more charged peptides were searched with a peptide tolerance of 10 ppm and a MS/MS tolerance of 0.6 Da for fragment ions. The MS/MS validation was performed by Scaffold 3.6.2 (Proteome Software, Inc.) as well as manual inspection of spectra.
Analysis of Glycosylated Proteins in A. pleuropneumoniae
Strain 4074 was grown at 37 °C in brain heart infusion (Difco) supplemented with 1 μg/ml β-NAD (Sigma) to an A600 of 1.2 and fractionated into periplasmic, cytoplasmic, inner, and outer membrane fractions as described previously (17). All fractions were prepared for mass spectrometric analysis. The cytoplasmic fraction was further enriched for glycopeptides using Oasis MAX resin (Waters)7 and analyzed as described above.
Expression and Purification of Autotransporter Proteins
E. coli 1-liter cultures of Dh5α cells harboring the expression plasmids for the relevant proteins were grown to an A600 of 0.5 and protein expression was induced by addition of arabinose to a final concentration of 0.2%. After 6 h of incubation at 37 °C, the cells were harvested, and an aliquot was analyzed by immunoblot. For protein purification, the cell were resuspended in Tris buffer (30 mm Tris-HCl, pH 8.0, 300 mm NaCl), supplemented with 0.1 mg/ml DNase I (Fermentas GmbH) and half a tablet of protease inhibitor mixture (EDTA-free; Roche Applied Science) and broken in a French pressure cell (three passes at 1000 p.s.i.). After centrifugation (30 min, 15,000 × g), the supernatant was loaded on a HisTrap HP 1-ml column (GE Healthcare), and the column was washed with 50 column volumes (CV) of Tris buffer containing 30 mm imidazole. Proteins were eluted with 200 mm imidazole and dialyzed against Tris buffer.
For further purification, the dialysate was brought to 6 m urea (Sigma), 10 mm DTT, incubated at 60 °C for 1 h, and loaded onto a 2-ml nickel-nitrilotriacetic acid-agarose column (Qiagen). The column was washed with increasing concentrations of imidazole (0, 30, and 40 mm in 30 mm Tris-HCl, pH 8.0, 300 mm NaCl, 6 m urea, 10 mm DTT, 20 CV each). The pure protein was finally eluted with 3 × 2 ml of 30 mm Tris-HCl, pH 8.0, 300 mm NaCl, 6 m urea, 10 mm DTT, 100 mm imidazole and dialyzed against 30 mm Tris-HCl, pH 8.0, 300 mm NaCl. 100 μg of each purified protein was analyzed by LC-ESI-MS/MS as described above.
In Vitro Glycosylation of AtaC1866–2428
ApNGT was purified as described previously (14). 20-μl reactions (0.5 μm AtaC1866–2428, 0.05 μm ApNGT, and 50 μm of nucleotide-activated hexose (UDP-Glc or UDP-Gal) in 20 μl of 25 mm Tris-HCl, pH 8.0, 150 mm NaCl) were incubated for 18 h at 30 °C. The reaction products were analyzed by SDS-PAGE with subsequent immunoblot using anti-His4 antibodies and N-Hex reactive human serum MS14.
Analysis of Monosaccharides on AtaC1866–2428
Analysis was essentially performed as described previously (18). In short, 250 μg of protein was precipitated by 25% TCA, and the pellet was washed in acetone and hydrolyzed for 5 h at 95 °C in 2.5 m TFA. After evaporation, the pellet was dissolved in 50 μl of 1% sodium acetate solution. Anthranilic acid labeling reagent was prepared by dissolving 30 mg of anthranilic acid (Sigma) and 20 mg of cyanoboroanhydride (Sigma) in 1 ml of 2.4% sodium acetate, 2% boric acid in methanol. 50 μl of labeling reagent was added to the hydrolyzed protein solution, and the mixture was incubated for 45 min at 80 °C. The samples were diluted to 5 ml in HPLC eluent A (0.3% 1-aminobutane, 0.5% phosphoric acid, and 1% THF in H2O), and 90 μl were analyzed by HPLC according to separation system I (18). The analysis was performed in three technical replicates.
Cytoplasmic Glycosylation of Heterologous Proteins
E. coli BL21(DE3) cells harboring the plasmids encoding the various substrate proteins in combination with an ApNGT-encoding plasmid or the empty vector were grown in LB supplemented with the appropriate antibiotics to an A600 of 0.5. Expression of the ApNGT and AcrA was induced by addition of 0.4% arabinose. In the case of EPO and CtxB, the expression of the substrate proteins was induced 2 h later by addition of 0.5 mm isopropyl β-d-thiogalactopyranoside. Cells were harvested 6 h after ApNGT induction, and an aliquot was analyzed by SDS-PAGE and immunoblot. For mass spectrometric analysis, cells were broken by passage through a French pressure cell (3 × 1000 p.s.i.), followed by centrifugation (20,000 × g, 30 min, 4 °C). The pellet was washed two times with 20 mm Tris-HCl, pH 8, 1% Triton X-100, 2 m urea, 1 mm EDTA. The soluble fraction of the AcrA samples as well as the washed pellets of the CtxB and EPO samples were analyzed by nanoLC-ESI-CID-MS/MS as described above.
RESULTS
Autotransporter Adhesins of A. pleuropneumoniae Are N-Glycosylated
ApNGT has been shown to possess glycosyltransferase activity (13, 14), but no proteins have been identified as endogenous substrates of this enzyme in vivo. To identify potential substrates of ApNGT in A. pleuropneumoniae (serotype 1 str. 4074), LC-ESI-MS/MS analysis of protein extracts was performed, and proteomic data were searched via Mascot engine to identify N-hexosylated peptides. In total, ten N-modified sites belonging to two proteins were identified (UniProt numbers E0E6S1 and E0E5Q8) (Table 1 and supplemental Fig. S1), and all glycosylated sites were confirmed based on the presence of peptide fragment ions containing hexose. Both proteins are annotated as belonging to the autotransporter adhesins protein family. The modified residues were all part of the consensus sequon NX(S/T). Among the modified sequons, we found a strong preference toward threonine at position +2 (nine sites, 90%) with only a minor fraction containing serine (one site, 10%). We also detected a dihexosylated peptide (TTVIN(Hex)N(Hex)STTITAKPTTVGTVTTYDLDLSK) where the two modified residues are directly adjacent, indicating that ApNGT is not blocked by glycosylation of a residue nearby.
TABLE 1.
Summary of LC-ESI-MS/MS analysis of the soluble fraction of A. pleuropneumoniae strain 4074
Modified residues are underlined and in bold. (Annotated spectra are shown in supplemental Fig. S1.)
| UniProt number | Protein name | Observed m/z | Error | Mascot score | Peptide sequence |
|---|---|---|---|---|---|
| ppm | |||||
| E0E6S1 | Autotransporter adhesin | 1073.2260 (3+) | 1.8 | 68.3 | K.TTVINNSTTITAKPTTVGTVTTYDLDLSK.T |
| 1127.2435 (3+) | 1.6 | 47.8 | K.TTVINNSTTITAKPTTVGTVTTYDLDLSK.T | ||
| 1489.0522 (3+) | 1.6 | 58.2 | K.NSTVIGNQNTINLTDVATNNVNTGYVNIQGDHNTINAAR.G | ||
| 1117.0408 (4+) | 1.4 | 33.8 | K.NSTVIGNQNTINLTDVATNNVNTGYVNIQGDHNTINAAR.G | ||
| 1053.4770 (3+) | 1.1 | 37.1 | K.SSYAQGDYSVAIGNAENTTNTASVNATK.V | ||
| 1214.9037 (3+) | 1.7 | 64.8 | R.NAYHLQSAGGGASNNTVSLGTNAYAGGEDSVAIGTR.A | ||
| E0E5Q8 | Autotransporter adhesin | 994.4711 (2+) | 2.3 | 57.9 | K.ATANGTNATAIGSSAQTK.G |
| 772.8460 (2+) | 1.6 | 45.9 | K.YDGTNNTQASGVR.A |
ApNGT Modifies Pasteurellaceae Autotransporter Adhesins in the Cytoplasm of E. coli
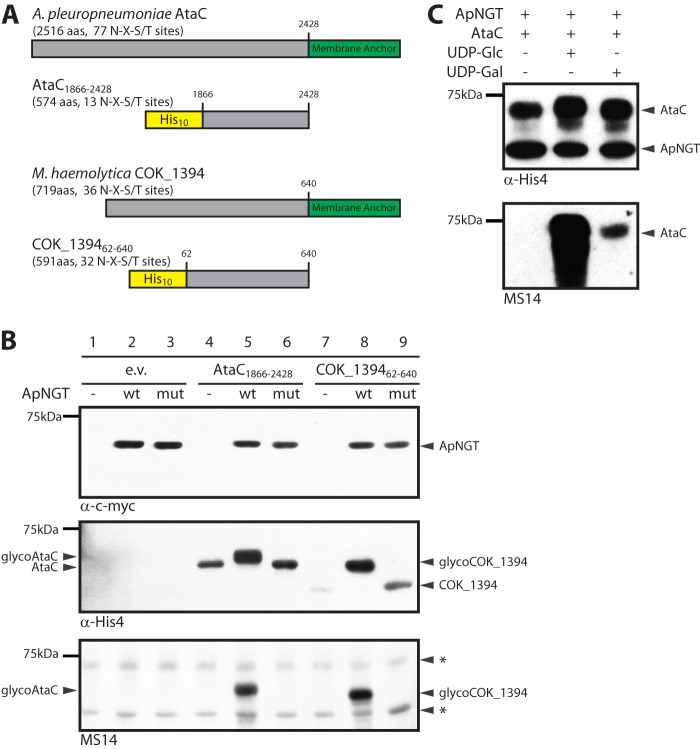
Having identified substrates of ApNGT, we set out to transfer this N-glycosylation system to Escherichia coli. Because autotransporter adhesins seem to be the preferred substrates of ApNGT, we mined the proteomes of Pasteurellaceae species for suitable autotransporter adhesins with a high density of NX(S/T) sites. The search yielded A. pleuropneumoniae AP76 autotransporter adhesin AtaC (APP7_0520, 2516 amino acids, 77 NX(S/T) sites) and the M. haemolytica autotransporter adhesin COK_1394 (719 amino acids, 36 NX(S/T) sites). However, expression of full-length adhesins in E. coli proved not to be successful. We therefore constructed truncated and tagged versions of the proteins that were missing the predicted β-barrel membrane anchor domain as well as the N terminus that includes the signal sequence (Fig. 1A, AtaC1866–2428 574 amino acids, 13 NX(S/T) sites and COK_139462–640 591 amino acids, 36 NX(S/T) sites). We co-expressed each of the truncated proteins together with ApNGT in the cytoplasm of E. coli and analyzed the tagged proteins in whole cell extracts by SDS-PAGE followed by immunoblot with tag-specific antibodies. The electrophoretic mobility of both AtaC1866–2428 and COK_139462–640 clearly shifted toward a higher molecular weight when co-expressed with ApNGT (Fig. 1B, middle panel, lanes 5 and 8), indicating modification of the proteins. Co-expression with ApNGT also increased the level of the two adhesin fragments in the cytoplasm of E. coli, as visualized by more intense bands on the immunoblot (Fig. 1B, middle panel, lanes 5 and 8). This stabilizing effect of the co-expression was particularly striking in the case of the truncated COK_1394 that was barely detectable in the absence of ApNGT (Fig. 1B, middle panel, lane 7). To assess the function of ApNGT in the potential glycosylation and the stabilization of the adhesin fragments, we took advantage of a catalytically inactive mutant of ApNGT. Based on the crystal structure of ApNGT, several residues that were required for UDP-glucose binding (Lys441, Asn521, and Asp525) were identified, and mutation of either one of them inactivates the enzyme (15). We co-expressed the triple mutant ApNGT-K441A, N521A,D525A together with the adhesin fragments AtaC1866–2428 and COK_139462–640 and analyzed the autotransporter fragments by SDS-PAGE and subsequent immunoblot (Fig. 1B, middle panel, lanes 6 and 9). As expected, co-expression of the mutant ApNGT did not result in a mobility shift (indicating glycosylation) but increased the levels of AtaC1866–2428 and COK_139462–640 significantly.
FIGURE 1.
A, schematic representation of truncated autotransporter constructs AtaC1866–2428 and COK_139462–640 used for expression in E. coli. The corresponding proteins are 574 and 591 amino acids in length and feature 13 and 32 potential glycosylation sites, respectively, as well as an N-terminal His10 tag for detection and purification. B, analysis of whole cell extracts of E. coli cells expressing the autotransporter constructs in the absence of ApNGT (−, lanes 4 and 7), in the presence of wild type ApNGT (wt, lanes 5 and 8), or with ApNGT-K441A,N521A,D525A (mut, lanes 6 and 9). Proteins were separated by SDS-PAGE on 7% acrylamide gels and analyzed by immunoblot. Lanes 1–3 show whole cell extracts of E. coli cells expressing the ApNGT constructs in the absence of substrate protein. The NGT proteins were detected via the c-Myc epitope (top panel), the substrate proteins were detected via the His10 tag (middle panel), and glycosylation was detected by the human serum MS14 (bottom panel). Unspecific bands are marked with asterisks. C, immunoblot analysis of in vitro glycosylated AtaC1866–2428. UDP-Glc and UDP-Gal served as sugar donors. ApNGT and AtaC1866–2428 were detected via the His6 tag (His10-tag resp.) (upper panel). Serum MS14 was analyzed for reactivity against the different N-linked hexoses (lower panel).
Human Sera Specifically React with N-Glycosylated Proteins
To directly detect glycosylated proteins in E. coli extracts, we took advantage of antisera from multiple sclerosis patients that were shown to react specifically with an N-glucosylated synthetic peptide (19, 20). We tested such sera for reactivity against our autotransporter constructs, expressed and modified by ApNGT in E. coli. In an immunoblot, serum MS14 reacted with remarkable specificity with the autotransporter protein fragments when they were co-expressed with wild type ApNGT but not with the proteins co-expressed with the catalytically inactive mutant (Fig. 1B, bottom panel). Similar reactivity was also achieved with other N-glucose-specific sera of multiple sclerosis patients (data not shown). Because ApNGT is able to transfer both glucose and galactose to proteins (13, 14), we performed in vitro glycosylation reactions of purified AtaC1866–2428 using UDP-Glc and UDP-Gal as sugar donor and probed the reaction products with serum MS14. Clear reactivity could be demonstrated against N-glucosylated AtaC1866–2428. Reactivity against N-galactosylated AtaC1866–2428 was significantly lower, supporting the specificity of this serum reported previously (20) (Fig. 1C). Most importantly, these results demonstrated the glycosylation of proteins in the cytoplasm of E. coli.
AtaC and COK_1394 Are Modified with Glucose at a Relaxed Consensus Sequon
The newly established N-glycosylation system in E. coli allowed the study of the glycosylation products in more detail. We purified glycosylated AtaC1866–2428 and COK_139462–640 using nickel-nitrilotriacetic acid affinity chromatography (Fig. 2A). During purification of the glycoproteins, we noted that ApNGT bound tightly to the autotransporter fragments and co-purified with them under native conditions (Fig. 2B, lane 1). We therefore included a second purification step under denaturing conditions (6 m urea, 10 mm DTT) to separate the substrate proteins from the enzyme (Fig. 2B). We digested the pure autotransporter protein fragments with trypsin and analyzed the peptides by LC-ESI-MS/MS. We achieved a sequence coverage of 95.9% for AtaC1866–2428 and detected a total of 26 residues modified with a hexose. COK_139462–640 analysis gave a sequence coverage of 39.3%, and we identified 16 modified positions. The modified sites were further confirmed using HCD or ETD fragmentation. We observed a relaxed peptide substrate specificity of ApNGT in vivo. In addition to the canonical NX(S/T) glycosylation consensus sites, asparagine residues in NXA, NXG, NXD, and NXV sequons were found to be modified as well. Furthermore, we detected a total of four glycosylated Gln residues in these two proteins (Table 2, Fig. 2C, and supplemental Fig. S2). These results did not allow us to define a consensus sequon (Fig. 2D). However, 67% of modified sequons had a hydroxyl amino acid (Ser/Thr) at position +2, with Thr being heavily favored over Ser (22 versus 6 modified sites). At the positions immediately adjacent to the glycosylated residue, small, uncharged amino acids, especially Ala, were highly enriched. Large, positively charged as well as aromatic residues were mostly absent from the stretch of protein surrounding the glycosylation sites. Most interestingly, our analysis also revealed the ApNGT-dependent O-glycosylation of the adhesin fragment AtaC1866–2428. Mass spectrometric analysis using the ETD method for fragmentation yielded two hexose-modified Ser residues (Table 2). These modifications were only found when the protein was co-expressed with ApNGT, supporting the hypothesis that ApNGT was able to act as protein O-glycosyltransferase as well.
FIGURE 2.
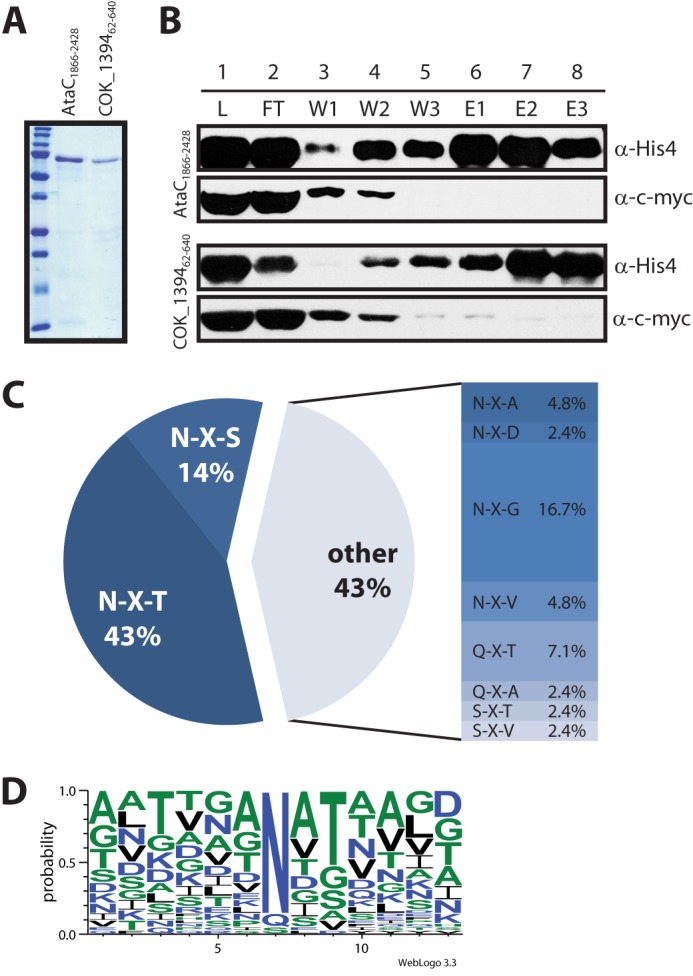
Glycosylation of autotransporter adhesin fragments co-expressed with ApNGT in E. coli. A, purified, glycosylated autotransporter adhesin fragments were analyzed by SDS-PAGE on a 15% acrylamide gel and stained by Coomassie Brilliant Blue R-250. B, fractions from the second purification step (under denaturing conditions) were separated by SDS-PAGE on a 7% acrylamide gel and analyzed by immunoblot. The top two panels show fractions from the purification of AtaC1866–2428, and the bottom two panels show fractions from the purification of COK_139462–640. The autotransporter fragments were detected via the His10 tag, the ApNGT protein via the c-Myc epitope. The fractions analyzed are: load (L, dialyzed eluate from first purification step under native conditions), flow-through (FT), wash fractions (W1–W3, 0, 30, and 40 mm imidazole), and elution fractions (E1–E3, 100 mm imidazole). Note that ApNGT co-purifies with the autotransporter fragments under native conditions (lane 1, L) and is subsequently removed by nickel-nitrilotriacetic acid purification under denaturing conditions. (For details see “Experimental Procedures.”) C, frequency of glycosylated sequons. D, consensus sequence for protein glycosylation by ApNGT in E. coli generated by Weblogo 3.3 (43) from an alignment of all modified sites (42 in total).
TABLE 2.
Summary of LC-ESI-MS/MS analysis of purified AtaC1866–2428 and COK_139462–640
Modified residues are underlined and in bold. The sequencing methods used were CID, HCD, and ETD. (Annotated spectra are shown in supplemental Fig. S2.)
| Site | Observed m/z | Error | Mascot Score | Peptide Sequence | Sequencing method |
|---|---|---|---|---|---|
| ppm | |||||
| AtaC | |||||
| Asn1 | 447.2411 | −1.2 | 62 | K.FVGNDGKVITK.E | ETD |
| Asn2 | 832.4755 | 4.45 | 75 | K.VITKELNETLTIK.G | CID |
| Asn3 | 799.0779 | 4.43 | 89 | K.ELNETLTIKGNLSTAADVTDK.N | ETD |
| Asn5 | 688.3727 | 4.8 | 50 | R.VDNVDNALIIK.M | CID |
| Asn9 or Asn10 | 927.4421 | 2.46 | 71 | K.GGNTVSLTTSGLDNGGNK.V | CID |
| Asn14 | 1084.8655 | −1.44 | 58 | K.VINVAAGDVNANSTDAVNGSQLYAVSEVANK.G | CID |
| Asn14 and Asn15 | 1138.8866 | 1.70 | 64 | K.VINVAAGDVNANSTDAVNGSQLYAVSEVANK.G | CID |
| Asn19 | 1322.9632 | 4.09 | 96 | K.GWNIQTNGNDTTNVKPGDTVNFVNGDNIAITNDGTK.V | CID |
| Asn19 and Asn24 | 1207.0965 | −0.19 | 56 | K.GWNIQTNGNDTTNVKPGDTVNFVNGDNIAITNDGTKVTVGLVK.N | CID |
| Asn27 | 975.9964 | −0.58 | 58 | K.VGDNVSLTKDGLTAGDVK.I | CID |
| Asn28 and Gln3 | 866.4425 | −0.87 | 63 | K.ISATTGINAGDKQITNVASGLGGK.K | CID |
| Gln3 | 1218.1327 | −1.75 | 101 | K.ISATTGINAGDKQITNVASGLGGK.K | CID |
| Asn30 | 1396.0009 | 2.10 | 60 | K.KLSEAEGDTLTNAANIGDLQTAVSSVTDASQGGGFGLADDK.G | CID |
| Asn31 | 1395.9998 | 1.31 | 61 | K.KLSEAEGDTLTNAANIGDLQTAVSSVTDASQGGGFGLADDK.G | CID |
| Asn32 | 1680.8085 | 1.38 | 182 | K.LSEAEGDTLTNAANIGDLQTAVSSVTDASQGGGFGLADDKGANVTQNLGK.T | HCD |
| Asn34 | 427.2275 | −4.73 | 48 | K.GDGKNISTVVK.G | ETD |
| Asn37 | 990.1605 | 2.60 | 94 | K.DGSLTIGNTTINSDQVKVGDVTVSSNGK.V | CID |
| Asn40 | 1477.3645 | 0.81 | 98 | K.VSGVADGDISPNSTEAINGSQLYDANQNIANYLGGGSKLDDK.G | CID |
| Asn46 | 1365.3445 | 0.71 | 95 | K.VDGNTTTANNVGDAITNLNNEVVKPLTFEGDTGVASKR.K | CID |
| Asn46 and Asn48 | 1367.33 | 1.88 | 91 | K.VDGNTTTANNVGDAITNLNNEVVKPLTFEGDTGVASK.R | CID |
| Asn53 | 688.3674 | 0.53 | 74 | K.LSENNIGVVSDGKGTLAVK.L | ETD |
| Asn54 | 754.3826 | −1.32 | 63 | K.TVNANTVNANTVK.A | CID |
| Asn54 and Asn55 | 835.4126 | 3.21 | 68 | K.TVNANTVNANTVK.A | CID |
| Asn55 | 960.8211 | 1.88 | 89 | K.TVNANTVNANTVKAGDTTINSDGVTIK.D | HCD |
| Asn62 | 1117.5781 | −0.95 | 120 | R.ITNVKAGQADTDAVNVSQLK.G | CID |
| Ser98 | 745.0231 | −4.65 | 74 | K.MARTLTDLTSATFTNAGGDK.S | ETD |
| Ser118 | 1251.3420 | −4.30 | 116 | K.VGDVTVSSNGKVSGVADGDISPNSTEAINGSQLYDANQNIANYLGGGSK.L | ETD |
| COK_1394 | |||||
| Asn1 and Asn3 | 1362.1475 | 0.88 | 105 | K.SVANATYSLALGSSANANATSTATR.G | CID |
| Asn22 | 885.1159 | 0.58 | 98 | K.NILGGNATVKPDGSVTYTNIGGTNK.N | CID |
| Asn33 | 900.9458 | −1.04 | 73 | K.AATAANTTNYEIGLTK.K | CID |
| Gln7 and Asn35 | 917.1111 | 1.42 | 43 | K.AIDDFTKDTQATVVSNDGTVTVK.S | CID |
| Gln7 and Asn35 | 1375.1627 | 1.14 | 42 | K.AIDDFTKDTQATVVSNDGTVTVK.S | CID |
| Gln7 | 898.9432 | 1.7 | 73 | K.DTQATVVSNDGTVTVK.S | CID |
| Gln7 | 1294.1369 | 1.69 | 47 | K.AIDDFTKDTQATVVSNDGTVTVK.S | CID |
| Gln12 | 1001.0058 | −1.1 | 48 | R.NSIAIGPNATATGGEQAAVALGTNSNANGNGALSLGVATVSK.G | CID |
| Asn43 | 1415.3826 | −1.33 | 159 | K.SRNSIAIGPNATATGGEQAAVALGTNSNANGNGALSLGVATVSK.G | CID |
| Gln13, Asn48, and Asn49 | 1313.1304 | 1.88 | 63 | K.GIQATAVGHSANATANGTTALGR.Q | CID |
| Asn48 and Asn49 | 1232.1031 | 1.28 | 78 | K.GIQATAVGHSANATANGTTALGR.Q | CID |
| Asn50 and Asn52 | 1194.0403 | 1.72 | 93 | R.QTNATAGDATAVGSNANATAEK.A | CID |
| Asn52 | 1113.0132 | 1.23 | 134 | R.QTNATAGDATAVGSNANATAEK.A | CID |
| Asn56 and Asn57 | 1139.897 | 2.19 | 79 | R.ANATAQFATALGMGAQATLNSSVALGSESVVR.A | CID |
| Asn58 | 1208.0884 | 0.87 | 99 | R.AATPTENATVGGITYNGFAGVNK.D | CID |
To investigate the nature of the hexoses that were transferred to AtaC1866–2428 by ApNGT in the cytoplasm of E. coli, we compared the monosaccharides attached to purified AtaC1866–2428 protein that was expressed in the presence or absence of ApNGT. The sugar residues were released from the purified proteins by acid hydrolysis, fluorescently labeled, and quantitatively analyzed by reverse phase HPLC (Fig. 3, A and B). The only sugar residue that significantly changed in abundance between the glycoprotein sample and the unmodified control was the one corresponding to glucose (retention time: 35.5 min). No significant amount of any of the other sugars that were analyzed (glucosamine, galactosamine, mannose, galactose, ribose, arabinose, xylose, and fucose) could be detected in either sample. When expressed in E. coli, ApNGT therefore predominantly transfers glucose to its substrate proteins. Comparison of the peak areas with a labeled standard allowed us to quantify the degree of glycosylation. Our data indicate that AtaC1866–2428 was modified with an average of nine glucose residues per protein.
FIGURE 3.
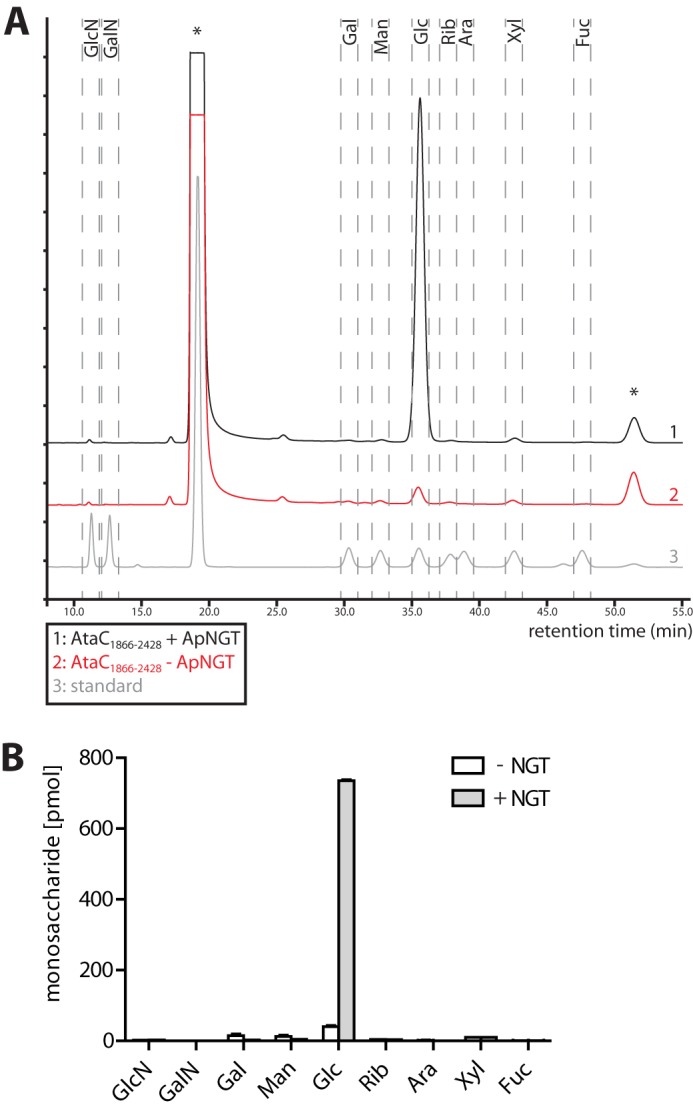
Monosaccharide analysis of purified glycosylated AtaC1866–2428. A, representative chromatograms from the analysis of monosaccharide composition of AtaC1866–2428. AtaC1866–2428 was expressed in the presence (black) and the absence (red) of ApNGT, purified, hydrolyzed, and its monosaccharides were labeled with anthranilic acid and analyzed by reverse phase HPLC. Each sample corresponds to the monosaccharide content of 4.5 μg (77.25 pmol) of purified AtaC1866–2428. The chromatogram of labeled monosaccharide standard is shown in gray. The analyzed sugars were GlcN, galactosamine (GalN), Gal, mannose (Man), Glc, Rib, arabinose (Ara), xylose (Xyl), and fucose (Fuc). Each peak in the standard corresponds to 40 pmol of labeled monosaccharide. Peaks from unreacted anthranilic acid labeling reagent are marked with an asterisk. B, quantification of the different monosaccharides. The peak areas were compared with the standard sample for absolute quantification of the level of each monosaccharide. Bars represent the mean values of three replicates, and error bars represent the S.E.
Glycosylation of Heterologous Proteins by ApNGT
To test whether the activity of ApNGT was restricted to autotransporter adhesins as substrates or whether it would also glycosylate other proteins, we tested our cytoplasmic glycosylation system on heterologous proteins. When co-expressed with ApNGT, both human erythropoietin (hEPO) and CtxB showed reactivity to the N-glucose-specific serum MS14 (Fig. 4). Analysis of the proteins by LC-ESI-MS/MS confirmed the modification of these proteins with N-linked hexose (Table 3 and supplemental Fig. S3). All of the three naturally N-glycosylated sites of hEPO (Asn51, Asn65, and Asn110) were found to be occupied, but no additional sites were modified. CtxB was glycosylated at Asn25. The Campylobacter jejuni efflux pump subunit AcrA is a substrate of the C. jejuni classical N-glycosylation machinery (21) and can also be glycosylated by ApNGT in vitro (14). Probing of whole cell extracts of E. coli cells co-expressing AcrA with ApNGT by immunoblot with serum MS14 did not conclusively demonstrate glycosylation, most likely because of low glycosylation efficiency (data not shown). LC-ESI-MS/MS analysis of these extracts, however, yielded a glycopeptide indicating glycosylation of AcrA at Asn274 (Table 3 and supplemental Fig. S3; AVFDN272N273N274(Hex)STLLPGAFATITSEGFIQK). In C. jejuni, AcrA is glycosylated at Asn273 (22), illustrating the different substrate requirements of the two glycosylation systems.
FIGURE 4.
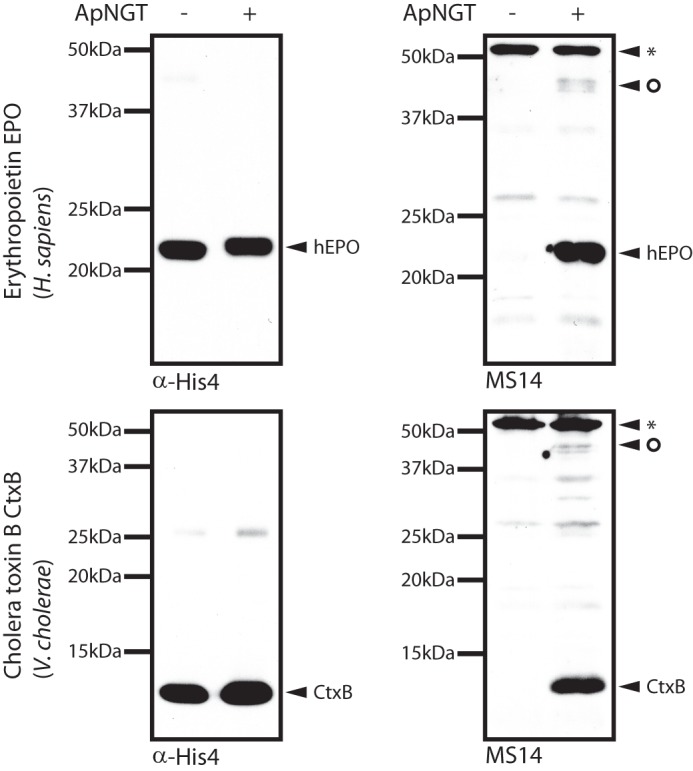
Immunoblots of whole cell extracts of E. coli cells expressing hEPO (top) and CtxB (bottom) in the presence (+) or absence (−) of ApNGT. Proteins were separated by SDS-PAGE on 12% (hEPO) and 15% (CtxB) acrylamide gels, respectively, followed by analysis via immunoblot. CtxB and EPO were detected via their His6 tags, and serum MS14 was used to detect glycosylation. Unspecific bands are marked with an asterisk. The circle marks potentially glycosylated E. coli proteins.
TABLE 3.
Summary of LC-ESI-MS/MS analysis of the glycosylation of human erythropoietin, AcrA, and CtxB co-expressed with ApNGT in the cytoplasm of E. coli
Modified residues are underlined and in bold. (Annotated spectra are shown in supplemental Fig. S3.)
| Site | Observed m/z | Error | Mascot Score | Peptide sequence | Sequencing method |
|---|---|---|---|---|---|
| ppm | |||||
| Human erythropoietin | |||||
| Asn51 or Asn65 | 1483.1589 (2+) | 2.29 | 37 | K.EAENITTGCAEHCSLNENITVPDTK.V | CID |
| K.EAENITTGCAEHCSLNENITVPDTK.V | |||||
| Asn51 and Asn65 | 1043.1256 (3+) | 1.82 | 22 | K.EAENITTGCAEHCSLNENITVPDTK.V | CID |
| Asn110 | 841.1046 (3+) | 2.32 | 70 | R.GQALLVNSSQPWEPLQLHVDK.A | CID |
| C. jejuni efflux pump subunit AcrA | |||||
| Asn254 | 973.1572 (3+) | 3.5 | 81 | K.AVFDNNNSTLLPGAFATITSEGFIQK.N | CID/HCD |
| Cholera toxin subunit B (CtxB) | |||||
| Asn25 | 1510.1835 (2+) | 3.8 | 101 | -MTPQNITDLCAEYHNTQIHTLNDK.I | CID |
When expressed in the cytoplasm of E. coli, ApNGT also came in contact with endogenous, cytoplasmic E. coli proteins, some of which contain sequons that could potentially be glycosylated. Immunoblots of E. coli whole cell extracts separated by SDS-PAGE and probed using N-Glc reactive sera indicate glycosylation of endogenous substrates (Fig. 4). To investigate these potential modifications on E. coli proteins, we analyzed the proteome of E. coli cells expressing ApNGT by LC-ESI-MS/MS and filtered for hexose-modified proteins (Table 4 and supplemental Fig. S4). We found 15 hexose-modified residues belonging to 8 different proteins. The sequons found to be modified were predominately canonical glycosylation sites (NX(S/T), 80%) with only a few alternative sequons modified (NXA and NXL, 20%). Among the glycosylated proteins, there were three that are annotated as outer membrane proteins (YuaO, Ag43, and MdtE). Much like AtaC and COK_1394, Ag43 is an autotransporter adhesin and is involved in autoaggregation (23). YuaO is so far uncharacterized but also shows sequence similarity to autotransporters. Together these two proteins contain more than half of all the modified sites identified, again indicating a preference of ApNGT toward this protein family. The rest of the glycosylated proteins we identified were only represented by a single glycopeptide each. These include four metabolic enzymes (GAPDH, PCKA, PurH, and ArgG) and a chaperone (DnaK). They are among the most abundant cytoplasmic proteins of E. coli (24). Taken together, these results show that although ApNGT favors autotransporter adhesins as peptide substrate in vivo, it glycosylates other protein classes from various origins and can be used as a general N-glycosylation system in the bacterial cytoplasm.
TABLE 4.
Summary of LC-ESI-MS/MS analysis of E. coli proteins glycosylated by ApNGT
Modified residues are underlined and in bold. (Annotated spectra are shown in supplemental Fig. S4.)
| UniProt# | Name | Description | Observed m/z | Error | Mascot score | Peptide sequence |
|---|---|---|---|---|---|---|
| ppm | ||||||
| P0A6Y8 | DnaK | Chaperone protein | 769.7334 (3+) | 1.10 | 71.9 | R.IINEPTAAALAYGLDKGTGNR.T |
| P15639 | PurH | Bifunctional purine biosynthesis protein | 806.3792 (2+) | 2.80 | 72.7 | K.NNMTIGIGAGQMSR.V |
| P0A9B2 | GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | 1085.1689 (3+) | 1.30 | 47 | K.GANFDKYAGQDIVSNASCTTNCLAPLAK.V |
| Q9JMS5 | YuaO | Uncharacterized protein | 809.8575 (2+) | 0.12 | 79.7 | K.GNNASATLDYTDSK.I |
| 834.4002 (2+) | 1.60 | 79.5 | K.VSGNMSIAQGNAASAK.V | |||
| 802.8511 (2+) | 1.60 | 74 | R.GNNASATLDYSDSK.I | |||
| 854.9266 (2+) | −5.30 | 73.5 | K.TQLELNNSALTASGK.V | |||
| 639.8131 (2+) | 1.30 | 50.9 | K.GNASIGVGDQAK.A | |||
| P39180 | Ag43 | Antigen 43 | 825.4345 (2+) | 1.30 | 96.6 | R.LQVDAGGTATNVTLK.Q |
| 654.8363 (3+) | 0.82 | 33 | K.TVNNDTLTIR.E | |||
| 878.9494 (2+) | 5.80 | 57.4 | K.SGSGTLTVSNTTLTQK.A | |||
| P22259 | PCKA | Phosphoenolpyruvate carboxykinase | 646.3407 (2+) | 0.09 | 42 | R.DALLENVTVR.E |
| P0A6E4 | ArgG | Argininosuccinate synthase | 665.3247 (2+) | 0.04 | 33 | K.DLEYLNSSVK.I |
| P37636 | MdtE | Multidrug resistance protein | 825.4197 (2+) | 5.7 | 71.4 | R.TQLNEAEANVTVAK.A |
DISCUSSION
We took advantage of A. pleuropneumoniae NGT to establish a general N-glycosylation system in the cytoplasm of E. coli. We found that ApNGT is able to glycosylate various different proteins and peptides in vitro (14) and in vivo. In A. pleuropneumoniae, two autotransporter adhesins were found to be glycosylated. However, because of the lack of an efficient enrichment method for the glycopeptides generated by ApNGT, our glycoproteome analysis was most likely incomplete, and we only observed the most abundant substrates. Further analysis might yield more glycosylated residues belonging to more substrate proteins. We cannot exclude the possibility that some of the glycosylated residues we detected were not modified by ApNGT but by another protein glycosyltransferase. There is, however, no evidence for such a second glycosylation activity in A. pleuropneumoniae. When ApNGT was expressed in E. coli, we noted that most proteins we found to be efficiently glycosylated were also outer membrane adhesins (AtaC, COK_1394, Antigen 43, and YuaO), pointing to a central role of the N-linked glycosylation for cell surface proteins. ApNGT glycosylated the endogenous E. coli autotransporters (YuaO and Antigen 43) that naturally are not N-glycoproteins but share the secretion pathway with AtaC. This indicated a preference of ApNGT toward this family of substrate proteins regardless of the origin. Interestingly, Ag43 as well as the related AIDA-I and TibA adhesins of E. coli are naturally O-heptosylated (25–28), and the fact that Ag43 can also be efficiently N-glycosylated points to an overlap in protein substrate specificity of these two glycosylation systems. In addition, CtxB, EPO, and AcrA, all secreted proteins, were glycosylated. We expressed these proteins without signal sequence so that they remained in the cytoplasm. Autotransporters have been shown to exhibit extremely slow folding kinetics (29, 30), whereas CtxB and EPO are insoluble and accumulate as inclusion bodies under these conditions. Therefore, our results might point to a requirement for a long residence time of the substrate in the cytoplasm in an unfolded state.
In H. influenzae, levels of HMW1A adhesin are reduced, and translocation to the outer membrane is impaired in the absence of the glycosyltransferase HMW1C (9). In our study, we noted an increased stability of the expressed autotransporter fragments in the presence of ApNGT. Because this was at least partially independent of glycosyltransferase activity of ApNGT, we propose that ApNGT has a chaperone activity for outer membrane proteins, especially adhesins. This is supported by the finding that ApNGT tightly binds to its substrate proteins AtaC and COK_1394 and co-purifies with them. In fact, it took harsh conditions (6 m urea, 10 mm DTT) to separate the adhesin fragments from the enzyme. Alternatively, ApNGT binds preferentially to unfolded proteins, a feature of extracellular proteins retained in the cytoplasm.
We report the use of human sera (19, 20) for detection of proteins modified with Asn-linked hexoses. The sera reacted with proteins modified with N-linked Glc and weakly with Gal residues. Our experimental setup did not allow us to elucidate the specificity of these polyclonal sera for the different N-linked hexoses, and we were unable to address the involvement of protein epitopes in antibody binding. Nevertheless, these sera are very useful tools to analyze NGT-type cytoplasmic N-linked protein glycosylation systems. Whether or not N-linked glucose represents the primary antigen that led to these specific antibodies remains unclear. However, the relatively high occurrence of bacteria harboring the same type of N-linked glycosylation system as A. pleuropneumoniae in the human population might offer an explanation for the presence of these antibodies: approximately 50% of children are colonized with nontypable H. influenzae strains (NTHi) (31), and depending on sample and assay method, between 20 and over 90% of subjects showed antibody reactivity against Y. enterocolitica epitopes (32–35).
Transfer of the cytoplasmic glycosylation system of A. pleuropneumoniae to E. coli allowed in depth analysis of ApNGT-modified glycoproteins and revealed a more relaxed peptide substrate specificity than previously reported (13, 14). Only ∼57% of residues that were found to be modified were in an NX(S/T) consensus sequon, with Thr being much more frequent than Ser. We also observed Ala, Gly, Val, Asp, and Leu at position +2 and detected the glycosylation of Gln and Ser side chains. Neither the hydroxyl amino acid at position +2 nor the Asn residue itself seems to be strictly necessary for glycosylation by ApNGT. This speaks against a role of the +2 amino acid in catalysis but might reflect a contribution to peptide substrate binding as has been observed for the Campylobacter lari oligosaccharyltransferase PglB (36, 37). However, the relatively high frequency of NX(S/T) sequons among the modified sites marks this sequon as the preferred substrate, consistent with the observation that we did not find any noncanonical sequons modified in A. pleuropneumoniae. Moreover, the fact that ApNGT acts both as an N- and O-glycosyltransferase reflects a unique property of this enzyme and demonstrates its remarkable flexibility. In contrast to its relaxed peptide substrate specificity, ApNGT is much more specific toward the sugar donor substrate. When expressed in E. coli, ApNGT predominately transfers glucose to its substrate proteins. No significant amount of any of the other sugars was observed on our model substrate. This matches very well with the observed in vitro sugar substrate specificities of ApNGT (13, 14) and H. influenzae HMW1C (11) but contrasts with the monosaccharide distribution observed on H. influenzae adhesin HMW1A, where galactose was the most abundant sugar, followed by glucose and marginal amounts of mannose (9). This might indicate additional regulation of N-glycosyltransferase activity in vivo or the presence of elongating enzymes adding other monosaccharides to the initial N-linked hexose.
A. pleuropneumoniae also encodes an α1,6-glucosyltransferase able to elongate the N-linked glucose that is the product of the ApNGT reaction (14). In our analysis of A. pleuropneumoniae glycosylation, we did not find any evidence for N-Glc elongation, and the sites we identified were modified only by a single hexose. However, because our methods were not fine-tuned to detect such possible elongated glycans, further analysis is needed to conclusively assess the extent of elaboration of A. pleuropneumoniae N-glycans by α1,6-glucosyltransferase or further enzymes.
As noted earlier (14), there is a convergent evolution of the use of the NX(S/T) sequon as substrate determinant for N-linked protein glycosylation. The molecular basis for NX(S/T) recognition by the classical N-glycosylation machinery is now understood (36, 37), but ApNGT belongs to a different class of glycosyltransferases, and substrate recognition must be different. Therefore we speculate that “molecular mimicry”—having glycan structures closely resembling the core of the host N-glycans—might be the driving force in the convergent evolution process that shaped the peptide substrate specificity of NGT in these pathogenic bacteria.
Our study also lays the basis for a number of potential biotechnological applications. The E. coli expression system provides an inexpensive way to produce glycosylated adhesins for diagnostics or vaccination purposes. The autotransporter adhesins of A. pleuropneumoniae mediate adhesion to the host cells and are therefore crucial for colonization and pathogenesis (38). All sequenced strains of A. pleuropneumoniae as well as a number of other pathogenic Pasteurellaceae species (such as H. influenzae or M. haemolytica), carry genes encoding a cytoplasmic glycosylation system. This type of glycosylation, because it is not found in the eukaryotic hosts and does not differ between various strains or even species, might be an interesting target for treatment or vaccination against porcine pleuropneumoniae and other diseases caused by Pasteurellaceae. ApNGT also glycosylates heterologous substrate proteins and can therefore be used as a preparative tool for glycoproteins. However, the relaxed substrate specificity of ApNGT as compared with eukaryotic oligosaccharyltransferase might lead to glycosylation of additional sites, which could cause problems in some applications. Furthermore, the fact that ApNGT acts in the cytoplasm and preferentially glycosylates autotransporters or unfolded proteins limits the number of applicable substrates. ApNGT is especially useful for the production of glycoproteins that form inclusion bodies (such as hEPO), which can be efficiently purified and folded in vitro (39). Unlike many other glycosylation systems that often yield many different glycoforms, such glycoproteins are homogeneously modified with glucose with no detectable amounts of other glycoforms. Such N-linked hexoses that are the product of the ApNGT reaction can be used as starting material to produce defined glycoproteins by transglycosylation (40), for glycoPEGylation (41), or for coupling of desired ligands using hydrazide chemistry (42).
Acknowledgments
We thank A. Essig, S. Müller, and the Functional Genomics Center Zurich for help with mass spectrometry and R. Gauss for help with HPLC. We also thank V. Perreten (University of Bern) and M. Wittenbrink (University of Zurich) for providing us with A. pleuropneumoniae and M. hemolytica strains, respectively.
The project was supported by Swiss National Science Foundation Grants CRSII3_12733 and 3100170-105541 (to M. A.) and by ETH Zürich. A patent application (“Pasteurellaceae vaccines,” PCT/EP2012/065480) covering the use of N-glycosylated Pasteurellaceae autotransporter adhesins as vaccines has been filed by ETH Zurich on behalf of A. N., F. S., and M. A. and was licensed to Malcisbo AG.

This article contains supplemental Table S1 and Figs. S1–S4.
S. Snovida, C. W. Kuo, C. W. Lin, and K. H. Khoo, manuscript in preparation.
- NGT
- N-glycosyltransferase
- ApNGT
- A. pleuropneumoniae NGT
- ESI
- electrospray ionization
- CID
- collision-induced dissociation
- HCD
- higher energy collisional dissociation
- ETD
- electron transfer dissociation
- Hex
- hexose
- Gal
- galactose
- hEPO
- human erythropoietin
- CtxB
- cholera toxin subunit B.
REFERENCES
- 1. Schwarz F., Aebi M. (2011) Mechanisms and principles of N-linked protein glycosylation. Curr. Opin. Struct. Biol. 21, 576–582 [DOI] [PubMed] [Google Scholar]
- 2. Kelleher D. J., Gilmore R. (2006) An evolving view of the eukaryotic oligosaccharyltransferase. Glycobiology 16, 47R–62R [DOI] [PubMed] [Google Scholar]
- 3. Aebi M., Bernasconi R., Clerc S., Molinari M. (2010) N-Glycan structures. Recognition and processing in the ER. Trends Biochem. Sci. 35, 74–82 [DOI] [PubMed] [Google Scholar]
- 4. Helenius A., Aebi M. (2004) Roles of N-linked glycans in the endoplasmic reticulum. Annu. Rev. Biochem. 73, 1019–1049 [DOI] [PubMed] [Google Scholar]
- 5. Sharon N., Lis H. (1995) Lectins. Proteins with a sweet tooth. Functions in cell recognition. Essays Biochem. 30, 59–75 [PubMed] [Google Scholar]
- 6. Kaneko Y., Nimmerjahn F., Ravetch J. V. (2006) Anti-inflammatory activity of immunoglobulin G resulting from Fc sialylation. Science 313, 670–673 [DOI] [PubMed] [Google Scholar]
- 7. Rudd P. M., Elliott T., Cresswell P., Wilson I. A., Dwek R. A. (2001) Glycosylation and the immune system. Science 291, 2370–2376 [DOI] [PubMed] [Google Scholar]
- 8. Apweiler R., Hermjakob H., Sharon N. (1999) On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim. Biophys. Acta 1473, 4–8 [DOI] [PubMed] [Google Scholar]
- 9. Grass S., Buscher A. Z., Swords W. E., Apicella M. A., Barenkamp S. J., Ozchlewski N., St Geme J. W., 3rd (2003) The Haemophilus influenzae HMW1 adhesin is glycosylated in a process that requires HMW1C and phosphoglucomutase, an enzyme involved in lipooligosaccharide biosynthesis. Mol. Microbiol. 48, 737–751 [DOI] [PubMed] [Google Scholar]
- 10. Gross J., Grass S., Davis A. E., Gilmore-Erdmann P., Townsend R. R., St Geme J. W., 3rd (2008) The Haemophilus influenzae HMW1 adhesin is a glycoprotein with an unusual N-linked carbohydrate modification. J. Biol. Chem. 283, 26010–26015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Grass S., Lichti C. F., Townsend R. R., Gross J., St Geme J. W., 3rd (2010) The Haemophilus influenzae HMW1C protein is a glycosyltransferase that transfers hexose residues to asparagine sites in the HMW1 adhesin. PLoS Pathog. 6, e1000919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009) The carbohydrate-active EnZymes database (CAZy). An expert resource for Glycogenomics. Nucleic Acids Res. 37, D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Choi K. J., Grass S., Paek S., St Geme J. W., 3rd, Yeo H. J. (2010) The Actinobacillus pleuropneumoniae HMW1C-like glycosyltransferase mediates N-linked glycosylation of the Haemophilus influenzae HMW1 adhesin. PLoS One 5, e15888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schwarz F., Fan Y. Y., Schubert M., Aebi M. (2011) Cytoplasmic N-glycosyltransferase of Actinobacillus pleuropneumoniae is an inverting enzyme and recognizes the NX(S/T) consensus sequence. J. Biol. Chem. 286, 35267–35274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kawai F., Grass S., Kim Y., Choi K. J., St Geme J. W., 3rd, Yeo H. J. (2011) Structural insights into the glycosyltransferase activity of the Actinobacillus pleuropneumoniae HMW1C-like protein. J. Biol. Chem. 286, 38546–38557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wisniewski J. R., Zougman A., Nagaraj N., Mann M. (2009) Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362 [DOI] [PubMed] [Google Scholar]
- 17. Thein M., Sauer G., Paramasivam N., Grin I., Linke D. (2010) Efficient subfractionation of gram-negative bacteria for proteomics studies. J. Proteome Res. 9, 6135–6147 [DOI] [PubMed] [Google Scholar]
- 18. Racaityte K., Kiessig S., Kálmán F. (2005) Application of capillary zone electrophoresis and reversed-phase high-performance liquid chromatography in the biopharmaceutical industry for the quantitative analysis of the monosaccharides released from a highly glycosylated therapeutic protein. J. Chromatogr. A 1079, 354–365 [DOI] [PubMed] [Google Scholar]
- 19. Lolli F., Mazzanti B., Pazzagli M., Peroni E., Alcaro M. C., Sabatino G., Lanzillo R., Brescia Morra V., Santoro L., Gasperini C., Galgani S., D'Elios M. M., Zipoli V., Sotgiu S., Pugliatti M., Rovero P., Chelli M., Papini A. M. (2005) The glycopeptide CSF114(Glc) detects serum antibodies in multiple sclerosis. J. Neuroimmunol. 167, 131–137 [DOI] [PubMed] [Google Scholar]
- 20. Lolli F., Mulinacci B., Carotenuto A., Bonetti B., Sabatino G., Mazzanti B., D'Ursi A. M., Novellino E., Pazzagli M., Lovato L., Alcaro M. C., Peroni E., Pozo-Carrero M. C., Nuti F., Battistini L., Borsellino G., Chelli M., Rovero P., Papini A. M. (2005) An N-glucosylated peptide detecting disease-specific autoantibodies, biomarkers of multiple sclerosis. Proc. Natl. Acad. Sci. U.S.A. 102, 10273–10278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wacker M., Linton D., Hitchen P. G., Nita-Lazar M., Haslam S. M., North S. J., Panico M., Morris H. R., Dell A., Wren B. W., Aebi M. (2002) N-Linked glycosylation in Campylobacter jejuni and its functional transfer into E. coli. Science 298, 1790–1793 [DOI] [PubMed] [Google Scholar]
- 22. Nita-Lazar M., Wacker M., Schegg B., Amber S., Aebi M. (2005) The N-X-S/T consensus sequence is required but not sufficient for bacterial N-linked protein glycosylation. Glycobiology 15, 361–367 [DOI] [PubMed] [Google Scholar]
- 23. Selkrig J., Mosbahi K., Webb C. T., Belousoff M. J., Perry A. J., Wells T. J., Morris F., Leyton D. L., Totsika M., Phan M. D., Celik N., Kelly M., Oates C., Hartland E. L., Robins-Browne R. M., Ramarathinam S. H., Purcell A. W., Schembri M. A., Strugnell R. A., Henderson I. R., Walker D., Lithgow T. (2012) Discovery of an archetypal protein transport system in bacterial outer membranes. Nat. Struct. Mol. Biol. 19, 506–510, S501 [DOI] [PubMed] [Google Scholar]
- 24. Ishihama Y., Schmidt T., Rappsilber J., Mann M., Hartl F. U., Kerner M. J., Frishman D. (2008) Protein abundance profiling of the Escherichia coli cytosol. BMC Genomics 9, 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Benz I., Schmidt M. A. (2001) Glycosylation with heptose residues mediated by the aah gene product is essential for adherence of the AIDA-I adhesin. Mol. Microbiol. 40, 1403–1413 [DOI] [PubMed] [Google Scholar]
- 26. Charbonneau M. E., Côté J. P., Haurat M. F., Reiz B., Crépin S., Berthiaume F., Dozois C. M., Feldman M. F., Mourez M. (2012) A structural motif is the recognition site for a new family of bacterial protein O-glycosyltransferases. Mol. Microbiol. 83, 894–907 [DOI] [PubMed] [Google Scholar]
- 27. Lindenthal C., Elsinghorst E. A. (1999) Identification of a glycoprotein produced by enterotoxigenic Escherichia coli. Infect. Immun. 67, 4084–4091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sherlock O., Dobrindt U., Jensen J. B., Munk Vejborg R., Klemm P. (2006) Glycosylation of the self-recognizing Escherichia coli Ag43 autotransporter protein. J. Bacteriol. 188, 1798–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Charbonneau M. E., Janvore J., Mourez M. (2009) Autoprocessing of the Escherichia coli AIDA-I autotransporter. A new mechanism involving acidic residues in the junction region. J. Biol. Chem. 284, 17340–17351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Junker M., Schuster C. C., McDonnell A. V., Sorg K. A., Finn M. C., Berger B., Clark P. L. (2006) Pertactin β-helix folding mechanism suggests common themes for the secretion and folding of autotransporter proteins. Proc. Natl. Acad. Sci. U.S.A. 103, 4918–4923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Faden H., Duffy L., Williams A., Krystofik D. A., Wolf J. (1995) Epidemiology of nasopharyngeal colonization with nontypeable Haemophilus influenzae in the first 2 years of life. J. Infect. Dis. 172, 132–135 [DOI] [PubMed] [Google Scholar]
- 32. Arscott P., Rosen E. D., Koenig R. J., Kaplan M. M., Ellis T., Thompson N., Baker J. R., Jr. (1992) Immunoreactivity to Yersinia enterocolitica antigens in patients with autoimmune thyroid disease. J. Clin. Endocrinol. Metab. 75, 295–300 [DOI] [PubMed] [Google Scholar]
- 33. Effraimidis G., Tijssen J. G., Strieder T. G., Wiersinga W. M. (2011) No causal relationship between Yersinia enterocolitica infection and autoimmune thyroid disease. Evidence from a prospective study. Clin. Exp. Immunol. 165, 38–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Resetkova E., Notenboom R., Arreaza G., Mukuta T., Yoshikawa N., Volpé R. (1994) Seroreactivity to bacterial antigens is not a unique phenomenon in patients with autoimmune thyroid diseases in Canada. Thyroid 4, 269–274 [DOI] [PubMed] [Google Scholar]
- 35. Wenzel B. E., Heesemann J., Wenzel K. W., Scriba P. C. (1988) Antibodies to plasmid-encoded proteins of enteropathogenic Yersinia in patients with autoimmune thyroid disease. Lancet 1, 56. [DOI] [PubMed] [Google Scholar]
- 36. Gerber S., Lizak C., Michaud G., Bucher M., Darbre T., Aebi M., Reymond J. L., Locher K. P. (2013) Mechanism of bacterial oligosaccharyltransferase. In vitro quantification of sequon binding and catalysis. J. Biol. Chem. 288, 8849–8861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lizak C., Gerber S., Numao S., Aebi M., Locher K. P. (2011) X-ray structure of a bacterial oligosaccharyltransferase. Nature 474, 350–355 [DOI] [PubMed] [Google Scholar]
- 38. Xiao L., Zhou L., Sun C., Feng X., Du C., Gao Y., Ji Q., Yang S., Wang Y., Han W., Langford P. R., Lei L. (2012) Apa is a trimeric autotransporter adhesin of Actinobacillus pleuropneumoniae responsible for autoagglutination and host cell adherence. J. Basic Microbiol. 52, 598–607 [DOI] [PubMed] [Google Scholar]
- 39. Wang Y. J., Liu Y. D., Chen J., Hao S. J., Hu T., Ma G. H., Su Z. G. (2010) Efficient preparation and PEGylation of recombinant human non-glycosylated erythropoietin expressed as inclusion body in E. coli. Int. J. Pharm. 386, 156–164 [DOI] [PubMed] [Google Scholar]
- 40. Lomino J. V., Naegeli A., Orwenyo J., Amin M. N., Aebi M., Wang L. X. (2013) A two-step enzymatic glycosylation of polypeptides with complex N-glycans. Bioorg. Med. Chem. 21, 2262–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. DeFrees S., Wang Z. G., Xing R., Scott A. E., Wang J., Zopf D., Gouty D. L., Sjoberg E. R., Panneerselvam K., Brinkman-Van der Linden E. C., Bayer R. J., Tarp M. A., Clausen H. (2006) GlycoPEGylation of recombinant therapeutic proteins produced in Escherichia coli. Glycobiology 16, 833–843 [DOI] [PubMed] [Google Scholar]
- 42. Wilchek M., Bayer E. A. (1987) Labeling glycoconjugates with hydrazide reagents. Methods Enzymol. 138, 429–442 [DOI] [PubMed] [Google Scholar]
- 43. Crooks G. E., Hon G., Chandonia J. M., Brenner S. E. (2004) WebLogo. A sequence logo generator. Genome Res. 14, 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]