Background: Tetraethylammonium is a broad potassium channel blocker applied in neuron research.
Results: Tetraethylammonium has the ability to lead cell apoptotic process; various biological and proteomics methods prove this result.
Conclusion: Novel biomarkers candidates are found to reveal mechanism of tetraethylammonium-induced apoptosis.
Significance: This provides new details on the new molecular pathway involved in apoptosis and gives a broader context in therapeutic strategies.
Keywords: Apoptosis, Bioinformatics, Flow Cytometry, Pharmacology, Proteomics, Western Blotting, HeLa Cell Line, Patch Clamp Technology, RT-qPCR, Tetraethylammonium
Abstract
Tetraethylammonium (TEA) is a potassium channel (KCh) blocker applied in the functional and pharmacological studies of the KChs. The MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay, a colorimetric assay to quantitatively measure living cells, demonstrated that TEA reduced the HeLa cell viability dose-dependently. Flow cytometry analysis indicated an increased apoptosis rate of the HeLa cell after exposing to TEA. The patch clamp technique revealed that the K+ current of the HeLa cell was inhibited up to 80% when exposed to TEA. In addition, quantitative real-time PCR approach set up cross-talk among the cytotoxicity of TEA, 4-aminopyridine, and anti-cancer drug such as cisplatin. Using comparative proteomics combined with MALDI-TOF MS/MS, 33 significantly changed proteins were found from TEA treatment group; among these proteins, 12 were up-regulated, and 21 were down-regulated. Here we indicated that these proteins were closely connected with many biological functions such as oxidative stress response, signal transduction, metabolism, protein synthesis, and degradation. Both Western blotting and quantitative real-time PCR approaches further verified these differential proteins. Ingenuity Pathways Analysis software, a tool to analyze “omics” data and model biological system, was applied to analyze the interaction pathways of these proteins. The subcellular locations of the differential proteins are also predicted from Uniprot. All results above can help in our understanding of the mechanism of TEA-induced cytotoxicity and provide potential cancer biomarkers. Various experimental results in this study (like those for cisplatin) indicated that TEA is not only a KCh blocker but also a potential anti-cancer drug.
Introduction
Apoptosis disorder is a result of a series of diseases, such as myocardial infarction (1), cancer (2), and autoimmune disease (3). Apoptotic volume decrease, cytochrome c release, apoptosome formation, and DNA fragmentation are common cell characteristics of apoptosis (4). Potassium channels (KChs),2 which participate in controlling membrane potential, are the most important and diverse ion channels on the cell membrane. Nearly every subfamily of KChs is correlated with various stages of cancer (5–7). Many KCh types are widely expressed and crucial in cellular processes and ionic-driving force across membranes (8). K+ current efflux and anion inflow help maintain cell electroneutrality during the apoptotic volume decrease process (9). Tetraethylammonium (TEA) is a non-selective KChs inhibitor that has been extensively used as a molecular probe to study their structure (10) and characteristics of the regulation pathways (11, 12). A four-peptide sequence containing Phe or Tyr residue on the loop surface of KCh proteins is a recognition region for TEA, and it is very conservative (13). Previous research reported that TEA induced apoptosis by inhibiting Bcl-2 and Bax expression, generating increased production of intracellular reactive oxygen species (14), up-regulating expression of p53 and p21 proteins, and arresting cells in the G1 phase (15). This research mainly focused on monitoring the K+ current using the patch clamp technique and alteration of some key cancer genes profiles in TEA-induced apoptosis (16, 17). Proteomics analysis integrating the potential response pathway to understand the mechanism of the various types of inhibitor-induced apoptosis is proposed as a more powerful tool to uncover useful biomarkers indicative of the cytotoxicity of the inhibitors and drugs.
In this study we report new evidence that TEA had the capacity for inhibiting HeLa proliferation and inducing apoptosis by a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay and flow cytometry analysis, respectively. In addition, proteomics combined with bioinformatics and other analytical methods was carried out to study differential proteins and potential networks of TEA cytotoxicity to HeLa cell. Cross-talk analysis on mRNA levels was set up among TEA, 4-aminopyridine (4-AP), and cisplatin (CDDP). 4-AP is another calcium-activated potassium channel blocker; it decreases cell viability and induces DNA fragmentation dose-dependently (18), whereas EGTA, which is a chelating agent of Ca2+, can mitigate the apoptosis rate induced by 4-AP (19). CDDP is a broad-spectrum antitumor drug in clinical use and has been widely applied in chemotherapy (20). In addition, CDDP is reported by Luo et al. (21) to lead to the apoptosis of HepG2 cells; some differential expressed proteins for the apoptosis were identified during CDDP treatment (21). The mRNA primers corresponding to these differential proteins found in the Luo et al. (21) study were amplified to compare the expression levels among TEA, 4-AP, and CDDP. Similar to most anticancer drugs such as CDDP, TEA also has a potential capacity for inducing the apoptosis of various cancer cells besides KChs blocking.
EXPERIMENTAL PROCEDURES
Culture of the HeLa Cell Line
The HeLa cell line was purchased from Cell Bank of the Chinese Academy of Sciences, Shanghai, China. It was cultured at 37 °C, with 5% CO2 in DMEM (high glucose) supplemented with 10% fetal calf serum, 100 units/ml penicillin, and 100 mg/ml streptomycin. A Chinese hamster ovary (CHO) cell line (22) (a kindly gift from Prof. Kaczmarek's laboratory, Yale University) was cultured at 37 °C with 5% CO2 in DMEM supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 mg/ml streptomycin.
Cytotoxicity Assay with MTT Assay
The toxicity of tetraethylammonium (Sigma) was evaluated in HeLa cells using an MTT reduction assay (23). Briefly, HeLa cells were seeded on a 96-well plate for 24 h to reach the density of 104 cells/ml. Various concentrations of TEA at three treating periods were conducted in parallel. Another 20-μl MTT solution (0.25 g/ml) was added, and cells were incubated at 37 °C for 4 h. Another 100 μl of dimethyl sulfoxide (DMSO) was added to each well to dissolve the purple formazon crystals after the removal of the MTT solution, and the plate was gently shaken at room temperature for 20 min. The optical density was measured at 570 nm using a plate reader (FLUOstar Omega). Cells without treatment were used as a control for 100% cellular viability. Each group included triplicate assays to confirm the results.
Flow Cytometry with Propidium Iodide (PI) and Annexin V-FITC/PI Dual Staining Assays
All experiments were performed using EPICS XL flow cytometry (Beckman) in triplicate as previous described (24). The cells were stained with PI, and around 10,000 dyed cells were captured for cell phase distribution analysis as described elsewhere (25). An annexin V-FITC apoptosis detection kit (Beyotime, China) was used for the dual staining assay. Briefly, 106 cells were harvested and washed twice with PBS. The cells were resuspended in binding buffer (10 mm HEPES, pH 7.5, 140 mm NaCl, 2.5 mm CaCl2), and annexin V- FITC and PI were added to a final concentration of 1 and 10 μg/ml, respectively, and they were incubated for 10 min in the dark at room temperature before flow cytometry analysis.
Patch Clamp Procedure
For the patch clamp experiment, a standard setup including an inverted microscope and an Axon amplifier was used as described before (15). A two-step procedure was applied for making glass capillaries. The pipette solution was made up as follows: 142 mm NaCl 5 mm KCl, 2 mm CaCl2, 2 mm MgCl2, 10 mm HEPES, 15 mm glucose, adjusted to pH 7.35. The bath medium was as follows: 114 mm gluconate, 17.5 mm KCl 2 mm MgCl2, 10 mm HEPES, 2 mm MgATP, adjusted with KOH to pH 7.3. Recording started while holding voltage at −80 mV, with step voltage from −40 to 60 mV. TEA was then applied for 5 min, and the current of the same cell was recorded. Data acquisition and analysis were carried out using pCLAMP 9 software (Axon Instruments).
HeLa Cell Lysate Preparation
HeLa cells were harvested and homogenized in lysis buffer (8 m urea, 2 m thiourea, 4% CHAPS, 1% Nonidet P-40, 65 mm DTT, 40 mm Tris, 0.5% ampholyte 3-10, and 1 mm PMSF) using an IKA® T10 basic disperser. After extraction at 4°C overnight, the homogenate was centrifuged for 1 h at 12,000 × g, the supernatants were collected, and their protein contents were measured using the Bradford assay respectively (26).
Two-dimensional Electrophoresis for Proteomic Analysis
150 μg of whole HeLa cell lysate was mixed with a rehydration solution containing 7 m urea, 2 m thiourea, 4% CHAPS (w/v), 1% DTT (w/v), and 0.5% IPG (immobilized pH gradient) buffer (Bio-Rad). The sample mixture was loaded onto pH 4–7, 17-cm ReadyStrip IPG strips (Bio-Rad). First-dimensional isoelectric focusing steps were conducted using the following protocol: active rehydration at room temperature for 12 h, voltage steps from 1 h at 100 V, 1 h at 500 V, then slowing ramping from 1,000 to 8,000 V to reach a total of 80,000 V h (PROTEAN IEF System, Bio-Rad). Equilibration steps were applied immediately after focusing with equilibration buffer (6 m urea, 50 mm Tris, 2% w/v SDS, 30% v/v glycerol, and a trace of bromphenol blue, pH 8.8), containing 1% w/v DTT for 20 min followed by incubation in the same buffer containing 2.5% w/v iodoacetamide for another 20 min. The equilibrated strip was applied directly to 12% SDS-polyacrylamide gels (SDS-PAGE) and separated at a constant current of 25 mA/gel until the bromphenol blue reached the bottom of the gel. For each protein sample (control or TEA treatment), three experimental triplicates were subjected to two-dimensional electrophoresis and visualized using silver-staining. The stained gel was captured by using the ChemiDocTX system (Bio-Rad) and analyzed to find any statistically different proteins with ImageMaster 2D Platinum software (27).
Protein Digestion and Identification Using MALDI-TOF MS/MS
Selected proteins spots were manually excised and digested with trypsin (Promega) following the manufacturer's instructions. The protein digest solution was extracted with ZipTip and washed with 0.1% TFA solution. An equal volume of peptide and matrix α-cyano-4-hydroxycinnamic acid saturated with 50% acetonitrile, 0.05% TFA were mixed, and 1 μl of the mixture was spotted onto a MALDI stainless steel target and evaporated at room temperature.
An ABI 4800 Proteomics Analyzer MALDI-TOF/TOF mass spectrometer (AB SCIEX, MA) was used for analysis, and spectra were calibrated using digested BSA. MALDI-TOF MS and TOF-TOF MS/MS were performed, and data were acquired in the positive MS reflector mode with a scan range from 800 to 4000 Da. The 10 most abundant precursors were selected for further MS/MS analysis. The MASCOT search engine was applied for interpretation of the mass spectra, allowing up to one missed cleavage, and variable modifications of carbamidomethylation and methionine oxidation, and maximal mass tolerances for precursor and fragments were set at 10 ppm and 0.05 Da, respectively. Results with % confidence interval values >95% were considered to be positive identifications.
Western Blotting Verification
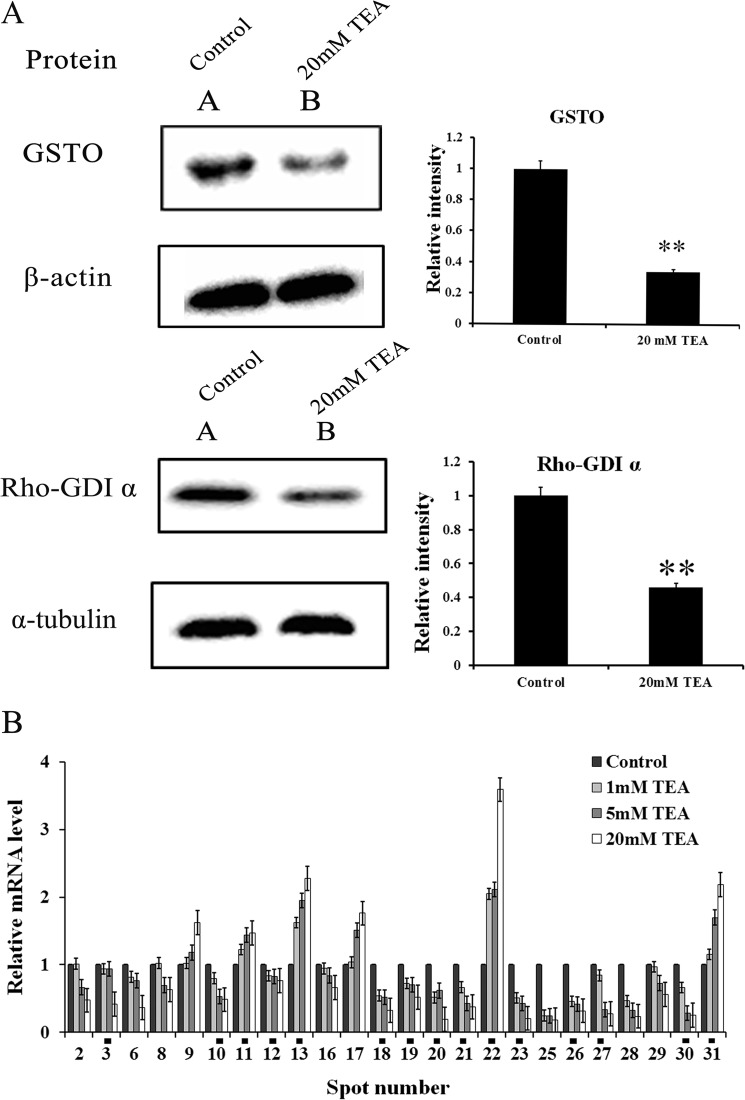
Proteins from HeLa whole cell lysate were separated on 12% gels. Briefly, after electrophoresis, the resolved proteins were transferred to a PVDF membrane and then blocked in 5% w/v nonfat milk TBST for 1 h (50 mmol/liter Tris-HCl, pH 7.6, 150 mmol/liter NaCl, and 0.1% Tween 20). The PVDF membrane was then incubated with the primary antibodies anti-glutathione S-transferase ω1 (GSTOl) or anti-Rho-GDPα (Santa Cruz, CA, 1:5000 dilution) and anti-β-actin or anti-α-tubulin (Santa Cruz, 1:5000 dilution) overnight at 4 °C. Secondary antibody HRP-conjugated goat anti-mouse IgG (Pierce, 1:10,000 dilution) was incubated with the PVDF membrane for 1 h after washing 3 times with TBST. The membrane was again washed 3 times with TBST, and the blot was developed using ECL.
Quantitative Real-time PCR
Quantitative real-time PCR analysis was performed as previously described (20). Total RNAs were extracted from the HeLa cell using the TRIzol reagent (Invitrogen), and cDNAs were synthesized using ImProm-IITM Reverse Transcription (Promega). A Rotor-GeneTM 6000 real-time rotary analyzer (Corbett Life Science) was used to amplify both target and internal control templates following the manufacturer's instructions (TaKaRa, Japan) (1 cycle at 95 °C for 5 min and 40 amplification cycles at 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 45 s). In brief, 1 μl of reverse-transcribed product template, 5 μl of SYBR Premix Ex TaqTM II, and the gene-specific primer pairs at a final concentration of 500 nm for each primer made 10 μl of reaction system. HeLa DNA was used to construct standard curves (CT versus log copy number) for target amplicons and normalized according to the β-actin gene expression.
Network Analysis
The molecular functions and subcellular locations of these proteins were searched in the Uniprot database based on accession numbers. The pathway maps containing regulated-proteins were constructed manually based on previous search outputs. To set up a signaling network model, which would be affected by TEA treatment in the HeLa cell, Ingenuity Pathways Analysis (IPA) software (Ingenuity Systems) was used to analyze each gene object corresponding to each modulated protein. Matched genes generated global molecular networks obtained from the pathway knowledge base. Functional analysis of each network was most significant for the genes involved in this network. The genes mapped to the biological networks were ranked by scores indicating the probability that a collection of genes equal to or greater than the number in the network could be achieved by chance alone, and three was set as the cutoff value for identifying significant gene networks affected by the TEA (28).
RESULTS
HeLa Cell Viability Assay with MTT Method
MTT assay is a colorimetric assay, and it is based on conversion of MTT into insoluble formazan crystals (purple) by mitochondrial succinate dehydrogenase, and the latter is dissolved by DMSO and measured spectrophotometrically. Because the reduction of MTT can only occur in metabolically active cells, this assay is broadly used to measure cell viability and evaluate cytotoxic effects of drugs on cell lines in vitro. To determine the level to which the TEA inhibited HeLa proliferation, HeLa cells were cultured in a 96-well plate at various TEA concentrations, and the viability rates were calculated after exposure to TEA. As summarized in Fig. 1, low concentrations of TEA ranging from 0.5 to 2.0 mm slightly decreased the viabilities of HeLa cells; in addition, with the increasing TEA concentrations from 2 to 30 mm, the viabilities decrease dramatically, especially between 36 and 48 h. When the TEA concentration was increased up to 20 mm at 36 h, the viability trend became stable. Based on these experimental data, 20 mm and 36 h were chosen as the experimental conditions for further studying the TEA cytotoxicity mechanism using proteomics tools.
FIGURE 1.
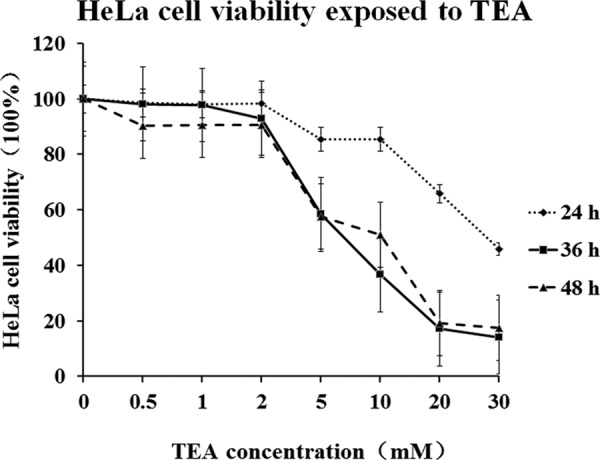
Dose-response curve between TEA concentration and cell viability of HeLa cells. Cells were treated with different concentrations of TEA for 24, 36, and 48 h. Cell viability rates were determined by using MTT assay. The results are expressed as the percentage of the viability rate of control cells. All values are represented as the means ± S.D. of three independent experiments.
The Capacity for Inducing HeLa Apoptosis of TEA
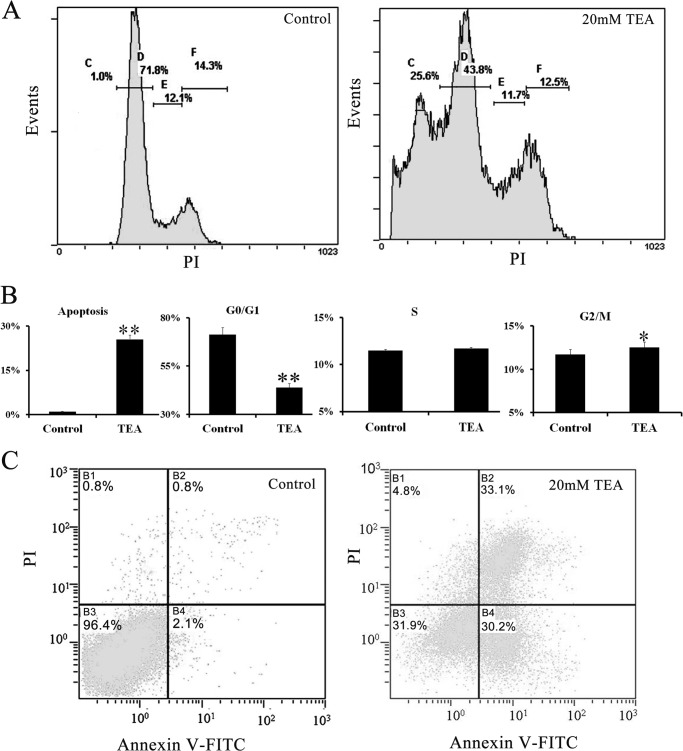
Using flow cytometry with PI staining, we analyzed changes of different cell growth phases after TEA exposure and further validated the drug effectiveness. Four growth phases both in the control and TEA treatment groups are clearly seen in Fig. 2. They are the apoptosis phase (Fig. 2, A–C), the G0/G1 phase (Fig. 2, A–D), the S phase (Fig. 2, A–E), and the G2/M phase (Fig. 2, A–F). The results show that TEA obviously changes cell phase distribution (Fig. 2B). The G0/G1 phase decreased from 71.3 to 43.8%, and there was a strong signal in the apoptosis phase in the TEA exposure group. Using annexin V-FITC/PI dual staining, the apoptosis rate increased from 2.1% in the control to 31.9% after exposure to 20 mm TEA for 36 h (Fig. 2C); this result is similar with that obtained from the PI staining method (Fig. 2A). The HeLa cells induced using TEA were then used to find differential proteins revealing the mechanism of cell apoptosis under conditions of 20 mm and a reactive time of 36 h.
FIGURE 2.
Flow cytometry analysis of cell cycle distribution and apoptosis rate by TEA inducement of HeLa cell. A, cell phase analysis of HeLa cells using flow cytometry with PI staining. B, quantitative analysis of the distribution of cells in cell phases in the bar graph (% of cells in each phase relative to the total population). C, assessment of TEA-induced apoptosis and necrosis using flow cytometry with annexin V-FITC/PI staining. Lower left, annexin V−/PI− cells (normal); lower right, annexin V+/PI− cells (early apoptosis); upper right, annexin V+/PI+ cells (necrosis). All values are represented as the means ± S.D. of three independent experiments. * represents p < 0.05, ** represents p < 0.01.
TEA Influences K+ Current on HeLa Cells and Kv2.1 CHO Cells
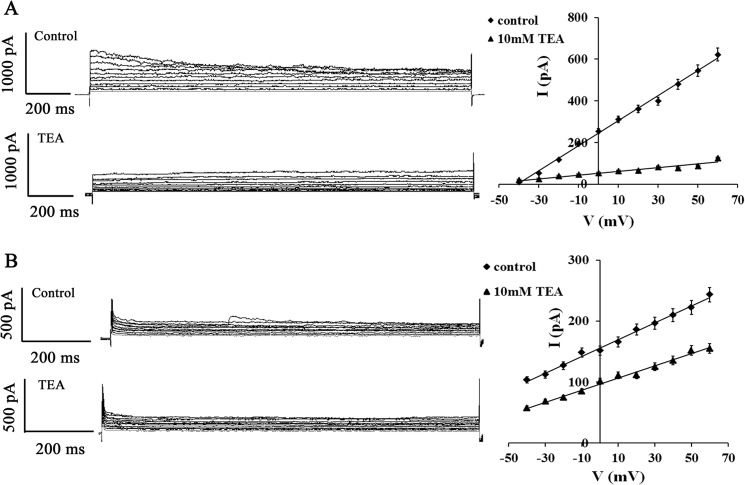
To determine the characteristics of KChs on the HeLa cell membrane, a whole-cell patch clamp was established to record outward K+ current (15). The results indicate that K+ current decreases up to 80.8% in the 10 mm TEA exposure group compared with the control (Fig. 3A). CHO cells were chosen to prove efficiency of TEA blocking. There is no KCh in the CHO cell, and after transfection with Kv2.1channel and steady expression of the Kv2.1 channel for 10 generations, it is named CHOKv2.1 (22). TEA was selected to block Kv2.1, and the K+ current was measured again after blocking. The data prove that the K+ current of CHOKv2.1 is blocked by TEA up to 30% (Fig. 3B). Because there is only one Kv2.1 channel on the CHO cell membrane, the current altitude and blocking percentage are lower than that of the HeLa cell, which has much more complex KChs.
FIGURE 3.
Voltage-activated outward K+ currents in HeLa cells and Kv2.1 current in CHO cells blocked by TEA. A, the whole-cell patch clamp was applied in recording HeLa cells of voltage-gated outward K+ currents. The current-voltage relation of K+ current was set up by depolarizing from −40 to 60 mV with 10-mV increment of each pulse. These outward K+ currents in the same cell were inhibited by external TEA (10 mm). B, the whole-cell patch clamp was applied in recording one CHOkv2.1 cell of Kv2.1 current. These Kv2.1 currents in the same cells were inhibited by external TEA (10 mm). All values are represented as the means ± S.D. of seven independent experiments.
A mRNA Expression Cross-talk Set Up to Analysis Toxicity of TEA, 4-AP, and CDDP
Luo et al. (21) successfully identified 19 differential expressed proteins associated with apoptosis under CDDP treatment in HepG2 cell line; there were protein #1 14-3-3, #2 53BP, #3 AKT, #4 CASP3, #5 DNA-PK, #6 GADD45A, #7 GRB2, #8 MDC, #9 MTOR, #10 p21, #11 p27, #12 p38, #13 p53, #14 p78, #15 PCNA, #16 PLAA, #17 RAD51, #18 RFC, and #19 XRCC. RT-qPCR was used to analyze the mRNA expression levels of corresponding differential proteins of HeLa cells under TEA (20 mm), 4-AP (5 mm), and CDDP (2.0 μg/ml) exposure (Fig. 4). #1 14-3-3, #2 53BP, and #3 AKT show lower mRNA expression levels under both TEA and 4-AP inducement; whereas for #8 MDC and #11 P27, the TEA group has higher expression levels compared with the control group, and another 4-AP group has the lower ones. #15 PCNA, #16 PLAA, and #17 RAD51 show opposite expression tendencies between these two groups. These results meant that TEA could not only induce HeLa apoptosis by blocking KChs but also caused some potential apoptosis pathways by regulating transcriptional levels of cancer genes, as did CDDP. TEA and CDDP share the similar expression tendencies up to nine mRNAs; these are #2 53BP, #3 AKT, #4 CASP3, #6 GADD45A, #10 p21, #12 p38, #13 p53, #15 PCNA, and #17 RAD51. Especially for #6 GADD45A, which has a >10-fold change in both groups, its transcript level increased under stressful growth arrest conditions and functioned as tumor and autoimmune suppressors (29). GADD45A, a p53- and BRCA1-regulated stress-inducible gene, might induce a p53-dependent apoptosis pathway in this experiment due to significant changes of p53 expression levels under these two chemicals stresses (30). Moreover, these pathways are somewhat different than to those of 4-AP, and this can be due to blocking different subfamilies of KChs.
FIGURE 4.
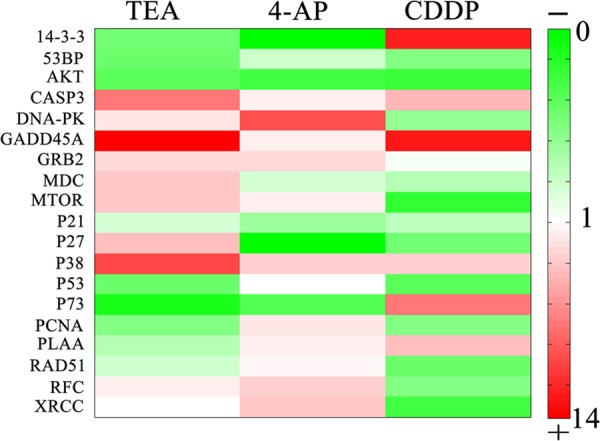
Heatmap visualization of RT-qPCR analysis of mRNA levels in the HeLa cell. RT-qPCR was performed on cDNA using gene-specific primers for differential proteins induced by CDDP of the HepG2 cell. Relative quantification of each gene expression level was normalized according to the β-actin gene expression. First column, TEA (20 mm); second column, 4-AP (5 mm); third column, CDDP (2 μg/ml). Red represents up-regulation, and green represents down-regulation. Color intensity represents the mRNA expression values. Rows, mRNA; column, treatment.
Proteomic Analysis HeLa Apoptosis Induced with TEA
Protein Profiles of HeLa Apoptosis
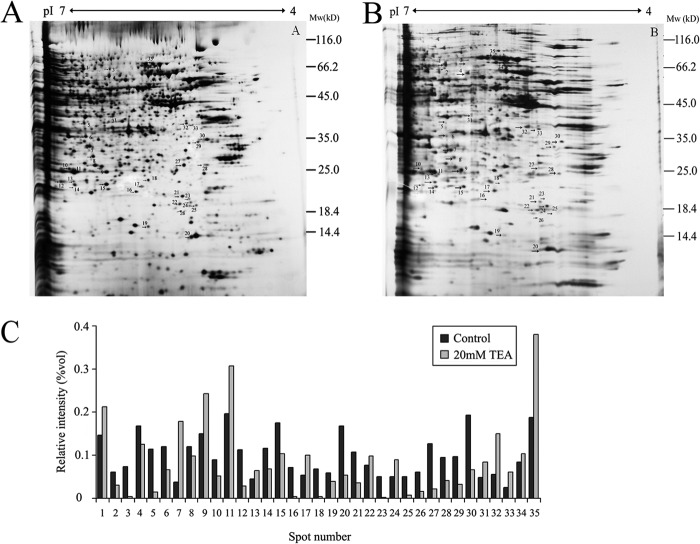
20 mm TEA was utilized to induce HeLa apoptosis for 36 h. The whole cell lysate extract from control and TEA treatment groups were separated using two-dimensional electrophoresis techniques. ImageMaster 2D Platinum software was used to analyze the differential proteins with the silver-staining method (Fig. 5, A and B). Quantitative image analysis using the proteomic software ImageMaster 2D Platinum Trail (V5.0) reveals 35 protein spots showing statistically significant changes (decrease or increase; p < 0.01) in TEA exposure group versus the control group (Fig. 5C). These spots were selected for further analysis using tandem mass spectrometry and mascot database search.
FIGURE 5.
Representative two-dimensional electrophoresis maps indicating protein spots that had significant change after 20 mm TEA exposure in HeLa cells. Whole cell lysate proteins (150 μg) were separated by two-dimensional electrophoresis and visualized using silver staining. Arrows indicate the proteins with a significantly modified expression level after TEA treatment. A, control; B, 20 mm TEA. C, relative quantification of the differential expressed proteins.
Protein Identification
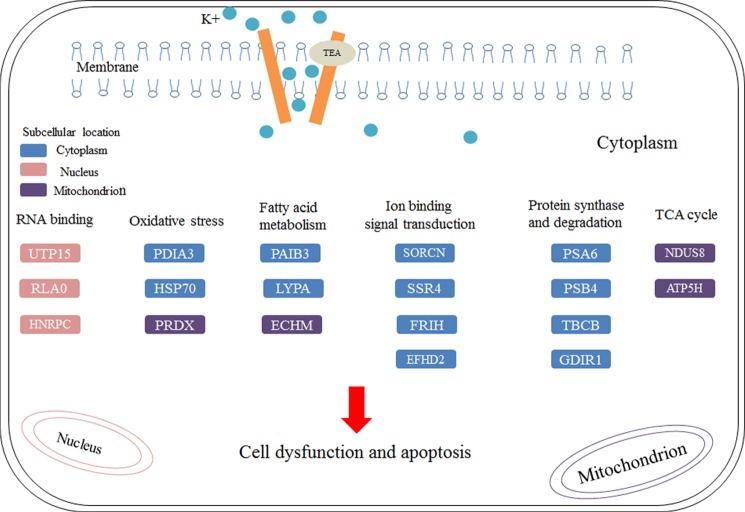
A total of 33 protein spots were identified using MALDI-TOF MS/MS and database searching. Of those identified, 12 (spots #1, 7, 9, 11, 13, 14, 17, 22, 32, 33, 34, and 35) are up-regulated, and the remaining proteins are identified (spots 2, 3, 4, 6, 8, 10, 12, 15, 16, 18, 19, 20, 21, 23, 25, 26, 27, 28, 29, 30 and 31) are down-regulated (Table 1). Enlarged comparisons of the differential proteins found in Fig. 5 are shown in Fig. 6. To better understand the biological functions of these up-regulated and down-regulated expressed proteins by TEA inducement, we searched the UniProt database to identify their molecular functions and predict their subcellular locations. Most of proteins are identified in cytoplasm (45%) and 23% in nucleus and mitochondrion. Subcellular locations and pathways of partial differential proteins are visualized in Fig. 7. The results uncover that these proteins are mainly involved in RNA binding, oxidative stress, fatty acid metabolism, ion binding signal transduction, protein synthesis and degradation, and TCA cycle.
TABLE 1.
Differential proteins selected under TEA inducement identified by using MALDI-TOF-MS/MS
Mass spectrometric identification of differentially regulated proteins of HeLa cell under TEA inducement is shown. The table shows the gel-based IDs, protein names, Uniprot accession numbers, predicted mass and pI, protein score, and TEA/control change -fold. Only significant hits are determined by Mascot are considered as positive identifications.
| Spot no. | Protein name | Accession no. | Mr (Da)/pI | Protein score | Relative intensity ratio | Change |
|---|---|---|---|---|---|---|
| 1 | POTE ankyrin domain family member E | Q6S8J3/POTEE | 122882.3/5.83 | 271 | 1.43 | ↑ |
| 2 | Seryl-tRNA synthetase, cytoplasmic | P49591/SYSC | dinner 59253.2/6.05 | 376 | 0.56 | ↓ |
| 3 | T-complex protein 1 subunit α | P17987/TCPA | 60818.8/5.8 | 644 | 0.11 | ↓ |
| 4 | Protein disulfide isomerase A3 | P30101/PDIA3 | 57145.9/5.98 | 831 | 0.76 | ↓ |
| 6 | 60 S acidic ribosomal protein P0 | P05388/RLA0 | 34422.9/5.71 | 120 | 0.56 | ↓ |
| 7 | Dienoyl-CoA isomerase, mitochondrial | Q13011/ECH1 | 36135/8.16 | 699 | 2.31 | ↑ |
| 8 | Glutathione S-transferase ω1 | P78417/GSTO1 | 27833.2/6.23 | 393 | 0.82 | ↓ |
| 9 | Enoyl-CoA hydratase, mitochondrial | P30084/ECHM | 31823.3/8.34 | 285 | 1.49 | ↑ |
| 10 | Platelet-activating factor acetylhydrolase IB subunit γ | Q15102/PA1B3 | 25832.2/6.33 | 160 | 0.58 | ↓ |
| 11 | Proteasome subunit α type-6 | P60900/PSA6 | 27838/6.34 | 380 | 1.38 | ↑ |
| 12 | Triosephosphate isomerase | P60174/TPIS | 26937.8/6.45 | 79 | 0.35 | ↓ |
| 13 | BAG family molecular chaperone regulator 2 | O95816/BAG2 | 23928.3/6.25 | 81 | 1.23 | ↑ |
| 14 | Acyl-protein thioesterase | O75608/LYPA | 24995.6/6.29 | 100 | 1.26 | ↑ |
| 15 | Protein DJ-1 | Q99497/PARK7 | 20049.6/6.33 | 250 | 0.62 | ↓ |
| 16 | Thioredoxin-dependent peroxide reductase, mitochondrial | P30048/PRDX3 | 28017.3/7.67 | 247 | 0.058 | ↓ |
| 17 | Proteasome subunit β type 4 | P28070/PSB4 | 29242.5/5.72 | 340 | 1.45 | ↑ |
| 18 | Heat shock protein β1 | P04792/HSPB1 | 22825.2/5.98 | 534 | 0.063 | ↓ |
| 19 | Translocon-associated protein subunit δ | P51571/SSRD | 19157.7/5.76 | 347 | 0.75 | ↓ |
| 20 | Prefoldin subunit 6 | I15212/PFD6 | 14573.8/8.83 | 59 | 0.34 | ↓ |
| 21 | ATP synthase subunit d, mitochondrial | O75947/ATP5H | 18536.5/5.21 | 194 | 0.37 | ↓ |
| 22 | Chromobox protein homolog 3 | Q13185/CBX3 | 20969.4/5.23 | 197 | 1.21 | ↑ |
| 23 | Ferritin heavy chain | P02794/ FRIH | 21383.4/5.3 | 137 | 0.074 | ↓ |
| 25 | Sorcin | P30626/SORCN | 21947.5/5.32 | 121 | 0.12 | ↓ |
| 26 | NADH dehydrogenase iron-sulfur protein 8, mitochondrial | O00217/NDUS8 | 24202.8/6 | 147 | 0.24 | ↓ |
| 27 | Ran-specific GTPase-activating protein | P43487/RANG | 23466.6/5.19 | 241 | 0.13 | ↓ |
| 28 | Rho GDP-dissociation inhibitor 1 | P52565/GDIR1 | 23249.7/5.02 | 367 | 0.52 | ↓ |
| 29 | EF-hand domain-containing protein D2 | Q96C19/EFHD2 | 26794.5/5.15 | 77 | 0.36 | ↓ |
| 30 | Tubulin-folding cofactor B | Q99426/TBCB | 27593.6/5.06 | 82 | 0.35 | ↓ |
| 31 | U3 small nucleolar RNA-associated protein 15 homolog | Q8TED0/UTP15 | 58660.6/9.18 | 44 | 0.33 | ↓ |
| 32 | Diphthine synthase | Q9H2P9/DPH5 | 31973.4/5.19 | 73 | 1.73 | ↑ |
| 33 | Heterogeneous nuclear ribonucleproteins C1/C2 | P07910/HNRPC | 33706.6/4.95 | 147 | 1.83 | ↑ |
| 34 | Stress-70 protein, mitochondrial | P38646/GRP75 | 73919.9/5.87 | 650 | 1.15 | ↑ |
| 35 | Heat shock 70-kDa protein 1A/1B | P08107/HSP71 | 70294.1/5.48 | 975 | 1.82 | ↑ |
FIGURE 6.
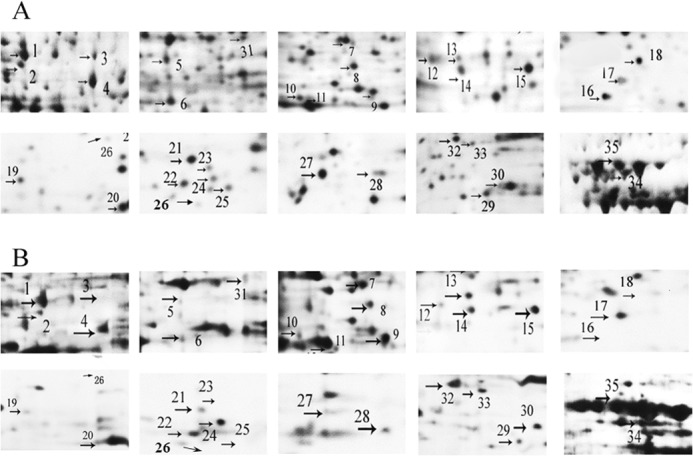
Magnified images of significantly different changed protein spots between the control and TEA-treated groups. A, differential proteins found in control group. B, differential proteins found in the TEA group.
FIGURE 7.
Pathway prediction of TEA induced HeLa cell cytotoxicity based on proteomic analysis. Differentially expressed proteins participate in RNA binding, oxidative stress, fatty acid metabolism, ion binding signal transduction, protein synthesis and degradation, and TCA cycle. Subcellular predictions and molecular functions of these proteins were constructed manually using UniProt database. The blue rectangles represent proteins located in cytoplasm, the pink rectangles represent proteins located in nucleus, and the purple rectangles represent proteins located in mitochondrion.
Validation of Differential Proteins Using Western Blotting
To validate the alterations of differential proteins identified after TEA exposure, GSTO1 and Rho GDP dissociation inhibitor 1 (GDIR1) were representatively selected and subjected to Western blotting (Fig. 8A). As a result, the abundances of GSTO1 and Rho-GDIα decrease significantly in the TEA exposure group, which show good consistency with that of two-dimensional electrophoresis result (Figs. 5 and 6), and they also show lower transcriptional levels in TEA group (Fig. 8B), thus confirming lower mRNA expression levels for GSTO1 and GDIR1 under TEA challenge.
FIGURE 8.
Western blotting and RT-qPCR analysis of representative proteins and their corresponding mRNAs identified by two-dimensional electrophoresis gel. A, Western blotting analysis of GSTO1 and Rho-GDIα expression levels in the HeLa cell. Left panel, 45 μg of protein was loaded onto a 12% SDS-PAGE and probed with antibody against GSTO and Rho-GDIα. β-Actin and α-tubulin were also measured as the loading control and used for data normalization. Right panel, the quantification of the signal has been analyzed with densitometric scanning. All values are represented as the means ± S.D. of three independent experiments. * represents p < 0.05; ** represents p < 0.01. B, RT-qPCR analysis of mRNA level in the HeLa cell. RT-qPCR was performed on cDNA using gene specific primers. Relative quantification of each gene expression level was normalized according to the β-actin gene expression. The PCR data of treatments was calibrated to the control values (control = 1). All the spots show significant difference (p < 0.05) in mRNA expression levels where the TEA concentrations are set at 20 and 5 mm. Spot numbers labeled with black squares are expressed as significantly different between control and TEA treated groups (all in 1, 5, and 20 mm).
The mRNA Expression Levels of Differential Expressed Proteins
The 35 differentially expressed proteins shown in Fig. 5 were analyzed by using Image Master 2D Platinum software (Version 5.0, GE Healthcare), obtaining partial significant enlarged maps (Fig. 6), respectively. In addition, 33 of these different proteins corresponding to the protein numbers shown in Fig. 6 have been successfully identified by PMF (MALDI-TOF MS/MS) techniques except for the sample numbers of 5 and 24 (Table 1). To examine whether the differential proteins in the two-dimensional electrophoresis gels were reliable and had similar patterns on the transcriptional levels, mRNA was extracted from each experimental group, and RT-qPCR analyses were carried out to calculate their mRNA relative expression levels using β-actin as the internal reference. 24 mRNA-amplified levels corresponding to the identified proteins shown in Table 1 were analyzed except for 1, 4, 7, 14, 15, 32, 33, 34, and 35. Compared to the results between control and the TEA exposure group, we found that all the spots show a significant difference in mRNA expression levels where the TEA concentrations are set at 20 mm (p < 0.01) and 5 mm (p < 0.05). Even when the concentration of TEA is lowered to 1 mm, most of the spots show similar tendencies. The spot numbers labeled with a black square (Fig. 8B) are expressed significantly differently between control and TEA treat groups (all in 1, 5, and 20 mm). # 9, 11, 13, 17, 22, and 31 have higher mRNA expression levels with higher concentrations of TEA, whereas the mRNA expression levels of the remaining proteins (#2, 3, 6, 8, 10, 12, 16, 18, 19, 20, 21, 23, 25, 26, 27, 28, 29, and 30) show different tendencies. The selected mRNA expression levels are consistent with the levels of protein expressions in the two-dimensional electrophoresis gels except for #31, which might due to post-translational modification process. Those mRNA results indicate that most identified proteins shown in Table 1 and Fig. 6 are credible.
Protein Networks Involved in TEA Exposure Treatment
To investigate whether the differentially expressed proteins interacted biologically, IPA software was also selected to generate systematic network analysis to elucidate integrated signaling pathways under TEA exposure. Counterparts of all these genes were imported to the IPA module, and a total of 33 proteins were located in two pathways for network generation (Fig. 9). One network (Fig. 9A) involves post-translational modification, protein folding, and hereditary disorder and has 35 genes (the maximum number of genes per network), among which 17 differential protein genes are involved in this network (score = 44, highly significant). Of these 17 proteins, 8 are up-regulated, whereas 9 are down-regulated. The other network (Fig. 9B) involves carbohydrate metabolism, small molecule biochemistry, and vitamin and mineral metabolism, and 16 focus genes are involved in this 35-gene network (score = 41, highly significant). Of these 16 proteins, 7 are up-regulated, whereas nine are found down-regulated in this network.
FIGURE 9.
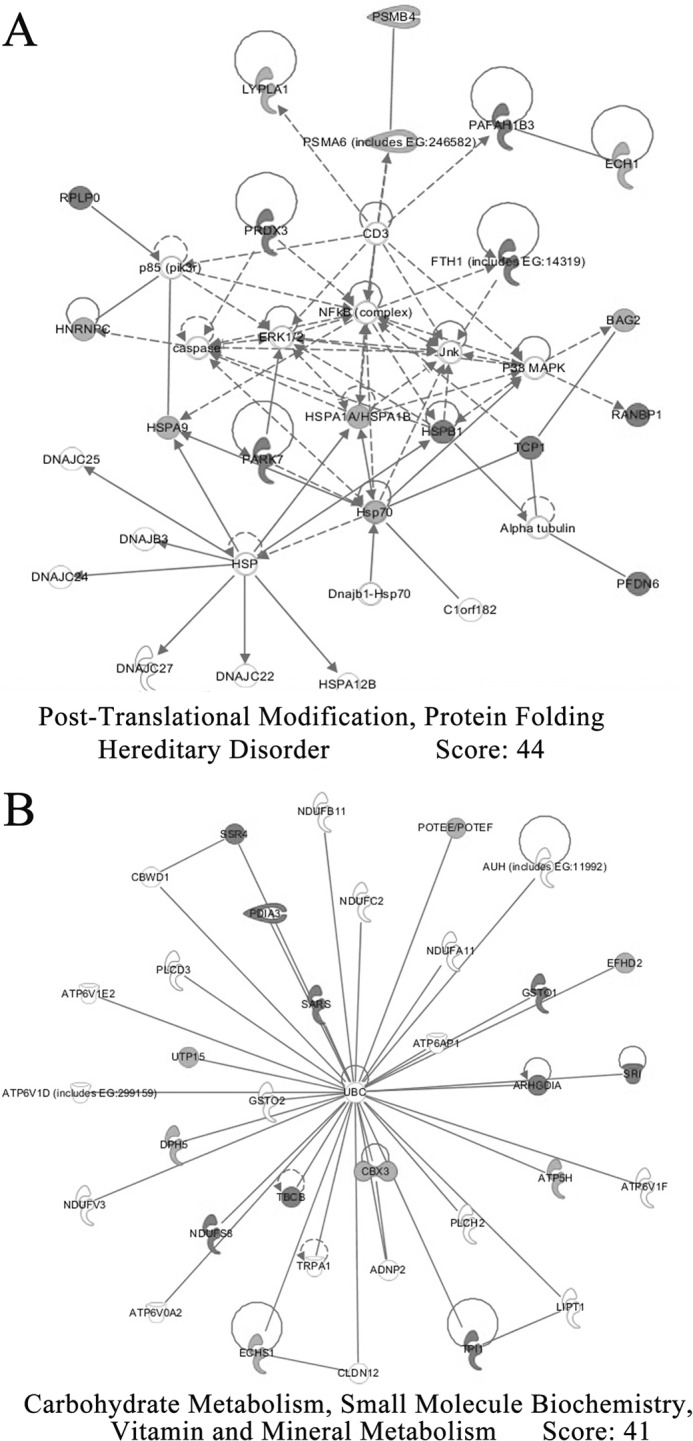
Network analysis of differentially expressed proteins performed using the IPA software. The networks describing proteins affecting gene expression, cell death, and cellular development are presented. Proteins are represented as nodes. Nodes in black represent up-regulated proteins, whereas nodes in gray represent down-regulated proteins. No proteins represented by white nodes were observed. Direct interactions or regulation are connected by solid lines, whereas indirect effects mediated by additional proteins are connected by dashed lines.
DISCUSSION
TEA Inhibits HeLa Cell Proliferation and Induces Cell Apoptosis
It is well understood that TEA can obviously block KChs and induce cell death in a clinic study. K+ is a key cation in the cell; it plays an important role in cell growth and proliferation. In this study we chose different concentrations and treatment periods of TEA exposed to HeLa cells. Both MTT assay and a morphological study (data not shown) reveal that HeLa cells are very sensitive to TEA (Fig. 1), especially when the concentration of TEA is larger than 2 mm, and the growth rates of HeLa cell are inhibited by TEA dose-dependently.
Apoptotic volume decrease is a hallmark of the apoptotic process, during which K+ effluxes and occurs to induce apoptotic body formation. K+ current efflux provokes substantial cellular shrinkage and affects the activation of caspases (31). PI-staining flow cytometry (Fig. 2, A and B) indicates that the TEA changes cell phases distribution. The G0/G1 phase percentage is greatly decreased (from 71.3 to 43.8%) under the TEA treatment, and the apoptosis phase percentage is significantly increased up to 25.6%, compared with only 1% in the control one. Annexin V-FITC/PI dual staining (Fig. 2C) reveals that the apoptosis phase percentage highly relies on the TEA concentration and is 9.6% in 10 mm TEA (data not shown) and 31.9% in 20 mm TEA, respectively. The results obtained from both staining methods are close to each other, supporting that the methods set up in flow cytometry are reliable.
Other Supporting Techniques to Verify Cytotoxicity of TEA
The whole-cell configuration of the patch clamp technique was used to examine K+ current in HeLa cells. It were found that the K+ current was obviously blocked up to 80.8% compared with that in the control group (Fig. 3A). Another cell line, CHO Kv2.1, which is capable of steady expressing the Kv2.1 channel after transfection with CHO, was blocked by TEA again around 30% (Fig. 3B). This block rate is lower than that in HeLa cells, and it is due to more diverse KChs on the HeLa cell membrane, and every subfamily of KChs has different response behaviors of step voltage stimuli, compared to only one Kv2.1 channel in CHO cell.
CDDP is a highly effective anticancer drug in clinic use. In total 19 significantly changed proteins in HepG2 apoptosis were identified by using proteomics techniques under CDDP and CDDP-transferrin treatment (21). Those proteins are common key regulators that are involved in the apoptotic process of various cell lines, such as AKT, p53, p38, and CASP3. RT-qPCR reveals that the mRNA levels of these 19 proteins are evidently changed during exposure to either TEA or 4-AP (Fig. 4), but the presence of both KChs blockers induce the cell to express somewhat different tendencies of mRNA levels, such as #9 MDC, #12 p27, #16 PCNA, #17 PLAA. In a rat cortical study, Chen et al. (32) isolated 4-AP-sensitive outward K+ currents by subtraction of the currents recorded in the presence of both TEA and 4-AP from those recorded in the presence TEA alone. The mRNAs, which have significantly different expression between the TEA and 4-AP group, might result exclusively from the TEA-sensitive outward K+ current and are closely related to TEA-relative apoptosis. On the other hand, the mRNA level of DNA-PK is much higher in 4-AP group and shows a slight change in TEA group. Previous research reported that DNA-PK failed to prevent expression of cyclooxygenase-2 expression; however, TEA attenuates expression of cyclooxygenase-2 (33). It provides evidence that TEA and DNA-PK mediate different pathways in COX-2 expression, and the same is true in the apoptotic process. DNA-PK might dominate in 4-AP-induced apoptosis, but more evidence is needed to support this hypothesis. Currently, the study of the comparison of TEA- and 4-AP-induced apoptosis is sparse. Caspases are crucial mediators of programmed cell death (apoptosis), and caspase-3 is a frequently activated death protease, catalyzing the specific cleavage of many key cellular proteins. Moreover, similar to CDDP as an efficient chemical cure of tumor diseases clinically, TEA is also a highly effective drug against cell apoptosis. It is suggested that TEA and CDDP take a similar or the same pathway for inducing cancer cell apoptosis, such as the caspase-3-dependent pathway (34).
Proteomics Techniques Uncover the Potential Mechanism of TEA-induced Apoptosis
A differential proteomics approach has been developed as an effective tool to identify the biological functions of proteins and, in addition, bottom-up proteomics has also been used widespread to find significant biomarkers in gel-based and gel-free methods. In our study we used this approach to find significant and differential expressed proteins under TEA treatment, and these proteins would aid our understanding of the molecular mechanism of HeLa cell apoptosis induced with TEA. These proteins were divided into several functional groups such as metabolism, oxidative stress response, signal transduction, protein degradation, and translation.
Our study demonstrated that TEA induced lipid metabolism disorder in the HeLa cell. Enoyl-CoA hydratase (ECH) facilitates a trans-2-enoyl-CoA thioester to form a β-hydroxyacyl-CoA thioester by the addition of water to its double bond (35). This is also the second step in the β-oxidation in fatty acid metabolism, and ECH found involved in tumor formation and proliferation, tumor cell adhesion, and migration capacities decreases (36, 37). Platelet-activating factor acetylhydrolase (PAIB3) hydrolyzes platelet-activating factor, and it is very conservative in platelets, lymphocytes, and neutrophils. It recognizes and scavenges fragmented phospholipids under oxidative stress and becomes antagonistic to a platelet-activating factor substrate (38). Acyl-protein thioesterase (LYPA) hydrolyzes fatty acid from serine residues, and overexpression of LYPA accelerates the turnover of palmitate bound to G protein (39). Thus, disorder of fatty acid metabolism associated with TEA induced apoptosis, especially when TEA blocks the K+ channels on the cell membrane, where abundant fatty acids exist.
TEA also exerts significant effects on some important oxidative stress proteins, such as protein disulfide isomerase A3 (PDIA3), thioredoxin-dependent peroxide reductase (PRDX3), and heat shock protein 70 1A/1B (HSP71). PDIA3 is a member of the PDI family and is responsible for disulfide bond oxidation, reduction, and isomerization and protein translation and folding before transport to the endoplasmic reticulum (40). Overexpression of PDI is a protective effect against apoptotic cell death in neurodegenerative disease (41). PDI is associated with ubiquitin and inhibits stress-induced cell apoptosis (42). Peroxiredoxins are a family of enzymes that catalyzes the reduction of hydrogen peroxide and hydroperoxide to water and alcohol, respectively. PRDX3, which is located in the mitochondrial, catalyzes two conserved cysteines to form a disulfide bond (43). PRDX3 reduces reactive oxygen species, and overexpression of this protein protects cells against oxidative stress and reduces the cell apoptotic process (44). In our study, PRDX3 and PDIA3 were down-regulated in the TEA treatment group, indicating unbalanced reactive oxygen species scavenge and formation within cells, leading to the apoptotic process.
HSP70 regulates the tumor apoptotic process and co-expresses with C-Myc and p53 and inhibits apoptosis by down-regulation of the associated genes, protein, or enzyme activities. HSP70 inhibits TNF-α-induced apoptosis by promoting proteasomal degradation of apoptosis signal-regulating kinase 1 (Ask1) (45). Moreover, HSP70 can co-express the BAG family molecular chaperone regulator (BAG2) by regulation of CHIP under oxidative stress. We also noted that BAG2 is another differential protein selected under TEA inducement in our study. Up-regulated HSP70 and BAG2 suggested that TEA itself would give rise to a protective mechanism to against apoptosis.
Our study also showed that TEA treatment affected signal transduction. Sorcin exists in cytoplasm to modulate calcium hemostasis, Ca2+-ATPase activation, and Ca2+-Na+ exchange within in the cell (46, 47). Overexpression of sorcin gives rise to an increase of Bcl-2 and decrease in Bax levels, whereas the knockdown of sorcin leads to the inhibition of cell apoptosis via dysfunction of caspase-3 (48, 49). In our study regulation of sorcin in the TEA treatment group promoted the apoptotic process, and this is in good accordance with previous research. Ca2+ is considered as an intracellular messenger playing signaling roles in all known cell types (50). We also noted that TEA blocked the calcium-activated potassium channel and that efflux of Ca2+ was accompanied with blocking of K+ outward movement. It was tempting to speculate that sorcin differential expression was directly associated with KChs blocking. Translocon-associated protein subunit δ (SSR4) functions as a calcium regulation protein and was also observed down-regulated under TEA treatment. Calcium binding EF-hand is a superfamily protein that participates in enzyme activity regulation, cellular skeleton formation, mitosis, and apoptosis (51). The EF-hand domain-containing protein D2, which was identified as another differential protein in this study, belongs to the EF-hand superfamily protein (52). It is co-expressed with actin and increases the TNF-α level. EFhd2 combines with the caspase-9 complex and induces apoptosis via its substrate during cellular skeleton development. EFhd2 is down-regulated and co-immunoprecipitates with caspase-9, which is involved in transduction in the apoptotic process (53). Similarly, we also observed the decreased abundance of EFhd2 in this study.
Our study showed that TEA treatment affected protein synthesis and degradation. We selected two subunits of proteasome from differential proteins: proteasome subunit α type-6 (PSA6) and proteasome subunit β type-4 (PSB4). They both belong to the 26 S proteasome multisubunit complex that is responsible for degradation of ubiquitinated substrates (54). Proteasome inhibitors induce p53-dependent cell apoptosis. Tubulin-folding cofactor B (TBCB) participates in microtubulin αβ complex formation (55); it is localized in the centrosomes and mid-body and is required for spindle organization, cell abscission, centriole formation, and ciliogenesis (56). However, little research has reported the relationship between TBCB and apoptosis. It is possible that TBCB could be a biomarker for TEA-induced apoptosis. Rho GDIR1 is a member of the GDI family, which keeps the GTPase inactive by binding and stabilizing the GDP-bound form. Rho-GDI is involved in tumor cell apoptosis, invasion, and metastases by post-translational modification (57). GDI is found in human tumors and chemo-resistant cancer cell lines, raising the possibility that RhoGDI might play a role in the development of drug resistance in cancer cells (58). Herein, a decreased GDIR1 level was observed in HeLa cells, indicating that TEA could induce apoptosis by decreasing its expression. It is inferred that GDIR1 could be an anti-apoptotic molecule that mediates cellular resistance to these chemotherapy agents.
Proteomics tools are helpful in uncovering potential biomarkers and the biological pathway of a drug. DJ-1 is first considered as putative oncogene and has been found in down-regulated tumor suppressor PTEN expression (59). In addition, DJ-1 was differentially down-regulated after K+ blocking by TEA treatment. CDDP resistance is associated with a high expression of DJ-1 in immunohistochemical experiments, and silencing DJ-1 decreases the cell proliferation rate effect of CDDP (60). Quantitative proteomics analysis reveals that PDIA3, DJ-1, and GSTO had significantly changed expression in the HeLa/CDDP-resistant group (61). Notably, these proteins also significant changed under CDDP inducement in rat primary hepatocytes (62), and in our study, they are considered as potential TEA biomarkers. It might be concluded that TEA induced HeLa apoptosis not only by blocking KChs but also by following previous and credible the mechanism, such as CDDP, initiating a similar or the same apoptotic biological pathway.
The IPA tool helps us to unlock the insights buried in cell, so as to quickly and easily build pathway models and to combine complex biological systems. One of the networks (Fig. 9B) closely relates to polyubiquitin C, which encodes polyubiquitin precursors. Ion channels are expressed ubiquitously in animal cells and are involved in numerous physiological processes (63). Abnormal expressions of ubiquitin C-associated proteins are due to dysfunction of KChs under TEA inducement. Export of differential protein profiles to IPA analysis would be helpful in identifying new proteins responsive to TEA treatment and would shed further light on the molecular mechanisms of TEA toxicity.
In conclusion, this study aims to reveal alterations in the differential proteins affected by TEA treatment so as to find novel biomarker candidates, and as a result, 33 proteins were found to be greatly modulated by TEA exposure. Identification and functional analysis of these proteins revealed that disturbance of lipid metabolism, oxidative stress damage, calcium binding signal transduction, protein synthesis, and degradation as well as dysfunction of cellular development were the major factors contributing to TEA cytotoxicity in the HeLa cell.
This study set up a cross-talk of TEA toxicity and CDDP by comparing mRNA expression levels of several apoptotic-related proteins found in HepG2 cell line. In addition, DJ-1, PDIA3, and GSTO were selected as differential proteins in TEA-treated groups; they are also regulated proteins in CDDP-resistant HeLa cells studies (61) as well as in a rat primary hepatocyte study of CDDP toxicity (62), and some important differential proteins are discovered in the p53-dependent apoptosis pathway as described above. These results provide new details on the new molecular pathway involved in apoptosis and give a broader context of apoptotic events in therapeutic strategies. However, it is worthy of note that further work is still required for validation before the proteins identified here can be accepted as reliable biomarkers of TEA in cancer clinic study.
Acknowledgments
We thank Dr. Leonard Kaczmarek (Yale University) for CHO cell lines and the experimental design. We also thank Professor John Hodgkiss of The University of Hong Kong for assistance with English.
This work was supported by State Natural Science Fund 30870515 and by PCSIRT (Program for Changjiang Scholars and Innovative Research Team in University) Project IRT0941, China.
- KCh
- potassium channel
- TEA
- tetraethylammonium
- CDDP
- cisplatin
- PI
- propidium iodide
- 4-AP
- 4-aminopyridine
- RT-qPCR
- quantitative real-time PCR
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- GSTO1
- GST Omega 1
- IPA
- Ingenuity Pathways Analysis software
- PCNA
- proliferating cell nuclear antigen
- GDIR1
- GDP dissociation inhibitor 1
- ECH
- enoyl-CoA hydratase
- PDIA3
- protein disulfide (PDI) isomerase
- PRDX3 A3
- thioredoxin-dependent peroxide reductase
- TBCB
- tubulin-folding cofactor B.
REFERENCES
- 1. Saraste A., Pulkki K., Kallajoki M., Henriksen K., Parvinen M., Voipio-Pulkki L.-M. (1997) Apoptosis in human acute myocardial infarction. Circulation 95, 320–323 [DOI] [PubMed] [Google Scholar]
- 2. Lowe S. W., Lin A. W. (2000) Apoptosis in cancer. Carcinogenesis 21, 485–495 [DOI] [PubMed] [Google Scholar]
- 3. Hanayama R., Tanaka M., Miyasaka K., Aozasa K., Koike M., Uchiyama Y., Nagata S. (2004) Autoimmune disease and impaired uptake of apoptotic cells in MFG-E8-deficient mice. Science 304, 1147–1150 [DOI] [PubMed] [Google Scholar]
- 4. Bortner C. D., Cidlowski J. A. (2007) Cell shrinkage and monovalent cation fluxes. Role in apoptosis. Arch. Biochem. Biophys. 462, 176–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Leanza L., Henry B., Sassi N., Zoratti M., Chandy K. G., Gulbins E., Szabò I. (2012) Inhibitors of mitochondrial Kv1.3 channels induce Bax/Bak-independent death of cancer cells. EMBO Mol. Med. 4, 577–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lee G. W., Park H. S., Kim E. J., Cho Y. W., Kim G. T., Mun Y. J., Choi E. J., Lee J. S., Han J., Kang D. (2012) Reduction of breast cancer cell migration via up-regulation of TASK-3 two-pore domain K+ channel. Acta Physiol. (Oxf.) 204, 513–524 [DOI] [PubMed] [Google Scholar]
- 7. Wallace J. L., Gow I. F., Warnock M. (2011) The life and death of breast cancer cells. Proposing a role for the effects of phytoestrogens on potassium channels. J. Membr. Biol. 242, 53–67 [DOI] [PubMed] [Google Scholar]
- 8. Park K.-S., Yang J.-W., Seikel E., Trimmer J. S. (2008) Potassium channel phosphorylation in excitable cells. Providing dynamic functional variability to a diverse family of ion channels. Physiology 23, 49–57 [DOI] [PubMed] [Google Scholar]
- 9. Wible B. A., Wang L., Kuryshev Y. A., Basu A., Haldar S., Brown A. M. (2002) Increased K+ efflux and apoptosis induced by the potassium channel modulatory protein KChAP/PIAS3β in prostate cancer cells. J. Biol. Chem. 277, 17852–17862 [DOI] [PubMed] [Google Scholar]
- 10. Tan Q., Ritzo B., Tian K., Gu L.-Q. (2012) Tuning the tetraethylammonium sensitivity of potassium channel Kcv by subunit combination. J. Gen. Physiol. 139, 295–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Patel S. K., Jackson L., Warren A. Y., Arya P., Shaw R. W., Khan R. N. (2013) A role for two-pore potassium (K2P) channels in endometrial epithelial function. J. Cell Mol. Med. 17, 134–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dilly S., Philippart F., Lamy C., Poncin S., Snyders D., Seutin V., Liégeois J.-F. (2013) The interactions of apamin and tetraethylammonium are differentially affected by single mutations in the pore mouth of small conductance calcium-activated potassium (SK) channels. Biochem. Pharmacol. 85, 560–569 [DOI] [PubMed] [Google Scholar]
- 13. Guidoni L., Carloni P. (2002) Tetraethylammonium binding to the outer mouth of the KcsA potassium channel. Implications for ion permeation. J. Recept. Signal. Transduct. Res. 22, 315–331 [DOI] [PubMed] [Google Scholar]
- 14. Yang K. B., Zhao S. G., Liu Y. H., Hu E. X., Liu B. X. (2009) Tetraethylammonium inhibits glioma cells via increasing production of intracellular reactive oxygen species. Chemotherapy 55, 372–380 [DOI] [PubMed] [Google Scholar]
- 15. Han X., Wang F., Yao W., Xing H., Weng D., Song X., Chen G., Xi L., Zhu T., Zhou J., Xu G., Wang S., Meng L., Iadecola C., Wang G., Ma D. (2007) Heat shock proteins and p53 play a critical role in K+ channel-mediated tumor cell proliferation and apoptosis. Apoptosis 12, 1837–1846 [DOI] [PubMed] [Google Scholar]
- 16. Shen Q.-J., Zhao Y.-M., Cao D.-X., Wang X.-L. (2009) Contribution of Kv channel subunits to glutamate-induced apoptosis in cultured rat hippocampal neurons. J. Neurosci. Res. 87, 3153–3160 [DOI] [PubMed] [Google Scholar]
- 17. Wang G., Zeng J., Shen C.-Y., Wang Z.-Q., Chen S.-D. (2008) Overexpression of Kir2.3 in PC12 cells resists rotenone-induced neurotoxicity associated with PKC signaling pathway. Biochem. Biophys. Res. Commun. 374, 204–209 [DOI] [PubMed] [Google Scholar]
- 18. Chin L. S., Park C. C., Zitnay K. M., Sinha M., DiPatri A. J., Jr., Perillán P., Simard J. M. (1997) 4-Aminopyridine causes apoptosis and blocks an outward rectifier K+ channel in malignant astrocytoma cell lines. J. Neurosci. Res. 48, 122–127 [PubMed] [Google Scholar]
- 19. Kim J. A, Kang Y. S., Jung M. W., Kang G. H., Lee S. H., Lee Y. S. (2000) Ca2+ influx mediates apoptosis induced by 4-aminopyridine, a K+ channel blocker, in HepG2 human hepatoblastoma cells. Pharmacology 60, 74–81 [DOI] [PubMed] [Google Scholar]
- 20. Ji X.-T., Huang L., Huang H.-Q. (2012) Construction of nanometer cisplatin core-ferritin (NCC-F) and proteomic analysis of gastric cancer cell apoptosis induced with cisplatin released from the NCC-F. J. Proteomics 75, 3145–3157 [DOI] [PubMed] [Google Scholar]
- 21. Luo L.-Z., Jin H.-W., Huang H.-Q. (2012) Transferrin-cisplatin specifically deliver cisplatin to HepG2 cells in vitro and enhance cisplatin cytotoxicity. J. Proteomics 77, 237–250 [DOI] [PubMed] [Google Scholar]
- 22. Zhang Y., McKay S. E., Bewley B., Kaczmarek L. K. (2008) Repetitive firing triggers clustering of Kv2.1 potassium channels in aplysia neurons. J. Biol. Chem. 283, 10632–10641 [DOI] [PubMed] [Google Scholar]
- 23. Mosmann T. (1983) Rapid colorimetric assay for cellular growth and survival. Application to proliferation and cytotoxicity assays. J. Immunol. Methods 65, 55–63 [DOI] [PubMed] [Google Scholar]
- 24. Huang Q.-Y., Huang H.-Q. (2011) Differential expression profile of membrane proteins in zebrafish (Danio rerio) brain exposed to methyl parathion. Proteomics 11, 3743–3756 [DOI] [PubMed] [Google Scholar]
- 25. Nicoletti I., Migliorati G., Pagliacci M. C., Grignani F., Riccardi C. (1991) A rapid and simple method for measuring thymocyte apoptosis by propidium iodide staining and flow cytometry. J. Immunol. Methods 139, 271–279 [DOI] [PubMed] [Google Scholar]
- 26. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 27. Huang Q.-Y., Huang L., Huang H.-Q. (2011) Proteomic analysis of methyl parathion-responsive proteins in zebrafish (Danio rerio) brain. Comp. Biochem. Physiol. C. Toxicol. Pharmacol. 153, 67–74 [DOI] [PubMed] [Google Scholar]
- 28. Chen D.-S., Chan K. M. (2009) Changes in the protein expression profiles of the Hepa-T1 cell line when exposed to Cu2+. Aquat. Toxicol. 94, 163–176 [DOI] [PubMed] [Google Scholar]
- 29. Salerno D. M., Tront J. S., Hoffman B., Liebermann D. A. (2012) Gadd45a and Gadd45b modulate innate immune functions of granulocytes and macrophages by differential regulation of p38 and JNK signaling. J. Cell Physiol. 227, 3613–3620 [DOI] [PubMed] [Google Scholar]
- 30. Zhan Q. (2005) Gadd45a, a p53- and BRCA1-regulated stress protein, in cellular response to DNA damage. Mutat. Res. 569, 133–143 [DOI] [PubMed] [Google Scholar]
- 31. Valencia-Cruz G., Shabala L., Delgado-Enciso I., Shabala S., Bonales-Alatorre E., Pottosin I. I., Dobrovinskaya O. R. (2009) Kbg and Kv1.3 channels mediate potassium efflux in the early phase of apoptosis in Jurkat T lymphocytes. Am. J. Physiol. Cell Physiol. 297, C1544–C1553 [DOI] [PubMed] [Google Scholar]
- 32. Chen L., Liu J., Xu C., Keblesh J., Zang W., Xiong H. (2011) HIV-1gp120 induces neuronal apoptosis through enhancement of 4-aminopyridine-sensitive outward K+ currents. PLoS ONE 6, e25994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sun H., Xu B., Sheveleva E., Chen Q. M. (2008) LY294002 inhibits glucocorticoid-induced COX-2 gene expression in cardiomyocytes through a phosphatidylinositol 3 kinase-independent mechanism. Toxicol. Appl. Pharmacol. 232, 25–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Porter A. G., Jänicke R. U. (1999) Emerging roles of caspase-3 in apoptosis. Cell Death Differ. 6, 99–104 [DOI] [PubMed] [Google Scholar]
- 35. Agnihotri G., Liu H.-W. (2003) Enoyl-CoA hydratase: Reaction, mechanism, and inhibition. Bioorg. Med. Chem. 11, 9–20 [DOI] [PubMed] [Google Scholar]
- 36. Zhang J., Song M., Wang J., Sun M., Wang B., Li R., Huang Y., Hou L., Jin Y., Wang M., Tang J. (2011) Enoyl coenzyme A hydratase 1 is an important factor in the lymphatic metastasis of tumors. Biomed. Pharmacother. 65, 157–162 [DOI] [PubMed] [Google Scholar]
- 37. Liu X., Feng R., Du L. (2010) The role of enoyl-CoA hydratase short chain 1 and peroxiredoxin 3 in PP2-induced apoptosis in human breast cancer MCF-7 cells. FEBS Lett. 584, 3185–3192 [DOI] [PubMed] [Google Scholar]
- 38. Thévenin A. F., Monillas E. S., Winget J. M., Czymmek K., Bahnson B. J. (2011) Trafficking of platelet-activating factor acetylhydrolase type II in response to oxidative stress. Biochemistry 50, 8417–8426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Duncan J. A., Gilman A. G. (1998) A cytoplasmic acyl-protein thioesterase that removes palmitate from G protein α subunits and p21RAS. J. Biol. Chem. 273, 15830–15837 [DOI] [PubMed] [Google Scholar]
- 40. Bardwell J. C. (1994) Building bridges. Disulphide bond formation in the cell. Mol. Microbiol. 14, 199–205 [DOI] [PubMed] [Google Scholar]
- 41. Tanaka S., Uehara T., Nomura Y. (2000) Up-regulation of protein-disulfide isomerase in response to hypoxia/brain ischemia and its protective effect against apoptotic cell death. J. Biol. Chem. 275, 10388–10393 [DOI] [PubMed] [Google Scholar]
- 42. Ko H. S., Uehara T., Nomura Y. (2002) Role of ubiquilin associated with protein-disulfide isomerase in the endoplasmic reticulum in stress-induced apoptotic cell death. J. Biol. Chem. 277, 35386–35392 [DOI] [PubMed] [Google Scholar]
- 43. Song I.-S., Kim H.-K., Jeong S.-H., Lee S.-R., Kim N., Rhee B. D., Ko K. S., Han J. (2011) Mitochondrial peroxiredoxin III is a potential target for cancer therapy. Int. J. Mol. Sci. 12, 7163–7185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nonn L., Berggren M., Powis G. (2003) Increased expression of mitochondrial peroxiredoxin-3 (thioredoxin peroxidase-2) protects cancer cells against hypoxia and drug-induced hydrogen peroxide-dependent apoptosis. Mol. Cancer Res. 1, 682–689 [PubMed] [Google Scholar]
- 45. Satou M., Nishi Y., Yoh J., Hattori Y., Sugimoto H. (2010) Identification and characterization of acyl-protein thioesterase 1/lysophospholipase I as a ghrelin deacylation/lysophospholipid hydrolyzing enzyme in fetal bovine serum and conditioned medium. Endocrinology 151, 4765–4775 [DOI] [PubMed] [Google Scholar]
- 46. Matsumoto T., Hisamatsu Y., Ohkusa T., Inoue N., Sato T., Suzuki S., Ikeda Y., Matsuzaki M. (2005) Sorcin interacts with sarcoplasmic reticulum Ca2+-ATPase and modulates excitation-contraction coupling in the heart. Basic Res. Cardiol. 100, 250–262 [DOI] [PubMed] [Google Scholar]
- 47. Fowler M. R., Colotti G., Chiancone E., Higuchi Y., Seidler T., Smith G. L. (2009) Complex modulation of L-type Ca2+ current inactivation by sorcin in isolated rabbit cardiomyocytes. Pflügers Arch. 457, 1049–1060 [DOI] [PubMed] [Google Scholar]
- 48. Kawakami M., Nakamura T., Okamura N., Komoto C., Markova S., Kobayashi H., Hashimoto N., Okumura K., Sakaeda T. (2007) Knock-down of sorcin induces up-regulation of MDR1 in HeLa cells. Biol. Pharm. Bull. 30, 1065–1073 [DOI] [PubMed] [Google Scholar]
- 49. Qi J., Liu N., Zhou Y., Tan Y., Cheng Y., Yang C., Zhu Z., Xiong D. (2006) Overexpression of sorcin in multidrug-resistant human leukemia cells and its role in regulating cell apoptosis. Biochem. Biophys. Res. Commun. 349, 303–309 [DOI] [PubMed] [Google Scholar]
- 50. Bhattacharjee A., Kaczmarek L. K. (2005) For K+ channels, Na+ is the new Ca2+. Trends Neurosci. 28, 422–428 [DOI] [PubMed] [Google Scholar]
- 51. Capozzi F., Luchinat C., Micheletti C., Pontiggia F. (2007) Essential dynamics of helices provide a functional classification of EF-hand proteins. J. Proteome Res. 6, 4245–4255 [DOI] [PubMed] [Google Scholar]
- 52. Xu Y., Liu W., Shen H., Yan J., Yang E., Wang H. (2010) Recombinant Mycobacterium bovis BCG expressing chimaeric protein of Ag85B and ESAT-6 enhances immunostimulatory activity of human macrophages. Microbes Infect. 12, 683–689 [DOI] [PubMed] [Google Scholar]
- 53. Checińska A., Giaccone G., Rodriguez J. A., Kruyt F. A., Jimenez C. R. (2009) Comparative proteomics analysis of caspase-9-protein complexes in untreated and cytochrome c/dATP stimulated lysates of NSCLC cells. J. Proteomics 72, 575–585 [DOI] [PubMed] [Google Scholar]
- 54. Wang X., Chen C.-F., Baker P. R., Chen P.-L., Kaiser P., Huang L. (2007) Mass spectrometric characterization of the affinity-purified human 26 S proteasome complex. Biochemistry 46, 3553–3565 [DOI] [PubMed] [Google Scholar]
- 55. Li H., DeRosier D. J., Nicholson W. V., Nogales E., Downing K. H. (2002) Microtubule structure at 8 Å resolution. Structure 10, 1317–1328 [DOI] [PubMed] [Google Scholar]
- 56. Fanarraga M. L., Carranza G., Castaño R., Jiménez V., Villegas J. C., Zabala J. C. (2010) Emerging roles for tublin folding cofactors at the centrosome. Commun. Integr. Biol. 3, 306–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cui J.-W., Li W.-H., Wang J., Li A.-L., Li H.-Y., Wang H.-X., He K., Li W., Kang L.-H., Yu M., Shen B.-F., Wang G.-J., Zhang X.-M. (2005) Proteomics-based identification of human acute leukemia antigens that induce humoral immune response. Mol. Cell Proteomics 4, 1718–1724 [DOI] [PubMed] [Google Scholar]
- 58. Zhang B., Zhang Y., Dagher M.-C., Shacter E. (2005) Rho GDP Dissociation inhibitor protects cancer cells against drug-induced apoptosis. Cancer Res. 65, 6054–6062 [DOI] [PubMed] [Google Scholar]
- 59. Lei Y., Huang K., Gao C., Lau Q. C., Pan H., Xie K., Li J., Liu R., Zhang T., Xie N., Nai H. S., Wu H., Dong Q., Zhao X., Nice E. C., Huang C., Wei Y. (2011) Proteomics identification of ITGB3 as a key regulator in reactive oxygen species-induced migration and invasion of colorectal cancer cells. Mol. Cell Proteomics 10.1074/mcp.M110.005397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zeng H.-Z., Qu Y.-Q., Zhang W.-J., Xiu B., Deng A.-M., Liang A.-B. (2011) Proteomic analysis identified DJ-1 as a cisplatin-resistant marker in non-small cell lung cancer. Int. J. Mol. Sci. 12, 3489–3499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chavez J. D., Hoopmann M. R., Weisbrod C. R., Takara K., Bruce J. E. (2011) Quantitative proteomic and interaction network analysis of cisplatin resistance in HeLa cells. PLoS ONE 6, e19892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cho Y.-E., Singh T. S. K., Lee H.-C., Moon P.-G., Lee J.-E., Lee M.-H., Choi E.-C., Chen Y.-J., Kim S.-H., Baek M.-C. (2012) In-depth identification of pathways related to cisplatin-induced hepatotoxicity through an integrative method based on an informatics-assisted label-free protein quantitation and microarray gene expression approach. Mol. Cell Proteomics 10.1074/mcp.M111.010884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Abriel H., Staub O. (2005) Ubiquitylation of ion channels. Physiology 20, 398–407 [DOI] [PubMed] [Google Scholar]