Background: Formyl peptide receptor 1 (FPR1) and FPR2 are highly homologous but bind fMet-Leu-Phe with very different affinities.
Results: Asp-281 provides a negative charge that renders FPR2 more sensitive to the length and composition of formyl peptides than FPR1.
Conclusion: Asp-281 is a major determinant for FPR2 binding.
Significance: This work provides a structural basis for differential interaction between formyl peptides and their receptors.
Keywords: Chemokines, Computer Modeling, G Protein-coupled Receptors (GPCR), Peptides, Receptor Structure-Function
Abstract
Unlike formyl peptide receptor 1 (FPR1), FPR2/ALX (FPR2) interacts with peptides of diverse sequences but has low affinity for the Escherichia coli-derived chemotactic peptide fMet-Leu-Phe (fMLF). Using computer modeling and site-directed mutagenesis, we investigated the structural requirements for FPR2 to interact with formyl peptides of different length and composition. In calcium flux assay, the N-formyl group of these peptides is necessary for activation of both FPR2 and FPR1, whereas the composition of the C-terminal amino acids appears more important for FPR2 than FPR1. FPR2 interacts better with pentapeptides (fMLFII, fMLFIK) than tetrapeptides (fMLFK, fMLFW) and tripeptide (fMLF) but only weakly with peptides carrying negative charges at the C terminus (e.g. fMLFE). In contrast, FPR1 is less sensitive to negative charges at the C terminus. A CXCR4-based homology model of FPR1 and FPR2 suggested that Asp-2817.32 is crucial for the interaction of FPR2 with certain formyl peptides as its negative charge may be repulsive with the terminal COO- group of fMLF and negatively charged Glu in fMLFE. Asp-2817.32 might also form a stable interaction with the positively charged Lys in fMLFK. Site-directed mutagenesis was performed to remove the negative charge at position 281 in FPR2. The D2817.32G mutant showed improved affinity for fMLFE and fMLF and reduced affinity for fMLFK compared with wild type FPR2. These results indicate that different structural determinants are used by FPR1 and FPR2 to interact with formyl peptides.
Introduction
Bacterial protein synthesis starts with an N-formyl methionine, hence producing N-formylated peptides and proteins. In mammals, mitochondrial protein synthesis shares this feature, and formyl peptides are released upon cell death. Mammalian phagocytes have cell surface receptors specialized in the detection of these formyl peptides that are strong chemoattractants for neutrophils (1, 2). In neutrophils, formyl peptide receptors (FPRs)2 are largely responsible for detection of invading bacteria, which guides these phagocytes to the site of infection and initiates a cascade of bactericidal activities including degranulation and superoxide generation (3, 4). In animal studies, mice lacking the Fpr1 gene are susceptible to certain bacterial infection (5), suggesting an important role for the receptor in host defense. Detection of mitochondrial formyl peptides by the FPRs is an important part of the inflammatory responses to endogenous damage-associated molecular patterns (6, 7).
The FPRs are G protein-coupled receptors (GPCRs) with a seven-membrane-span structure (4, 8). In humans, two FPRs are expressed on neutrophils, FPR1 and FPR2. FPR1 displays a high affinity (kd ∼ 1 nm) to the prototypic formyl peptide, N-formyl-Met-Leu-Phe (fMLF), which is an Escherichia coli-derived chemoattractant. FPR1 is well known for its ability to initiate Gαi-mediated signaling events including activation of the phosphatidylinositol 3-kinase, mitogen-activated protein kinases, and small GTPases that ultimately lead to cytoskeleton reorganization, cell migration, and release of highly toxic oxygen radicals as well as protein degradation enzymes. In contrast, FPR2 shares 69% sequence identity at the protein level yet displays low binding affinity for fMLF (kd ∼ 430 nm). Combined with its promiscuity in binding other peptides and nonpeptide ligands (4), it is questionable whether FPR2 is a bona fide formyl peptide receptor.
Numerous attempts were made to study the ligand binding pocket of FPR1 despite the lack of crystal structure for the receptor. Biochemical approaches including construction and analysis of receptor chimeras, point mutations, and cross-linking to fluorescent probes have been used in FPR1 binding characterization. Quehenberger et al. prepared and characterized chimera receptors consisting of FPR1 and FPR2 and identified three clusters of residues in the extracellular loops of FPR1 for high affinity interaction with fMLF (9). In addition, introduction of two positively charged residues (Arg and Lys) to positions 84 and 85 converted the low affinity FPR1/FPR2 chimera to a high affinity receptor for fMLF binding (10). Other studies employed point mutagenesis approaches and identified charged residues including Arg-842.63, Lys-852.64, Arg-2055.42, and Asp-2847.36 as well as Arg-1634.63 as being important for fMLF binding (11, 12). To further demonstrate the ligand binding properties, additional cross-link studies were conducted, and the results indicate that Lys-852.64 and Asp-2847.36 in FPR1 form an electrostatic interaction and formyl peptide binding serves to disrupt this interaction (13). Although these mutational and biochemical studies give insight into the interaction of formyl peptides with FPR1, studies of other GPCRs indicate that the interaction between receptors and a given ligand is complex and may vary in different receptors. Therefore, it is unclear whether the FPR1 ligand binding properties can be applied to FPR2.
In the present study we examined the ability of FPR2 to bind and respond to formyl peptides of different length and composition. We also constructed computer models based on available biochemical data from studies of the FPRs and on structural information of several recently crystallized GPCRs. This study follows a previous attempt in characterizing mitochondrial peptides that activate FPR2 (14) and our recent study of the mouse homologues of human FPRs (15). We report here the identification of structural features of formyl peptides required for full activation of FPR2 as well as constituents of the putative receptor binding site for these peptides. Our results suggest that FPR1 and FPR2 use different structural determinants for their interaction with formyl peptides.
EXPERIMENTAL PROCEDURES
Materials
The tripeptide fMLF (≥90% purity) was purchased from Sigma. Fluorescent peptides including WK(FITC)YMVm (m = D-Met; ≥95% purity), fMLFIK-FITC, and fMLFK-FITC were synthesized and conjugated by ChinaPeptides Co., Ltd (Shanghai, China). Other formyl and non-formyl peptides were synthesized at the Protein Research Laboratory at the University of Illinois at Chicago and purified to ≥90% homogeneity as indicated by mass spectrometry. FLIPR Calcium 5 reagent was purchased from Molecular Devices (Sunnyvale, CA). The KOD-plus-new PCR polymerase was purchased from TOYOBO Co., Ltd. (Osaka, Japan). Other chemicals were purchased from Sigma.
Computational Models of FPR1 and FPR2
Fig. 1A shows a phylogenetic tree of human FPR1 and FPR2 and all class A GPCRs with known structure. It can be seen that FPRs are located in a branch that includes opioid receptors (OPRX, OPRK, OPRD, and OPRM), the protease-activated receptor (PAR1), the neurotensin receptor (NTR1), and the chemokine CXCR4 and CCR5 receptors, with the latter being the most closely related receptor. This subfamily of peptide receptors is characterized by unique molecular signatures such as an ECL2 formed by two β-strands that maintain the binding site accessible from the extracellular environment and a closed helical segment (310 helix or tight turn) at the extracellular part of TM2 due to the conserved (S/T)×P2.58 motif (16). Thus, FPR1 and FPR2 were modeled using the structure of the CXCR4 chemokine receptor as template (PDB code 3ODU) (17) using Modeler 9v8 (18). The highly conserved Asn1.50 in TM 1, Asp2.50 in TM 2, Arg3.50 in TM 3, Trp4.50 in TM 4, Pro5.50 in TM 5, Pro6.50 in TM 6, and Pro7.50 in TM 7, which define the Ballesteros and Weinstein numbering scheme (19), were used as reference points in TM sequence alignments (see Fig. 1B). The extended conformation of fMLF was docked into the homology model of FPR1 using the Autodock Vina tool (20). All docking solutions were visually inspected and the pose in which the N-formyl group hydrogen bonds Arg-2055.42 and the negatively charged C terminus interacts with Arg-842.63 and Lys-852.64 was refined with energy minimization and molecular dynamics (MD) simulations. This computed peptide-receptor complex (fMLF-FPR1) was used as a template to model the binding of the studied peptides to FPR1 and FPR2.
FIGURE 1.
Sequence analysis of formyl peptide receptors. A, phylogenetic tree for all class A GPCRs with known structure plus human FPR1 and FPR2. Values at each ramification correspond to the bootstrap percentages. B, sequence alignment of CXCR4 (Uniprot ID: P61073) and human FPR1 (P21462) and FPR2 (P25090). The highly conserved Asn1.50 in TM 1, Asp2.50 in TM 2, Arg3.50 in TM 3, Trp4.50 in TM 4, Pro5.50 in TM 5, Pro6.50 in TM 6, and Pro7.50 in TM 7 used for the alignment of TM helices are shown in black, and conserved amino acids are shown in gray. Regions corresponding to TM helices or β-strands (in ECL2) of the CXCR4 crystal structure (PDB code 3ODU) are highlighted.
Molecular Dynamics Simulations of Peptide-Receptor Complexes
In a second step, these binding modes were refined using MD simulations of the peptide-receptor complexes. The complexes were embedded in a lipid bilayer (186 molecules of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine) with explicit solvent (14,300 water molecules) and counterions (65 Na+ and 75 Cl−). Model systems were energy-minimized and subsequently subjected to a 1-ns MD equilibration, with positional restraints on the Cα atoms of the receptor, to remove possible voids present in protein/lipids or protein/water interfaces. These restraints were released, and 100-ns MD trajectories were produced at constant pressure and temperature using the particle mesh Ewald method to evaluate electrostatic interactions with the GROMACS software v4.53 (21) using the protocol previously described (22). The stability of the peptide-receptor complexes was monitored by root mean square deviations.
Site-directed Mutagenesis
The mutation Asp-281 to Gly (D2817.32G) was introduced into the cDNA for FPR2 using a one-step PCR protocol with two back-to-back mutagenic primers. The full-length mutagenic fragment inside the plasmid vector pSFFV.neo was extended by KOD-plus-new polymerase (TOYOBO). The mutation was confirmed by DNA sequencing.
Cell Culture
The rat basophil leukemia cell line RBL-2H3, which does not respond to known formyl peptides, was transfected with an expression vector SFFV.neo containing the cDNAs for FPR1, FPR2, or its mutant D2817.32G as previously described (23). Stable transfectants were continuously selected with G418 (250 μg/ml) after initial transfectants were isolated with 500 μg/ml G418. Flow cytometry analysis of stable cell lines was conducted using anti-FPR2 antibodies (GENOVAC, Freiburg, Germany) to determine receptor expression. RBL-transfected cells were maintained in DMEM supplemented with 20% FBS.
Calcium Mobilization
Stable transfectants of RBL cells were grown to confluence in black/clear bottom 96-well assay plates. Prior to experiments cells were washed with 0.5% bovine serum albumin in Hanks' balanced saline solution (with Ca2+ and Mg2+) and incubated in the same buffer. Cells were loaded for 1 h at 37 °C with the FLIPR calcium-sensitive dye (Calcium 5) according to the manufacturer's protocols (Molecular Devices). The addition of agonists was robotically controlled, and samples were read in a FlexStation III (Molecular Devices). Cells were excited at 485 nm and detected with an emission wavelength of 525 nm. Maximal Ca2+ increase was ∼500 nm. For internal control, 1 μm ATP was added. Data were acquired by SoftMax® Pro 6 (Molecular Devices) and analyzed with Origin 8.5 software (Northampton, MA). The dose-response curves were plotted as the means ± S.E. based on at least 3 experiments using 7–9 different concentrations of agonists.
Degranulation
β-Hexosaminidase release was measured as described (24). RBL-2H3 cells (0.2 × 106/well) expressing hFPR1, hFPR2, or D2817.32G were used in degranulation assays, which included a preincubation with 10 μm cytochalasin B followed by agonist stimulation for 10 min (24). After termination of the reaction, absorbance was monitored at 405 nm in a FlexStation III Spectrometer (Molecular Devices). Values (means ± S.E.) were expressed as a percent of total β-hexosaminidase present in the cells.
cAMP Detection
RBL-FPR1, RBL-FPR2, and RBL-D2817.32G cells were cultured in 24-well plates for 24–48 h. The culture medium containing 20% FBS were removed and replaced with 500 μl of serum-free DMEM containing 10 μm forskolin plus agonists at different concentrations. After 30 min of incubation at 37 °C, the cells were washed 3 times in cold PBS and resuspended in 300 μl of lysis buffer. cAMP level was determined using a commercially available kit (R&D Systems, Minneapolis, MN).
Ligand Binding Assay
Cells for saturation and competitive binding assays were prepared as described (15). For saturation binding assays, the fluorescein isothiocyanate (FITC)-conjugated peptide WK(FITC)YMVm was used at concentrations from 10−10 to 10−7 m, whereas fMLFK-FITC and fMLFIK-FITC were used from 10−9 to 10−5 m. Total binding and nonspecific binding were measured in the absence and presence of unlabeled ligand in access (50 μm WKYMVm or 50 μm fMLFIK). The cells were equilibrated for 1 h on ice and then analyzed for mean fluorescent intensity on a FACSCalibur® flow cytometer (BD Biosciences). The maximum binding sites were estimated with FITC-conjugated bead standards of known fluorescein equivalents (QuantumTM FITC-5 Premix, Lot #9975, Bangs Laboratories, Fishers, IN). The relative affinity of non-fluorescent ligands were measured in competitive binding assays in which WK(FITC)YMVm of saturated concentration (2.5 nm) was added on ice for 1 h before the addition of increasing concentrations of competitors. Samples were incubated for another hour on ice, and mean fluorescent intensity values were obtained by flow cytometry. Data were analyzed with Origin 8.5 software (Northampton, MA) and the competitive binding curves were plotted as the means ± S.E. based on at least three independent experiments.
RESULTS
Staphylococcus aureus- and Listeria monocytogenes-derived Formyl Peptides Are Potent Agonists for FPR2
FPR2 is a promiscuous receptor activated by a number of different agonists from a variety of sources. However, the highly potent tripeptide for FPR1, fMLF (1), is a weak agonist for FPR2, as it requires micromolar concentrations of the tripeptide to active FPR2 (25). The discrepancy in the response to fMLF suggests that different structural determinants are used for fMLF binding by FPR2 despite sharing 69% identical sequence with FPR1 (Fig. 1). We sought to determine whether formyl peptides with amino acid sequences from other bacterial strains are potent agonists for FPR2. S. aureus and L. monocytogenes produce chemotactic peptides that are less well characterized. Of the six formyl peptides identified from S. aureus cultures, one with the sequence fMIFL had the greatest effect on neutrophil chemotaxis (26). Chemotactic peptides from L. monocytogenes, including fMIVIL, were found to be highly potent on human phagocytes (14). The S. aureus- and L. monocytogenes-derived formyl peptides were recently found to activate mouse Fpr1, which also displays low affinity for fMLF (15, 27). We tested these peptides (structures in Fig. 2A) in FPR2-expressing RBL cells (RBL-FPR2) for their ability to mobilize calcium. Both the tetrapeptide fMIFL and the pentapeptide fMIVIL induced calcium mobilization in nanomolar concentrations (EC50 = 89 and 400 nm, respectively; Table 1), whereas fMLF did not induce significant calcium release at this concentration (Fig. 2B). Dose-dependent Ca2+ mobilization was determined in RBL-FPR2 cells. fMLF induced calcium mobilization only when used at micromolar concentrations (EC50 = 6.7 μm, Fig. 2C). The other two peptides, fMIFL and fMIVIL, were ∼100- and 10-fold more potent than fMLF, respectively (Fig. 2, D and E).
FIGURE 2.
Calcium mobilization in RBL-FPR2 cells stimulated with formyl and non-formyl peptides. A, the structures of formyl peptides fMLF (E. coli), fMIFL (S. aureus), fMIVIL (L. monocytogenes), and unformylated peptides MLF, MIFL, MIVIL. B, time-dependent mobilization of Ca2+ in RBL-FPR2 cells stimulated with fMLF, fMIVIL, and fMIFL. Peptides (100 nm) were added at the time indicated by the arrow. Tracings are shown as the ratio of the reading at 525 nm over base line and are representative of three separate experiments. C–E, dose-dependent induction of Ca2+ mobilization in RBL-FPR2 cells stimulated with formyl peptides (fMLF, fMIFL, fMIVIL) and non-formyl peptides of the same sequence (MLF, MIFL, MIVIL). Peak values of Ca2+ mobilization at the indicated concentrations are shown as the means ± S.E., based on three separate experiments, each conducted in triplicate.
TABLE 1.
Comparison of various formyl peptides for their potency in RBL cells expressing FPR1, FPR2, and FPR2-D2817.32G
The EC50 values (m) in calcium mobilization assays and IC50 values (m) in forskolin-induced cAMP accumulation assays are shown as the means ±S.E. based on at least three independent experiments. For agonist-stimulated calcium flux assays, stably transfected cells were washed and incubated for 1 h at 37 °C with FLIPR calcium-sensitive dye. The addition of agonists was robotically controlled. Cells were excited at 485 nm and detected with an emission wavelength of 525 nm. For agonist-inhibited cAMP accumulation induced by forskolin, cells were incubated for 0.5 h at 37 °C with 10 μm forskolin plus agonists at different concentrations. cAMP level was then determined using a commercial available kit (Mouse/Rat cAMP Parameter Assay, R&D Systems).
| Formyl Peptide | Ca2+ mobilization (EC50) |
cAMP accumulation (IC50) |
||||
|---|---|---|---|---|---|---|
| FPR1 | FPR2 | FPR2-D2817.32G | FPR1 | FPR2 | FPR2-D2817.32G | |
| m | m | |||||
| fMLF | 3.5 (±0.6) × 10−9 | 6.7 (±3.0) × 10−6 | 8.8 (±2.6) × 10−7 | 3.4 (±1.6) × 10−9 | 2.5 (±1.2) × 10−6 | 2.1 (±0.4) × 10−7 |
| fMLFE | 2.8 (±0.9) × 10−9 | 4.8 (±10) × 10−5 | 1.8 (±2.4) × 10−6 | 5.3 (±0.9) × 10−9 | 2.6 (±0.9) × 10−4 | 1.4 (±0.6) × 10−6 |
| fMLFK | 3.9 (±1.6) × 10−9 | 8.6 (±1.0) × 10−8 | 9.4 (±1.8) × 10−7 | 4.0 (±0.6) × 10−9 | 5.1 (±0.8) × 10−8 | 8.4 (±0.7) × 10−7 |
| fMLFW | 4.3 (±0.5) × 10−9 | 7.0 (±0.6) × 10−7 | 1.1 (±0.1) × 10−6 | 5.5 (±1.8) × 10−9 | 6.4 (±0.6) × 10−7 | 9.4 (±0.5) × 10−7 |
| fMLFII | 1.4 (±0.4) × 10−12 | 3.8 (±0.8) × 10−9 | 4.1 (±7.5) × 10−9 | 4.9 (±0.5) × 10−11 | 6.9 (±1.3) × 10−9 | 4.1 (±1.7) × 10−9 |
| fMLFIK | 2.0 (±0.5) × 10−9 | 1.2 (±0.4) × 10−8 | 2.2 (±0.1) × 10−8 | 2.6 (±0.6) × 10−9 | 5.2 (±1.6) × 10−8 | 2.1 (±0.3) × 10−7 |
| fMIFL | 4.2 (±0.4) × 10−12 | 8.9 (±1.1) × 10−8 | 7.6 (±4.7) × 10−8 | 7.1 (±0.7) × 10−11 | 2.6 (±0.7) × 10−7 | 8.6 (±3.1) × 10−8 |
| fMIVIL | 2.7 (±0.5) × 10−11 | 4.0 (±0.1) × 10−7 | 4.8 (±0.1) × 10−7 | 4.8 (±1.1) × 10−10 | 9.0 (±3.1) × 10−7 | 4.8 (±0.6) × 10−7 |
Optimal Activation of FPR2 Requires an N-Formyl Group
Although FPR2 is a low affinity receptor for the prototypic formyl peptide fMLF, the fact that it is activated by differential agonists demonstrates a role in regulating inflammatory responses (4, 28). The agonists that activate FPR2 are mostly peptides, some of them without an N-formyl methionine. To determine whether FPR2 requires the N-formyl group in these peptides for activation, peptides were synthesized with and without the N-formyl group, and their potency at FPR2 was compared. Consistent with previous findings (29), fMLF demonstrated low potency to the FPR2 receptor, and removal of the N-terminal formyl group (MLF) abrogated activation of FPR2 (Fig. 2C). Similar effects were obtained with peptides derived from S. aureus (fMIFL) and L. monocytogenes (fMIVIL) (Fig. 2, D and E). These findings reinforce the notion that FPR2 is a formyl peptide receptor.
C-terminal Amino Acids Are Structural Determinants for the Potency and Efficacy of Formyl Peptides at FPR2
Based on previous characterization of FPR1 interaction with formyl peptides, the FPR1 binding pocket can accommodate peptides up to five amino acids (29, 30). Formyl peptides of five and six residues may be conjugated to their C termini with the bulky FITC without losing their potency and efficacy (30), suggesting that the length and composition of the C termini and its chemical modification may not be critical to FPR1 binding. We sought to determine whether the projected ligand binding pocket of FPR2 has similar properties. The low potency and efficacy of fMLF at FPR2, compared with the high potency and efficacy of longer formyl peptides such as fMIFL and fMIVIL (see above), suggests that peptide length could be an important determinant for FPR2 activation.
To investigate the C-terminal interaction of the ligands with FPR2, we synthesized formyl peptides with various C-terminal substitutions to determine the effect of length and charge on FPR2 activation (Fig. 3). The fMLFK and fMLFW tetrapeptides and the fMLFII and fMLFIK pentapeptides showed increased potency over fMLF with the order of fMLFII > fMLFIK > fMLFK > fMLFW > fMLF based on EC50 values from the calcium mobilization assay (Fig. 3). The pentapeptide fMLFII was 1500-fold more potent than fMLF in the induction of FPR2-dependent calcium mobilization. However, placing a negatively charged residue (Glu, Asp) at the C terminus resulted in a peptide with extremely low potency, as fMLFE did not show activity unless used at concentrations of 10 μm and above (Fig. 3). A similarly charged peptide fMLFD also lacked potency (data not shown). Similar results were obtained in other functional assays, including degranulation (Fig. 4) and agonist-induced inhibition of cAMP accumulation (see Fig. 9 and Table 1). Although fMLFK and fMLFIK showed higher potency on FPR2 (EC50 = 86 and 12 nm in calcium flux assays, IC50 = 51 and 52 nm in cAMP reduction, Table 1), conjugation of FITC to their C termini abrogated potency (data not shown) and affinity (Fig. 5A and Table 2). For this reason, a FITC-conjugated non-formyl peptide WKYMVm, which retained high affinity at FPR2 (Kd = 0.8 nm; Fig. 5B and Table 3), was used in FPR2 binding assays. Competition binding assays were performed in the presence of 2.5 nm WK(FITC)MVm. As shown in Fig. 5C, fMLFIK, fMLFII, and fMLFK were strong competitors. In contrast, fMLF and fMLFE were weak competitors, and fMLFE showed no ability to displace WK(FITC)YMVm. These binding properties were consistent with the potency of the peptides in functional assays (see Figs. 3, 4, and 9, Tables 1 and 3). Collectively, the results suggest involvement of the C-terminal amino acids in peptide interaction with FPR2. We concluded that the lengths of the peptides as well as their C-terminal charges are determinants for optimal agonistic activity at FPR2.
FIGURE 3.
Effects of length and amino acid substitution on the potency of formyl peptides in calcium flux assays. The structures of formyl peptides used are shown on top of each panel (A–F). The dose-response curves are shown in the lower part of panel of each group. Dose curves are shown as the means ± S.E. of peak Ca2+ values at the indicated concentrations based on three independent experiments, each conducted in triplicate.
FIGURE 4.
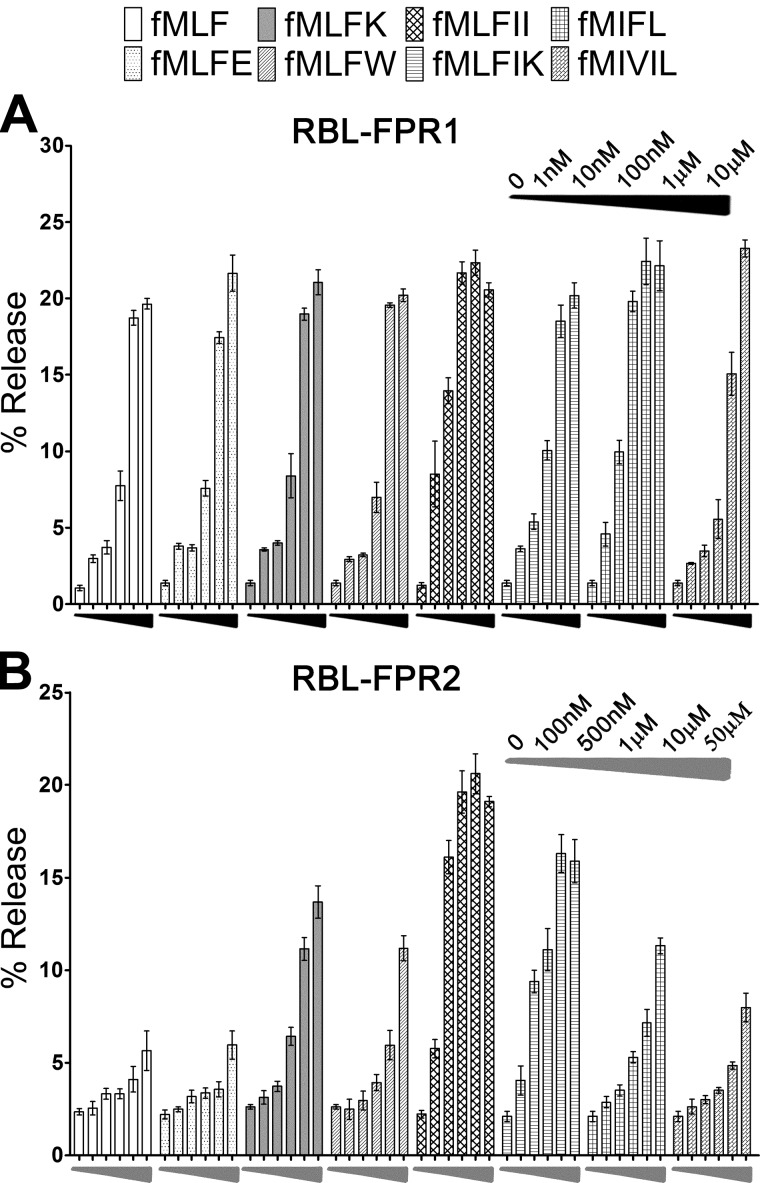
Comparison of degranulation induced by formyl peptides in RBL-FPR1 and RBL-FPR2 cells. RBL cells expressing FPR1 (A) or FPR2 (B) were stimulated with formyl peptides at indicated concentrations for 10 min. The β-hexosaminidase secreted into the culture medium was measured. Data are represented as the percentage of total cellular β-hexosaminidase released and are expressed as the means ± S.E. based on at least three separate experiments, each in duplicate.
FIGURE 9.
Effects of length and terminal charges of formyl peptides on cAMP accumulation. In the presence of 10 μm forskolin, formyl peptides including fMLF (A), fMLFE (B), fMLFK (C), fMLFW (D), fMLFII (E), fMLFIK (F), fMIFL (G), and fMIVIL (H) at different concentrations were incubated with RBL-2H3 cells expressing wild type FPR2 and D2817.32G, respectively. The dose curves for formyl peptides are shown as the means ± S.E. of cAMP concentrations detected at 450 nm after 30 min of incubation at 37 °C, calculated from cAMP standard curves (not shown). Three separate experiments were conducted in duplicate, and standard curves were generated for each set of samples.
FIGURE 5.
C-terminal modifications of formyl peptides differentially affect binding to FPR2 and FPR1. Total, nonspecific, and specific binding of fMLFIK-FITC (A and D) and WK(FITC)YMVm (B and E) to RBL-FPR2 cells (A and B) and RBL-FPR1 cells (D and E) are shown. Competition binding assays were performed on FPR2 (E) and FPR1 (F) in the presence of 2.5 nm WK(FITC)YMVm, which was competitively displaced by increasing concentrations of the indicated formyl peptides. Data were analyzed with the Origin 8.5 software, and the results are shown as the means ± S.E. from at least three experiments. MFI, mean fluorescent intensity.
TABLE 2.
Determination of fluorescent agonists binding to RBL cells expressing FPR1, FPR2, and the FPR2 mutant D2817.32G
For saturation binding assays, the FITC-conjugated peptides WK(FITC)YMVm, fMLFK-FITC, and fMLFIK-FITC were used, respectively. Total binding and nonspecific binding were measured in the absence and presence of unlabeled ligands in access (50 μm WKYMVm or fMLFIK). The cells were equilibrated for 1 h on ice and then analyzed for mean fluorescent intensity. The equilibrium dissociation constant Kd (nm) and Bmax (maximum binding capacity of the receptors) values of specific binding are shown in the table. The number of binding sites is calculated using FITC-coated standard beads. Data are the means ±S.E. (n ≥ 3). ND, not determined.
| Fluorescent agonist | Kd | Bmax | Binding sites |
|---|---|---|---|
| nm | |||
| RBL-FPR1 | |||
| WK(FITC)YMVm | 3.9 ± 1.1 | 96.7 ± 5.7 | 100,195 ± 13,592 |
| fMLFK-FITC | 0.3 ± 0.1 | 106 ± 5.9 | 109,44 ± 17,358 |
| fMLFIK-FITC | 0.9 ± 0.3 | 91.9 ± 5.4 | 89,315 ± 15,418 |
| RBL-FPR2 | |||
| WK(FITC)YMVm | 0.8 ± 0.1 | 125 ± 4.7 | 125,472 ± 16,562 |
| fMLFK-FITC | ND | ND | ND |
| fMLFIK-FITC | 1132 ± 680 | 112 ± 41.0 | 105,277 ± 96,172 |
| RBL-D2817.32G | |||
| WK(FITC)YMVm | 1.3 ± 0.3 | 143 ± 8.5 | 133,676 ± 41,085 |
| fMLFK-FITC | ND | ND | ND |
| fMLFIK-FITC | ND | ND | ND |
TABLE 3.
Competitive binding of various formyl peptides with WK(FITC)YMVm to RBL- FPR2 cells
The relative affinity of unconjugated ligands were measured in competitive binding assays in which a fixed amount of WK(FITC)YMVm (2.5 nm) was added to cells on ice for 1 h before the addition of increasing concentrations of the unconjugated peptides as competitors. Samples were incubated for another hour on ice, and mean fluorescent intensity values were obtained by flow cytometry. Shown are the means of IC50 values and maximum displacement (%) of unlabeled ligands in competition binding assays, based on at least three independent experiments. ND, not determined.
| Agonist | RBL-FPR2 |
RBL-D2817.32G |
||
|---|---|---|---|---|
| IC50 | Maximum displacement | IC50 | Maximum displacement | |
| m | % | m | % | |
| fMLF | 5.1 (±6.0) × 10−5 | 25 (±12) | 5.6 (±0.4) × 10−6 | 40 (±6.2) |
| fMLFE | ND | 10 (±1.8) | 8.3 (±3.5) × 10−6 | 34 (±6.6) |
| fMLFK | 1.9 (±0.6) × 10−6 | 90 (±0.9) | 8.5 (±5.9) × 10−6 | 80 (±6.3) |
| fMLFW | 3.1 (±11) × 10−6 | 45 (±8.3) | 9.1 (±1.7) × 10−6 | 46 (±9.6) |
| fMLFII | 2.9 (±8.2) × 10−6 | 82 (±3.5) | 2.1 (±0.7) × 10−6 | 89 (±8.7) |
| fMLFIK | 2.8 (±1.3) × 10−6 | 92 (±2.6) | 6.6 (±3.4) × 10−6 | 90 (±4.0) |
| fMIFL | 7.9 (±0.3) × 10−6 | 70 (±8.9) | 7.2 (±0.4) × 10−6 | 64 (±2.5) |
| fMIVIL | 7.8 (±0.4) × 10−6 | 38 (±3.5) | 4.2 (±0.2) × 10−6 | 35 (±6.9) |
Although FITC-conjugated fMLFIK displayed reduced binding to FPR2, fMLFIK-FITC retained high affinity for FPR1 (Kd = 0.9 nm, Fig. 5D, Table 2). This finding is consistent with the early observation that C-terminal residues of formyl peptides longer than five amino acids protrude to the open end of the putative binding pocket, as proposed by Sklar and coworkers (30). WK(FITC)YMVm was also tested on FPR1, and a Kd of 3.9 nm was obtained (Fig. 5E, Table 2). In competitive binding assays using WK(FITC)YMVm, the tripeptide fMLF and the tetrapeptides (fMLFE, fMLFK, fMLFW) displayed similar affinity to FPR1 despite charge difference in the C-terminal amino acids (Fig. 5F). In functional assays including degranulation (Fig. 4), calcium flux and cAMP accumulation assays (supplemental Figs. 1 and 2 and Table 1), fMLFE, which displayed low potency at FPR2 (Fig. 5C), performed as well as other formylated tetrapeptides when tested on FPR1.
Structural Models for FPR1 and FPR2 Peptide Binding
The above results indicate that, in contrast to FPR1, FPR2 binding is influenced by the C-terminal domain of formyl peptides. In an attempt to gain a better understanding of this effect, we constructed computer three-dimensional models of the complexes between N-formyl peptides and a CXCR4 chemokine-based model of FPR1 and FPR2 (see “Experimental Procedures”). In these models (Fig. 6), the N-formyl group hydrogen bonds Arg-2055.42, which ionic pairs with Asp1063.33 based on published reports (12). The crystal structure of the active state β2-adrenergic receptor in the active state bound to the BI-167107 agonist shows hydrogen bond interactions with the side chains at positions 5.42 and 5.46 (31). These interactions stabilize a receptor conformation that includes an inward movement of transmembrane helix TM 5, rotation of TM 3 that repositions a bulky hydrophobic side chain at position 3.40 (32), and relocation of the side chain at position 6.44 that contributes to a rotation and outward movement of TM6 for receptor activation (31). Thus, the interaction between the N-formyl group and Arg-2055.42 in TM 5 might trigger these movements, explaining the experimentally determined importance of the N-formyl group in receptor activation (Fig. 2). Importantly, Arg-2055.42 is conserved in both FPR1 (Fig. 6A) and FPR2 (Fig. 6, B–D); thus, the different structural requirements for peptide binding probably resides in residues located at the other side of the binding cavity. Sequence divergences are mainly observed in TM 2 and TM 7 (Fig. 7A). FPR1 contains Arg-842.63, Lys-852.64, and Asp2847.36, the last two forming an electrostatic interaction in the ligand-free receptor (Fig. 6A), in agreement with previous experimental data (13). There are Met and Asn in these positions, respectively, in FPR2 (Fig. 7A). Nevertheless, FPR2 contains the negative Asp-2817.32 side chain that is Gly in FPR1 but lacks any positive side chain in the TM 2 environment (Figs. 7A and 6, B–D). As a result, the molecular electrostatic potential on the inner surface of the binding cavity near TM 2 and TM 7 is positive (see the arrow in Fig. 7B) in FPR1, due to Arg-842.63 and Lys-852.64, and negative (see the arrow in Fig. 7C) in FPR2, due to Asp-2817.32. The C-terminal moiety of fMLF expands toward TM 2 in FPR1 in such a manner that the COO- group interacts with both Arg-842.63 and Lys-852.64, disrupting the Lys-852.64···Asp2847.36 ionic pair (Fig. 6A) that is an important event for receptor activation (13). In contrast, this COO- group of fMLF is repulsive with Asp-2817.32 of FPR2 (Fig. 6B), which impedes high affinity binding. According to the model, the fMLFII pentapeptide, which adds two Ile side chains to fMLF, moves the end-terminal COO- group away from the negatively charged environment of TM 2 in FPR2, thereby increasing its potency (Table 1). fMLFII (Fig. 6C) positions a hydrophobic Ile side chain in the hydrophobic environment of TM 2; thus, the much larger enhancement of potency in fMLFII (1763 times) than in fMLFIK (558 times) may be attributed to the fact that the end-terminal COO- group in fMLFII is closer to the extracellular side of TM 1, possibly interacting with Arg261.32 (Fig. 6C). In agreement with these modes of binding, the addition of a positive side chain to fMLF, forming the fMLFK tetrapeptide, enhances the potency by a factor of ∼10 relative to fMLF (Table 1) due to its interaction with Asp-2817.32 (Fig. 6D). In contrast, the addition of a negative side chain, forming the fMLFE tetrapeptide, is not tolerated in FPR2 (Table 1) because the Glu side chain is located in a negatively charged environment of TM 2 (not shown).
FIGURE 6.
Computational models of the complexes between N-formyl peptides and formyl peptide receptors (FPR1 and FPR2). CXCR4-based homology models of FPR1 (A) and FPR2 (B–D) in complex with formyl peptides (side chains are shown in light blue). In these models, the N-formyl group of peptides hydrogen bonds the Arg-2055.42··Asp1063.33 ionic pair. A, the end-terminal COO- group of fMLF interacts with both Arg-842.63 and Lys-852.64 of FPR1. B, the end-terminal COO- group of fMLF is repulsive with the negatively charged Asp-2817.32 of FPR2, which impedes the high affinity binding. C, the longer fMLFII pentapeptide moves the end-terminal COO- group away from Asp-2817.32 of FPR2, toward Arg261.32. D, the positive Lys side chain of fMLFK interacts with Asp-2817.32 of FPR2. Representative snapshots (4 structures collected every 35 ns), collected during the molecular dynamics simulations of the peptide-receptor complexes, are shown in the middle panels (0 ns in white, 35 ns in light gray, 70 ns in dark gray, and 100 ns in black). The key proposed interactions between the peptide and FPRs remain stable during the simulation. Root mean square deviations (rmsd) on receptor α-carbons (in black) and on peptide all-atoms (in gray) throughout the molecular dynamics simulations are shown in the bottom panels. The color code of the helices is: TM 1 (white), 2 (yellow), 3 (red), 4 (gray), 5 (green), 6 (blue), and 7 (brown).
FIGURE 7.

Structural comparison of FPR1 and FPR2 in selected regions. A, sequence alignment of FPR1 and FPR2. Residues referenced in the manuscript are boxed. Conserved side chains between both receptors are shown in black, and key sequence divergences are shown in green (for N-formyl peptide binding to FPR1) or gray (for N-formyl peptide binding to FPR2). B and C, molecular electrostatic potential on the inner surface of the binding cavity of FPR1 (B) and FPR2 (C). The orientation of the receptor is as in Fig. 5 above. Arrows show key positive areas in FPR1 due to Arg-842.63 and Lys-852.64 and a negative area in FPR2 due to Asp-2817.32.
Asp-2817.32 in FPR2 Is a Key Structural Determinant for Formyl Peptides Selectivity
To experimentally test the above models, we conducted site-directed mutagenesis and substituted Asp-2817.32 with glycine, the FPR1 counterpart at the 7.32 position (Fig. 1B). The mutant was stably expressed in RBL cells with an expression level similar to that of FPR2 and FPR1 (Table 2), based on flow cytometry analysis. The mutation did not significantly alter the equilibrium dissociation constant (Kd value) for WK(FITC)YMVm (1.3 nm for D2817.32G, compared with 0.8 nm for wild type FPR2; Table 2). Likewise, the mutation did not improve FPR2 binding of fMLFK-FITC and fMLFIK-FITC, which remained as poor agonists (data not shown). The ligand-receptor interaction was examined in both functional and binding assays (Figs. 8–10). The D2817.32G mutant and the wild type FPR2 showed no significant difference in response to the fMLFII and fMLFIK pentapeptides in calcium flux (Fig. 8 and Table 1), cAMP accumulation (Fig. 9 and Table 1), and competitive binding assays (Fig. 10 and Table 3). The longer peptide, fMLFIIK, could not discriminate between D2817.32G and wild type receptor (data not shown). These observations suggest that the D2817.32G mutation did not alter the global structure of the receptor.
FIGURE 8.
Substitution of Asp-2817.32 to Gly differentially affects the potency of formyl peptides of different length and terminal charges. The dose-response curves of wild type FPR2 and the mutant D2817.32G are shown, respectively. Dark lines represent the dose curves of wild type FPR2, and gray lines represent the curves of the D2817.32G mutant. The dose curves were based on peak Ca2+ mobilization at the indicated agonist concentrations and are shown as the means ± S.E. based on three separate experiments.
FIGURE 10.
Competitive binding of formyl peptides carrying different length and terminal charge to FPR2 and the D2817.32G mutant. Competition binding assays were performed in the presence of 2.5 nm WK(FITC)YMVm and increasing concentrations of the indicated peptides including fMLF (A), fMLFE (B), fMLFK (C), fMLFW (D), fMLFII (E), fMLFIK (F), fMIFL (G), and fMIVIL (H). Dark line, wild type hFPR2. Gray line, mutant D2817.32G. Data were analyzed with Origin 8.5 software, and the results are shown as means ± S.E. with >3 experiments.
In contrast, the D2817.32G mutation differentially altered FPR2-mediated response to, as well as binding affinity for, fMLF and all tetrapeptides tested in this work. As illustrated in Figs. 8–10 and summarized in Tables 1 and 3, fMLFK was ∼10-fold less potent on the D2817.32G mutant than on wild type FPR2, suggesting that removal of the negative charge of Asp-2817.32 reduced a favorable interaction with the positively charged lysine side chain, as predicted in the computer models. In comparison, the potency (Figs. 8 and 9) and affinity (Fig. 10) of fMLFE and fMLF improved by ∼10-fold when Asp-2817.32 was substituted with a Gly, providing experimental support to the computer models that predict Asp-2817.32 as being repulsive to the COO- group in fMLF and to the negative side chain of Glu in fMLFE. As a result of the removal of this negative charge at position 281, the difference in binding affinity and potency between the tested tripeptide and tetrapeptides was reduced (Tables 1 and 3). These results, summarized in Tables 1 and 3, suggested that Asp-2817.32 directly interacts with the C termini of formyl peptides of three to four amino acids due to proximity, but it is dispensable with peptides over five residues in length (Fig. 6).
DISCUSSION
Since its discovery in the early 1990s, FPR2 has been considered a controversial formyl peptide receptor largely due to its low affinity for fMLF (25, 33). In addition to its low affinity for fMLF, FPR2 displays promiscuous binding to various agonists, thus distinguishing it from the high affinity fMLF receptor, FPR1 (4). In our previously publications, we examined formyl peptides derived from L. monocytogenes and S. aureus for their activation of the mouse formyl peptide receptor, mouse Fpr1 (15, 27). We demonstrated the ability of formyl peptides from L. monocytogenes (fMIVIL) and S. aureus (fMIFL) to activate mouse Fpr1 with a >100-fold higher potency than fMLF (15, 27). In this study we showed similar results of fMIVIL and fMIFL in activating the low affinity human formyl peptide receptor, FPR2. Among the peptides tested, fMIVIL was previously shown to activate FPR2 (14), but the significance of this work was overlooked due to the higher potency of this peptide toward FPR1. In this study we not only demonstrated that FPR2 is activated by physiologically relevant formyl peptides such as fMIVIL from L. monocytogenes and fMIFL from S. aureus but also provided a potential mechanism for FPR2 interaction with formyl peptides of different length and composition.
FPR2 is activated by a number of endogenous ligands such as SAA as well as many exogenous ligands such as HIV envelope proteins and a number of synthetic peptides including MMK1 and WKYMVM or small molecules such as Quin C1 (4). Because of this high level of diversity, it was important to determine whether the formyl group of formyl peptides is necessary for FPR2 activation. Our results indicate that the Met-containing peptides without N-formyl group lacked FPR2-activating capability. Computational models of the complexes between these peptides and FPR1 and FPR2 showed that the N-formyl group interacts with Arg-2055.42 in TM 5 (Fig. 6), thus inducing or stabilizing relocation of the TM 5 extracellular side and the closely related TM 6, similarly to the mechanisms proposed for ligand interaction with the biogenic amine receptors (32, 34).
FPR1 and FPR2 as well as angiotensin, opioid, and chemokine families of GPCRs possess a conserved T2.56XP2.58 motif (TXP, X being a nonconserved residue) in TM 2, which plays a key role in receptor activation (35). Furthermore, it has been shown that a salt bridge linking the beginning of TM 7 and extracellular loop 2 of the β2-adrenergic receptor reorganizes upon agonist binding (22, 34). These data have suggested that both TM 2 and TM 7 play key roles during receptor activation. These findings are consistent with the experimental data obtained by Mills et al. (13), showing that formyl peptide binding to FPR1 disrupts the electrostatic interaction between Lys-852.64 in TM 2 and Asp2847.36 in TM 7. In our computational models, the fMLF tripeptide complexed with FPR1 disrupts this salt bridge through the interaction between the end-terminal COO- group of the peptide and Lys-852.64 (Fig. 6A), whereas the fMLFII pentapeptide places the hydrophobic Ile side chain between Lys-852.64 and Asp2847.36 (not shown). Notably, the amino acid composition in TM 2 and TM 7 of FPR2 is highly divergent from FPR1, supporting our prediction that longer peptides (e.g. fMLFII versus fMLF) as well as certain C-terminal side chains are contributing factors for FPR2 activation, as will be discussed below.
Quinn and co-workers (36) previously proposed, based on pharmacophore models developed for small molecule agonists, a more electronegative binding pocket in FPR2 compared with FPR1. We have shown that the negatively charged Asp-2817.32 in TM 7 provides the electrostatic negative potential in this domain of the receptor. Our computational models depict that the end-terminal COO- group of the fMLF tripeptide or the negatively charged Glu in the fMLFE tetrapeptide (not shown) is repulsive with this negatively charged area of FPR2, whereas the positive Lys side chain in the fMLFK tetrapeptide interacts favorably with Asp-2817.32. We were able to directly test these models using site-directed mutagenesis. Our results showed that substitution of the negatively charged Asp with Gly led to a decline in the interaction with fMLFK, whereas the substitution increased the potency and relative affinity for both fMLFE and fMLF. It is important to note that FPR1 lacks a negatively charged amino acid at the homologous position; hence, there is no favorable interaction with formyl peptides carrying a positive charge at the C terminus or repulsion with formyl peptides possessing a C-terminal negative charge. As a result, FPR1 interacts equally well with fMLFE and fMLFK.
Despite the above, Asp-2817.32 seems not to contribute to FPR2 interaction with the longer fMLFII pentapeptide (Figs. 8–10 and Tables 1 and 3). According to our computational models, fMLFII positions the hydrophobic Ile side chain between TMs 2 and 7 in a favorable manner and moves the end-terminal COO- group away from Asp-2817.32, toward the extracellular side of TM 1, where the molecular electrostatic potential is positive due to the FPR2-specific Arg261.32 (Fig. 6C). Therefore, formyl peptides of different lengths can interact with different structural features of a receptor. Some of these features, including the negatively charged Asp-2817.32, are found in FPR2, whereas others including the salt bridge between TM 2 and TM 7 are present only in FPR1. It should be pointed out that the salt bridge is important for high affinity interaction of formyl peptides with a receptor, as evidenced by the generally higher potency of all tested peptides at FPR1 compared with FPR2. Evidence supporting this notion also came from the observation that the D2817.32G mutation of FPR2 failed to restore the potency of fMLF and fMLFE to the level in FPR1. Thus, the salt bridge between TM 2 and TM 7 is indispensable for a strong interaction with fMLF, fMLFE, and other formyl peptides. The sequence of the mouse formyl peptide receptors, mouse Fpr1 and Fpr2, resembles that of human FPR2 in that they lack the salt bridge between TM 2 and TM 7. This is the probable reason for their low affinity binding to most of the formyl peptides tested, as compared with human FPR1 (15).
In summary, we have shown that selected formyl peptides from L. monocytogenes and S. aureus, which share certain C-terminal features such as one or two additional amino acids beyond the tripeptide fMLF and a positive charge, are potent agonists for FPR2. In addition, we have found that FPR1 and FPR2 interact with these peptides quite differently, and a negatively charged region in the binding pocket in FPR2 as well as the salt bridge between its TM 2 and TM 7 provide explanations to the difference. Further structural analysis is expected to delineate the mechanisms by which FPR2 interacts with various agonists in the forms of monomers and dimers, as shown in a recent report (37). The new information will expand our understanding of the FPRs as modulators of both proinflammatory and anti-inflammatory functions.
Acknowledgment
We thank Dr. Masakatsu Nanamori for helpful discussions.
This work was supported, in whole or in part, by National Basic Research Program of China (973 Program Grant 2012CB518000) and National Natural Science Foundation of China (Grant 31270941). This work was also supported by National Institutes of Health Grants R01 AI033503 and P01 HL077806, and by Specialized Research Fund for the Doctoral Program of Higher Education of China (Grant 20120073110069), the Ministerio de Ciencia e Innovacion (SAF2010-22198-C02-02) and the Instituto de Salud Carlos III (RD07/0067/0008) of Spain.

This article contains supplemental Figs. 1 and 2.
- FPR
- formyl peptide receptor
- GPCR
- G protein-coupled receptor
- TM
- transmembrane domain.
REFERENCES
- 1. Schiffmann E., Corcoran B. A., Wahl S. M. (1975) N-formylmethionyl peptides as chemoattractants for leucocytes. Proc. Natl. Acad. Sci. U.S.A. 72, 1059–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Niedel J. E., Kahane I., Cuatrecasas P. (1979) Receptor-mediated internalization of fluorescent chemotactic peptide by human neutrophils. Science 205, 1412–1414 [DOI] [PubMed] [Google Scholar]
- 3. Nauseef W. M. (2007) How human neutrophils kill and degrade microbes. An integrated view. Immunol. Rev. 219, 88–102 [DOI] [PubMed] [Google Scholar]
- 4. Ye R. D., Boulay F., Wang J. M., Dahlgren C., Gerard C., Parmentier M., Serhan C. N., Murphy P. M. (2009) International Union of Basic and Clinical Pharmacology. LXXIII. Nomenclature for the formyl peptide receptor (FPR) family. Pharmacol. Rev. 61, 119–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gao J. L., Lee E. J., Murphy P. M. (1999) Impaired antibacterial host defense in mice lacking the N-formylpeptide receptor. J. Exp. Med. 189, 657–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang Q., Raoof M., Chen Y., Sumi Y., Sursal T., Junger W., Brohi K., Itagaki K., Hauser C. J. (2010) Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature 464, 104–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. McDonald B., Pittman K., Menezes G. B., Hirota S. A., Slaba I., Waterhouse C. C., Beck P. L., Muruve D. A., Kubes P. (2010) Intravascular danger signals guide neutrophils to sites of sterile inflammation. Science 330, 362–366 [DOI] [PubMed] [Google Scholar]
- 8. Boulay F., Tardif M., Brouchon L., Vignais P. (1990) The human N-formylpeptide receptor. Characterization of two cDNA isolates and evidence for a new subfamily of G-protein-coupled receptors. Biochemistry 29, 11123–11133 [DOI] [PubMed] [Google Scholar]
- 9. Quehenberger O., Prossnitz E. R., Cavanagh S. L., Cochrane C. G., Ye R. D. (1993) Multiple domains of the N-formyl peptide receptor are required for high-affinity ligand binding. Construction and analysis of chimeric N-formyl peptide receptors. J. Biol. Chem. 268, 18167–18175 [PubMed] [Google Scholar]
- 10. Quehenberger O., Pan Z. K., Prossnitz E. R., Cavanagh S. L., Cochrane C. G., Ye R. D. (1997) Identification of an N-formyl peptide receptor ligand binding domain by a gain-of-function approach. Biochem. Biophys. Res. Commun. 238, 377–381 [DOI] [PubMed] [Google Scholar]
- 11. Lala A., Gwinn M., De Nardin E. (1999) Human formyl peptide receptor function role of conserved and nonconserved charged residues. Eur. J. Biochem. 264, 495–499 [DOI] [PubMed] [Google Scholar]
- 12. Mills J. S., Miettinen H. M., Cummings D., Jesaitis A. J. (2000) Characterization of the binding site on the formyl peptide receptor using three receptor mutants and analogs of Met-Leu-Phe and Met-Met-Trp-Leu-Leu. J. Biol. Chem. 275, 39012–39017 [DOI] [PubMed] [Google Scholar]
- 13. Mills J. S., Miettinen H. M., Barnidge D., Vlases M. J., Wimer-Mackin S., Dratz E. A., Sunner J., Jesaitis A. J. (1998) Identification of a ligand binding site in the human neutrophil formyl peptide receptor using a site-specific fluorescent photoaffinity label and mass spectrometry. J. Biol. Chem. 273, 10428–10435 [DOI] [PubMed] [Google Scholar]
- 14. Rabiet M. J., Huet E., Boulay F. (2005) Human mitochondria-derived N-formylated peptides are novel agonists equally active on FPR and FPRL1, while Listeria monocytogenes-derived peptides preferentially activate FPR. Eur. J. Immunol. 35, 2486–2495 [DOI] [PubMed] [Google Scholar]
- 15. He H. Q., Liao D., Wang Z. G., Wang Z. L., Zhou H. C., Wang M. W., Ye R. D. (2013) Functional characterization of three mouse formyl peptide receptors. Mol. Pharmacol. 83, 389–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gonzalez A., Cordomí A., Caltabiano G., Pardo L. (2012) Impact of helix irregularities on sequence alignment and homology modeling of G protein-coupled receptors. Chembiochem. 13, 1393–1399 [DOI] [PubMed] [Google Scholar]
- 17. Wu B., Chien E. Y., Mol C. D., Fenalti G., Liu W., Katritch V., Abagyan R., Brooun A., Wells P., Bi F. C., Hamel D. J., Kuhn P., Handel T. M., Cherezov V., Stevens R. C. (2010) Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science 330, 1066–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Martí-Renom M. A., Stuart A. C., Fiser A., Sánchez R., Melo F., Sali A. (2000) Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 29, 291–325 [DOI] [PubMed] [Google Scholar]
- 19. Ballesteros J. A., Weinstein H. (1995) Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G-protein coupled receptors. Methods Neurosci. 25, 366–428 [Google Scholar]
- 20. Trott O., Olson A. J. (2010) AutoDock Vina. Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput Chem. 31, 455–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Case D. A., Darden T. A., Cheatham T. E., III, Simmerling C. L., Wang J., Duke R. E., Luo R., Merz K. M., Pearlman D. A., Crowley M., Walker R. C., Zhang W., Wang B., Hayik S., Roitberg A., Seabra G., Wong K. F., Paesani F., Wu X., Brozell S., Tsui V., Gohlke H., Yang L., Tan C., Mongan J., Hornak V., Cui G., Beroza P., Mathews D. H., Schafmeister C., Ross W. S., Kollman P. A. (2006) AMBER9. University of California, San Francisco [Google Scholar]
- 22. Jongejan A., Bruysters M., Ballesteros J. A., Haaksma E., Bakker R. A., Pardo L., Leurs R. (2005) Linking agonist binding to histamine H1 receptor activation. Nat. Chem. Biol. 1, 98–103 [DOI] [PubMed] [Google Scholar]
- 23. He R., Tan L., Browning D. D., Wang J. M., Ye R. D. (2000) The synthetic peptide Trp-Lys-Tyr-Met-Val-d-Met is a potent chemotactic agonist for mouse formyl peptide receptor. J. Immunol. 165, 4598–4605 [DOI] [PubMed] [Google Scholar]
- 24. Nanamori M., Cheng X., Mei J., Sang H., Xuan Y., Zhou C., Wang M. W., Ye R. D. (2004) A novel nonpeptide ligand for formyl peptide receptor-like 1. Mol. Pharmacol. 66, 1213–1222 [DOI] [PubMed] [Google Scholar]
- 25. Ye R. D., Cavanagh S. L., Quehenberger O., Prossnitz E. R., Cochrane C. G. (1992) Isolation of a cDNA that encodes a novel granulocyte N-formyl peptide receptor. Biochem. Biophys. Res. Commun. 184, 582–589 [DOI] [PubMed] [Google Scholar]
- 26. Rot A., Henderson L. E., Copeland T. D., Leonard E. J. (1987) A series of six ligands for the human formyl peptide receptor. Tetrapeptides with high chemotactic potency and efficacy. Proc. Natl. Acad. Sci. U.S.A. 84, 7967–7971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Southgate E. L., He R. L., Gao J. L., Murphy P. M., Nanamori M., Ye R. D. (2008) Identification of formyl peptides from Listeria monocytogenes and Staphylococcus aureus as potent chemoattractants for mouse neutrophils. J. Immunol. 181, 1429–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Migeotte I., Communi D., Parmentier M. (2006) Formyl peptide receptors. A promiscuous subfamily of G protein-coupled receptors controlling immune responses. Cytokine Growth Factor Rev. 17, 501–519 [DOI] [PubMed] [Google Scholar]
- 29. Freer R. J., Day A. R., Muthukumaraswamy N., Pinon D., Wu A., Showell H. J., Becker E. L. (1982) Formyl peptide chemoattractants. A model of the receptor on rabbit neutrophils. Biochemistry 21, 257–263 [DOI] [PubMed] [Google Scholar]
- 30. Sklar L. A., Fay S. P., Seligmann B. E., Freer R. J., Muthukumaraswamy N., Mueller H. (1990) Fluorescence analysis of the size of a binding pocket of a peptide receptor at natural abundance. Biochemistry 29, 313–316 [DOI] [PubMed] [Google Scholar]
- 31. Swaminath G., Xiang Y., Lee T. W., Steenhuis J., Parnot C., Kobilka B. K. (2004) Sequential binding of agonists to the beta2 adrenoceptor. Kinetic evidence for intermediate conformational states. J. Biol. Chem. 279, 686–691 [DOI] [PubMed] [Google Scholar]
- 32. Schwartz T. W., Frimurer T. M., Holst B., Rosenkilde M. M., Elling C. E. (2006) Molecular mechanism of 7TM receptor activation. A global toggle switch model. Annu. Rev. Pharmacol. Toxicol. 46, 481–519 [DOI] [PubMed] [Google Scholar]
- 33. Murphy P. M., Ozçelik T., Kenney R. T., Tiffany H. L., McDermott D., Francke U. (1992) A structural homologue of the N-formyl peptide receptor. Characterization and chromosome mapping of a peptide chemoattractant receptor family. J. Biol. Chem. 267, 7637–7643 [PubMed] [Google Scholar]
- 34. Bokoch M. P., Zou Y., Rasmussen S. G., Liu C. W., Nygaard R., Rosenbaum D. M., Fung J. J., Choi H. J., Thian F. S., Kobilka T. S., Puglisi J. D., Weis W. I., Pardo L., Prosser R. S., Mueller L., Kobilka B. K. (2010) Ligand-specific regulation of the extracellular surface of a G-protein-coupled receptor. Nature 463, 108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Govaerts C., Blanpain C., Deupi X., Ballet S., Ballesteros J. A., Wodak S. J., Vassart G., Pardo L., Parmentier M. (2001) The TXP motif in the second transmembrane helix of CCR5. A structural determinant of chemokine-induced activation. J. Biol. Chem. 276, 13217–13225 [DOI] [PubMed] [Google Scholar]
- 36. Kirpotina L. N., Khlebnikov A. I., Schepetkin I. A., Ye R. D., Rabiet M. J., Jutila M. A., Quinn M. T. (2010) Identification of novel small-molecule agonists for human formyl peptide receptors and pharmacophore models of their recognition. Mol. Pharmacol. 77, 159–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cooray S. N., Gobbetti T., Montero-Melendez T., McArthur S., Thompson D., Clark A. J., Flower R. J., Perretti M. (2013) Ligand-specific conformational change of the G-protein-coupled receptor ALX/FPR2 determines proresolving functional responses. Proc. Natl. Acad. Sci. U.S.A. 110, 18232–18237 [DOI] [PMC free article] [PubMed] [Google Scholar]