Background: Expanded double negative T cells in systemic lupus erythematosus (SLE) originate from CD8+ T cells.
Results: cAMP responsive element modulator (CREM) α induces epigenetic remodeling of the CD8 cluster through DNMT3a and G9a.
Conclusion: CREMα centrally contributes to double negative T cell expansion in SLE pathogenesis.
Significance: CREMα governs T cell distribution in SLE.
Keywords: Chromatin, Chromatin Histone Modification, DNA Methylation, Epigenetics, Histone Methylation, Histone Modification, CD8, CREMα, SLE, Double Negative T Cells
Abstract
TCR-αβ+CD3+CD4−CD8− “double negative” T cells are expanded in the peripheral blood of patients with systemic lupus erythematosus (SLE) and lupus-prone mice. Double negative T cells have been claimed to derive from CD8+ cells that down-regulate CD8 co-receptors and acquire a distinct effector phenotype that includes the expression of proinflammatory cytokines. This, along with the fact that double negative T cells have been documented in inflamed organs, suggests that they may contribute to disease expression and tissue damage. We recently linked the transcription factor cAMP responsive element modulator (CREM) α, which is expressed at increased levels in T cells from SLE patients and lupus prone MRL/lpr mice, with trans-repression of a region syntenic to the murine CD8b promoter. However, the exact molecular mechanisms that result in a stable silencing of both CD8A and CD8B genes remain elusive. Here, we demonstrate that CREMα orchestrates epigenetic remodeling of the CD8 cluster through the recruitment of DNA methyltransferase (DNMT) 3a and histone methyltransferase G9a. Thus, we propose that CREMα is essential for the expansion of double negative T cells in SLE. CREMα blockade may have therapeutic value in autoimmune disorders with DN T cell expansion.
Introduction
Systemic lupus erythematosus (SLE)3 is an autoimmune disorder that can affect any organ or system and cause severe complications. Regardless of recent advances in the search for disease mechanisms, the molecular pathophysiology of SLE remains largely unknown. TCR-αβ+CD3+CD4−CD8− “double negative” (DN) T cells are expanded in the peripheral blood of SLE patients and lupus-prone MRL/lpr mice. We recently demonstrated that DN T cells in humans and MRL/lpr mice derive from CD8+ T cells by down-regulating CD8 surface-receptor expression (1).
The regulation of CD8 has been studied in humans and mice. In both species, mature CD4+ and CD8+ T cells derive from CD4−CD8− double negative thymocytes that convert into CD4+CD8+ double positive progenitor cells, which later on during their differentiation into mature T cells down-regulate either CD4 or CD8 (2, 3). Thymus-derived CD8+ T cells express heterodimers of CD8α and CD8β on their surface, whereas gut-derived CD8+ T cells or CD8+ dendritic cells express CD8α homodimers (2, 3). We have reported that during the TCR activation-induced transformation of CD8+ T cells into peripheral DN T cells, the transcription factor cAMP responsive element modulator (CREM) α trans-represses a region syntenic to the murine CD8b promoter in human CD8+ T cells. Trans-repression of this regulatory element results in transcriptional silencing and subsequently down-regulation of CD8 surface expression (1). This is of special interest, because CREMα is expressed at increased levels in T cells from SLE patients where it affects several T cell functions, including cytokine expression (1, 4). However, our findings did not completely explain the down-regulation of both CD8α and CD8β in response to TCR stimulation.
Phenotypes of specialized T cell subsets correlate with the exclusive expression of either CD4 or CD8, suggesting that the molecular mechanisms regulating CD4 or CD8 expression may also be involved in the definition of the phenotype of CD4+ helper or CD8+ cytotoxic T cells (5). Four clusters with increased DNase sensitivity have been identified within the murine CD8 locus, which are syntenic with six in the human cluster (2, 3). Transgenic reporter systems allowed the identification of several enhancer elements within the CD8 cluster (E8I–E8IV) (2, 3, 5–15). This enhancer network is required for lineage-specific regulation of CD8α and CD8β during T cell development and its elements undergo epigenetic remodeling during T cell development either allowing or prohibiting the expression of CD8A and/or CD8B (3). Epigenetic mechanisms regulate gene expression by influencing the accessibility of chromatin to transcription factors and RNA polymerases (16). The addition of methyl groups to the 5′-carbon end of cytidine residues in cytidine-phosphate-guanosine sequences of the genomic DNA, and post-translational modifications to the amino terminus of histone proteins represent the two main mechanisms during chromatin remodeling (16). It has been demonstrated that the CD8 cluster in mice undergoes epigenetic remodeling during T cell development in the thymus (4). Low degrees of DNA methylation in CD4+CD8+ double positive and CD8+ T cells allow the expression of murine CD8a and CD8b, whereas increased levels of DNA methylation around the CD8a and CD8b genes in CD4+ and DN T cells prohibit gene expression (4).
In this study we asked whether the CD8 cluster undergoes epigenetic remodeling in CD8+ T cells in response to TCR stimulation. We investigated whether the transcription factor CREMα, which is induced in response to TCR stimulation and expressed at increased levels in T cells from SLE patients induces chromatin remodeling of the CD8 cluster in response to TCR activation. We demonstrate that CREMα is recruited to several conserved non-coding regions within the human CD8 cluster, mediating epigenetic silencing of CD8A and CD8B. We conclude that the transcriptional regulator CREMα plays a central role in mature CD8+ T cell function and contributes to the expansion of DN T cells in patients with SLE.
MATERIALS AND METHODS
Cell Culture
Peripheral blood mononuclear cells were enriched for T lymphocytes by precipitation of non-T cells (Rosettesep, Stemcell Technologies) followed by density gradient centrifugation (Lymphoprep, Nycomed). From these T lymphocyte-enriched peripheral blood mononuclear cells, CD4−CD8+ T lymphocytes were isolated by negative selection (Dynabeads, Invitrogen). CD8+ T lymphocytes were cultured at a concentration of 1 × 106 cells/ml in RPMI1640 with 10% FCS in 12-well plates that had or had not been precoated with anti-CD3 and anti-CD28 antibodies (as indicated). Cells were collected after 120 h and harvested for quantitative RT-PCR, flow cytometry, methylated DNA immunoprecipitation (MeDIP), or chromatin immunoprecipitation (ChIP) as indicated.
Human Subjects
All SLE patients included in our studies were diagnosed according to the American College of Rheumatology classification criteria and recruited from the Division of Rheumatology at Beth Israel Deaconess Medical Center, Boston, MA, and gave written informed consent under protocol 2006-P-0298. Healthy age, gender, and ethnicity matched individuals were chosen as controls. Peripheral venous blood was collected in heparin-lithium tubes and total human T cells were purified as described before (1).
Mice
MRL/lpr mice were purchased from The Jackson Laboratory (Bar Harbor, ME) and housed in specific pathogen-free conditions. Experimental procedures were approved by the BIDMC Animal Care and Use Committee.
Flow Cytometry and Cell Sorting
Anti-CD4-PB, anti-CD8-PE, and anti-CD3-APC/Cy7 were purchased from BioLegend. Samples were acquired on a LSR II flow cytometer (BD Biosciences) and data were analyzed FlowJo version 7.2.2 (Tree Star). For the analysis of T lymphocyte populations, a first gate that included live cells was used. CD3+ T lymphocytes were then plotted in a CD4+ versus CD8+ graphic that allowed the identification of discrete CD4+, CD8+, and double negative T lymphocyte populations. For some experiments, stained cells were sorted in a FACSAria flow cytometer (BD Biosciences), post-sorting purity was >98%.
Semi-quantitative Real-time Polymerase Chain Reaction
Total RNA from control and SLE T lymphocytes was isolated, using the Qiagen RNeasy Mini Kit (Qiagen). cDNA was generated using a first strand cDNA synthesis kit (Invitrogen). For gene expression analyses, real-time PCR was performed using SYBR Green site-specific primers on an ABI OneStepPlus Real-time PCR System. Results were normalized to 18S. Primer sequences for quantitative RT-PCR, plasmid generation, MeDIP, and ChIP PCR will be provided upon request.
Gene Expression Plasmids
Expression plasmids for human CREMα, DNMT3a, and G9a have been described previously (17, 18, 19). Three million primary human CD8+ T lymphocytes were transfected with a total amount of 3 μg of the indicated expression plasmids using the Amaxa transfection system (Lonza) or Lipofectamine (Invitrogen) as indicated. After 24 (RNA, ChIP and MeDIP analyses) or 120 h (flow cytometry), cells were harvested and assayed.
Methylated CpG-DNA Immunoprecipitation (MeDIP)
The MeDIP assay was carried out according to the manufacturer's instructions (Zymo Research). Briefly, genomic DNA from T lymphocytes obtained from healthy individuals or SLE patients was purified using the AllPrep RNA/DNA/protein Mini kit (Qiagen), sheared to fragments of ∼200 bp using DNA shearase (Zymo Research). Subsequently, 100 ng of sheared genomic DNA were used for methylated CpG-DNA immunoprecipitation. The same amounts (100 ng) of (100%) methylated human DNA, and unmethylated human DNA (Zymo Research) were included as “input” and negative control. Methylated DNA was recovered and subjected to PCR analysis on an ABI OneStepPlus real-time PCR system.
Chromatin Immunoprecipitation (ChIP) Assays
Anti-H3K9m3e, anti-H3K27me3, and anti-G9a antibodies, nonspecific normal rabbit, and normal mouse IgG were obtained from Upstate (Millipore) and anti-DNMT3a antibody was from Abcam. Polyclonal anti-CREMα antibody detecting human CREMα has been described (20, 5).
Most ChIP experiments were carried out with the Upstate Biotechnology/Millipore Chip kit according to the manufacturer's instructions (Upstate Biotechnology/Millipore). For this assay, ChIP grade Protein A/G Plus-agarose was purchased from Pierce (ThermoScientific). For some experiments Forced CREMα expression combined with DNMT3a or G9a knock-down were carried out using Magnify ChIP assays (Invitrogen) according to the manufacturer's protocol. Briefly, 1–2 million cells were cross-linked with 1% formaldehyde, washed with cold phosphate-buffered saline, and lysed in buffer containing protease inhibitors (Roche Applied Science). Cell lysates were sonicated to shear DNA, sedimented, and diluted supernatants were immunoprecipitated with the indicated antibodies. A proportion (20% for Millipore, 10% for Invitrogen) of the diluted supernatants were kept as input (input represents PCR amplification of the total sample). The amount of immunoprecipitated DNA was subtracted by the amplified DNA that was bound by the nonspecific normal IgG and subsequently calculated as relative to the respective input DNA.
Co-immunoprecipitation of DNMT3a or G9a with CREMα
One million HEK293T cells were transfected with expression plasmids (on a pcDNA3 backbone) for CREMα, DNMT3a, G9a, or a combination as indicated (2 μg of each plasmid per transfection) using Lipofectamine (Invitrogen) according to the manufacturer's instructions. 24 h after transfection, cells were harvested and lysed in 400 μl of RIPA buffer including protease inhibitors (Roche Applied Science). Cell lysates were subjected to centrifugation (14,000 × g, 10 min, 4 °C) and 500 μg of total protein were incubated with anti-CREMα antibodies at 4 °C overnight and subjected to co-immunoprecipitation with the Pierce Co-IP kit, following the manufacturer's instructions (Pierce). Co-IP solutions were subjected to SDS-PAGE as described before (17, 18). Proteins were transferred to PVDF membranes and detected by anti-DNMT3a or G9a antibodies as indicated, applying suitable secondary peroxidase-linked anti-rabbit antibody (Santa Cruz) and ECL (Amersham Biosciences) as chemiluminescent. Input controls to confirm overexpression of the respective proteins was performed by immunblotting the non-immunoprecipitated cell lysates. Densitometry of protein-specific bands was performed on a Bio-Rad gel dock and quantified with Quantity One software (Bio-Rad).
Proximity Ligation Assay (PLA)
70 × 103 HEK 293T cells were cultured on 8-well cell culture slides (BD Falcon), and transfected with expression plasmids (on a pcDNA3 backbone) for CREMα, G9a, or a combination of CREMα and G9a (0.3 μg of each plasmid per transfection) using X-tremeGENE 9 DNA Transfection Reagent (Roche) according to the manufacturer's instructions. 24 h after transfection, cells were harvested and subjected to the Duolink proximity ligation assay following the manufacturer's instructions (Olink). Shortly thereafter, anti-CREM and anti-G9a antibodies were labeled with PLUS and MINUS oligonucleotide tails. Cells were fixed with 3.7% formaldehyde, washed with PBS, permeabilized with Triton X-100 (Invitrogen), and incubated with PLUS and MINUS oligonucleotide-labeled antibodies according to the manufacturer's instructions. Cells were washed several times and incubated with ligase (30 min) and polymerase solution (100 min). Then, cells were mounted with DAPI containing medium (Olink) and read on a fluorescence microscope (Zeiss). Experiments with primary human CD8+ T cells were performed using 5 × 106 T cells in Eppendorf tubes. After all the washing and incubations, cells were transferred to microscopy slides using a cytospin centrifuge (Shandon), mounted with DAPI containing medium (Olink), and read on a fluorescence microscope (Zeiss). The number of PLA signals per cell has been quantified using ImageJ software.
Forced Expression of CREMα and DNMT3a or G9a Knock-down
Three million primary human CD8+ T cells were transfected with a total amount of 3 μg of expression plasmid and 20 nm scrambled control siRNA or DNMT3a- or G9a-specific siRNA (OriGene) using Lipofectamine (Invitrogen) as indicated. Prior to these experiments, experimental conditions were optimized using Cy3-labeled control siRNA (OriGene) after transfection with Lipofectamine (Invitrogen). Transfection efficiency was >70%. Cells were collected after an overnight culture and processed for mRNA, MeDIP, or ChIP analysis as indicated. All experiments were repeated four to six times as indicated. Values in the bar diagrams are given as mean ± S.D.
Statistical Analysis
Paired two-tailed Student's t test was used for statistical analysis of all flow cytometry and transfection experiments as indicated. A p value of 0.05 was considered statistically significant. Results are indicated as the mean ± S.D., unless noted otherwise.
RESULTS
DNA Methylation of the CD8 Gene Cluster in CD8+ and CD8− T Cells
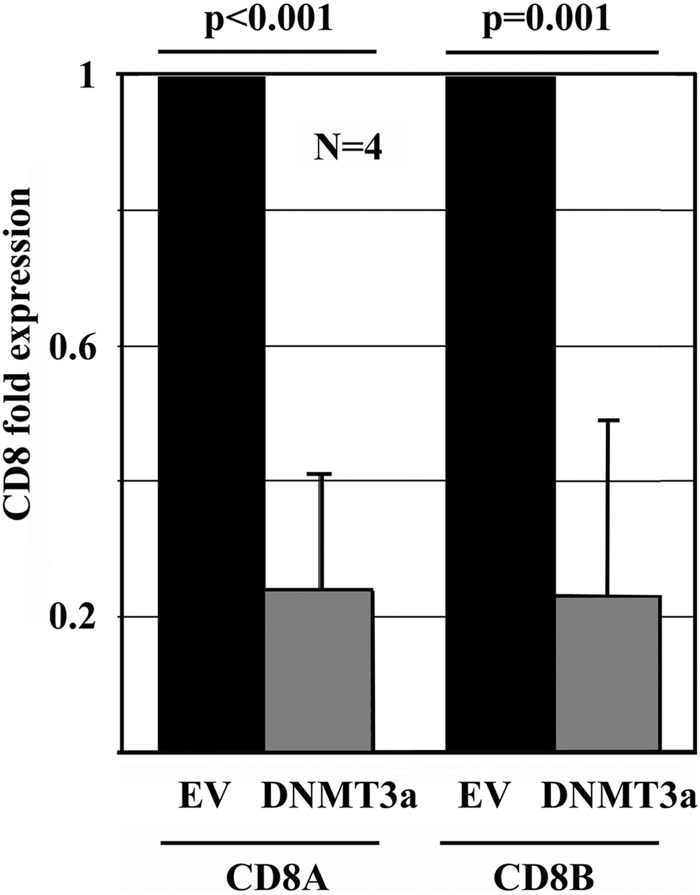
DNA methylation plays a well established role in the regulation of gene expression (16). To investigate whether DNA methyltransferase (DNMT)3a-mediated de novo DNA methylation affects CD8 expression, we over-expressed DNMT3a in primary human CD8+ T cells from healthy individuals (17, 18) and detected significantly reduced CD8A (p < 0.001) and CD8B (p = 0.001) transcription (Fig. 1).
FIGURE 1.
DNA methylation through DNMT3a regulates CD8 expression. CD8A and CD8B mRNA expression was assessed by quantitative RT-PCR after forced CpG-DNA methylation through DNMT3a (24 h).
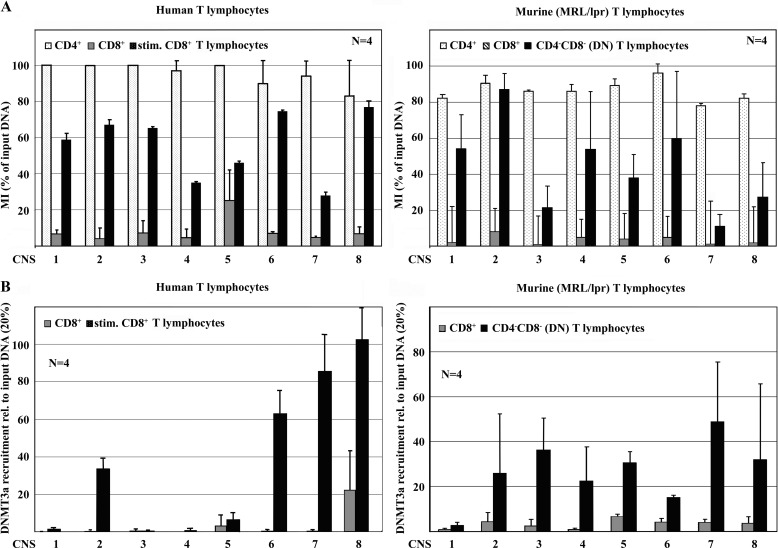
Thus, we aimed to determine DNA methylation of the human and murine CD8 genes in CD8+ T cells as compared with CD4+ and DN T cells. For the study of the human CD8 cluster, CD4+ and CD8+ T cells were sorted. Secondary to their low abundance in the peripheral blood, human DN T cells were in vitro induced by stimulating CD8+ T cells with anti-CD3 and anti-CD28 antibodies as reported previously (1). Murine CD4+, CD8+, and DN T cells were sorted from spleens of MRL/lpr mice on a FACSAria flow cytometer. Reflecting our gene expression data, human and murine CD4+ T cells displayed high levels of DNA methylation of the entire CD8 cluster, whereas CD8A and CD8B in CD8+ T cells were largely unmethylated (Fig. 2A). Murine and “induced” human DN T cells exhibited methylation levels that were between those in CD4+ and CD8+ T cells, suggesting that the CD8 genes are undergoing DNA methylation during the CD8 down-regulation process in DN T cell generation. Of note, DNA methylation of the human CD8 cluster was more homogenous when compared with murine CD8.
FIGURE 2.
CD8 expression is determined by DNA methylation. A, CD4+ and CD8+ T cells from healthy humans and MRL/lpr mice were sorted for the assessment of DNA methylation. Murine DN T cells were sorted; human DN T cells were induced by TCR stimulation. In human (left) and murine (right) CD4+ T cells, the CD8 cluster was methylated. In CD8+ T cells from both species, the CD8 cluster was largely unmethylated. Stimulated human CD8+ T cells and murine DN T cells exhibited increased DNA methylation of the CD8 cluster. B, recruitment of the DNMT3a to the CD8 cluster was assessed using ChIP. In human and murine DN T cells, DNMT3a is recruited to the CD8 cluster. DNA methylation of the human CD8 cluster is more homogenous when compared with mice. In both species DNMT3a recruitment is particularly strong at CNS2, -6, -7, and -8.
DNA Methylation of the CD8 Cluster Is Mediated by DNMT3a
We previously demonstrated that CREMα interacts with the de novo methyltransferase DNMT3a, mediating epigenetic remodeling (17, 18). Similar to the silencing of IL2 in effector T cells, the CD8 cluster undergoes de novo DNA methylation during generation of DN T cells. Thus, we wondered whether this could also be mediated by the interaction of CREMα with DNMT3a. We performed DNMT3a ChIP assays in human CD8+ T cells from healthy individuals and induced DN T cells as well in murine CD4+, CD8+, and DN T cells. As indicated in Fig. 2B, DNMT3a is recruited to the human and murine CD8 cluster in non-CD8 expressing T cells, particularly to CNS2, -7, and -8.
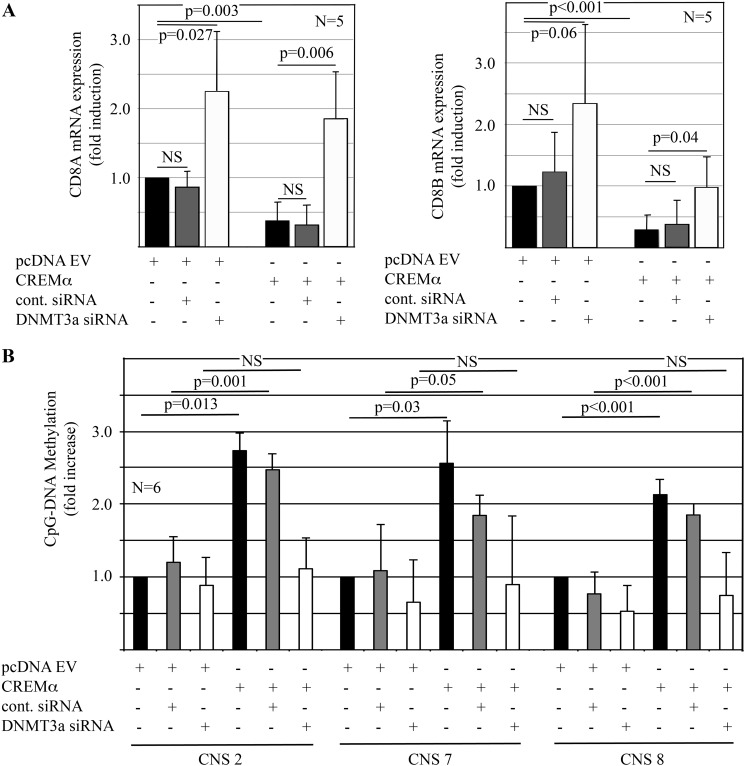
To establish the physiological relevance of the interaction between CREMα and DNMT3a, we over-expressed CREMα in primary human CD8+ T cells from healthy individuals, whereas knocking down DNMT3a (Fig. 3). Indeed, knock-down of DNMT3a resulted in increased CD8A (p = 0.027) and CD8B (p = 0.06) transcription, whereas forced CREMα expression mediated reduced CD8A (p = 0.003) and CD8B (p < 0.001) expression (Fig. 3, A and B). As suggested by previous studies and the finding that DNA methylation plays a role in the regulation of CD8, CREMα-mediated effects on CD8A and CD8B expression were reversible by DNMT3a knock-down (Fig. 3A). This was reflected by DNA methylation of CNS2, -7, and -8, which was increased in response to CREMα over-expression and reversible by DNMT3a knock-down (Fig. 3B).
FIGURE 3.
CREMα recruits DNMT3a to the CD8 cluster. A, DNMT3a knock-down results in increased CD8A and CD8B mRNA expression in human CD8+ T cells (24 h) when compared with controls (left: CD8A, right: CD8B, first to third lanes). CREMα reduces CD8A and CD8B mRNA expression (24h) (fourth and fifth lanes). DNMT3a knock-down reverses CREMα-mediated suppression of CD8A and CD8B mRNA (24 h) (sixth lane). B, CD8A and CD8B mRNA expression in response to CREMα with or without DNMT3a knock-down are reflected by DNA methylation.
Histone 3 Methylation (H3K9me3 and H3K27me3) Controls CD8 Expression
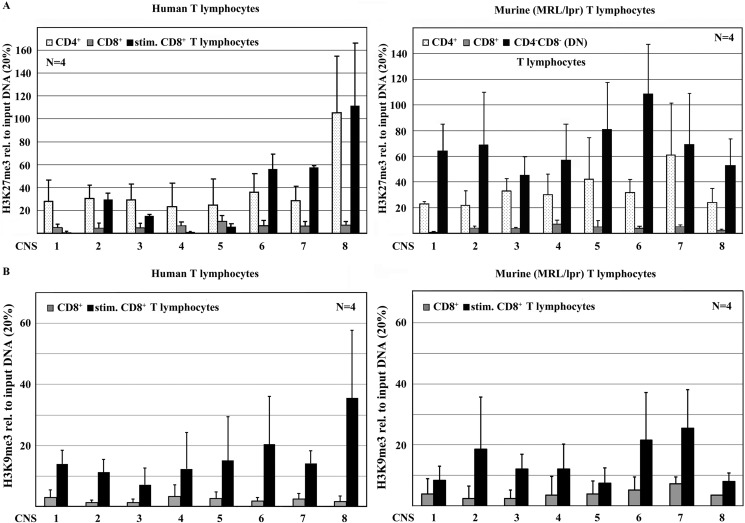
DNA methylation and histone methylation frequently follow the same patterns and can affect one another (21, 16). Thus, we asked whether the epigenetic silencing of CD8A and CD8B is facilitated also by histone modifications. Reflecting DNA methylation, human and murine CD4+ T cells displayed high levels of histone H3 tri-methylation at lysine residues 9 (H3K9me3) and 27 (H3K27me3) over the entire CD8 cluster, whereas CD8A and CD8B in CD8+ T cells were largely unmethylated. Furthermore, murine and induced human DN T cells exhibited increased H3K9me3 and H3K27me3 when compared with CD8+ T cells (Fig. 4). Interestingly, histone H3K9 and H3K27 methylation patterns were more consistent over the murine CD8 cluster, whereas histone H3 methylation peaks around CNS2, -6, -7, and -8 in human DN T cells. These differences may be the result of less “pure” DN T cell populations applied in the human experiments. As mentioned, human DN T cells were induced by TCR stimulation of CD8+ T cells over 5 days, reaching a DN T lymphocyte purity of usually around 50% (1).
FIGURE 4.
CD8 expression is reflected by H3K27 and H3K9 methylation. CD4+ and CD8+ T cells from healthy humans and MRL/LPR mice were sorted for the assessment of histone methylation of the CD8 cluster, using ChIP. Murine DN T cells were sorted; human DN T cells were induced by TCR stimulation. In human (left) and murine (right) CD4+ T cells, histone H3 was tri-methylated at residues lysine 27 (H3K27me3) (A) and at lysine 9 (H3K9me3) (B) over the entire CD8 cluster. In CD8+ T cells from both species, almost no H3K9me3 orH3K27me3 was detected, whereas induced human and murine DN T cells exhibited an increase in H3K27me3 and H3K9me3. Histone H3K9 and H3K27 methylation is more consistent over the murine CD8 cluster, whereas histone H3 methylation peaks around CNS2, -6, -7, and -8 in human DN T cells.
CREMα Recruits Histone Methyltransferase G9a to the CD8 Cluster
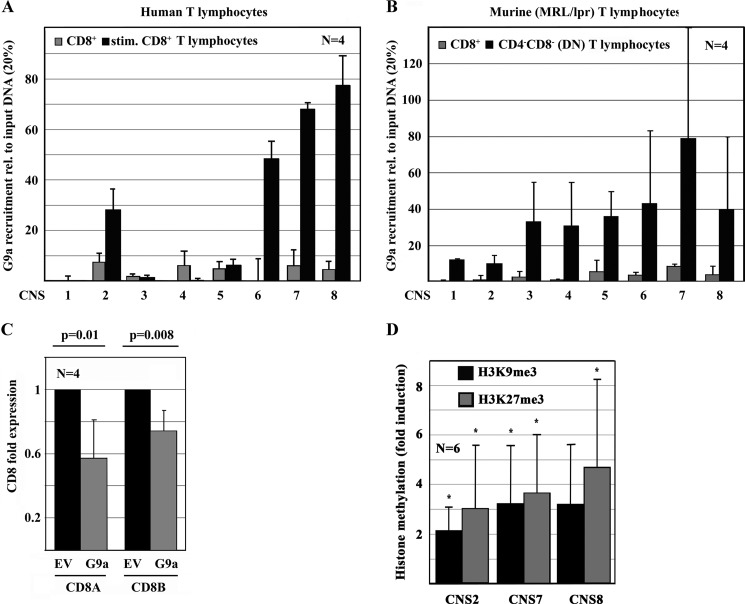
During the differentiation of cells and tissues, epigenetic remodeling is orchestrated by transcription factors, such as CREMα that interacts with DNMT3a (16, 21, 22). Because DNA and histone methylation patterns of the CD8 cluster follow the same patterns during peripheral DN T cell generation, we wondered which histone methyltransferase(s) were responsible for the methylation of histone H3K9 and H3K27 and whether they are mediated by CREMα, connecting DNA methylation and histone code. We performed ChIP assays, analyzing histone methyltransferase G9a recruitment to the CD8 cluster in CD8+, CD4+, and DN T cells from healthy humans and MRL/lpr mice. We chose G9a as a histone methyltransferase that has been reported relatively specific for histone methylation at H3K9 and H3K27 (23). As displayed in Fig. 5, G9a recruitment largely reflects H3K9 and H3K27 methylation of the CD8 cluster, suggesting these modifications are being mediated by G9a. To test this hypothesis, we forced G9a expression in primary human CD8+ T cells and monitored gene expression and histone H3K9 and H3K27 methylation (Fig. 5, C and D). Indeed, forced G9a expression resulted in a significant reduction of CD8A (p = 0.01) and CD8B (p = 0.008) mRNA expression that was mirrored by an increase in H3K9me and H3K27me3 around CNS2, -7, and -8, which exhibited the strongest recruitment of G9a.
FIGURE 5.
CREMα interacts with G9a mediating chromatin remodeling of the CD8 cluster. A and B, based on histone methylation patterns, the recruitment of G9a to the CD8 cluster has been tested, using ChIP. In both healthy humans (A) and MRL/lpr mice (B), the recruitment of G9a to the CD8 cluster in DN T cells is largely increased when compared with CD8+ T cells. C, forced expression of the histone methyltransferase G9a mediates a significant reduction of CD8A and CD8B mRNA expression through a significant increase of H3K9me3 and H3K27me3 (D) (*, indicates p < 0.05).
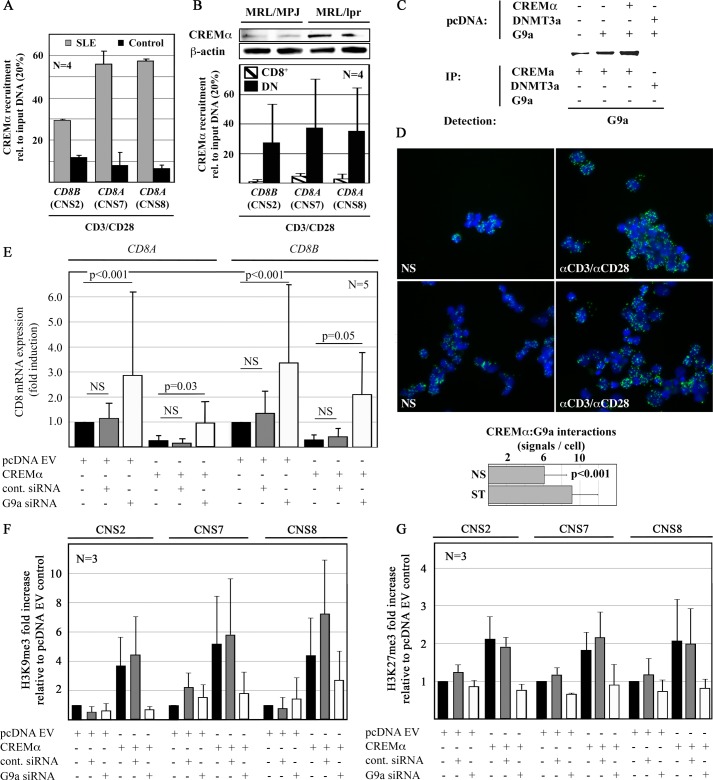
Because CREMα recruits DNMT3a to target genes in health and disease (1, 4, 5, 24, 25) and histone and DNA methylation usually follow the same patterns, we hypothesized that CREMα may also recruit G9a to the CD8 cluster, thus orchestrating epigenetic remodeling. To test our hypothesis, we investigated whether CREMα is differentially recruited to CNS2, -7, and -8 in CD8+ T cells from humans and MRL/lpr mice (Fig. 6, A and B). Indeed, CREMα is recruited to the CD8 cluster in both CD8+ T cells from healthy controls and SLE patients. However, T cells from SLE patients exhibit enhanced CREMα recruitment to CNS2, -7, and -8 when compared with controls, suggesting CREMα contributing to the enhanced generation of DN T cells in SLE (Fig. 6A). As SLE patients, MRL/lpr mice exhibit increased CREMα expression in T cells (Fig. 6B, upper panel) and exhibit enhanced recruitment of CREMα to CNS2, -7, and -8 was determined in DN T cells when compared with CD8+ T cells (Fig. 6B, lower panel).
FIGURE 6.
CREMα recruits G9a to the CD8 cluster. A, CREMα recruitment to CNS2, -7, and -8 is enhanced in CD8+ T cells from SLE patients. B, in analogy to SLE T cells, CREMα expression is increased in T cells from MRL/lpr mice (upper panel). In DN but not in CD8+ T cells, CREMα is recruited to CNS2, -7, and -8 suggesting CREMα is involved in the down-regulation of CD8 (lower panel). C, HEK293T cells were transfected with empty pcDNA3.1 plasmids. Proteins were co-immunoprecipitated with anti-CREMα or DNMT3a antibodies as indicated. Lysates were subjected to Western blotting with G9a antibodies. Representative results from one of three independent experiments are displayed. D, an interaction between CREMα and G9a has been established applying PLA. Ex vivo isolated CD8+ T cells exhibit interactions between CREMα and G9a that are enhanced after TCR stimulation (120 h). The number of PLA signals per unstimulated (NS) or stimulated (ST) CD8+ T cell was quantified using ImageJ software. Displayed is the number of signals per CD8+ T cell in 50 visual fields from 5 independent experiments (lower panel). E, G9a knock-down results in an increase of CD8A and CD8B mRNA expression in primary human CD8+ T cells (24 h) when compared with controls siRNA (left, CD8A; right, CD8B, first to third lanes). CREMα reduces CD8A and CD8B mRNA expression (24h) (fourth and fifth lanes). G9a knock-down reverses the CREMα effects on CD8A and CD8B (sixth lane). F and G, CD8A and CD8B expression patterns in response to CREMα with or without G9a knock-down are reflected by histone H3K9 (F) and H3K27 (G) methylation of CNS2, -7, and -8.
To investigate whether CREMα and G9a physically interact, we performed co-immunoprecipitation assays with anti-CREMα antibodies. In HEK293T cells that were transfected with an empty pcDNA3.1 plasmid we co-immunoprecipitated only small amounts of G9a with CREMα (Fig. 6C, first lane). Forced expression of G9a allowed increased amounts of G9a with CREMα to co-immunoprecipitate (Fig. 6C, second lane). Overexpression of both CREMα and G9a allowed strong co-immunoprecipitation of EHMT2/G9a with CREMα (Fig. 6C, third lane). This suggests a direct interaction between CREMα and G9a. To exclude the possibility of G9a interacting directly with DNMT3a, thus allowing co-immunoprecipitation with CREMα antibodies, we transfected cells with DNMT3a and G9a expression plasmids and tried to co-immunoprecipitate G9a with anti-DNMT3a antibodies (Fig. 6C, fourth lane). Failure to co-immunoprecipitate G9a with DNMT3a suggests that G9a interacts with CREMα rather than interacting with CREMα indirectly through DNMT3a. To reconfirm these findings with another technique, we forced the expression of CREMα and G9a in the same (HEK293T) cells, followed by PLA (Olink). Forced expression of either CREMα or G9a resulted in an increased signal when compared with controls. Forced expression of both CREMα and G9a resulted in the strongest signal, indicating in situ interaction (not shown). To translate our preliminary findings into the biological context of our study, we performed PLA in freshly isolated CD8+ T cells and after stimulation with anti-CD3 and anti-CD28 antibodies for 120 h. Indeed, CREMα and G9a also co-localized in primary human CD8+ T cells and the interaction was enhanced in response to stimulation for 120 h (Fig. 6D).
To determine the physiological relevance of the interaction between CREMα and G9a, we over-expressed CREMα in primary human CD8+ T cells while knocking down G9a (Fig. 6, E–Gs). G9a knock-down resulted in increased CD8A and CD8B mRNA expression, whereas reduced CD8A and CD8B expression in response to CREMα was reversible targeting G9a with siRNAs (Fig. 6E). This was reflected by increased H3K9me3 and H3K27me3 in response to CREMα, which was reversed by G9a knock-down (Fig. 6, F and G). Thus, our findings indicate that the interaction between CREMα and G9a plays a role in the regulation of CD8A and CD8B gene expression.
DISCUSSION
In the peripheral blood of SLE patients, DN T cells are expanded and contribute to disease expression and tissue damage (4, 5, 24, 25). DN T cells in SLE derive from CD8+ T cells by down-regulating CD8 surface expression and the acquisition of distinct effector phenotypes (4, 5, 24, 25). However, the molecular mechanisms instructing the transformation of CD8+ T cells into DN T cells remain largely unclear.
We previously demonstrated that the transcription factor CREMα trans-regulates a region syntenic to the murine CD8b promoter, thus contributing to the generation of DN T cells (1). Studies demonstrated that transcriptional silencing can be achieved by epigenetic remodeling of the murine CD8 cluster with a “closed” chromatin conformation prohibiting the recruitment of trans-activating signals (4, 5, 24, 25). However, the inducing stimuli and molecular mechanisms remained unknown. It has been shown that the presence of DNA methyltransferase (DNMT) 1 is essential for the silencing of CD8 in CD8− tissues. As a result, DNMT1-deficient mice exhibit enhanced CD8 expression secondary to a loss of inhibition (5, 26). Interestingly, this selectively occurred in TCR-γδ T cells and not in CD4+ TCR-αβ T cells that usually express the α-chain of the CD8αβ heterodimer, suggesting the chromatin conformation and resulting expression patterns of CD8 to be more variable when compared with CD4, which was constitutively repressed in non-CD4 expressing tissues (5, 26). This indicates that epigenetic remodeling of the CD8 cluster may be a central step in the transformation of CD8+ into DN T cells. We demonstrate that the CD8 cluster undergoes epigenetic remodeling in response to stimulation of CD8+ T cells, contributing to the generation of DN T cells. In both induced human and sorted MRL/lpr DN T cells DNA methylation along the entire CD8 cluster is mirrored by H3K9 and H3K27 tri-methylation, indicating that chromatin remodeling could be involved in the down-regulation of CD8 in mature DN T cells. Although the entire CD8 cluster in human DN T cells exhibits a consistently high methylation index, DNA methylation in murine DN T cells is region-specific with a low methylation index of CNS7 and -8. This is agreement with early findings of increased DNase hypersensitivity in DN T cells from MRL/lpr and CBA/CaH WEHI mice in the same regions (27, 28). Interestingly, histone H3K9 and H3K27 methylation is more consistent over the murine CD8 cluster, whereas histone H3 methylation peaks around CNS2, -6, -7, and -8 in human DN T cells. Taken together, CNS2, -6, -7, and -8 undergo the most striking epigenetic changes during the generation of DN T cells in both species and map to previously reported elements that are the target of chromatin remodeling during CD8+ lineage determination in the thymus of mice (4, 25). Species-specific characteristics in the distribution of DNA and histone H3 methylation remain to be understood. However, the discrete differences add up to a “common” epigenetic pattern that results in silencing of CD8 in DN T cells.
Transcription factors influence chromatin conformation through the recruitment of DNA and/or histone methyltransferases (16–18, 21). We and others demonstrated that CREMα and its counteractor CREB diametrically influence the transcriptional activity of cytokine genes through both trans-regulation and the recruitment of epigenetic modulators (22). The ATF transcription factor CREB has been demonstrated to interact with the p300 co-activator that has histone acetyltransferase activity (29). We recently reported that CREMα mediates epigenetic remodeling of cytokine genes during the priming of CD4+ T cells, including diametric effects on DNA methylation in effector T cells (18, 22). Because both CREB and CREMα orchestrate epigenetic remodeling and exhibit the aforementioned antithetic effects on CNS2, CREMα was a promising candidate in the search for regulators governing chromatin remodeling of the CD8 cluster during DN T cell generation. Indeed, we demonstrate that CREMα recruits both DNMT3a and the histone methyltransferase G9a to regulatory regions within the human and murine CD8 cluster (CNS2, 7, and 8), instructing chromatin remodeling and transcriptional silencing. Our finding that CREMα regulates CD8 expression through both trans-repression and the induction of epigenetic remodeling is in agreement with recent studies, targeting transcriptional regulation of murine CD4, CD8a, and CD8b during T cell development in the thymus. The observation that the transcription factor Runt-related transcription factor (RUNX) 1 is required for trans-repression of CD4 and that the closely related transcriptional regulator RUNX3 instructs chromatin remodeling of CD4 in DN thymocytes and CD8+ T cells provides further evidence that transcription factors orchestrate epigenetic priming of immune cells (4, 5, 22, 24, 30). Interestingly, RUNX3 and the Runx/core binding factor-β are necessarily required for CD8 co-receptor expression in activated CD8+ T cells through the recruitment to the E8I enhancer (31). The absence of E8I resulted in chromatin remodeling of the entire CD8 cluster with enhanced H3K27me3 and reduced histone H3 acetylation, both reflecting a “closure” of the murine CD8a gene (31). This is of special interest, because E8I maps to our CNS6 and -7, and is in close proximity to CNS8, all of which undergo CREMα-instructed epigenetic remodeling in response to TCR activation. Thus, reduced recruitment of RUNX3 to this region could also play a role in the transformation of CD8+ T cells into peripheral induced DN T cells and will be the focus of future studies.
Taken together, our data solidify the role of CREMα in the regulation of CD8. CREMα in addition to the previously reported trans-repression of CNS2 contributes to the down-regulation of CD8 through and the recruitment of DNMT3a and G9a. Because CREMα is increased in T cells from SLE patients and MRL/lpr mice, these mechanisms appear central for the generation of DN T cells in SLE and potentially other autoimmune diseases with increased numbers of DN T cells. This underlines the potential of CREMα as disease biomarker and putative therapeutic target in SLE. It remains to be determined whether CREMα instructs chromatin remodeling during the priming and differentiation of T cells in the thymus or if CREMα exclusively regulates CD8 in peripheral CD8+ T cells in response to activation.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 AI42269, R01 AI49954, and R01 AI85567 (to G. C. T.).
- SLE
- systemic lupus erythematosus
- DN
- double negative
- CREM
- cAMP responsive element modulator
- MeDIP
- methylated CpG-DNA immunoprecipitation
- PLA
- proximity ligation assay
- DNMT
- DNA methyltransferase
- CREB
- cAMP-response element-binding protein
- RUNX
- Runt-related transcription factor
- co-IP
- co-immunoprecipitation.
REFERENCES
- 1. Hedrich C. M., Rauen T., Crispin J. C., Koga T., Ioannidis C., Zajdel M., Kyttaris V. C., Tsokos G. C. (2013) cAMP responsive element modulator (CREM)α trans-represses the transmembrane glycoprotein CD8 and contributes to the generation of CD3+CD4−CD8− T cells in health and disease. J. Biol. Chem. 288, 31880–31887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hostert A., Tolaini M., Festenstein R., McNeill L., Malissen B., Williams O., Zamoyska R., Kioussis D. (1997) A CD8 genomic fragment that directs subset-specific expression of CD8 in transgenic mice. J. Immunol. 158, 4270–4281 [PubMed] [Google Scholar]
- 3. Kieffer L. J., Yan L., Hanke J. H., Kavathas P. B. (1997) Appropriate developmental expression of human CD8β in transgenic mice. J. Immunol. 159, 4907–4912 [PubMed] [Google Scholar]
- 4. Bilic I., Koesters C., Unger B., Sekimata M., Hertweck A., Maschek R., Wilson C. B., Ellmeier W. (2006) Negative regulation of CD8 expression via Cd8 enhancer-mediated recruitment of the zinc finger protein MAZR. Nat. Immunol. 7, 392–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kioussis D., Ellmeier W. (2002) Chromatin and CD4, CD8A and CD8B gene expression during thymic differentiation. Nat. Rev. Immunol. 2, 909–919 [DOI] [PubMed] [Google Scholar]
- 6. Ellmeier W., Sunshine M. J., Losos K., Hatam F., Littman D. R. (1997) An enhancer that directs lineage-specific expression of CD8 in positively selected thymocytes and mature T cells. Immunity 7, 537–547 [DOI] [PubMed] [Google Scholar]
- 7. Ellmeier W., Sunshine M. J., Losos K., Littman D. R. (1998) Multiple developmental stage-specific enhancers regulate CD8 expression in developing thymocytes and in thymus-independent T cells. Immunity 9, 485–496 [DOI] [PubMed] [Google Scholar]
- 8. Ellmeier W., Sunshine M. J., Maschek R., Littman D. R. (2002) Combined deletion of CD8 locus cis-regulatory elements affects initiation but not maintenance of CD8 expression. Immunity 16, 623–634 [DOI] [PubMed] [Google Scholar]
- 9. Garefalaki A., Coles M., Hirschberg S., Mavria G., Norton T., Hostert A., Kioussis D. (2002) Variegated expression of CD8α resulting from in situ deletion of regulatory sequences. Immunity 16, 635–647 [DOI] [PubMed] [Google Scholar]
- 10. Hostert A., Tolaini M., Roderick K., Harker N., Norton T., Kioussis D. (1997) A region in the CD8 gene locus that directs expression to the mature CD8 T cell subset in transgenic mice. Immunity 7, 525–536 [DOI] [PubMed] [Google Scholar]
- 11. Hostert A., Garefalaki A., Mavria G., Tolaini M., Roderick K., Norton T., Mee P. J., Tybulewicz V. L., Coles M., Kioussis D. (1998) Hierarchical interactions of control elements determine CD8α gene expression in subsets of thymocytes and peripheral T cells. Immunity 9, 497–508 [DOI] [PubMed] [Google Scholar]
- 12. Kieffer L. J., Bennett J. A., Cunningham A. C., Gladue R. P., McNeish J., Kavathas P. B., Hanke J. H. (1996) Human CD8α expression in NK cells but not cytotoxic T cells of transgenic mice. Int. Immunol. 8, 1617–1626 [DOI] [PubMed] [Google Scholar]
- 13. Zhang X. L., Seong R., Piracha R., Larijani M., Heeney M., Parnes J. R., Chamberlain J. W. (1998) Distinct stage-specific cis-active transcriptional mechanisms control expression of T cell coreceptor CD8α at double- and single-positive stages of thymic development. J. Immunol. 161, 2254–2266 [PubMed] [Google Scholar]
- 14. Zhang X. L., Zhao S., Borenstein S. H., Liu Y., Jayabalasingham B., Chamberlain J. W. (2001) CD8 expression up to the double-positive CD3(low/intermediate) stage of thymic differentiation is sufficient for development of peripheral functional cytotoxic T lymphocytes. J. Exp. Med. 194, 685–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Feik N., Bilic I., Tinhofer J., Unger B., Littman D. R., Ellmeier W. (2005) Functional and molecular analysis of the double-positive stage-specific CD8 enhancer E8III during thymocyte development. J. Immunol. 174, 1513–1524 [DOI] [PubMed] [Google Scholar]
- 16. Hedrich C. M., Tsokos G. C. (2011) Epigenetic mechanisms in systemic lupus erythematosus and other autoimmune diseases. Trends Mol. Med. 17, 714–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hedrich C. M., Rauen T., Tsokos G. C. (2011) cAMP-responsive element modulator (CREM)α protein signaling mediates epigenetic remodeling of the human interleukin-2 gene. Implications in systemic lupus erythematosus. J. Biol. Chem. 286, 43429–43436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hedrich C. M., Crispin J. C., Rauen T., Ioannidis C., Apostolidis S. A., Lo M. S., Kyttaris V. C., Tsokos G. C. (2012) cAMP response element modulator α controls IL2 and IL17A expression during CD4 lineage commitment and subset distribution in lupus. Proc. Natl. Acad. Sci. U.S.A. 109, 16606–16611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rauen T., Hedrich C. M., Juang Y. T., Tenbrock K., Tsokos G. C. (2011) cAMP-responsive element modulator (CREM)α protein induces interleukin 17A expression and mediates epigenetic alterations at the interleukin-17A gene locus in patients with systemic lupus erythematosus. J. Biol. Chem. 286, 43437–43446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Juang Y. T., Wang Y., Solomou E. E., Li Y., Mawrin C., Tenbrock K., Kyttaris V. C., Tsokos G. C. (2005) Systemic lupus erythematosus serum IgG increases CREM binding to the IL-2 promoter and suppresses IL-2 production through CaMKIV. J. Clin. Invest. 115, 996–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brenner C., Fuks F. (2007) A methylation rendezvous. Reader meets writers. Dev. Cell. 12, 843–844 [DOI] [PubMed] [Google Scholar]
- 22. Rauen T., Hedrich C. M., Tenbrock K., Tsokos G. C. (2013) cAMP responsive element modulator. A critical regulator of cytokine production. Trends Mol. Med. 19, 262–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tachibana M., Sugimoto K., Fukushima T., Shinkai Y. (2001) Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J. Biol. Chem. 276, 25309–25317 [DOI] [PubMed] [Google Scholar]
- 24. Sato T., Ohno S., Hayashi T., Sato C., Kohu K., Satake M., Habu S. (2005) Dual functions of Runx proteins for reactivating CD8 and silencing CD4 at the commitment process into CD8 thymocytes. Immunity 22, 317–328 [DOI] [PubMed] [Google Scholar]
- 25. Harker N., Garefalaki A., Menzel U., Ktistaki E., Naito T., Georgopoulos K., Kioussis D. (2011) Pre-TCR signaling and CD8 gene bivalent chromatin resolution during thymocyte development. J. Immunol. 186, 6368–6377 [DOI] [PubMed] [Google Scholar]
- 26. Lee P. P., Fitzpatrick D. R., Beard C., Jessup H. K., Lehar S., Makar K. W., Pérez-Melgosa M., Sweetser M. T., Schlissel M. S., Nguyen S., Cherry S. R., Tsai J. H., Tucker S. M., Weaver W. M., Kelso A., Jaenisch R., Wilson C. B. (2001) A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity 15, 763–774 [DOI] [PubMed] [Google Scholar]
- 27. Landolfi M. M., Van Houten N., Russell J. Q., Scollay R., Parnes J. R., Budd R. C. (1993) CD2−CD4−CD8− lymph node T lymphocytes in MRL lpr/lpr mice are derived from a CD2+CD4+CD8+ thymic precursor. J. Immunol. 151, 1086–1096 [PubMed] [Google Scholar]
- 28. Wu L., Pearse M., Egerton M., Petrie H., Scollay R. (1990) CD4−CD8− thymocytes that express the T cell receptor may have previously expressed CD8. Int. Immunol. 2, 51–56 [DOI] [PubMed] [Google Scholar]
- 29. Lee J. S., Zhang X., Shi Y. (1996) Differential interactions of the CREB/ATF family of transcription factors with p300 and adenovirus E1A. J. Biol. Chem. 271, 17666–17674 [PubMed] [Google Scholar]
- 30. Taniuchi I., Osato M., Egawa T., Sunshine M. J., Bae S. C., Komori T., Ito Y., Littman D. R. (2002) Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell 111, 621–633 [DOI] [PubMed] [Google Scholar]
- 31. Hassan H., Sakaguchi S., Tenno M., Kopf A., Boucheron N., Carpenter A. C., Egawa T., Taniuchi I., Ellmeier W. (2011) Cd8 enhancer E8I and Runx factors regulate CD8α expression in activated CD8+ T cells. Proc. Natl. Acad. Sci. U.S.A. 108, 18330–18335 [DOI] [PMC free article] [PubMed] [Google Scholar]