Background: Endothelial cells are well known inflammatory effectors, but TLR4 expression and function in human pericytes have not been addressed.
Results: Pericytes express TLR4, and stimulation with LPS or HMGB1 promotes cytokine and chemokine secretion and overexpression of adhesion molecules via NF-κB.
Conclusion: TLR4 activation in pericytes triggers a proinflammatory and proangiogenic program.
Significance: Pericytes are active players in inflammatory responses beyond their homeostatic role.
Keywords: Angiogenesis, Chemokines, Cytokine, Inflammation, Lipopolysaccharide (LPS), HMGB1, NF-κB, TLR4, Pericyte
Abstract
Pericytes and mesenchymal stem cells (MSCs) are ontogenically related, and in fact, no significant phenotypic differences could be observed by flow cytometry. Transcriptome analysis of human pericytes and MSCs revealed that 43 genes were up-regulated more than 10-fold in pericytes compared with MSCs. Identification of Toll-like receptor 4 (TLR4) as one of the most abundant RNA species in pericytes with respect to MSCs and confirmation of TLR4 expression on the cell surface led us to obtain a comprehensive overview of the expression program of lipopolysaccharide (LPS)-stimulated pericytes. Transcriptional profiling of LPS-treated cells revealed that 22 genes were up-regulated more than 5-fold. Of them, 10 genes encoded chemokines and cytokines (CXCL10, CCL20, IL8, CXCL1, IL6, CCL2, IL1B, CXCL2, IL1A, and CXCL6), and three genes encoded adhesion molecules (ICAM1, VCAM1, and SELE). LPS induced nuclear translocation of the transcription factor NF-κB in stimulated pericytes. Moreover, inhibition of NF-κB activation by SC-514 blocked LPS-induced up-regulation of a subset of chemokine genes, confirming the key role of NF-κB in LPS signaling in pericytes. At the protein level, we assessed the secretion of the proinflammatory cytokines and chemokines IL-6, IL-8, CXCL1, CXCL2, CXCL3, and CCL2 not only after LPS treatment but also in HMGB1-stimulated pericytes. Up-regulation of the adhesion molecules ICAM-1 and VCAM-1 resulted in an increased adhesion of peripheral blood leukocytes to an LPS-treated pericyte monolayer. The role of pericytes in the inflammatory context has been scarcely addressed; according to these results, pericytes should be considered as active players in the inflammatory cascade with potential physiopathological implications.
Introduction
Pericytes, the mural cells found surrounding capillaries in close association with endothelial cells (ECs),2 have a well known role in angiogenesis and vascular homeostasis, participating in vessel maturation, capillary stabilization, and regulation of flow rate (1). Pericyte number, morphology, and the degree of vascular pericyte coverage differ considerably depending on location and type of vessel (2). Pericytes not only provide structural support as classically assumed but also interact with ECs by direct cell-cell contacts and paracrine signals. Recently, several works have endowed pericytes with unexpected mesenchymal stem cell (MSC)-like properties. Pericytes can express MSC markers and behave like MSCs both in vitro and in vivo (3). In fact, pericytes of different sources have been shown to regenerate cartilage, bone, muscle, and skin (4–6). Conversely, MSCs have been attributed a perivascular origin (4) and can exhibit a pericyte-like behavior (7). In fact, it has been suggested that once the intimate relationship between ECs and pericytes is disrupted, the pericyte should be then considered an MSC (8). In agreement with their close ontogenetic relationship, pericytes and MSCs show a very similar phenotype. In an attempt to identify particular features at the genomic level, we compared the gene expression profiles of human brain vascular pericytes (HBVPs) and human umbilical cord-derived MSCs using Affymetrix Human Gene 1.0 ST microarrays. Interestingly, the second most highly expressed gene in HBVPs in comparison with MSCs was Toll-like receptor 4 (TLR4). TLRs are type I transmembrane glycoproteins that recognize pathogen-associated and damage-associated molecular patterns. TLR4 is part of the receptor complex that binds lipopolysaccharide (LPS) from the Gram-negative bacterial cell wall as well as endogenous ligands like the proinflammatory cytokine-like protein high mobility group box 1 (HMGB1). TLR4 is detected in monocytes, macrophages, dendritic cells, and several T cells populations, but its expression in non-immune cells is less documented, although some reports demonstrate that an LPS signaling system also exists in ECs (9–12). In a series of pioneering works by Edelman et al. (13–15), the authors demonstrated the expression of TLR4 by Western blotting in rat lung pericytes (13) as well as increased vessel permeability (14) and production of IL-1β (15) in LPS-treated pericytes, suggesting for the first time an active role of pericytes in the inflammatory cascade. More recently, the release of nitric oxide and several cytokines and chemokines by mouse brain vascular pericytes in response to LPS has been reported (16). Other interesting works studied the expression of immunologically relevant cell surface proteins by human pericytes after stimulation with TNF-α or IFN-γ (17, 18) but not with LPS. Moreover, no data can be found in the literature about the effect of HMGB1 on pericytes. Therefore and to our knowledge, neither TLR4 expression by human pericytes nor their responsiveness to LPS and HMGB1 has been documented yet. Here, we report comprehensive data of the transcriptional profile of LPS-treated HBVPs, cytokine and chemokine secretion after LPS and HMGB1 stimulation, and dissection of the signaling cascade beyond TLR4 activation.
EXPERIMENTAL PROCEDURES
Cell Culture and Reagents
Human MSCs from umbilical cord (19) were purchased from Inbiobank (Inbiomed, San Sebastian, Spain) and were cultured in low glucose DMEM (Lonza, Basel, Switzerland) supplemented with 10% MSC-qualified fetal calf serum (Promocell, Heidelberg, Germany). HBVPs were purchased from ScienCell Research Laboratories (Carlsbad, CA) and cultured in 2% fetal calf serum pericyte medium (ScienCell Research Laboratories). All primary cells were used between passages 2 and 5. LPS from Escherichia coli 026:B6 and anisomycin from Streptomyces griseolus were purchased from Sigma-Aldrich. Recombinant LPS-free HMGB1 in the redox form known to act as an inflammation mediator (20) was kindly provided by M. Bianchi (San Raffaele University and Scientific Institute, Milan, Italy). NF-κB inhibitor SC-514 and mitogen-activated protein kinase inhibitors SP600125 (JNK1, -2, and -3 inhibitor), PD98059 (MEK-1 inhibitor), and SB203580 (p38 inhibitor) were all purchased from Santa Cruz Biotechnology (Heidelberg, Germany).
Flow Cytometry
Cell surface HBVP and MSC antigen staining was performed by direct immunofluorescence using phycoerythrin-conjugated monoclonal antibodies against human PDGFRB (clone PR7212, R&D Systems), NG2/MCSP (clone LHM-2, R&D Systems), CD13 (clone WM15, BD Pharmingen), CD73 (clone AD2, BD Pharmingen), CD105 (clone SN6, eBioscience, Frankfurt, Germany), VCAM-1 (clone STA, ab33228, Abcam, Cambridge, UK), and ICAM-1 (clone 1H4, Immunotools, Friesoythe, Germany). Cell surface expression of TLR4 in HBVPs was analyzed using a mouse monoclonal antibody (clone HTA125, Abcam) followed by a goat anti-mouse phycoerythrin-conjugated secondary antibody (Jackson ImmunoResearch Laboratories, Suffolk, UK). Samples were analyzed with an EPICS XL flow cytometer (Coulter Electronics, Hialeah, FL).
Microarray Analysis
Total RNA from HBVPs treated with LPS for 4 h or untreated HBVPs and MSCs was extracted using the RNeasy Micro kit (Qiagen, Hilden, Germany). A DNase treatment step was included in the procedure (Qiagen). RNA was quantified with a NanoDrop ND-1000 spectrophotometer and checked for integrity on a Bioanalyzer 2100B (Agilent Technologies, Santa Clara, CA). For each cell type and condition, three independent RNA samples representing biological triplicates were obtained. Double-stranded cDNA was synthesized from 500 ng of total RNA using Ambion® WT Expression kit (Applied Biosystems, Carlsbad, CA). After cDNA purification, this DNA was used as template for the in vitro transcription. The obtained cRNA was fragmented and hybridized to the GeneChip® Human Gene 1.0 ST array (Affymetrix, Santa Clara, CA) for 16 h at 45 °C. Hybridized microarrays were washed and stained with a streptavidin-phycoerythrin conjugate in a GeneChipFluidics Station 450. All these procedures were carried out at the Universidad Complutense de Madrid Genomics and Proteomics Core Facility as suggested by the manufacturer. Hybridized cRNA was finally identified by the fluorescence signal in a GeneChip 3000 scanner. The CEL files generated from the scanning were converted to gene expression signals using the robust multichip average algorithm (21) in Affymetrix Expression Console. Subsequent analyses were performed with Babelomics gene expression and functional analysis suite version 4.2. Limma (22) was used for differential expression analysis. p values were corrected by calculating the false discovery rate (FDR). Genes with a signal ratio >2 or <0.5 and a p value <0.05 were considered significant. The microarray data produced in this analysis are deposited in the NCBI Gene Expression Omnibus (23) and are accessible through GEO Series accession numbers GSE46235 and GSE46236.
Bioinformatics
Grouping for ontologically similar genes was assessed with the Database for Annotation, Visualization and Integrated Discovery (DAVID 6.7) using the full human genome as reference background. For the contrast between LPS-treated and untreated HBVPs, a list of 76 genes was generated whose members passed the following threshold combination: p value <0.05 and -fold change >2. Data were analyzed in the “Functional Annotation Chart” tool using an Expression Analysis Systematic Explorer (EASE) threshold of 0.001 for molecular function, biological process, and cellular compartment gene ontology (GO) terms. An FDR value <0.05 indicated a significant enrichment.
Quantitative Real Time PCR
HBVPs were treated with different concentrations of LPS or HMGB1 for 1.5 or 4 h. Total RNA was extracted with the RNeasy Mini kit (Qiagen), and cDNA was synthesized from 500 ng of total RNA by random primer reverse transcription using a SuperScript VILO cDNA Synthesis kit (Invitrogen). Primer sequences for IL6 (24), IL8, CXCL1, -2, and -3 (25) as described previously were synthesized by Roche Diagnostics (Sant Cugat del Vallés, Spain). Primer pairs amplified products of 63–117 bp. Real time PCR was performed with a LightCycler 480 apparatus (Roche Diagnostics) using the LightCycler DNA Master SYBR Green kit (SABiosciences, Qiagen). The relative expression of each mRNA was calculated by the ΔCT method (where ΔCT is the value obtained by subtracting the CT value of the internal loading control gene SDHA mRNA from the CT value of the target mRNA). The amount of the target relative to the SDHA mRNA was expressed as 2−(ΔCT). -Fold expression changes were calculated using the equation 2−(ΔΔCT) where ΔΔCT is equal to ΔCT (experimental condition) − ΔCT (control condition). In some experiments, different concentrations of NF-κB and MAPK inhibitors were added to the culture medium 30 min before LPS treatment. Experiments were performed on three separate occasions. Experiments were performed in triplicate and on three separate occasions.
Western Blotting
Cells were treated with 1 μg/ml LPS for 4 h, lysed in Laemmli lysis buffer (Bio-Rad) for 10 min on ice, and collected by scraping. Equal amounts of proteins were resolved by 12% SDS-PAGE and transferred onto nitrocellulose membranes using the iBlot Dry Blotting System (Invitrogen). Membranes were incubated for 2 h with 2 μg/ml anti-human MyD88 (ab2064, Abcam) or 1 μg/ml anti-MD-2 (ab24182, Abcam) rabbit polyclonal antibody or 10 μg/ml anti-CD14 mouse monoclonal antibody (ab49755, Abcam). After that, membranes were incubated with IgG-IRDye800-conjugated anti-rabbit and anti-mouse antibodies diluted 1:15000 (Rockland Immunochemicals). During incubation with primary antibodies, membranes were simultaneously incubated with 500 ng/ml anti-human β-actin mouse monoclonal (ab8226, Abcam) or rabbit polyclonal (ab8227, Abcam) antibodies as a loading control followed by 1:15,000 diluted anti-mouse or anti-rabbit IgG-IRDye700-conjugated antibodies (Rockland Immunochemicals), respectively.
Visualization and quantitative analysis of protein bands were carried out with the Odyssey infrared imaging system (LI-COR Biosciences, Lincoln, NE). For the analysis of p38 activation, membranes were incubated with 1:1000 diluted anti-phospho-p38 MAPK (Thr-180/Tyr-182) rabbit monoclonal antibody (clone 3D7, Cell Signaling Technology) and anti-p38 rabbit polyclonal antibody (Santa Cruz Biotechnology) followed by anti-rabbit IgG-IRDye800.
Measurement of Cytokines and Chemokines
Concentrations of 23 cytokines and chemokines were measured semiquantitatively in conditioned media of treated (1 μg/ml LPS or 10 μg/ml HMGB1 for 4 h) or untreated HBVPs and MSCs with the human cytokine membrane-based antibody array kit I (RayBiotech). Instead of the HRP-conjugated streptavidin provided with the RayBiotech kit, 10 ng/ml IRDye800-labeled streptavidin (Rockland Immunochemicals) was used for detection with the Odyssey imaging system and quantification using the Odyssey application software (LI-COR Biosciences).
Additionally, levels of 11 cytokines in culture supernatants were quantified with the bead-based human Th1/Th2 FlowCytomix Multiplex kit (eBioscience, San Diego, CA) following the manufacturer's instructions. A FACSCalibur (BD Biosciences) flow cytometer device was used to analyze the samples.
Immunocytofluorescent Staining to Assess Nuclear Translocation of NF-κB
The NF-κB spatial translocation assay was performed according to Cellomics® NF-κB Activation HCS Reagent kit (Thermo Scientific Pierce, Rockford, IL) protocol. HBVPs were seeded onto cell chamber slides (Nunc) and stimulated with LPS (1 μg/ml) for 90 min. Then cells were fixed with 4% paraformaldehyde and permeabilized in 0.1% Triton X-100 for 10 min. After blocking, cells were incubated with a rabbit anti-NF-κB antibody followed by DyLight 488-conjugated anti-rabbit secondary antibody according to the manufacturer's instructions. Fluorescence images for NF-κB translocation were captured with a confocal laser-scanning microscope (TCS SP5, Leica Microsystems CMS GmbH, Mannheim, Germany).
Peripheral Blood Leukocyte Adhesion Assay
Adherence of peripheral blood leukocytes (PBLs) to LPS-stimulated HBVP monolayer was assessed. Confluent HBVPs seeded on 24-well plates were incubated with 1 μg/ml LPS for 24 h and washed three times with PBS. PBLs were isolated from healthy donors, labeled with a 10 μm concentration of the red fluorescent dye PKH26 (Sigma-Aldrich), and added to the HBVP monolayer (1.5 × 105 cells/well) for 2 h at 37 °C. Non-adherent PBLs were then removed by washing twice with PBS, and the remaining cells were photographed under an inverted epifluorescence microscope (Eclipse TS100-F, Nikon, Amstelveen, Netherlands). The number of adhered cells was quantified by direct visualization of four different fields of every independent experiment.
Statistics
Results were expressed as mean ± S.D. The data were evaluated using Student's t test and were considered to be statistically significant when the p value was ≤0.05.
RESULTS
Comparative Phenotypic Analysis of HBVPs and MSCs
The phenotypes of HBVPs and MSCs have been previously addressed separately by flow cytometry using combinations of positive and negative markers because none of them are absolutely specific for any cell type (26, 27). HBVPs and MSCs do not express endothelial and hematopoietic lineage markers such as CD31, CD34, and CD45. Here, we compared the cell surface expression of molecules characteristic of both MSCs (CD13, CD73, and CD105) and HBVPs (PDGFRB and NG2) (Fig. 1). Consistent with their close ontogenic relationship, we found that more than 95% of both HBVPs and MSCs were positive for NG2, CD13, CD73, and CD105. PDGFRB expression was slightly lower in MSCs with respect to HBVPs (81 versus 96% of positive cells); otherwise their phenotypes were hardly distinguishable.
FIGURE 1.
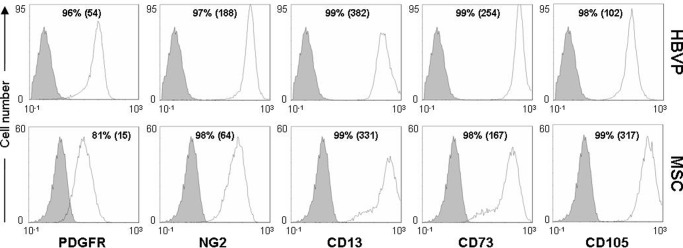
Comparative phenotypic analysis of HBVPs and MSCs. Cells were labeled with antibodies against the indicated antigens and analyzed by flow cytometry. Isotype-matched antibodies were used as control (in solid gray). Numbers indicate the percentage of positive cells together with the mean fluorescence intensity (in parentheses).
Differential Gene Expression Profiling between HBVPs and MSCs
Given that HBVPs may not be phenotypically distinguishable from MSCs, we attempted to identify differential gene expression profiles using global transcriptome analysis. The Human Gene 1.0 ST Array interrogates 28,869 well annotated genes. Among them, 607 genes (2.1%) were highly expressed (-fold change >2, p < 0.05) in HBVPs compared with MSCs, and 42 genes (0.14%) were expressed more than 10-fold, whereas 882 genes (3.05%) were significantly underexpressed in HBVPs (-fold change <0.5, p < 0.05), and 78 genes (0.27%) were expressed less than 10-fold (Table 1). According to the role of HBVPs in blood vessel homeostasis, the highly expressed genes in HBVPs included several molecules involved in angiogenesis: ANGPT2, ANGPTL4, EDN1, LAMA4, LAMA5, PGF, PLAU, PDGFA, and ROBO4. On the other hand, the highly expressed genes in MSCs included molecules involved in the biogenesis of extracellular matrix, such as collagens (COL1A1, COL1A2, COL3A1, COL5A1, COL11A1, and COL12A1), proteoglycans (HSPG2, VCAN, LUM, GPC1, DCN, and FMOD), matrix proteases (MMP-2, -3, -8, ADAMTS-1, -4, and -9), FBLN1, MATN1, and LOX. However, the most interesting finding in the differential expression pattern was the high relative expression of TLR4 in HBVPs versus MSCs (73.8-fold). Because TLR4 protein expression had not been documented in human pericytes, we decided to explore the functionality of this pathway triggered by LPS in HBVPs.
TABLE 1.
Genes up-regulated in HVBPs compared with MSCs (>10-fold). GEO accession number GSE46235
| Gene name | Accession no. | -Fold change | p value | False discovery rate |
|---|---|---|---|---|
| ZIC1 | NM_003412 | 84.347 | 2.81E − 16 | 1.17E − 12 |
| TLR4 | NR_024168 | 73.811 | 8.72E − 16 | 2.12E − 12 |
| SIX1 | NM_005982 | 63.233 | 5.26E − 16 | 1.39E − 12 |
| EPB41L3 | NM_012307 | 39.289 | 4.41E − 15 | 6.42E − 12 |
| EBF1 | NM_024007 | 23.831 | 1.85E − 14 | 1.68E − 11 |
| TFAP2C | NM_003222 | 22.265 | 4.14E − 13 | 1.47E − 10 |
| FAM49A | NM_030797 | 21.474 | 7.46E − 14 | 4.43E − 11 |
| C5orf23 | BC022250 | 21.317 | 1.23E − 15 | 2.50E − 12 |
| PTGS1 | NM_000962 | 19.319 | 5.14E − 14 | 3.44E − 11 |
| SEMA6D | NM_153618 | 18.987 | 4.35E − 12 | 9.05E − 10 |
| TMEM156 | NM_024943 | 18.525 | 3.62E − 13 | 1.37E − 10 |
| OLAH | NM_018324 | 17.606 | 1.05E − 10 | 1.09E − 08 |
| SULT1E1 | NM_005420 | 16.740 | 1.02E − 12 | 3.01E − 10 |
| GGT5 | NM_001099781 | 16.659 | 5.16E − 13 | 1.73E − 10 |
| TBX15 | NM_152380 | 16.610 | 1.84E − 12 | 4.64E − 10 |
| NPR3 | NM_000908 | 16.599 | 2.69E − 13 | 1.12E − 10 |
| MYO1D | NM_015194 | 16.139 | 1.45E − 12 | 3.90E − 10 |
| AOX1 | NM_001159 | 15.915 | 2.52E − 14 | 1.98E − 11 |
| ITGA10 | NM_003637 | 15.779 | 2.15E − 14 | 1.74E − 11 |
| LPAR4 | NM_005296 | 15.381 | 2.89E − 13 | 1.17E − 10 |
| AJAP1 | NM_018836 | 15.152 | 7.83E − 12 | 1.46E − 09 |
| IGFBP1 | NM_000596 | 14.912 | 4.48E − 12 | 9.18E − 10 |
| HTR1F | NM_000866 | 14.909 | 2.04E − 14 | 1.74E − 11 |
| SCUBE3 | NM_152753 | 14.131 | 3.03E − 09 | 1.72E − 07 |
| UNQ3104 | AY358109 | 13.360 | 4.39E − 12 | 9.06E − 10 |
| AFF3 | NM_002285 | 13.185 | 6.48E − 15 | 8.20E − 12 |
| RNF182 | NM_152737 | 13.088 | 6.05E − 11 | 7.24E − 09 |
| TRPC6 | NM_004621 | 12.990 | 3.01E − 12 | 6.83E − 10 |
| IGFBP2 | NM_000597 | 12.315 | 1.92E − 11 | 2.93E − 09 |
| KRT7 | NM_005556 | 12.245 | 1.88E − 11 | 2.90E − 09 |
| G0S2 | NM_015714 | 12.032 | 3.22E − 13 | 1.28E − 10 |
| MEOX2 | NM_005924 | 11.972 | 3.62E − 11 | 4.74E − 09 |
| ITGBL1 | NM_004791 | 11.935 | 8.38E − 15 | 9.37E − 12 |
| RARB | NM_000965 | 11.857 | 4.53E − 11 | 5.68E − 09 |
| PODXL | NM_001018111 | 11.472 | 4.13E − 12 | 8.64E − 10 |
| LY6K | NM_017527 | 11.442 | 9.48E − 13 | 2.90E − 10 |
| TNFRSF1B | NM_001066 | 10.565 | 9.31E − 10 | 6.50E − 08 |
| SORBS2 | NM_021069 | 10.411 | 5.48E − 11 | 6.68E − 09 |
| NNAT | NM_005386 | 10.338 | 5.31E − 11 | 6.55E − 09 |
| CYYR1 | NM_052954 | 10.314 | 6.42E − 11 | 7.59E − 09 |
| MAOA | NM_000240 | 10.219 | 2.60E − 10 | 2.24E − 08 |
| PTGER4 | NM_000958 | 10.009 | 1.43E − 12 | 3.88E − 10 |
Expression of TLR4 Signaling Complex in HBVPs
First of all, we assessed the expression of TLR4 on the cell surface of HBVPs and MSCs by flow cytometry (Fig. 2). TLR4 was expressed at low but detectable levels in HBVPs, but accordingly to the microarray data, it was undetectable in MSCs. We could not detect changes in TLR4 mRNA levels after LPS stimulation of HBVPs; however, there was an increase in its cell surface expression. On the contrary, LPS treatment did not increase TLR4 protein expression in MSCs. MD-2 and CD14 are components of the LPS recognition complex (28), and MyD88 is an adaptor protein that plays an important role as a signaling transducer in the TLR4 receptor pathway. To assess whether HBVPs were endowed with TLR4 signaling molecules, we examined the expression levels of these three proteins by Western blotting. HBVPs expressed both MyD88 and MD-2, but CD14 was undetectable. Furthermore, LPS stimulation did not increase the expression of any of these proteins (Fig. 3).
FIGURE 2.
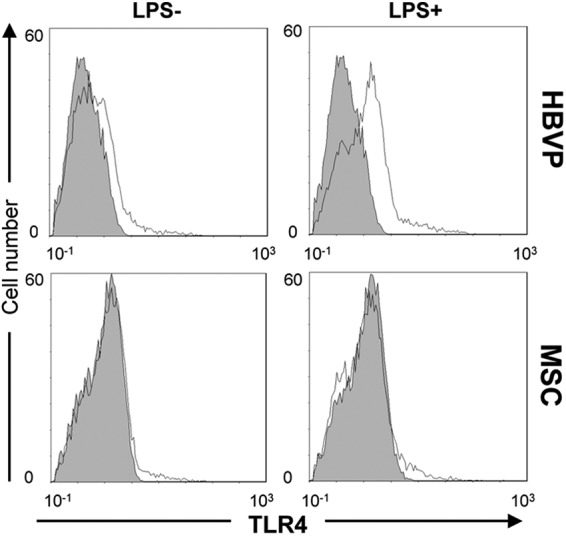
Cell surface expression of TLR4 detected by flow cytometry in HBVPs and MSCs stimulated or not with LPS. Filled histograms denote background fluorescence; line histograms denote TLR4 staining.
FIGURE 3.
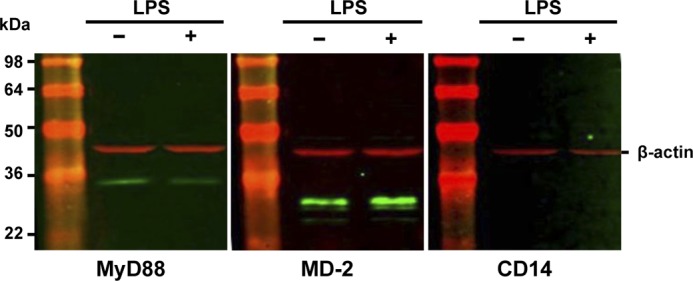
Expression of TLR4 signaling molecules in HBVPs. Immunoblot analysis of MyD88, MD-2, and CD14 is shown. β-Actin was used as a loading control.
Transcriptional Profile of LPS-stimulated HBVPs
To investigate the potential effect of LPS on pericyte gene expression, total RNA from unstimulated or stimulated (4 h with 1 μg/ml LPS) HBVPs was isolated and used for microarray analysis. Transcriptional profiling of LPS-treated HBVPs revealed that 76 annotated genes were differentially expressed >2-fold (p < 0.05), and 22 genes were up-regulated >5-fold (Table 2). Of them, 10 genes encoded chemokines and cytokines (CXCL10, CCL20, IL8, CXCL1, IL6, CCL2, IL1B, CXCL2, IL1A, and CXCL6), and three genes encoded adhesion molecules (ICAM1, VCAM1, and SELE). TNFAIP (TNF-α-induced protein)-2, -3, -5, and -6 were also included among the highly up-regulated genes as were PTGS2 (COX2) and SOD2. No annotated gene was significantly down-regulated. DAVID clustering analysis confirmed the enrichment in the biological process category of the terms “inflammatory response,” “defense response,” and “immune response” with FDR values of <5 × 10-14. Highly significant enrichments were also seen for the molecular function terms “cytokine activity” and “chemokine activity” (FDR < 5 × 10−12) (Table 3).
TABLE 2.
Genes up-regulated in HBVPs after LPS treatment (>5-fold). GEO accession number GSE46236
| Gene name | Accession no. | -Fold change | p value | False discovery rate |
|---|---|---|---|---|
| CXCL10 | NM_001565 | 59.39 | 3.66E − 04 | 4.82E − 01 |
| CCL20 | NM_004591 | 38.33 | 1.03E − 05 | 4.23E − 02 |
| IL8 | NM_000584 | 32.41 | 2.93E − 06 | 2.11E − 02 |
| CXCL1 | NM_001511 | 23.04 | 1.73E − 06 | 1.66E − 02 |
| IL6 | NM_000600 | 16.38 | 3.09E − 03 | 9.99E − 01 |
| CCL2 | NM_002982 | 15.52 | 2.47E − 08 | 7.11E − 04 |
| TNFAIP3 | NM_006290 | 15.08 | 5.19E − 05 | 1.38E − 01 |
| ICAM1 | NM_000201 | 14.18 | 6.46E − 05 | 1.43E − 01 |
| IL1B | NM_000576 | 12.66 | 4.10E − 05 | 1.31E − 01 |
| VCAM1 | NM_001078 | 11.71 | 4.77E − 06 | 2.75E − 02 |
| PTGS2 | NM_000963 | 10.99 | 1.02E − 03 | 8.85E − 01 |
| CXCL2 | NM_002089 | 8.53 | 7.61E − 06 | 3.65E − 02 |
| TNFAIP2 | NM_006291 | 8.52 | 1.27E − 03 | 9.99E − 01 |
| TNFAIP5 | NM_002852 | 6.56 | 1.09E − 04 | 2.08E − 01 |
| TNFSF18 | NM_005092 | 6.54 | 1.73E − 06 | 1.66E − 02 |
| TNFAIP6 | NM_007115 | 5.93 | 3.69E − 04 | 4.82E − 01 |
| SELE | NM_000450 | 5.83 | 4.36E − 02 | 9.99E − 01 |
| IL1A | NM_000575 | 5.64 | 2.37E − 02 | 9.99E − 01 |
| MX1 | NM_002462 | 5.36 | 9.46E − 02 | 9.99E − 01 |
| LIF | NM_002309 | 5.19 | 2.39E − 03 | 9.99E − 01 |
| SOD2 | NM_001024465 | 5.15 | 1.02E − 04 | 2.08E − 01 |
| CLDN1 | NM_021101 | 5.06 | 5.39E − 03 | 9.99E − 01 |
| CXCL6 | NM_002993 | 5.02 | 1.42E − 03 | 9.99E − 01 |
TABLE 3.
DAVID enriched GO terms of genes upregulated in LPS-treated HVBPs
MF, molecular function; BP, biological process; CC, cellular compartment.
| Category/term | Count/list total | Percentage | p value | Pop hits/total | -Fold enrichment | Bonferroni | Benjamini | FDR |
|---|---|---|---|---|---|---|---|---|
| GOTERM_BP_FAT | ||||||||
| GO:0006954, inflammatory response | 22/65 | 30.99 | 7.98E − 19 | 325/13,528 | 14.09 | 9.93E − 16 | 9.93E − 16 | 1.30E − 15 |
| GO:0006952, defense response | 27/65 | 38.03 | 8.29E − 19 | 615/13,528 | 9.14 | 1.03E − 15 | 5.16E − 16 | 1.35E − 15 |
| GO:0006955, immune response | 27/65 | 38.03 | 1.40E − 17 | 690/13,528 | 8.14 | 1.75E − 14 | 5.82E − 15 | 2.28E − 14 |
| GO:0009611, response to wounding | 24/65 | 33.80 | 8.35E − 17 | 530/13,528 | 9.42 | 1.38E − 13 | 3.45E − 14 | 1.78E − 13 |
| GO:0050900, leukocyte migration | 10/65 | 14.08 | 4.98E − 12 | 57/13,528 | 36.51 | 6.20E − 09 | 1.24E − 09 | 8.09E − 09 |
| GO:0042330, taxis | 13/65 | 18.31 | 9.55E − 12 | 160/13,528 | 16.91 | 1.19E − 08 | 1.98E − 09 | 1.55E − 08 |
| GO:0006935, chemotaxis | 13/65 | 18.31 | 9.55E − 12 | 160/13,528 | 16.91 | 1.19E − 08 | 1.98E − 09 | 1.55E − 08 |
| GO:0006916, antiapoptosis | 14/65 | 19.72 | 1.08E − 11 | 206/13,528 | 14.14 | 1.34E − 08 | 1.91E − 09 | 1.75E − 08 |
| GO:0043066, negative regulation of apoptosis | 16/65 | 22.54 | 6.85E − 11 | 354/13,528 | 9.41 | 8.53E − 08 | 1.07E − 08 | 1.11E − 07 |
| GO:0043069, negative regulation of programmed cell death | 16/65 | 22.54 | 8.35E − 11 | 359/13,528 | 9.28 | 1.04E − 07 | 1.16E − 08 | 1.36E − 07 |
| GO:0060548, negative regulation of cell death | 16/65 | 22.54 | 8.68E − 11 | 360/13,528 | 9.25 | 1.08E − 07 | 1.08E − 08 | 1.41E − 07 |
| GO:0007626, locomotory behavior | 14/65 | 19.72 | 3.80E − 10 | 274/13,528 | 10.63 | 4.73E − 07 | 4.30E − 08 | 6.17E − 07 |
| GO:0042981, regulation of apoptosis | 21/65 | 29.58 | 3.90E − 10 | 804/13,528 | 5.44 | 4.85E − 07 | 4.05E − 08 | 6.33E − 07 |
| GO:0043067, regulation of programmed cell death | 21/65 | 29.58 | 4.64E − 10 | 812/13,528 | 5.38 | 5.78E − 07 | 4.44E − 08 | 7.54E − 07 |
| GO:0010941, regulation of cell death | 21/65 | 29.58 | 4.95E − 10 | 815/13,528 | 5.36 | 6.17E − 07 | 4.40E − 08 | 8.04E − 07 |
| GO:0006915, apoptosis | 18/65 | 25.35 | 1.65E − 09 | 602/13,528 | 6.22 | 2.05E − 06 | 1.37E − 07 | 2.67E − 06 |
| GO:0012501, programmed cell death | 18/65 | 25.35 | 2.06E − 09 | 611/13,528 | 6.13 | 2.57E − 06 | 1.60E − 07 | 3.35E − 06 |
| GO:0008219, cell death | 19/65 | 26.76 | 3.33E − 09 | 719/13,528 | 5.50 | 4.14E − 06 | 2.44E − 07 | 5.40E − 06 |
| GO:0016265, death | 19/65 | 26.76 | 3.71E − 09 | 724/13,528 | 5.46 | 4.62E − 06 | 2.57E − 07 | 6.03E − 06 |
| GO:0002237, response to molecule of bacterial origin | 9/65 | 12.68 | 6.34E − 09 | 86/13,528 | 21.78 | 7.90E − 06 | 4.16E − 07 | 1.03E − 05 |
| GO:0030595, leukocyte chemotaxis | 7/65 | 9.86 | 1.83E − 08 | 37/13,528 | 39.37 | 2.28E − 05 | 1.14E − 06 | 2.97E − 05 |
| GO:0060326, cell chemotaxis | 7/65 | 9.86 | 2.55E − 08 | 39/13,528 | 37.36 | 3.17E − 05 | 1.51E − 06 | 4.14E − 05 |
| GO:0007610, behavior | 15/65 | 21.13 | 2.92E − 08 | 469/13,528 | 6.66 | 3.63E − 05 | 1.65E − 06 | 4.74E − 05 |
| GO:0031349, positive regulation of defense response | 8/65 | 11.27 | 4.83E − 08 | 73/13,528 | 22.81 | 6.01E − 05 | 2.61E − 06 | 7.84E − 05 |
| GO:0032496, response to lipopolysaccharide | 8/65 | 11.27 | 7.02E − 08 | 77/13,528 | 21.62 | 8.74E − 05 | 3.64E − 06 | 1.14E − 04 |
| GO:0042127, regulation of cell proliferation | 18/65 | 25.35 | 8.77E − 08 | 787/13,528 | 4.76 | 1.09E − 04 | 4.37E − 06 | 1.42E − 04 |
| GO:0048584, positive regulation of response to stimulus | 11/65 | 15.49 | 1.43E − 07 | 236/13,528 | 9.70 | 1.78E − 04 | 6.86E − 06 | 2.33E − 04 |
| GO:0002684, positive regulation of immune system process | 11/65 | 15.49 | 1.55E − 07 | 238/13,528 | 9.62 | 1.93E − 04 | 7.15E − 06 | 2.52E − 04 |
| GO:0009617, response to bacterium | 10/65 | 14.08 | 2.84E − 07 | 193/13,528 | 10.78 | 3.54E − 04 | 1.26E − 05 | 4.62E − 04 |
| GO:0019221, cytokine-mediated signaling pathway | 7/65 | 9.86 | 9.13E − 07 | 70/13,528 | 20.81 | 1.14E − 03 | 3.92E − 05 | 1.48E − 03 |
| GO:0034097, response to cytokine stimulus | 7/65 | 9.86 | 1.87E − 06 | 79/13,528 | 18.44 | 2.33E − 03 | 7.77E − 05 | 3.04E − 03 |
| GO:0016477, cell migration | 10/65 | 14.08 | 5.55E − 06 | 276/13,528 | 7.54 | 6.88E − 03 | 2.23E − 04 | 9.01E − 03 |
| GO:0006928, cell motion | 12/65 | 16.90 | 1.23E − 05 | 475/13,528 | 5.26 | 1.52E − 02 | 4.79E − 04 | 2.00E − 02 |
| GO:0048870, cell motility | 10/65 | 14.08 | 1.31E − 05 | 307/13,528 | 6.78 | 1.61E − 02 | 4.93E − 04 | 2.12E − 02 |
| GO:0051674, localization of cell | 10/65 | 14.08 | 1.31E − 05 | 307/13,528 | 6.78 | 1.61E − 02 | 4.93E − 04 | 2.12E − 02 |
| GO:0008284, positive regulation of cell proliferation | 11/65 | 15.49 | 2.23E − 05 | 414/13,528 | 5.53 | 2.74E − 02 | 8.18E − 04 | 3.63E − 02 |
| GO:0050727, regulation of inflammatory response | 6/65 | 8.45 | 2.88E − 05 | 76/13,528 | 16.43 | 3.53E − 02 | 0.0010253 | 4.68E − 02 |
| GOTERM_MF_FAT | ||||||||
| GO:0005125, cytokine activity | 17/57 | 23.94 | 8.58E − 17 | 195/12,983 | 19.86 | 1.92E − 14 | 1.92E − 14 | 1.33E − 13 |
| GO:0008009, chemokine activity | 11/57 | 15.49 | 3.42E − 15 | 46/12,983 | 54.47 | 5.95E − 13 | 2.98E − 13 | 4.21E − 12 |
| GO:0042379, chemokine receptor binding | 11/57 | 15.49 | 6.93E − 15 | 49/12,983 | 51.13 | 1.19E − 12 | 3.97E − 13 | 8.43E − 12 |
| GOTERM_CC_FAT | ||||||||
| GO:0005615, extracellular space | 23/53 | 32.39 | 4.66E − 15 | 685/12,782 | 8.10 | 4.34E − 13 | 4.34E − 13 | 5.11E − 12 |
| GO:0044421, extracellular region part | 23/53 | 32.39 | 4.47E − 12 | 960/12,782 | 5.78 | 4.16E − 10 | 2.08E − 10 | 4.90E − 09 |
| GO:0005576, extracellular region | 27/53 | 38.03 | 8.32E − 09 | 2,010/12,782 | 3.24 | 7.74E − 07 | 2.58E − 07 | 9.11E − 06 |
Validation of Microarray Results
To verify the gene expression profile determined by microarray analysis, the expression levels of IL6, IL8, CXCL1, CXCL2, and CXCL3 were analyzed by quantitative RT-PCR (qRT-PCR) using total RNA obtained from untreated or LPS-treated HBVPs (Fig. 4A). SHDA gene was used for data normalization, and primer specificity was assessed by sequencing. The direction of gene expression changes was in agreement by qRT-PCR and microarray; a result was considered valid if the -fold changes were greater than or equal to 2 (29). This criterion was fulfilled by all genes. For the majority of the genes, -fold differences were also of comparable magnitude with the exception of CXCL3.
FIGURE 4.
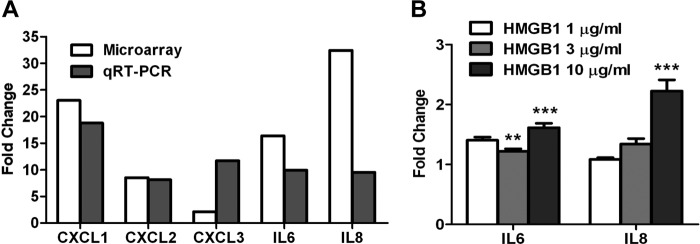
qRT-PCR analysis of genes selected from the microarray profile in LPS-stimulated HBVPs and effect of HMGB1 on gene expression. A, expression levels of five selected genes were evaluated by qRT-PCR to validate microarray data in HBVPs treated with 1 μg/ml LPS for 4 h. B, analysis of IL6 and IL8 levels by qRT-PCR in HBVPs stimulated with different concentrations of HMGB1 for 4 h.***, p < 0.001. Error bars represent S.D.
HMGB1 Also Induces Changes in HBVP Gene Expression
TLR4 is also one of the cognate receptors for HMGB1 (30). To study the potential role of HMGB1 as a proinflammatory mediator in pericytes, HBVPs were incubated for 4 h with different concentrations of recombinant HMGB1, and the expression levels of IL6 and IL8 were analyzed by qRT-PCR as in previous experiments. HMGB1 exposure of HBVPs led to statistically significant, dose-dependant up-regulation of both transcripts (p < 0.001) (Fig. 4B). However, -fold changes were consistently lower when compared with those following LPS stimulation.
LPS and HMGB1 Stimulate Cytokine and Chemokine Secretion by HBVPs but Not by MSCs
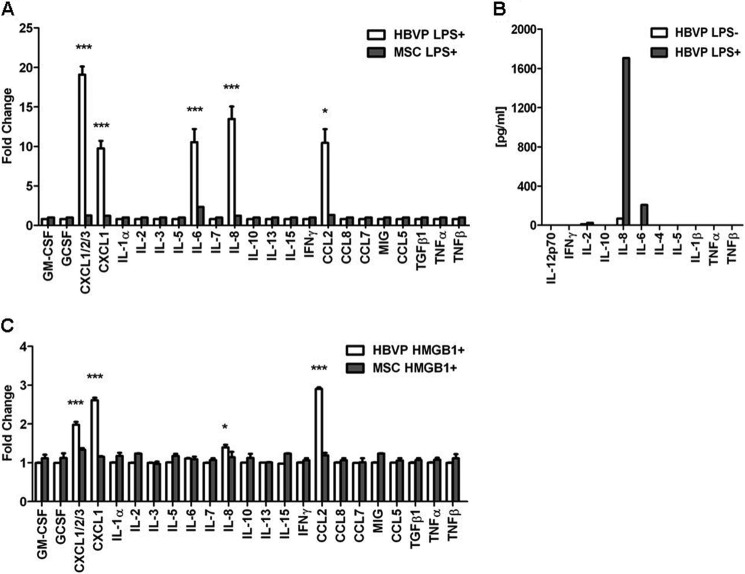
To assess whether modulation of gene expression by LPS had a correlation at the protein level, we studied the secretion of 23 cytokines and chemokines by LPS-treated HBVPs (Fig. 5A). HBVPs spontaneously released low levels of CXCL2, CXCL3, IL-8, and CCL2. Stimulation of HBVPs with LPS resulted in significant secretion of the proinflammatory cytokine IL-6 and increased production of IL-8, CXCL1, CXCL2, CXCL3, and CCL2 in comparison with unstimulated pericytes. In fact, we could demonstrate increased secretion of the corresponding protein for all the cytokine/chemokine genes represented in the antibody array whose expressions were up-regulated >5-fold in the microarray analysis except for IL-1α. Given that TLR4 expression in MSCs was undetectable on the cell surface, their capacity to respond to LPS should be expected to be limited. In fact, LPS treatment of MSCs did not significantly promote the secretion of any of the cytokines/chemokines included in the array, and only a slight increase in IL-6 could be detected. HBVP cytokine secretion data were confirmed using a quantitative flow cytometry, bead-based approach (Fig. 5B). Increased levels of IL-8 were produced by LPS-treated HBVPs (1706 pg/ml) compared with control cells (68.51 pg/ml). There was no detectable constitutive secretion of IL-6 by HBVPs, but stimulated pericytes secreted 206.42 pg/ml. No secretion of IL-1α, TNF-α, or TNF-β was detected by this method. On the other hand, HMGB1 stimulation of HBVPs led to secretion of CXCL1, CXCL2, CXCL3, IL-8, and CCL2 (Fig. 5C). As observed with RNA, protein levels in the conditioned medium were lower when compared with those following LPS stimulation. Treatment of MSCs with HMGB1 did not induce the release of any of the cytokines/chemokines tested.
FIGURE 5.
Secretion of cytokines and chemokines by HBVPs and MSCs after LPS and HMGB1 stimulation. A, semiquantitative detection of cytokines and chemokines in the conditioned media of HBVPs and MSCs treated with 1 μg/ml LPS for 4 h. B, cytokine quantification in the conditioned medium of LPS-stimulated HBVPs. C, semiquantitative detection of cytokines and chemokines in the conditioned media of HBVPs and MSCs treated with 10 μg/ml HMGB1 for 4 h. Data are representative of three independent experiments (mean ± S.D. of triplicates). *, p < 0.05; ***, p < 0.001. Error bars represent S.D.
NF-κB Signaling Pathway, but Not p38 MAPK, Is Activated by LPS in HBVPs
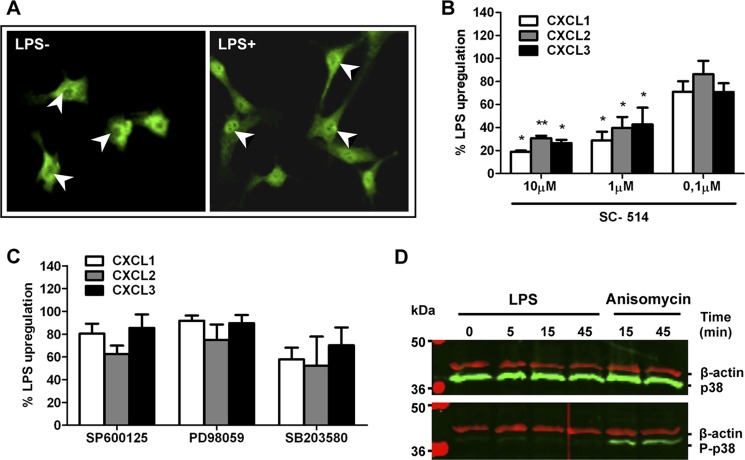
It has long been known that TLR4 can induce the activation of NF-κB and the expression of proinflammatory cytokines (31), but the functionality of this pathway in HBVPs has not been previously addressed. As NF-κB activation implies its translocation to the nucleus, we studied the subcellular location of NF-κB by immunofluorescent staining in unstimulated and LPS-stimulated HBVPs. As expected, in unstimulated control pericytes, NF-κB is localized mainly in the cytoplasm with only minor nuclear staining detected. Stimulation of HBVPs with LPS induced NF-κB translocation from the cytoplasm to the nucleus, which is highly indicative of the NF-κB activation (Fig. 6A). To test whether the NF-κB pathway is important for the production of proinflammatory mediators by pericytes, HBVPs were pretreated for 30 min with increasing concentrations of the inhibitor SC-514 and then stimulated with LPS. Treatment of HBVPs with SC-514 resulted in a concentration-dependent inhibition of LPS-stimulated CXCL1, CXCL2, and CXCL3 gene expression as assessed by qRT-PCR (Fig. 6B). At the highest concentration of SC-514 used (10 μm), the increase of chemokine mRNA levels after stimulation with LPS was significantly inhibited. LPS-induced TLR4 signaling has also been shown to activate a number of kinases, including MAPK, in different cell types (32). To further characterize the signal transduction pathway, we tested several MAPK inhibitors for their capacity to reduce gene up-regulation by pericytes. Preincubation of HBVPs with 10 μm SB203580 (p38 MAPK inhibitor), 10 μm SP600125 (JNK1 and -2 inhibitor), or 10 μm PD98059 (MEK-1 inhibitor) did not significantly inhibit up-regulation of CXCL1, CXCL2, and CXCL3 (Fig. 6C), although a trend toward inhibition of response could be observed with SB203580.To further clarify the role of p38 MAPK in LPS-mediated activation of pericytes, lysates of LPS-treated HBVPs were subjected to Western blotting to assess p38 MAPK phosphorylation status. As a positive control for p38 MAPK activation, HBVPs were treated with anisomycin. As shown in Fig. 6D, LPS treatment of HBVPs at different time points did not induce phosphorylation of p38 MAPK or any significant change in total p38 MAPK content.
FIGURE 6.
LPS mediates activation of the NF-κB signaling pathway in HBVPs but not phosphorylation of p38 MAPK. A, NF-κB p65 is detected in the cytoplasm of unstimulated cells (arrowheads show non-stained nuclei). Nuclear translocation of NF-κB p65 takes place after LPS stimulation for 90 min (arrowheads indicate stained nuclei). Original magnification, ×400. B, suppression of LPS-promoted CXCL1, -2, and -3 gene expression by an NF-κB inhibitor. HBVPs were incubated with different doses of the NF-κB inhibitor SC-514 for 30 min prior to LPS stimulation for 90 min. RNA was isolated and analyzed by qRT-PCR. *, p < 0.05; **, p < 0.005. C, LPS-promoted CXCL1, -2, and -3 up-regulation is not disrupted in HBVPs treated with different mitogen-activated protein kinase inhibitors, SP600125 (JNK1 and -2 inhibitor), PD98059 (MEK-1 inhibitor), and SB203580 (p38 inhibitor), prior to LPS stimulation. D, LPS treatment does not alter the expression level or phosphorylation status of p38 MAPK as assessed by Western blot at different time points. As a positive control, cells were treated with 10 μm anisomycin to induce p38 MAPK activation. Results shown in A and D represent those obtained in at least three independent experiments. Error bars represent S.D.
ICAM-1 and VCAM-1 Are Up-regulated in LPS-treated HBVPs
In ECs, regulation of the vascular proadhesive phenotype is tightly controlled by NF-κB. In HBVPs, we demonstrated the increased gene expression of adhesion molecules (SELE, ICAM1, and VCAM1) in response to stimulation with LPS (Table 1). To assess whether LPS treatment also resulted in up-regulation of their cell surface expression, HBVPs were treated with LPS for different times (4, 8, and 24 h) and analyzed by flow cytometry with antibodies against ICAM-1 and VCAM-1 (Fig. 7A). Unstimulated HBVPs exhibited basal expression of ICAM-1 and low levels of VCAM-1. Remarkably, HBVPs cultured with LPS showed a strong increase in the expression of both adhesion molecules that was already evident after 4 h of stimulation and peaked at 24 h when cell surface expression level and mean fluorescence intensity were similar for both ICAM-1 and VCAM-1. VCAM-1 was neither expressed nor induced in LPS-treated MSCs. MSCs exhibited low basal ICAM-1 expression, which only increased slightly after incubation with LPS for 24 h, with a profile similar to that of unstimulated HBVPs. Mean fluorescence intensities for ICAM-1 and VCAM-1 are depicted in Fig. 7, B and C, respectively.
FIGURE 7.
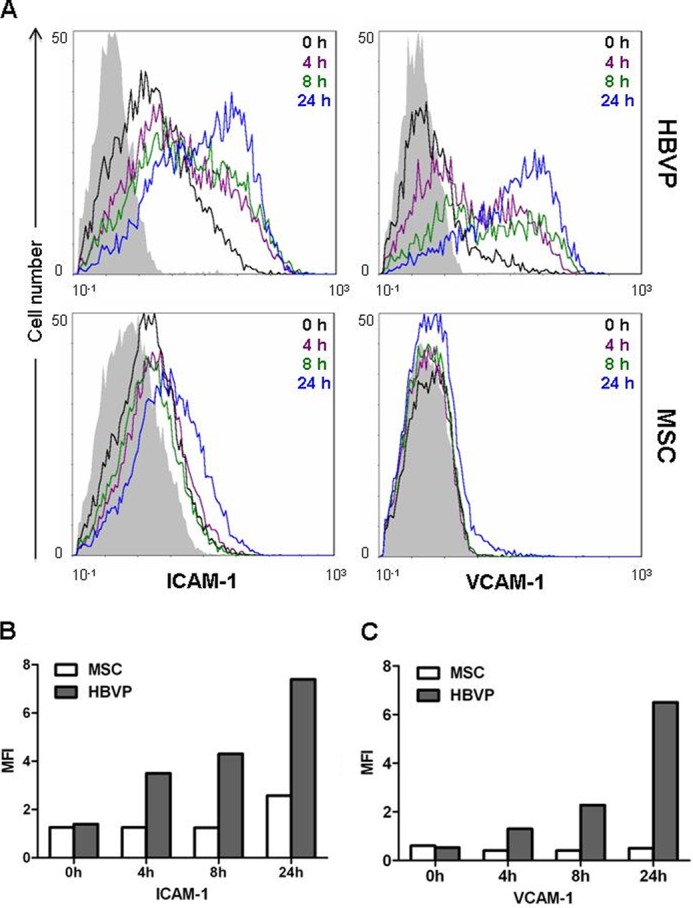
Expression of ICAM-1 and VCAM-1 on the surface of LPS-treated HBVPs and MSCs. A, ICAM-1 and VCAM-1 expression was analyzed by flow cytometry at different time points after LPS addition. Isotype-matched antibodies were used as control (in solid gray). B, bar graphs represent the mean fluorescence intensities (MFI) of each marker and time point. One representative result from three independent experiments is shown.
Peripheral Blood Leukocyte Adhesion to LPS-treated HBVPs
The biological relevance of ICAM-1 and VCAM-1 up-regulation was confirmed by the significantly increased adhesion of PBLs to LPS-treated HBVPs (24 h). PKH26-labeled PBLs were added on the top of confluent monolayers of untreated or LPS-treated HBVPs for 2 h followed by extensive washing (Fig. 8A). The number of adhered PKH26+ cells was quantified by direct visualization of four different fields of every independent experiment. Due to the basal expression of adhesion molecules by HBVPs, a greater number of PBLs adhered to control HBVPs than to plastic (57.3 ± 3.8 and 44.7 ± 6.6, respectively; p = 0.045) (Fig. 8B). However, preincubation of HBVPs with LPS significantly enhanced PBL adhesion (118 ± 19.7 cells) compared with non-treated HBVPs (p = 0.005).
FIGURE 8.
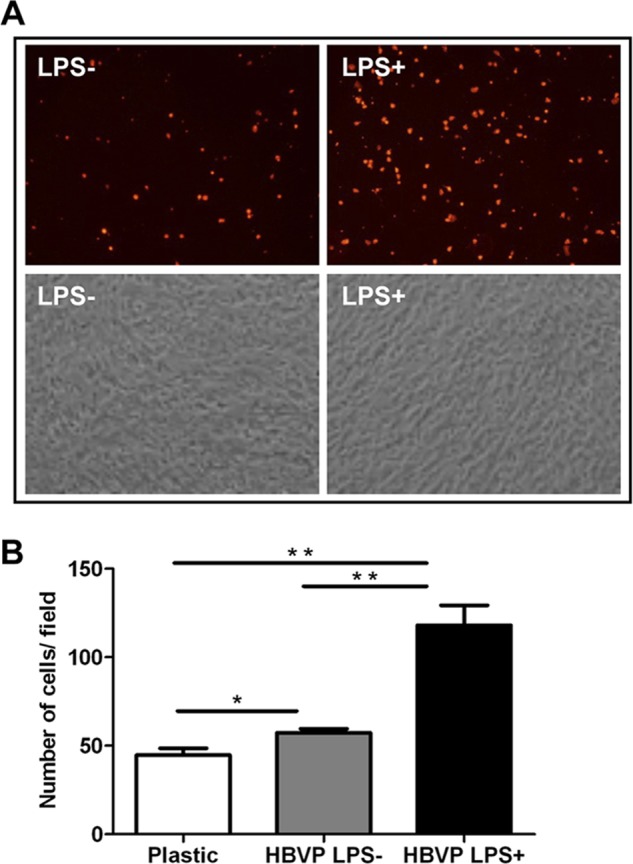
PBL adhesion to HBVPs treated with LPS. A, fluorescently labeled PBLs were added onto a confluent monolayer of LPS-treated HBVPs and washed after 2 h. Original magnification, ×200. B, quantification of adhered cells. Values correspond to the arithmetic mean ± S.D. of three different fields belonging to a representative experiment. *, p < 0.05; **, p < 0.005. Error bars represent S.D.
DISCUSSION
LPS from Gram-negative bacteria constitutes the best known pathogen-associated molecular pattern and is “sensed” by TLR4. It is well known that ECs express TLR4, and their activation by LPS exerts a critical role in the pathophysiology of sepsis and in mounting an innate response to control infection (11, 12). But blood vessels are not only constituted by ECs: mural cells envelop the endothelium in mature, stable vessels (33). However, their potential implication in inflammatory and innate immune responses has been scarcely addressed. The pericyte gene expression profile revealed a potential role for TLR4 in this context, and therefore, our next goals were to assess the expression of the protein on the HBVP cell surface and study their capacity to respond to LPS (pathogen-associated molecular pattern) and HMGB1 (damage-associated molecular pattern). TLR4 cell surface expression on HBVPs was apparently lower than expected in relation with transcript levels (similar to those of SDHA), and notwithstanding, it is enough for mounting a robust LPS response. Intracellular compartmentalization of TLR4 has been reported in human epithelial cells, which could nevertheless respond to LPS stimulation (34). We observed that LPS-treated HBVPs had increased surface expression of TLR4 in the absence of TLR4 mRNA up-regulation, suggesting that LPS may be able to induce TLR4 relocalization (35). It has been long known that the mechanism of TLR4 activation in monocytes and macrophages involves the auxiliary proteins MD-2 and CD14 (28). However, much concerning the function of TLR4 in non-immune cells remains to be studied. We have demonstrated the constitutive expression of MD-2 in HBVPs along with the lack of CD14. CD14 may be found in two distinct forms: as a glycosylphosphatidylinositol-linked membrane receptor on myeloid cells and as a soluble protein (sCD14). ECs do not express CD14 either, and it has been proposed that they require sCD14 found circulating in the plasma to respond to LPS (36). However, we treated HBVPs with LPS in the absence of serum in the culture medium, and therefore, the response to LPS that we observed could be considered CD14-independent. The proinflammatory effect of HMGB1 has been described in several types of cells, including monocytes (37), neutrophils (38), dendritic cells (39), ECs (40), and fibroblasts (41). Here, we have also demonstrated up-regulation of different chemokines and cytokines at both transcript and protein levels in HBVPs after exposure to HMGB1 albeit at lower levels than those observed with LPS as HMGB1 constitutes a comparatively weak TLR4 activator (40). In fact, it has been proposed that HMGB1 only exhibits full proinflammatory activity when complexed to LPS (42) because a combination of HMGB1 and LPS results in a higher increase in TNF-α production in human peripheral blood monocytes than LPS or HMGB1 treatment alone (43). Alternatively, HMGB1 signaling in HBVPs may be more dependent on CD14 expression than LPS signaling as optimal HMGB1-dependent TLR4 activation seems to require this coreceptor at least in murine macrophages (44). At the protein level, we have corroborated the secretion of CXCL1, CXCL2, CXCL3, CCL2, IL-6, and IL-8 by LPS and HMGB1-stimulated pericytes. Interestingly, when compared with previous results using LPS-treated mouse brain pericytes (16), the profile of cytokines and chemokines released by HBVPs is considerably different. Stimulation of mouse brain pericytes with LPS at the same concentration used in this study resulted in significant release of IL-1α, TNF-α, IL-3, IL-10, IL-12 (p70), IL-13, G-CSF, GM-CSF, and CCL5, which were undetectable in the conditioned medium of stimulated HBVPs. CXCL1 and CCL2 were produced by LPS-treated pericytes from both species, and there was no detectable production of IL-1α, IL-2, IL-4, and IL-5 by human or mouse brain pericytes. These data support major species differences in vascular cell immunological functions as highlighted by Pober and Tellides (45). On the other hand, the pattern of proteins released by LPS-treated HBVPs closely resembles that of LPS-stimulated ECs. LPS induced the secretion of IL-6, IL-8, and CCL2 but not of IL-1α, TNF-α, or IL-10 in human coronary artery ECs (9). Intriguingly, in human coronary artery ECs, LPS induces mRNA expression of IL1A, IL1B, and TNFA but no release of the proteins. Similarly, in HBVPs, we observed significant up-regulation of IL1A and IL1B mRNA (5.6-fold, p < 0.05 and 12.7-fold, p < 0.0001, respectively), but expression at the protein level could not be detected. CXCL1, CXCL2, CXCL3, and IL-8 belong to the CXC chemokine subgroup with glutamic acid-leucine-arginine (the “ELR” motif) immediately proximal to the CXC sequence. The specific receptors for these chemokines, CXCR1 and CXCR2, are expressed by ECs (46, 47). In fact, it is well known that ELR-positive CXC chemokines promote the migration and proliferation of ECs and are potent promoters of angiogenesis (48). In addition to the ELR+ CXC chemokine family, CCL2 is the best known CC chemokine mediator of neovascularization. This pattern of chemokine production described here may shed new light on the endothelial-pericyte interactions, suggesting a dual role for pericytes according to their activation status. In physiological conditions, pericytes are necessary to maintain a quiescent stable endothelium (49); however, activated pericytes could act as drivers of angiogenesis during inflammatory processes. When exposed to inflammatory mediators, the quiescent endothelium becomes activated and expresses additional proinflammatory factors and adhesion molecules that enable the extravasation of circulating leukocytes to sites of inflammation (12). Similarly, the chemokines secreted by HBVPs can potentially mediate leukocyte recruitment during inflammatory conditions as reported for ECs. CXCR1 and CXCR2 are expressed mainly by neutrophils but also by monocytes, T cell subsets, natural killer cells, basophils, and mast cells. In fact, IL-8 is a powerful neutrophil chemotactic factor. CCR2 is expressed mainly by monocytes but also by B cells, activated T cells, and dendritic cells. All these cell types are susceptible to be lured by activated pericytes. In melanoma, for example, CCL2 secreted by pericytes surrounding peritumoral vessels has been implied in inflammatory leukocyte recruitment (50). Monocytes and macrophages recruited by these chemokines could secrete additional soluble factors and release HMGB1 that could further stimulate pericytes in a positive feedback loop. But LPS-treated pericytes not only produce chemokines that could attract immune cells, they also overexpress adhesion molecules such as ICAM-1 and VCAM-1. Recent works have demonstrated that mouse pericytes facilitate neutrophil transmigration through postcapillary venular walls in an ICAM-1-dependent manner using a model of TNF-α- or IL-1β-stimulated mouse muscle in vivo (51, 52). In fact, capillary and arteriolar NG2+ pericytes can “instruct” extravasating neutrophils and monocytes with migratory cues (53). Moreover, pericytes can also interact with T cells and modulate adaptative immune responses. In fact, VCAM-1 up-regulation by TNF-α was proposed long ago to mediate interactions between T cells and human brain pericytes (17). More recently, human placental pericytes were shown to negatively regulate allogenic T cell responses (18), and human retinal pericytes were demonstrated to inhibit T cell proliferation (54). In a different context, tumor-derived pericytes have been shown to negatively influence CD4+ T cell activation and proliferation (55). Undoubtedly, pericytes share many characteristics with MSCs, which are known to display strong immunosuppressive functions (56), and the above cited studies constitute a good example. In fact, we have used MSCs as a source of mural cells to develop human functional blood vessels in immunodeficient mice (27), blurring the boundaries between pericytes and MSCs. However, the immunoregulatory effect of pericytes and MSCs might be differentially steered at least regarding inflammation. MSCs have been shown to be effective at improving survival of mice in induced sepsis models or after systemic LPS administration (57–59). On the contrary, we have demonstrated that pericytes can sense LPS through TLR4, overexpressing an array of soluble factors and membrane molecules that can fuel inflammation. To make the picture more complex, at least some MSCs seem to express TLR4 (60). The effect of TLR4 activation on the immunosuppressive properties of these cells is a controversial issue. In summary, LPS and HMGB1 activation of HBVPs may trigger a complex proinflammatory and proangiogenic program in the context of infection and sterile inflammation, respectively. Therefore, pericytes should be considered as active members in the inflammatory response beyond their homeostatic role. Modulating the activity of pericytes may provide a novel means of treating acute inflammatory conditions, especially in those tissues where they are especially abundant such as the central nervous system.
Acknowledgments
We thank Marco Bianchi, Maura Casalgrandi, and Ricardo Sánchez-Prieto for providing technical advice and reagents as well as for critical reading of the manuscript.
This work was supported by Fondo de Investigaciones Sanitarias/Instituto de Salud Carlos III Grants PI08/90856 and PS09/00227 and Fundación Investigación Biomédica Hospital Puerta de Hierro (to L. S.) and by Ministerio de Economía y Competitividad Grant BIO2011-22738 and Comunidad de Madrid Grant S2010/BMD-2312 (to L. A.-V.).
- EC
- endothelial cell
- TLR
- Toll-like receptor
- HMGB1
- high mobility group box 1
- MSC
- mesenchymal stem cell
- HBVP
- human brain vascular pericyte
- PDGFRB
- PDGF receptor β
- FDR
- false discovery rate
- GO
- gene ontology
- PBL
- peripheral blood leukocyte
- qRT-PCR
- quantitative real time-PCR.
REFERENCES
- 1. Armulik A., Genové G., Betsholtz C. (2011) Pericytes: developmental, physiological, and pathological perspectives, problems, and promises. Dev. Cell 21, 193–215 [DOI] [PubMed] [Google Scholar]
- 2. Díaz-Flores L., Gutiérrez R., Madrid J. F., Varela H., Valladares F., Acosta E., Martín-Vasallo P., Díaz-Flores L., Jr (2009) Pericytes. Morphofunction, interactions and pathology in a quiescent and activated mesenchymal cell niche. Histol. Histopathol. 24, 909–969 [DOI] [PubMed] [Google Scholar]
- 3. Corselli M., Chen C.-W., Crisan M., Lazzari L., Péault B. (2010) Perivascular ancestors of adult multipotent stem cells. Arterioscler. Thromb. Vasc. Biol. 30, 1104–1109 [DOI] [PubMed] [Google Scholar]
- 4. Crisan M., Yap S., Casteilla L., Chen C.-W., Corselli M., Park T. S., Andriolo G., Sun B., Zheng B., Zhang L., Norotte C., Teng P.-N., Traas J., Schugar R., Deasy B. M., Badylak S., Buhring H.-J., Giacobino J.-P., Lazzari L., Huard J., Péault B. (2008) A perivascular origin for mesenchymal stem cells in multiple human organs. Cell Stem Cell 3, 301–313 [DOI] [PubMed] [Google Scholar]
- 5. Dellavalle A., Maroli G., Covarello D., Azzoni E., Innocenzi A., Perani L., Antonini S., Sambasivan R., Brunelli S., Tajbakhsh S., Cossu G. (2011) Pericytes resident in postnatal skeletal muscle differentiate into muscle fibres and generate satellite cells. Nat. Commun. 2, 499. [DOI] [PubMed] [Google Scholar]
- 6. Paquet-Fifield S., Schlüter H., Li A., Aitken T., Gangatirkar P., Blashki D., Koelmeyer R., Pouliot N., Palatsides M., Ellis S., Brouard N., Zannettino A., Saunders N., Thompson N., Li J., Kaur P. (2009) A role for pericytes as microenvironmental regulators of human skin tissue regeneration. J. Clin. Investig. 119, 2795–2806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bexell D., Gunnarsson S., Tormin A., Darabi A., Gisselsson D., Roybon L., Scheding S., Bengzon J. (2009) Bone marrow multipotent mesenchymal stroma cells act as pericyte-like migratory vehicles in experimental gliomas. Mol. Ther. 17, 183–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. da Silva Meirelles L., Caplan A. I., Nardi N. B. (2008) In search of the in vivo identity of mesenchymal stem cells. Stem Cells 26, 2287–2299 [DOI] [PubMed] [Google Scholar]
- 9. Zeuke S., Ulmer A. J., Kusumoto S., Katus H. A., Heine H. (2002) TLR4-mediated inflammatory activation of human coronary artery endothelial cells by LPS. Cardiovasc. Res. 56, 126–134 [DOI] [PubMed] [Google Scholar]
- 10. Andonegui G., Bonder C. S., Green F., Mullaly S. C., Zbytnuik L., Raharjo E., Kubes P. (2003) Endothelium-derived Toll-like receptor-4 is the key molecule in LPS-induced neutrophil sequestration into lungs. J. Clin. Investig. 111, 1011–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mai J., Virtue A., Shen J., Wang H., Yang X. F. (2013) An evolving new paradigm: endothelial cells—conditional innate immune cells. J. Hematol. Oncol. 6, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Danese S., Dejana E., Fiocchi C. (2007) Immune regulation by microvascular endothelial cells: directing innate and adaptive immunity, coagulation, and inflammation. J. Immunol. 178, 6017–6022 [DOI] [PubMed] [Google Scholar]
- 13. Edelman D. A., Jiang Y., Tyburski J., Wilson R. F., Steffes C. (2006) Toll-like receptor-4 message is up-regulated in lipopolysaccharide-exposed rat lung pericytes. J. Surg. Res. 134, 22–27 [DOI] [PubMed] [Google Scholar]
- 14. Edelman D. A., Jiang Y., Tyburski J. G., Wilson R. F., Steffes C. P. (2007) Lipopolysaccharide activation of pericyte's Toll-like receptor-4 regulates co-culture permeability. Am. J. Surg. 193, 730–735 [DOI] [PubMed] [Google Scholar]
- 15. Edelman D. A., Jiang Y., Tyburski J. G., Wilson R. F., Steffes C. P. (2007) Cytokine production in lipopolysaccharide-exposed rat lung pericytes. J. Trauma 62, 89–93 [DOI] [PubMed] [Google Scholar]
- 16. Kovac A., Erickson M. A., Banks W. A. (2011) Brain microvascular pericytes are immunoactive in culture: cytokine, chemokine, nitric oxide, and LRP-1 expression in response to lipopolysaccharide. J. Neuroinflammation 8, 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Verbeek M. M., Westphal J. R., Ruiter D. J., de Waal R. M. (1995) T lymphocyte adhesion to human brain pericytes is mediated via very late antigen-4/vascular cell adhesion molecule-1 interactions. J. Immunol. 154, 5876–5884 [PubMed] [Google Scholar]
- 18. Maier C. L., Pober J. S. (2011) Human placental pericytes poorly stimulate and actively regulate allogeneic CD4 T cell responses. Arterioscler. Thromb. Vasc. Biol. 31, 183–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Covas D. T., Siufi J. L., Silva A. R., Orellana M. D. (2003) Isolation and culture of umbilical vein mesenchymal stem cells. Braz. J. Med. Biol. Res. 36, 1179–1183 [DOI] [PubMed] [Google Scholar]
- 20. Venereau E., Casalgrandi M., Schiraldi M., Antoine D. J., Cattaneo A., De Marchis F., Liu J., Antonelli A., Preti A., Raeli L., Shams S. S., Yang H., Varani L., Andersson U., Tracey K. J., Bachi A., Uguccioni M., Bianchi M. E. (2012) Mutually exclusive redox forms of HMGB1 promote cell recruitment or proinflammatory cytokine release. J. Exp. Med. 209, 1519–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Irizarry R. A., Hobbs B., Collin F., Beazer-Barclay Y. D., Antonellis K. J., Scherf U., Speed T. P. (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4, 249–264 [DOI] [PubMed] [Google Scholar]
- 22. Smyth G. K., Michaud J., Scott H. S. (2005) Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics 21, 2067–2075 [DOI] [PubMed] [Google Scholar]
- 23. Edgar R., Domrachev M., Lash A. E. (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Starkie R. L., Arkinstall M. J., Koukoulas I., Hawley J. A., Febbraio M. A. (2001) Carbohydrate ingestion attenuates the increase in plasma interleukin-6, but not skeletal muscle interleukin-6 mRNA, during exercise in humans. J. Physiol. 533, 585–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Facciabene A., Peng X., Hagemann I. S., Balint K., Barchetti A., Wang L.-P., Gimotty P. A., Gilks C. B., Lal P., Zhang L., Coukos G. (2011) Tumour hypoxia promotes tolerance and angiogenesis via CCL28 and Treg cells. Nature 475, 226–230 [DOI] [PubMed] [Google Scholar]
- 26. Guijarro-Muñoz I., Cuesta A. M., Alvarez-Cienfuegos A., Geng J. G., Alvarez-Vallina L., Sanz L. (2012) The axonal repellent Slit2 inhibits pericyte migration: potential implications in angiogenesis. Exp. Cell Res. 318, 371–378 [DOI] [PubMed] [Google Scholar]
- 27. Sanz L., Santos-Valle P., Alonso-Camino V., Salas C., Serrano A., Vicario J. L., Cuesta A. M., Compte M., Sánchez-Martín D., Alvarez-Vallina L. (2008) Long-term in vivo imaging of human angiogenesis: Critical role of bone marrow-derived mesenchymal stem cells for the generation of durable blood vessels. Microvasc. Res. 75, 308–314 [DOI] [PubMed] [Google Scholar]
- 28. Park B. S., Song D. H., Kim H. M., Choi B.-S., Lee H., Lee J.-O. (2009) The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature 458, 1191–1195 [DOI] [PubMed] [Google Scholar]
- 29. Rajeevan M. S., Ranamukhaarachchi D. G., Vernon S. D., Unger E. R. (2001) Use of real-time quantitative PCR to validate the results of cDNA array and differential display PCR technologies. Methods 25, 443–451 [DOI] [PubMed] [Google Scholar]
- 30. Park J. S., Svetkauskaite D., He Q., Kim J.-Y., Strassheim D., Ishizaka A., Abraham E. (2004) Involvement of toll-like receptors 2 and 4 in cellular activation by high mobility group box 1 protein. J. Biol. Chem. 279, 7370–7377 [DOI] [PubMed] [Google Scholar]
- 31. Medzhitov R. (2007) Recognition of microorganisms and activation of the immune response. Nature 449, 819–826 [DOI] [PubMed] [Google Scholar]
- 32. Carter A. B., Monick M. M., Hunninghake G. W. (1999) Both Erk and p38 kinases are necessary for cytokine gene transcription. Am. J. Respir. Cell Mol. Biol. 20, 751–758 [DOI] [PubMed] [Google Scholar]
- 33. Jain R. K. (2003) Molecular regulation of vessel maturation. Nat. Med. 9, 685–693 [DOI] [PubMed] [Google Scholar]
- 34. Guillot L., Medjane S., Le-Barillec K., Balloy V., Danel C., Chignard M., Si-Tahar M. (2004) Response of human pulmonary epithelial cells to lipopolysaccharide involves Toll-like receptor 4 (TLR4)-dependent signaling pathways: evidence for an intracellular compartmentalization of TLR4. J. Biol. Chem. 279, 2712–2718 [DOI] [PubMed] [Google Scholar]
- 35. Perros F., Lambrecht B. N., Hammad H. (2011) TLR4 signalling in pulmonary stromal cells is critical for inflammation and immunity in the airways. Respir. Res. 12, 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pugin J., Schürer-Maly C. C., Leturcq D., Moriarty A., Ulevitch R. J., Tobias P. S. (1993) Lipopolysaccharide activation of human endothelial and epithelial cells is mediated by lipopolysaccharide-binding protein and soluble CD14. Proc. Natl. Acad. Sci. U.S.A. 90, 2744–2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Andersson U., Wang H., Palmblad K., Aveberger A. C., Bloom O., Erlandsson-Harris H., Janson A., Kokkola R., Zhang M., Yang H., Tracey K. J. (2000) High mobility group 1 protein (HMG-1) stimulates proinflammatory cytokine synthesis in human monocytes. J. Exp. Med. 192, 565–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Park J. S., Arcaroli J., Yum H.-K., Yang H., Wang H., Yang K.-Y., Choe K.-H., Strassheim D., Pitts T. M., Tracey K. J., Abraham E. (2003) Activation of gene expression in human neutrophils by high mobility group box 1 protein. Am. J. Physiol. Cell Physiol. 284, C870–C879 [DOI] [PubMed] [Google Scholar]
- 39. Messmer D., Yang H., Telusma G., Knoll F., Li J., Messmer B., Tracey K. J., Chiorazzi N. (2004) High mobility group box protein 1: an endogenous signal for dendritic cell maturation and Th1 polarization. J. Immunol. 173, 307–313 [DOI] [PubMed] [Google Scholar]
- 40. Treutiger C. J., Mullins G. E., Johansson A.-S., Rouhiainen A., Rauvala H. M., Erlandsson-Harris H., Andersson U., Yang H., Tracey K. J., Andersson J., Palmblad J. E. (2003) High mobility group 1 B-box mediates activation of human endothelium. J. Intern. Med. 254, 375–385 [DOI] [PubMed] [Google Scholar]
- 41. Rossini A., Zacheo A., Mocini D., Totta P., Facchiano A., Castoldi R., Sordini P., Pompilio G., Abeni D., Capogrossi M. C., Germani A. (2008) HMGB1-stimulated human primary cardiac fibroblasts exert a paracrine action on human and murine cardiac stem cells. J. Mol. Cell. Cardiol. 44, 683–693 [DOI] [PubMed] [Google Scholar]
- 42. Bianchi M. E. (2009) HMGB1 loves company. J. Leukoc. Biol. 86, 573–576 [DOI] [PubMed] [Google Scholar]
- 43. Youn J. H., Oh Y. J., Kim E. S., Choi J. E., Shin J.-S. (2008) High mobility group box 1 protein binding to lipopolysaccharide facilitates transfer of lipopolysaccharide to CD14 and enhances lipopolysaccharide-mediated TNF-α production in human monocytes. J. Immunol. 180, 5067–5074 [DOI] [PubMed] [Google Scholar]
- 44. Kim S., Kim S. Y., Pribis J. P., Lotze M., Mollen K. P., Shapiro R., Loughran P., Scott M. J., Billiar T. R. (2013) Signaling of high mobility group box 1 (HMGB1) through toll-like receptor 4 in macrophages requires CD14. Mol. Med. 19, 88–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pober J. S., Tellides G. (2012) Participation of blood vessel cells in human adaptive immune responses. Trends Immunol. 33, 49–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Li A., Dubey S., Varney M. L., Singh R. K. (2002) Interleukin-8-induced proliferation, survival, and MMP production in CXCR1 and CXCR2 expressing human umbilical vein endothelial cells. Microvasc. Res. 64, 476–481 [DOI] [PubMed] [Google Scholar]
- 47. Singh S., Wu S., Varney M., Singh A. P., Singh R. K. (2011) CXCR1 and CXCR2 silencing modulates CXCL8-dependent endothelial cell proliferation, migration and capillary-like structure formation. Microvasc. Res. 82, 318–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Keeley E. C., Mehrad B., Strieter R. M. (2011) Chemokines as mediators of tumor angiogenesis and neovascularization. Exp. Cell Res. 317, 685–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Barlow K. D., Sanders A. M., Soker S., Ergun S., Metheny-Barlow L. J. (2013) Pericytes on the tumor vasculature: Jekyll or Hyde? Cancer Microenviron. 6, 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Samaniego R., Estecha A., Relloso M., Longo N., Escat J. L., Longo-Imedio I., Avilés J. A., del Pozo M. A., Puig-Kröger A., Sánchez-Mateos P. (2013) Mesenchymal contribution to recruitment, infiltration, and positioning of leukocytes in human melanoma tissues. J. Invest. Dermatol. 133, 2255–2264 [DOI] [PubMed] [Google Scholar]
- 51. Proebstl D., Voisin M.-B., Woodfin A., Whiteford J., D'Acquisto F., Jones G. E., Rowe D., Nourshargh S. (2012) Pericytes support neutrophil subendothelial cell crawling and breaching of venular walls in vivo. J. Exp. Med. 209, 1219–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang S., Cao C., Chen Z., Bankaitis V., Tzima E., Sheibani N., Burridge K. (2012) Pericytes regulate vascular basement membrane remodeling and govern neutrophil extravasation during inflammation. PLoS One 7, e45499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Stark K., Eckart A., Haidari S., Tirniceriu A., Lorenz M., von Brühl M.-L., Gärtner F., Khandoga A. G., Legate K. R., Pless R., Hepper I., Lauber K., Walzog B., Massberg S. (2013) Capillary and arteriolar pericytes attract innate leukocytes exiting through venules and “instruct” them with pattern-recognition and motility programs. Nat. Immunol. 14, 41–51 [DOI] [PubMed] [Google Scholar]
- 54. Tu Z., Li Y., Smith D. S., Sheibani N., Huang S., Kern T., Lin F. (2011) Retinal pericytes inhibit activated T cell proliferation. Invest. Ophthalmol. Vis. Sci. 52, 9005–9010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bose A., Barik S., Banerjee S., Ghosh T., Mallick A., Bhattacharyya Majumdar S., Goswami K. K., Bhuniya A., Banerjee S., Baral R., Storkus W. J., Dasgupta P. S., Majumdar S. (2013) Tumor-derived vascular pericytes anergize Th cells. J. Immunol. 191, 971–981 [DOI] [PubMed] [Google Scholar]
- 56. Nauta A. J., Fibbe W. E. (2007) Immunomodulatory properties of mesenchymal stromal cells. Blood 110, 3499–3506 [DOI] [PubMed] [Google Scholar]
- 57. Matthay M. A., Thompson B. T., Read E. J., McKenna D. H., Jr., Liu K. D., Calfee C. S., Lee J. W. (2010) Therapeutic potential of mesenchymal stem cells for severe acute lung injury. Chest 138, 965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Xu J., Woods C. R., Mora A. L., Joodi R., Brigham K. L., Iyer S., Rojas M. (2007) Prevention of endotoxin-induced systemic response by bone marrow-derived mesenchymal stem cells in mice. Am. J. Physiol. Lung Cell. Mol. Physiol. 293, L131–L141 [DOI] [PubMed] [Google Scholar]
- 59. Gonzalez-Rey E., Anderson P., González M. A., Rico L., Büscher D., Delgado M. (2009) Human adult stem cells derived from adipose tissue protect against experimental colitis and sepsis. Gut 58, 929–939 [DOI] [PubMed] [Google Scholar]
- 60. Nemeth K., Mayer B., Mezey E. (2010) Modulation of bone marrow stromal cell functions in infectious diseases by toll-like receptor ligands. J. Mol. Med. 88, 5–10 [DOI] [PMC free article] [PubMed] [Google Scholar]