Abstract
In the United States, biliary atresia (BA) is the most frequent indication for liver transplantation in pediatric patients. BA is a complex disease, with suspected environmental and genetic risk factors. A genome-wide association study in Chinese patients identified association to the 10q24.2 (hg18) genomic region. This signal was upstream of two genes, XPNPEP1 and ADD3, both expressed in intrahepatic bile ducts. We tested association to this region in 171 BA patients and 1,630 controls of European descent and found the strongest signal to be at rs7099604 (p = 2.5 × 10−3) in intron 1 of the ADD3 gene. Moreover, expression data suggest that ADD3, but not XPNPEP1, is differentially expressed in BA patients. The role of ADD3 in biliary development is unclear, but our findings suggest that this gene may be functionally relevant for the development of BA.
Introduction
Biliary atresia (BA) is a disease of the liver and bile ducts affecting neonates in the first months of life. BA is a progressive idiopathic, necroinflammatory process initially limited to the extrahepatic biliary tree, but the progressive obliteration of the extrahepatic bile duct lumen leads to obstruction of bile flow resulting in cholestasis and chronic liver damage. If left uncorrected, the mortality rate is 100 %. BA is the most common indication for liver transplantation in children. Fifty percent of affected individuals undergo transplantation by 2 years of age and 80 % undergo transplantation by 20 years of age (Erlichman et al. 2009; Mack and Sokol 2005; Shneider et al. 2006).
The incidence rate of BA varies across ethnic groups. It is more commonly found in Asian populations including those living in Asia as well as those living in the United States. The incidence rate is 0.6, 1.0, 1.7, 2.0, and 3.0 per 10,000 live births for the respective Caucasian, Japanese, Taiwanese, Filipinos in Hawaii, and Chinese in Hawaii populations (Muraji et al. 2009). While the etiology of BA is unknown, it is proposed to be multi-factorial, with infectious, inflammatory and genetic risk factors (Shneider et al. 2006).
A previous genome-wide association study (GWAS) of Chinese individuals (324 BA cases, 481 controls) identified a region on chromosome 10q as a potential susceptibility locus. The most significant finding was association with SNP rs17095355 (p = 6.94 × 10−9) on chromosome 10 at position 111735750 (hg19) (Garcia-Barcelo et al. 2010). This SNP is located upstream of two genes expressed in the hepatobiliary system, XPNPEP1, encoding X-Prolyl Aminopeptidase 1, and ADD3, encoding an F-actin binding protein called adducin 3 (Citterio et al. 2003; Ersahin et al. 2005). Association with the same SNP was replicated in a Thai cohort (p < 0.003) of 124 BA cases compared against 114 controls (Kaewkiattiyot et al. 2011). Both of these investigations were conducted on Asian subjects and there has been no report of this association in other ethnic groups. We investigated this region in our data from an ongoing GWAS study in a Caucasian cohort to test if this locus affects risk of BA in a non-Asian population.
Materials and methods
Sample collection
The BA patients in this studied were enrolled through two studies. The majority of the BA patients (n = 240) were enrolled under IRB-approved protocols in the Childhood Liver Disease Research and Education Network (ChiLDREN), an NIDDK-funded research network involving 16 pediatric centers across North America, including the Children’s Hospital of Philadelphia (CHOP). The remaining BA patients (n = 51) were followed at CHOP and directly enrolled into an IRB-approved study at this institution. These patients had a diagnosis of BA made by clinical presentation, liver histology and intraoperative cholangiogram. Most patients also had the diagnosis confirmed by examination of the biliary remnant from a Roux-en-Y hepatic portoenterostomy (Kasai operation). The DNA samples from patients enrolled in ChiLDREN were provided to us by the Rutgers University NIDDK biorepository (Shneider et al. 2012). The DNA samples of patients enrolled at CHOP were extracted from peripheral blood using the 5Prime DNA extraction kit or from lymphoblastoid cell lines transformed from peripheral blood. The genotypes of 3,000 healthy controls were supplied by the Center for Applied Genomics (CAG) at CHOP. These healthy individuals were enrolled from several different primary care clinics in the Greater Philadelphia region through an IRB-approved protocol (Shaikh et al. 2009). All samples were genotyped by the CAG on the HumanHap 610 Quad SNP BeadArray (Illumina, San Diego, CA), with the exception of 11 BA patients genotyped on the HumanHap 550v3 SNP BeadArray for a previous study (Cui et al. 2013; Leyva-Vega et al. 2010).
The liver biopsies and clinical data for the samples used in the quantitative PCR (qPCR) expression assays were obtained from infants with cholestasis enrolled into a prospective study of the ChiLDREN or from infants evaluated at Cincinnati Children’s Hospital Medical Center through the participating institution’s IRB-approved protocols. For subjects with BA, liver biopsies were obtained from 64 infants during the preoperative workup or at the time of intraoperative cholangiogram (age 66.1 ± 26.6 days). Subjects with intrahepatic cholestasis served as diseased controls (DC). Liver biopsy samples were obtained percutaneously or intraoperatively from 14 infants at the time of diagnostic evaluation (age 77.1 ± 48.7 days). This group consisted of children diagnosed with Alagille syndrome (n = 1), ABCB4 deficiency (n = 2), alpha-1-antitrypsin deficiency (n = 2) and cholestasis with unknown etiology (n = 9). A separate group of normal controls (NC) consisted of liver biopsy samples obtained from deceased-donor children aged 22–42 months (n = 7) as described previously (Li et al. 2011a). All samples were obtained after informed consent from patients’ parents or legal guardians.
SNP array quality control
The genotypes from the 550v3 and 610 Quad BeadArrays were merged together. The markers unique to the 550v3 were excluded from further analysis and the markers unique to the 610 Quad were set to missing in the 11 samples genotyped on the 550v3 array. There were 551,044 markers that passed quality control filtering criteria (see Sup. Fig. 1).
A genetically matched set of cases and controls was identified from the SNP array data (see Sup. Fig. 2 for detailed selection criteria). Of the 229 BA patients with self-reported ethnicity, 169 (74 %) were self-reported Caucasian, 33 (14 %) were African American, 31 (14 %) were Hispanic, 7 (3 %) were Asian, 1 was American Indian or Alaska Native, 1 was Native Hawaiian or Other Pacific Islander, and the remaining reported an unlisted ethnicity. For our analysis, we chose to focus on Caucasians because they were the largest ethnic group in our cohort. We clustered all the subjects using the multidimensional scaling (MDS) algorithm in PLINK (Purcell et al. 2007). The MDS plot (Sup. Figs. 3A and 3B) depicts the ethnic diversity of our patient and in-house control populations as compared to the HapMap 3 reference population (International HapMap 3 Consortium 2010). We controlled for the population structure of our cohort by first isolating individuals who were within the range of self-identified Caucasians for the first and second MDS components, and then by filtering for only individuals who had these two components within two standard deviations of the mean of this subpopulation (Sup. Fig. 3C). After quality control and filtering, the remaining 171 cases and 1,630 controls were used for our association study. We performed a second MDS analysis on this smaller cohort to look for finer population substructure and accounted for this by adjusting for the first two components in the association tests.
Linkage disequilibrium in 10q25.1
We investigated the linkage disequilibrium (LD) structure around rs17095355 on 10q24.2 (now referred to as 10q25.1 in hg19) in the HapMap 3 Release 2 Chinese and Caucasian SNP data. It is important to understand the population structure differences in this region between Chinese and Caucasians because the SNP association was detected in a Chinese cohort whereas we tested a Caucasian cohort. The confidence interval method in Haploview was used to define haplotype blocks (Barrett et al. 2005; Gabriel et al. 2002). Also, this method was used to calculate the r2 correlation between these blocks.
Imputation and association study
The logistic regression test adjusted for the first two MDS components was performed using PLINK on genome-wide genotyped SNPs to estimate the genomic inflation factor (Purcell et al. 2007). Results of this test were plotted as a quantile–quantile (Q–Q) plot in R (R Core Team 2012) to check for deviation from the expected distribution of p values.
Imputation of SNP genotypes was performed at and around rs17095355 to look for the strongest signal of association in this candidate region. We had 339 SNPs in the 2 Mb region around rs17095355 genotyped by the SNP array. We used these genotypes for imputation using IMPUTE2 (Howie et al. 2009). The 1000 Genomes Project (1000G) Phase I low coverage data from 1,092 individuals of diverse ethnic backgrounds served as our imputation reference panel (1000 Genomes Project Consortium 2012). SNPs that were imputed with low confidence (info <0.8), that were rare in our cases and controls (minor allele frequency or MAF <0.01), as well as those that appeared to be non-unique in hg19 were removed. The imputation procedure is summarized in Sup. Fig. 4. Association test of the imputed data adjusting for the first two MDS components was performed using SNPTEST (Marchini et al. 2007) on 333 SNPs that passed our quality control criteria in the region that spanned the Chinese LD block around rs17095355 and encompassed the two flanking genes, including a 5 kb region downstream of each gene (chr10:111619000–111901000 in hg19 coordinates) by logistic regression under an additive model. Using SNPT-EST, we also performed an association test of the same SNPs conditional on rs17095355 and rs7099604 by logistic regression analysis under a standard additive model.
Confirmation of imputation results
From imputation, a probability score was assigned to each of the three possible genotypes of a biallelic SNP. A confidently imputed genotype is considered one where the probability of the most likely genotype is greater or equal to 90 % (Marchini et al. 2007). Sanger sequencing confirmed the genotype at rs7099604 in 57 BA patients with confidently imputed genotypes. Sequencing was performed using a primer set consisting of 5′-TGG GCA TAA GTG CCT TGG ATG A-3′ (forward) and 5′-ACA AGC AGA CAG TTG TGG AGA-3′ (reverse). The primers were designed using PrimerQuest and ordered through Integrated DNA Technologies (Coralville, IA).
Immunohistochemistry
A control liver sample from an autopsied infant with no known liver disease and a BA sample (liver tissue from a wedge biopsy and biliary remnant from a Kasai operation) were provided by the Pathology Department at CHOP. These samples had been fixed by routine methods in 10 % buffered formalin and paraffin-embedded. The existing autopsy sample was obtained from a 2-month-old infant. The samples from the BA patient were collected at 1.5 months of age. Immunohistochemistry for P1/XPNPEP1 monoclonal antibody (R&D Systems, Minneapolis, MN) at a dilution of 1:400, and Adducin-gamma rabbit polyclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA) at a dilution of 1:100, was done on the Leica Bond-Max Autostainer using the Bond Polymer Refine Detection System (Leica Microsystems, Buffalo Grove, IL). Paraffin sections were cut onto charged glass slides at 5 μm, and dried overnight at 45 °C. Deparaffinization, epitope retrieval with ER2 buffer (pH 9) for 20 min for ADD3 and ER1 buffer (pH 6) for XPNPEP1 and detection were all performed on the Leica Bond-III stainer. The detection reagents include a horseradish peroxidase-linked polymer, which works for mouse and rabbit antibodies and DAB chromogen. The sections were counterstained with Gill’s hematoxylin, washed, dehydrated with graded concentrations of ethanol, cleared in xylene, mounted and examined microscopically.
Expression analysis
Quantitative PCR was performed using Brilliant III Ultra-Fast SYBR Green QPCR Master Mix and the Mx3005P qPCR System (Agilent Technologies, Santa Clara, CA) as previously reported (Shivakumar et al. 2004). The mRNA expression of target genes was normalized to the endogenous reference GAPDH gene. PCR primers used in the study are listed in Sup. Table 1.
The Kruskal–Wallis test was performed to test for a difference among the three groups (NC, DC, and BA). Then, pairwise Dunn’s multiple comparison tests were performed to determine the source of the difference. Statistical analysis was done using Prism 5.0c (GraphPad Software, San Diego, CA), and p < 0.05 was regarded as statistically significant. The correlation between gene expression and SNP genotype was tested by fitting a linear regression with the genotype expressed as number of minor alleles (0, 1, or 2) as a predictor of the expression.
Sanger Sequencing of ADD3
The ADD3 coding exons were sequenced in 60 patients to look for novel or rare mutations. Primers were designed to sequence all 14 coding exons of ADD3 (Sup. Table 2). PCR was performed with AmpliTaq Gold DNA polymerase (Life Technologies, Carlsbad, CA) using the optimized annealing conditions for each primer set. The quality of the PCR product was confirmed by running 5 μl of PCR product on a 1.5 % agarose gel at 120 V for 45 min. The unincorporated primers and dNTPs were removed from each sample using 2 μl of exonuclease 1 and 4 μl of fast thermosensitive alkaline phosphatase. Capillary electrophoresis was performed at the Nucleic Acid/Protein Core Facility (NAPCore) at CHOP. The sequences were aligned to the hg19 reference of ADD3 to identify variants.
Results
Association study of 10q25.1 from genotyped data
In order to test for the effects of population stratification, we calculated a genomic inflation factor (λ) based on the genome-wide association tests. The value of λ was estimated to be 1.01 and therefore no strong evidence for substructure was detected in our sample. A Q–Q plot of the observed p values did not indicate significant deviation from the expected values (Sup. Fig. 5).
Looking at association test results for the SNPs genotyped on the array in our Caucasian cohort, we searched for a signal at the 10q25.1 locus previously reported to be associated with BA in a Chinese study. We found no SNPs that achieved genome-wide significance at this locus. The significance of rs17095355 was unknown because it was not present on the array platform used to genotype our samples. However, SNPs strongly correlated with it based on the HapMap 3 samples of Chinese descent (CHB + CHD) (r2 = 0.99) achieved nominal significance (p < 0.03) (Table 1).
Table 1.
Regional genotyped and imputed SNPs achieving the highest significance
| SNP | Position | Alleles (minor/major) | MAF (cases) | MAF (controls) | Odds ratio (95 % CI) | p value | Conditional p value
|
r2
|
Annotation | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs17095355 | rs7099604 | rs17095355 (CHB + CHD) | rs7099604 (CHB + CHD) | rs17095355 (EUR) | rs7099604 (EUR) | ||||||||
| rs921348 | 111707498 | A/G | 0.208 | 0.169 | 1.29 (0.97, 1.70) | 0.026 | 0.927 | 0.953 | 0.99 | 0.51 | 1.00 | 0.41 | Intergenic |
| rs7906904 | 111708193 | A/G | 0.213 | 0.174 | 1.29 (0.98, 1.69) | 0.021 | 0.479 | 0.981 | 0.99 | 0.51 | 0.96 | 0.46 | Intergenic |
| rs17095355* | 111735750 | C/T | 0.204 | 0.166 | 1.29 (0.98, 1.71) | 0.026 | – | 0.917 | – | 0.49 | – | 0.41 | Intergenic |
| rs12569425 | 111758177 | G/A | 0.202 | 0.151 | 1.42 (1.07, 1.88) | 0.011 | 0.219 | 0.568 | 0.75 | 0.70 | 0.62 | 0.62 | Intergenic |
| rs7069128 | 111760075 | A/G | 0.202 | 0.151 | 1.42 (1.07, 1.88) | 0.011 | 0.219 | 0.568 | 0.75 | 0.70 | 0.62 | 0.62 | Intergenic |
| rs7099604 | 111774513 | A/G | 0.199 | 0.146 | 1.45 (1.09, 1.92) | 0.002 | 0.053 | – | 0.49 | – | 0.41 | – | ADD3 |
The most significant genotyped and imputed SNPs as well as the relevant SNPs from the Chinese study are included. The imputed SNPs are italicized. The most significant SNP (rs7099604) in this region is in boldface. The r2 correlation between the SNPs listed and rs17095355 as well as rs7099604 are listed separately. Linkage disequilibrium was determined with respect to two populations of 1000G data, the subjects of Chinese descent (CHB and CHD) and of European descent (EUR, comprised of CEU, FIN, GBR, IBS, TSI)
MAF minor allele frequency
SNP associated to BA in an Asian population
Fine-mapping of the 10q25.1 region following imputation
An exploratory analysis of the candidate region haplotype structures in HapMap 3 Chinese and Caucasian data was performed to better understand variability at this locus. The LD plots generated from the genotypes of the Han Chinese in Beijing, China (CHB) (Sup. Fig. 6A) and of the United States residents of western and northern European ancestry from Utah and Tuscans in Italy (CEU + TSI) (Sup. Fig. 6B) depict the diversity of this region between ethnic groups. The bars above the LD plots represent haplotype blocks defined according to the default algorithm in Haploview (Gabriel et al. 2002). In the Chinese population studied, the LD block containing rs17095355 is strongly correlated with a neighboring LD block (r2 = 0.91). Together, these two blocks partially encompass both XPNPEP1 and ADD3. To extensively cover the biologically relevant regions of these genes, we performed subsequent analysis on all SNPs included in the genomic region from 5 kb downstream of XPNPEP1 to 5 kb downstream of ADD3 (chr10:111619000–111901000 in hg19 coordinates).
Following SNP imputation, we performed an association test on 333 SNPs in the candidate region correcting for the first two MDS components. Results for the most significant imputed (rs7099604) and genotyped (rs12569425 and rs7069128) SNPs are listed in Table 1. Also, rs17095355 and the genotyped SNPs most highly correlated with it (rs921348 and rs7906904) are included. A comprehensive summary of all the genotyped and imputed SNPs tested can be found in Sup. Table 3. While rs17095355 was nominally significant (p = 0.026), it was not the most significant SNP in this region. The strongest signal came from rs7099604 (p = 0.002), a SNP that lies in the first intronic region of ADD3 (Table 1). This SNP was not reported in the published association study (Garcia-Barcelo et al. 2010), and it is only moderately correlated with rs17095355 (r2 = 0.49 in 197 1000G Phase I CHB + CHD samples). The SNPs most strongly correlated with rs7099604 in the 1000G Phase I CHB + CHD samples showed p values around 10−4 in the Chinese BA GWAS (Garcia-Barcelo et al. 2010).
Most markers achieving marginal significance (p ≤ 0.05) were highly correlated (r2 > 0.6) with rs7099604, but a handful had low correlation (r2 < 0.2) (Fig. 1a, Sup. Table 3). To explore if multiple independent SNPs in this region may be associated with BA, we performed additional association tests conditional on the genotype at rs7099604. The results are reported in Fig. 1b. Overall, the association was less significant for the SNPs in the intergenic region, including rs17095355 (conditional association p = 0.917), and in ADD3. Curiously, a few markers in XPNPEP1 that did not reach nominal significance or were barely significant (p = 0.04) have an increased significance with the conditional association test. Conversely, we also performed an association test of this region conditional on rs17095355 and found that rs7099604 remains borderline significant (conditional association test p = 0.053).
Fig. 1.
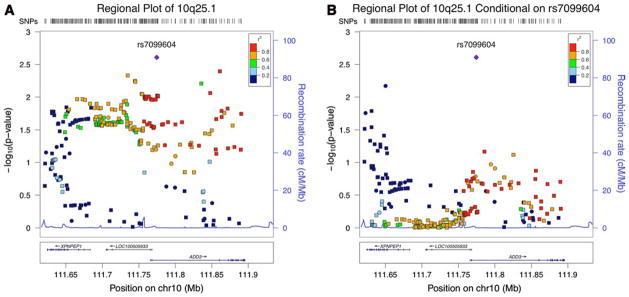
Regional association plot of 10q25.1. The most significant SNP in this region, rs7099604, is depicted as a purple diamond. The remaining colors correspond to each SNP’s r2 correlation to rs7099604 in the European population of the 1000 Genomes Phase I data. The circles represent the markers that were genotyped using the SNP array and squares represent the imputed markers. a The regional association plot of the p values from the additive genetic model. b The regional association plot of the p values conditional on rs7099604 (color figure online)
Confirmation of imputation results
We determined the genotype at rs7099604 of 58 patients with confidently imputed genotypes. These genotypes were determined by Sanger sequencing, and 5 of 5 GG, 28 of 29 AG, and 20 of 24 AA genotypes were correctly imputed. The 4 incorrectly predicted AA genotypes were determined to be either AG (n = 3) or GG (n = 1). The incorrectly predicted AG genotype was confirmed to be GG. Interestingly, the genotypes of 4 of the 8 patients genotyped on the older HumanHap 550v3 SNP arrays and experimentally checked via Sanger sequencing did not validate. However, only one of the 49 patients genotyped on the HumanHap 610 SNP array and experimentally checked was imputed incorrectly (2 % error rate). These experimental findings also show that the imputation algorithm was conservative when predicting a deviation from the major allele, as the error in genotypes underestimated the prevalence of the risk allele, G. From the experimental validation, we believe the imputed data is reliable and suitable for downstream analysis.
Localization of XPNPEP1 and ADD3
The function of X-prolyl aminopeptidases is to breakdown bradykinin, a peptide that participates in regulating blood pressure and this aminopeptidase can be inhibited by apstatin (Li et al. 2011b). XPNPEP1 is ubiquitously expressed throughout the body, with the most expression found in the small intestine. The role of XPNPEP1 in the liver and hepatobiliary tract has not been established, although an increased expression of serum leucine aminopeptidase has been associated with a several hepatobiliary diseases affecting intrahepatic or extrahepatic bile ducts (Kowlessar et al. 1961). From the immunohistochemistry stains of liver sample tissue, we showed that XPNPEP1 is present in both hepatocytes as well as the biliary epithelium in normal and diseased livers (Fig. 2a). Moreover, protein expression of XPNPEP1 was also present in the extrahepatic biliary epithelium, but not in the surrounding fibrous tissue.
Fig. 2.
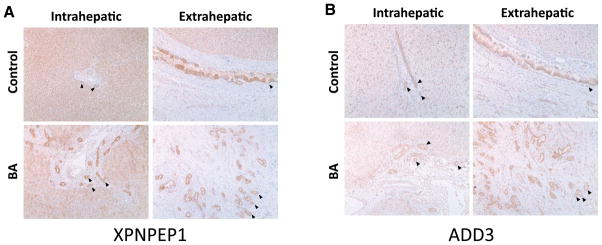
XPNPEP1 and ADD3 are expressed in intra- and extrahepatic bile ducts by immunohistochemistry. The black arrows represent bile ducts. a The staining of XPNPEP1 is present in the intrahepatic as well as the extrahepatic bile ducts. It seems that XPNPEP1 is ubiquitously expressed, as it also stains the hepatocytes. b ADD3 seems to be only expressed in the intra- and extrahepatic bile ducts as it faintly stains the hepatocytes and surrounding tissues
The adducin gene family has an established association with hypertension (Cusi et al. 1997). Although adducin proteins are known to be ubiquitously expressed membrane-skeletal proteins, the presence of ADD3 in the hepatobiliary system has not been described. By immunohistochemistry, we found evidence of ADD3 expression in the biliary epithelium (Fig. 2b), but there seemed to be little or no expression of ADD3 in the surrounding hepatic cells. ADD3 was also present in the extrahepatic biliary epithelium (Fig. 2b).
Expression of XPNPEP1 and ADD3
The expression of XPNPEP1 and ADD3 was measured in the liver of normal control (NC), diseased control (DC) and BA subjects by qPCR. There did not seem to be any apparent differences in XPNPEP1 expression between the three groups of patients (Fig. 3a). However, there was a significant difference in the level of expression of ADD3 between BA and normal control subjects (p < 0.001) (Fig. 3b). No significant difference was observed between the diseased control group and the normal controls as well as the BA group. We also investigated the possibility of the expression of these two genes being linked to the underlying genotype at the significantly associated SNP rs7099604 (Sup. Fig. 7). Although the sample size is small, there did not seem to be any correlation of rs7099604 genotype with expression of XPNPEP1 (p = 0.34) and ADD3 (p = 0.61).
Fig. 3.
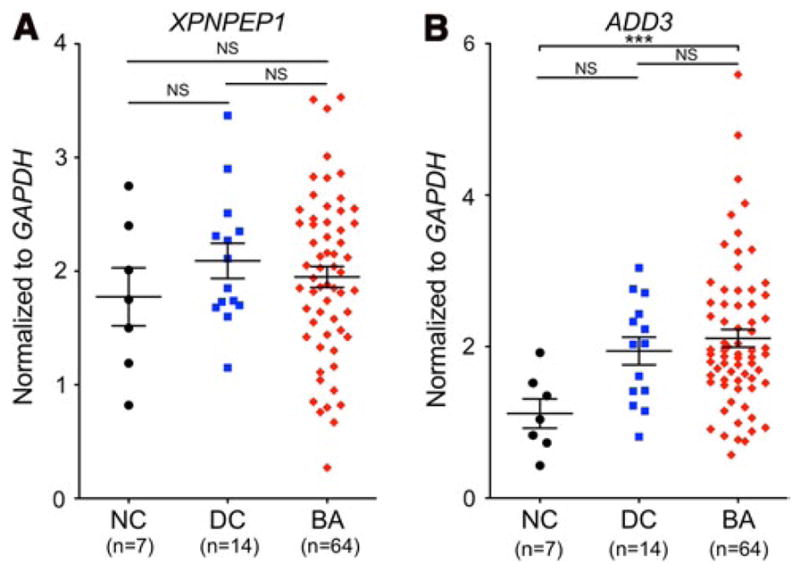
Relative expression of ADD3 and XPNPEP1. The expression levels of XPNPEP1 and ADD3 were measured in normal controls (NC), disease controls (DC), and BA patients (BA). The comparisons between these groups were not significant (NS) for XPNPEP1. A significant difference was detected (***p < 0.001) between the NC and BA for ADD3
ADD3 gene sequencing
We looked for coding changes in ADD3 by sequencing 60 patients. Sanger sequencing was carried out in a diverse representation of patients with rs7099604 genotypes (14 AA, 43 AG, 3 GG), but we did not identify any deviation from the human reference genome.
Discussion
Findings in 10q25.1
Two independent studies in Asian patients showed an association of 10q25.1 variants with BA (Garcia-Barcelo et al. 2010; Kaewkiattiyot et al. 2011). Our investigation of genotyped and imputed SNPs in this region indicates that the association to BA at this locus is also present in Caucasians. However, we find that while rs17095355, the most significant SNP in Asian samples, is also nominally significant in our cohort, rs7099604 in the first intron of ADD3 has a stronger signal in our Caucasian cohort. These differences may be attributed to regional differences in haplotype structure (Sup. Fig. 6) as well as variation in the minor allele frequency of these SNPs between the two ethnic groups. The dbSNP data on the frequency distribution of rs17095355, the candidate BA SNP reported in two Asian cohorts, show a large disparity in allele frequencies between Asians and Caucasians as well as within Caucasians (Sup. Fig. 8). The lower level of LD in this region combined with decreased prevalence of the polymorphism at rs17095355 in Caucasians may have decreased our power to detect an association with BA. Even so, the marginally significant p value from a conditional association test on rs17095355 indicates a residual association at rs7099604 that is not accounted for by the previously reported SNP. Our data suggest that while rs17095355 could be a good marker of the true signal in 10q25.1 in the Asian population, it may not necessarily be the best marker of the underlying signal in the European population. We propose rs7099604 as a better marker of the true association at 10q25.1 than rs17095355 in a Caucasian cohort, and its intronic location indicates ADD3 as the more relevant gene to the etiology of BA.
The negative findings from ADD3 exon sequencing in 60 BA patients suggest that the association signal is not from a direct change in its coding sequence. It is possible that the causal variant responsible for the association signal could alter the function of a regulatory element. The top two associated SNPs are intronic to ADD3, but the third most significant SNP, rs7067604, lies 33 kb upstream of ADD3. This SNP lies in a strong enhancer region in the HepG2 cell line from the ENCODE data (Rosenbloom et al. 2013).
Implication of ADD3 in BA
As the LD block containing the association signal on 10q25.1 spans XPNPEP1 and ADD3, it has been difficult to conclude where the signal arises. Both genes are expressed in the biliary epithelium and are downstream of the previously reported associated SNP. Interestingly, ADD3 seems to stain more specifically for the bile ducts, whereas XPN-PEP1 seems to be more ubiquitously expressed in the liver. Our most significant signal is an intronic polymorphism in ADD3, although the genotype at this SNP does not seem to directly correlate with the expression of ADD3. Examination of the intrahepatic biliary tree revealed that zebrafish with a gene knockdown of xpnpep1 exhibited little or no biliary abnormalities, but biliary defects were present in the add3a knockdown construct (R.P. Matthews, personal communication). These results seem to correspond well with the gene expression data, indicating no difference between XPNPEP1 expression in BA and the control cohorts and a significant difference between ADD3 expression in BA and the normal control cohort. While this manuscript was under revision, a publication by Cheng et al. also reported ADD3 as a likely candidate gene for BA. They found that the Chinese BA patients with the risk haplotype had significantly lower expression of ADD3 suggesting that the risk haplotype can contribute to BA by decreasing ADD3 expression (Cheng et al. 2013).
In conclusion, we demonstrated that the 10q25.1 region reported in Asian populations is also significantly associated with BA in the Caucasian population. While the original association signal lies upstream of XPNPEP1 and ADD3, SNP imputation in this region suggests a stronger association signal intronic to ADD3 and supports its role as a BA susceptibility gene.
Supplementary Material
Acknowledgments
We would like to thank the patients and their families who donated their samples for this study. We also would like to acknowledge R.P. Matthews for his preliminary data on the biliary phenotype of xpnpep1 and add3a knockdown constructs in zebrafish, and M. Leyva-Vega, A. Hutchinson, and L.D. Leonard for their help in obtaining the BA samples from ChiLDREN. This work was supported by The Fred and Suzanne Biesecker Liver Center at CHOP as well as R01-DK090045 to N.B.S, U01-DK062481 to K.M.L., and R01-DK083781 to J.A.B. E.A.T. was also supported by training grant T32-HG000046.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00439-013-1368-2) contains supplementary material, which is available to authorized users.
Contributor Information
Ellen A. Tsai, Email: eatsai@gmail.com, Genomics and Computational Biology Graduate Group, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA
Christopher M. Grochowski, Department of Pathology and Laboratory Medicine, The Children’s Hospital of Philadelphia, Philadelphia, PA, USA
Kathleen M. Loomes, Division of Gastroenterology, Hepatology and Nutrition, The Children’s Hospital of Philadelphia, Philadelphia, PA, USA. Department of Pediatrics, Perelman School of Medicine, University of Pennsylvania, Philadelphia 19104, PA, USA
Kazuhiko Bessho, Pediatric Liver Care Center and Division of Pediatric Gastroenterology, Hepatology and Nutrition, Cincinnati Children’s Hospital Medical Center, Cincinnati, USA. Department of Pediatrics, University of Cincinnati, Cincinnati, OH, USA.
Hakon Hakonarson, Department of Pediatrics, Perelman School of Medicine, University of Pennsylvania, Philadelphia 19104, PA, USA. Center for Applied Genomics, The Children’s Hospital of Philadelphia, Philadelphia, PA 19104, USA. Division of Human Genetics, The Children’s Hospital of Philadelphia, Philadelphia, PA, USA.
Jorge A. Bezerra, Pediatric Liver Care Center and Division of Pediatric Gastroenterology, Hepatology and Nutrition, Cincinnati Children’s Hospital Medical Center, Cincinnati, USA. Department of Pediatrics, University of Cincinnati, Cincinnati, OH, USA
Pierre A. Russo, Department of Pathology and Laboratory Medicine, The Children’s Hospital of Philadelphia, Philadelphia, PA, USA. Department of Pathology and Laboratory Medicine, Perelman School of Medicine, University of Pennsylvania, Philadelphia 19104, PA, USA
Barbara A. Haber, Hepatology, Infectious Diseases Clinical Research Department, Merck Research Laboratories, North Wales, PA, USA
Nancy B. Spinner, Department of Pathology and Laboratory Medicine, The Children’s Hospital of Philadelphia, Philadelphia, PA, USA. Department of Pathology and Laboratory Medicine, Perelman School of Medicine, University of Pennsylvania, Philadelphia 19104, PA, USA
Marcella Devoto, Email: devoto@email.chop.edu, Department of Pediatrics, Perelman School of Medicine, University of Pennsylvania, Philadelphia 19104, PA, USA. Division of Human Genetics, The Children’s Hospital of Philadelphia, Philadelphia, PA, USA. Department of Biostatistics and Epidemiology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA. Department of Molecular Medicine, University La Sapienza, Rome, Italy.
References
- 1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Cheng G, Tang CS, Wong EH, Cheng WW, So MT, Miao X, Zhang R, Cui L, Liu X, Ngan ES, Lui VC, Chung PH, Chan IH, Liu J, Zhong W, Xia H, Yu J, Qiu X, Wu XZ, Wang B, Dong X, Tou J, Huang L, Yi B, Ren H, Chan EK, Ye K, O’Reilly PF, Wong KK, Sham PC, Cherny SS, Tam PK, Garcia-Barcelo MM. Common genetic variants regulating ADD3 gene expression alter biliary atresia risk. J Hepatol. 2013 doi: 10.1016/j.jhep.2013.07.021. [DOI] [PubMed] [Google Scholar]
- Citterio L, Tizzoni L, Catalano M, Zerbini G, Bianchi G, Barlassina C. Expression analysis of the human adducin gene family and evidence of ADD2 beta 4 multiple splicing variants. Biochem Biophys Res Commun. 2003;309:359–367. doi: 10.1016/j.bbrc.2003.08.011. [DOI] [PubMed] [Google Scholar]
- Cui S, Leyva-Vega M, Tsai EA, Eauclaire SF, Glessner JT, Hakonarson H, Devoto M, Haber BA, Spinner NB, Matthews RP. Evidence from human and zebrafish that GPC1 is a biliary atresia susceptibility Gene. Gastroenterology. 2013;144(5):1107–1115. doi: 10.1053/j.gastro.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusi D, Barlassina C, Azzani T, Casari G, Citterio L, Devoto M, Glorioso N, Lanzani C, Manunta P, Righetti M, Rivera R, Stella P, Troffa C, Zagato L, Bianchi G. Polymorphisms of alpha-adducin and salt sensitivity in patients with essential hypertension. Lancet. 1997;349:1353–1357. doi: 10.1016/S0140-6736(97)01029-5. [DOI] [PubMed] [Google Scholar]
- Erlichman J, Hohlweg K, Haber BA. Biliary atresia: how medical complications and therapies impact outcome. Expert Rev Gastroenterol Hepatol. 2009;3:425–434. doi: 10.1586/egh.09.30. [DOI] [PubMed] [Google Scholar]
- Ersahin C, Szpaderska AM, Orawski AT, Simmons WH. Aminopeptidase P isozyme expression in human tissues and peripheral blood mononuclear cell fractions. Arch Biochem Biophys. 2005;435:303–310. doi: 10.1016/j.abb.2004.12.023. [DOI] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- Garcia-Barcelo MM, Yeung MY, Miao XP, Tang CS, Chen G, So MT, Ngan ES, Lui VC, Chen Y, Liu XL, Hui KJ, Li L, Guo WH, Sun XB, Tou JF, Chan KW, Wu XZ, Song YQ, Chan D, Cheung K, Chung PH, Wong KK, Sham PC, Cherny SS, Tam PK. Genome-wide association study identifies a susceptibility locus for biliary atresia on 10q24.2. Hum Mol Genet. 2010;19:2917–2925. doi: 10.1093/hmg/ddq196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap 3 Consortium. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaewkiattiyot S, Honsawek S, Vejchapipat P, Chongsrisawat V, Poovorawan Y. Association of X-prolyl aminopeptidase 1 rs17095355 polymorphism with biliary atresia in Thai children. Hepatol Res. 2011;41:1249–1252. doi: 10.1111/j.1872-034X.2011.00870.x. [DOI] [PubMed] [Google Scholar]
- Kowlessar OD, Haeffner LJ, Riley EM, Sleisenger MH. Comparative study of serum leucine aminopeptidase, 5-nucleotidase and non-specific alkaline phosphatase in diseases affecting the pancreas, hepatobiliary tree and bone. Am J Med. 1961;31:231–237. doi: 10.1016/0002-9343(61)90111-5. [DOI] [PubMed] [Google Scholar]
- Leyva-Vega M, Gerfen J, Thiel BD, Jurkiewicz D, Rand EB, Pawlowska J, Kaminska D, Russo P, Gai X, Krantz ID, Kamath BM, Hakonarson H, Haber BA, Spinner NB. Genomic alterations in biliary atresia suggest region of potential disease susceptibility in 2q37.3. Am J Med Genet A. 2010;152A:886–895. doi: 10.1002/ajmg.a.33332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Bessho K, Shivakumar P, Mourya R, Mohanty SK, Dos Santos JL, Miura IK, Porta G, Bezerra JA. Th2 signals induce epithelial injury in mice and are compatible with the biliary atresia phenotype. J Clin Invest. 2011a;121:4244–4256. doi: 10.1172/JCI57728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Bartlam M, Rao Z. Encyclopedia of inorganic and bioinorganic chemistry. Wiley; New York: 2011b. Human cytosolic X-Prolyl aminopeptidase. [Google Scholar]
- Mack CL, Sokol RJ. Unraveling the pathogenesis and etiology of biliary atresia. Pediatr Res. 2005;57:87R–94R. doi: 10.1203/01.PDR.0000159569.57354.47. [DOI] [PubMed] [Google Scholar]
- Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- Muraji T, Suskind DL, Irie N. Biliary atresia: a new immunological insight into etiopathogenesis. Expert Rev Gastroenterol Hepatol. 2009;3:599–606. doi: 10.1586/egh.09.61. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. R: a language and environment for statistical computing. 2012 http://www.R-project.org.
- Rosenbloom KR, Sloan CA, Malladi VS, Dreszer TR, Learned K, Kirkup VM, Wong MC, Maddren M, Fang R, Heitner SG, Lee BT, Barber GP, Harte RA, Diekhans M, Long JC, Wilder SP, Zweig AS, Karolchik D, Kuhn RM, Haussler D, Kent WJ. ENCODE data in the UCSC genome browser: year 5 update. Nucleic Acids Res. 2013;41:D56–D63. doi: 10.1093/nar/gks1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaikh TH, Gai X, Perin JC, Glessner JT, Xie H, Murphy K, O’Hara R, Casalunovo T, Conlin LK, D’Arcy M, Frackelton EC, Geiger EA, Haldeman-Englert C, Imielinski M, Kim CE, Medne L, Annaiah K, Bradfield JP, Dabaghyan E, Eckert A, Onyiah CC, Ostapenko S, Otieno FG, Santa E, Shaner JL, Skraban R, Smith RM, Elia J, Goldmuntz E, Spinner NB, Zackai EH, Chiavacci RM, Grundmeier R, Rappaport EF, Grant SF, White PS, Hakonarson H. High-resolution mapping and analysis of copy number variations in the human genome: a data resource for clinical and research applications. Genome Res. 2009;19:1682–1690. doi: 10.1101/gr.083501.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivakumar P, Campbell KM, Sabla GE, Miethke A, Tiao G, McNeal MM, Ward RL, Bezerra JA. Obstruction of extrahepatic bile ducts by lymphocytes is regulated by IFN-gamma in experimental biliary atresia. J Clin Invest. 2004;114:322–329. doi: 10.1172/JCI21153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shneider BL, Brown MB, Haber B, Whitington PF, Schwarz K, Squires R, Bezerra J, Shepherd R, Rosenthal P, Hoofnagle JH, Sokol RJ. A multicenter study of the outcome of biliary atresia in the United States, 1997 to 2000. J Pediatr. 2006;148:467–474. doi: 10.1016/j.jpeds.2005.12.054. [DOI] [PubMed] [Google Scholar]
- Shneider BL, Abel B, Haber B, Karpen SJ, Magee JC, Romero R, Schwarz K, Bass LM, Kerkar N, Miethke AG, Rosenthal P, Turmelle Y, Robuck PR, Sokol RJ. Portal hypertension in children and young adults with biliary atresia. J Pediatr Gastroenterol Nutr. 2012;55:567–573. doi: 10.1097/MPG.0b013e31826eb0cf. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


