Abstract
The unfolded protein response (UPR) is activated as a consequence of alterations to ER homeostasis. It upregulates a group of ER chaperones and cochaperones, as well as other genes that improve protein processing within the secretory pathway. The UPR effector ATF6α augments—but is not essential for—maximal induction of ER chaperones during stress, yet its role, if any, in protecting cellular function during normal development and physiology is unknown. A systematic analysis of multiple tissues from Atf6α−/− mice revealed that all tissues examined were grossly insensitive to loss of ATF6α. However, combined deletion of ATF6α and the ER cochaperone p58IPK resulted in synthetic embryonic lethality. These findings reveal for the first time that an intact UPR can compensate for the genetic impairment of protein folding in the ER in vivo. The also expose a role for p58IPK in normal embryonic development.
Keywords: ER stress, unfolded protein response, chaperones, protein folding
Introduction
Disruption of protein folding in the endoplasmic reticulum (ER) is sensed by ER-resident transmembrane proteins which initiate signaling cascades collectively known as the unfolded protein response (UPR) [1]. Three pathways, initiated by ATF6α, IRE1α, and PERK, characterize the canonical UPR, and each culminates in the production of a transcriptional activator [2]. IRE1α is conserved amongst eukaryotes and catalyzes splicing of an mRNA encoding the transcription factor XBP1 in vertebrates [3,4,5]. PERK is metazoan-specific, and phosphorylates the translation factor eIF2α to transiently depress general protein synthesis but also to stimulate translation of a subset of mRNAs, including that of the transcription factor ATF4 [6,7,8]. ATF6α is likewise metazoan-specific, and is cleaved by regulated intramembrane proteolysis to liberate an active transcription factor [9,10].
The IRE1α and PERK pathways have roles in embryonic or postnatal development. Deficiency of IRE1α or XBP1 compromises fetal liver development [11,12], and liver-specific rescue of XBP1 results in postnatal exocrine pancreas insufficiency [13]. PERK ablation leads to postnatal loss of insulin-producing pancreatic β cells [14]. While the IRE1α and PERK pathways converge upon transcriptional regulation of genes that improve ER protein folding and processing [3,5,6], both can also regulate genes involved in process unrelated to ER folding such as membrane biosynthesis and lipid metabolism [15,16,17,18].
In contrast to ablation of the IRE1α or PERK pathways, deletion of ATF6α does not result in prenatal or neonatal lethality and Atf6α−/− mice are fertile and grossly normal [19,20]. Microarray experiments have suggested that the direct targets of ATF6α are largely restricted to genes encoding ER chaperones and cochaperones, ER-associated degradation factors, and other genes with direct roles in protein folding and processing [19,21]. Its function is partially compensated by its paralog ATF6β, and chaperone insufficiency appears to underlie the embryonic lethality of Atf6α−/−; Atf6β−/− animals [20,22]. Perhaps because of this redundancy, to date Atf6α−/− mice have only yielded phenotypes in the presence of exogenous superphysiological challenges such as inhibition of N-linked glycosylation, exposure to neurotoxin, exercise, experimental colitis, or maintenance on a high fat diet [23,24,25,26,27]. Thus, the role of ATF6α, if any, in maintaining homeostasis during normal development and physiology is not clear.
The sensitivity of mice lacking ATF6α to inhibition of N-linked glycosylation or experimental colitis is phenocopied in animals lacking the ER-resident DnaJ protein p58IPK/ERDJ6 [23,24]. While this protein was originally identified as a negative regulator of the cytosolic eIF2α kinase PKR [28], and thereafter as a negative regulator of PERK [29,30], it has since been found to contain an N-terminal ER targeting sequence, and p58IPK is localized essentially quantitatively in the ER lumen; no detectable amounts of the protein were found in the cytosol under either basal or ER stress conditions [31]. In the ER lumen, p58IPK interacts with the HSP70 chaperone BiP/GRP78 [31,32,33], as would be expected for a DnaJ-family protein [34]. Mice lacking p58IPK are smaller than wild-type littermates, and develop diabetes attributed to compromised pancreatic β cell function [35]. The similar sensitivity to ER stress-inducing agents in animals lacking p58IPK or ATF6α suggests that a major function of p58IPK is to protect the protein folding capacity of the ER, and raises the possibility that p58IPK might serve in this capacity in tissues in addition to the endocrine pancreas, but that loss of p58IPK function in these tissues is masked by compensatory activation of the UPR.
Materials and Methods
All protocols for animal use were reviewed and approved by the University Committee on Use and Care of Animals at the University of Iowa. Animals were fed standard rodent chow and housed in a controlled environment with 12 hr light and dark cycles. Atf6α−/− [19] and p58IPK−/− [35] animals have been backcrossed into the C57BL/6J strain for >10 generations. For histological analysis of wild-type and Atf6α−/− mice, tissues were fixed in formalin, embedded in paraffin, sectioned, stained with hematoxylin and eosin, and examined in groups matched by age and gender for histological signs of pathology by a veterinary pathologist (A.K.O.) who was blinded to genotype. Multiple regions of each organ were examined. Numbers of mice examined were as follows: female wild-type, aged 7-17 months: 6; female Atf6α−/−, aged 7-17 months: 8; male wild-type, aged 17 months: 3; and male Atf6α−/−, aged 17 months: 5. Timed intercrosses were dated by detection of a mucus plug, which was considered as E0.5; staging was confirmed by embryo size and developmental features. ADRP immunostaining, injections with tunicamycin, and RNA analysis from liver were performed as described [36].
Results
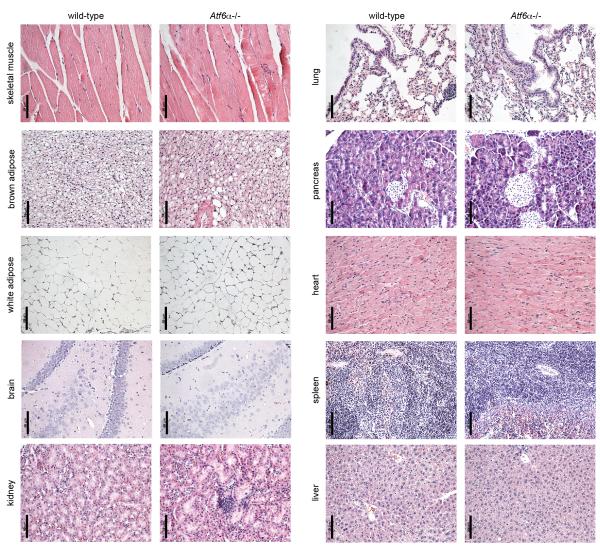
Atf6α−/− mice are histologically normal
To date, the phenotypes reported for Atf6α−/− mice or fish have been associated with exogenous challenges that are likely to compromise ER function and elicit bona fide ER stress [23,24,25,26,27]. While Atf6α−/− mice are viable, fertile, and grossly normal, it had not previously been reported whether deletion of ATF6α compromises the integrity of any tissue. Thus, we collected multiple tissues from wild-type and age-matched Atf6α−/− mice of both genders and of varying ages with an emphasis on older (>1 year) animals, in which a modest phenotype might be most likely to be manifested. These tissues—skeletal muscle, brown and white adipose, brain, kidney, lung, pancreas, heart, spleen, and liver—were fixed, paraffin-embedded, stained, and analyzed by a veterinary pathologist (A.K.O.) who was blinded to genotype. In this analysis, no systematic genotype-dependent abnormalities of any sort were observed in any tissue examined. Although multiple regions of each tissue were analyzed, here we show representative images demonstrating that the structural organization of each of these tissues is intact in Atf6α−/− mice (Figure 1). While these findings do not exclude the possibility of phenotypes for Atf6α−/− mice in tissues not examined, or by non-histological criteria, they are in clear contrast to the effects of manipulation of other UPR pathways on secretory tissues such as pancreas and liver [11,13,37]. Taken together with the susceptibility of Atf6α−/− animals to various exogenous challenges, this result suggests that ATF6α is necessary to protect against superphysiological stressors but is dispensable for normal physiological maintenance. Therefore, to whatever extent normal physiological processes elicit ER stress, the remaining limbs of the UPR are sufficient to protect tissue structure and function.
Figure 1.
No histological abnormalities in Atf6α−/− mice.
The indicated tissues were stained with hematoxylin and eosin and examined for histological signs of pathology by a veterinary pathologist who was blinded to genotype. Multiple regions of each organ were examined. Representative images of the indicated tissues are shown, all from 17 month-old female mice. Results from male mice and from younger female mice were similar. Scale bar = 50 μm.
Embryonic lethality upon deletion of ATF6α and p58IPK
Given that the most dramatic metabolic phenotypes in otherwise viable UPR-compromised animals are observed only upon challenge, we reasoned that a phenotype might be elicited in Atf6α−/− mice by intercrossing them with mice lacking the ER cochaperone p58IPK/ERDJ6. We predicted that deletion of p58IPK would be a sufficient stimulus for eliciting ER stress and hence a phenotype in Atf6α−/− animals in whatever tissues p58IPK is important during normal physiology.
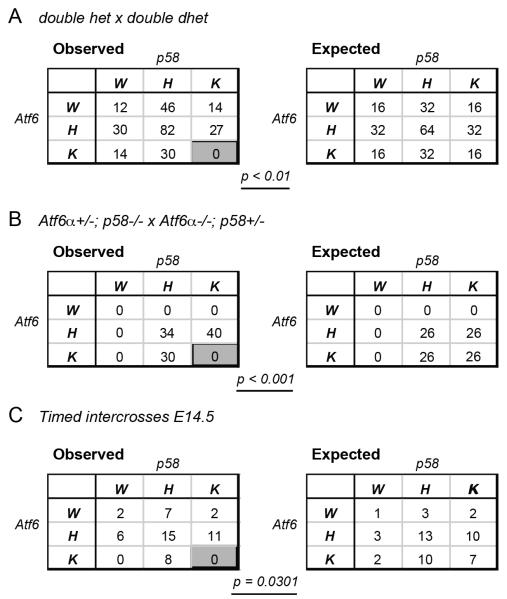
To our surprise, homozygous deletion of both genes resulted in synthetic embryonic lethality prior to embryonic day 14.5. Intercrosses among animals heterozygous for both genes yielded no Atf6α−/−; p58−/− animals among 255 weaned progeny (Figure 2A). Intercrosses among animals homozygous for deletion of one gene and heterozygous for the other similarly produced no Atf6α−/−; p58−/− animals at weaning (Figure 2B). To exclude the possibility that double knockout animals were born but then died prior to weaning, timed intercrosses were established using various combinations of heterozygous and homozygous animals (Table S1), with pups analyzed at embryonic day 14.5. We obtained a sufficient number of embryos to conclude that embryonic lethality of Atf6α−/−; p58−/− animals occurs prior to E14.5 (Figure 2C). We did recover one double-nullizygous pup from among two litters of E9.5 embryos (Table S1), which was in the process of being reabsorbed and which provided only enough embryonic material for genotyping.
Figure 2.
Combined deletion of ATF6α and p58IPK results in embryonic lethality.
(A) Atf6α+/−; p58IPK+/− animals were intercrossed, and the genotypes of the progeny from multiple litters were recorded. W = wild-type; H = heterozygous; K = knockout. The observed genotype frequencies are given in the table on the left, while the expected frequencies are in the table on the right. P-value was determined by Chi-square analysis. Total animal numbers do not correspond because of rounding for the expected group.
(B) Same as (A), except Atf6α+/−; p58−/− males were crossed to Atf6α−/−; p58+/− females and vice-versa.
(C) Animals of various combinations of Atf6α and p58 genotypes were subjected to timed intercross, and embryos were harvested and genotyped at embryonic day 14.5. The observed and expected preponderances of each genotype from all crosses are shown. The outcome of each cross is given in Table S1.
We next tested whether heterozygosity for p58IPK would be sufficient to elicit a phenotype in Atf6α−/− animals. We found that Atf6α−/−; p58+/− animals were grossly normal, were comparably fertile to wild-type animals, and displayed no overt tissue pathology. Histological examination of the pancreas, kidney, and liver—tissues in which a basal or stress-induced phenotype in p58−/− animals has been reported [24,35,38]—of these animals was also normal (data not shown).
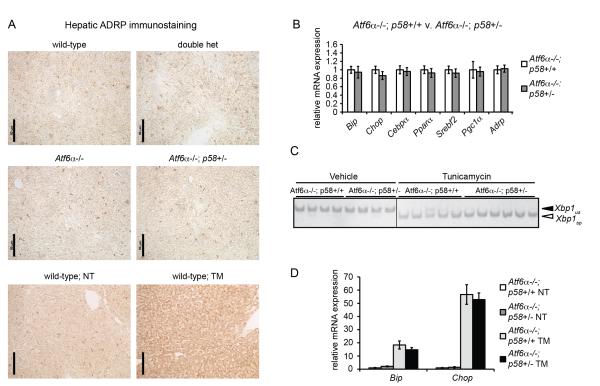
We then examined the consequence of p58IPK heterozygosity in an Atf6α−/− background in the liver. We chose the liver because p58IPK is expressed at the highest levels in that tissue [31,39] and because of our interest in the relationship between ER stress and fatty liver—also known as hepatic steatosis. As exogenous ER stress in the liver elicits dysregulation in hepatic lipid metabolism and consequent fatty liver that is dramatically exacerbated in Atf6α−/− mice [24,40], we hypothesized that loss of one allele of p58IPK might be a sufficient ER stress—in the sensitizing context of loss of ATF6α—to lead to hepatic lipid accumulation. However, in contrast to the stark steatosis seen upon challenge of even wild-type animals with the ER stress-inducing agent tunicamycin (TM), animals lacking both alleles of Atf6α and one allele of p58IPK did not show evidence for accumulated hepatic lipid droplets, as evidenced by immunostaining for the droplet marker protein ADRP (Figure 3A). Similarly, while exogenous ER stress causes a pronounced suppression of lipid metabolic genes in Atf6α−/− animals [24,40,41], Atf6α−/−; p58+/− animals showed no significant changes in the basal expression of such genes compared to Atf6α−/−; p58+/+ animals, nor in expression of the UPR target genes Bip and Chop (Figure 3B). Loss of one allele of p58IPK also did not further sensitize Atf6α−/− animals to ER stress in the liver; these animals did not evince altered Xbp1 mRNA splicing nor enhanced upregulation of Bip and Chop upon challenge with TM (Figure 3C, D).
Figure 3.
Heterozygosity for p58IPK does not sensitize Atf6α−/− animals to ER stress in the liver.
(A) Liver sections from animals of the indicated genotypes were examined for lipid droplet accumulation by immunostaining for ADRP. For comparative purposes, the increase in ADRP immunostaining seen upon challenge of wild-type animals with 1 mg/kg TM for 18h from a separate experiment is shown.
(B) Littermate Atf6α−/−; p58+/+ and Atf6α−/−; p58+/− animals of both genders were sacrificed and the expression of UPR target genes and metabolic genes in the liver was determined by qRT-PCR. None of the genes showed statistically significant differences in expression between genotypes.
(C) Atf6α−/−; p58+/+ and Atf6α−/−; p58+/− animals were challenged with either vehicle or 250 μg/kg TM. 16 hours after treatment RNA was prepared from liver samples and Xbp1 mRNA splicing was assessed by conventional RT-PCR. Unspliced (us) and spliced (spl) forms are indicated. Image is shown in black-to-white inverted form for greater visual clarity.
(D) Same as (C), except the expression of Bip and Chop mRNA was determined by qRT-PCR from liver samples. Differences in expression between genotypes upon TM treatment were not statistically significant.
Discussion
The embryonic lethality of Atf6α−/−; p58−/− animals suggests that p58IPK plays a role in embryonic development, the loss of which can be compensated by ATF6α. Several possibilities could account for this lethality: p58IPK might facilitate the folding of one or a small number of developmentally important proteins, and if its deletion induces ER stress, then ATF6α might compensate by upregulating other chaperoning pathways, including those mediated by other ER DnaJ proteins such as ERDJ3 and ERDJ4 [42,43]. Alternatively, p58IPK might contribute in a less specific way to the overall efficiency of protein folding in one or more tissues; and in the absence of ATF6α, loss of this contribution might result in unameliorated ER stress and either tissue malfunction (e.g., failure to secrete important developmental morphogens) or insurmountable levels of ER stress-induced apoptosis, which would be enhanced when the UPR is compromised. It is also possible that loss of ATF6α produces ER dysfunction and increases the load on the ER, and that an ER lacking p58IPK is ill-equipped to handle such increased load—i.e., that ATF6α acts upstream of p58IPK in inducing ER dysfunction rather than the other way around. However, given that there is as yet no evidence that loss of ATF6α alone induces ER dysfunction in any tissue or cell type yet studied, here or elsewhere, we think this possibility is unlikely. We also cannot formally exclude that the developmental role of p58IPK is exerted through putative regulation of PERK or PKR, but the absence of evidence for detectable cytosolic p58IPK militates against this possibility.
The site of p58IPK embryonic function might be in the nascent pancreas, the development of which takes place from E8.5-E14.5 [44]. However, the pattern of expression of p58IPK in the embryo is unknown, and in the adult it is ubiquitously expressed, suggesting that it might also function broadly in the embryo. Ultimately, a systematic series of timed intercrosses will be necessary to better understand the timing of the lethality, the tissue or tissues affected, and the molecular mechanisms likely at work.
This work is the first to our knowledge to combine deletion of an ER chaperone or cochaperone with that of a UPR signaling molecule in a vertebrate. A key conclusion from the synthetic embryonic lethality of Atf6α−/−; p58−/− animals is that the UPR in general, and the ATF6α pathway in particular, can compensate for ER chaperone insufficiency and reverse a protein folding defect. Although it is possible that ATF6α accomplishes this by regulating other processes that indirectly improve ER function and so reduce the requirement for p58-dependent folding, the fact that p58IPK functions as a cochaperone and that ATF6α is known to regulate chaperone and cochaperone expression makes ATF6α-dependent upregulation of compensatory folding pathways the most likely explanation for survival of Atf6α+/+; p58−/− animals. While this conclusion at first glance seems self-evident, both pharmacological (e.g., tunicamycin, thapsigargin…) and physiological (obesity, exercise…) inducers of ER stress perturb cellular function in many ways, and it is conceivable that the UPR could protect cellular function by other means—for example through regulation of protein synthesis rates, calcium homeostasis, glycosylation efficiency, etc. rather than by a UPR pathway that predominantly regulates the expression of chaperones. Thus, while Atf6α−/− animals have no apparent basal phenotype, compounding their deletion with that of p58IPK elucidates physiological roles for UPR-regulated protein folding pathways.
Supplementary Material
Highlights.
▪ The ER stress sensing molecule ATF6α is dispensable for normal tissue organization
▪ Heterozygosity for p58IPK does not sensitize Atf6α−/− animals to hepatic ER stress
▪ Combined homozygous deletion of ATF6α and the ER cochaperone p58IPK leads to embryonic lethality
▪ An intact UPR can compensate for genetic disruption of ER protein folding in vivo
Acknowledgments
We thank K. E. Berquam-Vrieze for assistance with timed mating experiments and the Tootle and Cornell labs in the Department of Anatomy and Cell Biology at the University of Iowa for sharing equipment. This work was funded by grant R01 DK084058 from the National Institute of Diabetes and Digestive and Kidney Disorders to D.T.R; D.D.M. was supported by an Institutional Postdoctoral Training Grant from the University of Iowa Cardiovascular Center, and J.A.G. was supported by an Institutional Predoctoral T32 Training Grant from the University of Iowa Department of Pharmacology. Funders had no role in study design, data collection or analysis, manuscript preparation, or decision to submit. J.A.G. and D.T.R. conceived and designed the experiments. J.A.G., H.M.T., and D.D.M. performed the experiments. A.K.O. analyzed tissue specimens for pathology. J.A.G. and D.T.R. analyzed data. D.T.R. wrote the paper.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Rutkowski DT, Kaufman RJ. A trip to the ER: coping with stress. Trends Cell Biol. 2004;14:20–28. doi: 10.1016/j.tcb.2003.11.001. [DOI] [PubMed] [Google Scholar]
- [2].Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- [3].Lee K, Tirasophon W, Shen X, Michalak M, Prywes R, Okada T, Yoshida H, Mori K, Kaufman RJ. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 2002;16:452–466. doi: 10.1101/gad.964702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Shen X, Ellis RE, Lee K, Liu CY, Yang K, Solomon A, Yoshida H, Morimoto R, Kurnit DM, Mori K, Kaufman RJ. Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell. 2001;107:893–903. doi: 10.1016/s0092-8674(01)00612-2. [DOI] [PubMed] [Google Scholar]
- [5].Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- [6].Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- [7].Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- [8].Shi Y, Vattem KM, Sood R, An J, Liang J, Stramm L, Wek RC. Identification and characterization of pancreatic eukaryotic initiation factor 2 alpha-subunit kinase, PEK, involved in translational control. Mol Cell Biol. 1998;18:7499–7509. doi: 10.1128/mcb.18.12.7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ye J, Rawson RB, Komuro R, Chen X, Dave UP, Prywes R, Brown MS, Goldstein JL. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell. 2000;6:1355–1364. doi: 10.1016/s1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- [10].Yoshida H, Okada T, Haze K, Yanagi H, Yura T, Negishi M, Mori K. ATF6 activated by proteolysis binds in the presence of NF-Y (CBF) directly to the cisacting element responsible for the mammalian unfolded protein response. Mol Cell Biol. 2000;20:6755–6767. doi: 10.1128/mcb.20.18.6755-6767.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Reimold AM, Etkin A, Clauss I, Perkins A, Friend DS, Zhang J, Horton HF, Scott A, Orkin SH, Byrne MC, Grusby MJ, Glimcher LH. An essential role in liver development for transcription factor XBP-1. Genes Dev. 2000;14:152–157. [PMC free article] [PubMed] [Google Scholar]
- [12].Zhang K, Wong HN, Song B, Miller CN, Scheuner D, Kaufman RJ. The unfolded protein response sensor IRE1alpha is required at 2 distinct steps in B cell lymphopoiesis. J Clin Invest. 2005;115:268–281. doi: 10.1172/JCI21848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Lee AH, Chu GC, Iwakoshi NN, Glimcher LH. XBP-1 is required for biogenesis of cellular secretory machinery of exocrine glands. Embo J. 2005;24:4368–4380. doi: 10.1038/sj.emboj.7600903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Harding HP, Zeng H, Zhang Y, Jungries R, Chung P, Plesken H, Sabatini DD, Ron D. Diabetes mellitus and exocrine pancreatic dysfunction in perk−/− mice reveals a role for translational control in secretory cell survival. Mol Cell. 2001;7:1153–1163. doi: 10.1016/s1097-2765(01)00264-7. [DOI] [PubMed] [Google Scholar]
- [15].Lee AH, Scapa EF, Cohen DE, Glimcher LH. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science. 2008;320:1492–1496. doi: 10.1126/science.1158042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Oyadomari S, Harding HP, Zhang Y, Oyadomari M, Ron D. Dephosphorylation of translation initiation factor 2alpha enhances glucose tolerance and attenuates hepatosteatosis in mice. Cell Metab. 2008;7:520–532. doi: 10.1016/j.cmet.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].So JS, Hur KY, Tarrio M, Ruda V, Frank-Kamenetsky M, Fitzgerald K, Koteliansky V, Lichtman AH, Iwawaki T, Glimcher LH, Lee AH. Silencing of Lipid Metabolism Genes through IRE1alpha-Mediated mRNA Decay Lowers Plasma Lipids in Mice. Cell Metab. 2012;16:487–499. doi: 10.1016/j.cmet.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Wang S, Chen Z, Lam V, Han J, Hassler J, Finck BN, Davidson NO, Kaufman RJ. IRE1alpha-XBP1s Induces PDI Expression to Increase MTP Activity for Hepatic VLDL Assembly and Lipid Homeostasis. Cell Metab. 2012;16:473–486. doi: 10.1016/j.cmet.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Wu J, Rutkowski DT, Dubois M, Swathirajan J, Saunders T, Wang J, Song B, Yau GD, Kaufman RJ. ATF6alpha optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev Cell. 2007;13:351–364. doi: 10.1016/j.devcel.2007.07.005. [DOI] [PubMed] [Google Scholar]
- [20].Yamamoto K, Sato T, Matsui T, Sato M, Okada T, Yoshida H, Harada A, Mori K. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev Cell. 2007;13:365–376. doi: 10.1016/j.devcel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- [21].Adachi Y, Yamamoto K, Okada T, Yoshida H, Harada A, Mori K. ATF6 Is a transcription factor specializing in the regulation of quality control proteins in the endoplasmic reticulum. Cell Struct Funct. 2008;33:75–89. doi: 10.1247/csf.07044. [DOI] [PubMed] [Google Scholar]
- [22].Ishikawa T, Okada T, Ishikawa-Fujiwara T, Todo T, Kamei Y, Shigenobu S, Tanaka M, Saito TL, Yoshimura J, Morishita S, Toyoda A, Sakaki Y, Taniguchi Y, Takeda S, Mori K. ATF6alpha/beta-mediated adjustment of ER chaperone levels is essential for development of the notochord in medaka fish. Mol Biol Cell. 2012;24:1387–1395. doi: 10.1091/mbc.E12-11-0830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Cao SS, Zimmermann EM, Chuang BM, Song B, Nwokoye A, Wilkinson JE, Eaton KA, Kaufman RJ. The Unfolded Protein Response and Chemical Chaperones Reduce Protein Misfolding and Colitis in Mice. Gastroenterology. 2013 doi: 10.1053/j.gastro.2013.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Rutkowski DT, Wu J, Back SH, Callaghan MU, Ferris SP, Iqbal J, Clark R, Miao H, Hassler JR, Fornek J, Katze MG, Hussain MM, Song B, Swathirajan J, Wang J, Yau GD, Kaufman RJ. UPR pathways combine to prevent hepatic steatosis caused by ER stress-mediated suppression of transcriptional master regulators. Dev Cell. 2008;15:829–840. doi: 10.1016/j.devcel.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Wu J, Ruas JL, Estall JL, Rasbach KA, Choi JH, Ye L, Bostrom P, Tyra HM, Crawford RW, Campbell KP, Rutkowski DT, Kaufman RJ, Spiegelman BM. The unfolded protein response mediates adaptation to exercise in skeletal muscle through a PGC-1alpha/ATF6alpha complex. Cell Metab. 2011;13:160–169. doi: 10.1016/j.cmet.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Egawa N, Yamamoto K, Inoue H, Hikawa R, Nishi K, Mori K, Takahashi R. The endoplasmic reticulum stress sensor, ATF6alpha, protects against neurotoxin-induced dopaminergic neuronal death. J Biol Chem. 2011;286:7947–7957. doi: 10.1074/jbc.M110.156430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Usui M, Yamaguchi S, Tanji Y, Tominaga R, Ishigaki Y, Fukumoto M, Katagiri H, Mori K, Oka Y, Ishihara H. Atf6alpha-null mice are glucose intolerant due to pancreatic beta-cell failure on a high-fat diet but partially resistant to diet-induced insulin resistance. Metabolism. 2012;61:1118–1128. doi: 10.1016/j.metabol.2012.01.004. [DOI] [PubMed] [Google Scholar]
- [28].Lee TG, Tang N, Thompson S, Miller J, Katze MG. The 58,000-dalton cellular inhibitor of the interferon-induced double-stranded RNA-activated protein kinase (PKR) is a member of the tetratricopeptide repeat family of proteins. Mol Cell Biol. 1994;14:2331–2342. doi: 10.1128/mcb.14.4.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].van Huizen R, Martindale JL, Gorospe M, Holbrook NJ. P58IPK, a novel endoplasmic reticulum stress-inducible protein and potential negative regulator of eIF2alpha signaling. J Biol Chem. 2003;278:15558–15564. doi: 10.1074/jbc.M212074200. [DOI] [PubMed] [Google Scholar]
- [30].Yan W, Frank CL, Korth MJ, Sopher BL, Novoa I, Ron D, Katze MG. Control of PERK eIF2alpha kinase activity by the endoplasmic reticulum stress-induced molecular chaperone P58IPK. Proc Natl Acad Sci U S A. 2002;99:15920–15925. doi: 10.1073/pnas.252341799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Rutkowski DT, Kang SW, Goodman AG, Garrison JL, Taunton J, Katze MG, Kaufman RJ, Hegde RS. The role of p58IPK in protecting the stressed endoplasmic reticulum. Mol Biol Cell. 2007;18:3681–3691. doi: 10.1091/mbc.E07-03-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Petrova K, Oyadomari S, Hendershot LM, Ron D. Regulated association of misfolded endoplasmic reticulum lumenal proteins with P58/DNAJc3. Embo J. 2008;27:2862–2872. doi: 10.1038/emboj.2008.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Tao J, Petrova K, Ron D, Sha B. Crystal structure of P58(IPK) TPR fragment reveals the mechanism for its molecular chaperone activity in UPR. J Mol Biol. 2010;397:1307–1315. doi: 10.1016/j.jmb.2010.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Qiu XB, Shao YM, Miao S, Wang L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell Mol Life Sci. 2006;63:2560–2570. doi: 10.1007/s00018-006-6192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Ladiges WC, Knoblaugh SE, Morton JF, Korth MJ, Sopher BL, Baskin CR, Macauley A, Goodman AG, Leboeuf RC, Katze MG. Pancreatic {beta}-Cell Failure and Diabetes in Mice With a Deletion Mutation of the Endoplasmic Reticulum Molecular Chaperone Gene P58IPK. Diabetes. 2005;54:1074–1081. doi: 10.2337/diabetes.54.4.1074. [DOI] [PubMed] [Google Scholar]
- [36].Chikka MR, McCabe DD, Tyra HM, Rutkowski DT. C/EBP homologous protein (CHOP) contributes to suppression of metabolic genes during endoplasmic reticulum stress in the liver. J Biol Chem. 2013;288:4405–4415. doi: 10.1074/jbc.M112.432344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Harding HP, Zhang Y, Bertolotti A, Zeng H, Ron D. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol Cell. 2000;5:897–904. doi: 10.1016/s1097-2765(00)80330-5. [DOI] [PubMed] [Google Scholar]
- [38].Datta R, Shah GN, Rubbelke TS, Waheed A, Rauchman M, Goodman AG, Katze MG, Sly WS. Progressive renal injury from transgenic expression of human carbonic anhydrase IV folding mutants is enhanced by deficiency of p58IPK. Proc Natl Acad Sci U S A. 2010;107:6448–6452. doi: 10.1073/pnas.1001905107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Korth MJ, Lyons CN, Wambach M, Katze MG. Cloning, expression, and cellular localization of the oncogenic 58-kDa inhibitor of the RNA-activated human and mouse protein kinase. Gene. 1996;170:181–188. doi: 10.1016/0378-1119(95)00883-7. [DOI] [PubMed] [Google Scholar]
- [40].Arensdorf AM, Dezwaan McCabe D, Kaufman RJ, Rutkowski DT. Temporal clustering of gene expression links the metabolic transcription factor HNF4alpha to the ER stress-dependent gene regulatory network. Front Genet. 2013;4:188. doi: 10.3389/fgene.2013.00188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Yamamoto K, Takahara K, Oyadomari S, Okada T, Sato T, Harada A, Mori K. Induction of liver steatosis and lipid droplet formation in ATF6alpha-knockout mice burdened with pharmacological endoplasmic reticulum stress. Mol Biol Cell. 2010;21:2975–2986. doi: 10.1091/mbc.E09-02-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Shen Y, Hendershot L. ERdj3, a Stress-inducible Endoplasmic Reticulum DnaJ Homologue, Serves as a CoFactor for BiP’s Interactions with Unfolded Substrates. Mol Biol Cell. 2005;16:40–50. doi: 10.1091/mbc.E04-05-0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Lai CW, Otero JH, Hendershot LM, Snapp E. ERdj4 protein is a soluble endoplasmic reticulum (ER) DnaJ family protein that interacts with ER-associated degradation machinery. J Biol Chem. 2012;287:7969–7978. doi: 10.1074/jbc.M111.311290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Jorgensen MC, Ahnfelt-Ronne J, Hald J, Madsen OD, Serup P, Hecksher-Sorensen J. An illustrated review of early pancreas development in the mouse. Endocr Rev. 2007;28:685–705. doi: 10.1210/er.2007-0016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.