Abstract
O-sulfotransferases (OSTs) are critical enzymes in the cellular biosynthesis of the biologically and pharmacologically important heparan sulfate and heparin. Recently, these enzymes have been cloned and expressed in bacteria for application in the chemoenzymatic synthesis of glycosaminoglycan-based drugs. OST activity assays have largely relied on the use of radioisotopic methods using [35S] 3'-phosphoadenosine-5'-phosphosulfate and scintillation counting. Herein, we examine alternative assays that are more compatible with a biomanufacturing environment. A high throughput microtiter-based approach is reported that relies on a coupled bienzymic colorimetric assay for heparan sulfate and heparin OSTs acting on polysaccharide substrates using arylsulfotransferase-IV and p-nitrophenylsulfate as a sacrificial sulfogroup donor. A second liquid chromatography-mass spectrometric assay, for heparan sulfate and heparin OSTs acting on structurally defined oligosaccharide substrates, is also reported that provides additional information on the number and positions of the transferred sulfo groups within the product. Together, these assays allow quantitative and mechanistic information to be obtained on OSTs that act on heparan sulfate and heparin precursors.
Keywords: enzymes, mass spectrometry, bioassays, sulfotransferases, coupled assay, heparin, heparan sulfate
Introduction
Glycosaminoglycans (GAGs) are linear polysaccharides that are found ubiquitously in intracellular and extracellular domains [1]. GAGs are divided among four unique families: heparan sulfate, chondroitin sulfate, keratan sulfate, and hyaluronan. These families are differentiated by composition, structure, and the number of sulfo groups that they carry [2]. These differences lead to highly specific roles in human biology, including but not limited to GAG-protein binding [3,4], structural GAGs [5,6], cell adhesion and cell-cell interactions [7], and morphogenesis [8].
Heparin is the most highly sulfated member of the heparan sulfate family of GAGs and is a GAG of substantial biological interest, primarily because of its binding to many proteins. One of the most important biological functions of heparin is its capacity to bind to the serine protease inhibitor, antithrombin III (AT) [9] and inhibit the coagulation pathway. In the coagulation cascade, prothrombin is enzymatically cleaved by Factor Xa to form thrombin, which in turn, acts on fibrinogen to form a fibrin clot. AT inhibits this pathway by binding to Factor Xa, preventing the formation of thrombin and ultimately the formation of fibrin. The heparin-AT complex binds to Factor Xa with very high affinity (107 M−1 s−1) making this complex a potent inhibitor of the coagulation pathway [10]. Heparin also forms a ternary complex with ATIII and thrombin, inhibiting thrombin activity and preventing blood coagulation [11].
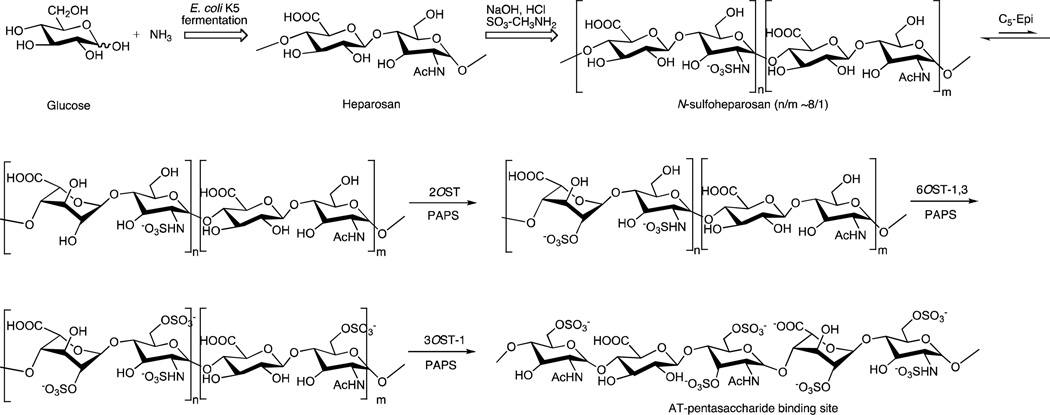
The in vivo biosynthesis of heparin requires enzymatic catalysis resulting in: (1) formation of the heparosan backbone on the serine-linked tetrasaccharide of the serglycin core protein through the action of polysaccharide synthases; (2) N-deacetylase, N-sulfotransferase (NDST)-1 and NDST-2 catalyzed N-deacetylation, N-sulfonation of the glucosamine residues; (3) the formation of iduronic acid residues from glucuronic acid residues by C5-epimerase (C5-epi); (4) the 2-O-sulfonation of iduronic acid residues by 2-O-sulfotransferase (2-OST); and (5) the 6-O-sulfonation by 6-O-sulfotransferases (6-OSTs) and 3-O-sulfonation by 3-O-sulfotransferase-1 (3-OST-1) of glucosamine residues. Commercial heparins are derived from a number of different animal tissues and can vary with regard to sulfonation extent and molecular weight [12].
There is a major concern about the conditions used to isolate and purify many commercial heparins. Heparin production involves steps that occur at the slaughterhouse that are not performed under current good manufacturing practices (cGMP). These steps greatly increase the risk of the introduction of impurities, contamination or adulteration, a clear hazard to the recipients of these heparin-containing products [13]. Such manufacturing pitfalls increase the demand for a safer heparin product produced entirely under cGMP protocols. Our laboratory is investigating a fermentative-chemoenzymatic approach for the preparation of bioengineered heparins.
Under a bioengineering approach to the production of heparin, the heparosan backbone, a repeating →1)-β-d-GlcA(1→4)-β-d-GlcNAc(1→ disaccharide, where GlcA is glucuronic acid and GlcNAc is N-acetylglucosamine, is produced by fermentation of E. coli K5 on glucose and ammonium chloride [14]. Heparosan is then converted to an N-sulfoheparosan, with a major repeating →1)-β-d-GlcA(1→4)-β-d-GlcNS(1→ (where S is sulfo) disaccharide structure, by enzymatic [15] or by chemical means [16]. Enzymatic conditions rely on treating heparosan with NDST enzymes, which N-deacetylate the GlcNAc residue and N-sulfonate the resulting amino group. Chemical conditions begin with harsh basic conditions for N-deacetylation of heparosan followed by chemical N-sulfonation using sulfur trioxide trimethylamine complex. The remaining enzymatic steps are believed to be identical to those in vivo: C5-epimerization of the N-sulfoheparosan converting most of the β-d-GlcA residues to α-l-iduronic acid (α-l-IdoA) residues, followed by the 2-O-sulfonation of the IdoA residues to IdoA2S, the 6-O-sulfonation of GlcNS to GlcNS6S residues, and finally 3-O-sulfonation of a small percentage of the GlcNS6S and GlcNAc to GlcNS3S6S and GlcNAc3S residues, respectively (Fig. 1). The primary differences between the in vivo biosynthesis and the plant scale bioengineering of heparin are in the elongation of the heparosan backbone and the N-sulfonation of the backbone. In vivo, the heparosan backbone is elongated by KfiA, KfiB, KfiC, and KfiD enzymes in Escherichia coli (17) and in animals by Ext enzymes, in which case the backbone is subsequently N-deacetylated and N-sulfonated enzymatically by NDST enzymes (18). In bioengineered heparin, the heparosan backbone is formed by E. coli K5 fermentation, then N-deacteylated with sodium hydroxide and N-sulfonated using sulfur trioxide methylamine. Finally, in both the biosynthesis and fermentative chemoenzymatic synthesis of heparin O-sulfonation is accomplished using O-sulfotransferases (OSTs).
Fig. 1.
Chemoenzymatic synthesis of heparin. Fermentation of E. coli K5 on glucose and ammonia affords heparosan which is then treated with sodium hydroxide to remove over 80% of its N-acetyl groups reduce its molecular weight to afford an N-sulfoheparosan. The N-sulfoheparosan is then converted through a series of enzymatic steps using C5-Epi, 2OST, 6OST-1 and −3 and 3OST-1 to form bioengineered heparin. The structure of an AT-pentasaccharide binding site is shown as the final product. These enzymatic steps closely parallel the final steps within heparin biosynthesis.
A major challenge to prepare a bioengineered heparin has been the development of assays to readily assess the activity of OSTs [19]. Commonly used radioisotopic assays pose problems for routine use in a quality control laboratory of a manufacturing facility [19]. The most accurate and reliable method of analyzing the bioengineered enzyme activity is nuclear magnetic resonance spectroscopy, which is time consuming, requiring added steps for product isolation and purification prior to analysis, and requires sophisticated instrumentation, making it potentially very useful [20, 21], but challenging for the routine assessment of enzyme activity.
Sulfonation reactions are also being increasingly recognized as important in the metabolism of drugs, hormones, carcinogens, neurotransmitters, lipids, and peptides [22]. Essential to understanding these metabolic sulfotransferases is the development of robust enzyme activity assays. Sulfotransferase assays have been the subject of a recent review that reported the most commonly used assay to be based on the measurement by scintillation counting of radioactive sulfate incorporated from PAP35S [19]. While these assays are widely used and very sensitive they are also hazardous, expensive, do not provide information on the position and number of incorporated sulfo groups, and have not been widely used to determine kinetic information.
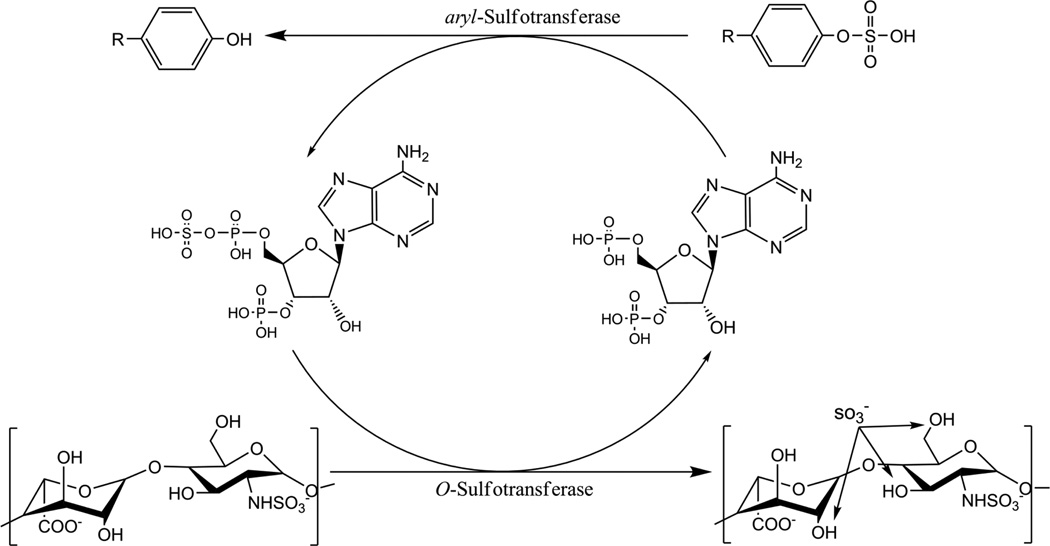
The current study offers two advances in OST assay method development. The first is a real-time colorimetric method for the evaluation of heparin OST activity using a coupled enzyme system, and the ability to obtain quantitative information on OST substrate specificities. In this assay, a recycling enzyme, aryl-sulfotransferase IV (AST-IV), takes the 3’-adenosine 5’-phosphate (PAP) product of the OST reaction and recycles it into the 3’-adenosine 5’-phosphosulfate (PAPS) cofactor (Fig. 2) using an additional sulfate donor, p-nitrophenyl sulfate (PNPS). One of the products of this reaction, p-nitrophenol (PNP), is visible and can be measured spectrophotometrically over time.
Fig. 2.
Schematic outlining the aryl-sulfotransferase/O-sulfotransferase coupled assay. In a system initialized with PAPS, the O-sulfotransferase converts PAPS to PAP and sulfates the GAG. PAP is then recycled to PAPS by aryl-sulfotransferase with p-nitrophenylsulfate (R = NO2) acting as the sulfate donor. The products of the arylsulfotransferase reaction are PAPS, which is reused with the OST enzyme, and p-nitrophenol, PNP, which is measureable spectrophotometrically.
Since this coupled systems offers an indirect measurement, a second method, relying on hydrophilic interaction liquid chromatography (HILIC)-Fourier transform mass spectrometry (FTMS), was developed to assess heparin OST enzyme activity directly based on measuring the products formed using defined substrates of various chain lengths. This lower throughput method is accurate, and does not require workup steps usually required by other mass spectrometric methods. The HILIC- FTMS method is also useful for obtaining mechanistic information about the OSTs and exact location and number of sulfo groups introduced.
Experimental procedures
Materials
Heparosan was produced through the large-scale fermentation of E. coli K5, a capsular K5 antigen corresponding to a polysaccharide comprised of GlcA and GlcNAc in a 1:1 molar ratio [23]. This product, with a molecular weight of nearly 70 kDa, was chemically de-N-acetylated using NaOH and chemically N-sulfonated using a sulfur trioxide trimethylamine complex (Sigma Aldrich, St. Louis, MO) [12]. The resultant N-sulfoheparosan typically has a molecular weight range of 10–16 kDa with 85% N-sulfo groups and 15% residual N-acetyl groups. PAP and PNPS were purchased from Sigma. PAPS was prepared according to previously published methods [24, 25].
The following enzymes and appropriate expression vectors and bacterial expression cells, prepared as previously described [26]. These enzymes include rat AST-IV (EC 2.8.2.1) in pET15 expression vector and BL21 expression cells; human C5-epimerase (NCBI: NM_015554.1), mouse 6OST isoform 1 (NCBI: NM_015818.2), and mouse 6OST isoform 3 (NCBI: NM_015820.3) in pMAL-c2x expression vector and Rosetta-gami-B with GroEL expression cells; hamster 2-OST (GenBank: D88811.1) in pMAL-c2x expression vector and Rosetta-gami-B expression cells; mouse 3OST-1 (NCBI: NM_010474.2) in pET28 expression vector and BL21(DE3)RIL expression cells. AST-IV and 3OST-1 possess N-terminal (His)6 tags. C5-epi, 2-OST, 6OST-1, and 6OST-3 possess N-terminal maltose binding protein tags. A recombinant 6OST-3 prepared in CHO cells was obtained from R&D Systems (Minneapolis, MN) and used where indicated for comparative purposes. Heparin lyases I, II, and III were cloned from the genomic DNA of Flavobacterium heparinum and the expression and purification of the recombinant heparin lyases was carried out in E. coli as previously described [27].
Enzyme preparation, purification, and quantification
Cells on agar plates were selected and grown for 16 h in 5 ml of lysogeny broth media in 14 ml BD Falcon tubes, supplemented with the appropriate antibiotics. The 5 ml culture was then transferred to a baffled 2.8-L Erlenmeyer flask containing 1-L of lysogeny broth media and the appropriate antibiotics. The cultures were incubated at 37°C and shaken at approx. 180 RPM until the solution optical density at 600 nm reached 0.7–0.9 absorbance units. At this point, the flask was transferred to an incubator shaker at 22°C and 180 RPM. After 30 min, the first inducer was added to the culture. If necessary, the second inducer was added after an additional 20 min. The flask was then shaken for 20 h.
After incubation was completed, the 1-L solution was centrifuged using a Sorvall centrifuge from Thermo Scientific (Rockland, IL) at 3500 × g for 30 min at 4°C. The supernatant was discarded and the cell pellet was re-suspended in 20 ml of buffer, containing 25 mM Tris (pH 7.4) and 500 mM NaCl for C5-epimerase, 2-OST, and 6-OST isoforms 1 and 3, or containing 25 mM Tris (pH 7.4), 500 mM NaCl, and 30 mM imidazole for AST-IV and 3-OST isoform 1. The re-suspended solution was sonicated using a Misonix 3000 (Farmingdale, NY) sonicator at 30 W and 50% cycle (15 s on, 15 s off) for a 3 min total on time. The sonicated solution was then centrifuged at 9400 × g for 60 min at 4°C. The supernatant was retained and the cell pellet was discarded.
The supernatant was passed through a Millipore (Billerica, MA) 0.45 µm sterile filter in preparation for affinity isolation and purification. A 20 ml gravity-flow column was washed 3-times with 20 ml distilled water, followed by the loading of 3 ml of Ni-NTA resin from GE Healthcare (Piscataway, NJ) or 5 ml of twice with 15 ml of washing buffer, containing 25 mM Tris (pH 7.4) and 500 mM NaCl for amylose, or containing 25 mM Tris (pH 7.4), 500 mM NaCl, and 30 mM imidazole for Ni-NTA. The solutions containing the free enzymes were then passed through the resin, followed by clearing of unbound material from the resin with an additional 10 ml of washing buffer. Finally, the bound enzymes were released from the resins using elution buffer, containing 25 mM Tris (pH 7.4), 500 mM NaCl, and 40 mM maltose for amylose, or containing 25 mM Tris (pH 7.4), 500 mM NaCl, and 300 mM imidazole for Ni-NTA.
Following elution, the enzymes were buffer-exchanged from Tris-buffer containing high concentrations of imidazole or maltose to standard phosphate buffered saline (pH 7.0) by centrifugation in Millipore (Ipswich, MA) Centrifugal Filter Units with a molecular weight cutoff of 3000 Da. Enzyme solutions were centrifuged at 3500 × g and re-suspended in phosphate buffered saline. Solution protein concentrations were determined by the bicinchoninic acid assay, purchased from Thermo Scientific (Rockland, IL), against a bovine serum albumin standard. Protein purity was determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis: enzyme solution was denatured at 100°C, mixed with loading dye in a 1:1 ratio, and loaded into 4–15% polyacrylamide gels purchased from Bio-Rad Life Sciences (Hercules, CA). Gels were run at 30 volts for 90 min in running buffer (25 mM Tris, 192 mM glycine, 0.1% sodium dodecyl sulfate). Molecular weights were verified by Precision Plus Protein™ All Blue Standards purchased from Bio-Rad.
Preparation of defined substrates for HILIC-MS method
A partial digestion of heparosan polysaccharide was carried out to ~ 40% completion over 17 min at room temperature [28, 29]. Heparosan (23 mg at 1 mg/ ml) was mixed with 0.28 units (1 ml) of purified heparin lyase III in 50 mM sodium phosphate (pH 7.6). The total reaction volume was 528 ml. The digestion was stopped by heating at 100°C for 10 min. The reaction was then repeated at a 600 mg scale of heparosan. The digested polysaccharide was resolved on a BioGel P-10 column, which was eluted with a buffer containing 0.2 M NaCl at a flow rate of 2 ml /h. The fractions were visualized by UV spectroscopy at 232 nm. The resultant oligosaccharides were desalted on a BioGel P-2 column. N-sulfoheparosan (250 mg), containing 100% GlcNS, was also digested following the same reaction protocol, and the reaction products were also resolved on a BioGel P-10 column and desalted on a BioGel P-2 column. The substrates were then characterized to confirm oligosaccharide structure and purity. Analysis was performed by mass spectrometry and nuclear magnetic resonance spectroscopy. The resulting heparosan and N-sulfoheparosan oligosaccharide substrates ranged from degree of polymerization (dp) 2–10.
Preparation of [34S]-PAPS for HILIC-MS method
[34S]PAPS was synthesized enzymatically using a modified method from similar previously described syntheses of [35S]PAPS by substituting Na234SO4 for Na2SO4 (from ISOFLEX USA) [30, 31]. The reaction included 90 mM adenosine triphosphate, 100 mM MgCl2, 1 M LiCl, 0.8 mg/ml pyrophosphatase, 0.8 mg/ml Kluyveromyces lactis expressed arylsulfotransferase, 0.8 mg/ml adenosine-5’-phosphosulfate-3’-phosphokinase, and 50 mM Tris-HCl at pH 8.0. The reaction was incubated at 30°C for 6 h. The [34S]PAPS product was analyzed using HPLC (polyamine II column, YMC America, Inc.) as follows: 100% water for 10 min, followed with a linear gradient of 0–100% of 1 M KH2PO4 for 30 min, followed by 100% 1 M KH2PO4 for 15 min at a flow rate of 1 ml min−1 with detection at 254 nm. Purification of PAPS was achieved on a DEAE-Sepharose fast flow column (GE Healthcare; 1.5 × 60 cm). The diethylaminoethyl column was washed with water, and PAPS was eluted with a gradient of 0– 500 mM NaCl at 5.0 ml min−1 for 200 min. Fractions containing PAPS as determined by HPLC on a polyamine II column were pooled and stored at −80°C. The purity of the [34S]PAPS product was assessed by MS analysis and determined to be 90%+ with less than 10% PAP contamination) (Fig. S1, Electronic Supplementary Material).
Colorimetric method for measuring enzyme activity
OST activity assays were carried out, unless otherwise specified, in transparent, U-bottom, 96-well plates purchased from Greiner Bio-One (Monroe, NC). A typical reaction volume of each well was 250 µL with the following conditions: 125 µl PNPS (10 mM in phosphate buffered saline, pH 7.0), 25 µl N-sulfoheparosan (1 mg/ml), 25 µl of an OST enzyme (400–800 µg/ml), 25 µl AST-IV (2–3 mg/ml), 25 µl PAPS or PAP (250–500 µM), and 25 µl C5-epi (250–500 µg/ml) or PBS. The concentrations of the substrates and the enzymes were then varied on a case-by-case basis. The 96-well plate was incubated at 37°C in a temperature controlled SpectraMax plate reader (Molecular Devices, Sunnyvale, CA). Kinetic plots of p-nitrophenol (PNP) formation were generated by sample measurements at 400 nm at even time intervals over at least 30 min. Using the extinction coefficient for PNP at pH 7.0 (at 400 nm, εPNP = 10,500 M−1cm−1), output absorbance readings from the plate reader were converted to µM concentrations of PNP and the data were re-plotted in terms of [PNP] as a function of time. Initial enzyme velocities (vi) were calculated as the change in [PNP] over a linear segment of the plot. Vi values, corresponding to varied PAP, PAPS, or enzyme concentrations, were fit to Michaelis-Menten and Eadie-Hofstee functions to yield the kinetic parameters.
Reaction conditions for HILIC method
The activities of OSTs were assessed by incubating 5.0 µg of purified 2-OST with 2.5 µg of N-sulfoheparosan derived decasaccharide and 2 µg PAPS in 22.5 µl of 50 mM MES buffer pH 7.0. The reaction occurred over 24 h with aliquots removed at 0, 1, 2, 3, 4, 7, and 24 h. Each aliquot was quenched by adding 50 µl of 100% acetonitrile.
MS analysis
The products were analyzed by HILIC using a 2.0 × 50 mm (Torrance, CA) The products were analyzed by HILIC using a 2.0 × 50 mm Luna HILIC column (Phenomenex, Torrance, CA) coupled to an electrospray ionization LTQ-Orbitrap XL FTMS (Thermo Fisher Scientific, San-Jose, CA). The mobile phase A was 5 mM ammonium acetate prepared with HPLC grade water. Mobile phase B was 5 mM ammonium acetate prepared in 98% HPLC grade acetonitrile with 2% HPLC grade water. An Agilent 1200 HPLC binary pump was used to deliver the gradient from 10% A to 80% A over 8 min at a flow rate of 250 µl/min after injecting the samples. The optimized MS parameters, used to prevent in-source fragmentation, included a spray voltage of 4.2 kV, a capillary voltage of −40 V, a tube lens voltage of −50 V, a capillary temperature of 275 °C, a sheath flow rate of 30, and an auxiliary gas flow rate of 6. External calibration of mass spectra routinely produced a mass accuracy of better than 3 ppm. All FT mass spectra were acquired at a resolution 60,000 within a 400–2000 Da mass range.
Results and discussion
Approach
Each OST has two substrates, the sulfo donor PAPS and a phenol or a carbohydrate hydroxyl acceptor. We initially examined the activity of the recycling enzyme, AST-IV, which is critical for transferring the sulfo group from PNPS to PAP to form the PAPS cofactor. Through the action of AST-IV, therefore, the bienzymic system can be used to assess conversion of PNPS into the visibly detectable PNP. AST coupled assays were first reported on NodST, an enzyme that catalyzes the formation of 6-O-sulfochitobiose [32, 33, 34]. These studies also provided an indirect measurement of NodST’s Michaelis-Menten kinetics. Assaying the heparin OSTs adds several additional levels of complexity. First, these enzymes act on various substrates, ranging from oligomers to polymers, which have multiple non-identical acceptor sites. For example, a polysaccharide substrate of molecular weight 10,000 might have ~20 glucosamine residues, some substituted with N-acetyl groups and some with N-sulfo groups, and ~20 uronic acid residues, some consisting of GlcA and some IdoA. In addition, the reactivity of these residues might differ whether they are in the center of the polysaccharide chain, at the non-reducing end or at the reducing end. Finally, the sequence context might have a profound effect on whether or not an OST acts and to what extent.
Coupling AST-IV to OST catalysis was then tested with all three classes of site-specific glycan sulfotransferases; 6OST-1 and 6OST-3, 2OST, and 3OST-1. The 6OSTs are structurally the simplest heparin biosynthetic enzymes, and are believed to act on unmodified chains of simple structures, i.e., heparosan or N-sulfoheparosan, or slightly more modified disaccharides, i.e., 2-O sulfonated N-sulfoheparosan, with different reactivities. The 6OST activity assay was coupled to AST-IV to recycle PAPS, maintaining the PAP concentration low to prevent product inhibition [35]. 2OST was examined on N-sulfoheparosan, C5-epi-treated N-sulfoheparosan, and several defined oligosaccharide substrates. Finally, 3OST-1 was studied on a highly modified substrate, heparan sulfate. Each heparin OST assay was first performed on polysaccharide substrates using an AST-IV coupled colorimetric assay to measure activity. This was then followed by the determination of OST activity on defined oligosaccharide substrates followed by HILIC-FTMS analysis. It is important to note that while all of the E. coli expressed recombinant OSTs used in this study show good specificity and are catalytic, a fairly high enzyme to substrate ratio is required. This may be the result of reduced activity do to their truncation (missing their transmembrane domain), the presence of fusion proteins (with either (His)6- or maltose-binding protein-tags), or the lack of glycosylation (present in the natural OSTs).
AST-IV kinetics
Rapid OST kinetic analysis requires the coupled, bienzymic recycling activity of AST-IV, which must not be rate limiting. Therefore, we first examined the kinetics of AST-IV using PNPS and PAP as substrates. At concentrations of PNPS and PAP of 5.0 and 0.25 mM, respectively, the rate of PNP release was linear up to 8.5 µg/ml AST-IV (Fig. S2 (A), Electronic Supplementary Material). The enzyme followed conventional Michaelis-Menten kinetics with a Km for PAP of 25.3 µM with 5 mM PNPS (Fig. S2 (B), Electronic Supplementary Material). This value is similar to that found in the literature [34, 36]. The Vmax for PAP as substrate was 165.5 nmol/min/mg enzyme at 5 mM PNPS, similar to literature values [37].
Assays for 6OST-1 and 6OST-3
Initial experiments revealed that 6OST-1 and 6OST-3 showed no measurable activity on heparosan using the coupled colorimetric assay, consistent with the literature [38] and unsurprising considering the low percentage of N-acetylated and 6-sulfonated disaccharides present in heparin [16]. However, both enzymes were active on heparan sulfate and N-sulfoheparosan, with activity on the former at least 2-fold higher than that on the latter, indicating that the presence of the 2-O-sulfate and IdoA residues are favorable for 6OST activity (Fig. S3, Electronic Supplementary Material). This reactivity was confirmed by 1H-nuclear magnetic resonance spectroscopy (Fig. S4 (A), Electronic Supplementary Material). Furthermore, disaccharide analysis of the action of 6OST-3 on a partially 2-O-sulfated heparan sulfate revealed that the enzyme strongly favored the N-sulfo, 2-O-sulfo disaccharides over the unsulfated and N-sulfo disaccharides (Fig. S4 (B), Electronic Supplementary Material).
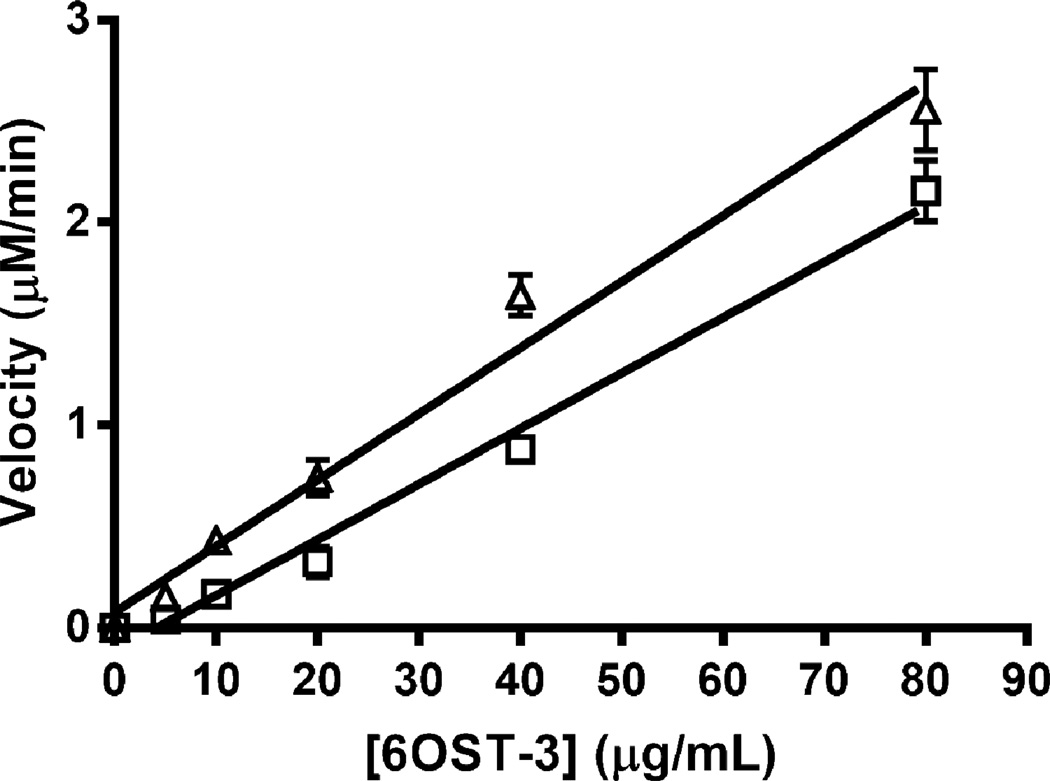
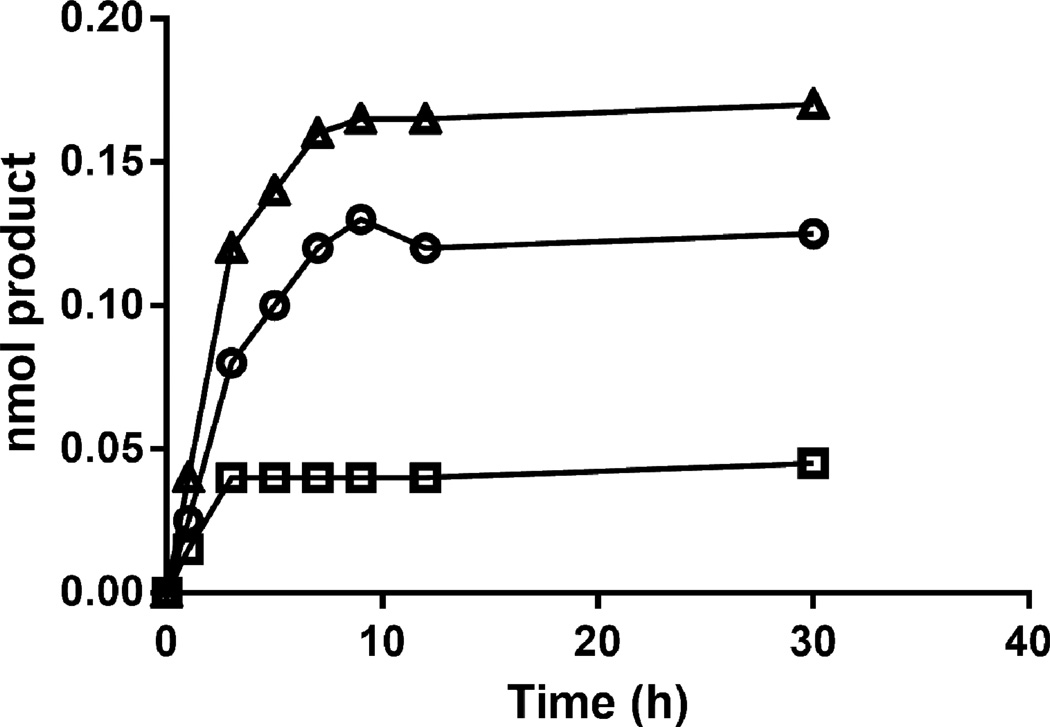
We assessed the rate-limiting step in the bienzymic assay with AST-IV to obtain more quantitative kinetics information on the 6-OSTs. At PAPS and N-sulfoheparosan concentrations of 75 µM and 100 µg/ml, respectively, increasing the 6OST-3 activity up to 80 µg/ml, the highest 6OST-3 concentration tested, resulted in a linear increase in reaction rate (Fig. 3). Hence, under conditions used in this work, AST-IV was not rate limiting. The activity of 6OST-3 on N-sulfoheparosan could be increased by pre-incubating the enzyme with C5-epi (Table 1 and Fig. 4), suggesting a preference for the action of this isoform at GlcNS residues in close proximity to IdoA residues.
Fig. 3.
Effect of 6OST-3 concentration on rate of 6-O-sulfonation with (Δ) and without (□) pre-incubation with C5-epi. The polysaccharide substrate concentration was kept between 0.1 and 0.25 mg/mL and the enzyme substrate ration was < 1:1.
Table 1.
Initial reaction rates for O-sulfotransferases on N-sulfoheparosan.
| Reaction Conditions | Velocity (nmol/min mg OST) |
|---|---|
| 2OST | 0.00 ± 0.85 |
| 2OST & C5-Epi | 1.63 ± 0.80 |
| 6OST3 | 45.1 ± 1.04 |
| 6OST3 & C5-Epi | 86.4 ± 0.89 |
Fig. 4.
Time course comparison of the action of 6OST-3 and 2OST on N-sulfoheparosan is shown in the presence and absence of C5-Epi. 2OST only (○); 2OST, C5-epimerase (□); 6OST-3 only (Δ); 6OST-3, C5-epimerase (∇).
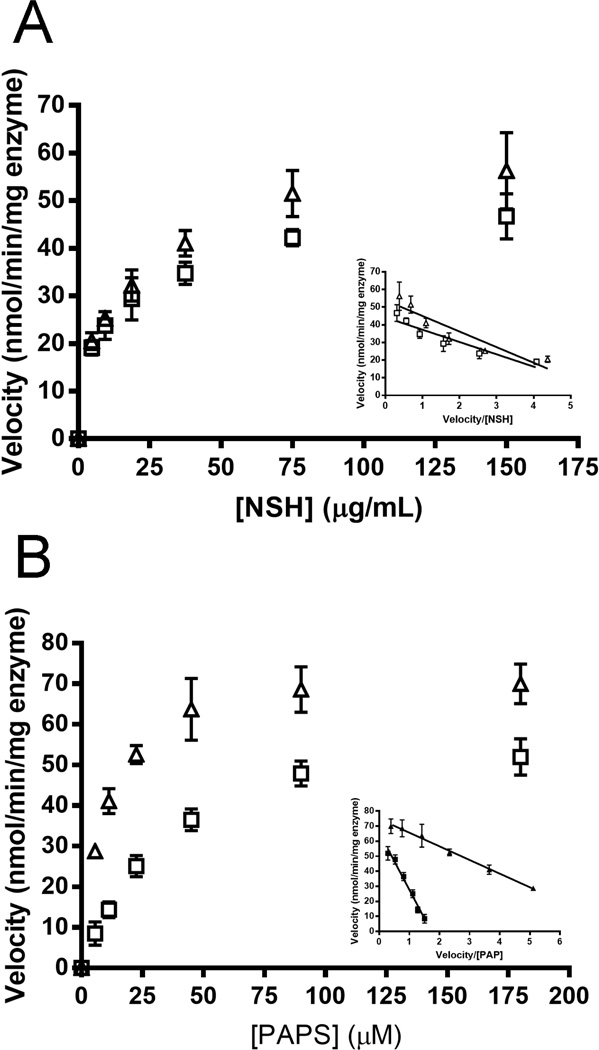
For polysaccharide substrate the enhanced reactivity of 6OST-3 due to the C5-epi was not evident (Fig. 5A). Specifically, in the presence of 80 µg/ml 6-OST 3, the KM and Vmax values for N-sulfoheparosan were 13.8 µg/ml and 51 nmol/(min mg enzyme), respectively. On the C5-epimerized N-sulfoheparosan as polysaccharide substrate, the Km and Vmax values were 14.1 µg/ml and 61.0 nmol/(min mg enzyme), respectively. While the influence of the C5-epi was marginal on the kinetics of 6OST-3 on the polysaccharide substrate, the kinetics with the PAPS substrate was strongly influenced by the C5-epi (Fig. 5B). Specifically, the Km of PAPS was approx. 4-fold lower in the presence of the C5-epi (8.3 vs. 33 µM, respectively), while the Vmax values for PAPS were similar at 73.5 nmol/(min mg enzyme) or 62.5 nmol/(min mg enzyme) with or without the C5-epi, respectively.
Fig. 5.
Michaelis-Menten kinetics (Eadie-Hofstee plot as Inset) of 6OST-3 on N-sulfoheparosan. (A) As a function of N-sulfoheparosan concentration with (Δ) and without (□) preincubation with C5-epi. (B) As a function PAPS concentration with (Δ) and without (□) preincubation with C5-epi.
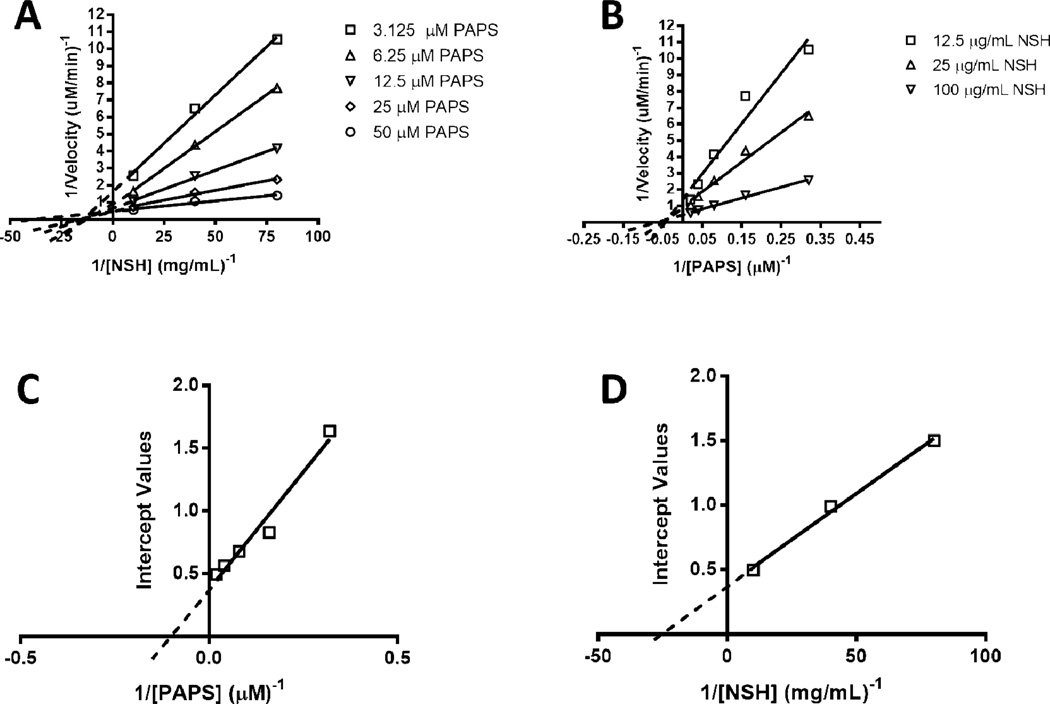
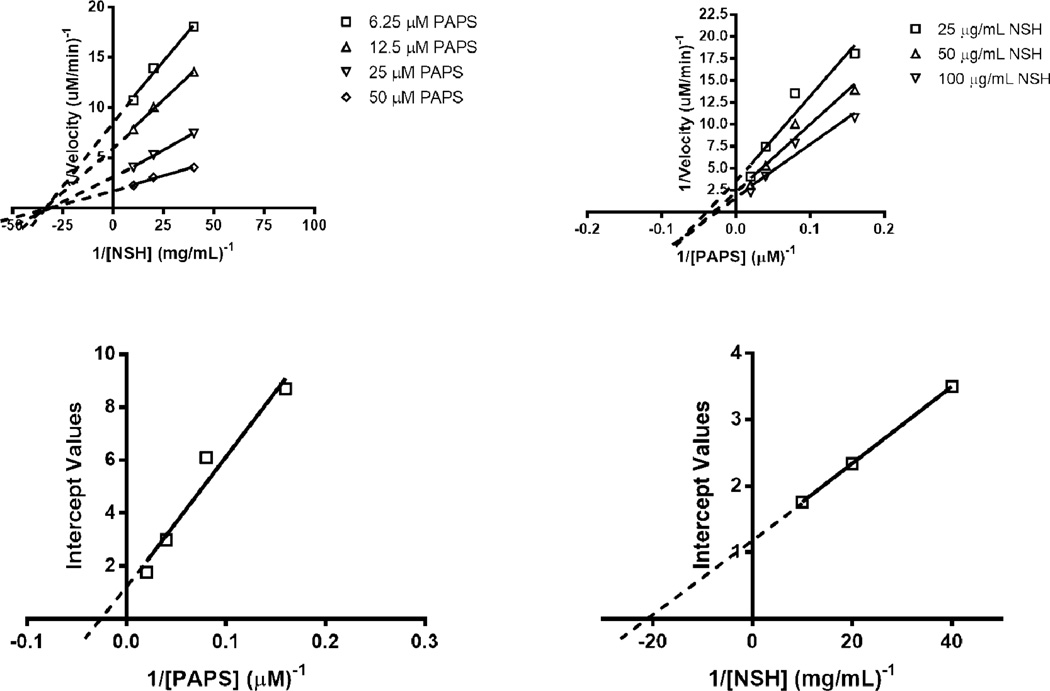
To further understand the influence of C5-epimerization on 6OST-3 activity, two-substrate Michaelis-Menten kinetic analysis was performed by varying PAPS (5–40 µM) and N-sulfoheparosan (12–100 µg/ml) concentrations, with and without 3 h pre-incubation of 6OST-3 with C5-epimerase. Under conditions where N-sulfoheparosan was preincubated with C5-epi, the primary Lineweaver-Burk plots suggested ternary complex kinetics, represented by coincident intersection (Figs. 6 A and B). The secondary replots against the intercept values (Figs. 6 C and D, Table 2) afforded , kcat = 0.2 s−1, and a kcat/KM,PAPS = 2.04 × 104 s−1 M−1 . Under conditions without C5-epi present, ternary complex mechanism kinetics was also observed (Figs. 7 A and B). Secondary replots against the intercept values (Figs. 7C and D, Table 2) afforded , kcat = 0.04 s−1 , and a kcat/KM,PAPS = 1.02×103 s−1 M−1.
Fig. 6.
Dual-substrate kinetics of 6OST-3 on C5-epi treated N-sulfoheparosan. (A) Primary Lineweaver-Burk plot with respect to [N-sulfoheparosan]−1. (B) Primary Lineweaver-Burk plot with respect to [PAPS]−1. (C) Secondary replot of intercepts against [PAPS]−1. (D) Secondary replot of intercepts against [N-sulfoheparosan]−1.
Table 2.
Detailed kinetic parameters of 6OST-3
| Substrate | KM,PAPS(µM) | KM,NSH (µg/mL) | kcat (s−1) | kcat/KM,PAPS (s−1M−1) | KS,PAPS(µM) | KS,NSH (µg/mL) |
|---|---|---|---|---|---|---|
| Epimerized N-sulfoheparosan | 9.8 | 37 | 0.2 | 2.04×104 | 17.5 | 66 |
| N-sulfoheparosan | 39.2 | 41 | 0.04 | 1.02×103 | 19.2 | 3 |
Fig. 7.
Dual-substrate kinetics of 6OST-3 in the absence of C5-epi treated N-sulfoheparosan. (A) Primary Lineweaver-Burk plot with respect to [N-sulfoheparosan]−1. (B) Primary Lineweaver-Burk plot with respect to [PAPS]−1. (C) Secondary replot of intercepts against [PAPS]−1. (D) Secondary replot of intercepts against [N-sulfoheparosan]−1.
Based on the intersection point of the primary plots (Figs. 6 A and B, Figs. 7 A and B), apparent dissociation constants for the PAPS and NSH substrates with 6OST-3 could be calculated. For PAPS, the apparent dissociation constant (Ks,paps) is nearly identical between both epimerized and non-epimerized N-sulfoheparosan (~ 15–20 µM), whereas the apparent dissociation constant for epimerized and non-epimerized N-sulfoheparosan are distinct (KS,EPINSH = ~66 µg/ml, KS,NSH = ~3 µg/ml). This suggests that 6OST-3 follows a kinetic mechanism wherein an ordered ternary complex is formed, with PAPS first binding to the enzyme followed by the binding of the N-sulfoheparosan. Following the transfer of the sulfo group, the 6-O sulfonated N-sulfoheparosan is released followed by the PAP product. In order to verify this order, the binding of 6OST-3 to N-sulfoheparosan was tested in the absence of PAPS using surface plasmon resonance (SPR). As shown in Fig. S5 (Electronic Supplementary Material), 6OST-3 was incapable of binding to N-sulfoheparosan in the absence of PAPS, which confirms the ordered ternary complex mechanism.
Structurally defined N-sulfoheparosan oligosaccharide substrates were employed to investigate further the reactivity and substrate specificity of 6OST-3. These experiments initially used the commercial recombinant mouse enzyme expressed in Chinese Hamster Ovary (CHO) cells (R&D Systems). The sequence comparison for the commercial 6OST-3 differed slightly from the E. coli expressed enzyme prepared in our laboratory (Fig. S6, Electronic Supplementary Material). In addition, this enzyme was glycosylated by the CHO cells in which it had been prepared. The CHO cell expressed 6OST-3 showed greater specific activity but identical specificity as the E. coli expressed 6OST-3 on the substrates tested. In these studies [34S]PAPS, including the stable sulfur isotope (Fig. S1, Electronic Supplementary Material), replaced previously used [32S]PAPS to improve the sensitivity of the method as the product masses would increase by 2.0159 amu for each sulfo group incorporated [39].
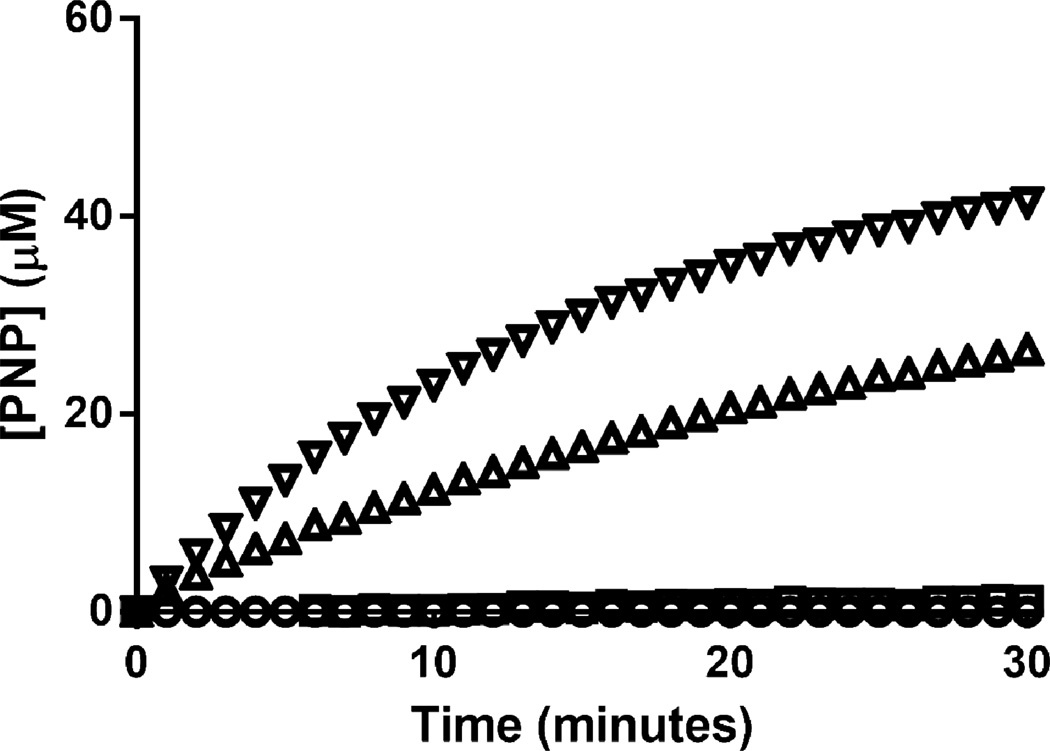
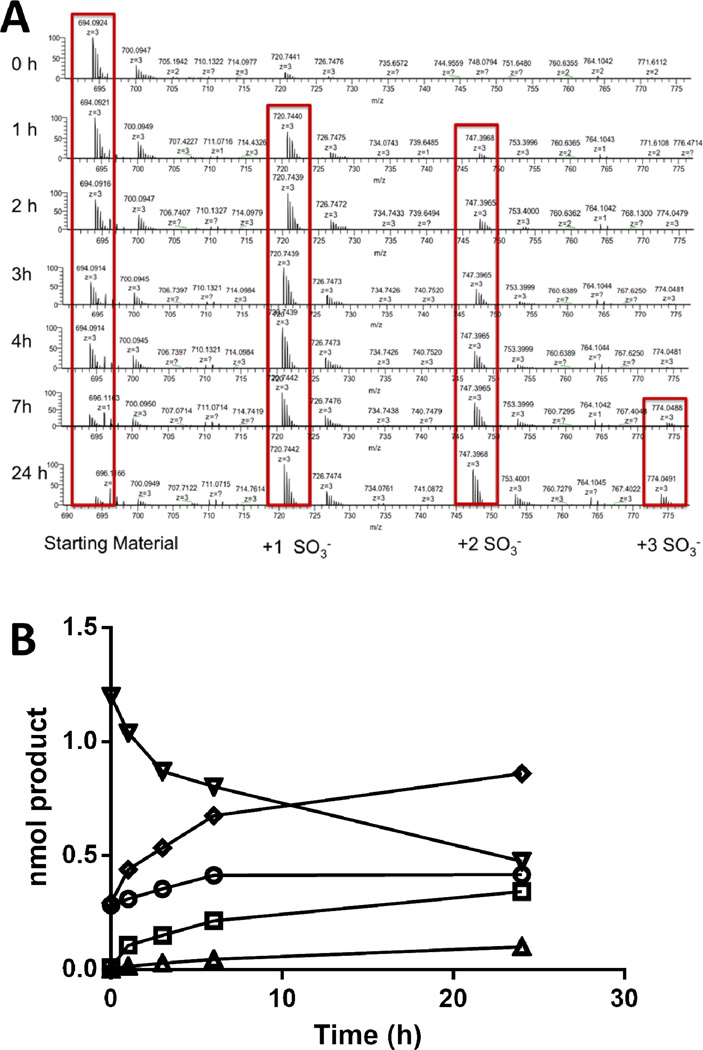
Activity on each individual substrate was assessed by incubating the commercial 6OST-3, or lab prepared 6OST-3, with 50 µM of heparosan hexasaccharide, N-sulfoheparosan tetrasaccharide, N-sulfoheparosan hexasacharide, N-sulfoheparosan octasaccharide, or N-sulfoheparosan decasaccharide, and 120 µM [34S]PAPS in 50 mM MES buffer pH 7.0 for a total reaction volume of 100 µl. Over 24 h, 6 µl aliquots were removed at specific time points. Each aliquot was added to 15 µl of 100% acetonitrile to quench the reaction. The samples were then analyzed and the sum of the sulfonated products was plotted. The sulfonation of an N-sulfoheparosan tetrasaccharide over time is presented in Fig. 8. The addition of one sulfo or two sulfo groups was monitored and the initial rates were converted into molar activity.
Fig. 8.
The 6OST-3 catalyzed sulfonation of N-sulfoheparosan to form one (○) or two (□) sulfo groups, total product formed (Δ), as determined by HILIC-MS.
A summary of the activities based on varying chain lengths of substrate is presented in Table 3. No activity was observed on the heparosan hexasaccharide, consistent with the aforementioned lack of 6OST activity on heparosan. The N-sulfo tetrasaccharide was a poor substrate, while the N-sulfo hexasaccharide, N-sulfo octasaccharide, and N-sulfo decasaccharide showed similar activities suggesting a minimum chain length requirement. The crystal structure of 6OST-3 has not been determined, and hence, the binding sites are unknown. However, these data might help to clarify its substrate specificity.
Table 3.
Substrate specificity of commercial 6OST-3 against NS oligosaccharides.
| Substrate | Specific Activity (nmol product/(min mg protein) |
|---|---|
| NS tetrasaccharide | 0.73 |
| NS hexasaccharide | 11.82 |
| NS hexasaccharide (Lab Prep. 6OST-3) | 10.94 |
| NS octasaccharide | 19.07 |
| NS decasaccharide | 14.01 |
| Heparosan hexasaccharide | No Reaction (<0.01) |
Assays for 2OST
As with 6OSTs, we determined 2OST to be rate limiting in the coupled reaction with AST-IV (data not shown). 2OST showed no measurable activity on either heparosan or N-sulfoheparosan. However, addition of C5-epimerase, resulted in the introduction of IdoA residues into N-sulfoheparosan, and formation of measurable activity of 2OST (Fig. S3, Electronic Supplementary Material), as confirmed using both disaccharide compositional analysis (Fig. S7, Electronic Supplementary Material) and nuclear magnetic resonance spectroscopy (Fig. S4 (A), Electronic Supplementary Material). Treatment of N-sulfoheparosan with C5-epimerase and 2OST afforded new peaks in the 1H-nuclear magnetic resonance spectroscopy at 3.9, 4.2, 4.3 and 5.2 ppm, corresponding to signals associated with IdoA2S [12]. These results are consistent with the reported preference of 2OST for IdoA containing substrates [40].
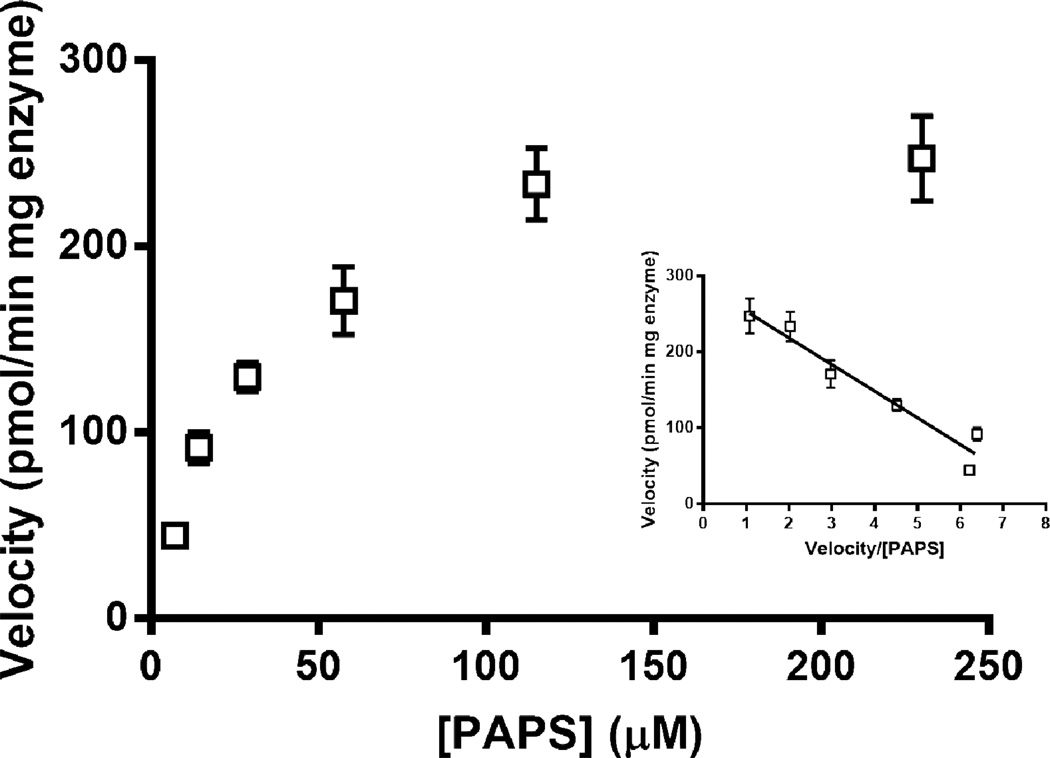
We proceeded to evaluate the activity of 2OST on an epimerized N-sulfoheparosan substrate. The concentrations of the GAG and OST were increased above those used in the 6OST assay to generate a robust response. Under varied PAPS concentrations, maintaining a constant epimerized N-sulfoheparosan concentration of 0.5 mg/ml and a 2-OST concentration of 0.25 mg/ml, the Vmax and KM values were calculated to be 35.1 µM and 288 pmol/min mg enzyme, respectively (Fig. 9). These values were similar to those reported in literature [41] for 2OST activity on completely de-O-sulfated heparin (CDSHP), a substrate that lack all O-sulfonations, but possesses the IdoA residues and N-sulfonations critical for 2OST activity.
Fig. 9.
Michaelis-Menten kinetics (Eadie-Hofstee plot as Inset) of 2OST on completely de-O-sulfonated heparin.
Finally, we examined 2OST activity on the structurally defined N-sulfoheparosan decasaccharide substrate to confirm 2OST activity on GlcA residues. The N-sulfoheparosan decasaccharide substrate showed a peak in its mass spectra at m/z 694.08 ([M-3H]3-, z = −3, MW 2085.2886) that disappeared over time with the appearance of a mono-2-O-sulfo-N-sulfoheparosan decasaccharide product peak at m/z 720.739 ([M-3H]3-, z = −3, MW 2165.2454) after 1 h, di-2-O-sulfo-N-sulfoheparosan decasaccharide product peak at m/z 747.397 ([M-3H]3-, z = −3, MW 22.45.2022) after 1 h, and tri-2-O-sulfo-N-sulfoheparosan decasaccharide product peak at m/z 774.049 ([M-3H]3-, z = −3, MW 2325.159) after 7 h (Fig. 10A). The peak area of each sulfonated product was determined and compared with the peak area of the known concentration of substrate to calculate the concentration of each product as a function of time (Fig. 10B), assuming similar ionization efficiencies. The sum of the sulfonated products was also plotted and its linear region (up to 5 min) was used to estimate an enzyme activity of 0.33-pmol product formed·min−1, with a specific activity of 131-pmol product formed·min−1·mg−1 protein. It is important to note that loss of 0.1–10% of sulfo groups can take place through in source fragmentation increasing with the level of sulfo group substitution. Since LC-MS analysis affords different retention times for oligosaccharides with differing levels of sulfation, in source fragmentation can be clearly distinguished from an analyte containing oligosaccharides having different sulfation levels.
Fig. 10.
(A) Time course of 2-O-sulfonation of N-sulfo decasaccharide. The increasing amounts of mono-, di- and tri-sulfonated products were calculated from the HILIC-MS data. (B) Time course of N-sulfoheparosan (∇) 2-O-sulfonation, from the addition of one sulfo (○), two sulfo (□), or three sulfo groups (Δ), total product formed (◊), as determined by HILIC-MS.
Assays for 3OST-1
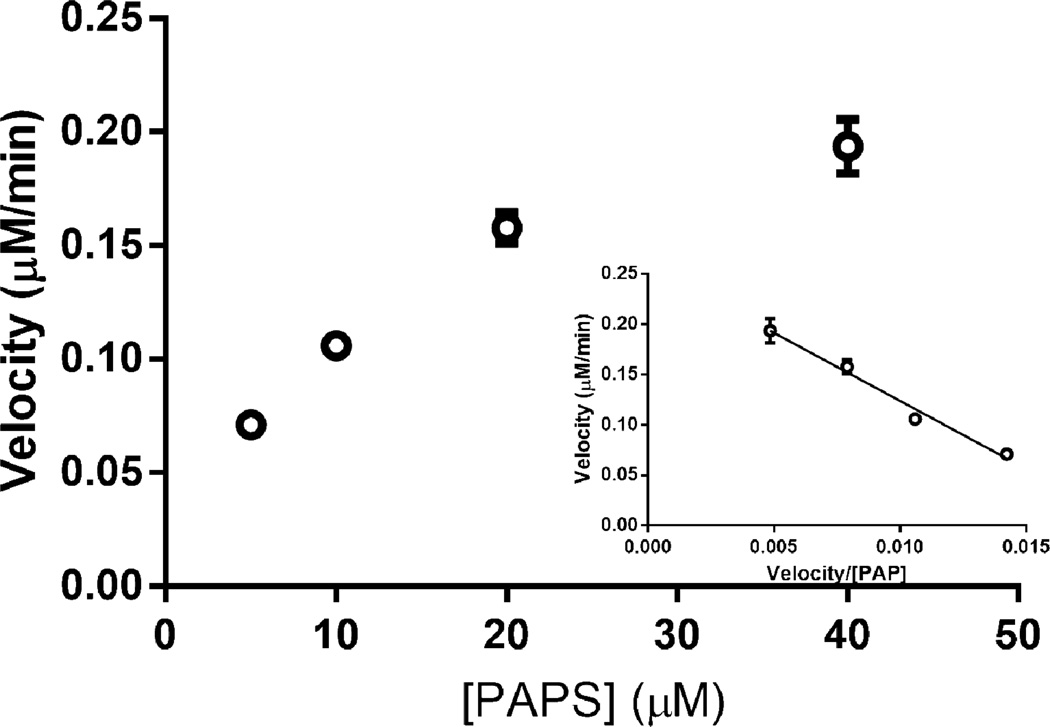
3OST-1 is responsible for installing the AT-pentasaccharide binding site into approx. one-third of the heparin chains [11], and thus, recognizes a rare sequence within the precursor substrate structure [42]. 3-OST-1 acts on a polysaccharide containing the sequence: →4) α-D-GlcNY (1→4) α-/β-D-GlcA/L-IdoA (1→4) α-D-GlcNY6X (1→4) β-D-GlcA (1→4) α-D-GlcNS6X (1→4) α-L-IdoA2S (1→4) α-D-GlcN6S(1→4)α/β-D-GlcA/L-IdoA2X (1→ (where Y = Ac or S and X = H or S, bold residues correspond to the AT-pentasaccharide binding site and bold/italicized residue is the one that will be sulfonated at the 3-position by 3-OST-1). No measurable activity was observed for 3OST-1 acting on heparosan or N-sulfoheparosan using the coupled colorimetric assay. The more highly modified heparan sulfate polysaccharide, however, is known to act as a substrate for 3OST-1 converting a low AT affinity heparan sulfate into a high affinity chain [11, 42]. A 3OST-1 assay, using coupled AST-IV (Fig. 11), afforded a PAPS KM for 3OST-1 of 13.5 µM, consistent with previously published literature using radioisotopic methods [43]. In addition, heparin lyase II treatment followed by LC-MS analysis of the resulting high AT affinity heparan sulfate confirmed the formation of a 3-O-sulfo group containing tetrasaccharide consistent with the generation of an AT pentasaccharide binding site (Fig. S8, Electronic Supplementary Material).
Fig. 11.
Michaelis-Menten kinetics (Eadie-Hofstee plot as Inset) of 3OST-1 on heparan sulfate.
Conclusions
We have outlined two assays for the real-time and near real-time analysis of heparan sulfate sulfotransferase enzyme activity. Under real-time conditions, a colorimetric methodology was used, coupling the OSTs with an AST-IV enzyme to produce a visibly measurable product. This method was also coupled with the C5-epi to gauge the importance of iduronic acid residues in OST bioactivity. Robust colorimetric assays were run with 6OST-3, to determine detailed kinetic parameters, and products were verified by LC-MS and nuclear magnetic resonance spectroscopy. Variations of PAPS concentration and epimerization of GlcA into IdoA residues had a significant effect on 6OST-3 and 2OST kinetics, but no obvious effect was seen under conditions of varied N-sulfoheparosan concentration. In the near real-time methodology, hydrophilic interaction chromatography was used with a defined substrate to measure the overall additions of sulfo groups to the substrate.
Supplementary Material
Abbreviations
- Ac
Acetyl
- AST-IV
Aryl-sulfotransferase
- AT
Antithrombin III
- CDSNS HP
Completely de-O-sulfonated heparin
- cGMP
Current good manufacturing process
- C5-epi
C5 Epimerase
- FTMS
Fourier transform mass spectrometry
- GAGs
Glycosaminoglycans
- GlcA
Glucuronic acid
- GlcN
Glucosamine
- HILIC
Hydrophilic interaction liquid chromatography
- HP
Heparin
- HS
Heparan sulfate
- IdoA
Iduronic acid
- LC-MS
Liquid chromatography – mass spectrometry
- NDST
N-deacetylase N-sulfotransferase
- PAP
3’-adenosine 5’-phosphate
- PAPS
3’-adenosine 5’-phosphosulfate
- PNP
para-nitrophenol
- PNPS
para-nitrophenylsulfate
- S
Sulfo
- 2-OST
2-O sulfotransferase
- 3-OST-1
3-O sulfotransferase isoform 1
- 6-OST-1
6-O sulfotransferase isoform 1
- 6-OST-3
6-O sulfotransferase isoform 3
References
- 1.Esko JD, Kimata K, Lindahl U. Chap. 16, Proteoglycans and Sulfated Glycosaminoglycans. In: Varki A, editor. Essentials of Glycobiology. 2nd. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 2008. [PubMed] [Google Scholar]
- 2.Ly M, Laremore TN, Linhardt RJ. Proteoglycomics: Recent progress and future challenges. OMICS. 2010;14:389–399. doi: 10.1089/omi.2009.0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Capila I, Linhardt RJ. Heparin-protein interactions. Angew Chem Int Edn. 2002;41:391–412. doi: 10.1002/1521-3773(20020201)41:3<390::aid-anie390>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 4.Kreuger J, Spillman D, Li J-P, Lindahl U. Interactions between heparan sulfate and proteins: the concept of specificity. J Cell Biol. 2006;174:323–327. doi: 10.1083/jcb.200604035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gallagher JT. The extended family of proteoglycans: social residents of the pericellular zone. Curr Opin Cell Biol. 1989;1:1201–1218. doi: 10.1016/s0955-0674(89)80072-9. [DOI] [PubMed] [Google Scholar]
- 6.Hardingham TE, Fosang AJ. Proteoglycans: many forms and many functions. FASEB J. 1992;6:861–870. [PubMed] [Google Scholar]
- 7.Bernfield M, Götte M, Park PW, Reizes O, Fitzgerald ML, Lincecum J, Zako M. Functions of cell surface heparan sulfate proteoglycans. Annu Rev Biochem. 1999;68:729–777. doi: 10.1146/annurev.biochem.68.1.729. [DOI] [PubMed] [Google Scholar]
- 8.Garner OB, Yamaguchi Y, Esko JD, Videm V. Small changes in lymphocyte development and activation in mice through tissue-specific alteration of heparan sulphate. Immunology. 2008;125:420–429. doi: 10.1111/j.1365-2567.2008.02856.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Islam T, Butler M, Sikkander SA, Toida T, Linhardt RJ. Further evidence that periodate cleavage of heparin occurs primarily through the antithrombin binding site. Carbohydr Res. 2002;337:2239–2243. doi: 10.1016/s0008-6215(02)00229-x. [DOI] [PubMed] [Google Scholar]
- 10.Jordan RE, Oosta GM, Gardner WT, Rosenberg RD. The binding of low molecular weight heparin to hemostatic enzymes. J Biol Chem. 1980;255:10081–10090. [PubMed] [Google Scholar]
- 11.Linhardt RJ. Heparin: Structure and activity. J Med Chem. 2003;46:2551–2554. doi: 10.1021/jm030176m. [DOI] [PubMed] [Google Scholar]
- 12.Zhang F, Yang B, Ly M, Solakyildirim K, Xiao Z, Wang Z, Beaudet JM, Torelli AY, Dordick JS, Linhardt RJ. Structural characterization of heparins from different commercial sources. Anal Bioanal Chem. 2011;401:2793–2803. doi: 10.1007/s00216-011-5367-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu H, Zhang Z, Linhardt RJ. Lessons learned from the contamination of heparin. Nat Prod Rep. 2009;26:313–321. doi: 10.1039/b819896a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Z, Zhang Z, McCallum SA, Linhardt RJ. NMR quantification for monitoring heparosan K5 capsular polysaccharide production. Anal Biochem. 2010;398:275–277. doi: 10.1016/j.ab.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei Z, Swiedler SJ, Ishihara M, Orellana A, Hirschberg CB. A single protein catalyzes both N-deacetylation and N-sulfation during the biosynthesis of heparan sulfate. Proc Natl Acad Sci USA. 1993;90:3885–3888. doi: 10.1073/pnas.90.9.3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Z, Yang B, Zhang Z, Ly M, Takieddin M, Mousa S, Liu J, Dordick JS, Linhardt RJ. Control of the heparosan N-deacetylation leads to an improved bioengineered heparin. Appl Microbiol Biotech. 2011;91:91–99. doi: 10.1007/s00253-011-3231-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeAngelis PL, Liu J, Linhardt RJ. Chemoenzymatic synthesis of glycosaminoglycans: Re-creating, re-modeling, and re-designing nature’s longest or most complex carbohydrate chains. Glycobiology. 2013;23:764–777. doi: 10.1093/glycob/cwt016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Esko JD, Selleck SB. Order out of chaos: assembly of ligand binding sites in heparan sulfate. Annu Rev Biochem. 2002;71:435–471. doi: 10.1146/annurev.biochem.71.110601.135458. [DOI] [PubMed] [Google Scholar]
- 19.Paul P, Suwan J, Liu J, Dordick JS, Linhardt RJ. Recent advances in sulfotransferase enzyme activity assays. Anal Bioanal Chem. 2012;403(6):1491–1500. doi: 10.1007/s00216-012-5944-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Limtiaco JFK, Beni S, Jones CJ, Langeslay DJ, Larive CK. NMR methods to monitor the enzymatic depolymerization of heparin. Anal Bioanal Chem. 2011;399:593–603. doi: 10.1007/s00216-010-4132-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nilsson M, Khajeh M, Botana A, Bernstein MA, Morris GA. Diffusion NMR and trilinear analysis in the study of reaction kinetics. Chem Commun. 2009:1252–1254. doi: 10.1039/b820813a. [DOI] [PubMed] [Google Scholar]
- 22.Gamage N, Barnett A, Hempel N, Duggleby RG, Windmill KF, Martin JL, McManus ME. Human sulfotransferases and their role in chemical metabolism. Toxicol Sci. 2006;90:p5–22. doi: 10.1093/toxsci/kfj061. [DOI] [PubMed] [Google Scholar]
- 23.Wang Z, Dordick JS, Linhardt RJ. E. coli K5 heparosan fermentation and improvement by genetic engineering. Bioengin Bugs. 2011;2:1–15. doi: 10.4161/bbug.2.1.14201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Satishchandran C, Markham GD. Adenosine-5'-phosphosulfate kinase from Escherichia coli K12. Purification, characterization, and identification of a phosphorylated enzyme intermediate. J Biol Chem. 1989;264:15012–15021. [PubMed] [Google Scholar]
- 25.Karamohamed S, Milsson J, Nourizad K, Ronaghi M, Pettersson B, Nyrén P. Production, purification, and luminometric analysis of recombinant Saccharomyces cerevisiae MET3 adenosine triphosphate sulfurylase expressed in Escherichia coli. Prot Expr Purif. 1999;15:381–388. doi: 10.1006/prep.1999.1032. [DOI] [PubMed] [Google Scholar]
- 26.Xu Y, Pempe EH, Liu J. Chemoenzymatic synthesis of heparin oligosaccharides with both anti-factor Xa and anti-factor IIa activities. J Biol Chem. 2012;287:29054–29061. doi: 10.1074/jbc.M112.358523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duncan M, Liu M, Fox C, Liu J. Characterization of the N-deacetylase domain from the heparan sulfate N-deacetylase/N-sulfotransferase 2. Biochem Biophys Res Commun. 2006;339:1232–1237. doi: 10.1016/j.bbrc.2005.11.142. [DOI] [PubMed] [Google Scholar]
- 28.Pervin A, Gallo C, Jandik KA, Han X-J, Linhardt RJ. Preparation and structural characterization of large heparin-derived oligosaccharides. Glycobiology. 1995;5:83–95. doi: 10.1093/glycob/5.1.83. [DOI] [PubMed] [Google Scholar]
- 29.Hileman RE, Smith AE, Toida T, Linhardt RJ. Preparation and structure of heparin lyase-derived heparan sulfate oligosaccharides. Glycobiology. 1997;7:231–239. doi: 10.1093/glycob/7.2.231. [DOI] [PubMed] [Google Scholar]
- 30.Liu R, Liu J. Enzymatic placement of 6-O-sulfo groups in heparan sulfate. Biochemistry. 2011;50:4382–4391. doi: 10.1021/bi200072t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shailubhai K, Singh RK, Schmuke JJ, Jacob GS. An enzymatic procedure for the preparation and purification of 3’-phosphoadenosine 5'-phospho-[35S]sulfate ([35S]PAPS): Applications in syntheses of 8-azido and 8-bromo derivatives of [35S]PAPS. Anal Biochem. 1996;243:165–170. doi: 10.1006/abio.1996.0496. [DOI] [PubMed] [Google Scholar]
- 32.Burkart MD, Wong C-H. A continuous assay for the spectrophotometric analysis of sulfotransferases using aryl sulfotransferase IV. Anal Biochem. 1999;274:131–137. doi: 10.1006/abio.1999.4264. [DOI] [PubMed] [Google Scholar]
- 33.Burkart MD, Izumi M, Chapman E, Lin C-H, Wong C-H. Regeneration of PAPS for the enzymatic synthesis of sulfated oligosaccharides. J Org Chem. 2000;65:5565–5574. doi: 10.1021/jo000266o. [DOI] [PubMed] [Google Scholar]
- 34.Pi N, Armstrong JI, Bertozzi CR, Leary JA. Kinetic analysis of NodST sulfotransferase using an electrospray ionization mass spectrometry assay. Biochemistry. 2002;41:13283–13288. doi: 10.1021/bi020457g. [DOI] [PubMed] [Google Scholar]
- 35.Xiong J, Bhaskar U, Li G, Fu L, Li L, Zhang F, Dordick JS, Linhardt RJ. Immobilized enzymes to convert N-sulfo, N-acetyl heparosan to a critical intermediate in the production of bioengineered heparin. J Biotechnol. 2013;167:241–247. doi: 10.1016/j.jbiotec.2013.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu D, Song D, Pederson LC, Liu J. Mutational study of heparan sulfate 2-O-sulfotransferase and chondroitin sulfate 2-O-sulfotransferase. J Biol Chem. 2007;282:8356–8367. doi: 10.1074/jbc.M608062200. [DOI] [PubMed] [Google Scholar]
- 37.Marshall AD, Darbyshire JF, Hunter AP, McPhie P, Jakoby WB. Control of activity through oxidative modification at the conserved residue Cys66 of aryl sulfotransferase IV. J Biol Chem. 1997;272:9153–9160. doi: 10.1074/jbc.272.14.9153. [DOI] [PubMed] [Google Scholar]
- 38.Habuchi H, Tanaka M, Habuchi O, Yoshida K, Suzuki H, Ban K, Kimata K. The occurrence of three isoforms of heparan sulfate 6-O-sulfotransferase having different specificities for hexuronic acid adjacent to the targeted N-sulfoglucosamine. J Biol Chem. 2000;275:2859–2868. doi: 10.1074/jbc.275.4.2859. [DOI] [PubMed] [Google Scholar]
- 39.Tran VM, Nguyen TKN, Raman K, Kuberan B. Applications of isotopes in advancing structural and functional heparanomics. Anal Bioanal Chem. 2011;399:559–570. doi: 10.1007/s00216-010-4166-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sheng J, Xu Y, Dulaney SB, Huang X, Liu J. Uncovering biphasic catalytic mode of C5-epimerase in heparan sulfate biosynthesis. J Biol Chem. 2012;287:20996–21002. doi: 10.1074/jbc.M112.359885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li K, Bethea HN, Liu J. Using engineered 2-O-sulfotransferase to determine the activity of heparan sulfate C5-epimerase and its mutants. J Biol Chem. 2010;285:11106–11113. doi: 10.1074/jbc.M109.081059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Linhardt RJ, Wang HM, Loganathan D, Bae JH. Search for the heparin antithrombin III-binding site precursor. J Biol Chem. 1992;267:2380–2387. [PubMed] [Google Scholar]
- 43.Edavettal SC, Lee KA, Negishi M, Linhardt RJ, Liu J, Pederson LC. Structural Analysis of the Sulfotransferase (3-OST-3) Involved in the Biosynthesis of an Entry Receptor for Herpes Simplex Virus 1. J Biol Chem. 2004;279:25789–25797. doi: 10.1074/jbc.M405013200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.