Abstract
Streptococcus mutans Antigen I/II (AgI/II) has been widely studied as a candidate vaccine antigen against human dental caries. In this report we follow up on prior studies that indicated that anti-AgI/II immunomodulatory monoclonal antibodies (MAbs) exerted their effects by destabilizing the native protein structure and exposing cryptic epitopes. We show here that similar results can be obtained by immunizing mice with truncated polypeptides out of the context of an intra-molecular interaction that occurs within the full-length molecule and that appears to dampen the functional response against at least two important target epitopes. Putative T cell epitopes that influenced antibody specificity were identified immediately upstream of the alanine-rich repeat domain. Adherence inhibiting antibodies could be induced against two discrete domains of the protein, one corresponding to the central portion of the molecule and the other corresponding to the C-terminus.
Keywords: Streptococcus, Monoclonal antibody, immune complex, immunodominance, vaccine design
1. Introduction
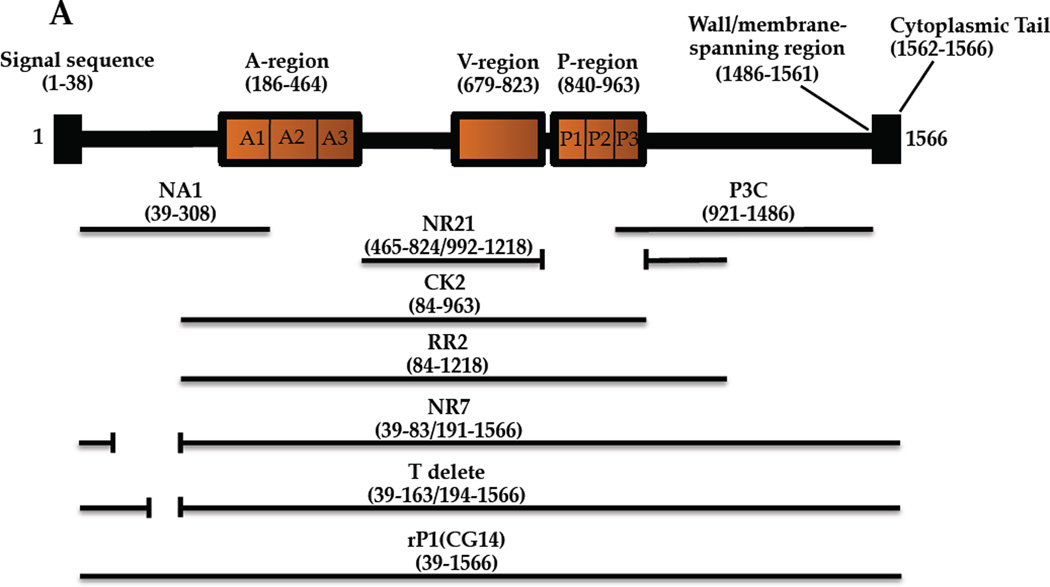
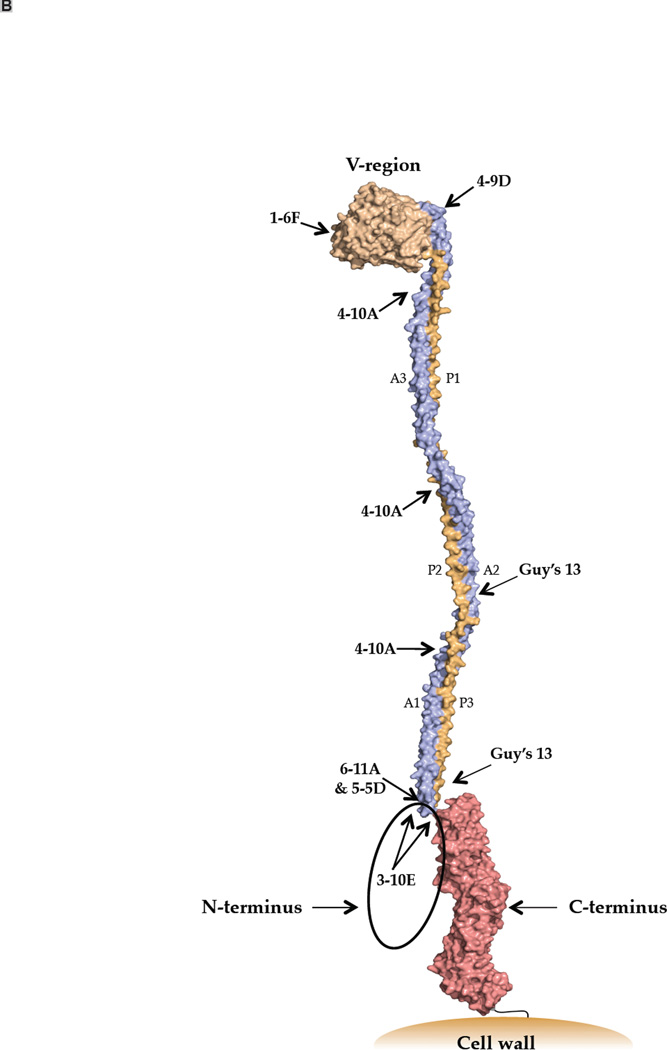
Due to its etiological association with dental caries [1], multiple antigens of Streptococcus mutans have been studied as vaccine candidates [2–6]. One such protein is the cell-surface localized Antigen I/II adhesin [7], also called P1 [8], Antigen B [9], or PAc [10]. AgI/II family members mediate interactions with host salivary constituents, cell matrix proteins, and other bacteria (reviewed in [11]). Until recently, a lack of high-resolution structural information hindered the design and interpretation of immunological studies. As deduced from the primary sequence, AgI/II has discontinuous alanine (A)- and proline (P)-rich tandem repeats that flank a variable (V) region where strain differences are clustered [10, 12, 13]. Recently, an unusual tertiary structure was discovered in which the A-repeats form an α-helix that intertwines with the polyproline II (PPII) P-region helix to form a long narrow stalk [14]. The intervening segment including the V-region comprises a β sandwich arranged in two sheets [15]. The crystal structure of the C-terminus also revealed β sheet structure with three consecutive domains adopting a DE-variant IgG fold [16]. Hence, two globular regions lie on either end of an extended stalk. A high affinity intra-molecular interaction between the N-terminus, which has not been crystalized, and the C-terminus increases stability of AgI/II and enhances adhesive function [17]. The primary and modeled tertiary structures of AgI/II are illustrated (Figure 1).
Figure 1.
Schematic representations of S. mutans Antigen I/II illustrating location of putative T cell epitopes and approximate antibody binding sites. (A) A representation of the primary structure of AgI/II and the recombinant polypeptides used in this study. (B) A three-dimensional model of Ag I/II. Approximate binding sites of monoclonal antibodies are indicated.
AgI/II’s interaction with salivary components is complex and involves two distinct adherence sites [16, 18]. The interaction differs depending on whether the major physiologic receptor, salivary agglutinin (SAG), is immobilized or is in fluid-phase. Monoclonal antibodies differ in their ability to inhibit adherence to SAG compared to SAG-mediated bacterial aggregation indicating that the determinants that mediate these two processes are not identical [19]. SAG is an oligomeric protein complex consisting primarily of the scavenger receptor glycoprotein gp340, and also containing amylase, sIgA and an 80 kDa protein [20, 21]. Different regions of both gp340 [22] and AgI/II [19] contribute to the different interactions. S. mutans adherence in vivo involves binding of AgI/II to immobilized SAG within the salivary pellicle coating the tooth surface [23]. Disruption of this interaction by antibodies is the focus of preventative therapeutic protocols. In contrast, interaction of fluid-phase SAG with cell surface AgI/II represents an innate host defense mechanism [24, 25], whereby aggregated are removed by swallowing. Hence it is desirable to elicit antibodies that disrupt SAG-mediated adherence, but not aggregation.
Numerous studies have demonstrated the relevance of an antibody response against AgI/II in protection against S. mutans colonization and cariogenicity (reviewed in [3, 11, 26, 27]). Both salivary and serum antibodies, that enter the oral cavity via transudation through the gingival crevice, have been reported to be protective [6, 28–33], or in some instances non-protective [34–36]. Subtle and potentially unapparent differences among immune responses can be crucial in determining the outcome of a host pathogen interaction. Naturally dominant epitopes are often not optimal for protection and pathogens can persist in the face of an immune response [37]. Therefore, it is fine specificity and functional activity, more so than total antibody amount, which likely determines whether colonization and cariogenicity is sufficiently inhibited to prevent disease by S. mutans.
Our laboratory has evaluated seven different anti-AI/II (P1) MAbs for immunomodulatory properties using an active immunization approach that incorporated them as part of immune complexes (IC) with whole bacterial cells [38–44]. Their approximate binding sites are illustrated in Figure 1B and were deduced based on reactivity with internal deletion constructs and combinations of truncated polypeptides [44–47]. MAbs 1–6F and 4–9D are influenced by overall conformation and bind within the region intervening the A- and P-repeats. 4–10A recognizes a repeated epitope formed by interacting A- and P-region sequences. Guy’s 13 also binds an epitope formed by interacting of A- and P-region sequences, but on a different part of the stalk. 6–11A, 5–5D, 3–10E bind epitopes that depend on the A-P interaction, but also involve a pre-A-post-P-region interaction. 3–10E’s binding is almost completely eliminated when this interaction is disrupted. MAbs 1–6F, 4–9D, and 4–10A inhibit bacterial adherence, while 6–11A, 5–5D, 3–10E, and Guy’s 13 do not [41, 44].
Previous studies showed that when incorporated within ICs, MAbs 6–11A, 5–5D, 3–10E, 4–10A, and Guy’s 13 redirected the adaptive immune response toward one of increased efficacy with regard to inhibition of bacterial adherence to SAG [38–44]. The presence of the MAbs within ICs altered the fine specificity and isotype composition of the elicited antibody response. These effects appeared to stem from a structural perturbation of the cell surface adhesin resulting in increased exposure of at least one normally cryptic or subdominant epitope, with the epitope recognized by 1–6F, an adherence-inhibiting MAb, shown to be affected [43, 44]. In the current study we sought to determine whether the effects of anti-Ag I/II MAbs could be mimicked by immunization with truncated and internal deletion variants of AgI/II in which important target epitopes might be better exposed. As a result, novel putative T helper cell and C-terminal epitopes were identified.
2. Materials and Methods
2.1 Bacterial strains, plasmids, expression and protein purification
S. mutans NG8 was grown aerobically for 16 hr in Todd-Hewitt broth with 0.3 % yeast extract (BBL, Cockeysville, MD). E. coli strains were grown aerobically at 37°C in Luria-Bertani broth (1 % [wt/vol] tryptone, 0.5 % [wt/vol] yeast extract, 1 % [wt/vol] NaCl) supplemented with ampicillin (50–100 µg/mL) or kanamycin (25–50 µg/mL). Construction of the CK1 and RR2 [45], NA1, P3C, and NR7 [17], and NR21 [43] polypeptides has been described. Recombinant proteins were purified on amylose or nickel resin. An additional in-frame deletion construct of AgI/II lacking two putative T-cell epitope sequences (aa 164–193) was generated by circle PCR-mutagenesis using primers 5’-GCTGCTCATGAGGCAGCTGCAAATGCTGC and 5’-GCAGCATTTGCAGCTGCCTCATGAGCAGC with pCG14 [48] as template. Amplified DNA was self-ligated using Quick T4 ligase (New England Biolabs) and transformed into E. coli Top10. Plasmid DNA [17] with the confirmed deletion, pPC303, was transformed into E. coli M15 (pRep4). The histidine-tagged T-delete protein was purified on nickel resin following induction of mid-exponential phase cells with 1 mM IPTG for 2–4 hours at 37 °C.
2.2 Mice
Six-eight week old female BALB/c mice were purchased from Charles River (Laboratories, Wilmington, MA) and housed in biosafety level 2 facilities under infectious disease conditions and fed a standard diet.
2.3 Source of antibodies
MAb 3–10E was obtained from a previously established hybridoma [49]. IgG was purified from murine ascites fluid using an ImmunoPure (A Plus) IgG Purification Kit (Pierce, Rockford, IL). Peroxidase-labeled and unlabeled secondary reagents were obtained from Southern Biotech (Birmingham, AL).
2.4 Immunizations and sample collections
AgI/II demonstrates numerous SDS-resistant discontinuous conformational epitopes that are preserved in Western blots [45]
Groups of five mice were immunized intraperitoneally with emulsified polyacrylamide gel slices containing recombinant full-length AgI/II (CG14), or recombinant AgI/II polypeptides NR7, NR21, CK2, RR2, T-delete, NA1, PC3, or a mixture of NA1 and P3C. Sham immunized groups received polyacrylamide only. Protein samples were loaded onto 7.5 % SDS-polyacrylamide preparative gels, negatively stained with 0.3 M cupric chloride for 5 min and rinsed with water. Negative stained bands of the correct size (∼1 □g total protein) were excised and de-stained with three 10 min washes with 0.25 M EDTA and 0.25 M Tris (pH 9) at room temperature with a final exchange into PBS. Gel slices were emulsified into 1 ml of PBS, and 100 µl was used to immunize individual mice. Mice were pre-bled 1 week before the first inoculation, immunized on days 0, 14, 21, and 35, and exsanguinated on day 50. Experiments were approved by the University of Florida Institutional Animal Care and Use Committee (Protocol # 201105486).
2.5 S. mutans adherence by BIAcore
Adherence of S. mutans to SAG immobilized on a CM5 sensor chip (GE Healthcare, Piscataway, NJ) was evaluated using the BIAcore 3000 machine (BIAcore AB, Uppsala, Sweden) [40]. SAG was prepared by adsorption to and desorption from S. mutans NG8 as described [22, 50]. Sera from mice within each group were pooled. Adherence of S. mutans reacted with sera (diluted 1:50) from mice immunized with full-length protein, versus those from mice immunized with truncated polypeptides, were compared. Each sample was run at least three times. Values were normalized to the delta RU of an S. mutans control sample run for each experiment in the absence of sera.
2.6 Biotin-labeling of MAb 3–10E and competition ELISA
Approximately 1 mg of purified 3–10E was biotinylated using EZ-Link™ Biotin-LC-Hydrazide (Pierce, Rockford, IL). ELISA plate wells were coated with S. mutans (∼107 cfu/well) [49] in carbonate-bicarbonate buffer, pH 9.6. Pooled antisera from each group were serially diluted two-fold in PBS containing 0.03% Tween-20 beginning at 1:25, 100 µl were added to the wells followed immediately by biotinylated 3–10E and incubated at 37 °C for two hours. Plates were washed and avidin-HRP conjugate (Pierce, IL) was applied to the wells for 30 minutes at room temperature. Plates were washed again and developed with 0.1 M o-phenylenediamine dihydrochloride containing 0.012 % hydrogen peroxide in 0.01 M phosphate citrate buffer. Percent inhibition was calculated as [OD450 direct binding of biotin-labeled 3–10E - OD450 experimental well / OD450 direct binding of biotin-labeled 3–10E] × 100. Positive and negative control wells contained unlabeled 3–10E, no inhibitor, or avidin-HRP only.
2.7 Quantitative subclass ELISA
ELISA plate wells were coated with S. mutans whole cells as described above or with 100 ng/well of purified recombinant NR21. Pooled sera from each group were serially diluted three-fold beginning at 1:50 and 100 µl were to the wells. Antibody reactivity was detected using affinity-purified peroxidase-labeled goat anti-mouse peroxidase conjugated IgG1, IgG2a, or IgG2b subclass specific antibodies (Southern Biotech) at a 1:2000 dilution. Plates were developed as above. Concentrations of anti-NR21 subclass antibodies were calculated by interpolation on standard curves generated using purified mouse subclass reagents (Southern Biotech, Birmingham, AL).
2.8 Prediction of T cell epitopes
We utilized RANKPEP (http://bio.dfci.harvard.edu/RANKPEP) to identify AgI/II peptides likely to interact with the class II I-Ad and I-Ed MHC alleles from Balb/c mice. The 1522 amino acid sequence representing the mature polypeptide was input and the 5 peptides predicted to bind the I-Ad and I-Ed molecules with the highest affinities, respectively, were identified. These are listed in order in Table 1 including their locations and the truncated polypeptides that contain them.
Table 1.
MHC class II P1 helper T cell epitopes predicted by RANKPEP*
| I-Ad peptides | Residue numbers | Polypeptides containing predicted epitopes |
|---|---|---|
| LLERGQSATATYTNL | 599–613 | CK1, CK2, RR2, NR21, NR7 |
| AYQKALAAYQAELKR | 227–241 | CK1, CK2, RR2, NR7 |
| NLPEAQGSASKEAEQ | 62–76 | NR7 |
| EKDMAAHKAEVERIN | 179–193 | CK2, RR2 |
| RVQEANAAAKAAYDT | 241–255 | CK1, CK2, RR2, NR7 |
| I-Ed peptides | Residue numbers | Polypeptides containing predicted epitopes |
| DRTLVAKQSVVKFQL | 1017–1031 | RR2, NR7 |
| QKALAAYQAELKRVQ | 229–243 | CK1, CK2, RR2, NR7 |
| NEEIRKRNATAKAEY | 271–285 | CK1, CK2, RR2, NR7 |
| ANEEIRKRNATAKAE | 270–284 | CK1, CK2, RR2, NR7 |
| VAKIKAKNQATKEQY | 164–178 | CK2 |
The rankpep prediction program (http://bio.dfci.harvard.edu/RANKPEP) was used to identify the top 5 potential peptides likely to be displayed to helper T cells by MHC Class II I-Ad and I-Ed molecules in BALB/c mice and their distribution among the polypeptide immunogens was tabulated.
2.9 Statistics
Statistically significant differences were determined by one- or two-way analysis of variance (ANOVA) using Graph Pad Prism 4.0. A p-value of less than 0.05 was considered significant. Tukey’s Multiple Comparison Test determined differences among the groups.
3. Results
3.1. P1 polypeptides vary in eliciting adherence-inhibiting antibodies
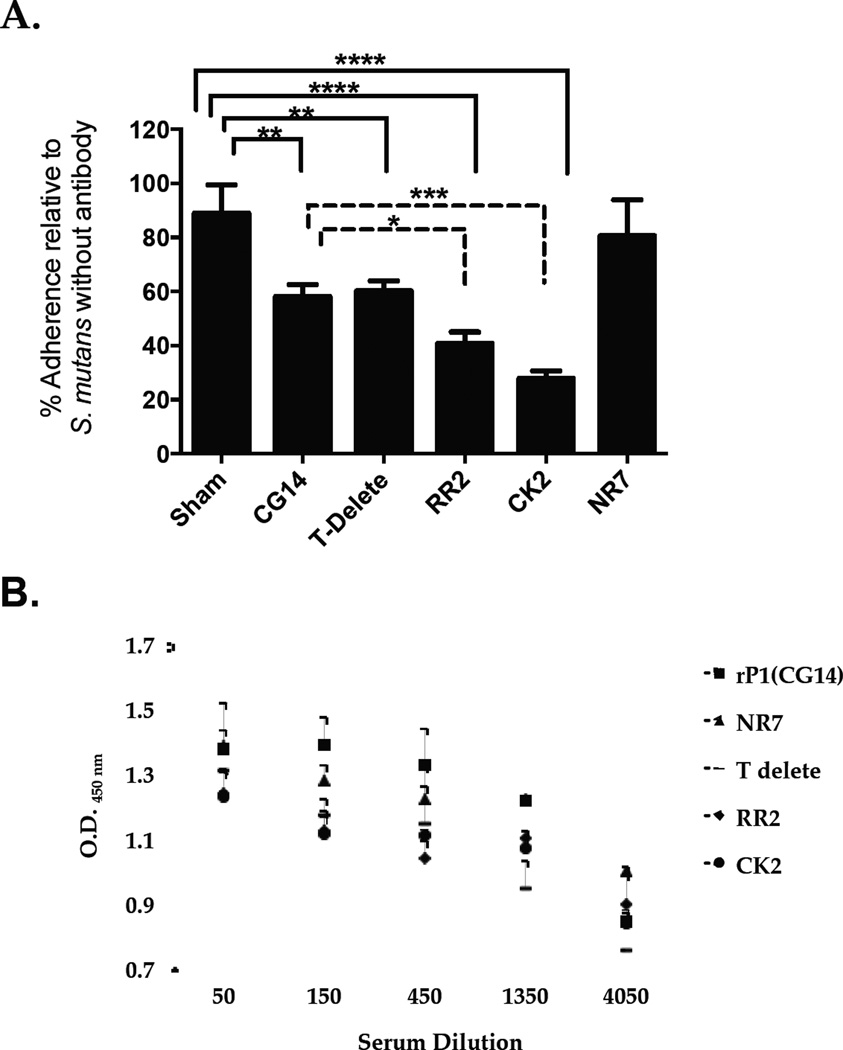
To evaluate the immunogenicity of truncated P1 polypeptides (Figure 1a), mice were immunized with full-length recombinant AgI/II (CG14), compared to RR2, CK2, and NR7. These were chosen because of their increased reactivity with MAb 1–6F [45], a strong inhibitor of S. mutans adherence [22, 41]. Inhibition of S. mutans adherence to immobilized SAG correlates with the level of 1–6F–like antibodies in the sera of mice immunized with IC of S. mutans and immunomodulatory MAbs [43]. As previously observed [43], pooled sera from mice that received CK2 was better able to inhibit adherence compared to that from mice that received full-length AgI/II (Figure 2A). This was also true of pooled sera from mice that received RR2. Surprisingly though, pooled sera from mice that received the internally-deleted NR7 polypeptide was no better at inhibiting adherence than pooled sera from mice immunized with CG14, despite the change in antigenicity and improved recognition of NR7 by MAb 1–6F [45]. Comparable levels of anti-S. mutans IgG were measured in all groups of mice that received any of the AgI/II constructs (Figure 2B), with no statistically significant differences detected among the groups.
Figure 2.
Evaluation of S. mutans adherence to SAG and detection of anti-S. mutans IgG. (A) BIAcore SPR analysis was used to evaluate bacterial adherence following incubation of S. mutans with sera collected from mice immunized with full-length P1 (CG14) and truncated derivatives of P1 (NA1, P3C). Values were normalized to the change in resonance units (ΔRU) for S. mutans NG8, in the absence of added serum, detected following the 60-second injection cycle. Immunogens are indicated on the X-axis. Tukey’s Multiple Comparison Test determined differences among the groups; *p≤0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001 (B) The presence of anti-S. mutans IgG in the sera of mice immunized with P1 versus truncated P1 polypeptides was evaluated by serial dilution and standard ELISA. Data are representative of at least three independent experiments. Statistical significance between groups was evaluated by two-way ANOVA and no significant differences were observed.
3.2. Predicted helper T cell epitopes affect antibody specificity and isotype
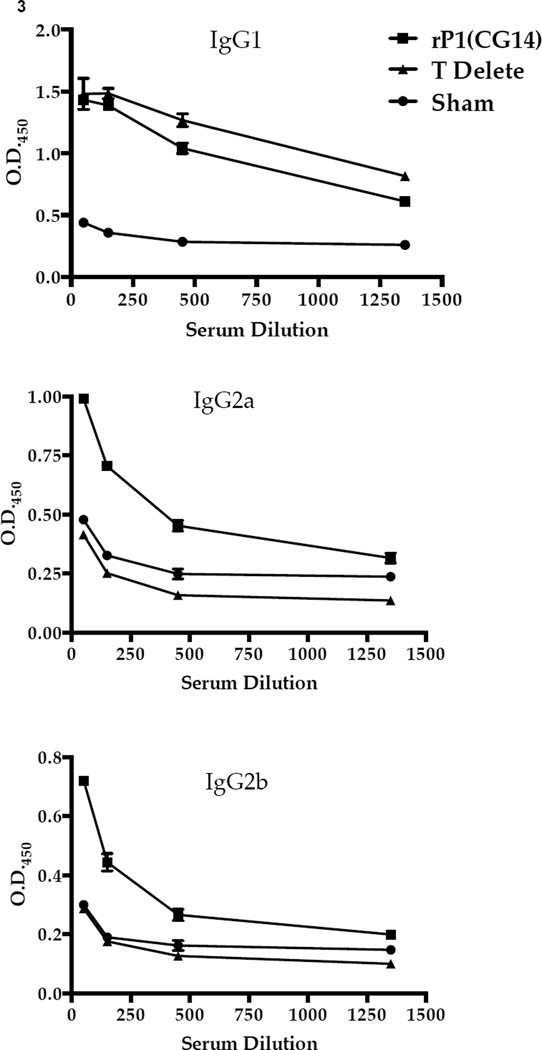
The top five I-Ad and five I-Ed AgI/II T helper cell epitopes in Balb/c mice were predicted using RANKPEP (Table 1). Since I-Ed peptide 5 and I-Ad peptide 4 were contiguous and contained within the segment that had been deleted from NR7 (aa 84–190), we constructed a more defined in-frame deletion (aa 164–192). This construct was no more effective than CG14 at eliciting an adherence-inhibiting response (Figure 2A). Next, we determined whether elimination of the two putative T cell epitopes affected formation of 1–6F–like antibodies, particularly those of the IgG2a and IgG2b isotypes, since these had correlated with improved adherence inhibition [41, 43]. We utilized polypeptide NR21 that corresponds to the central region of AgI/II and has an in-frame deletion of the P-region. It is recognized by MAb 1–6F, but no others in our panel; hence, it is a useful tool to evaluate 1–6F–like antibodies of particular isotypes contained within polyclonal sera. The levels of anti-NR21 antibodies in the sera of mice that had been immunized with the T-delete polypeptide, particularly the IgG2a and IgG2b isotypes, were significantly decreased compared to the full-length recombinant AgI/II group (Figure 3).
Figure 3.
Evaluation of the specificity and isotype of serum antibody responses by ELISA. The level of anti-NR21-specific IgG1, IgG2a, and IgG2b subclass antibody in the sera collected from sham-immunized mice, mice immunized with full-length P1, or the truncated protein lacking 2 putative T cell epitopes was evaluated by ELISA. All results are expressed as mean ± SEM and data are representative of at least three independent experiments. Statistically significant differences of the T-delete compared to CG14 were determined by two-way ANOVA. p≤0.05 (IgG1) , p≤0.0001 (IgG2a), p≤0.01 (IgG2b)
3.3 Relevant C-terminal epitopes are masked within AgI/II
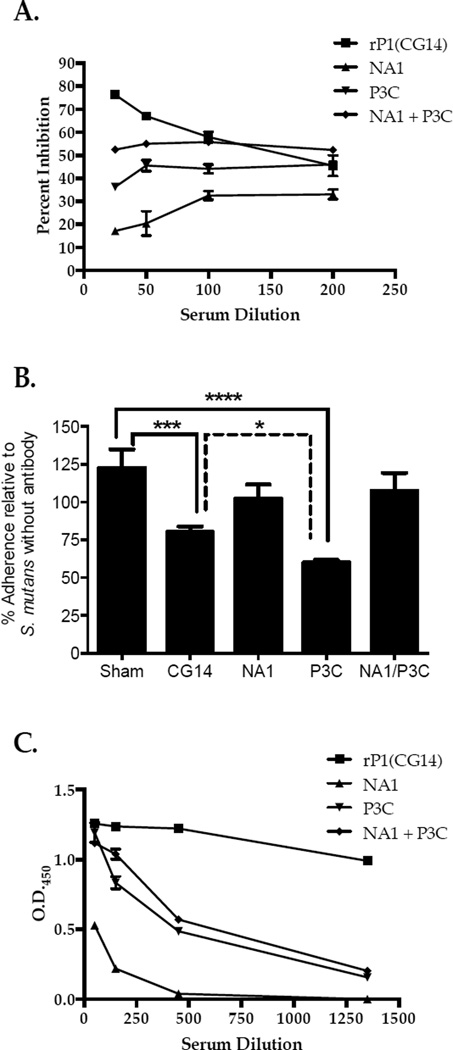
To evaluate potential masking of protective epitopes within the native protein resulting from the pre-A/post-P-region interaction additional murine immunizations using NA1 and P3C were performed (Figure 1A). These fragments form a stable complex that reconstitutes multiple discontinuous epitopes, including that recognized by MAb 3–10E. MAb 3–10E does not itself inhibit adherence [41], but serves as a marker for achievement of native structure [17]. To evaluate the ability of NA1 and P3C to maintain an interaction in vivo, a competition ELISA was employed in which the ability of pooled immune sera to inhibit binding of biotinylated 3–10E to S. mutans was tested. Pooled sera from the full-length CG14 group, followed by the NA1+P3C group, contained antibodies able to compete for MAb 3–10E binding, while pooled sera from P3C-, and particularly NA1-immunized mice, were less able to compete (Figure 4A). Pooled sera from mice immunized with P3C also were increased in their ability to inhibit S. mutans adherence to immobilized SAG compared to pooled sera from CG14 (p<0.05) or sham immunized (p<0.0001) groups (Figure 4B). This effect was not observed for pooled sera from the NA1 or NA1+P3C groups. Of note, total anti-S. mutans IgG measured in pooled sera from the P3C immunized mice was significantly less than that measured from mice immunized with CG14 and comparable to that from mice immunized with the NA1/P3C mixture (Figure 4C).
Figure 4.
Functional C-terminal epitopes are masked by intra-molecular interactions. (A) Sera from mice immunized with full-length P1 (CG14), NA1, P3C, or a mixture of NA1 and P3C were used in a competition ELISA to evaluate inhibition of binding of biotin-labeled MAb 3–10E to S. mutans. Serum dilution is indicated on the x-axis. (B) BIAcore SPR analysis was used to evaluate bacterial adherence following incubation of S. mutans with sera collected from immunized mice. Values were normalized to the change in resonance units (ΔRU) for S. mutans NG8, in the absence of added serum, detected following the 60-second injection cycle. Immunogens are indicated on the X-axis. Tukey’s Multiple Comparison Test determined differences among the groups; * p≤0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001. (C) The level of anti-S. mutans IgG in the sera from P1 versus truncated P1 -immunized mice was evaluated by serial dilution and standard ELISA. Data are representative of at least three independent experiments. Statistically significant differences compared to the CG14 group were determined by two-way ANOVA. p≤0.01 (P3C, NA1+P3C), p≤0.0001 (NA1).
4. Discussion
In proteins with complex conformations, formation of functional antibodies may be hampered by inaccessibility of cryptic, but protective, target epitopes. Binding of an antibody to its cognate antigen can induce a conformational change in that antigen [51–67]. Importantly, these conformational changes can lead to exposure of functional subdominant or neoepitopes [54, 60, 65, 68–87], and such determinants can represent important targets of functional or neutralizing antibodies [62, 68, 88–97]. This is most evident in viral systems in which the availability of well-resolved crystal structures has outpaced that of bacterial molecules.
A tertiary model for S. mutans AgI/II has now allowed the approximation of SAG binding domains [14, 16], and MAb binding sites [44]. In this study we capitalized on information from previous work that suggested destabilization of AgI/II structure was the basis of immunomodulatory effects by several MAbs [43]. Destabilization of protein by antibody can identify key internal segments that are integral to overall tertiary structure [63]. We had discovered that several anti-AgI/II MAbs recognize epitopes contributed to by aa 84–190 immediately upstream of the A-region [39, 98]. Circular dichroism, surface plasmon resonance, and differential scanning calorimetry confirmed this segment contributes to folding and function of the adhesin, and increases stability of its elongated hybrid helical stalk [17]. The NR7 polypeptide lacking this sequence demonstrates increased exposure of the 1–6F epitope [45]; therefore, we speculated that NR7 would represent a superior immunogen. Surprisingly, it did not. Therefore, we evaluated whether elimination of two putative T cell epitopes within the segment deleted from NR7 affected immunogenicity against the 1–6F epitope. Compared to unaltered AgI/II, the T-delete polypeptide resulted in the formation of significantly less antibody reactive with the NR21 polypeptide that is used as a tool for measurement of 1-6F-like antibodies. Unlike NR7, the T-delete polypeptide did not lose its ability to interact with MAb 3–10E or gain in reactivity with MAb 1–6F (data not shown), hence it appears to be structurally similar to the unaltered full-length adhesin. This suggests that its altered immunogenicity was a consequence of the elimination of the two putative T cell epitopes identified by RANKPEP rather than a structural alteration.
Two other constructs also demonstrate increased reactivity with MAb 1–6F compared to full-length AgI/II [45]. The longer, RR2, lacks a major portion of the C-terminus but retains an ability to form the 3–10E epitope [99]. RR2 was superior to CG14 in its ability to elicit an efficacious adherence-inhibiting response. The second shorter construct, CK2, lacks the entire C-terminus, and was also a more efficacious immunogen than full-length CG14. The interaction of AgI/II with SAG is multivalent and contains two distinct binding sites [18]. These have been localized within two discrete fragments. A3VP1, corresponds to the third alanine-rich repeat through the third proline-rich repeat, a segment contained within both RR2 and CK2. The second binding site is contained within the C-terminus [16]. Based on our current results, it appears that relevant target epitopes are associated with each of the binding sites, but are obscured within the context of the complete native structure. P3C, but not its intra-molecular binding partner NA1, was significantly better able to elicit adherence-inhibiting antibodies compared to the full-length adhesin or a mixture of P3C and NA1. The A3VP1-containing RR2/CK2 polypeptides, as well the P3C polypeptide, therefore retain important functional epitopes, yet expose them in such a way that a more desirable host response is achieved. It will be important in future studies to test whether co-immunization with a mixture of CK2 and P3C has an additive effect. Unlike RR2, CK2 lacks the entire N-terminus of AgI/II so would not be expected to bind to or influence the immunogenicity of the C-terminus.
While dental caries is associated with acidogenic and aciduric organisms such as S. mutans, oral health is associated with other oral streptococci that also express AgI/II family molecules [100–102]. Cross-reactivity of immune sera from this study was observed against S. oralis and to a lesser extent S. mitis; however, these organisms did not adhere to the SAG prepared using S. mutans (data not shown). Since the binding specificities of AgI/II homologs among species are known to differ [103], the cross-reactive antibodies should not be problematic. Minimal cross-reactivity was observed against S. salivarius and S. sanguinis. In a previous study [45] we found that anti-AgI/II monoclonal antibodies cross-reactive with S. gordonii were directed against epitopes that were subsequently mapped to the hybrid helical stalk rather than to the globular adherence domains lying at either end of it [44]. This is consistent with our current study in which more cross-reactivity was detected in sera from animals immunized with polypeptides that included the stalk. Unlike S. oralis and S. mitis, S. gordonii demonstrated substantial adherence to SAG, but this binding was not blocked by the anti-P3C sera that significantly inhibited S. mutans adherence (data not shown). Taken together, our results suggest that AgI/II-derived vaccine immunogens can be designed to selectively interfere with adherence of S. mutans. Ag I/II-mediated adhesion represents a sucrose-independent mechanism. Thus in conjunction with immunogens such as glycosyltransferase and glucan binding proteins that target sucrose-dependent adhesion [2] truncated AgI/II polypeptides such as CK2 and/or P3C would be useful adjuncts in an integrated immunoprophylactic approach.
The three-dimensional structure of an antigen clearly impacts immunogenicity against conformational epitopes, but stimulation of particular T cell subsets can also depend on minor alterations within an Ag [104]. Despite the presence of the newly identified predicted T cell epitopes in full-length CG14 (Table 1), its folded structure still appears to interfere with responses against important protective epitopes. Our results reiterate that in any system, optimal immunogenicity is a balance between structure, epitope exposure and availability of T cell epitopes following antigenic processing of any given conformer, and are consistent with an accumulating body of literature that points to an ability to capitalize on information regarding antibody-mediated changes in an antigen and to apply that information to mimic antibody-mediated destabilization of protein structure. These data will facilitate future studies regarding S. mutans AgI/II as a vaccine candidate. In addition, this approach can serve as a model for improvement of protective responses against pathogenic microbes for which the natural immunodominance of candidate antigens is less than optimal.
Highlights.
Streptococcus mutans adhesin AgI/II elicits adherence-inhibiting antibodies.
We utilize truncated proteins to identify targets of relevant antibodies.
Putative amino-terminal T cell epitopes impact formation of adherence-inhibiting antibodies.
The C-terminus was identified as a new target of adherence-inhibiting antibodies.
An intramolecular interaction appears to dampen the adherence-inhibiting immune response.
Acknowledgements
This work was supported by the National Institute for Dental and Craniofacial Research R01-DE13882 to L.J.B., training grant T32-DE07200 to R.A.R., and a University of Florida Alumni Fellowship and training grant T90-DE021990 to K.P.H.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hamada S, Slade HD. Biology, immunology, and cariogenicity of Streptococcus mutans . Microbiol Rev. 1980;44:331–384. doi: 10.1128/mr.44.2.331-384.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taubman MA, Nash DA. The scientific and public-health imperative for a vaccine against dental caries. Nat Rev Immunol. 2006;6:555–563. doi: 10.1038/nri1857. [DOI] [PubMed] [Google Scholar]
- 3.Michalek SM, Katz J, Childers NK. A vaccine against dental caries: an overview. BioDrugs. 2001;15:501–508. doi: 10.2165/00063030-200115080-00002. [DOI] [PubMed] [Google Scholar]
- 4.Smith DJ, Shoushtari B, Heschel RL, King WF, Taubman MA. Immunogenicity and protective immunity induced by synthetic peptides associated with a catalytic subdomain of mutans group streptococcal glucosyltransferase. Infect Immun. 1997;65:4424–4430. doi: 10.1128/iai.65.11.4424-4430.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith DJ, Taubman MA. Experimental immunization of rats with a Streptococcus mutans 59-kilodalton glucan-binding protein protects against dental caries. Infect Immun. 1996;64:3069–3073. doi: 10.1128/iai.64.8.3069-3073.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang P, Jespersgaard C, Lamberty-Mallory L, Katz J, Huang Y, Hajishengallis G, et al. Enhanced immunogenicity of a genetic chimeric protein consisting of two virulence antigens of Streptococcus mutans and protection against infection. Infect Immun. 2002;70:6779–6787. doi: 10.1128/IAI.70.12.6779-6787.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Russell MW, Lehner T. Characterisation of antigens extracted from cells and culture fluids of Streptococcus mutans serotype c . Arch Oral Biol. 1978;23:7–15. doi: 10.1016/0003-9969(78)90047-x. [DOI] [PubMed] [Google Scholar]
- 8.Forester H, Hunter N, Knox KW. Characteristics of a high molecular weight extracellular protein of Streptococcus mutans . J Gen Microbiol. 1983;129(Pt 9):2779–2788. doi: 10.1099/00221287-129-9-2779. [DOI] [PubMed] [Google Scholar]
- 9.Russell RR. Wall-associated protein antigens of Streptococcus mutans . J Gen Microbiol. 1979;114:109–115. doi: 10.1099/00221287-114-1-109. [DOI] [PubMed] [Google Scholar]
- 10.Okahashi N, Sasakawa C, Yoshikawa M, Hamada S, Koga T. Molecular characterization of a surface protein antigen gene from serotype c Streptococcus mutans, implicated in dental caries. Mol Microbiol. 1989;3:673–678. doi: 10.1111/j.1365-2958.1989.tb00215.x. [DOI] [PubMed] [Google Scholar]
- 11.Brady LJ, Maddocks SE, Larson MR, Forsgren N, Persson K, Deivanayagam CC, et al. The changing faces of Streptococcus antigen I/II polypeptide family adhesins. Mol Microbiol. 2010;77:276–286. doi: 10.1111/j.1365-2958.2010.07212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brady LJ, Crowley PJ, Ma JK, Kelly C, Lee SF, Lehner T, et al. Restriction fragment length polymorphisms and sequence variation within the spaP gene of Streptococcus mutans serotype c isolates. Infect Immun. 1991;59:1803–1810. doi: 10.1128/iai.59.5.1803-1810.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kelly C, Evans P, Bergmeier L, Lee SF, Progulske-Fox A, Harris AC, et al. Sequence analysis of the cloned streptococcal cell surface antigen I/II. FEBS Lett. 1989;258:127–132. doi: 10.1016/0014-5793(89)81632-1. [DOI] [PubMed] [Google Scholar]
- 14.Larson MR, Rajashankar KR, Patel MH, Robinette RA, Crowley PJ, Michalek S, et al. Elongated fibrillar structure of a streptococcal adhesin assembled by the high-affinity association of {alpha}- and PPII-helices. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.0912293107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Troffer-Charlier N, Ogier J, Moras D, Cavarelli J. Crystal structure of the V-region of Streptococcus mutans antigen I/II at 2.4 A resolution suggests a sugar preformed binding site. J Mol Biol. 2002;318:179–188. doi: 10.1016/S0022-2836(02)00025-6. [DOI] [PubMed] [Google Scholar]
- 16.Larson MR, Rajashankar KR, Crowley PJ, Kelly C, Mitchell TJ, Brady LJ, et al. Crystal structure of the C-terminal region of Streptococcus mutans antigen I/II and characterization of salivary agglutinin adherence domains. The Journal of biological chemistry. 2011;286:21657–21666. doi: 10.1074/jbc.M111.231100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heim KP, Crowley PJ, Brady LJ. An intramolecular interaction involving the N terminus of a streptococcal adhesin affects its conformation and adhesive function. The Journal of biological chemistry. 2013;288:13762–13774. doi: 10.1074/jbc.M113.459974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hajishengallis G, Koga T, Russell MW. Affinity and specificity of the interactions between Streptococcus mutans antigen I/II and salivary components. J Dent Res. 1994;73:1493–1502. doi: 10.1177/00220345940730090301. [DOI] [PubMed] [Google Scholar]
- 19.Brady LJ, Piacentini DA, Crowley PJ, Oyston PC, Bleiweis AS. Differentiation of salivary agglutinin-mediated adherence and aggregation of mutans streptococci by use of monoclonal antibodies against the major surface adhesin P1. Infect Immun. 1992;60:1008–1017. doi: 10.1128/iai.60.3.1008-1017.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oho T, Yu H, Yamashita Y, Koga T. Binding of salivary glycoprotein-secretory immunoglobulin A complex to the surface protein antigen of Streptococcus mutans . Infect Immun. 1998;66:115–121. doi: 10.1128/iai.66.1.115-121.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prakobphol A, Xu F, Hoang VM, Larsson T, Bergstrom J, Johansson I, et al. Salivary agglutinin, which binds Streptococcus mutans and Helicobacter pylori, is the lung scavenger receptor cysteine-rich protein gp-340. J Biol Chem. 2000;275:39860–39866. doi: 10.1074/jbc.M006928200. [DOI] [PubMed] [Google Scholar]
- 22.Loimaranta V, Jakubovics NS, Hytonen J, Finne J, Jenkinson HF, Stromberg N. Fluid- or surface-phase human salivary scavenger protein gp340 exposes different bacterial recognition properties. Infect Immun. 2005;73:2245–2252. doi: 10.1128/IAI.73.4.2245-2252.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Esberg A, Lofgren-Burstrom A, Ohman U, Stromberg N. Host and bacterial phenotype variation in adhesion of Streptococcus mutans to matched human hosts. Infection and immunity. 2012;80:3869–3879. doi: 10.1128/IAI.00435-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Madsen J, Mollenhauer J, Holmskov U. Review: Gp-340/DMBT1 in mucosal innate immunity. Innate Immun. 2010;16:160–167. doi: 10.1177/1753425910368447. [DOI] [PubMed] [Google Scholar]
- 25.Edwards AM, Manetti AG, Falugi F, Zingaretti C, Capo S, Buccato S, et al. Scavenger receptor gp340 aggregates group A streptococci by binding pili. Molecular microbiology. 2008;68:1378–1394. doi: 10.1111/j.1365-2958.2008.06220.x. [DOI] [PubMed] [Google Scholar]
- 26.Brady LJ. Antibody-mediated immunomodulation: a strategy to improve host responses against microbial antigens. Infect Immun. 2005;73:671–678. doi: 10.1128/IAI.73.2.671-678.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Michalek SM, Childers NK. Development and outlook for a caries vaccine. Crit Rev Oral Biol Med. 1990;1:37–54. doi: 10.1177/10454411900010010401. [DOI] [PubMed] [Google Scholar]
- 28.Challacombe SJ, Bergmeier LA, Rees AS. Natural antibodies in man to a protein antigen from the bacterium Streptococcus mutans related to dental caries experience. Arch Oral Biol. 1984;29:179–184. doi: 10.1016/0003-9969(84)90051-7. [DOI] [PubMed] [Google Scholar]
- 29.Lehtonen OP, Grahn EM, Stahlberg TH, Laitinen LA. Amount and avidity of salivary and serum antibodies against Streptococcus mutans in two groups of human subjects with different dental caries susceptibility. Infect Immun. 1984;43:308–313. doi: 10.1128/iai.43.1.308-313.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tenovuo J, Lehtonen OP, Aaltonen AS. Caries development in children in relation to the presence of mutans streptococci in dental plaque and of serum antibodies against whole cells and protein antigen I/II of Streptococcus mutans . Caries Res. 1990;24:59–64. doi: 10.1159/000261240. [DOI] [PubMed] [Google Scholar]
- 31.Ebersole JL. Humoral immune responses in gingival crevice fluid: local and systemic implications. Periodontol 2000. 2003;31:135–166. doi: 10.1034/j.1600-0757.2003.03109.x. [DOI] [PubMed] [Google Scholar]
- 32.Huang Y, Hajishengallis G, Michalek SM. Induction of protective immunity against Streptococcus mutans colonization after mucosal immunization with attenuated Salmonella enterica serovar typhimurium expressing an S. mutans adhesin under the control of in vivo-inducible nirB promoter. Infect Immun. 2001;69:2154–2161. doi: 10.1128/IAI.69.4.2154-2161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lehner T, Russell MW, Caldwell J, Smith R. Immunization with purified protein antigens from Streptococcus mutans against dental caries in rhesus monkeys. Infect Immun. 1981;34:407–415. doi: 10.1128/iai.34.2.407-415.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gregory RL, Hobbs LC, Kindle JC, VanTo T, Malmstrom HS. Immunodominant antigens of Streptococcus mutans in dental caries-resistant subjects. Hum Antibodies Hybridomas. 1990;1:132–136. [PubMed] [Google Scholar]
- 35.Kelly CG, Todryk S, Kendal HL, Munro GH, Lehner T. T-cell, adhesion, and B-cell epitopes of the cell surface Streptococcus mutans protein antigen I/II. Infect Immun. 1995;63:3649–3658. doi: 10.1128/iai.63.9.3649-3658.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hocini H, Iscaki S, Bouvet JP, Pillot J. Unexpectedly high levels of some presumably protective secretory immunoglobulin A antibodies to dental plaque bacteria in salivas of both caries-resistant and caries-susceptible subjects. Infect Immun. 1993;61:3597–3604. doi: 10.1128/iai.61.9.3597-3604.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Casadevall A, Pirofski LA. Exploiting the redundancy in the immune system: vaccines can mediate protection by eliciting 'unnatural' immunity. J Exp Med. 2003;197:1401–1404. doi: 10.1084/jem.20030637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brady LJ, van Tilburg ML, Alford CE, McArthur WP. Monoclonal antibody-mediated modulation of the humoral immune response against mucosally applied Streptococcus mutans . Infect Immun. 2000;68:1796–1805. doi: 10.1128/iai.68.4.1796-1805.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Isoda R, Robinette RA, Pinder TL, McArthur WP, Brady LJ. Basis of beneficial immunomodulation by monoclonal antibodies against Streptococcus mutans adhesin P1. FEMS Immunol Med Microbiol. 2007;51:102–111. doi: 10.1111/j.1574-695X.2007.00279.x. [DOI] [PubMed] [Google Scholar]
- 40.Oli MW, McArthur WP, Brady LJ. A whole cell BIAcore assay to evaluate P1-mediated adherence of Streptococcus mutans to human salivary agglutinin and inhibition by specific antibodies. J Microbiol Methods. 2005 doi: 10.1016/j.mimet.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 41.Oli MW, Rhodin N, McArthur WP, Brady LJ. Redirecting the humoral immune response against Streptococcus mutans antigen P1 with monoclonal antibodies. Infect Immun. 2004;72:6951–6960. doi: 10.1128/IAI.72.12.6951-6960.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rhodin NR. Immunomodulation by an anti-Streptococcus mutans monoclonal antibody [Ph.D.] Gainesville, Florida: University of Florida; 2003. [Google Scholar]
- 43.Robinette RA, Oli MW, McArthur WP, Brady LJ. Beneficial immunomodulation by Streptococcus mutans anti-P1 monoclonal antibodies is Fc independent and correlates with increased exposure of a relevant target epitope. J Immunol. 2009;183:4628–4638. doi: 10.4049/jimmunol.0803300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robinette RA, Oli MW, McArthur WP, Brady LJ. A therapeutic anti-Streptococcus mutans monoclonal antibody used in human passive protection trials influences the adaptive immune response. Vaccine. 2011;29:6292–6300. doi: 10.1016/j.vaccine.2011.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McArthur WP, Rhodin NR, Seifert TB, Oli MW, Robinette RA, Demuth DR, et al. Characterization of epitopes recognized by anti-Streptococcus mutans P1 monoclonal antibodies. FEMS Immunol Med Microbiol. 2007;50:342–353. doi: 10.1111/j.1574-695X.2007.00260.x. [DOI] [PubMed] [Google Scholar]
- 46.van Dolleweerd CJ, Chargelegue D, Ma JK. Characterization of the conformational epitope of Guy's 13, a monoclonal antibody that prevents Streptococcus mutans colonization in humans. Infect Immun. 2003;71:754–765. doi: 10.1128/IAI.71.2.754-765.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Dolleweerd CJ, Kelly CG, Chargelegue D, Ma JK. Peptide mapping of a novel discontinuous epitope of the major surface adhesin from Streptococcus mutans . J Biol Chem. 2004;279:22198–22203. doi: 10.1074/jbc.M400820200. [DOI] [PubMed] [Google Scholar]
- 48.Brady LJ, Cvitkovitch DG, Geric CM, Addison MN, Joyce JC, Crowley PJ, et al. Deletion of the central proline-rich repeat domain results in altered antigenicity and lack of surface expression of the Streptococcus mutans P1 adhesin molecule. Infect Immun. 1998;66:4274–4282. doi: 10.1128/iai.66.9.4274-4282.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ayakawa GY, Boushell LW, Crowley PJ, Erdos GW, McArthur WP, Bleiweis AS. Isolation and characterization of monoclonal antibodies specific for antigen P1, a major surface protein of mutans streptococci. Infect Immun. 1987;55:2759–2767. doi: 10.1128/iai.55.11.2759-2767.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rundegren J, Arnold RR. Differentiation and interaction of secretory immunoglobulin A and a calcium-dependent parotid agglutinin for several bacterial strains. Infect Immun. 1987;55:288–292. doi: 10.1128/iai.55.2.288-292.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Inagaki H, Katoh M, Kurosawa-Ohsawa K, Tanaka S. A new sandwich enzyme-linked immunosorbent assay (ELISA) for transforming growth factor alpha (TGF alpha) based upon conformational modification by antibody binding. Journal of immunological methods. 1990;128:27–37. doi: 10.1016/0022-1759(90)90460-d. [DOI] [PubMed] [Google Scholar]
- 52.Valenzuela CF, Dowding AJ, Arias HR, Johnson DA. Antibody-induced conformational changes in the Torpedo nicotinic acetylcholine receptor: a fluorescence study. Biochemistry. 1994;33:6586–6594. doi: 10.1021/bi00187a028. [DOI] [PubMed] [Google Scholar]
- 53.Mazza MM, Retegui LA. Monoclonal antibodies to human growth hormone induce an allosteric conformational change in the antigen. Immunology. 1989;67:148–153. [PMC free article] [PubMed] [Google Scholar]
- 54.Cao L, Yoshino T, Nishiuchi R, Yamadori I, Akagi T. Homotypic cell aggregation via conformational change of CD44 molecule induced by anti-CD44 monoclonal antibodies. Immunobiology. 1995;193:1–14. doi: 10.1016/S0171-2985(11)80152-X. [DOI] [PubMed] [Google Scholar]
- 55.Davies DR, Cohen GH. Interactions of protein antigens with antibodies. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:7–12. doi: 10.1073/pnas.93.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Towbin H, Erard F, van Oostrum J, Schmitz A, Rordorf C. Neoepitope immunoassay: an assay for human interleukin 1 beta based on an antibody induced conformational change. J Immunoassay. 1996;17:353–369. doi: 10.1080/01971529608005798. [DOI] [PubMed] [Google Scholar]
- 57.Frillingos S, Wu J, Venkatesan P, Kaback HR. Binding of ligand or monoclonal antibody 4B1 induces discrete structural changes in the lactose permease of Escherichia coli . Biochemistry. 1997;36:6408–6414. doi: 10.1021/bi970233b. [DOI] [PubMed] [Google Scholar]
- 58.Monaco-Malbet S, Berthet-Colominas C, Novelli A, Battai N, Piga N, Cheynet V, et al. Mutual conformational adaptations in antigen and antibody upon complex formation between an Fab and HIV-1 capsid protein p24. Structure Fold Des. 2000;8:1069–1077. doi: 10.1016/s0969-2126(00)00507-4. [DOI] [PubMed] [Google Scholar]
- 59.Ulrichts H, Harsfalvi J, Bene L, Matko J, Vermylen J, Ajzenberg N, et al. A monoclonal antibody directed against human von Willebrand factor induces type 2B-like alterations. J Thromb Haemost. 2004;2:1622–1628. doi: 10.1111/j.1538-7836.2004.00865.x. [DOI] [PubMed] [Google Scholar]
- 60.Ganesan R, Eigenbrot C, Kirchhofer D. Structural and mechanistic insight into how antibodies inhibit serine proteases. The Biochemical journal. 2010;430:179–189. doi: 10.1042/BJ20100634. [DOI] [PubMed] [Google Scholar]
- 61.Kitamura H, Shindo K, Sasabe M, Matsuura E, Sakairi N, Nishi N. Conformational change of DNA on the formation of DNA-anti-DNA antibody immune complex. Nucleic Acids Symp Ser. 1997:145–146. [PubMed] [Google Scholar]
- 62.Lazear E, Whitbeck JC, Ponce-de-Leon M, Cairns TM, Willis SH, Zuo Y, et al. Antibody-induced conformational changes in herpes simplex virus glycoprotein gD reveal new targets for virus neutralization. Journal of Virology. 2012;86:1563–1576. doi: 10.1128/JVI.06480-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cha K, Reeves PJ, Khorana HG. Structure and function in rhodopsin: destabilization of rhodopsin by the binding of an antibody at the N-terminal segment provides support for involvement of the latter in an intradiscal tertiary structure. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:3016–3021. doi: 10.1073/pnas.97.7.3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hernandez R, Paredes A, Brown DT. Sindbis virus conformational changes induced by a neutralizing anti-E1 monoclonal antibody. Journal of Virology. 2008;82:5750–5760. doi: 10.1128/JVI.02673-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hato T, Watanabe A, Nakatani S, Minamoto Y, Fujita S. A novel mechanism for exposure of fibrinogen binding sites on GPIIb-IIIa by a monoclonal antibody. Thromb Haemost. 1995;73:138–143. [PubMed] [Google Scholar]
- 66.Verhamme I, Kvassman JO, Day D, Debrock S, Vleugels N, Declerck PJ, et al. Accelerated conversion of human plasminogen activator inhibitor-1 to its latent form by antibody binding. The Journal of biological chemistry. 1999;274:17511–17517. doi: 10.1074/jbc.274.25.17511. [DOI] [PubMed] [Google Scholar]
- 67.Flynn DC, Olmsted RA, Mackenzie JM, Jr., Johnston RE. Antibody-mediated activation of Sindbis virus. Virology. 1988;166:82–90. doi: 10.1016/0042-6822(88)90149-3. [DOI] [PubMed] [Google Scholar]
- 68.Ros C, Gerber M, Kempf C. Conformational changes in the VP1-unique region of native human parvovirus B19 lead to exposure of internal sequences that play a role in virus neutralization and infectivity. Journal of Virology. 2006;80:12017–12024. doi: 10.1128/JVI.01435-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Paredes AM, Ferreira D, Horton M, Saad A, Tsuruta H, Johnston R, et al. Conformational changes in Sindbis virions resulting from exposure to low pH and interactions with cells suggest that cell penetration may occur at the cell surface in the absence of membrane fusion. Virology. 2004;324:373–386. doi: 10.1016/j.virol.2004.03.046. [DOI] [PubMed] [Google Scholar]
- 70.Lu C, Shimaoka M, Zang Q, Takagi J, Springer TA. Locking in alternate conformations of the integrin alphaLbeta2 I domain with disulfide bonds reveals functional relationships among integrin domains. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:2393–2398. doi: 10.1073/pnas.041618598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gilmore AP, Metcalfe AD, Romer LH, Streuli CH. Integrin-mediated survival signals regulate the apoptotic function of Bax through its conformation and subcellular localization. The Journal of cell biology. 2000;149:431–46. doi: 10.1083/jcb.149.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Takagi J, Isobe T, Takada Y, Saito Y. Structural interlock between ligand-binding site and stalk-like region of beta1 integrin revealed by a monoclonal antibody recognizing conformation-dependent epitope. J Biochem. 1997;121:914–921. doi: 10.1093/oxfordjournals.jbchem.a021673. [DOI] [PubMed] [Google Scholar]
- 73.Ugarova TP, Ljubimov AV, Deng L, Plow EF. Proteolysis regulates exposure of the IIICS-1 adhesive sequence in plasma fibronectin. Biochemistry. 1996;35:10913–10921. doi: 10.1021/bi960717s. [DOI] [PubMed] [Google Scholar]
- 74.Higazi AA, Upson RH, Cohen RL, Manuppello J, Bognacki J, Henkin J, et al. Interaction of single-chain urokinase with its receptor induces the appearance and disappearance of binding epitopes within the resultant complex for other cell surface proteins. Blood. 1996;88:542–551. [PubMed] [Google Scholar]
- 75.Honda S, Tomiyama Y, Pelletier AJ, Annis D, Honda Y, Orchekowski R, et al. Topography of ligand-induced binding sites, including a novel cation-sensitive epitope (AP5) at the amino terminus, of the human integrin beta 3 subunit. The Journal of biological chemistry. 1995;270:11947–11954. doi: 10.1074/jbc.270.20.11947. [DOI] [PubMed] [Google Scholar]
- 76.Ugarova TP, Zamarron C, Veklich Y, Bowditch RD, Ginsberg MH, Weisel JW, et al. Conformational transitions in the cell binding domain of fibronectin. Biochemistry. 1995;34:4457–4466. doi: 10.1021/bi00013a039. [DOI] [PubMed] [Google Scholar]
- 77.Niewiarowska J, Cierniewski CS. GPIIIa(90–102) and GPIIIa(631–653) epitopes as markers of conformational changes occurring during the activation of the glycoprotein IIb/IIIa complex. European journal of biochemistry / FEBS. 1994;224:803–809. doi: 10.1111/j.1432-1033.1994.00803.x. [DOI] [PubMed] [Google Scholar]
- 78.Di Simone C, Buchmeier MJ. Kinetics and pH dependence of acid-induced structural changes in the lymphocytic choriomeningitis virus glycoprotein complex. Virology. 1995;209:3–9. doi: 10.1006/viro.1995.1225. [DOI] [PubMed] [Google Scholar]
- 79.Meyer WJ, Gidwitz S, Ayers VK, Schoepp RJ, Johnston RE. Conformational alteration of Sindbis virion glycoproteins induced by heat, reducing agents, or low pH. Journal of Virology. 1992;66:3504–3513. doi: 10.1128/jvi.66.6.3504-3513.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Geng JG, Moore KL, Johnson AE, McEver RP. Neutrophil recognition requires a Ca(2+)-induced conformational change in the lectin domain of GMP-140. The Journal of biological chemistry. 1991;266:22313–22318. [PubMed] [Google Scholar]
- 81.Flynn DC, Meyer WJ, Mackenzie JM, Jr., Johnston RE. A conformational change in Sindbis virus glycoproteins E1 and E2 is detected at the plasma membrane as a consequence of early virus-cell interaction. Journal of Virology. 1990;64:3643–3653. doi: 10.1128/jvi.64.8.3643-3653.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nilsson B, Nilsson Ekdahl K, Avila D, Nilsson UR, Lambris JD. Neoantigens in complement component C3 as detected by monoclonal antibodies. Mapping of the recognized epitopes by synthetic peptides. The Biochemical journal. 1990;268:55–61. doi: 10.1042/bj2680055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Strandberg K, Kjellberg M, Erb EM, Persson U, Mosher DF, Villoutreix BO, et al. Activated protein C-protein C inhibitor complex formation: characterization of a neoepitope provides evidence for extensive insertion of the reactive center loop. Biochemistry. 2000;39:15713–15720. doi: 10.1021/bi001640h. [DOI] [PubMed] [Google Scholar]
- 84.Saunders DN, Buttigieg KM, Gould A, McPhun V, Baker MS. Immunological detection of conformational neoepitopes associated with the serpin activity of plasminogen activator inhibitor type-2. The Journal of biological chemistry. 1998;273:10965–10971. doi: 10.1074/jbc.273.18.10965. [DOI] [PubMed] [Google Scholar]
- 85.Medved L, Tsurupa G, Yakovlev S. Conformational changes upon conversion of fibrinogen into fibrin. The mechanisms of exposure of cryptic sites. Annals of the New York Academy of Sciences. 2001;936:185–204. doi: 10.1111/j.1749-6632.2001.tb03505.x. [DOI] [PubMed] [Google Scholar]
- 86.Li J, Smolyar A, Sunder-Plassmann R, Reinherz EL. Ligand-induced conformational change within the CD2 ectodomain accompanies receptor clustering: implication for molecular lattice formation. Journal of molecular biology. 1996;263:209–226. doi: 10.1006/jmbi.1996.0570. [DOI] [PubMed] [Google Scholar]
- 87.de Gasparo MM, Wood JM, Heusser CH. Mechanism of inhibition of human renin by monoclonal antibodies. Hypertension. 1988;11:209–216. doi: 10.1161/01.hyp.11.3.209. [DOI] [PubMed] [Google Scholar]
- 88.Elemer GS, Edgington TS. Monoclonal antibody to an activation neoepitope of alpha M beta 2 inhibits multiple alpha M beta 2 functions. Journal of immunology. 1994;152:5836–5844. [PubMed] [Google Scholar]
- 89.Joyner AS, Willis JR, Crowe JE, Jr., Aiken C. Maturation-induced cloaking of neutralization epitopes on HIV-1 particles. PLoS pathogens. 2011;7:e1002234. doi: 10.1371/journal.ppat.1002234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gray L, Sterjovski J, Ramsland PA, Churchill MJ, Gorry PR. Conformational alterations in the CD4 binding cavity of HIV-1 gp120 influencing gp120-CD4 interactions and fusogenicity of HIV-1 envelopes derived from brain and other tissues. Retrovirology. 2011;8:42. doi: 10.1186/1742-4690-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Moscoso CG, Sun Y, Poon S, Xing L, Kan E, Martin L, et al. Quaternary structures of HIV Env immunogen exhibit conformational vicissitudes and interface diminution elicited by ligand binding. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:6091–6096. doi: 10.1073/pnas.1016113108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pantophlet R. Antibody epitope exposure and neutralization of HIV-1. Current pharmaceutical design. 2010;16:3729–3743. doi: 10.2174/138161210794079182. [DOI] [PubMed] [Google Scholar]
- 93.Burastero SE, Figini M, Frigerio B, Lusso P, Mollica L, Lopalco L. Protective versus pathogenic anti-CD4 immunity: insights from the study of natural resistance to HIV infection. J Transl Med. 2009;7:101. doi: 10.1186/1479-5876-7-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li Q, Yafal AG, Lee YM, Hogle J, Chow M. Poliovirus neutralization by antibodies to internal epitopes of VP4 and VP1 results from reversible exposure of these sequences at physiological temperature. Journal of Virology. 1994;1968:3965–70. doi: 10.1128/jvi.68.6.3965-3970.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kwong PD, Doyle ML, Casper DJ, Cicala C, Leavitt SA, Majeed S, et al. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature. 2002;420:678–682. doi: 10.1038/nature01188. [DOI] [PubMed] [Google Scholar]
- 96.Hioe CE, Visciano ML, Kumar R, Liu J, Mack EA, Simon RE, et al. The use of immune complex vaccines to enhance antibody responses against neutralizing epitopes on HIV-1 envelope gp120. Vaccine. 2009;28:352–360. doi: 10.1016/j.vaccine.2009.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kumar R, Visciano ML, Li H, Hioe C. Targeting a Neutralizing Epitope of HIV Envelope Gp120 by Immune Complex Vaccine. J AIDS Clin Res. 2012:S8. doi: 10.4172/2155-6113.S8-002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rhodin NR, Cutalo JM, Tomer KB, McArthur WP, Brady LJ. Characterization of the Streptococcus mutans P1 epitope recognized by immunomodulatory monoclonal antibody 6–11A. Infect Immun. 2004;72:4680–4688. doi: 10.1128/IAI.72.8.4680-4688.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Crowley PJ, Seifert TB, Isoda R, van Tilburg M, Oli MW, Robinette RA, et al. Requirements for surface expression and function of adhesin P1 from Streptococcus mutans . Infect Immun. 2008;76:2456–2468. doi: 10.1128/IAI.01315-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gross EL, Beall CJ, Kutsch SR, Firestone ND, Leys EJ, Griffen AL. Beyond Streptococcus mutans: dental caries onset linked to multiple species by 16S rRNA community analysis. PLoS One. 2012;7:e47722. doi: 10.1371/journal.pone.0047722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nyvad B, Crielaard W, Mira A, Takahashi N, Beighton D. Dental caries from a molecular microbiological perspective. Caries research. 2013;47:89–102. doi: 10.1159/000345367. [DOI] [PubMed] [Google Scholar]
- 102.Peterson SN, Snesrud E, Liu J, Ong AC, Kilian M, Schork NJ, et al. The dental plaque microbiome in health and disease. PLoS One. 2013;8:e58487. doi: 10.1371/journal.pone.0058487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jakubovics NS, Stromberg N, van Dolleweerd CJ, Kelly CG, Jenkinson HF. Differential binding specificities of oral streptococcal antigen I/II family adhesins for human or bacterial ligands. Molecular Microbiology. 2005;55:1591–1605. doi: 10.1111/j.1365-2958.2005.04495.x. [DOI] [PubMed] [Google Scholar]
- 104.Antoniou AN, Blackwood SL, Mazzeo D, Watts C. Control of antigen presentation by a single protease cleavage site. Immunity. 2000;12:391–398. doi: 10.1016/s1074-7613(00)80191-0. [DOI] [PubMed] [Google Scholar]