Abstract
Based on our finding that the peroxisome proliferator-activated receptor (PPAR)γ agonist ciglitazone at high doses was able to mediate PPARγ-independent transcriptional repression of androgen receptor (AR) in a tumor cell-specific manner, we used Δ2CG, a PPARγ-inactive analogue of ciglitazone, to conduct lead optimization to develop a novel class of AR-ablative agents. Structure-activity analysis indicates a high degree of flexibility in realigning Δ2CG’s structural moieties without compromising potency in AR repression, as evidenced by the higher AR-ablative activity of the permuted isomer 9 [(Z)-5-(4-hydroxy-benzylidene)-3-(1-methyl-cyclohexylmethyl)-thiazolidine-2,4-dione]. Further modificiations of 9 gave rise to 12 [(Z)-5-(4-hydroxy-3-trifluoromethyl-benzylidene)-3-(1-methyl-cyclohexylmethyl)-thiazolidine-2,4-dione] which completely inhibited AR expression in LNCaP cells at low μM concentrations. This AR downregulation led to growth inhibition in LNCaP cells through apoptosis induction. Moreover, the role of AR repression in the antiproliferative effect of compound 12 was validated by the differential inhibition of cell viability between androgen-responsive and androgen-nonresponsive cells.
Introduction
Mounting evidence indicates that dysregulation of androgen receptor (AR) through gene amplification or mutations plays a key role in the development of androgen-refractory prostate cancer1–8, a hallmark of incurable and lethal prostate cancer progression. These molecular changes enhance AR sensitivity or permit AR activation by antiestrogen, thus allowing prostate cancer cells to become resistant to androgen ablation-induced apoptosis9, 10. From a clinical perspective, targeting AR expression represents an important strategy to improve the treatment of androgen-independent prostate cancer and ultimately to increase the survival of prostate cancer patients. A recent study indicates that knocking down the AR protein level by a small interfering RNA (siRNA) resulted in significant apoptotic cell death in LNCaP androgen-responsive prostate cancer cells, but not in the AR-null PC-3 cells11. Moreover, in a LNCaP tumor xenograft model, short hairpin RNA (shRNA)-mediated AR knockdown was effective in blocking tumor growth and delaying tumor progression12, which provides a proof-of-principle of this AR-targeted therapy.
Although a number of natural product-based, small-molecule agents exhibit the ability to suppress AR expression, including resveratrol13, vitamin E succinate14, genistein15, and curcumin6, their therapeutic use in humans is limited by unattainable therapeutic concentrations as a result of low potency. Thus, it is of urgency to develop potent AR-ablative agents in the pursuit of new strategies for prostate cancer treatment. During the course of our investigation of the effect of the thiazolidinedione family of peroxisome proliferator-activated receptor (PPAR)γ agonists on repressing prostate specific antigen (PSA)16, we demonstrated that troglitazone and ciglitazone at high doses mediated PPARγ-independent transcriptional repression of AR in a tumor cell-specific manner17. This PPARγ-independent suppression of AR expression might, in part, underlie the antiproliferative activity of troglitazone in prostate cancer cells, and is of translational value to the development of troglitazone and ciglitazone into potent ARablative agents. Our early optimization of troglitazone has generated STG28 (Fig. 1A) with moderate potency in suppressing AR expression in LNCaP prostate cancer cells17. In this study, the lead optimization of ciglitazone gave rise to compound 12, which exhibited the ability to mediate AR ablation at low μM concentrations.
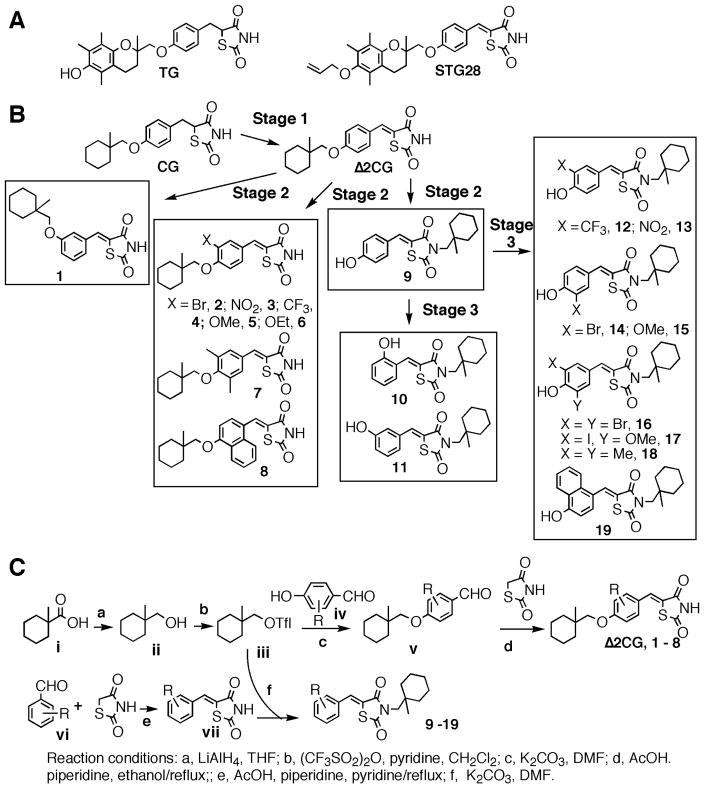
Figure 1.
(A) Representative structures of troglitazone (TG), STG28, ciglitazone (CG), and derivatives. (B) Schematic representation of the course of structural optimization of ciglitazone to develop AR-ablative agents. (C) The general synthetic procedure for ciglitazone derivatives.
Chemistry
The lead optimization of ciglitazone to develop compound 12 consisted of three stages (Fig. 1B). Stage 1 was to abrogate ciglitazone’s PPARγ agonist activity by introducing a double bond adjoining the terminal thiazolidinedione ring, leading to the PPARγ inactive analogue Δ2CG16. Stage 2 was to structurally modify Δ2CG via three distinct strategies: (a) regioisomerization of the (1-methylcyclohexyl)-methyl moiety to yield compound 1, (b) phenyl ring substitutions to give compounds 2 – 8, and (c) permutational rearrangement of the terminal cyclohexyl moiety to generate compound 9. In Stage 3, compound 9 underwent modifications at the terminal phenyl ring, generating two series of compounds, i.e., 10 and 11, and 12 – 19. These Δ2CG derivatives were synthesized according to general procedures described in Fig. 1C, and their ability to suppress AR expression in LNCaP cells was assessed by the AR promoter-luciferase reporter gene assay followed by Western blot analysis.
Results
Dissociation of the PPARγ activity does not affect the ability of Δ2CG to inhibit AR expression at both mRNA and protein levels in LNCaP cells
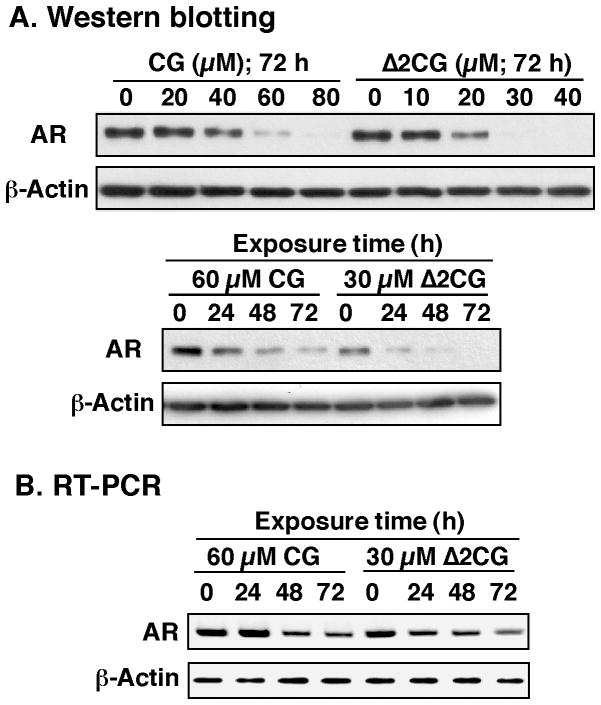
Dose- and/or time-dependent effects of ciglitazone and Δ2CG on suppressing AR expression were assessed in LNCaP cells in 10% fetal bovine serum (FBS) by Western blotting and reverse transcriptase-polymerase chain reaction (RT-PCR). These analyses indicate that Δ2CG, albeit lacking PPARγ agonist activity, exhibited modestly higher potency than ciglitazone in mediating transcriptional repression of AR. For example, the concentrations required for complete suppression of AR protein expression were approximately 30 μM and 60 μM for Δ2CG and ciglitazone, respectively (Fig. 2A).
Figure 2.
Effect of ciglitazone (CG) and Δ2CG on AR ablation in LNCaP cells. (A) Dose- and time-dependent effects of CG and Δ2CG on suppressing AR protein expression levels. Cells were exposed to CG or Δ2CG under the indicated conditions in 10% FBS-supplemented medium, and the lysates were subjected to Western blot analysis. (B) Time-dependent effect of CG (60 μM) and Δ2CG (30 μM) on suppressing the mRNA levels of AR. Cells were treated with either agent in 10% FBS-supplemented medium for the indicated times. Total RNA was isolated and subjected to RT-PCR analysis as described in the Experimental Section.
Furthermore, RT-PCR analysis indicates that the downregulation of AR expression occurred at the transcriptional level (Fig. 2B). Together, these findings confirmed the ability of ciglitazone to ablate AR independently of PPARγ activation, which provided a molecular rationale to use Δ2CG as a starting point for lead optimization to generate potent AR-ablative agents. To expedite the screening of AR-ablative agents, we used a luciferase reporter assay17 to analyze the effect of individual derivatives on suppressing AR transcription by using LNCaP cells transiently transfected with the AR promoter-linked luciferase reporter plasmid.
Lead optimization of Δ2CG
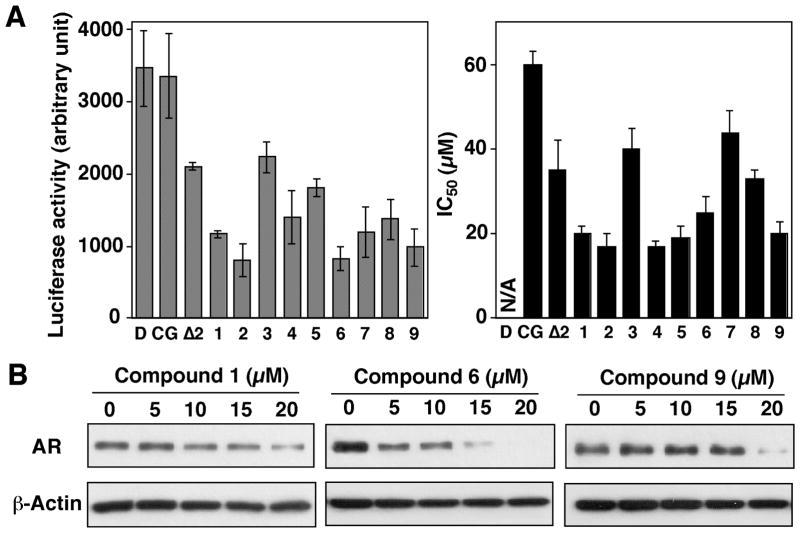
As aforementioned, Δ2CG underwent three types of structural modifications, leading to compounds 1 – 9. Individual derivatives at 10 μM, compared to ciglitazone and Δ2CG, each at 20 μM, were evaluated in the luciferase reporter assay in the transiently transfected LNCaP cells. Relative to Δ2CG, these derivatives showed improved potency in suppressing the activity of the AR promoter, suggesting that a high degree of flexibility existed in the structure-activity relationship. This premise was borne out by the regioisomer 1 and the permuted isomer 9, both of which showed enhanced AR-ablating activity despite substantial configuration changes (Fig. 3A, left panel). Moreover, examination of the IC50 values of individual derivatives in suppressing the viability of LNCaP cells after 48 h of treatment indicates a positive correlation between the ability to suppress AR mRNA transcription and that of inhibiting cell viability (Fig. 3A).
Figure 3.
Differential effects of ciglitazone, Δ2CG, and compounds 1 – 9 on suppressing AR expression in LNCaP cells. (A) Left panel: analysis of the effects of individual compounds on the transcriptional repression of the AR gene by the AR promoter-luciferase reporter assay. LNCaP cells were transiently transfected with an AR promoter-linked luciferase reporter plasmid and exposed to DMSO vehicle (D), ciglitazone (CG, 20 μM), Δ2CG (Δ2, 20 μM), or compounds 1 – 9 (10 μM) in 10% FBS-supplemented RPMI 1640 medium for 48 h. Analysis of luciferase activity was carried out as described in the Experimental Section. Columns, mean (n = 3); bars, standard deviation (SD). Right panel: The IC50 values of individual agents in inhibiting the cell viability of LNCaP cells. Cells were exposed to individual agents at various concentrations in 5% FBS-supplemented RPMI 1640 medium for 48 h, and cell viability was assessed by MTT assays. (B) Western blot analysis of the dose-dependent effect of compounds 1, 6, and 9 on reducing AR protein levels. Cells were exposed to individual agents at the indicated concentrations in 10% FBS-supplemented medium for 72 h, and the lysates were subjected to Western blot analysis.
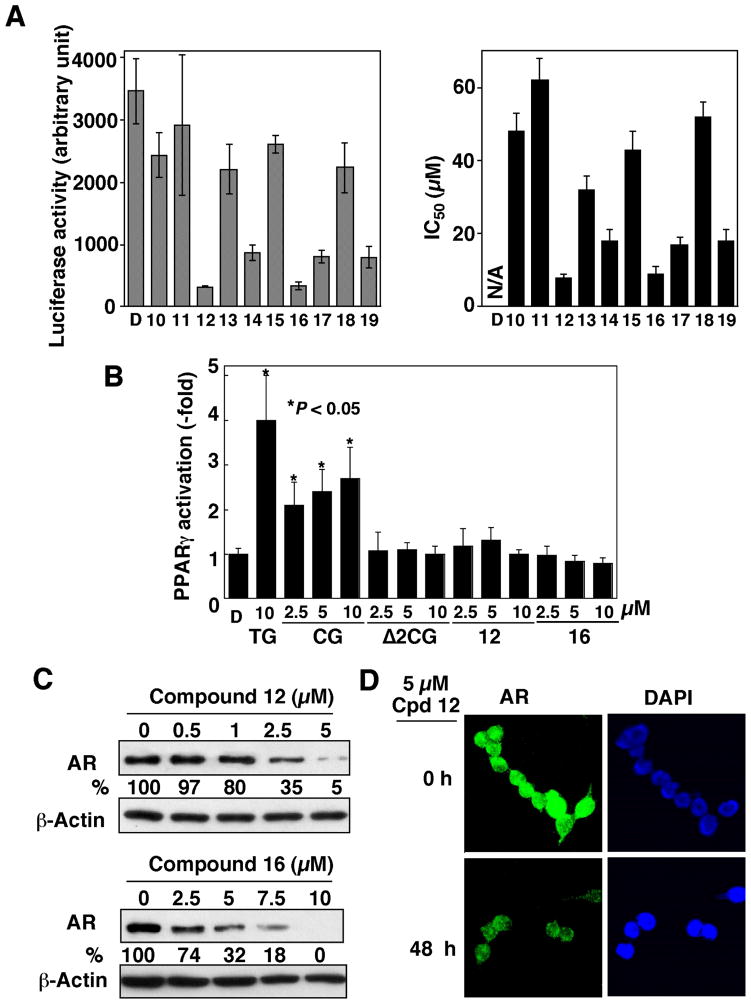
Of these derivatives, we chose compounds 1, 6, and 9 as representatives to conduct Western blot analysis. As shown in Fig. 3B, these three derivatives showed a dose-dependent effect on suppressing AR protein expression (Fig. 3B). In light of the unique structural feature of compound 9, we used this permuted derivative to carry out further structural optimization, generating compounds 10 – 19. The luciferase reporter analysis and cell viability assay indicated a subtle structure-activity relationship among these derivatives (Fig. 4A).
Figure 4.
Differential effect of compounds 10 – 19 on suppressing AR expression in LNCaP cells. (A) Left panel: analysis of the effects of DMSO vehicle (D) or individual compounds on the transcriptional repression of the AR gene by the AR promoter-luciferase reporter assay. LNCaP cells were transiently transfected with an AR promoter-linked luciferase reporter plasmid and exposed to compounds 10 – 19 (10 μM) in 10% FBS-supplemented RPMI 1640 medium for 48 h. Analysis of luciferase activity was carried out as described in the Experimental Section. Columns, mean (n = 3); bars, SD. Right panel: The IC50 values of individual agents in inhibiting the cell viability of LNCaP cells. Cells were exposed to individual agents at various concentrations in 5% FBS-supplemented RPMI 1640 medium for 48 h, and cell viability was assessed by MTT assays. (B) Dose-dependent effect of ciglitazone (CG), Δ2CG, and compounds 12 and 16, relative to that of 10 μM troglitazone (TG), on PPARγ activation in PC-3 cells. PC-3 cells were transiently transfected with PPRE-x3-TK-Luc reporter vector and then exposed to individual agents or DMSO vehicle (D) in 10% FBS-supplemented RPMI 1640 medium for 48 h. Analysis of luciferase activity was carried out as described in the Experimental Section. Columns, mean (n = 6); bars, SD. (C) Western blot analysis of the dose-dependent effect of compounds 12 and 16, on reducing AR protein levels. Cells were exposed to individual agents at the indicated concentrations in 10% FBS-supplemented medium for 72 h, and the lysates were subjected to Western blot analysis. (D) Immunocytochemical analysis of the effect of 5 μM compound 12 on suppressing AR expression after 24-h exposure. The nuclear counterstaining was achieved using a 4′,6-diamino-2-phenylindole (DAPI)-containing mounting medium.
For example, moving the terminal para-OH function to the ortho or meta position (compounds 10 and 11) abolished the ability to suppress AR promoter-luciferase activity and cell viability, indicating its important role in interacting with the target protein. Moreover, substitutions of the phenyl ring with CF3 or Br led to substantially higher potency in AR repression, while those with NO2 or electron-donating groups attenuated the activity (Fig. 4A). Of these derivatives, compounds 12 and 16 represented the optimal agents in inhibiting AR mRNA transcription and LNCaP cell viability.
To demonstrate that this drug-induced transcriptional repression of AR was independent of PPARγ, we examined the ability of compounds 12 and 16 versus troglitazone, ciglitazone, and Δ2CG to transactivate PPARγ by using the PPAR response element (PPRE)-luciferase reporter assay. In PC-3 cells transfected with a reporter construct (PPRE-x3-TK-Luc), troglitazone and ciglitazone at 10 μM exhibited a significant effect on increasing luciferase activity, ranging from 2.5-fold to 4-fold (P < 0.05). In contrast, compounds 12 and 16, like their parent compounds Δ2CG, lacked appreciable activity in PPARγ activation.
Western blot analysis indicates that the IC50 values for suppressing AR expression by compounds 12 and 16 after 72-h exposure were approximately 2 μM and 4 μM, respectively (Fig. 4C). The ability of compound 12 to suppress AR expression was further demonstrated by immunocytochemical analysis (Fig. 4D). As shown, exposure of LNCaP cells to 5 μM compound 12 for 48 h led to a substantial decrease in AR levels in the nucleus.
Antitumor effects of compound 12 in prostate cancer cells
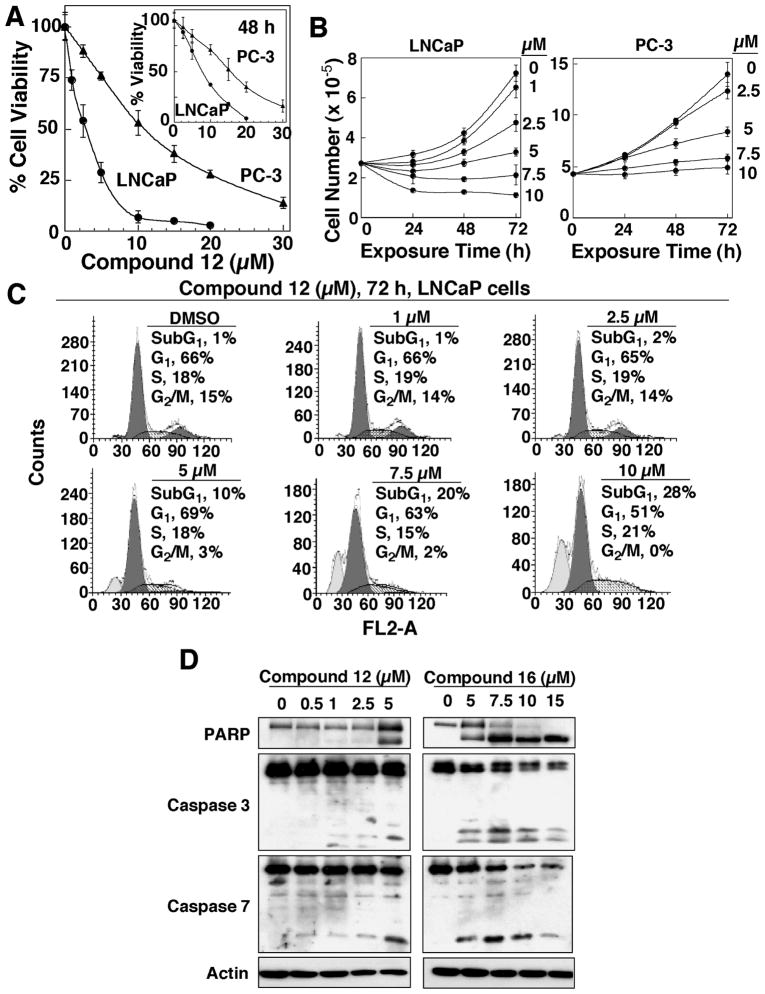
We assessed the antitumor effects of compound 12 in both LNCaP androgen-responsive and PC-3 androgen-nonresponsive prostate cancer cells via three different methods, including the MTT assay for cell viability, cell counting for cell proliferation, and flow cytometric analysis for cell cycle distribution. Due to lack of AR expression, PC-3 cells exhibited substantially lower sensitivity to the antiproliferative activities of compound 12 as compared to LNCaP cells. The IC50 values for suppressing cell viability were 8 μM and 3 μM, at 48 h and 72 h of drug treatment, respectively, in LNCaP cells, and 15 μM and 12 μM, respectively, in PC-3 cells (Fig. 5A).
Figure 5.
Antitumor effects of compound 12 in LNCaP cells. (A) Differential dose-dependent effects of compound 12 on the inhibition of cell viability of LNCaP versus PC-3 cells at 48 h (inset) and 72 h of treatment. Cells were exposed to the indicated concentrations of compound 12 in 5% FBS-containing RPMI 1640 medium for 48 and 72 h, and cell viability was determined by MTT assays. Points, mean (n = 6); bars, SD. (B) Dose- and time-dependent antiproliferative effects of compound 12 in LNCaP (left panel) and PC-3 (right panel) cells. Cells were seeded into six-well plates (250,000 cells/well), incubated for 24 h, and exposed to compound 12 at the indicated concentrations in 5% FBS-supplemented medium for different time intervals. Cells were harvested, and counted using a Coulter counter. (C) Flow cytometric analysis of LNCaP cells after treatment with DMSO or the indicated concentrations of compound 12 for 72 h. Percentages of cell cycle distribution represent the mean of two independent determinations. (D) Western blot analysis of the dose-dependent effects of compounds 12 and 16 on PARP cleavage, caspase 3 activation, and caspase 7 activation in LNCaP cells after 72 h of treatment.
This differential susceptibility was also manifest in the cell counting assay, in which compound 12 exhibited at least twofold higher potency in inhibiting the proliferation of LNCaP cells as compared to PC-3 cells (Fig. 5B). Moreover, cell cycle analysis was carried out after exposing LNCaP cells to different doses of compound 12 for 72 h (Fig. 5C). As shown, compound 12 caused a dose-dependent increase in the sub-G1 population, accompanied by decreases in the G2/M phase (Fig. 5C). Furthermore, the ability of compounds 12 and 16 to induce apoptotic death in LNCaP cells was demonstrated by their dose-dependent effects on modulating various apoptosis-related biomarkers, including PARP cleavage, and the proteolytic activation of caspase 3 and caspase 7 (Fig. 5D).
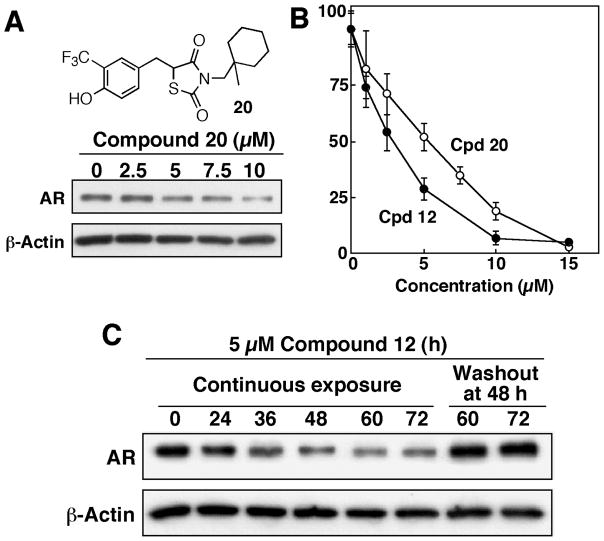
Our earlier study indicates that thiazolidinediones mediated the transcriptional repression of AR by facilitating the degradation of the transcription factor Sp117. However, it was reported that compounds containing a 5-arylidene-3-aryl-2,4-thiazolidinedione substructure underwent conjugation addition with p-thiocresol in the presence of piperidine upon heating18. This report raised a possibility that compound 12 and other derivatives might act as “Michael acceptors” by covalently modify the target enzyme/protein upon binding. Here, we obtained two lines of evidence to refute this possibility. First, compound 20, a saturated counterpart of compound 12, retained the ability to suppress AR expression and cell viability, though with slightly lower potency, in LNCaP cells (Fig. 6A and B). Second, the expression level of AR in drug-treated LNCaP cells would be rapidly restored once compound 12 was removed (or washed out) from the medium (Fig. 6C). This rapid restoration of the AR expression suggests a reversible nature of this ligand-protein interaction.
Figure 6.
Evidence that the ability of compound 12 to inhibit AR expression in LNCaP cells is not mediated through an irreversible mechanism. (A) Structure and the dose-dependent effect of compound 20 on suppressing AR expression in LNCaP cells. Cells were exposed to compound 20 at the indicated concentrations in 10% FBS-supplemented medium for 72 h, and the lysates were subjected to Western blot analysis. (B) Dose-dependent effects of compound 12 versus compound 20 in suppressing the viability of LNCaP cells. Cells were exposed to individual agents at various concentrations in 5% FBS-supplemented RPMI 1640 medium for 48 h, and cell viability was assessed by MTT assays. (C) Restoration of AR expression in LNCaP cells after compound 12 was washed out. The effect of 5 μM compound 12 on AR repression in LNCaP cells was examined at different intervals throughout a 72 h-period in two different manners. Continuous exposure: cells in T-25 flasks were incubated in drug-containing, 10% FBS-supplemented medium for 72 h, and washout at 48 h of treatment: cells in T-25 flasks were exposed to the agent for 44 h, followed by incubation in drug-free medium for additional 24 h. AR levels in cell lysates were analyzed by Western blot analysis.
Discussion
In light of the pivotal role of PPARγ in prostate cell proliferation and differentiation, the chemopreventive activity of thiazolidinediones in prostate cancer has been attributed to their ability to activate PPARγ signaling, leading to the terminal differentiation and growth arrest of tumor cells19–21. However, mounting evidence suggests that the antiproliferative ability of these agents is independent of their PPARγ agonist activity16, 17, 22–31. In our laboratory, we have identified several “off-target” mechanisms that might underlie the antitumor effects of thiazolidinediones, including Bcl-2/Bcl-xL inhibition, proteasomal degradation of cyclin D125, β-catenin32, and Sp117, and transcriptional repression of PSA16 and AR17. Separation of these pharmacological effects from PPARγ activation provides a mechanistic rationale for using thiazolidinediones as a scaffold to develop potent molecularly targeted agents. Considering the importance of AR in prostate tumorigenesis and tumor progression, we carried out lead optimization of ciglitazone and its PPARγ-inactive derivative Δ2CG to develop potent AR-ablative agents.
There existed a high degree of tolerance for the substructural rearrangement of Δ2CG without compromising the AR-ablative activity, as evidenced by the improved potency of compounds 1 and 9. In contrast, modifications of the phenyl ring exhibited a subtle effect on the AR-ablative potency. For example, changing the orientation of the terminal hydroxyl function of compound 9 completely abrogated the ability of the resulting compounds 10 and 11 to suppress AR expression, while the CF3- or di-Br-substitution led to enhanced potency. Together, these findings suggest that the benzylidene-thiazolidinedione substructure played a crucial role in interacting with the target protein.
Among all derivatives examined, compound 12 represented a structurally optimized derivative with an-order-of-magnitude higher potency than ciglitazone in suppressing AR expression. This AR downregulation led to growth inhibition in LNCaP cells through apoptosis induction, as evidenced by flow cytometry, PARP cleavage and caspase activation. The role of AR repression in the antiproliferative effect of compound 12 was supported by the differential inhibition of cell viability between LNCaP androgen-responsive and PC-3 androgen-nonresponsive cells. Because thiazolidinediones mediate AR repression through downregulation of Sp117, compound 12 also suppresses the transcription of many Sp1-targeted genes (data not shown), which accounts for the ability of compound 12 to inhibit PC-3 cell viability.
Relative to many natural product-based agents that suppress AR expression/function, such as resveratrol13, vitamin E succinate14, genistein15, and curcumin6, compound 12 is substantially more effective in downregulating AR expression. Thus, this AR-ablative agent has translational potential to foster new therapeutic strategies for prostate cancer treatment as a single agent or in combination with other molecularly targeted agents.
Conclusion
The in vivo efficacy of targeting AR expression to block tumor growth and delaying tumor progression has recently been demonstrated in a LNCaP tumor xenograft model by using shRNA-mediated AR knockdown12. This finding provides a proof-of-principle that inhibition of AR expression represents a therapeutically relevant strategy for prostate cancer treatment. Relative to shRNAs, small-molecule agents prove to be advantageous in many aspects of cancer therapy. Consequently, in vivo testing of compound 12 to suppress prostate tumor growth is currently underway.
Experimental Section
Chemical reagents and organic solvents were purchased from Sigma-Aldrich unless otherwise mentioned. Nuclear magnetic resonance spectra (1H NMR) were measured on a Bruker DPX 300 model spectrometer. Chemical shifts (δ) were reported in parts per million (ppm) relative to the TMS peak. Electrospray ionization mass spectrometry analyses were performed with a Micromass Q-Tof II High-Resolution electrospray mass spectrometer. Elemental analyses were performed by the Atlantic Microlab, Inc. (Norcross, GA), and were reported within 0.4% of calculated values. Flash column chromatography was performed with silica gel (230–400 mesh). Δ2CG and the two series of compounds, 1 – 8 and 9 – 19, were synthesized according to the general methods described in Figure 1B, which are illustrated by the synthesis of compounds 1 and 9 as examples.
(Z)-5-[3-(1-Methyl-cyclohexylmethoxy)-benzylidene]-thiazolidine-2,4-dione (1)
Step a
To a stirring solution of LiAlH4 (20 mmol) in anhydrous THF (10 mL) at 4°C was added 1-methyl-cyclohexanecarboxylic acid (i, 7.0 mmol) in 50 mL of THF dropwise over a period of 1 hour. The solution was stirred at refluxing temperature under N2 for 6 hours. The solution was cooled to 4°C by ice bath, and 1 mL of 1 N NaOH (1 mL) followed by H2O (2 mL) was slowly added to the solution to quench the reaction. The solution was stirred at 23 °C for 1 hour and then filtered to remove solid material. The solution was concentrated. Purification by flash silica gel chromatography (ethyl acetate/hexanes, 1:2) gave the product, (1-methyl-cyclohexyl)-methanol (ii), in 82% yield.
Step b
A solution of compound ii (1 mmol) in dry CH2Cl2 (5 mL) was cooled to 4°C, to which was added pyridine (1.1 mmol) and triflate anhydride (1.1 mmol). After stirring at 4°C for 2 h, the solution was concentrated, and the residue was purified by flash silica gel column chromatography (ethyl acetate/hexanes, 1:10) to afford trifluoro-methanesulfonic acid 1-methyl-cyclohexylmethyl ester (iii) in 35% yield.
Step c
A mixture of compound iii (0.5 mmol), 3-hydoxy-benzaldehyde (iv, 0.6 mmol) and K2CO3 (0.7 mmol) were dissolved in DMF (3 mL). The solution was heated to 80°C for 4 hr. The solution was poured into water, extracted with ethyl acetate (10 mL) three times, and concentrated. The residue was purified by chromatography and resulted in 0.22 mmol of 3-(1-methyl-cyclohexylmethoxy)-benzaldehyde (v) with a 44% yield.
Step d
A mixture consisting of compound v (0.5 mmol), 2, 4-thiazolidinedione (0.6 mmol) and catalytic amounts of piperidine was refluxed in EtOH (5 mL) for 24 h and then concentrated. The oily product was dissolved in ethyl acetate, poured into water and acidified with AcOH. The solution was extracted with ethyl acetate, dried and concentrated. The residue was purified by silica gel chromatography, providing compound 1 in 67% yield. 1H NMR (300 MHz, CDCl3) δ 1.04 (s, 3H), 1.46–1.56 (m, 10H), 3.69 (s, 2H), 6.78–7.28 (m, 2H), 7.08 (d, J = 8.40 Hz, 1H), 7.39 (dt, J = 2.10, 8.40 Hz, 1H), 7.84 (s, 1H), 8.21–8.78 (br, 1H); HRMS exact mass of (M + Na)+, 354.1140 amu; observed mass of (M + Na)+, 354.113 amu.
(Z)-5-[3-Bromo-4-(1-methyl-cyclohexylmethoxy)-benzylidene]-thiazolidine-2,4-dione (2)
1H NMR (300 MHz, CDCl3) δ 1.11 (s, 3H), 1.40–1.61 (m, 10H), 3.77 (s, 2H), 6.95 (d, J = 8.42 Hz, 1H), 7.41 (dd, J = 2.10, 8.42 Hz, 1H), 7.69 (d, 1H, J = 2.10), 7.74 (s, 1H), 8.38 (s, 1H); HRMS exact mass of (M + Na)+, 432.0245 amu; observed mass of (M + Na)+, 432.0247 amu.
(Z)-5-[4-(1-Methyl-cyclohexylmethoxy)-3-nitro-benzylidene]-thiazolidine-2,4-dione (3)
1H NMR (300 MHz, CDCl3) δ 1.08 (s, 3H), 1.42–1.59 (m, 10H), 3.79 (s, 2H,), 7.23 (d, J = 8.40 Hz, 1H), 7.85 (d, J = 8.42 Hz, 1H), 7.92 (s, 1H), 8.10 (s, 1H), 8.33 (s, 1H); HRMS exact mass of (M + Na)+, 399.0991 amu; observed mass of (M + Na)+, 399.0995 amu.
(Z)-5-[4-(1-Methyl-cyclohexylmethoxy)-3-trifluoromethyl-benzylidene]-thiazolidine-2,4-dione (4)
1H NMR (300 MHz, CDCl3) δ 1.08 (s, 3H), 1.42–1.59 (m, 10H), 3.79 (s, 2H), 7.10 (d, J = 8.40 Hz, 1H), 7.63 (d, J = 8.42 Hz, 1H), 7.71 (s, 1H), 7.80 (s, 1H), 8.09–8.12 (br, 1H), HRMS exact mass of (M + Na)+, 422.1014 amu; observed mass of (M + Na)+, 422.1019 amu.
(Z)-5-[3-Methoxy-4-(1-methyl-cyclohexylmethoxy)-benzylidene]-thiazolidine-2,4-dione (5)
1H NMR (300 MHz, CDCl3) δ 1.09 (s, 3H), 1.40–1.58 (m, 10H), 3.75 (s, 2H,), 3.96(s, 3H), 6.96 (d, J = 8.40 Hz, 1H), 7.00 (s, 1H), 7.11 (d, J = 8.42 Hz, 1H), 7.79 (s, 1H), 8.55 (s, 1H); HRMS exact mass of (M + Na)+, 384.1245 amu; observed mass of (M + Na)+, 384.1239 amu.
(Z)-5-[3-Ethoxy-4-(1-methyl-cyclohexylmethoxy)-benzylidene]-thiazolidine-2,4-dione (6)
1H NMR (300 MHz, CDCl3) δ 1.08 (s, 3H), 1.40–1.58 (m, 13H), 3.74 (s, 2H), 4.10 (q, J = 6.9 Hz, 2H), 6.95 (d, J = 8.40 Hz, 1H), 7.01 (d, J = 2.10 Hz, 1H), 7.11 (dd, J = 8.40, 2.10 Hz, 1H), 7.79 (s, 1H), 8.42 (s, 1H); HRMS exact mass of (M + Na)+, 389.1402 amu; observed mass of (M + Na)+, 389.1402 amu.
(Z)-5-[3,5-Dimethyl-4-(1-methyl-cyclohexylmethoxy)-benzylidene]-thiazolidine-2,4-dione (7)
1H NMR (300 MHz, CDCl3) δ 1.13 (s, 3H), 1.32–1.59 (m, 10H), 2.42 (s, 6H), 3.48 (s, 2H), 7.17 (s, 2H), 7.76(s, 1H), 8.26 (s, 1H); HRMS exact mass of (M + Na)+, 382.1453 amu; observed mass of (M + Na)+, 382.1448 amu.
(Z)-5-[4-(1-Methyl-cyclohexylmethoxy)-naphthalen-1-ylmethylene]-thiazolidine-2,4-dione (8)
1H NMR (300 MHz, CDCl3) δ 1.18 (s, 3H), 1.51–1.59 (m, 10H), 3.91 (s, 2H), 6.915 (d, J = 8.70 Hz, 1H), 7.55–7.69 (m, 3H), 8.12 (d, J = 8.70, 1H), 8.39 (d, J = 8.40, 1H), 8.59 (s, 1H); HRMS exact mass of (M + Na)+, 404.1296 amu; observed mass of (M + Na)+, 404.1299 amu.
(Z)-5-(4-Hydroxy-benzylidene)-3-(1-methyl-cyclohexylmethyl)-thiazolidine-2,4-dione (9)
Step e
A mixture of p-hydroxybenzaldehyde (vi; 0.5 mmol), 2,4-thiazolidinedione, (0.6 mmol), and catalytic amounts of piperidine and AcOH was refluxed in toluene (5 mL) for 24 h. The precipitated product was filtered, washed with toluene (3 × 10 mL), and dried in vacuo at 60°C overnight, yielding 5-(4-hydroxybenzylidene)-thiazolidine-2,4-dione (vii) in a 85% yield.
Step f
A solution of compound vii (0.5 mmol), compound iii (0.6 mmol) and K2CO3 (0.65 mmol) were stirred in DMF (3 mL) at 80°C for 4 hr, poured into water, extracted with ethyl acetate (3 × 10 mL), dried and concentrated. The residue was purified by chromatography, affording compound 9 in 42% yield. 1H NMR (300 MHz, CDCl3) δ 0.94 (s, 3H), 1.14–1.86 (m, 10H), 3.63 (s, 2H), 5.69 (s, 1H), 6.94(d, J = 8.40 Hz, 2H), 7.43(d, J = 8.40 Hz, 2H), 7.83 (s, 1H); HRMS exact mass of (M + Na)+, 354.1140 amu; observed mass of (M + Na)+, 354.1141 amu.
(Z)-5-(3-Hydroxy-benzylidene)-3-(1-methyl-cyclohexylmethyl)-thiazolidine-2,4-dione (10)
1H NMR (300 MHz, CDCl3) δ 0.96 (s, 3H), 1.24–1.67 (m, 10H), 3.65 (s, 2H), 5.24 (s, 1H), 6.70 (d, J = 1.5 Hz, 1H), 6.93(dd, J = 8.10, 1.5 Hz, 1H), 7.10(d, J = 7.80 Hz, 1H), 7.36 (dd, J = 7.80, 7.50 Hz, 1H), 7.84 (s, 1H); HRMS exact mass of (M + Na)+, 354.1140 amu; observed mass of (M + Na)+, 354.1143 amu.
(Z)-5-(2-Hydroxy-benzylidene)-3-(1-methyl-cyclohexylmethyl)-thiazolidine-2,4-dione (11)
1H NMR (300 MHz, CDCl3) δ 0.95 (s, 3H), 1.22–1.65 (m, 10H), 3.66 (s, 2H), 6.44 (d, J = 0.9 Hz, 1H), 6.91 (dd, J = 8.10, 0.9 Hz, 1H), 7.04 (td, J = 7.2, 0.6 Hz, 1H), 7.32 (tdd, J = 7.5, 1.5, 0.6Hz, 1H), 7.46 (dd, J = 7.80, 1.5Hz, 1H), 8.42 (s, 1H); HRMS exact mass of (M + Na)+, 354.1140 amu; observed mass of (M + Na)+, 354.1145 amu.
(Z)-5-(4-Hydroxy-3-trifluoromethyl-benzylidene)-3-(1-methyl-cyclohexylmethyl)-thiazolidine-2,4-dione (12)
1H NMR (300 MHz, CDCl3) δ 0.95 (s, 3H), 1.46–1.56 (m, 10H), 3.64 (s, 2H), 6.08–6.38 (br, 1H), 7.09 (d, J = 8.40 Hz, 1H), 7.59 (d, J = 8.40 Hz, 1H), 7.69 (s, 1H), 7.83 (s, 1H); HRMS exact mass of (M + Na)+, 422.1014 amu; observed mass of (M + Na)+, 422.1012 amu. Anal. (C19H20F3NO3S) C, H, N, S, O, F.
(Z)-5-(4-Hydroxy-3-nitro-benzylidene)-3-(1-methyl-cyclohexylmethyl)-thiazolidine-2,4-dione (13)
1H NMR (300 MHz, CDCl3) δ 0.96 (s, 3H), 1.23–1.57 (m, 10H), 3.68 (s, 2H), 7.31(d, J = 8.40 Hz, 1H), 7.74 (dd, J = 8.40, 2.1Hz, 1H), 7.81 (s, 1H), 8.29 (d, J = 2.1 Hz, 1H); HRMS exact mass of (M + Na)+, 399.0991 amu; observed mass of (M + Na)+, 399.0991 amu.
(Z)-5-(3-Bromo-4-hydroxy-benzylidene)-3-(1-methyl-cyclohexylmethyl)-thiazolidine-2,4-dione (14)
1H NMR (300 MHz, CDCl3) δ 0.79 (s, 3H), 1.17–1.46 (m, 10H), 3.36 (s, 2H), 7.01 (d, J = 8.40 Hz, 1H), 7.37 (d, J = 8.40 Hz, 1H), 7.73 (s, 2H); HRMS exact mass of (M + Na)+, 432.0245 amu; observed mass of (M + Na)+, 432.0245 amu. Anal. (C18H20BrNO3S) C, H, N, O
(Z)-5-(4-Hydroxy-3-methoxy-benzylidene)-3-(1-methyl-cyclohexylmethyl)-thiazolidine-2,4-dione (15)
1H NMR (300 MHz, CDCl3) δ 0.92 (s, 3H), 1.21–1.58 (m, 10H), 3.62 (s, 2H), 3.97 (s, 3H), 5.95 (br, 1H), 6.90–7.03 (m, 2H), 7.10 (d, J = 7.80 Hz, 1H), 7.82 (s, 1H), HRMS exact mass of (M + Na)+, 384.1245 amu; observed mass of (M + Na)+, 384.1245 amu. Anal. (C19H23NO4S) C, H, N, O.
(Z)-5-(3,5-Dibromo-4-hydroxy-benzylidene)-3-(1-methyl-cyclohexylmethyl)-thiazolidine-2,4-dione (16)
1H NMR (300 MHz, CDCl3) δ 0.94 (s, 3H), 1.32–1.56 (m, 10H), 3.63 (s, 2H), 6.22(s, 1H), 7.62 (s, 2H), 7.68(s, 1H); HRMS exact mass of (M + Na)+, 511.9330 amu; observed mass of (M + Na)+, 511.9329 amu. Anal. (C18H19Br2NO3S) C, H, N, S, O, Br.
(Z)-5-(4-Hydroxy-3-iodo-5-methoxy-benzylidene)-3-(1-methyl-cyclohexylmethyl)-thiazolidine-2,4-dione (17)
1H NMR (300 MHz, CDCl3) δ 0.94 (s, 3H), 1.22–1.62 (m, 10H), 3.63 (s, 2H), 3.96 (s, 3H), 6.44 (s, 1H), 6.97 (s, 1H), 7.50 (s, 1H), 7.73(s, 1H), HRMS exact mass of (M + Na)+, 510.0212 amu; observed mass of (M + Na)+, 510.0213 amu.
(Z)-5-(4-Hydroxy-3,5-dimethyl-benzylidene)-3-(1-methyl-cyclohexylmethyl)-thiazolidine-2,4-dione (18)
1H NMR (300 MHz, CDCl3) δ 0.94 (s, 3H), 1.22–1.66 (m, 10H), 2.30 (s, 6H), 3.62 (s, 2H), 5.06 (s, 1H), 7.17 (s, 2H), 7.78 (s, 1H); HRMS exact mass of (M + Na)+, 382.1453 amu; observed mass of (M + Na)+, 382.1454 amu.
(Z)-5-(4-Hydroxy-naphthalen-1-ylmethylene)-3-(1-methyl-cyclohexylmethyl)-thiazolidine-2,4-dione (19)
1H NMR (300 MHz, CDCl3) δ 0.98 (s, 3H), 1.20–1.66 (m, 10H), 3.67 (s, 2H), 5.91 (s, 1H), 6.91(d, J = 7.80 Hz, 1H), 7.56–7.67(m, 3H), 8.15 (d, J = 8.40 Hz, 1H), 8.29(d, 1H, J = 7.20 Hz), 8.60 (s, 1H); HRMS exact mass of (M + Na)+, 404.1296 amu; observed mass of (M + Na)+, 404.1296 amu.
5-(4-Hydroxy-3-trifluoromethyl-benzyl)-3-(1-methyl-cyclohexylmethyl)-thiazolidine-2,4-dione (20)
A mixture of compound 12 (20 mg) and Pd-C (40 mg) in methanol (5 mL) was stirred under hydrogen (50 psi) overnight, filtered, and concentrated to dryness under vacuum. The residue was purified by silica gel flash chromatography and re-crystallized with ethyl acetate-hexane (1:8), giving compound 20 (14 mg). 1H NMR (250 MHz, CDCl3) 0.81 (s, 3H), 1.16–1.59 (m, 10H), 3.13(dd, 1H, J = 9.3Hz, 8.7Hz), 3.45 (s, 2H), 3.51 (dd, 1H, J = 9.3Hz, 3.6Hz), 4.44 (dd, 1H, J = 3.6Hz, 8.7Hz), 5.53 (s, 1H), 6.92 (d, 1H, J = 8.40Hz), 7.33 (d, 1H, J = 8.40Hz), 7.39 (s, 1H), HRMS exact mass of (M + Na)+, 422.1170 amu; observed mass of (M + Na)+, 422.1173 amu.
Cell Culture
LNCaP androgen-responsive (p53+/+) and PC-3 androgen-nonresponsive (p53−/−) prostate cancer cells were obtained from the American Type Culture Collection (Manassas, VA), and were maintained in RPMI 1640 supplemented with 10% fetal bovine serum at 37°C in a humidified incubator containing 5% carbon dioxide.
Cell Counting and Cell Viability Assay
LNCaP or PC-3 cells were placed in six-well plates (2.5 × 105 cells/well) in 10% FBS-supplemented RPMI 1640 for 24 h, and treated with various concentrations of compound 12 for additional 24, 48 and 72 h. Cells were then trypsinized and counted by using a Coulter counter (Model Z1 D/T, Beckman Coulter, Fullerton, CA). Cell viability was assessed by using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay in six replicates in 96-well plates. LNCaP or PC-3 cells were seeded at 6000 cells per well in 10% FBS-supplemented RPMI 1640 for 24 h, followed by treatments with various compounds in 5% FBS-supplemented RPMI 1640 at the indicated concentrations. Controls received DMSO at a concentration equal to that in drug-treated cells. After the end of incubation, MTT (0.5 mg/ml) in 10% FBS-supplemented RPMI 1640 was added to each well, and cells were incubated at 37°C for 2 hours. Medium was removed and the reduced MTT dye was solubilized in DMSO (200 μl/well). Absorbance was determined at 570 nm by a 96-well plate reader.
Transfection and Luciferase assay
The 3.6-kilobase AR promoter-linked reporter plasmid p-3600ARCAT was kindly provided by Dr. Chawnshang Chang (University of Rochester Medical Center, Rochester, NY). The AR promoter gene (−3600 to +550) encompassing the transcription start site was isolated by using PCR to generate hAR-luc with the following primers: 5′-TACAGGTACCGGTATCTCGACCTGCAGGTC-3′ and 5′-TGTTAGATCTTGCTGAAGCCGCTCCCCAGT-3′. The fragment was subcloned into the pGL3 luciferase reporter vector (Promega, Madison, WI) at KpnI and BglII in the multiple cloning site. The PPRE-x3-TK-Luc reporter vector contains three copies of the PPAR-response element (PPRE) upstream of the thymidine kinase promoter-luciferase fusion gene and was kindly provided by Dr. Bruce Spiegelman (Harvard University, Cambridge, MA). The pCMVSp1 plasmid was purchased from Origene Technologies, Inc. (Rockville, MD). LNCaP or PC3 cells were transfected with 5 μg of individual plasmids in an Amaxa Nucleofector using a cell line-specific nucleofector kit according to the manufacturer’s protocol (Amaxa Biosystems, Cologne, Germany), and then seeded in 6-well plates at 5 × 105 cells per well for 48 h. The transfection efficiency was determined to be 70 – 80% by transfecting cells with 2 μg of pmaxGFP plasmid, followed by fluorescence microscopy to measure GFP expression. For each transfection, herpes simplex virus thymidine kinase promoter-driven Renilla reniformis luciferase was used as an internal control for normalization.
For the reporter gene assay, after transfection, cells were cultured in 24-well plates in 10% FBS-supplemented RPMI 1640 medium for 48 h, subject to different treatments for the indicated times, collected, and lysed with passive lysis buffer (Promega). Fifty-μl aliquots of the lysates were added to 96-well plates, and luciferase activity was monitored after adding 100 μl of luciferase substrate (Promega) each well by using a MicroLumatPlus LB96V luminometer (Berthold Technologies, Oak Ridge, TN) with the WinGlow software package. All transfection experiments were carried out in six replicate.
Cell Cycle Analysis
LNCaP cells were seeded in 6-well plates (2.5 × 106 cells/well) and treated with different concentrations of compound 12 for 72 h. After extensive washing with PBS, cells were trypsinized followed by fixation in ice-cold 80% ethanol at 4°C overnight. Cells were then centrifuged for 5 min at 1500 × g at room temperature, and stained with propidium iodide (50 μg/ml) and RNase A (100 units/ml) in PBS. Cell cycle phase distributions were determined on a FACScort flow cytometer and analyzed by the ModFitLT V3.0 program.
RT-PCR and Immunoblotting
LNCaP cells were cultured in T25 flasks at an initial density of 1×106 cells/flask. After exposure to various compounds at the indicated conditions, cells were subject to total RNA isolation by using an RNeasy mini-kit (QIAGEN, Valencia, CA). RNA concentrations were determined by measuring absorption at 260 nm in a spectrophotometer. Aliquots of 6 μg of total RNA from each sample were reverse-transcribed to cDNA using an Omniscript RT Kit (QIAGEN) according to the manufacturer’s instructions. The PCR primers used were as follows.
-
AR: 5′-ACACATTGAAGGCTATGAATGTC-3′ (forward)
5′-TCACTGGGTGTGGAAATAGATGGG-3′ (reverse)
-
β-actin: 5′-TCTACAATGAGCTGCGTGTG-3′ (forward)
5′-GGTCAGGATCTTCATGAGGT-3′ (reverse)
PCR reaction products were separated electrophoretically in 1.5% agarose gels. For immunoblotting, protein extracts were prepared by M-PER Mammalian Protein Extraction Reagent (Pierce, Rockford, IL) with freshly added 1% phosphatase and protease inhibitor cocktails (Calbiochem) followed by centrifugation at 13,000 × g for 10 min. Supernatant was collected and protein concentration was determined by protein assay reagent (Bio-Rad, CA). Protein extracts were then suspended in 2x SDS sample buffer, and subject to 10% SDS-polyacrylamide gels. After electrophoresis, proteins were transferred to nitrocellulose membranes using a semidry transfer cell. The transblotted membrane was washed twice with Tris-buffered saline containing 0.1% Tween 20 (TBST). After blocking with TBST containing 5% nonfat milk for 1 h, the membrane was incubated with mouse monoclonal anti-AR (Santa Cruz) or anti-β-actin (MP Biomedicals) antibodies (diluted 1:1000) in 1% TBST nonfat milk at 4°C overnight. After incubation with the primary antibody, the membrane was washed three times with TBST for a total of 30 min, followed by incubation with horseradish peroxidase conjugated goat anti-mouse IgG (diluted 1:2500) for 1 h at room temperature. After three thorough washes with TBST for a total of 30 min, the immunoblots were visualized by enhanced chemiluminescence.
Immunocytochemical Analysis
Cells were seeded onto coverslips in six-well plates (2.5 × 105 cells/well) for 24 h followed by exposure to 5 μM compound 12 for an additional 48 h. After extensive washing with PBS, cells were fixed and permeabilized with PBS containing 0.1% Triton X-100 for 1 h, and then incubated with anti-AR (1:100 dilution) in PBS containing 0.1% Triton X-100, 0.2% bovine serum albumin, 0.5 mM PMSF, and 1 mM DTT at room temperature for 12 h followed by Alexa Fluor 488-conjugated goat anti-mouse IgG (1:100; Molecular Probes) for 2 h. Nuclear counterstaining was performed by mounting with 4,6-diamidino-2-phenylindole (DAPI)-containing medium. Images of immunocytochemically labeled samples were observed using a Nikon microscope (Eclipse TE300).
Acknowledgments
This work is supported by National Institutes of Health Grant CA112250, Department of Defense Prostate Cancer Research Program Grant W81XWH-05-1-0089 and grants from William R. Hearst Foundation and Prostate Cancer Foundation, and the Lucius A. Wing Endowed Chair Fund at the Ohio State University.
Abbreviations
- PPARγ
peroxisome proliferator-activated receptor γ
- AR
androgen receptor
- shRNA
short hairpin RNA
- PSA
prostate specific antigen
- FBS
fetal bovine serum
- RT-PCR
reverse transcriptase-polymerase chain reaction
- PPRE
PPAR response element
- TBST
Tris-buffered saline containing 0.1% Tween 20
Footnotes
Supporting Information Available. Elemental analysis data are available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Chen CD, Welsbie DS, Tran C, Baek SH, Chen R, Vessella R, Rosenfeld MG, Sawyers CL. Molecular determinants of resistance to antiandrogen therapy. Nat Med. 2004;10:33–9. doi: 10.1038/nm972. [DOI] [PubMed] [Google Scholar]
- 2.Isaacs JT, Isaacs WB. Androgen receptor outwits prostate cancer drugs. Nat Med. 2004;10:26–7. doi: 10.1038/nm0104-26. [DOI] [PubMed] [Google Scholar]
- 3.Debes JD, Tindall DJ. Mechanisms of androgen-refractory prostate cancer. N Engl J Med. 2004;351:1488–90. doi: 10.1056/NEJMp048178. [DOI] [PubMed] [Google Scholar]
- 4.Grossmann ME, Huang H, Tindall DJ. Androgen receptor signaling in androgen-refractory prostate cancer. J Natl Cancer Inst. 2001;93:1687–97. doi: 10.1093/jnci/93.22.1687. [DOI] [PubMed] [Google Scholar]
- 5.Huang H, Tindall DJ. The role of the androgen receptor in prostate cancer. Crit Rev Eukaryot Gene Expr. 2002;12:193–207. doi: 10.1615/critreveukaryotgeneexpr.v12.i3.30. [DOI] [PubMed] [Google Scholar]
- 6.Lee DK, Chang C. Endocrine mechanisms of disease: Expression and degradation of androgen receptor: mechanism and clinical implication. J Clin Endocrinol Metab. 2003;88:4043–54. doi: 10.1210/jc.2003-030261. [DOI] [PubMed] [Google Scholar]
- 7.Roy-Burman P, Tindall DJ, Robins DM, Greenberg NM, Hendrix MJ, Mohla S, Getzenberg RH, Isaacs JT, Pienta KJ. Androgens and prostate cancer: are the descriptors valid? Cancer Biol Ther. 2005;4:4–5. doi: 10.4161/cbt.4.1.1563. [DOI] [PubMed] [Google Scholar]
- 8.Taplin ME, Balk SP. Androgen receptor: a key molecule in the progression of prostate cancer to hormone independence. J Cell Biochem. 2004;91:483–90. doi: 10.1002/jcb.10653. [DOI] [PubMed] [Google Scholar]
- 9.Koivisto P, Visakorpi T, Kallioniemi OP. Androgen receptor gene amplification: a novel molecular mechanism for endocrine therapy resistance in human prostate cancer. Scand J Clin Lab Invest Suppl. 1996;226:57–63. [PubMed] [Google Scholar]
- 10.Santos AF, Huang H, Tindall DJ. The androgen receptor: a potential target for therapy of prostate cancer. Steroids. 2004;69:79–85. doi: 10.1016/j.steroids.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 11.Liao X, Tang S, Thrasher JB, Griebling TL, Li B. Small-interfering RNA-induced androgen receptor silencing leads to apoptotic cell death in prostate cancer. Mol Cancer Ther. 2005;4:505–15. doi: 10.1158/1535-7163.MCT-04-0313. [DOI] [PubMed] [Google Scholar]
- 12.Cheng H, Snoek R, Ghaidi F, Cox ME, Rennie PS. Short hairpin RNA knockdown of the androgen receptor attenuates ligand-independent activation and delays tumor progression. Cancer Res. 2006;66:10613–20. doi: 10.1158/0008-5472.CAN-06-0028. [DOI] [PubMed] [Google Scholar]
- 13.Mitchell SH, Zhu W, Young CY. Resveratrol inhibits the expression and function of the androgen receptor in LNCaP prostate cancer cells. Cancer Res. 1999;59:5892–5. [PubMed] [Google Scholar]
- 14.Zhang Y, Ni J, Messing EM, Chang E, Yang CR, Yeh S. Vitamin E succinate inhibits the function of androgen receptor and the expression of prostate-specific antigen in prostate cancer cells. Proc Natl Acad Sci U S A. 2002;99:7408–13. doi: 10.1073/pnas.102014399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bektic J, Berger AP, Pfeil K, Dobler G, Bartsch G, Klocker H. Androgen receptor regulation by physiological concentrations of the isoflavonoid genistein in androgen-dependent LNCaP cells is mediated by estrogen receptor beta. Eur Urol. 2004;45:245–51. doi: 10.1016/j.eururo.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 16.Yang CC, Ku CY, Wei S, Shiau CW, Chen CS, Pinzone JJ, Ringel MD, Chen CS. Peroxisome Proliferator-Activated Receptor {gamma}-Independent Repression of Prostate-Specific Antigen Expression by Thiazolidinediones in Prostate Cancer Cells. Mol Pharmacol. 2006;69:1564–70. doi: 10.1124/mol.105.018333. [DOI] [PubMed] [Google Scholar]
- 17.Yang CC, Wang YC, Wei S, Lin LF, Chen CS, Lee CC, Lin CC, Chen CS. Peroxisome proliferator-activated receptor gamma-independent suppression of androgen receptor expression by troglitazone mechanism and pharmacologic exploitation. Cancer Res. 2007;67:3229–38. doi: 10.1158/0008-5472.CAN-06-2759. [DOI] [PubMed] [Google Scholar]
- 18.Mustafa A, Asker W, El-Din Sobhy ME. On the reactivity of the exocyclic double bond in 5-arylidene-3-aryl-2,4-thiazolidinediones; their reaction with diazoalkanes, p-thiacresol and piperidine. J Am Chem Soc. 1960;82:2597–601. [Google Scholar]
- 19.Jiang M, Shappell SB, Hayward SW. Approaches to understanding the importance and clinical implications of peroxisome proliferator-activated receptor gamma (PPARgamma) signaling in prostate cancer. J Cell Biochem. 2004;91:513–27. doi: 10.1002/jcb.10770. [DOI] [PubMed] [Google Scholar]
- 20.Koeffler HP. Peroxisome proliferator-activated receptor gamma and cancers. Clin Cancer Res. 2003;9:1–9. [PubMed] [Google Scholar]
- 21.Lieberman R. Chemoprevention of prostate cancer: current status and future directions. Cancer Metastasis Rev. 2002;21:297–309. doi: 10.1023/a:1021267128567. [DOI] [PubMed] [Google Scholar]
- 22.Bae MA, Song BJ. Critical role of c-Jun N-terminal protein kinase activation in troglitazone-induced apoptosis of human HepG2 hepatoma cells. Mol Pharmacol. 2003;63:401–8. doi: 10.1124/mol.63.2.401. [DOI] [PubMed] [Google Scholar]
- 23.Baek SJ, Wilson LC, Hsi LC, Eling TE. Troglitazone, a peroxisome proliferator-activated receptor gamma (PPAR gamma) ligand, selectively induces the early growth response-1 gene independently of PPAR gamma. A novel mechanism for its anti-tumorigenic activity. J Biol Chem. 2003;278:5845–53. doi: 10.1074/jbc.M208394200. [DOI] [PubMed] [Google Scholar]
- 24.Gouni-Berthold I, Berthold HK, Weber AA, Ko Y, Seul C, Vetter H, Sachinidis A. Troglitazone and rosiglitazone induce apoptosis of vascular smooth muscle cells through an extracellular signal-regulated kinase-independent pathway. Naunyn Schmiedebergs Arch Pharmacol. 2001;363:215–21. doi: 10.1007/s002100000352. [DOI] [PubMed] [Google Scholar]
- 25.Huang JW, Shiau CW, Yang YT, Kulp SK, Chen KF, Brueggemeier RW, Shapiro CL, Chen CS. Peroxisome proliferator-activated receptor gamma-independent ablation of cyclin D1 by thiazolidinediones and their derivatives in breast cancer cells. Mol Pharmacol. 2005;67:1342–8. doi: 10.1124/mol.104.007732. [DOI] [PubMed] [Google Scholar]
- 26.Motomura W, Okumura T, Takahashi N, Obara T, Kohgo Y. Activation of peroxisome proliferator-activated receptor gamma by troglitazone inhibits cell growth through the increase of p27KiP1 in human. Pancreatic carcinoma cells. Cancer Res. 2000;60:5558–64. [PubMed] [Google Scholar]
- 27.Okura T, Nakamura M, Takata Y, Watanabe S, Kitami Y, Hiwada K. Troglitazone induces apoptosis via the p53 and Gadd45 pathway in vascular smooth muscle cells. Eur J Pharmacol. 2000;407:227–35. doi: 10.1016/s0014-2999(00)00758-5. [DOI] [PubMed] [Google Scholar]
- 28.Palakurthi SS, Aktas H, Grubissich LM, Mortensen RM, Halperin JA. Anticancer effects of thiazolidinediones are independent of peroxisome proliferator-activated receptor gamma and mediated by inhibition of translation initiation. Cancer Res. 2001;61:6213–8. [PubMed] [Google Scholar]
- 29.Shiau CW, Yang CC, Kulp SK, Chen KF, Chen CS, Huang JW. Thiazolidenediones mediate apoptosis in prostate cancer cells in part through inhibition of Bcl-xL/Bcl-2 functions independently of PPARgamma. Cancer Res. 2005;65:1561–9. doi: 10.1158/0008-5472.CAN-04-1677. [DOI] [PubMed] [Google Scholar]
- 30.Sugimura A, Kiriyama Y, Nochi H, Tsuchiya H, Tamoto K, Sakurada Y, Ui M, Tokumitsu Y. Troglitazone suppresses cell growth of myeloid leukemia cell lines by induction of p21WAF1/CIP1 cyclin-dependent kinase inhibitor. Biochem Biophys Res Commun. 1999;261:833–7. doi: 10.1006/bbrc.1999.1049. [DOI] [PubMed] [Google Scholar]
- 31.Takeda K, Ichiki T, Tokunou T, Iino N, Takeshita A. 15-Deoxy-delta 12,14-prostaglandin J2 and thiazolidinediones activate the MEK/ERK pathway through phosphatidylinositol 3-kinase in vascular smooth muscle cells. J Biol Chem. 2001;276:48950–5. doi: 10.1074/jbc.M108722200. [DOI] [PubMed] [Google Scholar]
- 32.Wei S, Lin LF, Yang CC, Wang YC, Chang GD, Chen H, Chen CS. Thiazolidinediones Modulate the Expression of beta-Catenin and Other Cell-Cycle Regulatory Proteins by Targeting the F-Box Proteins of Skp1-Cul1-F-box Protein E3 Ubiquitin Ligase Independently of Peroxisome Proliferator-Activated Receptor {gamma} Mol Pharmacol. 2007;72:725–33. doi: 10.1124/mol.107.035287. [DOI] [PubMed] [Google Scholar]