Abstract
Synthetic kappa-opioid receptor (KOR) agonists induce dysphoric and pro-depressive effects, and variations in the KOR (OPRK1) and prodynorphin (PDYN) genes have been shown to be associated with alcohol dependence. We genotyped 23 single nucleotide polymorphisms (SNPs) in the PDYN and OPRK1 genes in 816 alcohol dependent subjects and investigated their association with (1) negative craving measured by a subscale of the Inventory of Drug Taking Situations (IDTS); (2), a self-reported history of depression; and, (3) the intensity of depressive symptoms measured by the Beck Depression Inventory-II (BDI). In addition, 13 of the 23 PDYN and OPRK1 SNPs, which were previously genotyped in a set of 1248 controls, were used to evaluate association with alcohol dependence. SNP and haplotype tests of association were performed. Analysis of a haplotype spanning the PDYN gene (rs6045784, rs910080, rs2235751, rs2281285) revealed significant association with alcohol dependence (p=0.00079) and with negative craving (p=0.0499). A candidate haplotype containing the PDYN rs2281285-rs1997794 SNPs that was previously associated with alcohol dependence was also associated with negative craving (p=0.024) and alcohol dependence (p=0.0008) in this study. A trend for association between depression severity and PDYN variation was detected. No associations of OPRK1 gene variation with alcohol dependence or other studied phenotypes were found. These findings support the hypothesis that sequence variation in the PDYN gene contributes to both alcohol dependence and the induction of negative craving in alcohol dependent subjects.
Keywords: OPRK1, PDYN, Alcohol dependence, Craving, Depression
INTRODUCTION
Animal studies suggest that dynorphins play a role in alcohol dependence (Shippenberg et al., 2007; Walker et al., 2011; Wee and Koob, 2010), whereas blockade of KOR decreases ethanol self-administration in ethanol-dependent but not in nondependent animals (Walker et al., 2011). This finding implies that the dynorphin/KOR regulated neurotransmission is persistently activated in dependent animals and, thus, contributes to increased ethanol self-administration. PDYN mRNA and dynorphins were found to be upregulated in the dorsolateral prefrontal cortex of alcohol dependent subjects (Bazov et al., 2011). This upregulation, which may be influenced by PDYN polymorphisms or epigenetic mechanisms, could contribute to impairment in cognitive control over alcohol drinking behavior (Shippenberg et al., 2007; Taqi et al., 2011a; Taqi et al., 2011b).
Evidence also demonstrates that the brain dynorphin/KOR system plays a role in the motivational aspects of stress by mediating pro-depressive-like states that involve elements of anhedonia, dysphoria, and aversion in humans and laboratory animals (Knoll et al., 2007; Todtenkopf et al., 2004; Wadenberg, 2003; Walsh et al., 2001). Synthetic KOR agonists produce dysphoria in humans (Pfeiffer et al., 1986; Wadenberg, 2003) and a wide variety of depressive-like effects in animal models (Carlezon et al., 2006; Dinieri et al., 2009; Mague et al., 2003; Todtenkopf et al., 2004; Tomasiewicz et al., 2008). Stress elevates the expression of dynorphins in animals (Shirayama et al., 2004) while KOR antagonists block the effects of stress and produce antidepressant-like effects (Knoll et al., 2007; Mague et al., 2003; Newton et al., 2002). Consistently, PDYN mRNA is elevated in the patch compartment of the striatum of suicide subjects, an effect presumably attributed to depression (Hurd et al., 1997). Collectively these findings suggest that stress-related activation of KOR by dynorphins can lead to depression-like syndrome in animal models and depression in humans.
Clinical research also indicates that negative affect contributes to relapse along with alcohol craving (Zywiak et al., 2006; Zywiak et al., 2003). The complex interplay between craving and an emotional/behavioral context associated with alcohol use is conceptualized by a three-pathway psychobiological model (Verheul et al., 1999). According to this model, the desire for drinking (craving) may present in the context of tension or negative emotions (negative or “relief” craving), a desire for the rewarding properties of alcohol (positive or “reward” craving), or obsessive thoughts about drinking (obsessive or “temptation” craving) (Verheul et al., 1999). The propensity to use alcohol in the context of these situations can be reliably identified by the Inventory of Drug Taking Situations (IDTS) (Annis et al., 1997; Turner et al., 1997). Data indicates that negative craving seems to be a more important determinant for drinking compared to reward craving or obsessive craving (Victorio-Estrada and Mucha, 1997; Victorio-Estrada et al., 1996). Thus, negative craving may be an important marker predictive of the severity of alcohol dependence and/or treatment response.
Genetic studies have demonstrated association between the OPRK1 and PDYN variation and alcohol dependence in humans (Edenberg et al., 2008; Williams et al., 2007; Xuei et al., 2006). Although no associations with PDYN or OPRK1 SNPs have been reported from genome-wide association studies of alcohol dependence ((Treutlein et al., 2009); (Bierut et al., 2010); (Edenberg et al., 2010)), the stringent corrections for multiple testing that need to be applied in these studies result in low power to detect small effects of individual SNPs. Thus, both genes remain strong candidates for alcohol dependence-related phenotypes based on evidence from candidate gene studies and evidence of functional importance of the dynorphin/KOR system in stress and alcohol self administration in model studies. However, the role of PDYN and OPRK1 variation in negative craving or in the presence of comorbid depression in alcohol dependent human subjects has not been investigated. To examine potential overlap in genetic factors predisposing to negative craving, intensity of depressive symptoms and/or self-reported history of depression in alcohol dependent subjects, we investigated the association of these phenotypes with OPRK1 and PDYN sequence variation. We hypothesized that variations in OPRK1 and PDYN are associated with increased negative craving (reflected by elevated score of the negative IDTS scale), increased level of depressive symptoms (reflected by elevated Beck Depression Inventory-II, or BDI, score), increased frequency of reported history of depression and increased risk for alcohol dependence. To test this hypothesis, we evaluated the association of each phenotype with individual SNPs, haplotypes covering the PDYN and OPRK1 genes, and candidate haplotypes previously reported to be associated with alcohol or cocaine dependence (Beardsley et al., 2005; Xuei et al., 2006; Yuferov et al., 2009).
METHODS
Study subjects and data collection
This study was approved by the Institutional Review Board of Mayo Clinic Rochester. All subjects provided informed consent and permission to use their information for future studies of alcohol dependence and related phenotypes. A total of 936 alcohol dependent cases and 1302 non-alcohol dependent controls were genotyped. After quality control procedures 816 cases and 1248 controls remained for analysis.
The case group included alcohol dependent (DSM-IV-TR) subjects participating in ongoing and completed studies of clinical and genetic predictors of alcohol dependence, treatment outcomes and related phenotypes (Boykoff et al., 2010; Karpyak et al., 2010; Karpyak et al., 2009; Kolla et al., 2011; Schneekloth et al., accepted for publication 2012). For this study, available information on the intensity of negative craving (measured by the negative subscale of IDTS) as well as a state-dependent measure of depressive symptomatology (measured by BDI) was extracted from the research database.
A subset of cases had participated in the Mayo Clinic Intensive Addictions Program (IAP) and completed the IDTS and BDI questionnaires as part of treatment-related assessments. Because the treatment-related assessments have evolved over time, not all IAP patients were evaluated with both the IDTS and BDI. Therefore, data for the negative craving subscale of IDTS was available for 196 of the 816 cases, while BDI data was available for 292 cases. The negative IDTS scores were approximately symmetrically distributed, with a mean of 47.94 (SD = 20.96). The BDI data were right skewed with the mean on the border between mild and moderate intensity of depression (18.74+/−11.61). In addition, self-reported information regarding the presence (n=193, 52.9%) or absence (n=172, 47.1%) of lifetime history of depression was available for a subset of 365 cases who participated in studies aimed at determining the genetic predictors of severe alcohol withdrawal (Karpyak et al., 2010; Karpyak et al., 2009).
Control subjects were selected from a group of controls that had previously participated in a genome-wide association study (GWAS) of venous thrombosis carried out at Mayo Clinic and gave consent for additional research studies (Heit et al., 2011).
In an effort to replicate the strongest SNP association detected in the discovery sample, we genotyped one PDYN SNP in an independent replication sample of 474 alcohol dependent subjects and 432 controls recruited at the Ludwigs-Maximilians-University of Munich, Germany. Detailed information about the discovery and replication samples is available in the Supplemental materials.
SNP selection and genotyping
The list of selected PDYN and OPRK1 SNPs is presented in Table 1. Non-alcoholic controls were genotyped previously as part of a study of venous thrombosis (Heit et al., 2011). For genotyping of cases, tag SNPs and candidate SNPs were selected to achieve gene coverage and to replicate previously reported associations. Tag SNPs were selected to match, whenever possible, the 13 PDYN and OPRK1 SNPs that had been previously genotyped in the controls (Heit et al., 2011). Candidate SNPs in PDYN and OPRK1 were chosen based on previously reported associations with alcohol, opiate and/or cocaine dependence (Table 1). The selected candidate and tag SNPs were genotyped in alcohol dependent subjects to evaluate potential associations with negative craving and depressive symptomatology. For SNPs that had been genotyped in the controls, association with alcohol dependence was also investigated.
Table 1.
Candidate SNPs genotyped in cases and controls
| Gene | SNP† | Major allele |
Minor allele |
MAF†† cases |
MAF†† controls |
Genotyped in controls |
Previously reported associations with alcohol, cocaine or opiate dependence |
|---|---|---|---|---|---|---|---|
| OPRK1 | rs963549^* | G | A | 0.1255 | 0.1259 | Y | |
| rs7817710 | C | A | 0.0808 | 0.0870 | Y | ||
| rs997917^ | A | G | 0.2953 | 0.2847 | Y | (Xuei et al., 2006; Yuferov et al., 2009) | |
| rs6473797^ | A | G | 0.2515 | (Xuei et al., 2006; Yuferov et al., 2009) | |||
| rs6473799* | A | G | 0.2515 | 0.2460 | Y | ||
| rs12548098 | A | G | 0.1017 | (Xuei et al., 2006; Yuferov et al., 2009) | |||
| rs7836120^* | A | G | 0.1691 | 0.1540 | Y | ||
| rs16918941^* | A | G | 0.0863 | 0.0794 | Y | (Xuei et al., 2006; Yuferov et al., 2009) | |
| rs6985606^* | A | G | 0.4907 | 0.4984 | Y | (Xuei et al., 2006; Yuferov et al., 2009) | |
| rs12056414 | G | A | 0.0821 | 0.0742 | Y | ||
| PDYN | rs6045784^ | A | G | 0.1017 | Xuei et al, 2006 | ||
| rs6132153 | A | G | 0.1628 | 0.1434 | Y | ||
| rs2235749 | G | A | 0.2711 | (Xuei et al., 2006; Yuferov et al., 2009) | |||
| rs910080^ | A | G | 0.2644 | (Yuferov et al., 2009) | |||
| rs10485703 | A | G | 0.1010 | Xuei et al, 2006 | |||
| rs6045819 | A | G | 0.1077 | (Xuei et al., 2006; Yuferov et al., 2009) | |||
| rs6045824 | A | G | 0.1028 | 0.1066 | Y | ||
| rs6035222 | G | A | 0.1114 | (Xuei et al., 2006) | |||
| rs6045868* | G | A | 0.2546 | 0.2464 | Y | (Xuei et al., 2006) | |
| rs2235751^* | A | G | 0.2543 | 0.2392 | Y | (Xuei et al., 2006) | |
| rs2281285^* | A | G | 0.1726 | 0.1422 | Y | (Xuei et al., 2006) | |
| rs1997794 | A | G | 0.3554 | (Clarke et al., 2009; Xuei et al., 2006) | |||
| rs10854244 | A | T | 0.2632 | Xuei et al, 2006 |
SNP = Single Nucleotide Polymorphism
MAF = Minor Allele Frequency
tag SNPs selected for gene-level haplotype analysis in cases only to assess for association with negative craving and depression-related outcomes
tag SNPs selected for gene-level case/control haplotype analysis comparing alcohol dependent cases with non-alcoholic controls to assess for association with alcohol dependence
Genotyping was performed separately in cases and controls. Twenty-three candidate SNPs were genotyped in cases using the Illumina BeadXpress platform with Illumina GoldenGate SNP assay. Thirteen of these PDYN and OPRK1 SNPs were also previously genotyped with the Illumina 660 genome-wide SNP array in the controls (Heit et al., 2011). In addition, 43 ancestry informative markers were genotyped to verify self-reported race (Supplemental Table 1). Genotyping details and quality control analysis of the discovery and replication samples are described in the Supplemental materials.
Data analysis and statistics
To avoid confounding effects of population stratification, subjects with self-reported race other than “white, non-Hispanic” were excluded from analyses, resulting in 817 cases and 1249 controls. To verify genetic ancestry of the remaining subjects, 43 ancestry informative markers were analyzed using STRUCTURE (Pritchard et al., 2000). Details are available in Supplemental materials. As a result of this analysis one case and one control subject were excluded, leaving 816 cases and 1248 controls for analysis.
Analyses of genetic association with negative craving and depression measures
Associations between the 23 SNPs in PDYN and OPRK1 and negative craving in alcohol dependent subjects were first evaluated using single SNP association tests. For each SNP, association with the raw score of the negative IDTS subscale was evaluated with a linear regression model, including age and gender as covariates. SNP genotypes were coded 0, 1, or 2 representing the number of copies of the minor allele. For the top SNP associations, permutation analysis (10,000 permutations) was used to correct for multiple testing of the 23 SNPs. For SNPs that showed significant association at the 0.05 level, we report both the uncorrected and corrected p-values. All statistical analyses were performed in R Statistical Software, version 2.13.0 (R Development Core Team, 2011) unless otherwise noted.
We also investigated the association of negative craving with previously reported (candidate) PDYN and OPRK1 haplotypes as well as haplotypes spanning the full gene. Xuei et al (2006) reported the association of PDYN haplotypes composed of rs2235749 and rs10485703 (Xuei Block 1) and rs1883723, rs2281285, and rs1997794 (Xuei Block 2) with alcohol dependence, while Yuferov et al (2009) reported the association of a haplotype composed of rs2235749, rs910080, and rs910079 with cocaine dependence. We investigated the potential association of these candidate haplotypes with negative craving, with the exclusion of rs1883723 (Xuei Block 2) and rs910079 (Yuferov) SNPs, which were not genotyped in our study. However, we note that rs910079 is in perfect linkage disequilibrium (LD; r2 = 1.0) with rs910080, and thus our rs2235749-rs910080 haplotype is equivalent to Yuferov’s rs2235749-rs910080-rs910079 haplotype.
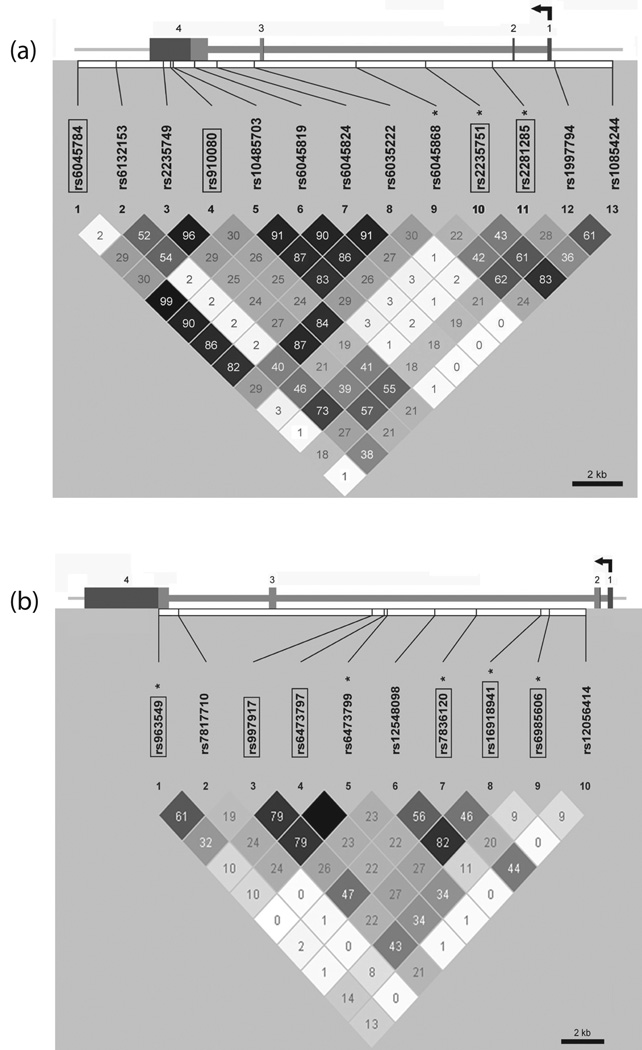
In addition to candidate haplotypes, we also considered haplotypes spanning each gene to examine association at the gene level. These full-gene haplotypes do not span the gene in the physical sense, but cover the genetic variation that is present across each gene by including tag SNPs based on patterns of linkage disequilibrium. Due to frequency limitations, rather than using all genotyped SNPs, a smaller set of tag SNPs was selected to cover each gene. LD in cases was estimated across each gene with Haploview software (Barrett et al., 2005), and a set of tag SNPs was identified with Tagger (de Bakker et al., 2005) to capture the genetic variation across each gene (Fig 1). Four tag SNPs were identified for PDYN (rs6045784, rs910080, rs2235751, rs2281285) and six tag SNPs were identified for OPRK1 (rs6473797, rs7836120, rs6985606, rs997917, rs963549, rs1691894). Haplotype association tests were performed using the score statistic proposed by Schaid et al (2002), adjusting for age and gender. Reported p-values for the global haplotype tests are not corrected for multiple testing; however corrections are reported for haplotype-specific tests.
Figure 1. LD plots showing r2 values and haplotype block structure for PDYN SNPs (a) and OPRK1 SNPs (b).
LD plots are based on 23 SNPs genotyped in alcohol dependent subjects only (similar results were observed for a subset of SNPs genotyped in the control subjects). Boxed SNPs denote tag SNPs chosen with Tagger for the case-only haplotype analysis. SNPs denoted with an asterisk are tag SNPs chosen with Tagger for the case-control analysis. SNPs marked in bold were genotyped in both cases and controls.
Single SNP, candidate haplotype and full-gene haplotype analyses were also performed to investigate association with severity of depressive symptomatology, measured by BDI at admission (state-dependent measure) and lifetime history of depression (trait-related measure) in alcohol dependent cases. Adjustments were made for gender but not age, which was not significantly associated with BDI scores at admission or lifetime history of depression. Because the BDI scores were right-skewed, a square-root transformation was applied prior to linear regression analysis. Associations with BDI at admission were evaluated with linear regression models, whereas associations with lifetime history of depression were evaluated with logistic regression models.
Analyses of genetic association with alcohol dependence
In addition to association analysis with negative craving and depression, we evaluated association of 13 PDYN and OPRK1 SNPs with alcohol dependence in a case-control analysis of 816 alcohol dependent subjects and 1248 controls. The association of each SNP with alcohol dependence was evaluated using a logistic regression model, including age and gender as covariates. Tests of haplotype association with alcohol dependence were also performed. Because only 13 of the 23 SNPs genotyped in alcohol dependent cases were also genotyped in controls, analyses of the same haplotypes that were investigated in the case-only analyses were not possible. Only one SNP involved in the reported candidate haplotypes from the studies by (Yuferov et al., 2009) and (Xuei et al., 2006) was also genotyped in the controls (rs2281285), eliminating the possibility to study the association of these haplotypes with alcohol dependence. Instead, we investigated the association of alcohol dependence with full-gene haplotypes constructed using tag SNPs identified with Tagger (de Bakker et al., 2005) to capture the genetic variation across each gene. For the case-control analyses, three tag SNPs were identified for PDYN (rs6045868, rs2235751, rs2281285) and five tag SNPs were identified for OPRK1 (rs6473799, rs7836120, rs6985606, rs963549, rs1691894).
LD patterns were consistent with reports from previous studies and with data available from HapMap, and were also similar between case and control subjects, with the exception of rs2281285. This SNP displayed reduced LD among case subjects, while the LD pattern in controls matched that of HapMap.
Replication of single SNP association findings
The PDYN rs2281285 SNP was genotyped in 915 subjects recruited from Ludwigs-Maximilians-University in Munich, Germany using a TaqMan® SNP Genotyping Assay (Applied Biosystems, Foster City, CA) in order to replicate an association detected in this study. Nine subjects failed genotyping, and data from 474 alcohol dependent subjects and 432 non-alcoholic controls were analyzed. Logistic regression analysis was conducted in the replication sample to examine association between alcohol dependence and rs2281285 as described for the original Mayo Clinic sample.
RESULTS
Association of PDYN and OPRK1 SNPs with negative craving
Results of the tests for association between negative craving and individual PDYN and OPRK1 SNPs are presented in Table 2. A nominally significant association was detected with rs2281285 (uncorrected p=0.012, multiple-testing corrected p = 0.14). The estimated effect size was 6.95 (SE=2.75), indicating that for each inherited copy of the minor G allele, the IDTS negative score was almost 7 points higher on average (Cohen’s D = 0.33). There were no statistically significant associations between negative craving and individual SNPs in the OPRK1 gene (Table 2). However, two OPRK1 SNPs (rs12548098 and rs16918941) and PDYN rs6132153 SNP revealed trends for association at the p < 0.07 level (uncorrected).
Table 2.
Association of individual PDYN and OPRK1 SNPs with negative craving subscale of IDTS, depressive symptoms at admission (BDI) and self-reported history of depression
| IDTS† (N=196) | BDI† (N=292) | History of Depression (N=365) |
||||||
|---|---|---|---|---|---|---|---|---|
| Gene | SNP | MAF (cases) |
Effect Size* |
P- value† |
Effect Size* |
P-value‡ | Odds Ratio |
P-value‡ |
| PDYN | rs6045784 | 0.10 | −4.47 | 0.18 | −0.24 | 0.21 | 1.06 | 0.83 |
| rs6132153 | 0.16 | 5.49 | 0.057 | −0.02 | 0.91 | 1.01 | 0.95 | |
| rs2235749 | 0.27 | 2.07 | 0.40 | −0.13 | 0.33 | 0.99 | 0.95 | |
| rs910080 | 0.26 | 1.53 | 0.54 | −0.14 | 0.30 | 1.04 | 0.84 | |
| rs10485703 | 0.10 | −4.95 | 0.15 | −0.28 | 0.17 | 1.06 | 0.83 | |
| rs6045819 | 0.11 | −5.28 | 0.11 | −0.39 | 0.038‡‡ | 1.00 | 1.0 | |
| rs6045824 | 0.10 | −4.76 | 0.17 | −0.39 | 0.050 | 1.17 | 0.57 | |
| rs6035222 | 0.11 | −2.90 | 0.38 | −0.34 | 0.078 | 1.01 | 0.96 | |
| rs6045868 | 0.25 | 1.29 | 0.60 | −0.15 | 0.27 | 1.01 | 0.96 | |
| rs2235751 | 0.25 | 3.52 | 0.14 | 0.01 | 0.95 | 0.85 | 0.36 | |
| rs2281285 | 0.17 | 6.95 | 0.012** | 0.07 | 0.63 | 1.13 | 0.57 | |
| rs1997794 | 0.36 | 1.70 | 0.45 | −0.04 | 0.73 | 0.94 | 0.69 | |
| rs10854244 | 0.26 | 3.89 | 0.094 | 0.08 | 0.53 | 0.91 | 0.57 | |
| OPRK1 | rs963549 | 0.13 | −0.45 | 0.89 | 0.10 | 0.59 | 0.80 | 0.34 |
| rs7817710 | 0.081 | 0.01 | 1.0 | 0.09 | 0.68 | 0.70 | 0.18 | |
| rs997917 | 0.30 | −0.50 | 0.84 | −0.02 | 0.91 | 0.77 | 0.13 | |
| rs6473797 | 0.25 | −0.26 | 0.92 | −0.03 | 0.80 | 0.74 | 0.092 | |
| rs6473799 | 0.25 | −0.26 | 0.92 | −0.03 | 0.80 | 0.74 | 0.092 | |
| rs12548098 | 0.10 | −6.90 | 0.068 | 0.38 | 0.066 | 0.70 | 0.17 | |
| rs7836120 | 0.17 | −2.71 | 0.39 | 0.16 | 0.36 | 0.85 | 0.45 | |
| rs16918941 | 0.086 | −7.42 | 0.064 | 0.28 | 0.20 | 0.58 | 0.055 | |
| rs6985606 | 0.49 | −2.22 | 0.30 | −0.09 | 0.49 | 0.81 | 0.20 | |
| rs12056414 | 0.082 | 5.06 | 0.24 | −0.03 | 0.88 | 1.27 | 0.40 | |
IDTS = Inventory of Drug Taking Situations
BDI = Beck Depression Inventory-II
P-values < 0.05 are presented in bold and underlined.
For IDTS and BDI, the reported effect size is the estimated coefficient for the minor allele effect from the regression model.
Presented p-values are not corrected for multiple testing except as noted below.
Corrected p-value = 0.14.
Corrected p-value = 0.36.
Haplotype association test results are shown in Table 3. A global test of full-gene haplotype association revealed a significant association of negative craving with variation in PDYN (p=0.0499) but not OPRK1 (p=0.32). Haplotype-specific frequencies and corresponding p-values are presented in Supplemental Table 2. We also found significant evidence of association (global test p=0.024) between negative craving and the candidate rs2281285-rs1997794 haplotype (a subset of Xuei Block 2). The combination of G-A alleles in this haplotype was estimated to be a risk haplotype associated with negative craving (G-A haplotype specific p=0.007; maximum statistic corrected p=0.039). Haplotype-specific frequencies and corresponding p-values are presented in Supplemental Table 3. We also found a marginally significant association (global test p=0.055) between negative craving and the other candidate Xuei Block 1 haplotype rs2235749-rs10485703; (Xuei et al., 2006). In this haplotype, combination of A-A alleles was associated with increased negative craving (A-A haplotype specific p=0.019, maximum statistic corrected p=0.054). There was no evidence of association between negative craving and the third candidate haplotype consisting of rs2235749 and rs910080 (equivalent to the rs2235749-rs910080-rs910079 haplotype described to be associated with cocaine dependence by Yuferov et al, 2009).
Table 3.
Association of PDYN and OPRK1 haplotypes with negative craving subscale of IDTS, depressive symptoms at admission (BDI) and self-reported history of depression
| Haplotype Analyses |
Gene name | SNPs included | P-value IDTS* (N=196) |
P-value BDI* (N=292) |
P-value History of Depression* (N=365) |
|---|---|---|---|---|---|
| Full-gene | PDYN | rs6045784, rs910080, rs2235751, rs2281285 | 0.0499 | 0.087 | 0.95 |
| OPRK1 | rs6473797, rs7836120, rs6985606, rs997917, rs963549, rs1691894 | 0.32 | 0.40 | 0.22 | |
| Candidate haplotype | PDYN Block 1 (Xuei et al | rs2235749, rs10485703 | 0.055 | 0.66 | 0.96 |
| PDYN Block 2 (Xuei et al ) | rs2281285, rs1997794 | 0.024 | 0.082 | 0.82 | |
| PDYN (Yuferov et al) | rs2235749, rs910080 | 0.30 | 0.74 | 0.52 |
P-values are based on the global score test for haplotype association proposed by Schaid et al., 2002 and are not corrected for multiple testing; P-values < 0.05 are presented in bold and underlined. Additional results for the statistically significant haplotypes, including specific haplotype frequencies, are included in Supplemental Tables 2–4.
Association of PDYN and OPRK1 SNPs with BDI score and depression history
We observed association between PDYN rs6041859 and BDI score at the 0.05 significance level (uncorrected p=0.038) but not after correction for multiple testing (corrected p=0.36). We also observed a trend for association of BDI with PDYN candidate haplotype (Xuei Block 2) rs2281285-rs1997794 (p=0.082). Haplotype-specific frequencies and corresponding p-values are presented in Supplemental Table 2. A global test of haplotype association also revealed a trend for association of BDI score with variation in PDYN (p=0.087) with risk haplotypes (rs6045784 A - rs910080 A - rs2235751 A - rs2281285 G) consistent with the results for association with the negative craving subscale of IDTS. Haplotype-specific frequencies and corresponding p-values are presented in Supplemental Table 3. No association was observed between BDI scores and OPRK1 (Table 3). Our data did not provide evidence for association of lifetime history of depression with any SNP (Table 2) or haplotype (Table 3).
Association of PDYN and OPRK1 variations with alcohol dependence
Results of analyses of association between alcohol dependence and PDYN and OPRK1 SNPs are presented in Table 4. Consistent with the association findings for negative craving, PDYN rs2281285 SNP was also found to be associated with alcohol dependence (uncorrected p=0.0083, corrected p=0.076). The estimated odds ratio for association of rs2281285 with alcohol dependence was 1.30 (95% CI 1.07–1.58), indicating a higher risk of dependence associated with the minor G allele.
Table 4.
Analyses of association between PDYN and OPRK1 SNPs and alcohol dependence.
| Gene | SNP | MAF cases | MAF controls | Odds Ratio | P-value* |
|---|---|---|---|---|---|
| PDYN | rs6132153 | 0.16 | 0.14 | 1.22 | 0.047† |
| rs6045824 | 0.10 | 0.11 | 0.93 | 0.55 | |
| rs6045868 | 0.25 | 0.25 | 1.06 | 0.46 | |
| rs2235751 | 0.25 | 0.24 | 1.12 | 0.19 | |
| rs2281285 | 0.17 | 0.14 | 1.30 | 0.0083** | |
| OPRK1 | rs963549 | 0.13 | 0.13 | 0.96 | 0.67 |
| rs7817710 | 0.081 | 0.087 | 0.90 | 0.39 | |
| rs997917 | 0.30 | 0.28 | 1.04 | 0.61 | |
| rs6473799 | 0.25 | 0.25 | 1.03 | 0.75 | |
| rs7836120 | 0.17 | 0.15 | 1.13 | 0.22 | |
| rs16918941 | 0.086 | 0.079 | 1.12 | 0.39 | |
| rs6985606 | 0.49 | 0.50 | 0.96 | 0.54 | |
| rs12056414 | 0.082 | 0.074 | 1.10 | 0.44 |
Age and gender were included as covariates in the models.
P-values < 0.05 are presented in bold and underlined.
Presented p-values are not corrected for multiple testing except as noted below.
Corrected p-value = 0.36.
Corrected p-value = 0.076.
Consistent with results found for negative craving, gene-level haplotype associations with alcohol dependence were statistically significant for PDYN (rs6045868-rs2235751-rs2281285 haplotype, p=0.0008), but not for OPRK1 (rs6473799-rs7836120-rs6985606-rs963549-rs1691894 haplotype, p=0.86). Haplotype-specific frequencies and corresponding p-values are presented in Supplemental Table 4. As explained in the methods, only 13 of the 23 SNPs genotyped in alcohol dependent cases were also genotyped in controls and these SNPs did not include the previously reported candidate haplotypes. Therefore, we were not able to test for association of alcohol dependence with these haplotypes.
Replication of Association of rs2281285 with Alcohol Dependence
The rs2281285 G allele frequency in the replication sample was 0.161 in cases (as compared to 0.173 in the discovery sample), and 0.148 in controls (as compared to 0.142 in the discovery sample). In the replication sample the estimated odds ratio for the effect of the variant rs2281285 G allele was OR=1.18 (95% CI=0.90–1.54) as compared to OR=1.30 (95% CI = 1.07–1.58) in the discovery sample. Association with alcohol dependence was not statistically significant in the replication sample (p=0.22).
DISCUSSION
This study investigated the association of variability in PDYN and OPRK1 genes with negative craving, alcohol dependence, depression history and intensity of depressive symptoms. Examination of a haplotype reflecting the variation within the PDYN gene, consisting of rs6045868, rs2235751, and rs2281285, provided evidence for association of the PDYN gene with alcohol dependence (p = 0.00079). Similarly, we found a statistically significant association of the PDYN gene haplotype consisting of rs6045784, rs910080, rs2235751, and rs2281285 with negative craving (p = 0.0499). To the best of our knowledge, this is the first report supporting association of the PDYN gene with negative craving. Haplotype analysis also revealed a trend for association of the same PDYN gene haplotype with the intensity of depressive symptoms measured by BDI. No association between a history of comorbid depression and PDYN was observed in our sample.
We also found a significant association between negative craving and the candidate PDYN rs2281285-rs1997794 haplotype (p=0.024), which is a subset of a haplotype that was previously reported to be associated with alcohol dependence (Xuei et al., 2006). Only one of three SNPs (rs2281285) involved in the previously reported candidate haplotypes (Xuei Block 1, Xuei Block 2, and Yuferov) was also genotyped in our control subjects, precluding analysis of the association of those haplotypes with alcohol dependence. Within the alcohol dependent sample, the same haplotype rs2281285 G-rs1997794 A that was associated with increased negative craving (elevated negative scale of IDTS) also exhibited a trend for association with increased intensity of depressive symptoms (elevated BDI score, p=0.082), which is consistent with the original hypothesis. We also identified consistent associations of individual SNPs with alcohol dependence and negative craving as well as a trend for association with state-dependent intensity of depressive symptoms within alcohol dependent cases. Specifically, we detected association between alcohol dependence and the rs2281285 SNP (uncorrected p=0.0083; corrected p=0.076). Association with this SNP has not been previously described, but association with a haplotype that includes this SNP has been reported (Xuei et al., 2006). We also identified a nominal association between the negative craving subscale of the IDTS and rs2281285 (uncorrected p=0.012, corrected p=0.14). Although this result does not hold up after correction for multiple testing, the finding that the same SNP is associated with both alcohol dependence and the negative subscale of IDTS strengthens the results.
The rs2281285 SNP is located in intron B of PDYN - 769 bp downstream of exon 2 / intron B border (Nikoshkov et al., 2005). This area may be involved in the regulation of transcription initiation from multiple sites giving rise to one of PDYN transcripts, FL2-PDYN mRNA. Two dominant transcripts, the canonical FL1-PDYN and FL2-PDYN mRNAs, allow translation of the full-length PDYN protein that is further processed to dynorphin peptides (Nikoshkov et al., 2005). FL1- and FL2-PDYN mRNAs display different brain region-specific distributions, which suggests that their expression is controlled by different promoters. Whereas FL1 transcript shows high expression in limbic-related structures, FL2 transcript is expressed in hypothalamus and claustrum (Nikoshkov et al., 2005). Allelic variability in this location may contribute to differential regulation of these two transcripts. The low risk rs2281285 A allele resides within the TAAAT sequence that represents DNA-binding element for several homeobox transcription factors of the POU family, whereas the high risk rs2281285 G allele destroys this sequence. The members of the POU family of transcription factors are known to regulate growth hormone and prolactin genes and are involved in the specification of the lactotrope, somatotrope, and thyrotrope phenotypes in the pituitary (http://www.uniprot.org/uniprot/P28069) and hypothalamus {Yamanaka, 2010 #1982}. PDYN mRNA and dynorphins are present at high levels in the pituitary gland and hypothalamus (Roman et al., 2006; Schafer et al., 1994; Wylot et al., 2008). Dynorphins expressed in these tissues could have an important role in hormonal stress responses relevant for alcohol dependence {Bilkei-Gorzo, 2008 #1983}. In this scenario, additional levels of PDYN regulation involving genetic polymorphisms of the POU factors and / or their cellular content may be envisaged.
We did not replicate the association between alcohol dependence and rs2281285 SNP in the German sample, which may be attributed to several factors. The discovery sample included 816 cases and 1248 controls, while the replication sample included 474 cases and 432 controls, providing less power to identify an association. The odds ratio estimate in the replication sample of OR=1.18 (95% CI=0.90–1.54) is consistent in magnitude and direction with the odds ratio in the discovery sample (OR=1.30, 95% CI = 1.07–1.58). The frequency of rs2281285 G allele in the control groups in the discovery and replication samples were very similar, whereas the rs2281285 G allele frequency for cases within the replication sample was lower compared to discovery sample. This may be due to differences between case subjects enrolled at Mayo and in Munich in terms of severity of alcohol dependence or presence of certain comorbidities. For instance, in a subset (n=365) of alcohol dependent subjects from the discovery sample with available information, 53% reported a history of depression as compared to 32% of alcohol dependent subjects in the replication sample. Although the differences were not statistically significant, in both samples, the minor allele was more frequent in subjects with a lifetime history of depression than their non-depressed alcohol-dependent counterparts (Mayo 0.178 vs. 0.167; German 0.183 vs. 0.164). This suggests that the association observed between rs2281285 and alcohol dependence may reflect a subtype of alcohol dependence with comorbid depression, which may be related to the association with negative craving. However, the data set available for this study was not sufficiently large to examine subtype associations. Nevertheless, these post-hoc comparisons provide interesting hypotheses for further research.
Contrary to our results for the PDYN gene, we detected no evidence for associations of OPRK1 haplotypes with negative craving, alcohol dependence or depression-related phenotypes. We were also not able to replicate an association between OPRK1 rs1691891 SNP and alcohol dependence and did not test for association of OPRK1 rs12548098 SNP, as it was not genotyped in controls. Although we found a trend for association of negative craving with two OPRK1 SNPs (rs16918941 p=0.064, rs12548098 p=0.068) that were previously reported to be associated with alcohol dependence (Xuei et al 2006), these findings are not statistically significant after correction for multiple testing.
Our findings should be considered in the context of the following limitations. Genotyping was performed separately in cases and controls, which may potentially bias study results due to differential genotyping errors. However, our QC analysis indicates low levels of genotyping failures and errors, which makes such a bias unlikely. On the other hand, use of “samples of convenience” limited our ability to comprehensively investigate association of the phenotypes of interest with candidate genetic variations. It also precluded assessment of control subjects for presence of heavy drinking, alcohol abuse, substance dependence or psychotic disorders to verify that control subjects were in fact disease free. However, the use of unselected controls representing the general population rather than selected, disease-free controls is a cost-efficient strategy for genetic association studies {Manolio, 2009 #1987}. Although effect estimates may be attenuated and power may be reduced due to the inclusion of control subjects who are not disease free {, 2007 #1986}, we used medical records to exclude subjects with a documented history of alcohol dependence to minimize this loss of power. Furthermore, the use of convenience controls limited our ability to consider the potential confounding effects of comorbid substance dependence (e.g. nicotine) on associations with alcohol dependence, and hindered determination of causality. Future studies should examine the role of PDYN and OPRK1 in other psychiatric comorbidities in a sample from a well-defined population to better understand the association and to investigate potential pleiotropic effects.
In addition, the data set available for this study contained information for each phenotype of interest (negative craving, depression history, BDI) only in a limited number of subjects, precluding subgroup analyses or the assessment of potential associations among these phenotypes. Moreover, the self-reported history of depression used in this study may not be the most appropriate phenotype for genetic association analysis due to reasons including, but not limited to, recall bias and lack of consistency in diagnostic approaches in each case. Based on these considerations and given the trend for associations revealed in this study, a more comprehensive investigation of the associations between PDYN and OPRK1 and depression history should be considered in a rigorously phenotyped sample of alcohol dependent subjects.
In conclusion, we have demonstrated an association of the PDYN gene variation with alcohol dependence and negative craving at the gene and haplotype levels. The role of individual variations, including rs2281285, requires further investigation. We did not find evidence of association of the OPRK1 gene with alcohol dependence or negative craving. In this study, the association between studied genetic variations and severity of depressive symptoms or self-reported history of depression was not statistically significant. Further research is necessary to validate these findings, determine genetic markers associated with alcohol dependence and negative cravings, and investigate molecular mechanisms behind the observed associations.
Supplementary Material
ACKNOWLEDGEMENTS
This study was supported by grants from the St. Marys Hospital Sponsorship Award (VMK), Samuel C. Johnson Genomics of Addiction Program (VMK, JMB, DAM,), NIH/NIAAA P20 AA17830Z (VMK, JMB, MAF, DAM), and the Swedish Council for Working Life and Social Research, Swedish Science Research Council and Swedish Research Council FORMAS(GB). Controls were recruited and genotyped as part of the GWAS of Venous Thrombosis study (NIH/NHGRI grant HG04735, PI J.A. Heit). We thank the Mayo Clinic Cancer Center for the use of the Genotyping Core, which provided genotyping services. Mayo Clinic Cancer Center is supported in part by an NCI Cancer Center Support Grant 5P30 CA15083-37. This project was also supported by NIH/NCRR/NCATS CTSA Grant Number UL1 RR024150. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Footnotes
STATEMENT OF INTEREST
Dr. Preuss has received research support, consultancy, or lecture fees from Pfizer, Astra-Zeneca, Eli-Lilly, Janssen-Cilag, Novartis and Probiodrug in the past 3 years. Dr. Frye has received grant support from Pfizer. Dr. Mrazek has an interest in intellectual property that has been licensed from the Mayo Clinic by AssureRx, which is a clinical decision support company. The remaining authors have no conflicts of interest.
Supplementary information is available at The International Journal of Neuropsychopharmacology website.
REFERENCES
- Annis HM, Turner NE, Sklar SM. Addiction Research Foundation, Centre for Addiction and Mental Health. Toronto, Canada: 1997. Inventory of Drug-Taking Situations: User’s Guide. [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Bazov I, Kononenko O, Watanabe H, Kuntic V, et al. The endogenous opioid system in human alcoholics: molecular adaptations in brain areas involved in cognitive control of addiction. Addiction biology. 2011 doi: 10.1111/j.1369-1600.2011.00366.x. [DOI] [PubMed] [Google Scholar]
- Beardsley PM, Howard JL, Shelton KL, Carroll FI. Differential effects of the novel kappa opioid receptor antagonist, JDTic, on reinstatement of cocaine-seeking induced by footshock stressors vs cocaine primes and its antidepressant-like effects in rats. Psychopharmacology. 2005;183(1):118–126. doi: 10.1007/s00213-005-0167-4. [DOI] [PubMed] [Google Scholar]
- Bierut LJ, Agrawal A, Bucholz KK, Doheny KF, et al. A genome-wide association study of alcohol dependence. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(11):5082–5087. doi: 10.1073/pnas.0911109107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boykoff N, Schneekloth TD, Hall-Flavin D, Loukianova L, et al. Gender differences in the relationship between depressive symptoms and cravings in alcoholism. The American journal on addictions. 2010;19(4):352–356. doi: 10.1111/j.1521-0391.2010.00057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlezon WA, Jr, Beguin C, DiNieri JA, Baumann MH, et al. Depressive-like effects of the kappa-opioid receptor agonist salvinorin A on behavior and neurochemistry in rats. Journal of Pharmacology and Experimental Therapeutics. 2006;316(1):440–447. doi: 10.1124/jpet.105.092304. [DOI] [PubMed] [Google Scholar]
- Clarke TK, Krause K, Li T, Schumann G. An association of prodynorphin polymorphisms and opioid dependence in females in a Chinese population. Addiction biology. 2009;14(3):366–370. doi: 10.1111/j.1369-1600.2009.00151.x. [DOI] [PubMed] [Google Scholar]
- de Bakker PI, Yelensky R, Pe'er I, Gabriel SB, et al. Efficiency and power in genetic association studies. Nature Genetics. 2005;37(11):1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- Dinieri JA, Nemeth CL, Parsegian A, Carle T, et al. Altered sensitivity to rewarding and aversive drugs in mice with inducible disruption of cAMP response element-binding protein function within the nucleus accumbens. Journal of Neuroscience. 2009;29(6):1855–1859. doi: 10.1523/JNEUROSCI.5104-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Koller DL, Xuei X, Wetherill L, et al. Genome-wide association study of alcohol dependence implicates a region on chromosome 11. Alcoholism, Clinical and Experimental Research. 2010;34(5):840–852. doi: 10.1111/j.1530-0277.2010.01156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Wang J, Tian H, Pochareddy S, et al. A regulatory variation in OPRK1, the gene encoding the kappa-opioid receptor, is associated with alcohol dependence. Human Molecular Genetics. 2008;17(12):1783–1789. doi: 10.1093/hmg/ddn068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heit JA, Cunningham JM, Petterson TM, Armasu SM, et al. Genetic variation within the anticoagulant, procoagulant, fibrinolytic and innate immunity pathways as risk factors for venous thromboembolism. Journal of thrombosis and haemostasis. 2011;9(6):1133–1142. doi: 10.1111/j.1538-7836.2011.04272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurd YL, Herman MM, Hyde TM, Bigelow LB, et al. Prodynorphin mRNA expression is increased in the patch vs matrix compartment of the caudate nucleus in suicide subjects. Molecular Psychiatry. 1997;2(6):495–500. doi: 10.1038/sj.mp.4000319. [DOI] [PubMed] [Google Scholar]
- Karpyak VM, Biernacka JM, Weg MW, Stevens SR, et al. Interaction of SLC6A4 and DRD2 polymorphisms is associated with a history of delirium tremens. Addiction biology. 2010;15(1):23–34. doi: 10.1111/j.1369-1600.2009.00183.x. [DOI] [PubMed] [Google Scholar]
- Karpyak VM, Kim JH, Biernacka JM, Wieben ED, et al. Sequence Variations of the Human MPDZ Gene and Association With Alcoholism in Subjects With European Ancestry. Alcoholism, Clinical and Experimental Research. 2009;33(4):712–721. doi: 10.1111/j.1530-0277.2008.00888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoll AT, Meloni EG, Thomas JB, Carroll FI, et al. Anxiolytic-like effects of kappa-opioid receptor antagonists in models of unlearned and learned fear in rats. Journal of Pharmacology and Experimental Therapeutics. 2007;323(3):838–845. doi: 10.1124/jpet.107.127415. [DOI] [PubMed] [Google Scholar]
- Kolla BP, Schneekloth TD, Biernacka JM, Frye MA, et al. Trazodone and alcohol relapse: a retrospective study following residential treatment. American Journal on Addictions. 2011;20(6):525–529. doi: 10.1111/j.1521-0391.2011.00172.x. [DOI] [PubMed] [Google Scholar]
- Mague SD, Pliakas AM, Todtenkopf MS, Tomasiewicz HC, et al. Antidepressant-like effects of kappa-opioid receptor antagonists in the forced swim test in rats. Journal of Pharmacology and Experimental Therapeutics. 2003;305(1):323–330. doi: 10.1124/jpet.102.046433. [DOI] [PubMed] [Google Scholar]
- Newton SS, Thome J, Wallace TL, Shirayama Y, et al. Inhibition of cAMP response element-binding protein or dynorphin in the nucleus accumbens produces an antidepressant-like effect. Journal of Neuroscience. 2002;22(24):10883–10890. doi: 10.1523/JNEUROSCI.22-24-10883.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikoshkov A, Hurd YL, Yakovleva T, Bazov I, et al. Prodynorphin transcripts and proteins differentially expressed and regulated in the adult human brain. FASEB journal. 2005;19(11):1543–1545. doi: 10.1096/fj.05-3743fje. [DOI] [PubMed] [Google Scholar]
- Pfeiffer A, Brantl V, Herz A, Emrich HM. Psychotomimesis mediated by kappa opiate receptors. Science. 1986;233(4765):774–776. doi: 10.1126/science.3016896. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155(2):945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2011. [Google Scholar]
- Roman E, Ploj K, Gustafsson L, Meyerson BJ, et al. Variations in opioid peptide levels during the estrous cycle in Sprague-Dawley rats. Neuropeptides. 2006;40(3):195–206. doi: 10.1016/j.npep.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Schafer MK, Bette M, Romeo H, Schwaeble W, et al. Localization of kappa-opioid receptor mRNA in neuronal subpopulations of rat sensory ganglia and spinal cord. Neuroscience Letters. 1994;167(1–2):137–140. doi: 10.1016/0304-3940(94)91046-4. [DOI] [PubMed] [Google Scholar]
- Schneekloth TD, Biernacka JM, Hall-Flavin DK, Karpyak VM, et al. Alcohol Craving as a Predictor of Relapse. American Journal on Addictions. doi: 10.1111/j.1521-0391.2012.00297.x. (accepted for publication 2012). [DOI] [PubMed] [Google Scholar]
- Shippenberg TS, Zapata A, Chefer VI. Dynorphin and the pathophysiology of drug addiction. Pharmacology and Therapeutics. 2007;116(2):306–321. doi: 10.1016/j.pharmthera.2007.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama Y, Ishida H, Iwata M, Hazama GI, et al. Stress increases dynorphin immunoreactivity in limbic brain regions and dynorphin antagonism produces antidepressant-like effects. Journal of Neurochemistry. 2004;90(5):1258–1268. doi: 10.1111/j.1471-4159.2004.02589.x. [DOI] [PubMed] [Google Scholar]
- Taqi MM, Bazov I, Watanabe H, Nyberg F, et al. Prodynorphin promoter SNP associated with alcohol dependence forms noncanonical AP-1 binding site that may influence gene expression in human brain. Brain Research. 2011a;1385:18–25. doi: 10.1016/j.brainres.2011.02.042. [DOI] [PubMed] [Google Scholar]
- Taqi MM, Bazov I, Watanabe H, Sheedy D, et al. Prodynorphin CpG-SNPs associated with alcohol dependence: elevated methylation in the brain of human alcoholics. Addiction biology. 2011b;16(3):499–509. doi: 10.1111/j.1369-1600.2011.00323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todtenkopf MS, Marcus JF, Portoghese PS, Carlezon WA., Jr Effects of kappa-opioid receptor ligands on intracranial self-stimulation in rats. Psychopharmacology. 2004;172(4):463–470. doi: 10.1007/s00213-003-1680-y. [DOI] [PubMed] [Google Scholar]
- Tomasiewicz HC, Todtenkopf MS, Chartoff EH, Cohen BM, et al. The kappa-opioid agonist U69,593 blocks cocaine-induced enhancement of brain stimulation reward. Biological Psychiatry. 2008;64(11):982–988. doi: 10.1016/j.biopsych.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutlein J, Cichon S, Ridinger M, Wodarz N, et al. Genome-wide Association Study of Alcohol Dependence. Archives of General Psychiatry. 2009;66(7):773–784. doi: 10.1001/archgenpsychiatry.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner NE, Annis HM, Sklar SM. Measurement of antecedents to drug and alcohol use: psychometric properties of the Inventory of Drug-Taking Situations (IDTS) Behaviour Research and Therapy. 1997;35(5):465–483. doi: 10.1016/s0005-7967(96)00119-2. [DOI] [PubMed] [Google Scholar]
- Verheul R, van den Brink W, Geerlings P. A three-pathway psychobiological model of craving for alcohol. Alcohol and Alcoholism. 1999;34(2):197–222. doi: 10.1093/alcalc/34.2.197. [DOI] [PubMed] [Google Scholar]
- Victorio-Estrada A, Mucha RF. The Inventory of Drinking Situations (IDS) in current drinkers with different degrees of alcohol problems. Addictive Behaviors. 1997;22(4):557–565. doi: 10.1016/s0306-4603(96)00061-5. [DOI] [PubMed] [Google Scholar]
- Victorio-Estrada A, Mucha RF, Stephan ER. Excessive drinking situations in German alcoholics: replication of a three-factor model used for North Americans. Drug and Alcohol Dependence. 1996;41(1):75–79. doi: 10.1016/0376-8716(96)01241-0. [DOI] [PubMed] [Google Scholar]
- Wadenberg ML. A review of the properties of spiradoline: a potent and selective kappa-opioid receptor agonist. CNS drug reviews. 2003;9(2):187–198. doi: 10.1111/j.1527-3458.2003.tb00248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BM, Zorrilla EP, Koob GF. Systemic kappa-opioid receptor antagonism by nor-binaltorphimine reduces dependence-induced excessive alcohol self-administration in rats. Addiction biology. 2011;16(1):116–119. doi: 10.1111/j.1369-1600.2010.00226.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh SL, Strain EC, Abreu ME, Bigelow GE. Enadoline, a selective kappa opioid agonist: comparison with butorphanol and hydromorphone in humans. Psychopharmacology. 2001;157(2):151–162. doi: 10.1007/s002130100788. [DOI] [PubMed] [Google Scholar]
- Wee S, Koob GF. The role of the dynorphin-kappa opioid system in the reinforcing effects of drugs of abuse. Psychopharmacology. 2010;210(2):121–135. doi: 10.1007/s00213-010-1825-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams TJ, LaForge KS, Gordon D, Bart G, et al. Prodynorphin gene promoter repeat associated with cocaine/alcohol codependence. Addiction biology. 2007;12(3–4):496–502. doi: 10.1111/j.1369-1600.2007.00069.x. [DOI] [PubMed] [Google Scholar]
- Wylot B, Staszkiewicz J, Okrasa S. The expression of genes coding for opioid precursors, opioid receptors, beta-LH subunit and GnRH receptor in the anterior pituitary of cyclic gilts. Journal of Physiology and Pharmacology. 2008;59(4):745–758. [PubMed] [Google Scholar]
- Xuei X, Dick D, Flury-Wetherill L, Tian HJ, et al. Association of the kappa-opioid system with alcohol dependence. Molecular Psychiatry. 2006;11(11):1016–1024. doi: 10.1038/sj.mp.4001882. [DOI] [PubMed] [Google Scholar]
- Yuferov V, Ji F, Nielsen DA, Levran O, et al. A functional haplotype implicated in vulnerability to develop cocaine dependence is associated with reduced PDYN expression in human brain. Neuropsychopharmacology. 2009;34(5):1185–1197. doi: 10.1038/npp.2008.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zywiak WH, Stout RL, Trefry WB, Glasser I, et al. Alcohol relapse repetition, gender, and predictive validity. Journal of Substance Abuse Treatment. 2006;30(4):349–353. doi: 10.1016/j.jsat.2006.03.004. [DOI] [PubMed] [Google Scholar]
- Zywiak WH, Westerberg VS, Connors GJ, Maisto SA. Exploratory findings from the Reasons for Drinking Questionnaire. Journal of Substance Abuse Treatment. 2003;25(4):287–292. doi: 10.1016/s0740-5472(03)00118-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.