Abstract
Significance: Human apurinic/apyrimidinic endonuclease 1 (APE1, also known as REF-1) was isolated based on its ability to cleave at AP sites in DNA or activate the DNA binding activity of certain transcription factors. We review herein topics related to this multi-functional DNA repair and stress-response protein. Recent Advances: APE1 displays homology to Escherichia coli exonuclease III and is a member of the divalent metal-dependent α/β fold-containing phosphoesterase superfamily of enzymes. APE1 has acquired distinct active site and loop elements that dictate substrate selectivity, and a unique N-terminus which at minimum imparts nuclear targeting and interaction specificity. Additional activities ascribed to APE1 include 3′–5′ exonuclease, 3′-repair diesterase, nucleotide incision repair, damaged or site-specific RNA cleavage, and multiple transcription regulatory roles. Critical Issues: APE1 is essential for mouse embryogenesis and contributes to cell viability in a genetic background-dependent manner. Haploinsufficient APE1+/− mice exhibit reduced survival, increased cancer formation, and cellular/tissue hyper-sensitivity to oxidative stress, supporting the notion that impaired APE1 function associates with disease susceptibility. Although abnormal APE1 expression/localization has been seen in cancer and neuropathologies, and impaired-function variants have been described, a causal link between an APE1 defect and human disease remains elusive. Future Directions: Ongoing efforts aim at delineating the biological role(s) of the different APE1 activities, as well as the regulatory mechanisms for its intra-cellular distribution and participation in diverse molecular pathways. The determination of whether APE1 defects contribute to human disease, particularly pathologies that involve oxidative stress, and whether APE1 small-molecule regulators have clinical utility, is central to future investigations. Antioxid. Redox Signal. 20, 678–707.
Introduction
Shortly after determination of the DNA structure, it became appreciated that our genetic material is susceptible to spontaneous hydrolytic decay, as well as reactions with endogenous and exogenous physical and chemical agents. Such events can lead to modification of the DNA composition and alter the coding content of the genome, potentially driving mutagenesis or activation of cell death responses. Through many decades of research, it has become apparent that there exist enzymatic processes which recognize DNA damage and restore genetic integrity. Importantly, defects in the efficiency or accuracy of these so-called DNA repair or DNA damage tolerance pathways have been found to result in developmental failings, immunological deficiencies, cancer predisposition, neurological abnormalities, and premature aging characteristics, to name a few.
One of the most common forms of DNA damage is the loss of the base moiety from the intact sugar phosphate backbone (Fig. 1). Early studies from Lindahl estimated that roughly 10,000 depurination/depyrimidination events occur spontaneously per mammalian genome per day [reviewed in Lindahl (122)]. Since the base residue provides the instructional information, loss of this component of DNA can lead to error-prone bypass synthesis or polymerase arrest, and, thus, problems during replication or transcription. Such events can cause mutagenesis, chromosome instability, and gene expression defects, which underlie the cellular dysfunction and pathologies commonly associated with a DNA repair defect.
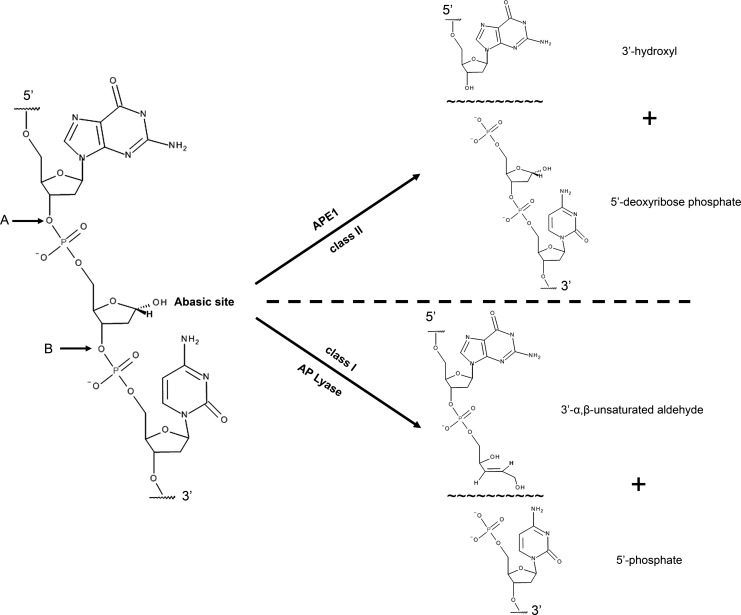
FIG. 1.
Chemical structure of a hydrolytic abasic site and the cleavage position for the major classes of AP site incision enzymes. The phosphodiester bond cleavage site, immediately adjacent to an abasic lesion (see arrows), is shown for a class I AP lyase (site B) and a class II AP endonuclease (site A). Class I AP lyases cleave via β-elimination, generating a 3′-α,β-unsaturated aldehyde and a 5′-phosphate. Class II AP endonucleases, for example, APE1, incise the DNA backbone hydrolytically, leaving behind 5′-deoxyribose phosphate and 3′-hydroxyl termini. For simplicity, just the strand containing the AP site is shown, with two “random” flanking bases. Images were created using the Accelrys Draw 4.1 software (Accelrys, San Diego, CA). AP, apurinic/apyrimidinic; APE1, apurinic/apyrimidinic endonuclease 1.
Given the high frequency of apurinic/apyrimidinic (AP) sites in DNA, and their potential for promoting deleterious outcomes, investigators searched for possible repair activities specific for these lesions. Reports in the 1970s [reviewed in Lindahl (121)], indeed, described protein fractions of varying purity from a range of organisms, including mammalian cell and tissue extracts, that exhibited the ability to cleave at abasic sites, which had been introduced into DNA by acid/heat hydrolysis or genotoxin-induced base release. Subsequent work by Linn and colleagues (147) revealed that there are two main classes of AP endonucleases: those that cleave 3′ to the abasic residue (class I) and those which cleave 5′ (class II) (Fig. 1). It was presumed that these incision activities represented an important step toward the eventual removal of abasic sites from chromosomal DNA.
Around the same time, a separate class of enzymes, termed DNA glycosylases, was discovered that had the capacity to release abnormal base residues from DNA in free form, producing an AP site intermediate. We now appreciate that there exist a collection of conserved DNA glycosylases that exhibit specificity for a broad range of substrate bases, including the deamination product of cytosine, uracil, the alkylation product 3-methyladenine, and the oxidation product 8-oxoguanine, to name a few (16, 177). Piecing together the information at the time, it was clear that organisms had evolved a DNA repair pathway which (i) excised a modified substrate base by breaking the N-glycosylic bond, (ii) cleaved the DNA backbone 5′ to the resulting AP site (class II incision), (iii) removed the 5′-abasic fragment via some form of nuclease degradation, (iv) carried out repair replication, and (v) sealed the remaining nick via DNA ligation. This pathway was designated base excision repair (BER), because unlike nucleotide excision repair, which involved the release of an oligonucleotide fragment, BER was initiated by direct base release (121).
Today, BER is considered the primary mechanism for coping with most forms of spontaneous hydrolytic, alkylative and oxidative DNA damage. BER has been sub-divided into two major pathways based on the number of nucleotides incorporated during repair synthesis, that is, single nucleotide and long patch, which involves incorporation of 2–13 nucleotides and strand-displacement polymerization. Single-strand break repair, which is a specialized process that is used for dealing with abnormal 3′- or 5′-termini that block DNA polymerase or ligase activity, also engages many of the core BER proteins. For a more detailed discussion of BER, its protein components, and its relationship to disease and other processes, the readers are directed to (39, 109) and the accompanying review by Izumi and colleagues.
Discovery and Cloning of Human AP Endonuclease 1
As alluded to earlier, early work from the Linn and Grossman laboratories described the purification of an endodeoxyribonuclease from HeLa cervical cancer cells and human placenta that acted on AP sites in substrate DNA molecules (64, 95, 187). This protein, which represented the predominant AP site incision activity in human cell extracts, was roughly 32–41 kDa in molecular weight and monomeric, was most active at a pH around 7.6 to 7.8, required the divalent metal Mg2+ (or to a lesser degree Mn2+) for efficient cleavage, and exhibited class II endonuclease specificity, generating 3′-hydroxyl and 5′-deoxyribose-5-phosphate strand break termini (Fig. 1). Nearly a decade later, in the early 1990s, the transcript encoding the human AP endonuclease (at the time, termed APE, HAP1, and APEX, since named apurinic/apyrimidinic endonuclease 1 [APE1]/APEX1) was cloned by the Demple, Hickson, and Seki groups using one of two approaches: (i) a screen of a lambda phage expression library with an antibody against the purified protein (37) or (ii) a cross-hybridization screen of a human cDNA library using either the bovine or mouse AP endonuclease cDNA as a probe (171, 182). Strikingly, around the same time, human APE1 was independently identified (and the gene subsequently cloned) by Curran and colleagues as the major nuclear protein (termed REF-1) to simulate the DNA-binding activity of the AP-1 (Fos/Jun) transcription factor complex (233). This activation was shown to be mediated through reduction of a conserved cysteine residue within the DNA binding domain of the target protein, and was also observed with factors such as NF-κB, Myb, and members of the ATF/CREB family. Thus, it was recognized early on that APE1/REF-1 (from here on referred to as APE1) was a multi-functional protein, with roles in both DNA repair and transcriptional regulation. Figure 2 depicts a linear schematic of human APE1, identifying several of the key elements of the protein that will be discussed throughout.
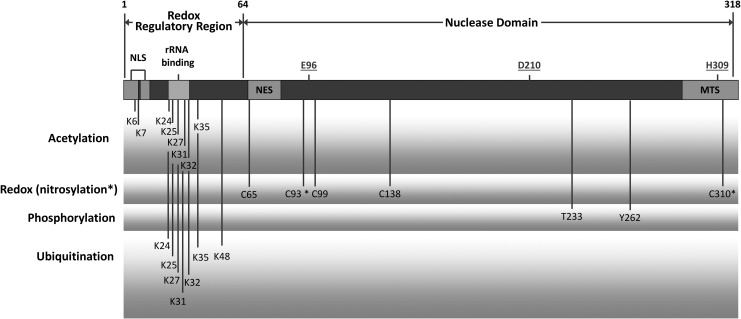
FIG. 2.
Schematic of key elements in the human APE1 protein. The nuclease domain of APE1 spans roughly residues 64 to 318, with the N-terminal portion of the protein encompassing much of the transcriptional regulatory functions of APE1 (with some overlap between the two regions). Key active site residues for the nuclease activities of APE1 (i.e., E96, D210, and H309) are identified via underline, and several PTM sites are depicted. A recently identified, lysine-rich, rRNA binding site is also designated. See text for further details. NLS, nuclear localization signal; NES, nuclear export signal; MTS, mitochondrial targeting sequence; PTM, post-translational modification.
APE1 Protein
The human APE1 gene is composed of five exons, spans roughly 2.5 to 3 kb of DNA, and is located on chromosome 14q11.2 (3, 72, 73, 172, 247). The main mRNA transcript is ∼1.5 kb in length, and appears to be ubiquitously expressed in all tissue and cell types from a housekeeping-like promoter that lacks a consensus TATA box, but harbors a CCAAT-like sequence and several putative CpG regulatory elements. The open reading frame is located within the last four exons, and it encodes a protein of 318 amino acids (theoretical molecular weight of 35.5 kDa). From residue ∼60 through the end of the protein, human APE1 shares sequence homology with Escherichia coli exonuclease III (xth), but harbors a unique N-terminal region (Fig. 3A). Exonuclease III had been shown earlier to exhibit a range of nuclease activities consistent with what had been described for the mammalian protein, including AP endonuclease, 3′-repair diesterase, and RNase H functions [reviewed in Demple and Harrison (36)]. Supportive of a critical biological role for these proteins, exonuclease III orthologs have been conserved throughout evolution and are present in many organisms (for examples, see Fig. 3A).
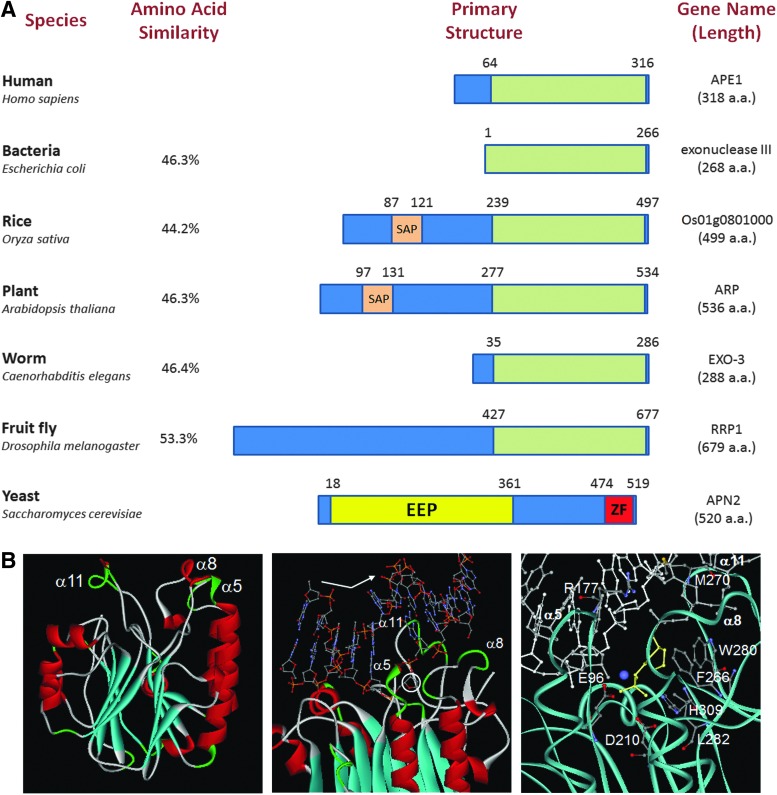
FIG. 3.
Human APE1 orthologs and key protein structural elements. (A) APE1 orthologs. The particular species (left) and gene name (right) are indicated. Shown is the primary structure, with the AP endonuclease domain designated as a green box. Total amino-acid sequence length and percent similarity with human APE1 are specified. SAP, SAF-A/B, Acinus and PIAS motif, which is a putative DNA/RNA binding domain; EEP, Exonuclease-Endonuclease-Phosphatase domain; ZF, zinc finger. (B) APE1 structural features. All images were created using the DS ViewerPro software (Accelrys, San Diego, CA). Left: ribbon diagram of human APE1 [coordinates from 1BIX (63)]. The major recognition loops are designated. Middle: diagram of APE1 bound to un-incised AP-DNA [coordinates from 1DE8 (145)]. Recognition loops are indicated, and the abasic site is circled. Note the 35° kinking of the DNA backbone (emphasized by arrow). Right: diagram of the binding surface and active site of APE1 in complex with incised AP-DNA and Mn2+ [coordinates from 1DE9 (145)]. DNA strands are shown in white, with the 5′-abasic residue designated in yellow. The protein is shown as a cyan ribbon. The recognition pocket (comprising F266, W280, and L282) and several key active site residues (E96, D210, and H309) are highlighted. The recognition loops, and two key binding residues (R177 and M270), are also indicated. The catalytic metal ion is depicted as a purple sphere. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
An initial analysis of the primary amino-acid sequence of human APE1 identified consensus nuclear localization signals (NLSs) within the acquired N-terminus (Fig. 2), which have since been confirmed to function in sub-cellular targeting (see “Intra-cellular targeting and mitochondrial function” section), as well as a number of putative phosphorylation sites throughout the length of the protein (see later). More recent, a comprehensive bioinformatic sequence analysis has revealed that APE1 is a part of a divalent cation-dependent phosphoesterase superfamily of proteins, which includes nucleases (e.g., DNase I, RNase H, and the LINE-1 or L1 retrotransposon endonuclease), inositol (polyphosphate) and, possibly, protein phosphatases, and sphingomyelinases (40). These proteins, likely arising via divergent evolution, share a conserved four-layered α/β-sandwich structural core, and exhibit a wide range of substrate specificities, while maintaining a generally conserved catalytic mechanism for cleaving phosphoester bonds in nucleic acids, proteins, and phospholipids [see for example (225)]. X-ray crystallography studies indicate that the superfamily of enzymes, while preserving the common α/β fold, possess specific loop regions and active site elements that likely dictate substrate specificity [see for example (63, 209, 223)].
Crystal structures solved by Tainer and colleagues of human APE1 bound to both substrate (intact) and product (incised) forms of AP-DNA indicate that the protein uses a rigid, pre-formed positively charged surface to kink the DNA helix and engulf the AP site strand to facilitate targeted complex formation (145). Stabilization of the kinked abasic substrate is further mediated by residues emanating from four loops and one α-helix (e.g., α5, α8, α11), with the “flipped-out” extra-helical AP site positioned within a pocket (comprising F266, W280, and L282) that excludes DNA bases (Fig. 3B). The importance of the α8 loop and the active site pocket residues in determining substrate specificity has been demonstrated by a biochemical analysis of site-specific mutant APE1 proteins (70, 188). A noteworthy feature of APE1 is that R177, which is located within the α5 loop, inserts into the major groove to provide a hydrogen bond to the AP site 3′ phosphate, and, in doing so, seems to lock the protein onto the incised DNA product. Since mutation of R177 improves the turnover rate of APE1, it would appear that the enzyme has evolved not for catalytic efficiency, but to facilitate coordination with the next enzyme in the BER pathway (145). This model of “passing the baton,” while supported by biochemical and structural data, however, lacks biological evidence.
Despite our sound understanding of how APE1 locates, scanning DNA in a quasi-processive manner (24), and specifically recognizes AP sites, there remains controversy around how APE1 cleaves the phosphodiester backbone. Nonetheless, there is general consensus that E96 plays a role in divalent metal coordination and that D210 and H309 have critical functions in the hydrolytic reaction chemistry (Fig. 3B). Other important residues include N68, D70, Y171, N212, D283, and D308, which are generally conserved throughout the diverse members of the phosphoesterase superfamily. Still, further studies are needed to delineate the precise catalytic reaction mechanism, which is debated in several recent publications (123, 148, 152, 210).
Before proceeding with our more detailed discussion of human APE1, it is worth mentioning that in E. coli there are two prominent AP endonucleases: exonuclease III and endonuclease IV (nfo) [reviewed in Demple and Harrison (36) and Mol et al. (145)]. However, in mammals, there is no obvious functional ortholog of endonuclease IV. Instead, there is a second protein with homology to exonuclease III, termed APE2 (also known as APEX2) (71, 208). The AP endonuclease and 3′-damage excision activities of the human APE2 protein are generally weak, and, thus, its function as a DNA repair enzyme remains in question (18, 19, 70). It is not our intent to describe the current picture of APE2 in detail, but we point out that, while APE2-null mice exhibit growth retardation and dyshematopoiesis, APE2-deficient mouse embryonic stem cells or immortalized fibroblasts do not display increased sensitivity to several genotoxic agents, including hydrogen peroxide, bleomycin, or x-ray irradiation (86). Moreover, deletion of APE2 in the CH12F3 mouse B cell line does not affect sensitivity to methylmethane sulfonate (MMS) (136). These data support the notion that APE2 is not a major player in DNA damage repair, and contend that APE1 is the predominant, if not sole, AP endonuclease in mammals. Given that APE2 is a member of the α/β-sandwich superfamily of enzymes that exhibit diverse signaling functions, it would be valuable to explore other substrate specificities of APE2, such as against proteins and phospholipids.
APE1 Biochemical Activities
3′-repair diesterase
The initial work on the purified mouse, bovine, and human AP endonuclease found that the mammalian protein exhibited not only a powerful class II AP site incision activity, but also the ability to excise 3′-damages, such as 3′-phosphates, 3′-phosphoglycolate esters, and 3′-deoxyribose fragments (27, 173, 182, 183). These 3′-end blocking groups prevent primer extension by a DNA polymerase or nick ligation by a DNA ligase, and, thus, have the potential to promote chromosome instability as unrepaired strand breaks. Such non-conventional groups arise as products of hydroxyl radical attack of the sugar phosphate backbone or as repair intermediates during DNA processing. Experiments using human whole cell extracts have shown that APE1 is rate limiting for the repair of DNA strand breaks induced by hydrogen peroxide (presumably 3′-phosphates) and bleomycin (3′-phosphoglycolates) (89), and is the predominant enzyme for the excision of 3′-phosphoglycolate residues from a single nucleotide gap (160). Biochemical studies also indicate that APE1 plays a prominent role in the removal of 3′-deoxyribose fragments that are generated by AP lyase β-elimination reactions, which are catalyzed by certain multi-functional DNA glycosylases (e.g., 8-oxoguanine DNA glycosylase [OGG1] and endonuclease III homolog 1 [NTH1]) or are promoted “non-specifically” by basic polypeptides, such as polyamines or histones [reviewed in Doetsch and Cunningham (41) and Hegde et al. (75)]. It should be pointed out that the ability of human APE1 to excise 3′-damages appears to be largely restricted to single-strand breaks or 3′-recessed double strand break ends, as the enzyme is highly inefficient at removing 3′-phosphoglycolates from blunt or 3′-overhang (single-stranded) termini (199). Indeed, there exist other proteins; for example, polynucleotide kinase 3′-phosphatase (PNKP) and tyrosyl-DNA phosphodiesterase 1 (TDP1), which provide complementary 3′-damage repair activities [reviewed in Caldecott (23) and Wilson (227)].
3′ to 5′ exonuclease
Work by Seki et al. found that the mouse and human proteins possess 3′ to 5′ exonuclease activity, which is a prominent function of the E. coli exonuclease III protein (182, 183). The exonuclease activity of human APE1, however, is poorly processive and ≥100-fold less efficient than its AP endonuclease activity, although the former activity is somewhat influenced by reaction conditions, sequence/positional context (see comments in previous section), and thermal stability of the duplex (28, 44, 45, 226, 231). Notably, APE1 displays the capacity to excise certain 3′-mispaired nucleotides with increased proficiency, and, in this setting, may act as an autonomous proofreading enzyme for DNA polymerases which lack editing activity (28, 226). Moreover, Chou et al. (29) isolated APE1 as the major protein that excises the clinically relevant, chain-terminating nucleoside analog, β-L-dioxolane-cytidine (also known as, troxacitabine), from the 3′-end of a 3′-recessed oligonucleotide substrate (see more in “APE1 as a Therapeutic Target”). We note that, at present, the crucial, biological role of the 3′ to 5′ exonuclease activity for the exonuclease III orthologs remains largely a mystery.
RNA cleavage
Similar to E. coli exonuclease III, human APE1 exhibits RNase H activity, albeit to a lesser extent than its bacterial counterpart (9). As with the 3′ to 5′ exonuclease activity, the biological significance of the ability of APE1 (or exonuclease III for that matter) to degrade an RNA molecule hybridized to a complementary DNA strand is unknown. We note that this activity in RNase H is used to remove the RNA primers during DNA replication, yet there is no evidence for a role of APE1 in this process. More recent work has found that APE1 can cleave abasic sites in RNA, and it has been proposed that the protein operates in RNA quality control, removing damaged RNA templates to prevent error-prone translation, although this model has not been sufficiently validated in cells (10, 215).
Lee and colleagues identified APE1 as a major endoribonuclease able to cleave the coding region determinant of the c-myc mRNA (preferentially in between UA and CA dinucleotides), thereby affecting transcript turnover (8). Subsequent in vitro experiments surprisingly found that APE1 catalyzes RNA incision in the absence of divalent metals (107). Since earlier work had demonstrated that APE1 can also incise acyclic AP sites in an Mg2+-independent manner (47), the results suggest that flexibility/dynamics of the phosphodiester linkage, which is greater adjacent to an acyclic lesion, plays an important role in the establishment of the transition state intermediate. More extensive structure-function analyses revealed that APE1 engages several of the same active site residues used for AP-DNA cleavage to promote RNA incision (excluding D283), requires a 2′-hydroxyl group on the sugar moiety for catalysis, and generates products with a 3′-phosphate, indicating both conserved and distinct mechanisms for AP-DNA and RNA cleavage (106). Although the specific activity of APE1 to cleave target mRNAs is much lower than its AP endonuclease function in vitro, Barnes et al. (8) found that transient knock-down (KD) of APE1 in HeLa cells results in an increased half-life and steady-state level of c-myc mRNA, providing evidence for a biological role for its RNA cleavage function. The identification of potential separation-of-function mutations in APE1 [see the D283N mutant alluded to earlier, (106)] should permit additional studies to determine whether the enzyme contributes to mRNA turnover in vivo.
Nucleotide incision repair
Strikingly, APE1, thought to be limited to cleavage of abasic sites in intact, double-stranded DNA, has been reported to recognize and incise at certain base damages (e.g., 5,6‐dihydro‐2′‐deoxyuridine, 5,6‐dihydrothymidine, 5‐hydroxy‐2′‐deoxyuridine, α‐2′‐deoxyadenosine, and α‐thymidine adducts), generating single-strand break ends with a 3′-hydroxyl and a 5′-dangling modified nucleotide (65). Distinct from its AP endonuclease function, this activity—proposed to initiate a corrective response termed nucleotide incision repair (NIR) that would serve as a back-up for the more classical glycosylase-initiated BER pathway—is most active at low MgCl2 and KCl concentrations, at a pH range of 6.4 to 6.8. Such a nuclease activity is seemingly consistent with earlier work which found that APE1 can incise at benzene-derived base adducts, similarly creating 3′-hydroxyl and 5′-dangling modified nucleotide ends (67). Molecular modeling of a duplex harboring the benzetheno exocyclic adduct of cytosine (pBQ-C), and subsequent molecular dynamics simulations with APE1, implied that the pBQ-C adduct can be accommodated in the enzyme's active site on specific structural rearrangements of both the DNA and protein, and that APE1 would be able to execute a similar reaction mechanism for phosphodiester bond cleavage as used on AP-DNA. However, a precise understanding of how APE1 specifically binds DNA base adducts requires high resolution structural information, and further experiments are necessary to substantiate the biological role of this activity in human cells.
Redox regulation
As mentioned earlier, APE1 was independently purified based on its ability to stimulate the DNA-binding activity of several transcription factors through a redox regulatory mechanism (233). Since then, APE1 has been reported to modulate the redox status of both ubiquitous (e.g., AP-1, Egr-1, NF-κB, p53, CREB, and HIF-1α) and tissue-specific transcription factors (e.g., PEBP-2, Pax-5, and -8, TTF-1) with functions in stress responses and other cellular processes [reviewed in Kelley et al. (101)]. Although the redox regulatory and DNA repair nuclease activities can be disabled separately by site-specific mutagenesis (219, 234), the precise molecular details of the protein-facilitated activation step remain elusive (103). In particular, none of the cysteine residues within APE1 are found in a C-X-X-C motif that is common to most redox regulatory factors, such as thioredoxin, a cellular component which appears to be involved with APE1 in a redox regulatory cascade (81). In addition, disulfide bond formation is considered a necessary step in the resolving activity of redox factors, and while most evidence supports a role for C65 in the thiol-mediated redox reaction (61, 219), none of the other six cysteine residues in APE1 are located appropriately in the 3-dimensional structure to facilitate disulfide bond formation with C65. Finally, although some data imply a role for C93 in the redox reaction, both C65 and C93 are buried within the protein structure, while being positioned at a distance (9 Å) that is not compatible with disulfide bond formation. Based on the current experimental evidence, Georgiadis and colleagues have proposed a model in which (i) C65 serves as the nucleophilic residue for reduction of the disulfide bond in the target transcription factor; (ii) C93 operates as the resolving residue; and (iii) APE1 undergoes a significant conformational change (possibly unfolding) to reveal a third cysteine residue (possibly C99) to facilitate the redox reaction (130). While studies are accumulating which indicate that the redox function of APE1 is important to processes such as cell growth and differentiation (see more in “Determination of cell fate” and “Small molecule inhibitors” sections), further experiments are needed to delineate the precise molecular mechanism of the reaction.
Trans-acting modulation
In addition to functioning as a transcriptional regulator through post-translational modification (PTM) of a target protein (see previous section), APE1 is also a component of nuclear protein complexes that bind to negative Ca2+ response elements (nCaREs) and modulate gene expression. In particular, on a rise in extracellular Ca2+, Okazaki et al. observed an increase in APE1 transcript and protein levels that resulted in augmented complex assembly at the nCaREs (nCaRE-A and nCaRE-B) of the parathyroid hormone (PTH) gene that promoted transcriptional suppression (153). Subsequent work found that APE1 binds to the nCaRE PTH sequences in cooperation with, minimally, the two subunits of the Ku antigen, p70 and p80 (30), and that complex formation of APE1 with the promoter elements is controlled by lysine (K6 and K7) acetylation (11). A role for APE1 in nCaRE-dependent transcriptional suppression was also observed for the human renin gene in chorio-decidual cells as a part of a response to elevated intra-cellular Ca2+ levels (54) and for the BAX gene in gastric epithelial cells as a part of an elaborate response to Helicobacter pylori infection that involves APE1 acetylation and poly(ADP)ribose polymerase 1 (PARP1) (12). Interestingly, the upstream region of the APE1 promoter contains three nCaRE-like sequences, at least one of which (nCaRE-B2) appears to be bound by an APE1-containing protein complex that negatively auto-regulates gene expression (90). A follow-up analysis revealed that heterogenous nuclear ribonucleoprotein L (hnRNP-L) was a component of the nCaRE-B2 binding complex (113). It would be worthwhile to determine the technical or biological (cell-dependent?) reasons for why an independent study did not identify these negative regulatory elements in the APE1 promoter (72), as well as whether APE1 has a broader role in modulating gene expression via binding CaREs or other promoter sequence elements throughout the genome. A more exhaustive characterization of the composition of the APE1-specific, multi-protein transcriptional complexes would also be worthwhile.
We close this section by noting that Gillespie et al. have proposed a model in which “controlled” oxidative DNA damage in hypoxic regulatory elements in specific promoters (e.g., of VEGF) may lead to BER processing events, such as binding and subsequent strand cleavage by APE1, which can induce changes in local sequence topology and DNA flexibility that could drive productive transcription [reviewed in Gillespie et al. (62)]. However, how such oxidative DNA damage is targeted and how repair may be strategically manipulated at the promoter site to foster transcription factor assembly and, ultimately, gene expression requires further clarification.
APE1 Biological Roles
Shortly after cloning of the APE1 gene, studies were undertaken to elucidate the cellular functions of the protein. In particular, Walker et al. (218) showed that HeLa cells stably expressing antisense APE1 RNA exhibited a similar doubling time, plating efficiency, and gross morphology, but were hyper-sensitive to killing by a range of DNA-damaging agents, including MMS, hydrogen peroxide, menadione, and paraquat, but not ultraviolet irradiation. Ono et al. (155) similarly found that C6 rat glioma cells expressing antisense APE1 RNA displayed normal growth rates, but reduced survival when challenged with MMS or hydrogen peroxide. These data are consistent with a role for APE1 in repairing alkylative and oxidative DNA lesions, which are likely to include abasic sites and dirty 3′-strand break ends, respectively, that engage the AP endonuclease and 3′-repair diesterase activities of the enzyme. A more comprehensive discussion of the phenotypes of APE1-defective cells, specifically in the context of clinical DNA-damaging agent sensitivity, is presented later in “APE1 as a Therapeutic Target.”
Complementation strategies were also employed to determine the potential biological functions of APE1. In particular, trans-complementation of E. coli strains which were deficient in the main AP endonucleases (exonuclease III and endonuclease IV) revealed that human APE1 is able to fully complement MMS sensitivity, but only partially correct hydrogen peroxide sensitivity (37, 171). A similar damaging agent rescue pattern was observed when the human protein was expressed in yeast-deficient in the major AP endonuclease, APN1 (72). Chinese hamster ovary cells designed to inducibly express a dominant-negative form of APE1 (termed ED, due to E96Q and D210N mutations), which displays better than wild-type binding affinity for substrate DNA, but is devoid of nuclease activity, exhibited pronounced hyper-sensitivity to MMS, yet a more mild increase in sensitivity to hydrogen peroxide (138). The combined data are consistent with APE1 maintaining a powerful AP endonuclease function that participates in the repair of AP sites formed by the direct and indirect action of MMS, and a lesser, albeit significant, 3′-repair activity for removal of 3′-blocking groups (e.g., 3′-phosphates) generated by the oxidizing agent hydrogen peroxide. More extensive trans-complementation has not been undertaken to our knowledge, but could be a powerful strategy if employed creatively in bacteria or yeast to assess the other reported biochemical activities of APE1 (see “APE1 Biochemical Activities” section), such as its exonuclease function.
Recent work by Demple and colleagues has shown that strong, chronic down-regulation of APE1 in multiple human cell lines using a siRNA approach results in accumulation of total genomic abasic sites, inhibition of proliferation, a dramatic increase in the proportion of cells with sub-G1 DNA content, and activation of apoptotic cell death (55). Successful rescue of these cellular phenotypes via trans-complementation with the yeast AP endonuclease APN1, which has no sequence or structural homology to APE1 and lacks a redox regulatory function, suggests that the repair nuclease activities shared between these two proteins are required for cell survival (consistent with the observation of endogenous DNA damage accumulation). Studies by the Mitra group using nullizygous embryonic fibroblasts from APE1−/− mice that are transgenic with a “floxed” human APE1 gene also found an essential role for the protein in cell viability, as Cre-mediated excision of the complementing gene resulted in rapid apoptosis (88). Trans-complementation experiments with various human APE1 gene mutations revealed that the DNA repair nuclease activity (an H309N mutant) or its acetylation-mediated transcriptional regulatory function (a K6R/K7R mutant), but not its redox regulatory function (a C65S mutant), was necessary for cell survival, although these studies should be interpreted with caution as expression of the complementing protein was not documented. More recently, the Tell laboratory created a stable, inducible KD HeLa cell line, in which APE1 expression is decreased to <5% after 10 days of doxycycline treatment (214, 215). Although the KD cells eventually die, exhibiting the typical growth arrest and apoptosis, this model has been a valuable tool for delineating the contribution of specific APE1 functions. Consistent with the previous work, KD cells complemented with a nuclease-deficient human APE1 cDNA harboring an H309N mutation did not survive, confirming the essential nature of the DNA repair activities of the enzyme (213). However, in contrast to what the Mitra laboratory observed (see above), the C65S redox regulatory mutant did not rescue the inviability of the KD cells; whereas the non-acetylatable K6R/K7R mutant provided near wild-type complementation for cell growth. While it remains unclear why opposite results were obtained in the two studies (possibly reflecting differences in strategies, cell types, etc.), the experiments provide evidence for biologically important roles for the redox regulatory function and acetylation of APE1.
Though it had generally become accepted that APE1 is required for mammalian cell viability, a recent report by Masani et al. (136) described the generation of an APE1 knock-out (KO) CH12F3 mouse B cell line that displays normal cell proliferation, while exhibiting the expected hyper-sensitivity to MMS. Although the precise reason(s) for the normal growth of the APE1-deficient cells is uncertain, it is likely that a natural or induced up-regulation of a compensatory pathway, such as nucleotide excision repair, homologous recombination, or translesion DNA synthesis, mediates survival in the face of the high levels of endogenous DNA damage. Thus, by extension, we anticipate that in mammalian cells in which APE1 is deficient, the genetic and biological background will ultimately determine the cellular attributes, potentially explaining the contrasting viability results mentioned in the preceding paragraph with the different site-directed mutants. Of course, variability in technical aspects of the approaches cannot be excluded from contributing to the disparate outcomes. Addressing these issues would seem to be a worthwhile pursuit going forward. We close this section by pointing out that both E. coli and S. cerevisiae which lack their respective major AP endonuclease genes are able to survive, although they exhibit mild growth defects.
Intra-cellular targeting and mitochondrial function
APE1 has long been recognized as a prominent nuclear protein, where it would perform its DNA repair and transcriptional regulatory activities. It is, in fact, its nuclear functions that are often presumed to be essential for cell or organismal viability, although given its alternative compartmentalization (introduced below), this cannot be stated with absolute certainty at present. Using GFP-tagged truncated and mutated APE1 expression constructs, Izumi and colleagues found that APE1 harbors two independent segments within the first 20 N-terminal residues that direct its strong nuclear localization (Fig. 2): (i) residues 2–7, which contain a classic pat7 type NLS (PKRGKK), and (ii) residues 8–13, which harbor two critical acidic amino acids (E12 and D13), but lack a signature NLS (91). These authors also reported that the N-terminal portion of APE1 mediates a physical interaction with the nuclear importins, KAPα1 and KAPα2, which likely facilitate APE1 nuclear internalization. Notably, a normally cytoplasmic, mutant form of APE1 was found to be sequestered in the nucleus on treatment with leptomycin B, a nuclear export inhibitor, suggesting that the protein possesses a nuclear export signal (NES), although this sequence was not explicitly identified at the time (see more in next section).
Despite the strong nuclear targeting elements within APE1, the protein has been reported to be cytoplasmic in some cell types, such as those with high metabolic or proliferative rates, displaying co-localization with mitochondria and the endoplasmic reticulum [reviewed in Tell et al. (204)]. Using both standard and rigorous mitochondrial purification techniques, several groups have, indeed, reported the existence of AP endonuclease activity, and an APE1-like protein species, in the mitochondrial fraction (57, 203, 207). Mitochondria possess an extra-nuclear genome (mtDNA), which due to its proximity to the electron transport chain (ETC), is highly susceptible to attack by reactive oxygen species (ROS) generated as by-products of oxidative phosphorylation. Persistent mtDNA damage can lead to problems in expression of the essential ETC proteins, leading to a greater dysfunction in oxidative phosphorylation that drives further ROS production, a vicious cycle fundamental to the mitochondrial theory of aging [reviewed in Miquel et al. (141)]. Due to the high frequency of potentially deleterious oxidative modification, mitochondria have acquired their own protective DNA repair mechanisms to preserve genome integrity and maintain mitochondrial function. We now recognize that BER is the major DNA repair system in mitochondria, with many of the mitochondrial BER (mtBER) proteins arising from either alternative splicing or translation [reviewed in de Souza-Pinto et al. (35), Liu and Demple (125), and Van Houten et al. (212)].
How APE1 is targeted to mitochondria is still not completely clear, but several studies have shed insights on this issue. One report found that a 33 residue N-terminal truncated form of APE1 is present in mitochondria from both bovine liver and mouse NIH3T3 cells (26). The authors proposed that APE1 is directed to this organelle by removal of the N-terminus, which harbors the prominent NLS (Fig. 2), via a site-specific cleavage event that might engage a mitochondrial matrix peptidase. However, given the many studies which have reported full-length APE1 in the mitochondrial fraction [see for instance (203, 213)] and the evidence that the N-terminal portion of APE1 is susceptible to degradation (182, 184), it seems likely that non-specific proteolysis occurred during the fractionation steps, leading to the creation of the 33 amino-acid N-terminal truncated form. Since there is no consensus mitochondrial targeting sequence (MTS) in APE1 based on bioinformatics scrutiny, Li et al. (118) screened for interactions between a series of peptides that span the length of the APE1 protein and three translocases of the outer mitochondrial membrane. These binding assays uncovered a putative MTS in the C-terminus of APE1, encompassing residues 289–318. Site-directed mutagenesis of residues K299 and R301 confirmed the importance of this region in directing either the 289–318 peptide or full-length APE1 to the mitochondrial compartment. Moreover, a recent study found that full-length APE1 resides mainly in the mitochondrial inter-membrane space (213), a localization pattern that appears distinct from other mtBER proteins, which are associated with mtDNA in the matrix (198). In this work (213), APE1 was shown to interact, in a proximity ligation assay, with the oxidoreductase Mia40, which plays a key role in oxidative protein folding within the mitochondrial inter-membrane space. Thus, the current data suggest a model in which a presently unknown PTM of full-length APE1 is necessary to reveal the “masked” MTS within the C-terminus and enable mitochondrial-specific protein interactions that direct mitochondrial localization.
Relatively few studies have assessed the specific contributions of APE1 to the maintenance of mitochondrial integrity. This fact is due, in part, to the difficulty in separately examining the mitochondrial and nuclear functions of the protein. To determine the effects of increased APE1 in mitochondria, Li et al. expressed a recombinant fusion protein composed of the strong MTS of manganese superoxide dismutase and the repair domain of APE1 (residues 34–318) in human umbilical vein endothelial cells (119). Targeted APE1 mitochondrial over-expression was shown to enhance cell viability and suppress apoptosis after hydrogen peroxide-induced oxidative stress, presumably by enhancing mtDNA repair. This observation suggests that APE1 is rate limiting in mtBER, although this finding appears to be cell-type specific, as a separate study found that mitochondrial targeting of E. coli exonuclease III in the human malignant breast epithelial cell line, MDA-MB-231, results in impaired mtDNA repair and a decrease in long-term survival after oxidative stress (192). One message taken from these studies is that additional work is needed to better understand the coordination between the components of mtBER.
As noted earlier, Vascotto et al. showed that the redox mutant C65S is unable to rescue the inviability of the KD HeLa cell line (213). However, they also found that this mutant was unable to counteract the decreased mitochondrial membrane potential, that is, the mitochondrial dysfunction, of the APE1-deficient cells. A more comprehensive analysis revealed that the C65A mutation affected not only the global gene expression pattern, presumably due to defects in redox regulation of nuclear transcription factor binding, but also its redox-assisted protein folding, which adversely affected its protein interactions, AP endonuclease activity, and mitochondrial localization. Thus, the C65 residue, which has been proposed to play an important role in the redox chemistry (see “Redox regulation” section), also appears to mediate intra-cellular trafficking, such that reduced APE1 mitochondrial accumulation results in mitochondrial impairment and apoptotic cell death. Li et al. independently confirmed that reduced overall APE1 levels correlate with loss of mitochondrial membrane potential and apoptosis, yet presented evidence that much of this effect was mediated indirectly through reduced expression of several nuclear genes which encode mitochondrial proteins, including those that facilitate mitochondrial trans-membrane transportation (117). The studies go on to identify the transcription factor NRF1, which plays an important role in modulating the expression of nuclear genes involved in respiration and mtDNA replication and transcription, as a key target of APE1-mediated redox regulation. Thus, in light of the complexity of APE1's functions, it is evident that more extensive studies employing separation-of-function, site-specific mutants will need to be performed to better clarify the cellular consequence(s) of disrupting a particular APE1 function, including its role in mitochondria.
Intra-cellular trafficking
Some of the early work indicating that APE1 localized to compartments other than the nucleus was reported by Kelley and colleagues. Using immunohistochemistry, they found that APE1 had both nuclear and cytoplasmic staining in certain brain and liver cells within paraffin-embedded autopsy specimens from a 70 year-old man (43). Since then, alternative localization patterns have been observed for APE1 that appear to be influenced by cell-type, environmental exposures, or disease status (see sections on “Cancer” and “Neuropathology” later). It would also appear that the intra-cellular distribution of APE1 can be regulated via an active process, although the precise molecular mechanisms are unclear [reviewed in Tell et al. (204)]. For example, Mitra and colleagues were one of the first to show that APE1 can translocate to the nucleus on the introduction of oxidative stress (168). Much research since has discovered that the APE1 distribution pattern can change on pro-oxidant injury, heavy metal or DNA-damaging agent exposure, or hormone or cytokine stimulation, to name a few [reviewed in Tell et al. (204, 205)].
While still in its infancy of understanding, there are some insights regarding the potential mechanisms for regulating APE1 intra-cellular distribution. For example, as described in the previous section, Vascotto et al. found that the redox state of C65 can control the mitochondrial localization of APE1 (213). In addition, one study showed that S-nitrosoglutathion, a nitric oxide donor and S-nitrosating agent, can induce APE1 nuclear export, with this nuclear to cytoplasmic translocation being mediated by S-nitrosation of residues C93 and C310 (165). The translocation process was reversible, could not be mimicked by hydrogen peroxide-induced oxidative stress, and involved, to some degree, reduced nuclear import. Deletion analysis revealed that residues 64–80 of APE1 likely harbor the NES and that there exists a “hidden” MTS in the final 69 amino acids of the protein [consistent with the results of (118); see the previous section]. The authors propose that S-nitrosation of C93 and C310 induces a protein comformational change that unmasks the NES, promoting cytosolic translocation. Collectively, the current data suggest a model in which unmodified, full-length APE1 is mainly targeted to the nucleus via its N-terminal elements, but on PTM (not fully defined to this point), the typically inaccessible NES or MTS is revealed (Fig. 2), driving re-distribution of the protein. A major emphasis of future work will be to unravel how protein interactions (see for example nucleophosmin below) and PTMs regulate APE1 activities and intra-cellular localization. We direct the readers to a recent review describing in greater detail potential PTMs of APE1 (21), and provide Table 1 as a summary of these alterations and their reputed effects, which are likely influenced in a cell type- or response-specific manner.
Table 1.
Post-Translational Modification of Human APE1
| PTM | Modifying enzyme | Site(s) of modification | Impact on activity | Cellular aspects | Reference |
|---|---|---|---|---|---|
| Phosphorylation | Cdk5 complex | T233 (T232 in mouse) | Nuclease: Decreased | Increased cell vulnerability to DNA damage | (84) |
| Was elevated in post-mortem brain tissue of PD and AD patients | |||||
| ND | Y262 | ND | Was detected in anaplastic large-cell lymphoma SU-DHL-1 cells | (175) | |
| CK II | ND | Nuclease: abolished | ND | (240) | |
| CK II | ND | Nuclease: normal | Was protective against MMS cytotoxicity | (53) | |
| Redox: increased | |||||
| CK I | ND | Nuclease: normal | ND | (240) | |
| PKC | ND | Nuclease: normal | Was elevated in response to oxidative stress | (82, 240) | |
| Redox: increased | |||||
| Acetylation | p300 & SIRT1 | K6/K7 | Nuclease: decreased | Increased binding of APE1 to PTH nCaRE-B promoter sequence | (11, 185, 241) |
| Affected YB-1 binding to MDR1 promoter | |||||
| Decreased the interaction between APE1 and XRCC1 | |||||
| Promoted anti-cancer drug resistance | |||||
| SIRT1 | K24/K25/K27/K31/K32 | Nuclease: decreased | Stabilized the binding of APE1 to mRNA | (49) | |
| SIRT1 | K27/K31/K32/K35 | Nuclease: decreased | Promoted APE1 nucleolar accumulation | (124) | |
| Regulated K6/K7 acetylation | |||||
| ND | K35 | ND | Was increased in triple negative breast cancer samples | (162) | |
| Ubiquitination | MDM2 | K24/K25/K27 | ND | Promoted nuclear exclusion | (20) |
| MDM2(Mono) | K48(Poly) | ND | Poly-ubiquitination reduced APE1 protein stability | (22) | |
| K24/K25/K27(Mono) | Was enhanced by T233 phosphorylation | ||||
| UBR3 | K24/K25/K27/K31/K32/K35 | ND | Regulated APE1 protein level and MMS sensitivity | (140) | |
| Nitrosylation | ND | C93/C310 | ND | Regulated intra-cellular localization | (165) |
| Was increased by nitrosative stress | |||||
| Glutathionylation | None | C99 | Nuclease: decreased | Was increased by oxidative stress | (108) |
AD, Alzheimer disease; APE1, apurinic/apyrimidinic endonuclease 1; MMS, methylmethane sulfonate; ND, not determined; PD, Parkinson disease; PTM, post-translational modification.
Participation in specific cellular processes
To this point, we have reviewed the major reported biochemical activities and the more general biological roles of APE1, but there are a few specific molecular processes that appear to engage the enzyme worth detailing:
First, APE1 appears to play a broad role in various aspects of RNA metabolism (see also section entitled “RNA cleavage”). Using a HeLa cell line in which the endogenous APE1 protein was depleted and a comparable amount of a Flag-tagged version of APE1 was re-introduced, Vascotto et al. discovered that many of the co-immunoprecipitating partners of the fusion protein operate in RNA processing and ribosome biogenesis (215). Most of these interactions, including one with nucleophosmin (NPM1), are mediated by the N-terminal 33 amino acids of APE1 and may involve stabilization by RNA molecules. In addition, a significant portion of nuclear APE1 was found to be concentrated within the nucleoli, co-localizing with both NPM1 and nucleolin. This nucleolar distribution was influenced by the cell cycle and dependent on active rRNA synthesis, supporting a role for APE1 in RNA metabolism. Based on a series of biochemical and cellular experiments, the authors argue that the NPM1-APE1 interaction functions to regulate the ability of the endonuclease to remove damaged RNA molecules from the transcript pool. Future investigations are needed to better define (i) the role of the interactions of APE1 with RNA and NPM1 in influencing intra-cellular distribution and (ii) the biological importance of APE1 in RNA quality control, transcript turnover, and RNA biogenesis (see accompanying review by Tell and colleagues for further discussion).
Second, APE1, and other components of BER, operate in the programmed processes of somatic hypermutation (SHM) and class switch recombination (CSR), which introduce genetic variability within the immunoglobulin genes of B cells [reviewed in Keim et al. (99)]. During the adaptive humoral immune response to foreign antigen, SHM introduces mutations into the variable region exons to promote antibody affinity maturation, while CSR promotes genetic re-arrangements in the constant region of the heavy chain to drive isotype switching and optimization of interactions between antibody and different effector molecules. Both SHM and CSR require the targeted action of activation-induced cytidine deaminase, a protein that is expressed restrictively in activated B cells and deaminates cytosine to uracil in DNA. These uracils serve as a critical launching point for the mutagenic and recombination events that take place during antibody diversification, and are excised by uracil-DNA glycosylase (UNG), which generates an abasic site product. Guikema et al., using APE1 haploinsufficient mice (APE1+/−), reported reduced CSR in the S region of splenic B cells due to a decrease in DNA double-strand break formation, consistent with a role for APE1 in the AP site cleavage step (66). The authors also found that CSR was reduced in APE2-deficient mice, implicating both exonuclease III-like proteins in the process of isotype switching and suggesting a possible specialized DNA processing function for APE2 in activated B cells. As noted earlier, Yu and colleagues were successful in generating an APE1 KO CH12F3 mouse B cell line (136). Employing this resource, they found that CSR was drastically reduced in APE1-null cells, whereas the process was unaffected in CH12F3 cells deleted for APE2, whether APE1 was present or not. This research provides the strongest evidence to date of an essential role for APE1 in CSR, while implying that APE2 is not involved in this phenomenon, even as a back-up enzyme. Consistent with APE1 operating in CSR, purified recombinant APE1 protein can incise at AP sites in a synthetic oligonucleotide substrate designed to mimic the R-loop structure that is presumably needed for the recombination event (10). It is noteworthy that, to our knowledge, the contribution of APE1, or APE2 for that matter, in SHM has not been explicitly addressed.
Third, APE1 functions as a member of a 270–420 kDa multi-protein complex, termed the SET complex, to aid in the execution of the granzyme A (GzmA)-activated cell death response [reviewed in Lieberman (120)]. GzmA is an abundant serine protease in the granules (i.e., specialized secretory lysosomes) of natural killer cells and cytotoxic T lymphocytes. On delivery to the target cell cytosol, GzmA interacts with the SET complex to initiate a caspase-independent cell death pathway that is characterized by DNA nicking and the production of large DNA fragments. This process is thought to be important in the immune defense against cancers and viruses that evade caspase-mediated apoptosis. Although the members of the SET complex have not been fully characterized, most of the functional components have been identified and include the nucleosome assembly protein SET, the DNA-binding protein HMGB2, the tumor suppressor protein pp32, the 5′–3′ exonuclease TREX1, the GzmA-activated DNase NM23-H1, and APE1. While the normal function of the SET complex is unknown, based on its protein composition, it likely operates to regulate chromatin structure, genomic integrity, and gene expression. The SET complex, in response to GzmA entry into the cell, translocates rapidly from the endoplasmic reticulum, where it usually resides, to the nucleus in a process that depends on mitochondrial damage and consequent ROS accumulation (135). Once in the nucleus, the complex presumably activates expression of key early response genes and carries out relevant DNA processing activities, such as DNA nicking. Lieberman and colleagues reported that a fraction of the total APE1 protein is a part of the SET complex, and that GzmA interacts with and cleaves APE1 at residue K31 (48). In addition, they found that GzmA inactivates both the AP endonuclease and redox regulatory functions of APE1. While the latter is consistent with the N-terminal region of APE1 playing a critical role in this activity, deletion of 61 residues from the N-terminus has little effect on its repair activity (89), implying an unknown mechanism for inactivation of its endonuclease function. Although the precise molecular task of APE1 in the caspase-independent cell death pathway remains unclear, given that silencing of APE1 enhances GzmA-induced cell death, the protein appears to play an important cellular role in the process. Moreover, over-expression of a non-cleavable version of APE1 had a protective effect against GzmA-activated cell death, indicating that degradation of the protein is important in the response. Recent studies also implicate APE1 in an analogous GzmK-mediated cell death pathway (68). Finally, Yan et al. reported that KD of any of the nucleases in the SET complex (i.e., APE1, NM23-H1, or TREX1) increases auto-integration, while reducing chromosomal integration of the human immunodeficiency virus 1 (HIV-1), suggesting that this multi-protein complex can positively affect HIV-1 infection (242).
Determination of cell fate
As evident from the earlier presentation, APE1 can play a critical role in protecting cells from apoptotic cell death. Consistent with the findings of experiments using KD or genetic approaches (see “APE1 Biological Roles” section), expression of the dominant-negative form of APE1, that is, ED, results in genomic AP site accumulation, G1 arrest, and apoptosis in Chinese hamster ovary cells (137). Nevertheless, APE1 also appears to regulate the activation of cellular senescence, a phenomenon by which normal diploid cells lose their capacity to divide (typically around 50 doublings in culture) and achieve a state of irreversible growth arrest, termed the “Hayflick Phenomenon,” after its discoverer Leonard Hayflick. This outcome is thought to have evolved in certain organisms, such as mammals, to prevent the onset of cancer, with evidence suggesting that senescent cells accumulate with age and promote tissue aging. Heo et al. found that in vitro replicative or oxidative stress-induced senescence of bone marrow-derived human mesenchymal stem cells is closely related to a decrease in endogenous APE1 expression (78). Significantly, exogenous adenoviral-mediated APE1 over-expression was able to suppress the elevated superoxide levels and associated senescent phenotype. In addition, Karimi-Busheri et al. found that tumor-initiating cells isolated as mammospheres from MCF-7 breast cancer cells have approximately twofold higher APE1 expression and reduced cellular senescence in comparison with the bulk population of MCF-7 cells; they found no difference in the expression of other BER-related genes, namely DNA polymerase β (POLβ) and X-ray cross-complementing 1 (XRCC1), in mammospheres (97). Finally, Krutá et al. reported that prolonged maintenance of human embryonic stem cells in culture leads to significantly reduced APE1 expression, as well as a decreased overall BER capacity, with no changes in OGG1 or POLβ protein levels (112). Seeing that the data implicating APE1 in mediating cellular senescence is primarily corollary, experiments are needed to determine whether APE1 has a direct role in regulating this outcome, and if so, via what mechanism.
While exploring for a role of APE1 in hematopoiesis, Zou et al. discovered that KD of APE1 in mouse embryonic stem cells results in a significant decrease in the formation of hemangioblast and in primitive and definitive hematopoietic colony frequencies (248). Moreover, inhibition of the redox regulatory role of APE1 via the small molecule (E)-3-(2-(5,6-dimethoxy-3-methyl-1,4-benzoquinonyl))-2-nonyl propenoic acid (also known as, E3330; see also section “Small molecule inhibitors”) caused impaired hemangioblast development, whereas the indirect AP site repair inhibitor, methoxyamine, had no effect. These data indicate a role for APE1 in positively regulating embryonic hematopoiesis, specifically through its redox function. Interestingly, Zou et al. found that KD of APE1 via siRNA in mouse embryoid bodies induced G1 arrest, but did not activate apoptotic cell death. Thus, APE1 has specific roles in regulating cellular responses, as well as cell fate, that involve both its DNA repair and transcriptional regulatory functions, and are likely dictated by cell type, environment, and genetic background.
APE1 in Disease
Similar to other core participants in mammalian BER, such as POLβ and XRCC1, deletion of both alleles of APE1 (also known as, Apex1) in mice leads to early lethality. The initial APE1 KO mouse model employed a gene targeting strategy that deleted a 3.6 kb genomic fragment encompassing most of exon 1 (which is a non-coding exon) and all of exons 2–4, which cover the entire protein coding region (235). No homozygous KO (APE1−/−) offspring were generated (out of 464 live births) when breeding APE1+/− parents, whereas heterozygous APE1 KO mice (APE1+/−) were reported to be normal in size, fertility, and behavior for approximately 9 months of age. APE1−/− blastocysts were obtained from the uteral lumen of pregnant females at around day E3.5, but no null embryos were explanted from the uterine tissue of heterozygous matings at day E6.5, indicating that embryonic death occurred between implantation and E6.5. At E5.5, a higher-than-expected percentage of deciduae from heterozygous matings contained embryos that appeared severely necrotic, characterized by disorganized patches of pyknotic cells, suggesting a severe consequence of APE1 absence at this stage of development. Since the redox status of pre-implantation embryos is altered dramatically during development (58), it would appear that a sufficient amount of APE1 is required to protect embryos against physiological situations of oxidative stress.
In a separate study, Chen and colleagues, using a targeting strategy that deleted critical coding exons 2 and 3, yet left intact expression elements of neighboring genes, likewise did not observe homozygous KO pups; while heterozygous KO mice appeared generally healthy for at least 12 months (128). Genotyping revealed that no APE1−/− embryos survived beyond E9.5. However, unlike the original mouse model, embryonic epiblasts did not become progressively disorganized, pyknotic, and growth retarded until E7.5 to E9.5, well after implantation. The variability in survival relative to the previous study (<E6.5) could be explained by the nature of the genomic deletion, or differences in the exact mouse genetic background or the general breeding environment. Notably, explanted APE1−/− blastocysts showed increased sensitivity to γ-irradiation, which is consistent with a defect in DNA repair.
It is worth emphasizing that none of the mouse studies to date have determined which function of APE1 is required for animal viability. Toward this goal, Curran and colleagues created a C64A homozygous transgenic mouse model (C64 is equivalent to the C65 residue in the human protein thought to be critical for redox activity; see “Redox regulation” section), which was found to exhibit normal viability and life expectancy, and no overt phenotypic abnormalities (156). However, detailed analyses uncovered that these animals were normal for redox activity, raising uncertainty about the precise redox reaction mechanism and concerns about the relevance of this model for addressing the biological importance of this function. We note that in a separate attempt to identify the critical role of APE1 in organism development, no targeted APE1 null mice, which harbored a human APE1 genomic transgene that was mutated to determine the contribution of a specific function and flanked by loxP elements to permit eventual excision, were obtained, presumably due to improper ectopic expression of the complementing transgene during embryogenesis (88).
Since APE1 is essential for animal development, the relationship between APE1 deficiency and mammalian disease has been largely extrapolated from haploinsufficient mouse model studies. Consistent with the earlier work, Meira et al. found that heterozygous matings generated no viable homozygous KO progeny, but these authors observed a reduction in the number of heterozygous mutant embryos and young pups compared with that predicted by normal Mendelian inheritance (139). This trend could be countered by the addition of anti-oxidants in the diet, suggesting involvement of oxidative stress in the reduced survival. Moreover, a detailed histopathological analysis found that APE1+/− mice, while having a normal life expectancy, displayed increased cardiac abnormalities (not defined) and spontaneous tumors, including lymphomas (two cases) and an adenocarcinoma and sarcoma (a single case each). APE1+/− embryonic fibroblasts and cerebellar granule cell neurons also exhibited increased sensitivity to oxidizing agents in culture, supporting the fundamental concept that reduced APE1 function can associate with elevated exposure-dependent susceptibility.
Huamani et al. found that heterozygous KO animals were similar to wild-type littermates for approximately 9 months for body and testis weight, and for testis, spleen, and liver histology (83). Consistent with the increased risk of carcinogenesis noted earlier, these authors observed a greater than twofold increase in spontaneous mutant frequencies in mixed spermatogenic cells from 9-month-old APE1+/− mice, and an approximately twofold increase in mutation frequency in the liver and spleen of 3-month-old APE1+/− mice, relative to wild-type littermates. A follow-up analysis by Walter and colleagues revealed that the heterozygous KO animals also accumulate more mtDNA damage with age relative to the wild-type control mice (217). In a separate study, Unnikrishnan et al. reported that the liver tissue of APE1+/− mice had increased genomic DNA damage in the form of aldehydic lesions and slightly increased apoptosis after treatment with the oxidizing agent and well-known hepato-carcinogen, 2-nitropropane (211). These data, in total, support the hypothesis that impaired function APE1 alleles, should they exist in the human population, will give rise to exposure-dependent disease risk. However, as with the viability issue (see above), the studies to date have not clearly discerned whether the defects observed in the heterozygous animals are the result of a deficiency in DNA repair or a more general stress response. It is noteworthy that most studies conducted to date have concentrated mainly on haploinsufficient mice that are 9 months or younger, and it would be worthwhile to examine older animals, with a focus on tissues and organs which exhibit high oxygen consumption, such as the brain, incorporating relevant exposures where valuable.
Genetic variants
In humans, a firm linkage between an APE1 defect and the development of disease has not been documented. However, research initially pioneered by Mohrenweiser and colleagues, and since expanded by several investigative teams, indicates that there exist a large number of DNA repair gene nucleotide sequence variants within the normal and disease population (143, 144, 189). In fact, it has been extrapolated that for a representative DNA repair pathway comprising 20 genes, a typical individual will be variant for at least 5 of those genes. Thus, individuals who are wild type for all alleles in a single pathway will be rare, as will individuals with the same genotype. While in most instances it is unclear how the sequence variation affects DNA repair capacity, it has been hypothesized that the reported synonymous and non-synonymous (missense) genetic variants can lead to changes in transcript production, mRNA stability, translation efficiency, or protein structure function, which can ultimately affect DNA repair efficiency/efficacy and disease susceptibility. We focus our discussion here on APE1 genetic variation that has been shown to alter the protein coding sequence or promoter activity.
Several years ago, studies began to identify and examine the consequence(s) of missense mutations in the APE1 gene. The most common single nucleotide polymorphism (SNP) T1349G (rs1130409), which is present in the population at a roughly 45% frequency (dependent on race, ethnicity, etc.) and changes residue D148 to E148, did not alter the in vitro AP endonuclease efficiency of the encoded protein (69). D148E, as well as two other polymorphic variants (frequency of appearance >3%), Q51H (rs1048945) and I64V (rs2307486), have since been shown to exhibit normal thermodynamic stability of protein folding, abasic endonuclease, 3′–5′ exonuclease and REF-1 activities, coordination during the early steps of BER in a simple reconstituted assay, and intra-cellular distribution when expressed exogenously in HeLa cells (87). A few missense variants, some of which were reported to be uniquely associated with amyotrophic lateral sclerosis (ALS), such as L104R, E126D, and D283G, were estimated or experimentally demonstrated to exhibit 40 to 90% reductions in AP endonuclease activity (69); however, as is discussed later, the validity of these variants is in question. In addition to the identified population variants, a separate study found novel somatic APE1 mutations in 3 out of 20 endometrial cancer cases; whereas no unique mutations were observed in 43 ovarian tumor samples (161). Two of the endometrial cancer-associated mutations resulted in an amino-acid substitution (P112L or R237C), with the other nucleotide change introducing a premature stop codon at residue W188 that would give rise to, minimally, a nuclease-inactive APE1 protein fragment. The P112L variant was found to exhibit normal activities in a range of biochemical and cell-based assays, whereas the R237C variant displayed reduced in vitro 3′ to 5′ exonuclease and 3′-damage processing activities (87), which could have contributed to the genome instability and carcinogenic process by reducing overall proofreading capacity and strand-break repair. While studies are ongoing to identify a causal relationship between an APE1 defect and human disease, we provide in Table 2 a summary of the current list of APE1 missense variants reported to date, many of which are rare and have not been functionally characterized. Lastly, we note that re-sequencing of the APE1 exons in the 60 cancer cell lines within the NCI-60 cell line panel uncovered no novel non-synonymous mutations, suggesting that APE1 variants are not frequent (87).
Table 2.
APE1 Missense Variants
| Variant | rsa | Allele frequency (%) | Submitter |
|---|---|---|---|
| G8R | rs202001645 | N/A | 1000Genome/EXOME_CHIP |
| P21L | rs150934075 | Once | 1000Genome |
| S26R | rs200630518 | N/A | 1000Genome |
| K35Q | rs61757709 | Once | Multiple |
| G39E | rs34632023 | 1.3 | APPLERA_GI|hCV25628237 |
| A43V | rs146439344 | N/A | 1000Genome |
| Q51Hb | rs1048945 | 2.5a | Multiple |
| I64T | rs61730854 | 1.3 | CORNELL |
| I64Vb | rs2307486 | 0.6a | Multiple |
| A88T | rs146768400 | Once | NHLBI-ESP|ESP2500-chr14-20924842 |
| E110G | rs200702900 | N/A | 1000Genome |
| C138Y | rs150356603 | Once | NHLBI-ESP|ESP2500-chr14-20924993 |
| I146V | rs201190560 | N/A | EXOME_CHIP |
| D148Eb | rs1130409 | 42.4 | Multiple |
| D163E | rs149168435 | 0.1 | NHLBI-ESP|ESP2500-chr14-20925199 |
| R187H | rs148298598 | Once | NHLBI-ESP|ESP2500-chr14-20925270 |
| R221C | rs147110862 | Once | NHLBI-ESP|ESP2500-chr14-20925371 |
| N222H | rs201945833 | 0.1 | EXOME_CHIP/CLINSEQ_SNP |
| R237H | rs189916038 | N/A | 1000Genome |
| G241Rb | rs33956927 | 0.5 | Multiple |
| P248L | rs201100630 | N/A | EXOME_CHIP |
| P311Sb | rs1803120 | N/A | Multiple |
| T313A | rs113056798 | Once | BUSHMAN|BUSHMAN-chr14-19995486 |
| A317Vb | rs1803118 | N/A | Multiple |
Frequency varies between populations.
Functionally characterized (see text).
N/A, not available (or unknown).
Though most research thus far has been focused on nucleotide substitutions which alter the coding sequence of DNA repair genes, one study found that an A-to-C SNP (rs1760944, ∼35% frequency) in the APE1 promoter associates with a decreased risk of lung cancer and results in reduced reporter gene expression in human embryonic lung fibroblasts and the H1299 non-small cell lung carcinoma cell line (127). The authors went on to demonstrate impaired binding of a transcription factor, presumed to be OCT-1, to the variant allele relative to the wild-type A allele. However, the mechanism by which lower APE1 expression would provide disease protection is unclear. In a separate study, Lo et al. (126) also found that the variant genotypes (A/C or C/C) associated with decreased lung cancer risk, yet they observed increased luciferase reporter gene expression with the C allele in comparison to the wild-type A allele in a panel of human lung adenocarcinoma cell lines (i.e., A549, H1355, CL1-0 and H928). While the differences in the promoter response likely stem from disparate cellular backgrounds (i.e., transcription factor constellations) and/or construct designs, as with most epidemiology association analysis involving DNA repair SNPs, a causative role for the variant in disease development has not been firmly established. The issue of individual genetic variability and disease susceptibility is obviously highly complex, and will take several more years to unravel in the arena of APE1 and BER [reviewed in Simonelli et al. (195), Wallace et al. (220), and Wilson et al. (228)].
Cancer
There is accumulating evidence that indicates a possible relationship between APE1 alterations and cancer etiology. First, APE1 deficiency leads to increased mutagenesis and genotoxin/carcinogen susceptibility (see, for example, the mouse studies described earlier in this section), a cellular phenotype that is common to a DNA repair defect. Second, as discussed in the preceding section, one study reported unique somatic mutations in APE1 in endometrial tumor samples that might have contributed to disease initiation or progression. Finally, although corollary, APE1 has often been seen to be over-expressed or to exhibit an atypical sub-cellular distribution pattern (with predominantly cytoplasmic localization) in many cancer types that is not observed in normal pre-cancerous tissue. Since the topic of APE1 in cancer has been recently reviewed in detail (1), we only discuss the issue briefly here, and provide Table 3 as a glimpse of the current field.
Table 3.
Studies of APE1 in Cancer
| Cancer type | Study highlights | Reference |
|---|---|---|
| NSCLC | More cytoplasmic APE1 expression in tumor cells | (93) |
| Higher APE1 nuclear staining associated with better overall survival | ||
| Predominantly nuclear APE1 expression in tumor cells | (163) | |
| Cytoplasmic staining of APE1 associated with poor prognosis for adenocarcinoma or lymph node metastasis | ||
| Increased APE1 expression in tumor cells | (222) | |
| Nuclear/cytoplasmic APE1 expression more prominent in tumor cells | ||
| Mainly nuclear expression in normal cells | ||
| High expression associated with cisplatin resistance, and poor disease-free and overall survival | ||
| Nuclear/cytoplasmic APE1 expression more prominent in tumor cells | (232) | |
| Higher cytoplasmic APE1 expression associated with adenocarcinoma, stage II+III, and HPV E6-positive tumors | ||
| High cytoplasmic APE1 is an independent, poor prognostic factor for NSCLC | ||
| Nuclear/cytoplasmic APE1 expression more prominent in tumor cells | (96) | |
| Increased cytoplasmic expression associated with recurrence of adenocarcinoma | ||
| Ovarian | Nuclear/cytoplasmic APE1 expression more prominent in tumor cells | (157) |
| High APE1 expression associated with advanced tumor (>grade 3) and poor prognosis | ||
| Both nuclear and cytoplasmic APE1 expression patterns in tumor cells | (4) | |
| Nuclear expression associated with cancer type, optimal debulking, and overall survival | ||
| Increased cytoplasmic APE1 expression in tumor cells | (146) | |
| Mainly nuclear APE1 expression in normal cells | ||
| Cytoplasmic and nuclear/cytoplasmic expression patterns in tumor cells | (190) | |
| Cytoplasmic expression associated with low differentiation, poor overall, and disease-free survival | ||
| Breast | More cytoplasmic expression in tumor cells | (94) |
| Mainly nuclear APE1 expression in normal cells | ||
| Nuclear staining associated with low angiogenesis, negative LN status | ||
| Cytoplasmic staining associated with high angiogenesis, positive LN status | ||
| Predominantly nuclear APE1 expression in tumor cells | (164) | |
| Cytoplasmic staining associated with percentage of positive p53 staining | ||
| Nuclear/cytoplasmic staining associated with poor survival | ||
| APE1 is an independent prognostic factor for survival | ||
| Increased nuclear APE1 expression in tumor cells | (162) | |
| Increased cytoplasmic APE1K27–35Ac in TNBC | ||
| Exclusively nuclear APE1K27–35Ac in normal cells | ||
| High overall expression and acetylation of APE1 are associated with TNBC | ||
| Cervical | Increased overall APE1 expression in tumor cells | (79) |
| High expression associated with radio-resistance | ||
| Prostate | More cytoplasmic APE1 observed in PIN and malignant cancer | (100) |
| Both nuclear and cytoplasmic staining increased in PIN and malignant cancer | ||
| Predominantly nuclear APE1 expression in normal cells | ||
| Bladder | Predominantly nuclear APE1 expression in tumor cells | (176) |
| High expression associated with radio-sensitivity, improved survival | ||
| Rectal | Nuclear/cytoplasmic APE1 expression more prominent in tumor cells | (105) |
| Cytoplasmic expression associated with poor disease-free survival with pre-operative radio/chemotherapy | ||
| Gastro-oesophageal | Nuclear/cytoplasmic APE1 expression more prominent in tumor cells | (4) |
| Nuclear expression inversely associated with overall survival with neo-adjuvant chemotherapy | ||
| Cytoplasmic expression associated with cancer differentiation | ||
| Pancreatico-biliary | Nuclear/cytoplasmic APE1 expression more prominent in tumor cells | (4) |
| Subcellular distribution varied among cancer types | ||
| Absence of cytoplasmic expression associated with perineural invasion, vascular invasion and impaired differentiation | ||
| Hepatocellular | Increased cytoplasmic APE1 expression in tumor cells | (38) |
| Cytoplasmic expression associated with poor differentiation, poor survival | ||
| Osteosarcoma | Increased overall APE1 expression in tumor cells | (221) |
| High expression associated with poor survival | ||
| Multiple myeloma | High overall expression associated with recurrence and malignancy | (238) |
| Head & neck | Cytoplasmic and nuclear/cytoplasmic expression pattern more prominent in tumor cells | (111) |
| Nuclear/cytoplasmic expression pattern in normal cells | ||
| Nuclear staining associated with differentiation, positive LN status, and poor survival | ||
| Nuclear APE1 staining inversely related to nuclear accumulation of p53 and response to chemotherapy | ||
| Higher APE1 mRNA in advanced tumor (Stage III/IV) and large tumor (T3/T4) with LN metastasis | (132) | |
| Gliomas | High AP endonuclease activity associated with malignancy | (13) |
| Pediatric ependymoma | Exclusively nuclear APE1 expression in tumor cells | (15) |
| Low AP endonuclease activity associated with better disease-free survival with radiotherapy | ||
| Germ cell | Nuclear/cytoplasmic expression pattern more prominent in tumor cells | (170) |
| Increased overall APE1 expression in tumor cells | ||
| Predominantly nuclear APE1 expression in normal cells | ||
| High expression associated with resistance to bleomycin and radiation |
NSCLC, non-small cell lung cancer; HPV, human papillomavirus; LN, lymph node; TNBC, triple negative breast cancer; PIN, prostatic intraepithelial neoplasia.
APE1 expression has been primarily measured in fixed, paraffin-embedded, biopsy tissue sections from patients and matched controls using immunohistochemical techniques. Collectively, the studies have indicated that, in general, high expression or a cytoplasmic or cytoplasmic/nuclear distribution is associated with DNA-damaging agent resistance, tumor aggressiveness, or poor prognosis (summarized in Table 3). What causes the increase in protein levels or leads to the distinct sub-cellular localization pattern is largely unknown, but presumably involves changes in the intra-cellular environment and PTMs to the protein (see also “Intra-cellular trafficking” section). Since cancer cells are typically vigorously growing, they have an elevated demand for energy production and, thus, generate a high level of ROS as a result of enhanced mitochondrial oxidative phosphorylation. Moreover, in solid tumors, due to changes in angiogenesis, there are cycles of hypoxia/re-oxygenation that contribute to a state of oxidative stress. Thus, one hypothesis put forward is that the cytoplasmic localization of APE1 is a feature of the response to increased ROS, where the APE1 protein re-distributes to the cytoplasm to maintain mtDNA integrity and function (see “Intra-cellular targeting and mitochondrial function” section) and/or to suppress Rac1-mediated oxidative stress and injury (6, 158). Elucidating the mechanisms and reasons for, as well as the consequences of, the altered APE1 expression patterns represents a critical step toward devising better treatment or management strategies for specific cancer patients.
While APE1 staining is a potential tool in cancer diagnosis, prognosis, and treatment prediction, it should be emphasized that not all disease samples exhibit distinct APE1 expression patterns, stressing the importance of developing individualized treatment plans. Moreover, in an effort to reduce experimental variability from lab to lab, as well as any artifacts, standardized methods for sample preparation and handling, and for immunodetection (e.g., immunohistochemistry) that employ the same antibodies and quantitative procedures to assess APE1 expression, are needed. The implementation of a high-volume tissue micro-array platform that spans multiple cancer types and controls represents a paradigm for future translational evaluations [see, for example, (4)], and hopefully will help provide a more definitive understanding of the molecular roles of APE1 in the carcinogenic process. Finally, it is noteworthy that elevated serum auto-antibodies against APE1 have recently been associated with the auto-immune disease, systemic lupus erythematosus, particularly those patients with psychiatric manifestations, and non-small cell lung carcinoma (33, 98). Future investigations will need to determine the contribution, if any, of the APE1 auto-antibodies to disease etiology and, more generally, whether APE1 auto-antibodies have utility in basic serological examinations.
Neuropathology
The central nervous system (CNS), consisting of the brain and spinal cord, is composed of some of the most metabolically active cells in the human body. As such, careful maintenance of ROS homeostasis and associated oxidative damage in cells of the CNS is vital for the well being of the organism. APE1 is, in general, highly expressed in the CNS, with some variability among the different cell types and regions of the human brain, including the existence of distinct nuclear and cytoplasmic distribution patterns [reviewed in Wilson and McNeill (229)]. Early investigations demonstrated that APE1 depletion by siRNA in primary rat hippocampal or sensory neuronal cell cultures results in reduced cell viability, as well as increased apoptosis and DNA damage, after hydrogen peroxide treatment (216). Jiang et al. found that inhibition of AP site repair using the inhibitor, methoxyamine, and not inhibition of the redox activity of APE1, induced hyper-sensitivity of differentiated (post-mitotic) SH-SY5Y neural cells to a range of oxidizing agents (92). APE1 protein levels were also shown to be correlated with the degree of neuro-protection against the deleterious effects of both transient focal and global cerebral ischemia in rodent models (104, 197), observations which are consistent with the emerging evidence that BER plays an important role in the defense against stroke [reviewed in Sykora et al. (201)]. In total, the experiments support the hypothesis that APE1, apparently in a DNA repair-dependent mechanism, and presumably other proteins which process oxidative DNA lesions, contributes to the maintenance of brain cell function, particularly when challenged with conditions of oxidative stress.
One of the first studies to suggest a relationship between an APE1 defect and human neuropathology found significantly lower APE1 protein and AP endonuclease activity in tissue extracts prepared from the frontal cortex of 11 patients suffering from sporadic ALS in comparison to six age-matched controls (110). ALS is a motor neuron disease that has been associated with oxidative stress, as pathogenic mutations have been identified in the radical scavenger superoxide dismutase gene (SOD1) in familial cases of the disorder. Consistent with APE1 contributing to the development of ALS, mutations, including the missense variants L104R, E126D, D148E, D283G, and G306A, were found in eight out of 11 patients with sporadic or familial ALS; while no mutations were detected in five healthy control subjects (154). In a separate, larger re-sequencing effort involving 153 ALS patients and 58 controls, Hayward et al. (74) observed three APE1 nucleotide substitutions common among patients and controls (including the D148E polymorphism), as well as a silent base change (at residue Y315) unique to three sporadic ALS patients and a 4-bp deletion in a single sporadic ALS patient. The deletion mutation introduces a stop codon at amino acid 145, producing a truncated protein product that, if stable, would minimally lack nuclease activity; whether this variant was inherited or sporadic, and whether it contributed to the disease, was unfortunately unable to be assessed. The fact that far fewer mutations were observed in this study was the first indication that APE1 plays a much smaller role in the etiology of ALS than insinuated by the apparent “jack-pot” of APE1 sequence variants reported in the earlier work (154). Consistent with this notion, Tomkins et al. (206) found no novel APE1 mutations in genomic DNA isolated from the cerebral cortex of 84 ALS patients, other than a 3′-UTR 4-bp deletion in a single case of ALS. They also observed that the SNPs related to Q51H and D148E are present at roughly equal frequencies among the ALS patients and controls. Coppede et al. (31) similarly observed no significant difference in the D148E allele frequency between 134 sporadic ALS patients and 129 matched controls, seemingly consistent with the normal biochemical properties of the variant enzyme (see “Genetic variants” section). In a separate study, however, APE1 protein was found to be increased or re-localized in CNS tissues of ALS patients, particularly within regions that contain motor neurons (186). It seems likely that these alterations are a consequence of the oxidative stress and increased DNA damage associated with ALS, and are not directly connected to disease initiation. While further studies to explore a possible role for APE1 in the different pathological stages of ALS might be worthwhile, it would seem to be most relevant to evaluate whether rare genetic mutations in APE1 (or BER genes for that matter), such as the deletion mutants described earlier, contribute directly to disease manifestation in a sub-set of patients.
A similar, generally inconclusive picture has emerged regarding the potential contribution of APE1 to Alzheimer disease (AD), and, to a lesser extent, Parkinson disease (PD). AD is a degenerative dementia that involves cognitive and functional impairments due to the loss of neurons in the cerebral cortex, whereas PD is characterized by motor deficits and cell death in the substantia nigra. While the exact cause of either disorder is unclear, studies have suggested the involvement of oxidative stress in the disease pathology, prompting an interest in specific DNA repair systems, such as BER, as risk factors. Several studies have, indeed, found that oxidative DNA damage is higher in AD brain tissue [reviewed in Santos et al. (179)], and Weissman et al. (224) discovered that reduced BER activity, namely uracil DNA glycosylase and POLβ functions (although APE1 function was unchanged in AD samples), correlates with AD progression in a small set of patients. In addition, Tan et al. reported that APE1 immunostaining was relatively high in the hippocampus and surrounding temporal cortex of some AD sufferers relative to matched controls, a phenomenon that again may simply reflect a response to disease-induced oxidative stress (202). Further analysis revealed that APE1 was also present in a subset of senile plaques and plaque-like structures, suggesting a role for the protein in β-amyloid deposition, a marker for AD. Generally consistent with this earlier work, Davydov et al. (34) reported that APE1 protein levels tended to be higher in mid-frontal cortex nuclear extracts of AD patients, and Marcon et al. (134) observed more intense nuclear immunostaining of APE1 in the pathogenic regions of both sporadic and familial AD brains. Molecular epidemiology association studies, however, have indicated that the polymorphic variant of APE1 (D148E) as well as common SNPs in OGG1 and XRCC1 are not independent risk factors of AD (159). With regard to PD, one recent study involving 60 case subjects and 108 normal controls found that the D148E variant of APE1 had a positive association as a risk factor for the disease (59), although it was acknowledged by the authors that more extensive follow-up experiments are needed to validate this observation. Finally, Huang et al. (84), in addition to confirming the importance of APE1 in neuronal cell viability, possibly in a mechanism involving Cdk5 phosphorylation of residue T232 (in the mouse protein, T233 in the human protein), which inactivates the enzyme's AP endonuclease activity (see Table 1), detected an increase in the T233-phosphorylated form of APE1 in post-mortem brain samples from PD and AD patients, uncovering a possible molecular connection between APE1 inactivation and neurodegenerative disease.
Despite the fact that strong evidence exists for a role of APE1 in protecting neuronal cells from conditions of oxidative stress, there is no firm evidence at present that indicates an involvement of the protein in human neurological disease. While global changes in protein expression or re-localization may be observed, these descriptive alterations likely reflect a response to the disease environment, presumably the state of oxidative stress, as also seen in cancer patient samples (see previous section). Moreover, it is important to point out that the large genome-wide association studies conducted on ALS, AD, and PD patients, to our knowledge, have not identified genes related to DNA repair pathways as major risk factors in any of these neurological disorders. Nevertheless, it remains plausible that rare APE1 mutations, either inherited (which allow for organism survival) or sporadic, could directly contribute to disease initiation or susceptibility. In addition, it is possible that reduced BER capacity, particularly in a setting of oxidative stress that is typical of neurological disease, could exacerbate the pathologic process.
It is worth emphasizing that a set of inherited disorders with neurological deficits have been genetically linked to single-strand break DNA repair defects [reviewed in Reynolds and Stewart (169)]. In particular, spinocerebellar ataxia with axonal neuropathy-1 (SCAN1) arises from mutations in TDP1; ataxia oculomotor apraxia-1 (AOA1) results from mutations in APTX (Aprataxin); and microcephaly, early-onset, intractable seizures, and developmental delay (MCSZ) are caused by pathogenic mutations in PNKP. Since APE1 maintains a significant strand break 3′-repair diesterase activity, it will be of interest to examine whether APE1 deficiency can adversely affect end-points related to brain development and function, such as by using conditional KO mouse models, as has been done for other BER genes, including XRCC1 (116). Clearly, future studies are needed to more directly evaluate the contribution of APE1 and BER to various pathologies involving oxidative stress [reviewed in Hegde et al. (76)].
APE1 as a Therapeutic Target
Since APE1 has roles in both disease suppression and therapeutic agent resistance, it is presumed that if APE1 activities can be strategically regulated, the protein would be a favorable target in both preventative and curative treatment paradigms. Although the concept of DNA repair activators is of growing interest as a prospective mechanism for improving “healthspan” (i.e., counter-acting the cumulative effects seen during aging and in age-related disease), efforts to this point have focused mainly on strategies to reduce or block DNA repair function. In particular, the current emphasis has been on the design of small-molecule DNA repair inhibitors, pursuing the hypothesis that inactivation of a DNA damage response will increase the efficacy of a relevant anti-cancer agent, many of which induce cell death by introducing lethal DNA lesions. Since APE1 plays a central and critical role in the BER process, the protein has received significant attention as an attractive target. As will be expounded in the next sections, APE1 could be targeted in (i) combinatorial treatment schemes to enhance the cytotoxicity of a germane clinical DNA-damaging agent or (ii) mono-therapies, in which inhibition of APE1 function would be synthetically lethal with a genetic defect present uniquely in the disease (e.g., cancer) tissue.
Protein depletion
As introduced earlier (see “APE1 Biological Roles” section), anti-sense-mediated depletion of APE1 increased cellular sensitivity to the alkylating agent MMS and the oxidizing agent hydrogen peroxide, providing an initial picture of the role of APE1 in resistance to DNA-damaging agent challenges (155, 218). On the advent of siRNA technologies, several groups began to look more exhaustively at the effects of APE1 depletion on sensitivity to a range of clinical DNA-interactive compounds. Some of the earliest work, published by Kelley and colleagues, revealed that APE1 provides resistance against not only MMS and hydrogen peroxide, but also the anti-cancer agents thiotepa, etoposide, and ionizing radiation (221). Through the efforts of many labs since, there is a general consensus emerging that indicates a role for APE1 in dictating cellular responsiveness to clinically relevant alkylators, most notably temozolomide (7, 14, 137, 142, 193), and a subset of chain-terminating nucleoside analogs, most prominently troxacitabine (114, 137, 180). Such a role is consistent with the biochemical activities of APE1, as alkylating agents, particularly mono-functional versions, induce a high number of abasic sites through both direct and indirect mechanisms, and certain anti-metabolites generate exonuclease 3′-end substrates. In addition, expression of the dominant-negative ED protein (introduced in “APE1 Biological Roles” section) in Chinese hamster ovary cells increases the toxicity of the anti-cancer agents, 5-fluorouracil and 5-fluorodeoxyuridine, with the latter causing greater cell death, presumably because its effects are targeted explicitly to DNA, and not to RNA as well (138). This observation is generally consistent with the recent findings that PARP1 and other BER enzymes, including APE1, play a role in resistance to 5-fluorodeoxyuridine more so than 5-fluorouracil, likely due to the higher levels of BER substrate base lesions, that is, 5-fluorouracil and uracil, introduced into genomic DNA by the former compound (60).
Hyper-sensitivity of APE1 KD cells has been reported for other clinical DNA-damaging agents, including ionizing radiation (32, 150, 237), bleomycin (56, 170, 218), gemcitabine (239), cisplatin (222, 246), etoposide (185, 221), and photodynamic therapy (236, 243). While in some cases it is not obvious what role APE1 would play in repairing the direct DNA damage (e.g., for the crosslinking agent cisplatin), some level of oxidative stress is involved in most genotoxin exposures. Thus, APE1 may be required for oxidative DNA lesion processing or for a more general stress response. Moreover, Chattopadhyay et al. (25) have reported that acetylated APE1, through an interaction with the transcription factor YB-1, can activate expression of the multi-drug resistance gene MDR1. Increased levels of the MDR1 P-glycoprotein membrane transporter are associated with enhanced drug efflux and resistance in cancer cells, and could explain the sensitizing effect of APE1 depletion to compounds such as cisplatin. Hence, APE1 may have multiple roles in providing DNA-damaging agent protection, engaging its DNA repair and/or transcriptional regulatory functions. We note that for many of the anti-cancer agents mentioned earlier, an APE1-dependent influence on sensitivity has not been observed in all studies, suggesting presumably cell type- or experimental assay-specific responses/outcomes.
In addition to the pre-clinical KD studies performed in culture, the effects of APE1 down-regulation on cancer cell growth or sensitivity to therapeutic agents have been investigated in xenograft or tumor-bearing mouse models as well. Fishel et al. reported that human SKOV-3 ovarian cancer cells treated with APE1 siRNA ex vivo and then implanted into female nude mice exhibit a dramatic reduction in tumor volume and cell proliferation during the period of APE1 depletion (51). Moreover, Xiang et al. found that LOVO colon cancer cells injected subcutaneously into nude mice and subsequently treated with APE1-specific siRNA adenoviral particles display reduced tumor growth, particularly when challenged with X-ray irradiation, relative to the controls (237). Using this established approach, the Wang group has since shown that in vivo APE1 depletion sensitizes A549 non-small cell lung cancer xenografts to hematoporphyrin derivative-mediated photodynamic therapy (243) and human hepatocellular carcinoma cell lines to radio-therapy (32). The compilation of the studies strongly indicates that transient suppression of APE1 activity can have a beneficial effect when treating cancer with certain chemo- or radio-therapies. It is important to emphasize, however, that since KD strategies result in general suppression of target gene expression, it is uncertain in this scenario which activity of the multi-functional APE1 protein determines sensitivity to the different DNA-damaging agents.
Small-molecule inhibitors
Since DNA repair inhibitors have potential utility in combinatorial treatment paradigms with clinically relevant DNA-interactive agents and in mono-therapies under the premise of synthetic lethality (see more in next section), several investigators have focused on the development of small-molecule DNA repair protein inhibitors. In addition, function-specific inhibitors represent potentially powerful tools to address the contribution of the targeted activity to specific cellular end points, including DNA-damaging agent sensitivity. Since the topic of APE1 inhibitors has been extensively reviewed recently (5, 230), it is not our intent to revisit this issue in depth. Instead, we provide Table 4 as a summary of the efforts to design and apply APE1 inhibitors, and touch on the current state of the mission to develop a clinically useful, small-molecule inactivator of the AP endonuclease or redox regulatory function of APE1.
Table 4.
Studies of Direct APE1 Nuclease Inhibitors
| Feature compound(s) | Brief summary | Reference |
|---|---|---|
| CRT0044876 (7-Nitroindole-2carboxylic acid) | First reported direct nuclease inhibitor, identified in a high-throughput screen of 5000 small molecules | (131) |
| Inhibits AP endonuclease (IC50 ∼3 μM), as well as 3′-phosphodiesterase (IC50 ∼5 μM) and 3′-phosphatase, activities | ||
| Inhibits AP endonuclease activity in cell extracts | ||
| Sensitizes cells to MMS and TMZ | ||
| Several compounds (most potent contain two carboxylate functional groups in a 3D arrangement with hydrophobic groups) | Identified in an in silico screen of 365,000 compounds using 3D pharmacophore models | (244) |
| IC50 in the low μM range | ||
| Not evaluated in cell-based experiments | ||
| NCI-13755 NCI-13793 |
Arylstibonic acid compounds identified from a screen of the NCI 2000 Diversity Set | (181) |
| IC50 4–17 nM | ||
| No effect on cellular sensitivity to DNA-damaging agents | ||
| 6-Hydroxy-DL-DOPA Myricetin Reactive Blue 2 |
Identified from a screen of the Sigma LOPAC | (194) |
| IC50 0.11–0.32 μM | ||
| Inhibit AP endonuclease activity in cell extracts | ||
| Sensitize cells to MMS | ||
| Multiple known biological targets | ||
| AR03 ([2,4,9-trimethylbenzo[b][1,8]-naphthyridin-5-amine) | Identified from a screen of 60,000 small molecules | (7) |
| IC50 ∼2 μM (no effect on redox function) | ||
| Inhibits AP endonuclease activity in cell extracts | ||
| Sensitizes glioblastoma cells to MMS and TMZ | ||
| Lucanthone Hycanthone |
IC50 5 μM and 80 nM, respectively (no effect on redox function) | (129, 149) |
| Sensitize cancer cells to MMS and TMZ | ||
| Currently in clinical trials as an adjuvant in brain tumor radiotherapy | ||
| Compounds containing 2-methyl-4-amino-6,7-dioxolo-quinoline structure | Identified from in silico screen | (196) |
| IC50 in the μM range (no effect on redox function) | ||
| Inhibits AP endonuclease activity in nuclear lysates | ||
| Sensitize cancer cells to the methylating agent, MeLex | ||
| 3-Carbamoylbenzoic acid and derivatives | SAR study around 3-carbamoylbenzoic acid scaffold | (2) |
| IC50 in the low μM range | ||
| Sensitize cancer cells to MMS, 5-fluorouracil and 5-fluorodeoxyuridine | ||
| Several compounds of different structures | Identified from the NIH MLSMR and additional public collections (352,498 total compounds) | (42) |
| IC50<30 μM | ||
| Sensitize HeLa cells to MMS | ||
| Compound 3 (N-(3-(benzo[d]thiazol-2-yl)-6-isopropyl-4,5,6,7-tetrahydrothieno[2,3-c]pyridin-2-yl)acetamide) and its analogue, compound 52 | SAR study around compound 3 | (167) |
| IC50 2 and 3.3 μM, respectively | ||
| Inhibit AP endonuclease activity in cell extracts | ||
| Sensitize HeLa cells to MMS and TMZ | ||
| Initial ADME profile: compound 3 crosses the blood-brain barrier more efficiently; compound 52 has a better general cytotoxicity profile, higher exposure levels, and a more favorable t(1/2) in the plasma |
TMZ, temozolomide; SAR, structure-activity relationship.
A number of laboratories have taken on the effort to identify and design potent inhibitors against the repair nuclease function(s) of APE1. These studies have generally relied on a high-throughput, fluorescence-based screening assay, in which inhibition of APE1 AP site incision activity prevents release of a 5′-fluorophore-containing DNA fragment from its strand-opposite 3′-attached quench, and, thus, an associated increase in quantifiable signal [originally described in (131)]. As summarized in Table 4, quite a few reports have appeared detailing the identification and validation of initial hits that inactivate the AP endonuclease activity of APE1 in vitro. However, most of the reported inhibitors have affinities (typically μM) that are not compatible with a suitable pharmaceutical agent, are not amendable to extensive chemical modification, or have multiple biological targets. The “low affinity” attribute may stem from the fact that small molecules are unable to replicate the “high affinity” binding contacts seen in the APE1/AP-DNA complex, which spans roughly 7–8 bp in length. Indeed, in an extensive structure-function activity relationship study, the introduced modifications only slightly increased the effectiveness of the analogs against APE1, either (i) supporting the notion that small molecules do not make sufficient contact with the protein for high affinity binding or (ii) indicating that the initial chemotype was a poor starting substrate. It is also noteworthy that none of the nuclease-targeted compounds identified to date have been demonstrated to have utility in pre-clinical animal cancer models. Nevertheless, in pre-clinical cell culture systems, APE1 repair inhibitors have been shown to enhance the cytotoxicity of temozolomide (7, 131, 142, 167), confirming that the nuclease function(s) of the protein plays a critical role in repairing alkylative DNA damage, presumably abasic sites. Future studies will need to examine the utility of the most promising compounds in both combinatorial and synthetic lethal treatment paradigms, and may need to employ covalently linked, small molecules that bind simultaneously multiple functional sites of APE1.
The design of APE1 redox inhibitors has been more limited, presumably due to the lack of a high-throughput screening assay to identify molecules that inactivate the redox function of the protein. Nevertheless, using a bead-based approach and nuclear cell extracts, Shimizu et al. (191) identified APE1 as a major binding protein for the quinone derivative, E3330 (introduced in “APE1 Biological Roles” section). E3330 had previously been shown to suppress NF-κB transactivational activity, and, in doing so, to promote an anti-inflammatory state, exhibiting potential utility in the treatment of various forms of hepatitis [(80) and references therein]. The APE1-E3330 interaction was validated using multiple biochemical and biological approaches, and was determined to have an apparent binding constant of 1.6 nM by surface plasmon resonance (191). Since this discovery, the ability of E3330 to selectively inactivate the redox activity of APE1 and suppress the activity of several transcription factor targets has been confirmed in multiple studies [reviewed in Kelley et al. (101)]. Moreover, E3330, and, in some instances, its synthesized chemical analogs, has been demonstrated to inhibit proliferation of human pancreatic and ovarian cancer cell lines in culture; angiogenesis or migration of cancer cells in in vitro models; and tumor growth of pancreatic cancer xenografts in mice (52, 102, 151, 249). Thus, despite the fact that there is controversy regarding the precise functionality of E3330 (133, 245), the findings with this compound and its analogs underscore the importance of further analyses around the molecular mechanisms and clinical value of targeting APE1 redox function in therapeutic paradigms. Indeed, an exciting development in recent years has been the observation that APE1 redox activity, through modulation of transcription factors such as HIF-1α (46, 85, 115), plays a significant role in regulating angiogenesis, and, as such, could represent a promising target in the control of tumor proliferation and metastasis. We close this section by mentioning that dietary agents, such as soy isoflavones (found in plants) and resveratrol (found in red wine), have been reported to have complex effects on the expression or the repair/redox activities of APE1, implying the potential for natural (intended or un-intended) regulation of APE1 cellular functions [reviewed in Kelley et al. (101) and Raffoul et al. (166)].
Synthetic lethality
One of the most significant advances in recent years regarding DNA repair as a potential target in cancer therapeutics was the observation that PARP inhibitors can lead to reduced survival of cells which lack BRCA1 or BRCA2 (17, 50). PARP1 is a DNA damage sensor that facilitates efficient repair of DNA single-strand breaks. BRCA1 and BRCA2 are tumor suppressor gene products associated with hereditary breast/ovarian cancer that participate in resolving DNA double-strand breaks. Through a mechanism that engages protein and DNA interactions reviewed elsewhere (174), BRCA1 and BRCA2 facilitate RAD51-mediated homologous recombination, a molecular process which enables the faithful exchange of genetic information from an intact sister chromatid to resolve one-ended, DNA double-strand breaks formed on replication fork collapse. The fact that PARP inhibitors promote lethality of cells deficient in homologous recombination suggested that the pathways engaging PARP1 and the BRCA proteins are synthetically lethal (i.e., simultaneous inactivation of both leads to cell death). Specifically, it was thought that inhibition of PARP1-mediated single-strand break repair would lead to increased levels of DNA double-strand breaks during copying of the genome, causing lethality in cells unable to efficiently resolve these damage-induced, replication intermediates. While the mechanism driving cell death has been modified in recent years, now involving potential trapping of PARP1 on DNA by the inhibitor (77), the enthusiasm for employing synthetic lethality in the treatment of repair-defective cancers has remained. Indeed, although issues and optimization phases remain, PARP inhibitors have shown promising results in select clinical trials [reviewed in Sandhu et al. (178)].
With regard to APE1, Sultana et al. observed synthetic lethality of BRCA1, BRCA2, or ATM double-strand break repair-deficient Chinese hamster lung or human cancer cell lines on treatment with an APE1 DNA repair inhibitor (200). In particular, exposure of these mutant cell lines to a direct or indirect endonuclease inhibitor resulted in reduced colony formation, elevated chromosomal AP sites and DNA single- and double-strand breaks, and G2/M cell cycle arrest. In the reverse direction, Chinese hamster ovary cells expressing the dominant-negative ED protein or human MDA-MB-231 breast cancer cells depleted for APE1 exhibited reduced survival in the presence of inhibitors (NU7441, KU55933, or Wortmannin) against the double-strand break repair signaling kinases, namely ATM and DNA-PKcs. Since the redox inhibitor E3330 did not induce synthetic lethality, it would appear that a defect in AP site repair induces replication fork collapse and the accumulation of DNA strand break intermediates that drive cell death. A more exhaustive search of the cellular pathways that are synthetically lethal with the disparate functions of APE1 would seem to be a worthwhile endeavor. Moreover, assuming effective and sufficiently validated APE1 inhibitors can be developed, future studies will need to assess the value of these small molecules in formal clinical trials.
Summary, Conclusions, and Future Perspectives
Abasic sites are a common form of DNA damage that are generated by spontaneous or damage-induced hydrolysis of the N-glycosylic bond, or via the action of DNA glycosylases that remove modified or substrate bases from DNA as a part of BER. In addition, single-strand breaks are created frequently by free radical attack of DNA or as intermediates of a DNA repair response. APE1 possesses a powerful AP endonuclease activity (around two orders of magnitude greater than its other nuclease activities), which is generally agreed to be a major biological function of the enzyme. Based on current work, its 3′-repair diesterase activity is also likely an important cellular function, minimally contributing to clean-up of DNA termini generated by β-elimination reactions at AP sites, and its 3′ to 5′ exonuclease activity appears to at least aid in the removal of certain chain-terminating nucleoside analogs in cells. Moreover, a great deal of biochemical and cell-based evidence supports the importance of the redox regulatory function of APE1 in moderating a range of cellular responses. The biological significance of the other functions, namely RNA cleavage and NIR, in our opinion, still requires further substantiation. Studies using genetically manipulated cell lines may help define which of the many activities of APE1 are important to the various biological end points discussed here. However, the difficulty in creating clean, separation-of-function mutants in APE1, particularly those that effectively distinguish the various nuclease activities of the enzyme, is likely to be a constraint.
Figure 4 provides an overview of the many proposed functions of APE1. Besides having prominent roles in the nucleus, APE1 also has important roles in mitochondria, at minimum in protecting the mitochondrial genome, and presumably in the endoplasmic reticulum as a part of the SET complex. As an extension of its formal biochemical activities, APE1 has general roles in a range of biological processes, including proliferation and growth, apoptosis, senescence, stress responses, angiogenesis, metabolism, and differentiation, to name a few. How the sub-compartmentalization and these various functions of APE1 are manipulated is just beginning to be understood, and presumably involve protein–protein interactions and PTMs. The unstructured, newly acquired N-terminal region of the protein also appears to play a key role in directing the participation of APE1 to specific molecular pathways. Teasing out the complexities that regulate the intra-cellular localization and the activities of this multi-functional protein will prove to be a real challenge, and should keep the APE1 aficionados out of trouble for many years to come.
FIG. 4.
The APE1 galaxy. As highlighted within, APE1 has multiple biochemical activities that impart multiple biological roles. These functions affect various cellular processes that can ultimately impact disease susceptibility, therapeutic response, and prognosis. In addition, complex mechanisms that involve protein interactions and PTMs regulate not only the activities of APE1, but also its intra-cellular compartmentalization. See text for details. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
It seems striking that at present there is no established link between an APE1 defect and human disease. This presumably stems from the fact that a severe APE1 deficit would be incompatible with life. Nevertheless, there is a collection of cell-based and animal model studies indicating that APE1 deficiency imparts reduced cell survival, increased sensitivity to DNA-damaging agent exposures, elevated mutagenesis, and an enhanced risk of disease development, to name a few. While studies continue to progress to explore the hypothesis that reduced BER capacity associates with increased disease susceptibility, the ongoing whole exome and whole genome re-sequencing efforts may reveal rare mutations in BER genes that contribute to disease manifestation. Those pathologies that are most likely to be caused or exacerbated by a BER defect are ones which involve oxidative stress and its associated DNA damage.
We close with an apology and a thank you. We have attempted to provide a comprehensive summary of the APE1 literature, and undoubtedly have either missed relevant publications or misinterpreted some of the reported data. While we aimed at delivering the content in an unbiased manner, there will always be some partiality in the presentation. For this we are sorry, but we hope that the review spurs on new ideas and identifies weaknesses in the APE1 field, and, of course, we thank the many investigators, both past and present, who have contributed to our understanding of this critical multi-functional protein.
Abbreviations Used
- AD
Alzheimer disease
- ALS
amyotrophic lateral sclerosis
- APE1
apurinic/apyrimidinic endonuclease 1
- BER
base excision repair
- CNS
central nervous system
- CSR
class switch recombination
- ETC
electron transport chain
- KD
knock-down
- KO
knock-out
- MMS
methylmethane sulfonate
- mtBER
mitochondrial BER
- mtDNA
mitochondrial DNA
- MTS
mitochondrial targeting sequence
- nCaREs
negative Ca2+ response elements
- NES
nuclear export signal
- NIR
nucleotide incision repair
- NLS
nuclear localization signal
- PARP1
poly(ADP)ribose polymerase 1
- PD
Parkinson disease
- PTM
post-translational modification
- ROS
reactive oxygen species
- SHM
somatic hypermutation
- SNP
single nucleotide polymorphism
Acknowledgments
This effort was supported by the Intramural Research Program at NIH, National Institute on Aging. The authors thank Peter Sykora and Jennifer Illuzzi for their critical reading of the article.
References
- 1.Abbotts R. and Madhusudan S. Human AP endonuclease 1 (APE1): from mechanistic insights to druggable target in cancer. Cancer Treat Rev 36: 425–435, 2010 [DOI] [PubMed] [Google Scholar]
- 2.Aiello F, Shabaik Y, Esqueda A, Sanchez TW, Grande F, Garofalo A, and Neamati N. Design and synthesis of 3-carbamoylbenzoic acid derivatives as inhibitors of human apurinic/apyrimidinic endonuclease 1 (APE1). Chem Med Chem 7: 1825–1839, 2012 [DOI] [PubMed] [Google Scholar]
- 3.Akiyama K, Seki S, Oshida T, and Yoshida MC. Structure, promoter analysis and chromosomal assignment of the human APEX gene. Biochim Biophys Acta 1219: 15–25, 1994 [DOI] [PubMed] [Google Scholar]
- 4.Al-Attar A, Gossage L, Fareed KR, Shehata M, Mohammed M, Zaitoun AM, Soomro I, Lobo DN, Abbotts R, Chan S, and Madhusudan S. Human apurinic/apyrimidinic endonuclease (APE1) is a prognostic factor in ovarian, gastro-oesophageal and pancreatico-biliary cancers. Br J Cancer 102: 704–709, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Al-Safi RI, Odde S, Shabaik Y, and Neamati N. Small-molecule inhibitors of APE1 DNA repair function: an overview. Curr Mol Pharmacol 5: 14–35, 2012 [PubMed] [Google Scholar]
- 6.Angkeow P, Deshpande SS, Qi B, Liu YX, Park YC, Jeon BH, Ozaki M, and Irani K. Redox factor-1: an extra-nuclear role in the regulation of endothelial oxidative stress and apoptosis. Cell Death Differ 9: 717–725, 2002 [DOI] [PubMed] [Google Scholar]
- 7.Bapat A, Glass LS, Luo M, Fishel ML, Long EC, Georgiadis MM, and Kelley MR. Novel small-molecule inhibitor of apurinic/apyrimidinic endonuclease 1 blocks proliferation and reduces viability of glioblastoma cells. J Pharmacol Exp Ther 334: 988–998, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barnes T, Kim WC, Mantha AK, Kim SE, Izumi T, Mitra S, and Lee CH. Identification of Apurinic/apyrimidinic endonuclease 1 (APE1) as the endoribonuclease that cleaves c-myc mRNA. Nucleic Acids Res 37: 3946–3958, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barzilay G, Walker LJ, Robson CN, and Hickson ID. Site-directed mutagenesis of the human DNA repair enzyme HAP1: identification of residues important for AP endonuclease and RNase H activity. Nucleic Acids Res 23: 1544–1550, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berquist BR, McNeill DR, and Wilson DM., 3rd.Characterization of abasic endonuclease activity of human Ape1 on alternative substrates, as well as effects of ATP and sequence context on AP site incision. J Mol Biol 379: 17–27, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhakat KK, Izumi T, Yang SH, Hazra TK, and Mitra S. Role of acetylated human AP-endonuclease (APE1/Ref-1) in regulation of the parathyroid hormone gene. EMBO J 22: 6299–6309, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhattacharyya A, Chattopadhyay R, Burnette BR, Cross JV, Mitra S, Ernst PB, Bhakat KK, and Crowe SE. Acetylation of apurinic/apyrimidinic endonuclease-1 regulates Helicobacter pylori-mediated gastric epithelial cell apoptosis. Gastroenterology 136: 2258–2269, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bobola MS, Blank A, Berger MS, Stevens BA, and Silber JR. Apurinic/apyrimidinic endonuclease activity is elevated in human adult gliomas. Clin Cancer Res 7: 3510–3518, 2001 [PubMed] [Google Scholar]
- 14.Bobola MS, Finn LS, Ellenbogen RG, Geyer JR, Berger MS, Braga JM, Meade EH, Gross ME, and Silber JR. Apurinic/apyrimidinic endonuclease activity is associated with response to radiation and chemotherapy in medulloblastoma and primitive neuroectodermal tumors. Clin Cancer Res 11: 7405–7414, 2005 [DOI] [PubMed] [Google Scholar]
- 15.Bobola MS, Jankowski PP, Gross ME, Schwartz J, Finn LS, Blank A, Ellenbogen RG, and Silber JR. Apurinic/apyrimidinic endonuclease is inversely associated with response to radiotherapy in pediatric ependymoma. Int J Cancer 129: 2370–2379, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brooks SC, Adhikary S, Rubinson EH, and Eichman BF. Recent advances in the structural mechanisms of DNA glycosylases. Biochim Biophys Acta 1834: 247–271, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, and Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434: 913–917, 2005 [DOI] [PubMed] [Google Scholar]
- 18.Burkovics P, Hajdu I, Szukacsov V, Unk I, and Haracska L. Role of PCNA-dependent stimulation of 3′-phosphodiesterase and 3′-5′ exonuclease activities of human Ape2 in repair of oxidative DNA damage. Nucleic Acids Res 37: 4247–4255, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burkovics P, Szukacsov V, Unk I, and Haracska L. Human Ape2 protein has a 3′-5′ exonuclease activity that acts preferentially on mismatched base pairs. Nucleic Acids Res 34: 2508–2515, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Busso CS, Iwakuma T, and Izumi T. Ubiquitination of mammalian AP endonuclease (APE1) regulated by the p53-MDM2 signaling pathway. Oncogene 28: 1616–1625, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Busso CS, Lake MW, and Izumi T. Posttranslational modification of mammalian AP endonuclease (APE1). Cell Mol Life Sci 67: 3609–3620, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Busso CS, Wedgeworth CM, and Izumi T. Ubiquitination of human AP-endonuclease 1 (APE1) enhanced by T233E substitution and by CDK5. Nucleic Acids Res 39: 8017–8028, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caldecott KW. Single-strand break repair and genetic disease. Nat Rev Genet 9: 619–631, 2008 [DOI] [PubMed] [Google Scholar]
- 24.Carey DC. and Strauss PR. Human apurinic/apyrimidinic endonuclease is processive. Biochemistry 38: 16553–16560, 1999 [DOI] [PubMed] [Google Scholar]
- 25.Chattopadhyay R, Das S, Maiti AK, Boldogh I, Xie J, Hazra TK, Kohno K, Mitra S, and Bhakat KK. Regulatory role of human AP-endonuclease (APE1/Ref-1) in YB-1-mediated activation of the multidrug resistance gene MDR1. Mol Cell Biol 28: 7066–7080, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chattopadhyay R, Wiederhold L, Szczesny B, Boldogh I, Hazra TK, Izumi T, and Mitra S. Identification and characterization of mitochondrial abasic (AP)-endonuclease in mammalian cells. Nucleic Acids Res 34: 2067–2076, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen DS, Herman T, and Demple B. Two distinct human DNA diesterases that hydrolyze 3′-blocking deoxyribose fragments from oxidized DNA. Nucleic Acids Res 19: 5907–5914, 1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chou KM. and Cheng YC. The exonuclease activity of human apurinic/apyrimidinic endonuclease (APE1). Biochemical properties and inhibition by the natural dinucleotide Gp4G. J Biol Chem 278: 18289–18296, 2003 [DOI] [PubMed] [Google Scholar]
- 29.Chou KM, Kukhanova M, and Cheng YC. A novel action of human apurinic/apyrimidinic endonuclease: excision of L-configuration deoxyribonucleoside analogs from the 3′ termini of DNA. J Biol Chem 275: 31009–31015, 2000 [DOI] [PubMed] [Google Scholar]
- 30.Chung U, Igarashi T, Nishishita T, Iwanari H, Iwamatsu A, Suwa A, Mimori T, Hata K, Ebisu S, Ogata E, Fujita T, and Okazaki T. The interaction between Ku antigen and REF1 protein mediates negative gene regulation by extracellular calcium. J Biol Chem 271: 8593–8598, 1996 [DOI] [PubMed] [Google Scholar]
- 31.Coppede F, Lo Gerfo A, Carlesi C, Piazza S, Mancuso M, Pasquali L, Murri L, Migliore L, and Siciliano G. Lack of association between the APEX1 Asp148Glu polymorphism and sporadic amyotrophic lateral sclerosis. Neurobiol Aging 31: 353–355, 2010 [DOI] [PubMed] [Google Scholar]
- 32.Cun Y, Dai N, Xiong C, Li M, Sui J, Qian C, Li Z, and Wang D. Silencing of APE1 enhances sensitivity of human hepatocellular carcinoma cells to radiotherapy in vitro and in a xenograft model. PLoS One 8: e55313, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Dai N, Cao XJ, Li MX, Qing Y, Liao L, Lu XF, Zhang SH, Li Z, Yang YX, and Wang D. Serum APE1 autoantibodies: a novel potential tumor marker and predictor of chemotherapeutic efficacy in non-small cell lung cancer. PLoS One 8: e58001, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davydov V, Hansen LA, and Shackelford DA. Is DNA repair compromised in Alzheimer's disease? Neurobiol Aging 24: 953–968, 2003 [DOI] [PubMed] [Google Scholar]
- 35.de Souza-Pinto NC, Wilson DM, 3rd, Stevnsner TV, and Bohr VA. Mitochondrial DNA, base excision repair and neurodegeneration. DNA Repair (Amst) 7: 1098–1109, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Demple B. and Harrison L. Repair of oxidative damage to DNA: enzymology and biology. Annu Rev Biochem 63: 915–948, 1994 [DOI] [PubMed] [Google Scholar]
- 37.Demple B, Herman T, and Chen DS. Cloning and expression of APE, the cDNA encoding the major human apurinic endonuclease: definition of a family of DNA repair enzymes. Proc Natl Acad Sci U S A 88: 11450–11454, 1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Di Maso V, Avellini C, Croce LS, Rosso N, Quadrifoglio F, Cesaratto L, Codarin E, Bedogni G, Beltrami CA, Tell G, and Tiribelli C. Subcellular localization of APE1/Ref-1 in human hepatocellular carcinoma: possible prognostic significance. Mol Med 13: 89–96, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dianov GL. and Hubscher U. Mammalian base excision repair: the forgotten archangel. Nucleic Acids Res 41: 3483–3490, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dlakic M. Functionally unrelated signalling proteins contain a fold similar to Mg2+-dependent endonucleases. Trends Biochem Sci 25: 272–273, 2000 [DOI] [PubMed] [Google Scholar]
- 41.Doetsch PW. and Cunningham RP. The enzymology of apurinic/apyrimidinic endonucleases. Mutat Res 236: 173–201, 1990 [DOI] [PubMed] [Google Scholar]
- 42.Dorjsuren D, Kim D, Vyjayanti VN, Maloney DJ, Jadhav A, Wilson DM, 3rd, and Simeonov A. Diverse small molecule inhibitors of human apurinic/apyrimidinic endonuclease APE1 identified from a screen of a large public collection. PLoS One 7: e47974, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Duguid JR, Eble JN, Wilson TM, and Kelley MR. Differential cellular and subcellular expression of the human multifunctional apurinic/apyrimidinic endonuclease (APE/ref-1) DNA repair enzyme. Cancer Res 55: 6097–6102, 1995 [PubMed] [Google Scholar]
- 44.Dyrkheeva NS, Khodyreva SN, Sukhanova MV, Safronov IV, Dezhurov SV, and Lavrik OI. 3′-5′ exonuclease activity of human apurinic/apyrimidinic endonuclease 1 towards DNAs containing dNMP and their modified analogs at the 3 end of single strand DNA break. Biochemistry (Mosc) 71: 200–210, 2006 [DOI] [PubMed] [Google Scholar]
- 45.Dyrkheeva NS, Lomzov AA, Pyshnyi DV, Khodyreva SN, and Lavrik OI. Efficiency of exonucleolytic action of apurinic/apyrimidinic endonuclease 1 towards matched and mismatched dNMP at the 3′ terminus of different oligomeric DNA structures correlates with thermal stability of DNA duplexes. Biochim Biophys Acta 1764: 699–706, 2006 [DOI] [PubMed] [Google Scholar]
- 46.Ema M, Hirota K, Mimura J, Abe H, Yodoi J, Sogawa K, Poellinger L, and Fujii-Kuriyama Y. Molecular mechanisms of transcription activation by HLF and HIF1alpha in response to hypoxia: their stabilization and redox signal-induced interaction with CBP/p300. EMBO J 18: 1905–1914, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Erzberger JP. and Wilson DM., 3rd.The role of Mg2+ and specific amino acid residues in the catalytic reaction of the major human abasic endonuclease: new insights from EDTA-resistant incision of acyclic abasic site analogs and site-directed mutagenesis. J Mol Biol 290: 447–457, 1999 [DOI] [PubMed] [Google Scholar]
- 48.Fan Z, Beresford PJ, Zhang D, Xu Z, Novina CD, Yoshida A, Pommier Y, and Lieberman J. Cleaving the oxidative repair protein Ape1 enhances cell death mediated by granzyme A. Nat Immunol 4: 145–153, 2003 [DOI] [PubMed] [Google Scholar]
- 49.Fantini D, Vascotto C, Marasco D, D'Ambrosio C, Romanello M, Vitagliano L, Pedone C, Poletto M, Cesaratto L, Quadrifoglio F, Scaloni A, Radicella JP, and Tell G. Critical lysine residues within the overlooked N-terminal domain of human APE1 regulate its biological functions. Nucleic Acids Res 38: 8239–8256, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, Martin NM, Jackson SP, Smith GC, and Ashworth A. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434: 917–921, 2005 [DOI] [PubMed] [Google Scholar]
- 51.Fishel ML, He Y, Reed AM, Chin-Sinex H, Hutchins GD, Mendonca MS, and Kelley MR. Knockdown of the DNA repair and redox signaling protein Ape1/Ref-1 blocks ovarian cancer cell and tumor growth. DNA Repair (Amst) 7: 177–186, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fishel ML, Jiang Y, Rajeshkumar NV, Scandura G, Sinn AL, He Y, Shen C, Jones DR, Pollok KE, Ivan M, Maitra A, and Kelley MR. Impact of APE1/Ref-1 redox inhibition on pancreatic tumor growth. Mol Cancer Ther 10: 1698–1708, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fritz G. and Kaina B. Phosphorylation of the DNA repair protein APE/REF-1 by CKII affects redox regulation of AP-1. Oncogene 18: 1033–1040, 1999 [DOI] [PubMed] [Google Scholar]
- 54.Fuchs S, Philippe J, Corvol P, and Pinet F. Implication of Ref-1 in the repression of renin gene transcription by intracellular calcium. J Hypertens 21: 327–335, 2003 [DOI] [PubMed] [Google Scholar]
- 55.Fung H. and Demple B. A vital role for Ape1/Ref1 protein in repairing spontaneous DNA damage in human cells. Mol Cell 17: 463–470, 2005 [DOI] [PubMed] [Google Scholar]
- 56.Fung H. and Demple B. Distinct roles of Ape1 protein in the repair of DNA damage induced by ionizing radiation or bleomycin. J Biol Chem 286: 4968–4977, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fung H, Kow YW, Van Houten B, Taatjes DJ, Hatahet Z, Janssen YM, Vacek P, Faux SP, and Mossman BT. Asbestos increases mammalian AP-endonuclease gene expression, protein levels, and enzyme activity in mesothelial cells. Cancer Res 58: 189–194, 1998 [PubMed] [Google Scholar]
- 58.Gardiner CS. and Reed DJ. Status of glutathione during oxidant-induced oxidative stress in the preimplantation mouse embryo. Biol Reprod 51: 1307–1314, 1994 [DOI] [PubMed] [Google Scholar]
- 59.Gencer M, Dasdemir S, Cakmakoglu B, Cetinkaya Y, Varlibas F, Tireli H, Kucukali CI, Ozkok E, and Aydin M. DNA repair genes in Parkinson's disease. Genet Test Mol Biomarkers 16: 504–507, 2012 [DOI] [PubMed] [Google Scholar]
- 60.Geng L, Huehls AM, Wagner JM, Huntoon CJ, and Karnitz LM. Checkpoint signaling, base excision repair, and PARP promote survival of colon cancer cells treated with 5-fluorodeoxyuridine but not 5-fluorouracil. PLoS One 6: e28862, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Georgiadis MM, Luo M, Gaur RK, Delaplane S, Li X, and Kelley MR. Evolution of the redox function in mammalian apurinic/apyrimidinic endonuclease. Mutat Res 643: 54–63, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gillespie MN, Pastukh VM, and Ruchko MV. Controlled DNA “damage” and repair in hypoxic signaling. Respir Physiol Neurobiol 174: 244–251, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gorman MA, Morera S, Rothwell DG, de La Fortelle E, Mol CD, Tainer JA, Hickson ID, and Freemont PS. The crystal structure of the human DNA repair endonuclease HAP1 suggests the recognition of extra-helical deoxyribose at DNA abasic sites. EMBO J 16: 6548–6558, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Grafstrom RH, Shaper NL, and Grossman L. Human placental apurinic/apyrimidinic endonuclease. Mechanism of action. J Biol Chem 257: 13459–13464, 1982 [PubMed] [Google Scholar]
- 65.Gros L, Ishchenko AA, Ide H, Elder RH, and Saparbaev MK. The major human AP endonuclease (Ape1) is involved in the nucleotide incision repair pathway. Nucleic Acids Res 32: 73–81, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guikema JE, Linehan EK, Tsuchimoto D, Nakabeppu Y, Strauss PR, Stavnezer J, and Schrader CE. APE1- and APE2-dependent DNA breaks in immunoglobulin class switch recombination. J Exp Med 204: 3017–3026, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guliaev AB, Hang B, and Singer B. Structural insights by molecular dynamics simulations into specificity of the major human AP endonuclease toward the benzene-derived DNA adduct, pBQ-C. Nucleic Acids Res 32: 2844–2852, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guo Y, Chen J, Zhao T, and Fan Z. Granzyme K degrades the redox/DNA repair enzyme Ape1 to trigger oxidative stress of target cells leading to cytotoxicity. Mol Immunol 45: 2225–2235, 2008 [DOI] [PubMed] [Google Scholar]
- 69.Hadi MZ, Coleman MA, Fidelis K, Mohrenweiser HW, and Wilson DM., 3rd.Functional characterization of Ape1 variants identified in the human population. Nucleic Acids Res 28: 3871–3879, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hadi MZ, Ginalski K, Nguyen LH, and Wilson DM., 3rd.Determinants in nuclease specificity of Ape1 and Ape2, human homologues of Escherichia coli exonuclease III. J Mol Biol 316: 853–866, 2002 [DOI] [PubMed] [Google Scholar]
- 71.Hadi MZ. and Wilson DM., 3rd.Second human protein with homology to the Escherichia coli abasic endonuclease exonuclease III. Environ Mol Mutagen 36: 312–324, 2000 [PubMed] [Google Scholar]
- 72.Harrison L, Ascione AG, Wilson DM, 3rd, and Demple B. Characterization of the promoter region of the human apurinic endonuclease gene (APE). J Biol Chem 270: 5556–5564, 1995 [DOI] [PubMed] [Google Scholar]
- 73.Harrison L, Ascione G, Menninger JC, Ward DC, and Demple B. Human apurinic endonuclease gene (APE): structure and genomic mapping (chromosome 14q11.2–12). Hum Mol Genet 1: 677–680, 1992 [DOI] [PubMed] [Google Scholar]
- 74.Hayward C, Colville S, Swingler RJ, and Brock DJ. Molecular genetic analysis of the APEX nuclease gene in amyotrophic lateral sclerosis. Neurology 52: 1899–1901, 1999 [DOI] [PubMed] [Google Scholar]
- 75.Hegde ML, Izumi T, and Mitra S. Oxidized base damage and single-strand break repair in mammalian genomes: role of disordered regions and posttranslational modifications in early enzymes. Prog Mol Biol Transl Sci 110: 123–153, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hegde ML, Mantha AK, Hazra TK, Bhakat KK, Mitra S, and Szczesny B. Oxidative genome damage and its repair: implications in aging and neurodegenerative diseases. Mech Ageing Dev 133: 157–168, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Helleday T. The underlying mechanism for the PARP and BRCA synthetic lethality: clearing up the misunderstandings. Mol Oncol 5: 387–393, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Heo JY, Jing K, Song KS, Seo KS, Park JH, Kim JS, Jung YJ, Hur GM, Jo DY, Kweon GR, Yoon WH, Lim K, Hwang BD, Jeon BH, and Park JI. Downregulation of APE1/Ref-1 is involved in the senescence of mesenchymal stem cells. Stem Cells 27: 1455–1462, 2009 [DOI] [PubMed] [Google Scholar]
- 79.Herring CJ, West CM, Wilks DP, Davidson SE, Hunter RD, Berry P, Forster G, MacKinnon J, Rafferty JA, Elder RH, Hendry JH, and Margison GP. Levels of the DNA repair enzyme human apurinic/apyrimidinic endonuclease (APE1, APEX, Ref-1) are associated with the intrinsic radiosensitivity of cervical cancers. Br J Cancer 78: 1128–1133, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hiramoto M, Shimizu N, Sugimoto K, Tang J, Kawakami Y, Ito M, Aizawa S, Tanaka H, Makino I, and Handa H. Nuclear targeted suppression of NF-kappa B activity by the novel quinone derivative E3330. J Immunol 160: 810–819, 1998 [PubMed] [Google Scholar]
- 81.Hirota K, Matsui M, Iwata S, Nishiyama A, Mori K, and Yodoi J. AP-1 transcriptional activity is regulated by a direct association between thioredoxin and Ref-1. Proc Natl Acad Sci U S A 94: 3633–3638, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hsieh MM, Hegde V, Kelley MR, and Deutsch WA. Activation of APE/Ref-1 redox activity is mediated by reactive oxygen species and PKC phosphorylation. Nucleic Acids Res 29: 3116–3122, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Huamani J, McMahan CA, Herbert DC, Reddick R, McCarrey JR, MacInnes MI, Chen DJ, and Walter CA. Spontaneous mutagenesis is enhanced in Apex heterozygous mice. Mol Cell Biol 24: 8145–8153, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Huang E, Qu D, Zhang Y, Venderova K, Haque ME, Rousseaux MW, Slack RS, Woulfe JM, and Park DS. The role of Cdk5-mediated apurinic/apyrimidinic endonuclease 1 phosphorylation in neuronal death. Nat Cell Biol 12: 563–571, 2010 [DOI] [PubMed] [Google Scholar]
- 85.Huang LE, Arany Z, Livingston DM, and Bunn HF. Activation of hypoxia-inducible transcription factor depends primarily upon redox-sensitive stabilization of its alpha subunit. J Biol Chem 271: 32253–32259, 1996 [DOI] [PubMed] [Google Scholar]
- 86.Ide Y, Tsuchimoto D, Tominaga Y, Nakashima M, Watanabe T, Sakumi K, Ohno M, and Nakabeppu Y. Growth retardation and dyslymphopoiesis accompanied by G2/M arrest in APEX2-null mice. Blood 104: 4097–4103, 2004 [DOI] [PubMed] [Google Scholar]
- 87.Illuzzi JL, Harris NA, Manvilla BA, Kim D, Li M, Drohat AC, and Wilson DM., III Functional assessment of population and tumor-associated APE1 protein variants. PLoS One 8: e65922, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Izumi T, Brown DB, Naidu CV, Bhakat KK, Macinnes MA, Saito H, Chen DJ, and Mitra S. Two essential but distinct functions of the mammalian abasic endonuclease. Proc Natl Acad Sci U S A 102: 5739–5743, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Izumi T, Hazra TK, Boldogh I, Tomkinson AE, Park MS, Ikeda S, and Mitra S. Requirement for human AP endonuclease 1 for repair of 3′-blocking damage at DNA single-strand breaks induced by reactive oxygen species. Carcinogenesis 21: 1329–1334, 2000 [PubMed] [Google Scholar]
- 90.Izumi T, Henner WD, and Mitra S. Negative regulation of the major human AP-endonuclease, a multifunctional protein. Biochemistry 35: 14679–14683, 1996 [DOI] [PubMed] [Google Scholar]
- 91.Jackson EB, Theriot CA, Chattopadhyay R, Mitra S, and Izumi T. Analysis of nuclear transport signals in the human apurinic/apyrimidinic endonuclease (APE1/Ref1). Nucleic Acids Res 33: 3303–3312, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jiang Y, Guo C, Fishel ML, Wang ZY, Vasko MR, and Kelley MR. Role of APE1 in differentiated neuroblastoma SH-SY5Y cells in response to oxidative stress: use of APE1 small molecule inhibitors to delineate APE1 functions. DNA Repair (Amst) 8: 1273–1282, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kakolyris S, Giatromanolaki A, Koukourakis M, Kaklamanis L, Kanavaros P, Hickson ID, Barzilay G, Georgoulias V, Gatter KC, and Harris AL. Nuclear localization of human AP endonuclease 1 (HAP1/Ref-1) associates with prognosis in early operable non-small cell lung cancer (NSCLC). J Pathol 189: 351–357, 1999 [DOI] [PubMed] [Google Scholar]
- 94.Kakolyris S, Kaklamanis L, Engels K, Fox SB, Taylor M, Hickson ID, Gatter KC, and Harris AL. Human AP endonuclease 1 (HAP1) protein expression in breast cancer correlates with lymph node status and angiogenesis. Br J Cancer 77: 1169–1173, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kane CM. and Linn S. Purification and characterization of an apurinic/apyrimidinic endonuclease from HeLa cells. J Biol Chem 256: 3405–3414, 1981 [PubMed] [Google Scholar]
- 96.Kang MW, Kang SK, Choi S, Lee CS, Jeon BH, and Lim SP. Upregulation of APE/ref-1 in recurrence stage I, non small cell lung cancer. Asian Cardiovasc Thorac Ann 20: 36–41, 2012 [DOI] [PubMed] [Google Scholar]
- 97.Karimi-Busheri F, Rasouli-Nia A, Mackey JR, and Weinfeld M. Senescence evasion by MCF-7 human breast tumor-initiating cells. Breast Cancer Res 12: R31, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Katsumata Y, Kawaguchi Y, Baba S, Hattori S, Tahara K, Ito K, Iwasaki T, Yamaguchi N, Oyama M, Kozuka-Hata H, Hattori H, Nagata K, Yamanaka H, and Hara M. Identification of three new autoantibodies associated with systemic lupus erythematosus using two proteomic approaches. Mol Cell Proteomics 10: M110 005330, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Keim C, Kazadi D, Rothschild G, and Basu U. Regulation of AID, the B-cell genome mutator. Genes Dev 27: 1–17, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kelley MR, Cheng L, Foster R, Tritt R, Jiang J, Broshears J, and Koch M. Elevated and altered expression of the multifunctional DNA base excision repair and redox enzyme Ape1/ref-1 in prostate cancer. Clin Cancer Res 7: 824–830, 2001 [PubMed] [Google Scholar]
- 101.Kelley MR, Georgiadis MM, and Fishel ML. APE1/Ref-1 role in redox signaling: translational applications of targeting the redox function of the DNA repair/redox protein APE1/Ref-1. Curr Mol Pharmacol 5: 36–53, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kelley MR, Luo M, Reed A, Su D, Delaplane S, Borch RF, Nyland RL, 2nd, Gross ML, and Georgiadis MM. Functional analysis of novel analogues of E3330 that block the redox signaling activity of the multifunctional AP endonuclease/redox signaling enzyme APE1/Ref-1. Antioxid Redox Signal 14: 1387–1401, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kelley MR. and Parsons SH. Redox regulation of the DNA repair function of the human AP endonuclease Ape1/ref-1. Antioxid Redox Signal 3: 671–683, 2001 [DOI] [PubMed] [Google Scholar]
- 104.Kim HW, Cho KJ, Park SC, Kim HJ, and Kim GW. The adenoviral vector-mediated increase in apurinic/apyrimidinic endonuclease inhibits the induction of neuronal cell death after transient ischemic stroke in mice. Brain Res 1274: 1–10, 2009 [DOI] [PubMed] [Google Scholar]
- 105.Kim JS, Kim JM, Liang ZL, Jang JY, Kim S, Huh GJ, Kim KH, and Cho MJ. Prognostic significance of human apurinic/apyrimidinic endonuclease (APE/Ref-1) expression in rectal cancer treated with preoperative radiochemotherapy. Int J Radiat Oncol Biol Phys 82: 130–137, 2012 [DOI] [PubMed] [Google Scholar]
- 106.Kim WC, Berquist BR, Chohan M, Uy C, Wilson DM, 3rd, and Lee CH. Characterization of the endoribonuclease active site of human apurinic/apyrimidinic endonuclease 1. J Mol Biol 411: 960–971, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kim WC, King D, and Lee CH. RNA-cleaving properties of human apurinic/apyrimidinic endonuclease 1 (APE1). Int J Biochem Mol Biol 1: 12–25, 2010 [PMC free article] [PubMed] [Google Scholar]
- 108.Kim YJ, Kim D, Illuzzi JL, Delaplane S, Su D, Bernier M, Gross ML, Georgiadis MM, and Wilson DM., 3rd.S-glutathionylation of cysteine 99 in the APE1 protein impairs abasic endonuclease activity. J Mol Biol 414: 313–326, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kim YJ. and Wilson DM., 3rd.Overview of base excision repair biochemistry. Curr Mol Pharmacol 5: 3–13, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kisby GE, Milne J, and Sweatt C. Evidence of reduced DNA repair in amyotrophic lateral sclerosis brain tissue. Neuroreport 8: 1337–1340, 1997 [DOI] [PubMed] [Google Scholar]
- 111.Koukourakis MI, Giatromanolaki A, Kakolyris S, Sivridis E, Georgoulias V, Funtzilas G, Hickson ID, Gatter KC, and Harris AL. Nuclear expression of human apurinic/apyrimidinic endonuclease (HAP1/Ref-1) in head-and-neck cancer is associated with resistance to chemoradiotherapy and poor outcome. Int J Radiat Oncol Biol Phys 50: 27–36, 2001 [DOI] [PubMed] [Google Scholar]
- 112.Krutá M, Balek L, Hejnova R, Dobsakova Z, Eiselleova L, Matulka K, Barta T, Fojtik P, Fajkus J, Hampl A, Dvorak P, and Rotrekl V. Decrease in abundance of apurinic/apyrimidinic endonuclease causes failure of base excision repair in culture-adapted human embryonic stem cells. Stem Cells 31: 693–702, 2012 [DOI] [PubMed] [Google Scholar]
- 113.Kuninger DT, Izumi T, Papaconstantinou J, and Mitra S. Human AP-endonuclease 1 and hnRNP-L interact with a nCaRE-like repressor element in the AP-endonuclease 1 promoter. Nucleic Acids Res 30: 823–829, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lam W, Park SY, Leung CH, and Cheng YC. Apurinic/apyrimidinic endonuclease-1 protein level is associated with the cytotoxicity of L-configuration deoxycytidine analogs (troxacitabine and beta-L-2′,3′-dideoxy-2′,3′-didehydro-5-fluorocytidine) but not D-configuration deoxycytidine analogs (gemcitabine and beta-D-arabinofuranosylcytosine). Mol Pharmacol 69: 1607–1614, 2006 [DOI] [PubMed] [Google Scholar]
- 115.Lando D, Pongratz I, Poellinger L, and Whitelaw ML. A redox mechanism controls differential DNA binding activities of hypoxia-inducible factor (HIF) 1alpha and the HIF-like factor. J Biol Chem 275: 4618–4627, 2000 [DOI] [PubMed] [Google Scholar]
- 116.Lee Y, Katyal S, Li Y, El-Khamisy SF, Russell HR, Caldecott KW, and McKinnon PJ. The genesis of cerebellar interneurons and the prevention of neural DNA damage require XRCC1. Nat Neurosci 12: 973–980, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Li M, Vascotto C, Xu S, Dai N, Qing Y, Zhong Z, Tell G, and Wang D. Human AP endonuclease/redox factor APE1/ref-1 modulates mitochondrial function after oxidative stress by regulating the transcriptional activity of NRF1. Free Radic Biol Med 53: 237–248, 2012 [DOI] [PubMed] [Google Scholar]
- 118.Li M, Zhong Z, Zhu J, Xiang D, Dai N, Cao X, Qing Y, Yang Z, Xie J, Li Z, Baugh L, Wang G, and Wang D. Identification and characterization of mitochondrial targeting sequence of human apurinic/apyrimidinic endonuclease 1. J Biol Chem 285: 14871–14881, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Li MX, Wang D, Zhong ZY, Xiang DB, Li ZP, Xie JY, Yang ZZ, Jin F, and Qing Y. Targeting truncated APE1 in mitochondria enhances cell survival after oxidative stress. Free Radic Biol Med 45: 592–601, 2008 [DOI] [PubMed] [Google Scholar]
- 120.Lieberman J. Granzyme A activates another way to die. Immunol Rev 235: 93–104, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lindahl T. DNA glycosylases, endonucleases for apurinic/apyrimidinic sites, and base excision-repair. Prog Nucleic Acid Res Mol Biol 22: 135–192, 1979 [DOI] [PubMed] [Google Scholar]
- 122.Lindahl T. Instability and decay of the primary structure of DNA. Nature 362: 709–715, 1993 [DOI] [PubMed] [Google Scholar]
- 123.Lipton AS, Heck RW, Primak S, McNeill DR, Wilson DM, 3rd, and Ellis PD. Characterization of Mg2+ binding to the DNA repair protein apurinic/apyrimidic endonuclease 1 via solid-state 25Mg NMR spectroscopy. J Am Chem Soc 130: 9332–9341, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lirussi L, Antoniali G, Vascotto C, D'Ambrosio C, Poletto M, Romanello M, Marasco D, Leone M, Quadrifoglio F, Bhakat KK, Scaloni A, and Tell G. Nucleolar accumulation of APE1 depends on charged lysine residues that undergo acetylation upon genotoxic stress and modulate its BER activity in cells. Mol Biol Cell 23: 4079–4096, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Liu P. and Demple B. DNA repair in mammalian mitochondria: much more than we thought? Environ Mol Mutagen 51: 417–426, 2010 [DOI] [PubMed] [Google Scholar]
- 126.Lo YL, Jou YS, Hsiao CF, Chang GC, Tsai YH, Su WC, Chen KY, Chen YM, Huang MS, Hu CY, Chen CJ, and Hsiung CA. A polymorphism in the APE1 gene promoter is associated with lung cancer risk. Cancer Epidemiol Biomarkers Prev 18: 223–229, 2009 [DOI] [PubMed] [Google Scholar]
- 127.Lu J, Zhang S, Chen D, Wang H, Wu W, Wang X, Lei Y, Wang J, Qian J, Fan W, Hu Z, Jin L, Shen H, Huang W, Wei Q, and Lu D. Functional characterization of a promoter polymorphism in APE1/Ref-1 that contributes to reduced lung cancer susceptibility. FASEB J 23: 3459–3469, 2009 [DOI] [PubMed] [Google Scholar]
- 128.Ludwig DL, MacInnes MA, Takiguchi Y, Purtymun PE, Henrie M, Flannery M, Meneses J, Pedersen RA, and Chen DJ. A murine AP-endonuclease gene-targeted deficiency with post-implantation embryonic progression and ionizing radiation sensitivity. Mutat Res 409: 17–29, 1998 [DOI] [PubMed] [Google Scholar]
- 129.Luo M. and Kelley MR. Inhibition of the human apurinic/apyrimidinic endonuclease (APE1) repair activity and sensitization of breast cancer cells to DNA alkylating agents with lucanthone. Anticancer Res 24: 2127–2134, 2004 [PubMed] [Google Scholar]
- 130.Luo M, Zhang J, He H, Su D, Chen Q, Gross ML, Kelley MR, and Georgiadis MM. Characterization of the redox activity and disulfide bond formation in apurinic/apyrimidinic endonuclease. Biochemistry 51: 695–705, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Madhusudan S, Smart F, Shrimpton P, Parsons JL, Gardiner L, Houlbrook S, Talbot DC, Hammonds T, Freemont PA, Sternberg MJ, Dianov GL, and Hickson ID. Isolation of a small molecule inhibitor of DNA base excision repair. Nucleic Acids Res 33: 4711–4724, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mahjabeen I, Baig RM, Sabir M, and Kayani MA. Genetic and expressional variations of APEX1 are associated with increased risk of head and neck cancer. Mutagenesis 28: 213–218, 2013 [DOI] [PubMed] [Google Scholar]
- 133.Manvilla BA, Wauchope O, Seley-Radtke KL, and Drohat AC. NMR studies reveal an unexpected binding site for a redox inhibitor of AP endonuclease 1. Biochemistry 50: 10540–10549, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Marcon G, Tell G, Perrone L, Garbelli R, Quadrifoglio F, Tagliavini F, and Giaccone G. APE1/Ref-1 in Alzheimer's disease: an immunohistochemical study. Neurosci Lett 466: 124–127, 2009 [DOI] [PubMed] [Google Scholar]
- 135.Martinvalet D, Zhu P, and Lieberman J. Granzyme A induces caspase-independent mitochondrial damage, a required first step for apoptosis. Immunity 22: 355–370, 2005 [DOI] [PubMed] [Google Scholar]
- 136.Masani S, Han L, and Yu K. Apurinic/apyrimidinic endonuclease 1 is the essential nuclease during immunoglobulin class switch recombination. Mol Cell Biol, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.McNeill DR, Lam W, DeWeese TL, Cheng YC, and Wilson DM., 3rd.Impairment of APE1 function enhances cellular sensitivity to clinically relevant alkylators and antimetabolites. Mol Cancer Res 7: 897–906, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.McNeill DR. and Wilson DM., 3rd.A dominant-negative form of the major human abasic endonuclease enhances cellular sensitivity to laboratory and clinical DNA-damaging agents. Mol Cancer Res 5: 61–70, 2007 [DOI] [PubMed] [Google Scholar]
- 139.Meira LB, Devaraj S, Kisby GE, Burns DK, Daniel RL, Hammer RE, Grundy S, Jialal I, and Friedberg EC. Heterozygosity for the mouse Apex gene results in phenotypes associated with oxidative stress. Cancer Res 61: 5552–5557, 2001 [PubMed] [Google Scholar]
- 140.Meisenberg C, Tait PS, Dianova II, Wright K, Edelmann MJ, Ternette N, Tasaki T, Kessler BM, Parsons JL, Kwon YT, and Dianov GL. Ubiquitin ligase UBR3 regulates cellular levels of the essential DNA repair protein APE1 and is required for genome stability. Nucleic Acids Res 40: 701–711, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Miquel J, de Juan E, and Sevila I. Oxygen-induced mitochondrial damage and aging. EXS 62: 47–57, 1992 [DOI] [PubMed] [Google Scholar]
- 142.Mohammed MZ, Vyjayanti VN, Laughton CA, Dekker LV, Fischer PM, Wilson DM, 3rd, Abbotts R, Shah S, Patel PM, Hickson ID, and Madhusudan S. Development and evaluation of human AP endonuclease inhibitors in melanoma and glioma cell lines. Br J Cancer 104: 653–663, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Mohrenweiser HW, Wilson DM, 3rd, and Jones IM. Challenges and complexities in estimating both the functional impact and the disease risk associated with the extensive genetic variation in human DNA repair genes. Mutat Res 526: 93–125, 2003 [DOI] [PubMed] [Google Scholar]
- 144.Mohrenweiser HW, Xi T, Vazquez-Matias J, and Jones IM. Identification of 127 amino acid substitution variants in screening 37 DNA repair genes in humans. Cancer Epidemiol Biomarkers Prev 11: 1054–1064, 2002 [PubMed] [Google Scholar]
- 145.Mol CD, Izumi T, Mitra S, and Tainer JA. DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination [corrected]. Nature 403: 451–456, 2000 [DOI] [PubMed] [Google Scholar]
- 146.Moore DH, Michael H, Tritt R, Parsons SH, and Kelley MR. Alterations in the expression of the DNA repair/redox enzyme APE/ref-1 in epithelial ovarian cancers. Clin Cancer Res 6: 602–609, 2000 [PubMed] [Google Scholar]
- 147.Mosbaugh DW. and Linn S. Further characterization of human fibroblast apurinic/apyrimidinic DNA endonucleases. The definition of two mechanistic classes of enzyme. J Biol Chem 255: 11743–11752, 1980 [PubMed] [Google Scholar]
- 148.Mundle ST, Delaney JC, Essigmann JM, and Strauss PR. Enzymatic mechanism of human apurinic/apyrimidinic endonuclease against a THF AP site model substrate. Biochemistry 48: 19–26, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Naidu MD, Agarwal R, Pena LA, Cunha L, Mezei M, Shen M, Wilson DM, 3rd, Liu Y, Sanchez Z, Chaudhary P, Wilson SH, and Waring MJ. Lucanthone and its derivative hycanthone inhibit apurinic endonuclease-1 (APE1) by direct protein binding. PLoS One 6: e23679, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Naidu MD, Mason JM, Pica RV, Fung H, and Pena LA. Radiation resistance in glioma cells determined by DNA damage repair activity of Ape1/Ref-1. J Radiat Res 51: 393–404, 2010 [DOI] [PubMed] [Google Scholar]
- 151.Nyland RL, Luo M, Kelley MR, and Borch RF. Design and synthesis of novel quinone inhibitors targeted to the redox function of apurinic/apyrimidinic endonuclease 1/redox enhancing factor-1 (Ape1/ref-1). J Med Chem 53: 1200–1210, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Oezguen N, Mantha AK, Izumi T, Schein CH, Mitra S, and Braun W. MD simulation and experimental evidence for Mg(2)+ binding at the B site in human AP endonuclease 1. Bioinformation 7: 184–198, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Okazaki T, Chung U, Nishishita T, Ebisu S, Usuda S, Mishiro S, Xanthoudakis S, Igarashi T, and Ogata E. A redox factor protein, ref1, is involved in negative gene regulation by extracellular calcium. J Biol Chem 269: 27855–27862, 1994 [PubMed] [Google Scholar]
- 154.Olkowski ZL. Mutant AP endonuclease in patients with amyotrophic lateral sclerosis. Neuroreport 9: 239–242, 1998 [DOI] [PubMed] [Google Scholar]
- 155.Ono Y, Furuta T, Ohmoto T, Akiyama K, and Seki S. Stable expression in rat glioma cells of sense and antisense nucleic acids to a human multifunctional DNA repair enzyme, APEX nuclease. Mutat Res 315: 55–63, 1994 [DOI] [PubMed] [Google Scholar]
- 156.Ordway JM, Eberhart D, and Curran T. Cysteine 64 of Ref-1 is not essential for redox regulation of AP-1 DNA binding. Mol Cell Biol 23: 4257–4266, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ouellet V, Le Page C, Guyot MC, Lussier C, Tonin PN, Provencher DM, and Mes-Masson AM. SET complex in serous epithelial ovarian cancer. Int J Cancer 119: 2119–2126, 2006 [DOI] [PubMed] [Google Scholar]
- 158.Ozaki M, Suzuki S, and Irani K. Redox factor-1/APE suppresses oxidative stress by inhibiting the rac1 GTPase. FASEB J 16: 889–890, 2002 [DOI] [PubMed] [Google Scholar]
- 159.Parildar-Karpuzoglu H, Dogru-Abbasoglu S, Hanagasi HA, Karadag B, Gurvit H, Emre M, and Uysal M. Single nucleotide polymorphisms in base-excision repair genes hOGG1, APE1 and XRCC1 do not alter risk of Alzheimer's disease. Neurosci Lett 442: 287–291, 2008 [DOI] [PubMed] [Google Scholar]
- 160.Parsons JL, Dianova II, and Dianov GL. APE1 is the major 3′-phosphoglycolate activity in human cell extracts. Nucleic Acids Res 32: 3531–3536, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Pieretti M, Khattar NH, and Smith SA. Common polymorphisms and somatic mutations in human base excision repair genes in ovarian and endometrial cancers. Mutat Res 432: 53–59, 2001 [DOI] [PubMed] [Google Scholar]
- 162.Poletto M, Di Loreto C, Marasco D, Poletto E, Puglisi F, Damante G, and Tell G. Acetylation on critical lysine residues of Apurinic/apyrimidinic endonuclease 1 (APE1) in triple negative breast cancers. Biochem Biophys Res Commun 424: 34–39, 2012 [DOI] [PubMed] [Google Scholar]
- 163.Puglisi F, Aprile G, Minisini AM, Barbone F, Cataldi P, Tell G, Kelley MR, Damante G, Beltrami CA, and Di Loreto C. Prognostic significance of Ape1/ref-1 subcellular localization in non-small cell lung carcinomas. Anticancer Res 21: 4041–4049, 2001 [PubMed] [Google Scholar]
- 164.Puglisi F, Barbone F, Tell G, Aprile G, Pertoldi B, Raiti C, Kelley MR, Damante G, Sobrero A, Beltrami CA, and Di Loreto C. Prognostic role of Ape/Ref-1 subcellular expression in stage I-III breast carcinomas. Oncol Rep 9: 11–17, 2002 [PubMed] [Google Scholar]
- 165.Qu J, Liu GH, Huang B, and Chen C. Nitric oxide controls nuclear export of APE1/Ref-1 through S-nitrosation of cysteines 93 and 310. Nucleic Acids Res 35: 2522–2532, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Raffoul JJ, Kucuk O, Sarkar FH, and Hillman GG. Dietary agents in cancer chemoprevention and treatment. J Oncol 2012: 749310, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Rai G, Vyjayanti VN, Dorjsuren D, Simeonov A, Jadhav A, Wilson DM, 3rd, and Maloney DJ. Synthesis, biological evaluation, and structure-activity relationships of a novel class of apurinic/apyrimidinic endonuclease 1 inhibitors. J Med Chem 55: 3101–3112, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Ramana CV, Boldogh I, Izumi T, and Mitra S. Activation of apurinic/apyrimidinic endonuclease in human cells by reactive oxygen species and its correlation with their adaptive response to genotoxicity of free radicals. Proc Natl Acad Sci U S A 95: 5061–5066, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Reynolds JJ. and Stewart GS. A single strand that links multiple neuropathologies in human disease. Brain 136: 14–27, 2013 [DOI] [PubMed] [Google Scholar]
- 170.Robertson KA, Bullock HA, Xu Y, Tritt R, Zimmerman E, Ulbright TM, Foster RS, Einhorn LH, and Kelley MR. Altered expression of Ape1/ref-1 in germ cell tumors and overexpression in NT2 cells confers resistance to bleomycin and radiation. Cancer Res 61: 2220–2225, 2001 [PubMed] [Google Scholar]
- 171.Robson CN. and Hickson ID. Isolation of cDNA clones encoding a human apurinic/apyrimidinic endonuclease that corrects DNA repair and mutagenesis defects in E. coli xth (exonuclease III) mutants. Nucleic Acids Res 19: 5519–5523, 1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Robson CN, Hochhauser D, Craig R, Rack K, Buckle VJ, and Hickson ID. Structure of the human DNA repair gene HAP1 and its localisation to chromosome 14q 11.2–12. Nucleic Acids Res 20: 4417–4421, 1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Robson CN, Milne AM, Pappin DJ, and Hickson ID. Isolation of cDNA clones encoding an enzyme from bovine cells that repairs oxidative DNA damage in vitro: homology with bacterial repair enzymes. Nucleic Acids Res 19: 1087–1092, 1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Roy R, Chun J, and Powell SN. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat Rev Cancer 12: 68–78, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Rush J, Moritz A, Lee KA, Guo A, Goss VL, Spek EJ, Zhang H, Zha XM, Polakiewicz RD, and Comb MJ. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat Biotechnol 23: 94–101, 2005 [DOI] [PubMed] [Google Scholar]
- 176.Sak SC, Harnden P, Johnston CF, Paul AB, and Kiltie AE. APE1 and XRCC1 protein expression levels predict cancer-specific survival following radical radiotherapy in bladder cancer. Clin Cancer Res 11: 6205–6211, 2005 [DOI] [PubMed] [Google Scholar]
- 177.Sampath H, McCullough AK, and Lloyd RS. Regulation of DNA glycosylases and their role in limiting disease. Free Radic Res 46: 460–478, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Sandhu SK, Yap TA, and de Bono JS. The emerging role of poly(ADP-Ribose) polymerase inhibitors in cancer treatment. Curr Drug Targets 12: 2034–2044, 2011 [DOI] [PubMed] [Google Scholar]
- 179.Santos RX, Correia SC, Zhu X, Smith MA, Moreira PI, Castellani RJ, Nunomura A, and Perry G. Mitochondrial DNA oxidative damage and repair in aging and Alzheimer's disease. Antioxid Redox Signal 18: 2444–2457, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Schild LJ, Brookman KW, Thompson LH, and Wilson DM., 3rd Effects of Ape1 overexpression on cellular resistance to DNA-damaging and anticancer agents. Somat Cell Mol Genet 25: 253–262, 1999 [DOI] [PubMed] [Google Scholar]
- 181.Seiple LA, Cardellina JH, 2nd, Akee R, and Stivers JT. Potent inhibition of human apurinic/apyrimidinic endonuclease 1 by arylstibonic acids. Mol Pharmacol 73: 669–677, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Seki S, Hatsushika M, Watanabe S, Akiyama K, Nagao K, and Tsutsui K. cDNA cloning, sequencing, expression and possible domain structure of human APEX nuclease homologous to Escherichia coli exonuclease III. Biochim Biophys Acta 1131: 287–299, 1992 [DOI] [PubMed] [Google Scholar]
- 183.Seki S, Ikeda S, Watanabe S, Hatsushika M, Tsutsui K, Akiyama K, and Zhang B. A mouse DNA repair enzyme (APEX nuclease) having exonuclease and apurinic/apyrimidinic endonuclease activities: purification and characterization. Biochim Biophys Acta 1079: 57–64, 1991 [DOI] [PubMed] [Google Scholar]
- 184.Seki S, Takata A, Nakamura T, Akiyama K, and Watanabe S. A possible cause of heterogeneity of mammalian apurinic/apyrimidinic endonuclease. Int J Biochem 25: 53–59, 1993 [DOI] [PubMed] [Google Scholar]
- 185.Sengupta S, Mantha AK, Mitra S, and Bhakat KK. Human AP endonuclease (APE1/Ref-1) and its acetylation regulate YB-1-p300 recruitment and RNA polymerase II loading in the drug-induced activation of multidrug resistance gene MDR1. Oncogene 30: 482–493, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Shaikh AY. and Martin LJ. DNA base-excision repair enzyme apurinic/apyrimidinic endonuclease/redox factor-1 is increased and competent in the brain and spinal cord of individuals with amyotrophic lateral sclerosis. Neuromol Med 2: 47–60, 2002 [DOI] [PubMed] [Google Scholar]
- 187.Shaper NL, Grafstrom RH, and Grossman L. Human placental apurinic/apyrimidinic endonuclease. Its isolation and characterization. J Biol Chem 257: 13455–13458, 1982 [PubMed] [Google Scholar]
- 188.Shen JC. and Loeb LA. Mutations in the alpha8 loop of human APE1 alter binding and cleavage of DNA containing an abasic site. J Biol Chem 278: 46994–47001, 2003 [DOI] [PubMed] [Google Scholar]
- 189.Shen MR, Jones IM, and Mohrenweiser H. Nonconservative amino acid substitution variants exist at polymorphic frequency in DNA repair genes in healthy humans. Cancer Res 58: 604–608, 1998 [PubMed] [Google Scholar]
- 190.Sheng Q, Zhang Y, Wang R, Zhang J, Chen B, Wang J, Zhang W, and Xin X. Prognostic significance of APE1 cytoplasmic localization in human epithelial ovarian cancer. Med Oncol 29: 1265–1271, 2012 [DOI] [PubMed] [Google Scholar]
- 191.Shimizu N, Sugimoto K, Tang J, Nishi T, Sato I, Hiramoto M, Aizawa S, Hatakeyama M, Ohba R, Hatori H, Yoshikawa T, Suzuki F, Oomori A, Tanaka H, Kawaguchi H, Watanabe H, and Handa H. High-performance affinity beads for identifying drug receptors. Nat Biotechnol 18: 877–881, 2000 [DOI] [PubMed] [Google Scholar]
- 192.Shokolenko IN, Alexeyev MF, Robertson FM, LeDoux SP, and Wilson GL. The expression of Exonuclease III from E. coli in mitochondria of breast cancer cells diminishes mitochondrial DNA repair capacity and cell survival after oxidative stress. DNA Repair (Amst) 2: 471–482, 2003 [DOI] [PubMed] [Google Scholar]
- 193.Silber JR, Bobola MS, Blank A, Schoeler KD, Haroldson PD, Huynh MB, and Kolstoe DD. The apurinic/apyrimidinic endonuclease activity of Ape1/Ref-1 contributes to human glioma cell resistance to alkylating agents and is elevated by oxidative stress. Clin Cancer Res 8: 3008–3018, 2002 [PubMed] [Google Scholar]
- 194.Simeonov A, Kulkarni A, Dorjsuren D, Jadhav A, Shen M, McNeill DR, Austin CP, and Wilson DM., 3rd.Identification and characterization of inhibitors of human apurinic/apyrimidinic endonuclease APE1. PLoS One 4: e5740, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Simonelli V, Mazzei F, D'Errico M, and Dogliotti E. Gene susceptibility to oxidative damage: from single nucleotide polymorphisms to function. Mutat Res 731: 1–13, 2012 [DOI] [PubMed] [Google Scholar]
- 196.Srinivasan A, Wang L, Cline CJ, Xie Z, Sobol RW, Xie XQ, and Gold B. Identification and characterization of human apurinic/apyrimidinic endonuclease-1 inhibitors. Biochemistry 51: 6246–6259, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Stetler RA, Gao Y, Zukin RS, Vosler PS, Zhang L, Zhang F, Cao G, Bennett MV, and Chen J. Apurinic/apyrimidinic endonuclease APE1 is required for PACAP-induced neuroprotection against global cerebral ischemia. Proc Natl Acad Sci U S A 107: 3204–3209, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Stuart JA, Mayard S, Hashiguchi K, Souza-Pinto NC, and Bohr VA. Localization of mitochondrial DNA base excision repair to an inner membrane-associated particulate fraction. Nucleic Acids Res 33: 3722–3732, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Suh D, Wilson DM, 3rd, and Povirk LF. 3′-phosphodiesterase activity of human apurinic/apyrimidinic endonuclease at DNA double-strand break ends. Nucleic Acids Res 25: 2495–2500, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Sultana R, McNeill DR, Abbotts R, Mohammed MZ, Zdzienicka MZ, Qutob H, Seedhouse C, Laughton CA, Fischer PM, Patel PM, Wilson DM, 3rd, and Madhusudan S. Synthetic lethal targeting of DNA double-strand break repair deficient cells by human apurinic/apyrimidinic endonuclease inhibitors. Int J Cancer 131: 2433–2444, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Sykora P, Wilson DM, 3rd, and Bohr VA. Base excision repair in the mammalian brain: implication for age related neurodegeneration. Mech Ageing Dev pii: S0047-6374(13)0005 1-1, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Tan Z, Sun N, and Schreiber SS. Immunohistochemical localization of redox factor-1 (Ref-1) in Alzheimer's hippocampus. Neuroreport 9: 2749–2752, 1998 [DOI] [PubMed] [Google Scholar]
- 203.Tell G, Crivellato E, Pines A, Paron I, Pucillo C, Manzini G, Bandiera A, Kelley MR, Di Loreto C, and Damante G. Mitochondrial localization of APE/Ref-1 in thyroid cells. Mutat Res 485: 143–152, 2001 [DOI] [PubMed] [Google Scholar]
- 204.Tell G, Damante G, Caldwell D, and Kelley MR. The intracellular localization of APE1/Ref-1: more than a passive phenomenon? Antioxid Redox Signal 7: 367–384, 2005 [DOI] [PubMed] [Google Scholar]
- 205.Tell G, Quadrifoglio F, Tiribelli C, and Kelley MR. The many functions of APE1/Ref-1: not only a DNA repair enzyme. Antioxid Redox Signal 11: 601–620, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Tomkins J, Dempster S, Banner SJ, Cookson MR, and Shaw PJ. Screening of AP endonuclease as a candidate gene for amyotrophic lateral sclerosis (ALS). Neuroreport 11: 1695–1697, 2000 [DOI] [PubMed] [Google Scholar]
- 207.Tomkinson AE, Bonk RT, and Linn S. Mitochondrial endonuclease activities specific for apurinic/apyrimidinic sites in DNA from mouse cells. J Biol Chem 263: 12532–12537, 1988 [PubMed] [Google Scholar]
- 208.Tsuchimoto D, Sakai Y, Sakumi K, Nishioka K, Sasaki M, Fujiwara T, and Nakabeppu Y. Human APE2 protein is mostly localized in the nuclei and to some extent in the mitochondria, while nuclear APE2 is partly associated with proliferating cell nuclear antigen. Nucleic Acids Res 29: 2349–2360, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Tsujishita Y, Guo S, Stolz LE, York JD, and Hurley JH. Specificity determinants in phosphoinositide dephosphorylation: crystal structure of an archetypal inositol polyphosphate 5-phosphatase. Cell 105: 379–389, 2001 [DOI] [PubMed] [Google Scholar]
- 210.Tsutakawa SE, Shin DS, Mol CD, Izumi T, Arvai AS, Mantha AK, Szczesny B, Ivanov IN, Hosfield DJ, Maiti B, Pique ME, Frankel KA, Hitomi K, Cunningham RP, Mitra S, and Tainer JA. Conserved structural chemistry for incision activity in structurally non-homologous apurinic/apyrimidinic endonuclease APE1 and endonuclease IV DNA repair enzymes. J Biol Chem 288: 8445–8455, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Unnikrishnan A, Raffoul JJ, Patel HV, Prychitko TM, Anyangwe N, Meira LB, Friedberg EC, Cabelof DC, and Heydari AR. Oxidative stress alters base excision repair pathway and increases apoptotic response in apurinic/apyrimidinic endonuclease 1/redox factor-1 haploinsufficient mice. Free Radic Biol Med 46: 1488–1499, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Van Houten B, Woshner V, and Santos JH. Role of mitochondrial DNA in toxic responses to oxidative stress. DNA Repair (Amst) 5: 145–152, 2006 [DOI] [PubMed] [Google Scholar]
- 213.Vascotto C, Bisetto E, Li M, Zeef LA, D'Ambrosio C, Domenis R, Comelli M, Delneri D, Scaloni A, Altieri F, Mavelli I, Quadrifoglio F, Kelley MR, and Tell G. Knock-in reconstitution studies reveal an unexpected role of Cys-65 in regulating APE1/Ref-1 subcellular trafficking and function. Mol Biol Cell 22: 3887–3901, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Vascotto C, Cesaratto L, Zeef LA, Deganuto M, D'Ambrosio C, Scaloni A, Romanello M, Damante G, Taglialatela G, Delneri D, Kelley MR, Mitra S, Quadrifoglio F, and Tell G. Genome-wide analysis and proteomic studies reveal APE1/Ref-1 multifunctional role in mammalian cells. Proteomics 9: 1058–1074, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Vascotto C, Fantini D, Romanello M, Cesaratto L, Deganuto M, Leonardi A, Radicella JP, Kelley MR, D'Ambrosio C, Scaloni A, Quadrifoglio F, and Tell G. APE1/Ref-1 interacts with NPM1 within nucleoli and plays a role in the rRNA quality control process. Mol Cell Biol 29: 1834–1854, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Vasko MR, Guo C, and Kelley MR. The multifunctional DNA repair/redox enzyme Ape1/Ref-1 promotes survival of neurons after oxidative stress. DNA Repair (Amst) 4: 367–379, 2005 [DOI] [PubMed] [Google Scholar]
- 217.Vogel KS, Perez M, Momand JR, Acevedo-Torres K, Hildreth K, Garcia RA, Torres-Ramos CA, Ayala-Torres S, Prihoda TJ, McMahan CA, and Walter CA. Age-related instability in spermatogenic cell nuclear and mitochondrial DNA obtained from Apex1 heterozygous mice. Mol Reprod Dev 78: 906–919, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Walker LJ, Craig RB, Harris AL, and Hickson ID. A role for the human DNA repair enzyme HAP1 in cellular protection against DNA damaging agents and hypoxic stress. Nucleic Acids Res 22: 4884–4889, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Walker LJ, Robson CN, Black E, Gillespie D, and Hickson ID. Identification of residues in the human DNA repair enzyme HAP1 (Ref-1) that are essential for redox regulation of Jun DNA binding. Mol Cell Biol 13: 5370–5376, 1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Wallace SS, Murphy DL, and Sweasy JB. Base excision repair and cancer. Cancer Lett 327: 73–89, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Wang D, Luo M, and Kelley MR. Human apurinic endonuclease 1 (APE1) expression and prognostic significance in osteosarcoma: enhanced sensitivity of osteosarcoma to DNA damaging agents using silencing RNA APE1 expression inhibition. Mol Cancer Ther 3: 679–686, 2004 [PubMed] [Google Scholar]
- 222.Wang D, Xiang DB, Yang XQ, Chen LS, Li MX, Zhong ZY, and Zhang YS. APE1 overexpression is associated with cisplatin resistance in non-small cell lung cancer and targeted inhibition of APE1 enhances the activity of cisplatin in A549 cells. Lung Cancer 66: 298–304, 2009 [DOI] [PubMed] [Google Scholar]
- 223.Weichenrieder O, Repanas K, and Perrakis A. Crystal structure of the targeting endonuclease of the human LINE-1 retrotransposon. Structure 12: 975–986, 2004 [DOI] [PubMed] [Google Scholar]
- 224.Weissman L, Jo DG, Sorensen MM, de Souza-Pinto NC, Markesbery WR, Mattson MP, and Bohr VA. Defective DNA base excision repair in brain from individuals with Alzheimer's disease and amnestic mild cognitive impairment. Nucleic Acids Res 35: 5545–5555, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Whisstock JC, Romero S, Gurung R, Nandurkar H, Ooms LM, Bottomley SP, and Mitchell CA. The inositol polyphosphate 5-phosphatases and the apurinic/apyrimidinic base excision repair endonucleases share a common mechanism for catalysis. J Biol Chem 275: 37055–37061, 2000 [DOI] [PubMed] [Google Scholar]
- 226.Wilson DM., 3rd.Properties of and substrate determinants for the exonuclease activity of human apurinic endonuclease Ape1. J Mol Biol 330: 1027–1037, 2003 [DOI] [PubMed] [Google Scholar]
- 227.Wilson DM., 3rd.Processing of nonconventional DNA strand break ends. Environ Mol Mutagen 48: 772–782, 2007 [DOI] [PubMed] [Google Scholar]
- 228.Wilson DM, 3rd, Kim D, Berquist BR, and Sigurdson AJ. Variation in base excision repair capacity. Mutat Res 711: 100–112, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Wilson DM, 3rd, and McNeill DR. Base excision repair and the central nervous system. Neuroscience 145: 1187–1200, 2007 [DOI] [PubMed] [Google Scholar]
- 230.Wilson DM, 3rd, and Simeonov A. Small molecule inhibitors of DNA repair nuclease activities of APE1. Cell Mol Life Sci 67: 3621–3631, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Wilson DM, 3rd, Takeshita M, Grollman AP, and Demple B. Incision activity of human apurinic endonuclease (Ape) at abasic site analogs in DNA. J Biol Chem 270: 16002–16007, 1995 [DOI] [PubMed] [Google Scholar]
- 232.Wu HH, Chu YC, Wang L, Tsai LH, Lee MC, Chen CY, Shieh SH, Cheng YW, and Lee H. Cytoplasmic Ape1 expression elevated by p53 aberration may predict survival and relapse in resected non-small cell lung cancer. Ann Surg Oncol 2012. [Epub ahead of print] DOI: 10.1245/s10434-012-2431-2 [DOI] [PubMed] [Google Scholar]
- 233.Xanthoudakis S. and Curran T. Identification and characterization of Ref-1, a nuclear protein that facilitates AP-1 DNA-binding activity. EMBO J 11: 653–665, 1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Xanthoudakis S, Miao GG, and Curran T. The redox and DNA-repair activities of Ref-1 are encoded by nonoverlapping domains. Proc Natl Acad Sci U S A 91: 23–27, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Xanthoudakis S, Smeyne RJ, Wallace JD, and Curran T. The redox/DNA repair protein, Ref-1, is essential for early embryonic development in mice. Proc Natl Acad Sci U S A 93: 8919–8923, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Xia L, Guan W, Wang D, Zhang YS, Zeng LL, Li ZP, Wang G, and Yang ZZ. Killing effect of Ad5/F35-APE1 siRNA recombinant adenovirus in combination with hematoporphrphyrin derivative-mediated photodynamic therapy on human nonsmall cell lung cancer. Biomed Res Int 2013: 957913, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Xiang DB, Chen ZT, Wang D, Li MX, Xie JY, Zhang YS, Qing Y, Li ZP, and Xie J. Chimeric adenoviral vector Ad5/F35-mediated APE1 siRNA enhances sensitivity of human colorectal cancer cells to radiotherapy in vitro and in vivo. Cancer Gene Ther 15: 625–635, 2008 [DOI] [PubMed] [Google Scholar]
- 238.Xie JY, Li MX, Xiang DB, Mou JH, Qing Y, Zeng LL, Yang ZZ, Guan W, and Wang D. Elevated expression of APE1/Ref-1 and its regulation on IL-6 and IL-8 in bone marrow stromal cells of multiple myeloma. Clin Lymphoma Myeloma Leuk 10: 385–393, 2010 [DOI] [PubMed] [Google Scholar]
- 239.Xiong GS, Sun HL, Wu SM, and Mo JZ. Small interfering RNA against the apurinic or apyrimidinic endonuclease enhances the sensitivity of human pancreatic cancer cells to gemcitabine in vitro. J Dig Dis 11: 224–230, 2010 [DOI] [PubMed] [Google Scholar]
- 240.Yacoub A, Kelley MR, and Deutsch WA. The DNA repair activity of human redox/repair protein APE/Ref-1 is inactivated by phosphorylation. Cancer Res 57: 5457–5459, 1997 [PubMed] [Google Scholar]
- 241.Yamamori T, DeRicco J, Naqvi A, Hoffman TA, Mattagajasingh I, Kasuno K, Jung SB, Kim CS, and Irani K. SIRT1 deacetylates APE1 and regulates cellular base excision repair. Nucleic Acids Res 38: 832–845, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Yan N, Cherepanov P, Daigle JE, Engelman A, and Lieberman J. The SET complex acts as a barrier to autointegration of HIV-1. PLoS Pathog 5: e1000327, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Yang ZZ, Li MX, Zhang YS, Xiang DB, Dai N, Zeng LL, Li ZP, Wang G, and Wang D. Knock down of the dual functional protein apurinic/apyrimidinic endonuclease 1 enhances the killing effect of hematoporphrphyrin derivative-mediated photodynamic therapy on non-small cell lung cancer cells in vitro and in a xenograft model. Cancer Sci 101: 180–187, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Zawahir Z, Dayam R, Deng J, Pereira C, and Neamati N. Pharmacophore guided discovery of small-molecule human apurinic/apyrimidinic endonuclease 1 inhibitors. J Med Chem 52: 20–32, 2009 [DOI] [PubMed] [Google Scholar]
- 245.Zhang J, Luo M, Marasco D, Logsdon D, Lafavers KA, Chen Q, Reed A, Kelley MR, Gross ML, and Georgiadis MM. Inhibition of apurinic/apyrimidinic endonuclease I's redox activity revisited. Biochemistry 52: 2955–2966, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Zhang Y, Wang J, Xiang D, Wang D, and Xin X. Alterations in the expression of the apurinic/apyrimidinic endonuclease-1/redox factor-1 (APE1/Ref-1) in human ovarian cancer and indentification of the therapeutic potential of APE1/Ref-1 inhibitor. Int J Oncol 35: 1069–1079, 2009 [PubMed] [Google Scholar]
- 247.Zhao B, Grandy DK, Hagerup JM, Magenis RE, Smith L, Chauhan BC, and Henner WD. The human gene for apurinic/apyrimidinic endonuclease (HAP1): sequence and localization to chromosome 14 band q12. Nucleic Acids Res 20: 4097–4098, 1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Zou GM, Luo MH, Reed A, Kelley MR, and Yoder MC. Ape1 regulates hematopoietic differentiation of embryonic stem cells through its redox functional domain. Blood 109: 1917–1922, 2007 [DOI] [PubMed] [Google Scholar]
- 249.Zou GM. and Maitra A. Small-molecule inhibitor of the AP endonuclease 1/REF-1 E3330 inhibits pancreatic cancer cell growth and migration. Mol Cancer Ther 7: 2012–2021, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]