Abstract
Significance: The well-studied sequences in the human genome are those of protein-coding genes, which account for only 1%–2% of the total genome. However, with the advent of high-throughput transcriptome sequencing technology, we now know that about 90% of our genome is extensively transcribed and that the vast majority of them are transcribed into noncoding RNAs (ncRNAs). It is of great interest and importance to decipher the functions of these ncRNAs in humans. Recent Advances: In the last decade, it has become apparent that ncRNAs play a crucial role in regulating gene expression in normal development, in stress responses to internal and environmental stimuli, and in human diseases. Critical Issues: In addition to those constitutively expressed structural RNA, such as ribosomal and transfer RNAs, regulatory ncRNAs can be classified as microRNAs (miRNAs), Piwi-interacting RNAs (piRNAs), small interfering RNAs (siRNAs), small nucleolar RNAs (snoRNAs), and long noncoding RNAs (lncRNAs). However, little is known about the biological features and functional roles of these ncRNAs in DNA repair and genome instability, although a number of miRNAs and lncRNAs are regulated in the DNA damage response. Future Directions: A major goal of modern biology is to identify and characterize the full profile of ncRNAs with regard to normal physiological functions and roles in human disorders. Clinically relevant ncRNAs will also be evaluated and targeted in therapeutic applications. Antioxid. Redox Signal. 20, 655–677.
Introduction
In all organisms, faithful propagation of genetic material and transmission into daughter cells is essential to avoid propagation of mutations that could lead to genomic instability and aberrant cell activities (4). A wide variety of DNA lesions may occur by extrinsic insults, such as ultraviolet (UV) radiation, ionizing radiation (IR), and numerous genotoxic chemicals. Meanwhile, the by-products of cellular metabolism, such as reactive oxygen species (ROS), also threaten the integrity of genomic DNA. In order to maintain genomic stability, eukaryotes have evolved a highly coordinated cellular system to recognize and repair damaged DNA. Collectively, this system is named DNA damage response (DDR), encompassing DNA damage signal transduction, DNA repair, cell-cycle checkpoints, and apoptosis. Of the many forms of DNA damage, double-stranded DNA break (DSB) is one of the most lethal to the cell if left unrepaired. The DSB is repaired by nonhomologous end joining (NHEJ) in all phases of the cell cycle and homologous recombination (HR) when a template is available in the S or G2 phase. Other types of DNA damage are processed through nucleotide excision repair (NER), base excision repair (BER), and mismatch repair (MMR) (39).
The DDR is composed of a set of tightly regulated steps: initial detection of DNA damage, recruitment of repair factors to damage sites, and the final repair of DNA lesions. Therefore, all the components in the DDR are functionally categorized into DNA damage sensors, signal transducers, and effectors. In addition to previously identified protein-coding genes, noncoding RNAs (ncRNAs) have been shown to join the DDR. Accumulating evidence suggests that various types of ncRNAs play an important role in DNA repair and safeguarding genome integrity (29, 115, 212, 225). The ncRNAs are highly abundant and functionally important although they are not translated into proteins (5, 43). Except for those housekeeping ribosomal and transfer RNAs (rRNAs and tRNAs), ncRNAs are divided into two major groups based on their size: small ncRNAs and long noncoding RNAs.
MicroRNAs
The most well-studied group of ncRNAs are microRNAs (miRNAs), which are ∼19- to 24-nucleotide (nt) small ncRNAs with post-transcriptional regulatory functions that control the translation of mRNA into protein (10). First identified in Caenorhabditis elegans, miRNAs are universally expressed in nearly all metazoans, plants, and even DNA viruses (70). To date, almost 2000 mature miRNAs have been annotated in the human genome that are involved in many cellular processes such as proliferation, differentiation, stress responses, apoptosis, and development (10). Classically, the biogenesis of miRNAs is a tightly regulated multistep process. In the nucleus, primary miRNAs (pri-miRNAs), which bear one or more imperfect stem-loop hairpin-like structures, are first processed by the RNaseIII enzyme Drosha and its cofactor DGCR8 (DiGeorge Syndrome Critical Region Gene 8). The processed products, which are called precursor miRNAs (pre-miRNAs), have a hairpin structure of ∼70-nt and are exported to the cytoplasm by the Ran GTPase Exportin-5. In the cytoplasm, the stem loops of pre-miRNAs are cleaved off by another RNaseIII, Dicer, and its cofactor, Tar RNA-binding protein (TRBP), resulting in the production of mature ∼22-nt RNA duplexes. Most often, one strand of the duplex is preferentially incorporated into a member of the Argonaute protein subfamily to form the RNA-induced silencing complex (RISC). The other strand, known as the miRNA star or passenger strand, is often degraded. RISCs loaded with mature miRNAs are subsequently guided by miRNAs to pair with target transcripts at their 3′ untranslated region (3′-UTR) and induce mRNA degradation or inhibition of translation (10). Since a single miRNA may potentially target more than 300 different mRNA transcripts, almost 60% of human protein-coding genes are predicted to be targets of miRNAs. In addition, several types of miRNAs may regulate the same pathway cooperatively by targeting the same or different targets.
Long noncoding RNAs
The other most common ncRNAs are mRNA-like long noncoding RNAs (lncRNAs), which range in length from 200 nt to ∼100 kilobases and lack significant open reading frames. Similar to their protein-coding counterpart, most lncRNAs are transcribed by RNA polymerase II (RNA pol II) and polyadenylated, and their expression is tightly associated with cell and tissue specificity (51, 64). However, compared with protein-coding genes or miRNAs, lncRNAs are usually expressed at lower levels and poorly conserved between species. With the advent of RNA sequencing and computational methods for transcriptome reconstruction, thousands of lncRNAs have been identified in various cell types and tissues in mammals (22). Since the number of characterized long noncoding transcripts has increased, so has the uncertainty regarding their putative functions. How do lncRNAs exert their functions? At present, only a handful of lncRNAs have been well characterized with distinctive biological roles, such as XIST and TSIX, in X-chromosome inactivation (20, 119), H19 and AIR in genomic imprinting (18, 189), NRON in cytoplasmic-to-nuclear trafficking of NFAT transcriptional factor (224), and HOTAIR and HOTTIP in trans-acting regulation of the HOX gene family (173, 216). Although only a minority have been studied in detail, it is now becoming evident that lncRNAs provide important transcriptional outputs of the genome.
LncRNAs include a heterogeneous group of noncoding transcripts. These RNAs can be classified according to their proximity to protein-coding genes: sense, antisense, bidirectional, intronic, and intergenic (168). Antisense lncRNAs are transcribed from the opposite DNA strand of a protein-coding gene and overlap, in part, with sense mRNA. Both ends of protein-coding genes may be able to encode antisense transcripts. Antisense transcription is a widespread phenomenon of mammalian genomes. More than 70% of transcription units sequenced in the mouse genome are antisense transcripts, and a majority of them are lncRNAs (99). However, only a few antisense lncRNAs have been functionally validated. As an example, BACE1-AS is a conserved lncRNA that is implicated in Alzheimer's disease–associated pathophysiology. As an antisense transcript for β-secretase-1 (BACE1), a crucial enzyme in Alzheimer's disease, BACE-AS has been found to regulate the stability of BACE1 mRNA and, subsequently, BACE1 protein expression in vitro and in vivo (52).
More recently, another group of lncRNAs, termed large or long intergenic ncRNAs (lincRNAs), has been identified by searching the distinctive “K4–K36” chromatin signature, which is indicative of active transcription. These lincRNAs are exclusively intergenic and are marked by trimethylation of lysine 4 of histone H3 (H3K4me3) at the promoter region and trimethylation of lysine 36 of histone H3 (H3K36me3) along the transcribed region. By searching for K4–K36 domains that do not overlap with known protein-coding genes, the Rinn laboratory has revealed approximately 1600 regions in the mouse genome and approximately 2500 regions in the human genome that show higher evolutionary conservation across mammals in contrast to other lncRNAs (72, 103). The biological functions of most lincRNAs are poorly defined, although they are presumably involved in transcription regulation by physical association with different chromatin regulatory proteins. For instance, using loss-of-function studies, dozens of lincRNAs have been found to play key roles in the circuitry controlling embryonic stem cell state in trans. Knockdown of these lincRNAs causes either exit from the pluripotent state or up-regulation of lineage commitment programs, comparable to knockdown of well-known embryonic stem cell regulators, such as Oct4 and Nanog (73).
Targets of ncRNAs in the DDR and ROS Signaling Pathways
Genotoxic stress is a life-threatening event for humans, as it changes the content and organization of DNA. In addition to many types of exogenous DNA-damaging agents, another important determinant of genomic integrity and cellular response to DNA damage is the level of intracellular ROS, which is tightly regulated through the coordinated activities of cellular pro-oxidants and antioxidants. Intracellular ROS can act as a cellular toxicant or a signaling molecule, depending on its concentration and localization. Endogenous ROS produced by cellular metabolism account for the major oxidative lesions that induce DNA damage (149, 163). A mechanistic link between ROS and DDR pathways has not been clearly elucidated. A recent study demonstrated that ROS induction after treatment of cells with neocarzinostatin (NCS) is partly mediated by increasing histone H2AX, a biomarker for the DDR (98). Therefore, ROS is not only a causative factor to generate DNA damage, but is also regulated by the DNA damage-signaling pathways.
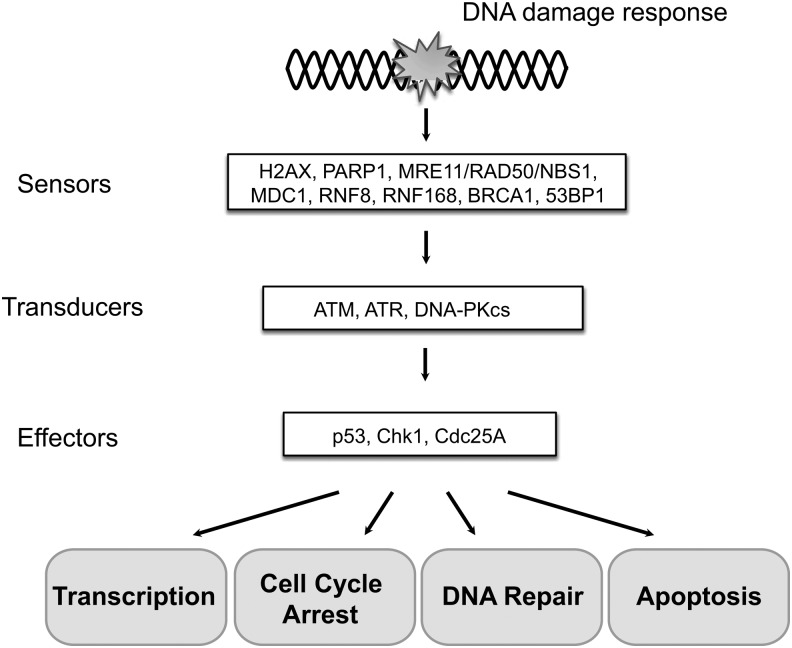
The DDR is a complex signal transduction pathway that detects DNA damage signals and transduces those signals to execute proper cellular responses (39). The signals collected and transmitted by sensors, mediators, and transducers are ultimately used to make a cell fate determination: Either the cell cycle is arrested to allow repair of damaged DNA or, if the damage is beyond repair, programs are initiated that instruct the cell to undergo apoptosis (Fig. 1).
FIG. 1.
The components of the canonical DNA damage response (DDR). The key proteins in the DDR pathway are summarized in the order of sensors, transducers, and effectors; while the effectors are involved in transcription activation, cell-cycle arrest, DNA repair, and apoptosis.
miRNA targets in the DDR
miRNAs modulate the initiation, maintenance, and activity of the DDR by targeting and modulating key genes in the pathway. These miRNAs are summarized in Table 1. The DDR is initiated at DNA damage sites by a number of sensors that recognize damaged DNA and recruit signaling proteins to the damage site. One of the most important sensors, H2AX, is extensively phosphorylated after DNA damage by two phosphatidylinositide 3-kinase-like kinases: ataxia telangiectasia mutated (ATM) and ataxia telangiectasia and Rad3 related (ATR). Overexpression of miR-24, the first miRNA found to target H2AX, down-regulates the level of H2AX, resulting in higher sensitivity of cells to IR (114). ATM is responsible for the activation of a number of signaling pathways after DSBs, the most lethal DNA damage in the cell. It is estimated that ATM may phosphorylate as many as 700 proteins in human cells (139). Homeostatic regulation of ATM activity in the DDR is primarily mediated by the Wip1 phosphatase (135). Recently, several miRNAs, including miR-18a, miR-100, miR-101, miR-181, and miR-421, have been identified as novel regulators to control the protein level of ATM (86, 188, 221, 232, 242). BRCA1, a critical tumor suppressor, BRCA1, is also recruited to DNA damage lesions, where it facilitates DNA repair. The level of BRCA1 is regulated by miR-182, miR-146a, and 146b-5p (60, 150).
Table 1.
Regulatory miRNAs in the DNA Damage Response
| Targets | Function in the DDR | MiRNAs | Refs. |
|---|---|---|---|
| ATM | Transducer | miR-18a, miR-100, miR-101, miR-181, miR-421 | (86, 188, 221, 232, 242) |
| BAX | Apoptosis | miR-886-5p | (122) |
| Bcl-2 | Apoptosis | miR-15a, miR-16-1, miR34a, miR-195 | (15, 41, 81) |
| BRCA1 | Mediator, DNA repair | miR-182, miR-146a, 146b-5p | (60, 150) |
| Cdc25A | Cell cycle checkpoint | miR-21, miR-424, miR-449a, miR-503, miR-449a/b | (26, 34, 46, 178, 217) |
| Cdc25B | Cell cycle checkpoint | miR-148a, miR-449a, miR-449b | (123) |
| Cdc25C | Cell cycle checkpoint | miR-129b | (117) |
| Cdc42 | Cell cycle checkpoint | miR-29, miR-137, miR-185, miR-206 | (36, 116, 124, 126, 127, 160) |
| CDK2 | Cell cycle | miR-124a, miR-372, miR-376a, miR-885-5p | (1, 152, 200, 214) |
| CDK6 | Cell cycle | miR-16, miR-22, miR-34, miR-107, miR-195, miR-124a, miR-29, miR-449a/b | (34, 53, 128, 166, 193, 228, 229) |
| Cyclin D | Cell cycle | miR-15, miR-16, miR-195 | (16, 128, 229) |
| Cyclin E | Cell cycle | miR-16, miR-29c, miR-195, miR-424 | (48, 154, 181, 213) |
| Cyclin G1 | Cell cycle | miR-122 | (68) |
| DNA-PK | DNA repair | miR-101 | (232) |
| E2F | Transcription factor | miR-11, miR-15, miR-16, miR-17, miR-20a, miR-106b | (159, 202, 203) |
| H2AX | Sensor/mediator | miR-24, miR-136 | (114, 220) |
| MDM2 | Cell cycle checkpoint | miR-143, miR-145, miR-605 | (227, 246) |
| MSH2 | DNA mismatch repair | miR-21, miR-155 | (205, 242) |
| p21 | Cell cycle checkpoint | miR-17, miR-20a/b, miR-106a/b, miR-663, miR-93, miR-215, miR-192 | (65, 84, 93, 239) |
| p27 | Cell cycle | miR-221/222, miR-181 | (54, 66) |
| p53 | Cell cycle checkpoint, apoptosis | miR-125b, miR-504 | (87, 117) |
| P57 | Cell cycle | miR-221/222 | (54) |
| p63 | Transcription factor | miR-92, miR-302 | (179) |
| PLK1 | Cell cycle checkpoint | miR-100 | (164) |
| PUMA | Apoptosis | miR-125b | (184) |
| RAD23B | DNA repair | miR-373 | (45) |
| RAD52 | DNA repair | miR-210, miR-373 | (45) |
| Wee1 | Cell cycle checkpoint | miR-195 | (81, 172) |
| Wip1 | Cell cycle checkpoint | miR-16 | (251) |
The tumor suppressor p53 has a central role in the activation of genes in multiple pathways, including cell-cycle regulation, tumor suppression, and apoptosis. miR-125b and miR-504 have been identified as negative regulators of p53 in several types of human cells (87, 117). Interestingly, miR-605 (227), and miR-143/miR-145 (246) are post-transcriptionally activated by p53 and, subsequently, target Mdm2, leading to rapid accumulation of p53. These findings reveal a miRNA/Mdm2/p53-positive feedback loop that ensures rapid activation of p53 after DNA damage (76).
miRNA targets in the ROS signaling pathways
ROS are chemically reactive molecules containing oxygen, including the superoxide anion radical (O2−), hydrogen peroxide (H2O2), hydroxyl radical (OH−), and nitric oxide (NO). ROS are constantly produced in the living cells and cause a significant portion of DNA damage. ROS not only are by-products of cellular metabolic processes, but also are generated by specific plasma membrane oxidases in response to growth factors and cytokines and serve as secondary messengers in specific signaling pathways. Recently, miRNAs are found to target the central regulators of the ROS signaling pathway (Nrf2, Keap1, and TNFα) and the ROS scavenger (superoxide dismutase [SOD], catalase, and thioredoxin reductase [TXNRD]), representing a key role in the regulation of antioxidant response (Table 2).
Table 2.
miRNAs That Regulate the Reactive Oxygen Species Pathway
| Targets | Function in the ROS | MiRNAs | Refs. |
|---|---|---|---|
| Catalase | Scavenger | miR-30b | (78) |
| GPX2 | Scavenger | miR-17* | (230) |
| Keap1 | Regulator | miR-141, miR-200a | (49, 208) |
| Nrf2 | Regulator | miR132, miR-155, miR-144, miR-153, miR-27a, miR-142-5p, miR-28 | (155, 177, 190, 210, 237) |
| SOD2 | Scavenger | miR-222, miR-382, miR-335, miR-23a, miR-17* | (7, 111, 131, 215, 230) |
| SOD3 | Scavenger | miR-21 | (249) |
| TNFα | Regulator | miR-21 | (249) |
| TXNRD2 | Scavenger | miR-34a, miR-17* | (7, 230) |
Nuclear factor-erythroid 2-related factor 2 (Nrf2) is an important transcription factor for cell survival during oxidative stress. On oxidative stress, Nrf2 binds to the antioxidant response element and activates the expression of cellular antioxidant enzymes, such as SOD, catalase, and glutathione peroxidase (GSHPX). Recently, Sangokoya et al. discovered that miR-144 modulated oxidative stress tolerance in sickle cell disease through reducing Nrf2 levels and increased anemia severity in patients homozygous for sickle cell disease (177). Subsequent studies showed that Nrf2 can be regulated by other miRNAs, including miR132, miR-155, miR-153, miR-27a, miR-142-5p, and miR-28, to trigger the redox homeostasis within cells (155, 190, 210, 237). miRNAs also regulate Kelch-like ECH-associated protein 1 (Keap1) that mediate the degradation of cytoplasmic Nrf2. For example, miR-200a binds with the Keap1 3′-UTR, leading to Keap1 mRNA degradation (49). A reduction in Keap1 level was implied to be associated with Nrf2 nuclear translocation and transcriptional activation of Nrf2-dependent antioxidant enzymes.
Some miRNAs directly inhibit ROS scavengers that modulate ROS signaling. Considered the first line of defense against ROS, SOD catalyzes the dismutation of superoxide into oxygen and hydrogen peroxide. The Chen group reported that miR-335 suppressed SOD2 expression with a concomitant increase in ROS and induced premature senescence of young mesangial cells (7). The reduction of SOD2 by miRNAs also links the antioxidant response to the epithelial-mesenchymal transition process in that it indirectly regulates matrix metalloproteinase 1 (MMP1) and transforming growth factor beta 1 (TGFβ1) (111, 131). Other crucial ROS scavengers believed to be targeted by miRNA are GSHPX and catalase. GSHPX is responsible for catalyzing the reduction of H2O2 and reduces lipid hydroperoxides to their corresponding alcohols and free H2O2 to water, whereas catalase catalyzes the conversion of H2O2 to water and oxygen, preventing the generation of hydroxyl radicals in cells (67). Recent studies have also suggested that miR-17 suppresses GSHPX2 via directly binding its 3′UTR sequence (230) and that miR-30b targets catalase and inhibits its expression at the transcript and protein levels under an oxidant environment (78). In summary, miRNA fine-tunes the ROS signaling pathway by modulating regulators and scavengers that function in the ROS signaling pathway (Fig. 2).
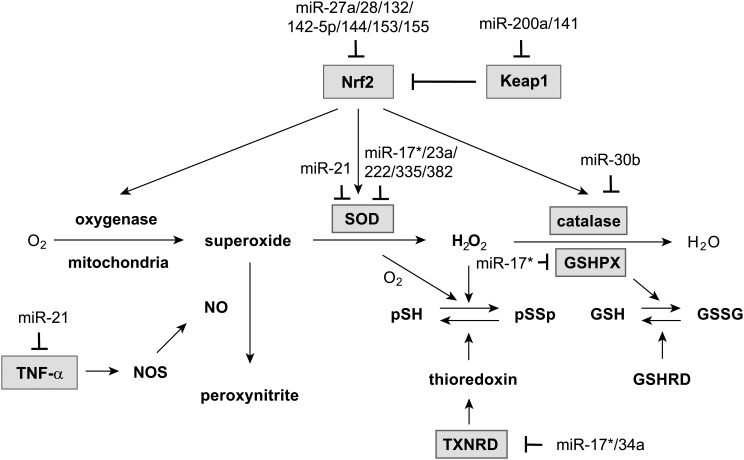
FIG. 2.
miRNAs in the reactive oxygen species (ROS) signaling pathway. ROS are constantly produced as superoxides, originating as by-products of cellular metabolism, or by oxygenases from specific signaling pathways. On oxidative stress, the nulcear factor-erythroid 2-related factor 2 (Nrf2)-mediated antioxidant response is activated to maintain intracellular redox homeostasis and limit oxidative damage. miR-200a and miR-141 form the Nrf2 positive feedback loop by targeting Kelch-like ECH-associated protein 1 (Keap1), a negative regulator of Nrf2. Other miRNAs target the central regulators Nrf2, transforming growth factor (TNF)-α, and the ROS scavengers superoxide dismutatese (SOD), catalase, glutathione peroxidase (GSHPX), and thioredoxin reductase (TXNRD).
Mitochondrial miRNAs and their roles in DNA repair and ROS signaling pathways
miRNAs are known to be encoded by the nuclear genome and to exert their influence by modulating gene expression mainly in the cytosol. Interestingly, recent studies using microarray analysis have found that a unique set of miRNAs are enriched in the rigorously purified mitochondria (8, 9, 13, 110), Such findings are beginning to shed light on how miRNAs are involved in the regulation of mitochondria. However, these miRNAs that are enriched in mitochondria (mitomiRs) are species specific and cell-type dependent. At least 15 nuclear-encoded miRNAs have been uniquely and reproducibly identified in mitochondria isolated from adult rat livers (110). Computational target prediction analysis (TargetScan and miRanda) for potential mRNA targets revealed that apoptosis, cell death, cell cycle/cell division, development, and neurogenesis were the most significant processes, but found no interaction with nucleus-derived transcripts expressed in the mitochondria. An examination of the 3′-UTR of mitochondria-derived mRNAs found a single potential interaction between miR-130a and mitochondria-encoded cytochrome c oxidase III (COX3) (110). A subsequent study in mice reported that 20 miRNAs were specifically and abundantly restrained in liver mitochondria with regard to the whole liver tissue, such as miR-122, miR-805, and miR-609 (13). Notably, the expression profile of mitochondria-associated miRNAs is significantly altered in mouse livers after treatment with streptozotocin, an antibiotic often used to induce experimental type 1 diabetes in mice. These results suggest that mitochondria-associated miRNAs may be correlated with mitochondria dysfunction (13).
More recently, a signature of 13 nuclear-encoded miRNAs was reproducibly enriched in mitochondria of human Hela cells, including hsa-miR-494, hsa-miR-1275, and hsa-miR-1974 (9). In comparison to cytosol-enriched miRNAs, these mature mitomiRs were smaller than 19 nt and exhibited unique thermodynamic features. A computational target analysis of the 13 mitomiRs in parallel with canonical miRNAs revealed that the former appeared to lack any preferential predicted targeting of the mitochondrial genes encoded by the nuclear genome, but 10 out of 13 mitomiRs were predicted to target 120 mtDNA sites (9). Intriguingly, three mitomiRs (hsa-miR-1974, hsa-miR-1977, and hsa-miR-1978) also showed a perfect match in the human mtDNA genome, but it remains to be ascertained whether the transcription of those three mitomiRs could also occur from the mitochondrial genome. Not only miRNAs are enriched in mitochondria, but also other components of miRNA machinery are also detectable in mitochondria. For example, it was shown that purified mitochondria contain Argonaute 2 (AGO2), a key active protein of the RISC complex, suggesting a potential function of miRNAs in mitochondria (8, 13)
Mitochondria are essential organelles in all cells, as they provide cellular energy by generating ATP via respiration, accounting for approximately 85%–90% of the oxygen consumed by the cell (106). Incomplete processing of oxygen or release of free electrons results in the production of ROS. The environment of the mitochondria is highly oxidative. Thus, mtDNA is believed to be more susceptible than in nuclear DNA to DNA damage caused by the oxidative effects of ROS than is nuclear DNA. Although mtDNA was originally thought to lack DNA repair activity, after four decades of research on mitochondria, it has become clear that they possess multiple mtDNA repair pathways, including BER, single-strand break repair, MMR, and, possibly, HR (102). All the mtDNA repair machinery identified so far in mammals are similar in activity to those operating in the nucleus, encoded by nuclear genes, and subsequently transported into the mitochondria (207). In the last decade, numerous studies revealed that miRNAs regulate multiple aspects of the nuclear DDR, including directly regulating the expression of diverse components of the DDR pathway and indirectly fine-tuning the expression of master regulatory proteins, such as p53, through cross-talking with other signaling pathways (132). However, the physiological and pathological roles of these miRNAs identified in mitochondria remain elusive. Whether they function similarly to regulate DNA repair and ROS signaling pathways needs to be further studied.
LncRNA targets in the DDR
The DDR is dependent on sequentially post-transcriptional and post-translational events that occur on the protein components of the pathway. Phosphorylation is a primary event that modulates the activity and stability of proteins in the DDR. In addition, chromatin remodeling and epigenetic factors can alter transcriptional status and thereby influence the DDR (206).
Recent large-scale genomic transcriptome analyses have revealed that transcription in mammals is not restricted to protein-coding regions, but pervades throughout the genome, giving rise to a complex network of transcripts, most of which have no protein-coding capacity (14). Although the functions of this class of transcripts (ncRNAs) are largely unknown, several studies have elucidated extensive signatures of lncRNAs within cell-cycle promoters and have found that these lncRNAs can control cell cycle or apoptosis during the DDR. One such lncRNA, PANDA (p21-associated lncRNA DNA damage activated), is induced after DNA damage and regulates apoptosis (89). Global gene expression analysis demonstrated that 224 genes were induced, and 193 genes were repressed by at least two-fold after PANDA knockdown. Genes induced by PANDA knockdown were significantly enriched for those involved in apoptosis. Somewhat surprisingly, PANDA does not affect the expression of p21, even though it is antisense to the p21 gene. PANDA is transcribed in a p53-dependent manner. Chromatin immunoprecipitation analysis showed that PANDA regulates apoptosis via modulation of the transcription factor NF-YA. An lncRNA from the promoter region of CCND1 (encoding cyclin D1) is induced by IR and regulates transcription of CCND1 in cis by forming a ribonucleoprotein complex repressor complex (219). This lncRNA, named lncRNA-CCND1, binds to and allosterically activates the RNA-binding protein TLS (translated in liposarcoma), which inhibits histone acetyltransferases and, thus, results in repression of CCND1 transcription. The lncRNA-ANRIL (antisense ncRNA in the INK4 locus) is transcribed in an opposite direction of p15Ink4b and regulates its expression directly or by recruitment of polycomb proteins (108, 162). It is transcriptionally up-regulated by the transcription factor E2F1 in an ATM-dependent manner after DNA damage, and elevated levels of ANRIL suppress the expression of INK4a, INK4b, and ARF at the late stage of the DDR, allowing the cell to return to normal at the completion of the DNA repair. A recent study showed that ANRIL splicing variants play a role in coordinating tissue remodeling, by modulating the expression of genes involved in cell proliferation, apoptosis, and extra-cellular matrix remodeling (42).
LncRNA expression is context specific. As an example, the DDR caused by the topoisomerase inhibitor camptothecin was shown to increase the cellular levels of two antisense lncRNAs at the 5′ (5′aHIF-1α) and 3′ (3′aHIF-1α) ends of the human antisense RNA of the HIF gene in human cancer cell lines and kidney tumor specimens (12). The observed induction of lncRNAs was dependent on DNA damaging agents and cells. In HCT116 cells, camptothecin treatment induces the transcription of both 5′ and 3′ antisense RNA, whereas desferoxamine only increases the transcription of 3′ antisense RNA. However, camptothecin could not induce the transcription of 3′ antisense RNA transcription in HeLa cells. Alternatively, the two antisense transcripts might, alternatively, be involved in mRNA degradation or chromatin inactivation of the HIF-1α gene locus after DNA damage and prevent hypoxic induction of HIF-1α protein (12, 133).
Transcribed from noncoding DNA sequences between protein-coding genes, lincRNAs are also involved in regulation of the DDR. The first studied lincRNA, lincRNA-p21, runs in an antisense direction opposite to the p21 gene. It regulates the expression of p53-repressed genes that are involved in apoptosis and cell-cycle regulation. Knockdown of either p53 or lincRNA-p21 increases the viability of cells, suggesting a direct physiological role of lincRNA-p21 in the DDR. A marked reduction in apoptosis was observed in doxorubicin-treated cells with depleted lincRNA-p21, leading to reduced levels of the proapoptotic genes NOXA and PERP. A biotin-labeled RNA pull-down experiment coupled with protein tandem mass spectroscopy analysis identified the RNA-binding protein hnRNP-K as a lincRNA-p21-associated protein, which possibly mediates the regulatory function of lincRNA-p21 in p53-dependent transcription (88).
Chromatin remodeling at the site of DNA breaks prepares for the consequent gene transcription, which is a key step in the DDR. Although exact details have not yet been worked out, several lncRNAs appear to modulate the chromatin state directly or through modifications of proteins involved in the DDR. A recent model delineated that lncRNAs provide a scaffold for the binding of polycomb protein complex PRC1/2 and direct it in a sequence-specific manner to regulate the expression of genes required during several physiological processes, including stress response. In agreement with their potential importance in gene regulation, many lncRNAs have been shown to be altered in human cancer (64). Along with their functional identification, it is expected that more lncRNAs will be identified in the DDR.
Potential roles of other small ncRNAs in the DDR
Although miRNAs and the recently identified lncRNAs are the most studied ncRNAs, other ncRNAs have been identified with deep sequencing technologies followed by extensive bioinformatics-based characterization of genomic locations and predictions of RNA secondary structures. These ncRNAs are also implicated in the DDR, including ncRNAs that regulate gene expression (such as small nucleolar RNAs [snoRNAs] and transfer RNAs) (6) and genes that mediate gene silencing (such as Piwi-interacting RNAs [piRNAs] and endogenous small interfering RNAs) (63).
DDRNAs
Most recently, a group of Dicer- and Drosha-dependent small RNAs (∼21 nt) were found to be induced by DSBs and involved in HR-mediated DSB repair. RNA deep sequencing revealed that these small RNAs arise from the sequences in the vicinity of DSB sites. These RNAs have been referred to as DDRNAs. Wei et al. proposed that DDRNAs function as guide molecules to direct either chromatin modifications or the recruitment of protein complexes to DSB sites to facilitate repair (222). Moreover, Francia et al. reported that DDR foci formation was sensitive to RNase A treatment and that DDRNAs, either chemically synthesized or generated in vitro by Dicer cleavage, were sufficient to restore DDR in the RNase A-treated cells (55).
SnoRNAs
SnoRNAs are a class of small RNA molecules that primarily mediate modifications of rRNAs, tRNAs, and small nuclear RNAs. Several snoRNAs have been suggested to be involved in the DDR. Through their analyses of snoRNA TIF-IA conditional knockout mice, Yuan et al. showed that TIF-IA plays a key role in cell-cycle arrest, up-regulation of p53, and induction of apoptosis (243). TIF-IA mediates growth-dependent regulation of ribosomal RNA synthesis, and its depletion may increase binding of ribosomal proteins, such as L11, to MDM2 and decrease the interaction of MDM2 with p53 and p19ARF. In another study, Michel et al. reported that loss of three box C/D snoRNAs in the L13a(rpL13a) locus was sufficient to render cells resistant to lipotoxic and oxidative stress in vitro and prevented the propagation of oxidative stress in vivo (148). Cells with disruption of one rpL13a allele have intact cell death pathways but are resistant to apoptosis induced by lipotoxic conditions.
tRNAs
Before fulfilling their functions, all tRNA transcripts should be extensively processed in multiple steps, including the removal of the 5′ leader by RNase P, removal of the 3′ trailer sequence by endonucleases and exonucleases, addition of CCA, splicing of introns, and modifications at multiple residues (85). Trm9 catalyzes conversion of cm5U to mcm5U at position 34 of substrate tRNAs (97). Lack of the mcm5U moiety of tRNAs can affect apoptosis, as shown in a study in which silencing of human Trm9 homolog ABH8 led to apoptosis of urothelial carcinoma lines and down-regulation of NOX-1-dependent ROS, as well as suppression of angiogenesis and invasion (185). Both mitochondrial and cytosolic tRNAs bind to cytochrome c. This binding prevents cytochrome c interaction with Apaf-1, thereby blocking Apaf-1 oligomerization and caspase activation. tRNA hydrolysis in cells enhances apoptosis and caspase activation, whereas introducing tRNA into living cells blocks apoptosis. These findings suggest that in addition to its well-established role in gene expression, tRNA may determine cellular responsiveness to apoptotic stimuli (142).
pi-RNAs
piRNAs are the largest class of small ncRNA molecules that are expressed in animal cells. They are distinct from miRNAs in that they are larger (26–31 nt rather than 21–24 nt), lack sequence conservation, and are more complex. Loss of Piwi proteins leads to germline-specific apoptosis, which may be triggered by DNA damage. In the miwi2 (piwi homologue in mouse) mutant mouse, the level of phosphorylated histone H2AX (γ-H2AX), a biomarker for the DDR, is significantly increased in zygotene-stage spermatocytes compared with wild-type cells (27). Failure to repair DSBs and/or defective synapses prevents germ cells from entering the pachytene stage of spermatogenesis and leads to apoptosis. Increased levels of apoptosis have also been observed in the testes of ziwi (piwi homologue in zebrafish) mutant zebrafish, (30). These findings suggest that piRNA biogenesis is involved in the maintenance of genome integrity. By sequencing small RNAs (RNA-seq) in primary marrow cells of patients with low- and high-grade myelodysplastic syndromes, Beck et al. found that piRNAs were enriched in only the small RNAome in low-grade myelodysplastic syndromes and potentially protected DNA from the accumulation of mutations (11).
qiRNAs
RNA interference pathways use small RNAs such as small interfering RNAs (siRNA) to mediate gene silencing in eukaryotes. It was recently reported that in the filamentous fungus, Neurospora DNA damage induces expression of the Argonaute protein QDE-2 and a novel class of small RNAs termed qiRNAs because of their association with QDE-2 (118). qiRNA biogenesis also requires DNA-damage-induced pre-siRNAs as precursors. Neurospora RNA interference mutants show increased sensitivity to DNA damage, suggesting a role for qiRNAs in the DDR by inhibiting protein translation.
Other ncRNAs
Despite recent progress in our understanding of ncRNAs, the findings to date represent only the tip of the iceberg. The transcription landscape in higher eukaryotes has turned out to be more complex than expected, suggesting that the RNA transcripts are not restricted to well-defined functional features. More recently, scientists have identified various chromatin-associated ncRNAs such as transcription initiation RNAs (180), 5′ capped promoter-associated small and large RNAs (2), and splice-site RNAs (196). However, the roles of these ncRNAs in the DDR need to be clarified.
Transcriptional Regulation of ncRNA Expression in the DDR
The first step for ncRNA expression is RNA polymerase-mediated transcription, which also involves transcription factors or cofactors and their interaction with the RNA polymerase complex and regulatory DNA elements in the ncRNA genes. Increasing evidence has shown that the DDR markedly alters the expression of ncRNAs and, thus, post-transcriptionally modulates DDR proteins such as sensors, mediators, transducers, and effectors to orchestrate a number of distinct actions in detecting DNA damage, arresting the cell cycle, mediating DNA repair, and inducing apoptosis (212). Here, we focus on the role of the DDR in the biogenesis of two major classes of ncRNAs, miRNAs and lncRNAs.
Transcriptional regulation of miRNAs after DNA damage
Various DNA damage stresses alter miRNA expression profiles. Pothof et al. first identified the differential expression of miRNAs in cell-cycle checkpoints and DNA repair in UV-treated cells (170). miRNA expression profiles were later analyzed in cells treated with other DNA damaging agents, including cisplatin, doxorubicin, IR, and NCS (59, 176, 195, 250). Different doses of DNA damage appear to activate unique as well as common sets of miRNAs, suggesting that miRNAs regulate the DDR by a mechanism based on the nature and intensity of the DNA damage.
Similar to regular protein-coding genes, miRNA genes are transcribed to pri-miRNAs. For some miRNAs, this transcription process is regulated in the DDR. The tumor suppressor p53 is a well-known transcription factor that is induced after DNA damage, which itself induces cell growth arrest, promotes apoptosis, blocks angiogenesis, and mediates DNA repair on DNA damage signals (141). Global miRNA expression analyses revealed that a cohort of miRNAs are up-regulated in a p53-dependent manner after DNA damage and identified the miRNA components of p53 transcriptional pathways. The miR-34 family (miR-34a and miR-34b/c) was the first identified transcriptional target of p53. When ectopically expressed, the miR-34 family causes a cell-cycle arrest in the G1 phase and inhibits cell-cycle progression by targeting numerous cell cycle regulators, suggesting their tumor-suppressing potential. For example, the miR-34 family directly targets and down-regulates cyclin-dependent kinase 4 (CDK4), CDK6, E2F3, Myc, and NMYC (33, 82). In addition to the miR-34 family, p53 directly transactivates miR-15a/16-1, miR-29, miR-107, miR-145, miR-192, miR-194, miR-215, and miR-605 (Fig. 3). These p53-induced miRNAs contribute to cell-cycle arrest by inhibiting the transcripts of several genes that regulate cell-cycle checkpoints or metabolism (19, 62, 83, 107, 126, 192). miR-16 and miR-29 target and repress Wip1 phosphatase, a master inhibitor in the DDR that inhibits the activation and stabilization of p53, leading to p53 induction (204, 251). Ectopic expression of miR-192/215 induces cell-cycle arrest through targeting a number of transcripts that regulate G1/S and G2/M checkpoints (21). miR-145 directly targets the oncogene c-Myc, suggesting that p53 represses c-Myc functions through regulation of miRNA expression (175, 192). Interestingly, p53-induced miRNAs regulate p53 activity in a positive feedback loop (76, 83). miR-34 inhibition of SIRT1 results in increased p53 acetylation and activation (231). miR-192, miR-194, miR-215, and MiR-605 directly inhibit Mdm2 expression, and miR-29 inhibits Wip1, leading to an increased p53 level and activity (19, 165, 227).
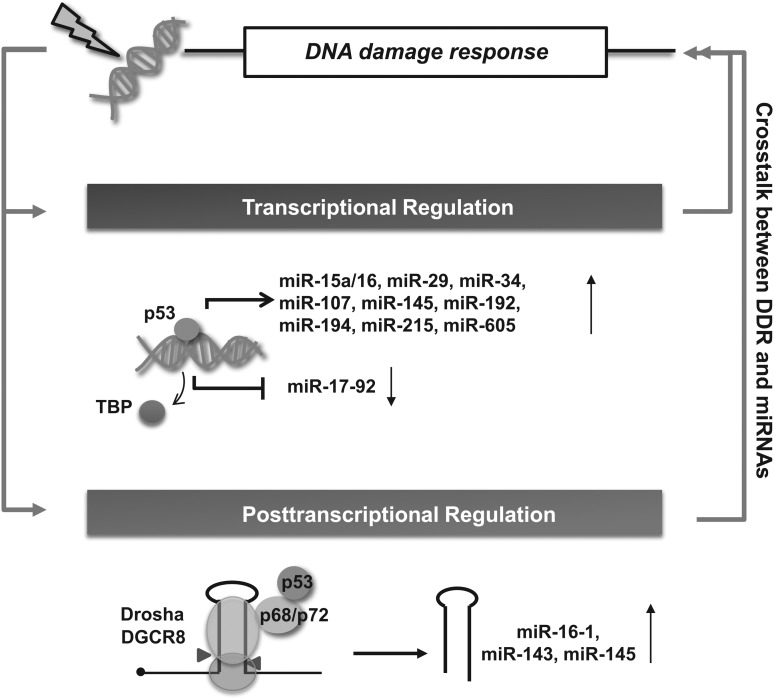
FIG. 3.
Transcriptional and post-transcriptional regulation of p53 in miRNA biogenesis. p53 modulates miRNA maturation at not only the transcription step but also the processing step of primary miRNA transcripts and results in production of crosstalk between the p53 network and the miRNA biogenesis machinery in the DDR. Many miRNAs regulated by p53 modulate the DDR by targeting proteins in the DDR, such as cell-cycle progression and apoptosis.
The p53 protein also functions as a transcriptional repressor by binding to miRNA promoters and preventing the recruitment of transcriptional activators. As an example, p53 suppresses the transcription of the miR-17-92 cluster gene by preventing recruitment of the TATA-binding protein to the TAATA site in the promoter (Fig. 3). The miR-17-92 cluster is repressed under hypoxic conditions via a p53-dependent mechanism, leading to sensitization to hypoxia-induced apoptosis (233). Thus, fine-tuning between the p53 signaling pathway and the miRNA network enables cells to amplify the p53 signal, which enhances cell sensitivity to external signals.
DNA damage-responsive transcription factors other than p53, such as NF-κB, E2F, and Myc, are also involved in the transcriptional control of miRNA expression. Both E2F and Myc activate the transcription of the miR-17-92 cluster, which, in turn, inhibits E2F and Myc expression, forming an autoregulatory negative feedback loop (3). In addition, Myc-induced miRNAs influence Myc-mediated cell proliferation and cell fate (104). For example, Myc-induced miR-20a targets cdkn1a, a gene encoding a negative regulator of cell-cycle progression, and Myc-induced miR-221 and miR-222 target the CDKN1b and CDKN1c genes, which initiate cell-cycle arrest. Owing to a lack of basic information regarding miRNA gene structure, not much is currently known about how miRNA gene expression is transcriptionally regulated. Global prediction and verification of promoter regions of miRNA genes would allow us to further explore the functional interaction of transcriptional machinery and epigenetic miRNA regulation.
Transcriptional regulation of lncRNAs after DNA damage
LncRNAs are an important class of pervasive genes that are involved in a variety of biological functions (74). They act to integrate contextual and environmental cues such as DNA damage stress. The majority of lncRNAs are transcribed by RNA polymerase II, as evidenced by Pol II occupancy, 5′ caps, histone modifications associated with Pol II transcriptional elongation, and polyadenylation (72). LncRNAs show cell type-specific expression and respond to diverse stimuli, suggesting that their expression is under considerable transcriptional control. However, only a small number of functional lncRNAs have been well characterized, and little is known about how lncRNA expression is controlled in different contexts.
The tumor suppressor protein p53 coordinates the cellular response to DNA damage through regulation of gene expression. In order to identify specific lncRNAs that operate within the p53 pathway, several research groups have designed tiling microarrays to detect their expression in mouse cell lines engineered with controllable expression of p53 or in various human samples treated with doxorubicin (89, 103). Hung et al. used an ultrahigh-density array that tiles the promoters of 56 cell-cycle genes to interrogate 108 samples representing diverse perturbations. They observed that DNA damage induced five lncRNAs from the CDKN1A promoter, one of which was PANDA, which is induced in a p53-dependent manner (89). PANDA was found to interact with transcription factor NF-YA and to prevent its binding to chromatin, leading to the repression of pro-apoptotic genes and thus facilitating cell-cycle arrest. In a subsequent study, Huarte et al. showed that one of the direct p53 targets in response to DNA damage, lincRNA-p21, which is located upstream of the p21 gene, acted as a transcriptional repressor mediating p53-induced apoptosis (88). p53 regulates lincRNA-p21 by directly activating its expression, through direct binding to the lincRNA-p21 promoter. Another p53-regulated lncRNA in the DDR is TUG1. TUG1 was originally identified as a lncRNA that is up-regulated by taurine in developing retinal cells and was found to repress the p53-dependent cell-cycle regulation (103). Recently, Yang et al. showed that TUG1 served as a scaffold RNA that binds with methylated Pc2 in Polycomb bodies, which are an important antimitogenic signal and, under certain stimuli, a stress-induced modification mark required for growth-control gene repression and senescence (236). Other than p53, transcription factor E2F1 regulates the expression of lncRNAs in the DDR. Our group found that ANRIL is induced by E2F1 after DNA damage in an ATM-dependent manner. Elevated ANRIL regulates cell proliferation and cellular senescence (211). Notably, ANRIL has been shown to interact with PRC2 and PRC1 complex to repress the expression of INK4B–ARF–INK4A locus (108). These findings showed that lncRNAs can play key regulatory roles in the p53 or E2F1 transcriptional response after DNA damage (Fig. 4).
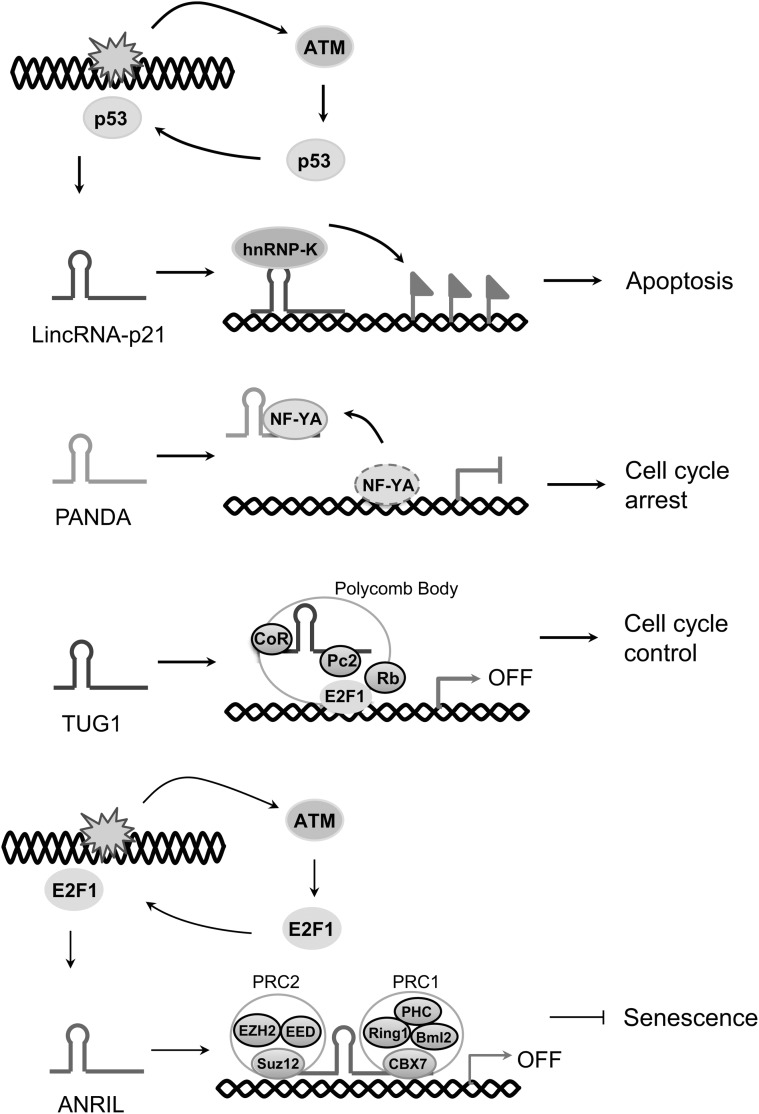
FIG. 4.
Regulation of lncRNA expression in the DDR. On DNA damage, ATM-induced transcription factors p53 and E2F1 transcriptionally activate the expression of, lincRNA-p21, PANDA, TUG1, and ANRIL. Up-regulated lincRNA-p21 functions as a guide to the hnRNP-K repressor complex, leading to p53-mediated apoptosis. Up-regulated PANDA acts as a decoy to interact with NF-YA, resulting in cell-cycle arrest. Induced TUG1 functions as a scaffold in the Polycomb body, repressing E2F1 target genes in cell-cycle control. Up-regulated ANRIL interacts with PRC1 and PRC2 complexes and represses the INK4b-ARF-INK4a locus, inhibiting cellular senescence.
While lincRNA-p21, PANDA, and several other lncRNAs may function in an important p53-dependent pathway in the DDR, it is tempting to speculate that other lncRNAs also play key roles in numerous other tumor suppressor and oncogenic pathways, representing a hitherto unknown paradigm in tumorigenesis.
Nuclear-cytoplasmic distribution and nuclear export/import of miRNAs in the DDR
Post-transcriptional processing has been shown to play a key role in the regulation of miRNA levels in the DDR (76, 212). While the processing of pri-miRNAs into pre-miRNAs is increased by the Drosha-p68/p72/p53 complex or the Drosha-KSRP complex in the nucleus, accumulated pre-miRNAs should be exported from the nucleus to the cytoplasm for further processing by the Dicer complex. Thus, XPO5-mediated nuclear export of pre-miRNAs represents a rate-limiting step for miRNA maturation (240). Previous studies have shown that depletion of Xpo5 resulted in a global reduction of miRNAs (138). Recently, Melo et al. identified a genetic defect on the Xpo5 gene in a subset of colorectal cancers with microsatellite instability, which traps pre-miRNAs in the nucleus, thereby reducing miRNA processing (143). They further found that haploinsufficient XPO5 failed to sustain the mature miRNA levels required to control cell differentiation and proliferation, and promoted tumorigenesis. Our group demonstrated that XPO5-mediated pre-miRNA nuclear export is promoted in an ATM-dependent manner after DNA damage (data unpublished). On DNA damage, we observed enhanced interaction between the nucleopore and XPO5 complex. This interaction accelerated the nuclear export of pre-miRNAs, thus facilitating more production of mature miRNAs in response to DNA damage. It is noted that some mature miRNAs are required to function in the nucleus. Therefore, import of these mature miRNAs to the nucleus is necessary in the DDR. CRM1 (or XPO1) was recently shown to interact with the Argonaute complex and to mediate nuclear-cytoplasmic shuttling of mature miRNAs (28). Inhibition of CRM1 resulted in an increased accumulation of miRNA guide sequences in the nucleus. It would be of great interest to investigate the modulation of CRM1-mediated miRNA transport after DNA damage.
Post-Transcriptional Regulation of ncRNA Expression in the DDR
miRNA genes are initially transcribed to yield a primary, long transcript that is sequentially processed in both the nucleus and cytoplasm. Nuclear processing is mediated by the RNase III enzyme Drosha–DGCR8 microprocessor complex to generate pre-miRNAs of ∼70-nt in length. Pre-miRNAs are subsequently transported to the cytoplasm by export 5-Ran-GTP, where they are cleaved by the RNase III enzyme Dicer to generate mature miRNAs (105). Accumulating evidence shows that post-transcriptional processing may play a major role in the regulation of miRNA levels. Recent studies have shown that DNA damage leads to increased levels of some pre-miRNAs and mature miRNAs without producing significant changes in their primary transcripts (195, 250). These findings suggest that the DDR is involved in this post-transcriptional processing of miRNAs by modulating the activity or stability of the processing machines and that there are functional connections between the DDR and miRNA maturation (76, 132, 212).
Major components in the Dicer and Drosha complexes and their regulation in the DDR
Drosha and Dicer plays a central role in miRNA biogenesis, and their expression levels directly influence the clinical outcomes of malignant cancer (147). Drosha interacts with DGCR8, a double-stranded RNA-binding domain protein, to form a complex called the microprocessor. They regulate each other post-transcriptionally. The Drosha–DGCR8 complex destabilizes DGCR8 mRNA by cleaving its hairpin structure. DGCR8, in turn, stabilizes Drosha through protein–protein interaction (77). This mutual regulation between Drosha and DGCR8 may contribute to the homeostatic control of miRNA biogenesis. Other major components in the miRNA maturation machinery are also under control in the cell. A recent study reported that oxidative stress-responsive heme oxygenase-1 (HMOX1) can modulate miRNA processing by down-regulating DGCR8 protein levels (109). Pothof et al. observed that UV damage triggered a cell cycle-dependent relocalization of Ago2 into stress granules and promoted miRNA expression in a partially ATM/ATR-independent manner (170). The p38 mitogen-activated protein kinase (MAPK) pathway and MAPK/Erk signaling regulate the stability of Ago2 and the Dicer-TRBP complex, respectively (161, 244). Phosphorylated TRBP stabilizes the Dicer-TRBP complex and increases mature miRNA production. Remarkably, Erk and MAPKs are phosphorylated and activated after DNA damage (47), which suggests a connection between the DDR and Dicer complex activity. TAp63, the transactivation isoform of p63, binds to the promoter of Dicer and transactivates its expression (191), revealing a direct regulation of Dicer expression.
Regulatory factors in the Dicer and Drosha complexes and their regulation in the DDR
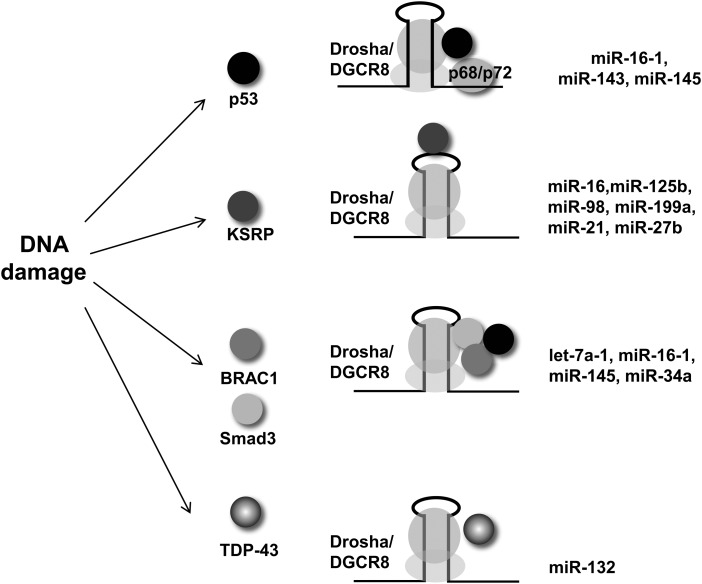
In addition to the major components Drosha and DGCR8, the microprocessor contains a variety of cofactors in this large complex around RNA. These cofactors have been suggested to promote the fidelity, specificity, and activity of the Drosha-mediated cleavage of pri-miRNAs (187). In particular, many of these accessory factors are also involved in the DDR. DEAD box RNA helicases p68 (DDX5) and p72 (DDX17) were identified in the Drosha complex and found to contribute to the efficient processing of a subset of pri-miRNAs into the corresponding mature miRNAs such as miR-16-1, miR-143, and miR-145 (56, 69). In addition to regulating transcription, p53 regulates the processing of miRNAs by binding to Drosha. A direct interaction between p53 and p68/p72 facilitates p53's promotion of miRNA processing under conditions of DNA damage (195) (Fig. 5). Recent studies from our group provide direct evidence that as many as a quarter of miRNAs are significantly induced on DNA damage in an ATM-dependent manner (250). Among these induced miRNAs, a cohort of miRNAs associated with KH-type splicing regulatory protein (KSRP), a key component of both the Drosha and Dicer complexes, was identified (201). Trabucchi et al. presented compelling evidence that KSRP promotes maturation of this selected group of miRNA precursors (201). KSRP binds with a high affinity to the terminal loop of these miRNAs and interacts with both Drosha and Dicer. As a key kinase for the initiation of the DNA damage signaling cascade, ATM directly binds to and phosphorylates KSRP, leading to enhanced interaction between KSRP and pri-miRNAs and increased KSRP activity in miRNA processing (250). These findings strongly support the hypotheses that ATM functions as a major switch for the activity of KSRP in miRNA biogenesis, and that KSRP acts as a molecular gatekeeper which accelerates the production of a subset of miRNAs that regulate cell activities in response to DNA damage.
FIG. 5.
Components in the DDR regulate the function of the Drosha complex in miRNA biogenesis. Many cofactors in the Drosha microprocessor promote its processing activity. DNA damage activates/induces these regulatory proteins that translate DNA damage signal to miRNA expression.
Other regulatory molecules that respond to DNA damage signals have recently been identified in the post-transcriptional regulation of miRNA expression. RNA-binding-motif protein 38 (RBM38), which is regulated by p53, may affect miRNA target gene selection by competing with miRNAs for binding to 3′-UTRs of target mRNAs. RBM38 is required for optimal induction of G1 cell-cycle arrest after DNA damage by protecting the 3′-UTR of the p53 target, p21, from binding by miRNAs. This study suggested that RBM38 decreases miRNA accessibility in a number of p53-induced transcripts, allowing an optimal target gene induction and cell-cycle control (121). BRCA1, a human tumor suppressor protein, interacts with the Drosha microprocessor complex and Smad3/p53/DHX9 and accelerates the processing of primary miRNA transcripts such as let-7a-1, miR-16-1, miR-145, and miR-34a (101). TAR DNA-binding protein 43 (TDP-43) facilitates the processing of a subset of pri-miRNAs and pre-miRNAs through its interaction with Drosha and Dicer complexes and the relevant miRNAs (100). Interestingly, TRP-43 also regulates Cdk6 messenger RNA level by interacting with a DNA damage-induced lncRNA, gadd7. Interaction of gadd7 with TRP-43 blocks the interaction of TDP-43 with Cdk6 mRNA at the post-transcriptional stage (129). This study demonstrated a unique type of regulation: TDP-43 modulates the DDR through miRNA biogenesis and DNA damage-induced lncRNA.
Although we now know that miRNA expression is regulated transcriptionally and post-transcriptionally in the DDR, many important questions remain to be addressed. In particular, it remains largely unknown how miRNA biogenesis responds to DNA damage for p53- or KSRP-independent miRNAs. There could be other mechanisms that account for the induction of those miRNAs. Further studies will provide insights into the molecular mechanisms by which DNA damage signaling is linked to miRNA biogenesis.
miRNA-lncRNA interaction in the DDR
Recent evidence suggests that some lncRNAs might potentially interact with other classes of ncRNAs, including miRNAs, and modulate their regulatory role. LncRNAs may regulate gene expression post-transcriptionally by directly binding to miRNAs, acting as a “sponge” to prevent specific miRNAs from binding to their target mRNAs (31, 96). These findings have revealed a novel layer of regulation in that distinct classes of ncRNAs interplay with each other and cooperate to regulate gene expression. A transcriptome scale analysis on miRNA-lncRNA interactions was recently conducted to study the role of lncRNA as a layer of regulatory interactions with miRNAs (94). However, extensive and accumulated data will be needed to identify crosstalk between miRNAs and lncRNAs in the DDR signaling pathway.
ncRNAs in Genome Instability and Human Cancers
miRNA and lncRNA gene variation in human cancers
Cancer is a disease involving multistep changes in the genome. Increasing evidence shows that expression of miRNAs and lncRNAs is deregulated in human cancer (51, 91). The first direct evidence for an involvement of miRNAs in human cancer derived from studies on chronic lymphocytic leukemia (CLL), in an attempt to identify tumor suppressor genes at chromosome 13q14, which was frequently deleted in about 60% of the cases. By examining this recurring deletion, Dr. Croce's group found that two miRNAs, miR-15a and miR-16-1, other than protein-coding genes, were located within the smallest minimal common region of deletion. Thus, this result provided the evidence that deletion of miRNAs could be involved in the pathogenesis of human cancers. Following these initial observations, several groups reported that many miRNAs in regions of chromosomal instability (amplification, translocation, or deletion) or nearby chromosomal breakpoints are prone to deletions or amplifications (24, 247). These cancer-associated miRNA variations include amplification of the miR-17-92 cluster in lymphoma (197), translocation of miR-17-92 in T-cell acute lymphoblastic leukaemia (T-ALL) (140), and amplification of miR-26a in glioblastoma (90). Overall, about half of miRNA genes are located in cancer-associated genomic regions or in fragile sites (24). In addition, recent studies suggest that epigenetic modifications of miRNA genes cause their dysregulation in human cancers (248). Finally, several key proteins in the miRNA biogenesis pathway, such as Dicer and Ago2, may be dysfunctional or dysregulated in cancer, which may promote tumorigenesis (247). Therefore, variations in DNA copy numbers and epigenetic markers or defects in the miRNA biogenesis machinery might contribute to miRNA dysregulation in human cancer.
Somatic mutation of lncRNAs in cancer is not well explored either. Several initial studies showed that lncRNAs may be a target for genomic amplification/deletion, somatic point mutations, or other targeted aberrations in cancer. For example, the co-amplification of MYC and the lncRNA gene PVT1 in the chromosome region of 8q24 contributes to serous ovarian adenocarcinoma and breast cancer pathophysiology. This amplification has also been associated with reduced survival. However, inhibition of PVT1, but not MYC, induces an amplification/overexpression-specific apoptotic response, suggesting that PVT1 exerts its pathophysiologic influence independently as an ncRNA (71). In addition, a GAS5-BCL6 gene fusion, resulting from a chromosomal translocation and retaining the full coding sequence of BCL6, has been reported in a patient with B-cell lymphoma (153). As another example, Poliseno and colleagues reported that the PTEN pseudogene, PTENP1, is selectively lost in prostate and colon cancers, which leads to dysregulated expression levels of these genes (167).
Roles of the DDR-associated ncRNAs in tumorigenesis
The importance of miRNAs in cancer is underscored by the observation that around half of miRNA genes are located in cancer-associated genomic regions or fragile sites (24, 182), which are frequently amplified or deleted, mutated, or epigenetically silenced in tumorigenesis. Global repression of miRNA processing machinery (Drosha, DGCR8, Exportin-5, Dicer1, and Ago2) promotes cellular transformation, and miRNA processing-impaired cells formed tumors with accelerated kinetics in a mouse model, suggesting a role for mature miRNAs in cancer-associated processes (113, 143, 144). Large-scale miRNA expression profiling of human cancers has revealed that miRNA dysregulation is associated with many types of cancer, including those originating from the blood, brain, thyroid, breast, lung, tongue, nose and pharynx, liver, gastro-intestinal system (esophageal, gastric, pancreatic, and colorectal cancers), and genitourinary system (cervical, ovarian, and prostate cancers) (44, 50, 75, 158). Aberrations in miRNA expression are closely linked to tumorigenesis. A single miRNA can target a variety of mRNAs and can play either an anti-apoptotic or a pro-apoptotic role depending on the cancer type. These characteristics make functional assessment of individual miRNAs difficult. Interestingly, those aforementioned DDR-associated miRNAs show a considerable overlap with frequently dysregulated miRNAs in human solid tumors, suggesting that DNA damage-responsive miRNAs play a role in cancer etiology. Some examples of DDR-associated ncRNAs involved in cancer are summarized in Table 3.
Table 3.
DNA Damage Response-Associated ncRNAs and Their Targets in Human Cancer
| Name | Function | Target/interactor | Cancer phenotype | Cancer type | Refs. |
|---|---|---|---|---|---|
| let-7 | TS | H-RAS, HMGA2 | Induces apoptosis | Lung, breast | (198, 241) |
| miR-15/16 | TS | Bcl-2, Wt-1 | Induces apoptosis and decreases tumorigenicity | Chronic lymphocytic leukemia, pituitary adenoma | (17, 23, 41) |
| miR-155 | OG | c-maf | Induces lymph proliferation | Chronic lymphocytic leukemia, lung, breast | (61, 92, 235) |
| miR-17∼92 | OG | E2F1, Bim, PTEN | Co-operates with c-myc to induce lymphoma in mice, promotes lymph proliferative disorder, and enhances cell proliferation | Lymphoma, lung | (80, 145, 226) |
| miR-21 | OG | PTEN, PDCD4, TPM1 | Induces apoptosis and decreases tumorigenicity | Breast, cholangiocarcinoma, chronic lymphocytic leukemia, lung | (57, 146, 186, 252) |
| miR-221 | OG | P27, p57 | Promotes aggressive growth and cell proliferation | Hepatocellular carcinoma, glioblastoma | (38, 54, 66) |
| miR-29 | TS | TCL-1, MCL1, DNMT3s | Induces apoptosis and decreases tumorigenicity | Non-small cell lung cancer, cholangiocarcinoma, lung | (151, 235) |
| miR-34 | TS | E2F, CDK4, CDK6, cyclin E2 | Induces apoptosis | Gastric, pancreatic, lung, neuroblastoma | (33, 95, 223) |
| miR-372/373 | OG | LATS2 | Promotes tumorigenesis in co-operation with RAS | Testicular tumors | (209) |
| ANRIL | OG | PRC1, PRC2 | Suppresses senescence via INK4A | Prostate, leukemia | (108, 162, 238) |
| LincRNA-p21 | TS | hnRNP-k | Mediates p53 signaling, induces apoptosis | Mouse models of lung, sarcoma, lymphoma | (88) |
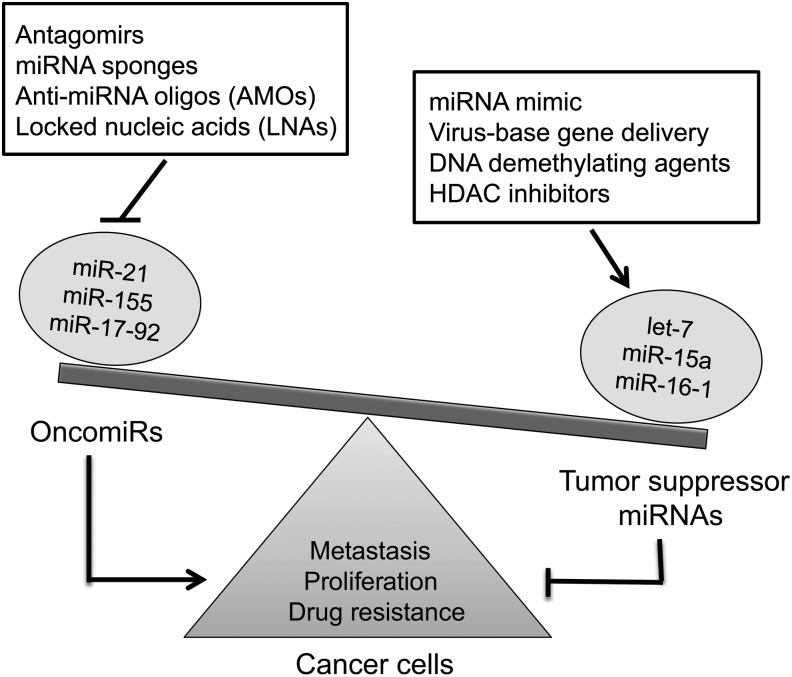
DDR-associated miRNAs have been characterized as tumor suppressors (such as miR-15/16, miR-29, miR-34, and the let-7 family) or oncogenes (such as, miR-155, miR-21, miR-221, and the miR-17-92 cluster) (Table 3 and Fig. 6). The miR-15/16 cluster induces apoptosis by targeting the important anti-apoptotic factor BCL2 at the post-transcriptional level (41). This miRNA cluster is frequently deleted in B-cell CLL, resulting in its down-regulation in the majority of cases of CLL (23). The miR-15-16 cluster has also been reported to be down-regulated in pituitary adenoma (17) and prostate carcinoma (169). Therefore, in these cancers, miR-15/16 expression is preferentially down-regulated to promote tumor progression by inhibiting apoptosis. The oncogenic miR-21 is the most consistently up-regulated miRNA across many cancer types (44, 50, 75, 158). miR-21 was first implicated as an antiapoptotic factor because of the finding that knockdown of miR-21 increased apoptotic cell death in human glioblastoma cells (32) and in a mouse model of breast cancer (186). miR-21 directly targets the tumor suppressor PTEN, which inhibits apoptosis in cancer cells (32). miR-21 was also reported to target PDCD4 (137), a pro-apoptotic gene frequently down-regulated in hepatocellular carcinoma (245), and other important tumor suppressor genes, including tropomyosin 1 (252) and serine peptidase inhibitor, clade B (ovalbumin), and member 5 (SERPINB5) (253). These observations suggested that miR-21 may also play a role in tumor invasion and metastasis. Hypoxia-regulated miRNAs, such as miR-210, are induced in response to low oxygen levels and play a role in cell survival by decreasing the activation of caspases, the central components of apoptotic signaling (112). Since hypoxia is an important feature of the cancer microenvironment, it is interesting that miR-210 is also overexpressed in many cancer types, indicating that hypoxia contributes to miRNA dysregulation in certain cancers. A recent study demonstrated that miR-210 is a good prognostic biomarker for breast cancer (25). In light of the roles of miRNA in apoptosis and cancer, miRNAs in tumorigenesis may not be merely either pro-apoptotic or anti-apoptotic. Rather, the coordination and perhaps synergism between several deregulated miRNAs and their protein-coding counterparts likely facilitate a favorable environment for cancer formation. Further studies that provide a detailed characterization of miRNA target and function will be necessary to improve our understanding of the role of miRNAs in tumorigenesis and to facilitate the design of appropriate therapies targeting this novel group of molecules.
FIG. 6.
miRNA-based therapeutic strategies against cancer. One strategy is to inhibit the function of oncomirs by the use of antigomirs, miRNA sponges, anti-miRNA oligonucleotides, or locked nucleic acids. Another strategy is to restore the activity of tumor suppressor miRNAs through miRNA mimic, virus-based gene delivery, or treatment with DN-demethylating agents and histone deacetylase inhibitors.
DDR-associated lncRNAs function as tumor suppressors and are deregulated in cancer. For example, lincRNA-p21 and MEG3 were found to mediate p53 signaling in the DDR and to induce apoptosis, thus inhibiting cell proliferation in a spectrum of tumors, including lung tumors, sarcoma, lymphoma, meningioma, hepatocellular tumors, leukemia, and pituitary tumors (171). Aberrant expression of the oncogenic lncRNAs uc.73a and PCGEM1 was found to inhibit apoptosis and to promote cell proliferation in leukemia and prostate cancer, respectively (171). An lncRNA suppressed in breast cancer, GAS5, was able to induce apoptosis and growth arrest, and to prevent GR-induced gene expression (171). ANRIL predisposes patients for hereditary cutaneous malignant melanoma (162). Altered ANRIL activity might result in dysregulated silencing of the INK4b/ARF/INK4a locus, thereby contributing to cancer initiation.
piRNA-pathway mutations often lead to a significant overexpression of retrotransposons and a high level of DNA damage. piRNAs and piRNA-like transcripts are involved in tumorigenesis in testicular tissue and a variety of other cancers (40, 136, 234), although their specific functions in tumorigenesis are unknown. Recent studies have implicated Piwi proteins in tumor development. For example, PIWIL1 and PIWIL2 are overexpressed in various cancer types (125, 194, 199). PIWIL1 overexpression is closely related to cell-cycle arrest (130), whereas PIWIL2 overexpression is related to anti-apoptotic signaling and cell proliferation (120). Furthermore, PIWI proteins are associated with stem cell self-renewal (183) and are restored in precancerous stem cells that have the potential for malignant differentiation (35). For some cancers, PIWIL2 overexpression may lead to cisplatin resistance, which might arise due to increasing chromatin condensation that prevents the normal process of DNA repair (218). piRNAs were also found to be aberrantly expressed in human cancer cells (37), implying that the Piwi pathway has a more important function outside germline cells than was originally thought.
Application of ncRNAs as diagnostic biomarkers and therapeutic targets
Oncogenic stress often activates the DDR in premalignant or early-stage tumors. While a large number of ncRNAs are induced or suppressed in the DDR, it is of great interest to identify the roles of these ncRNAs in tumorigenesis. The growing body of literature shows that ncRNAs play fundamental roles in development, differentiation, and malignancy, suggesting that this class of molecules contains potential diagnostic markers and novel targets for therapies. So far, most work in this field has been in the context of the roles of miRNAs in cancer, but there is also considerable interest in applying similar approaches to other ncRNAs.
Recent identification of stable miRNAs in serum, plasma, and other body fluids has paved the way for their use as a promising class of blood-based biomarkers for diagnosing human cancer. In addition to their stability in various body fluids, miRNAs possess several features that make them attractive diagnostic biomarkers. The expression patterns of miRNAs are frequently dysregulated in a variety of diseases, including cancers and appear to be tissue specific, showing signatures related to tumor classification, diagnosis, and progression. For example, a systematic expression analysis by Lu et al. of 217 mammalian miRNAs in various human cancers clearly reflected the developmental lineage and differentiation state of the tumors, and a general down-regulation of miRNAs was observed in tumors compared with normal tissues (134). In addition, Aharonov and coworkers applied the miRNA expression profiles to trace the tissue of origin of cancers of unknown primary origin (174). These findings highlight the potential of miRNA profiling in cancer diagnosis. Another advantage of using secretory miRNAs as biomarkers is that the level of miRNAs can be easily detected and quantified with next-generation sequencing technologies. A number of methods are currently available for the detection and quantification of miRNAs, such as Northern blot analysis, microarray analysis, quantitative real-time polymerase chain reaction, and in situ hybridization.
An important feature of almost all human tumors is the genome instability, which is a condition exacerbated by defects in the DDR. In general, we may exploit this inherent weakness to attack cancer cells, potentially without causing excessive damage to the surrounding normal cells. However, development of drug resistance is a persistent problem in cancer therapies using DNA damaging agents. Given the active roles of ncRNAs in the DDR and their ability to target components of DNA repair, ncRNAs are promising agents to improve efficacy of conventional cancer therapy. Indeed, several studies have shown that aberrant expression of ncRNAs, especially miRNAs, is associated with drug responsiveness, and that manipulation of endogenous ncRNAs expression may overcome chemoresistance in anticancer treatment. For example, restoration of the expression of miR-34a, a direct downstream target of p53, in human gastric cancer cells, restored p53-induced functions and increased chemotherapeutic sensitivity (95). Moreover, miRNA-mediated inhibition of the key sensors and transducers in the DDR, (for examples, ATM is inhibited by miR-101, miR-100 and miR-421 and DNA-PKcs is inhibited by miR-101) sensitized tumor cells to IR in vitro and in vivo (86, 157, 232). Using a mimetic miRNA screening approach, Ashworth and coworkers identified that several miRNAs, including miR-107 and miR-222, regulate the DDR and sensitize tumor cells to PARP inhibitor olaparib by repressing expression of RAD51, thus impairing DSB repair by HR. In addition, elevated expression of miR-107 has been observed in a subset of ovarian clear cell carcinomas, which correlates with PARP inhibitor sensitivity and reduced RAD51 expression, raising the possibility to use these miRNAs as biomarkers to identify patients who may benefit from treatment with PARP inhibitors (156).
A number of approaches have been developed to achieve systematic depletion or restoration of miRNA functions. Suppression of endogenous miRNAs can be reached with multiple strategies, such as locked nucleic acids, anti-miRNA oligonucleotides (AMOs), “miRNA sponges,” and antagomirs (58, 79). In contrast, the restoration of miRNAs can be achieved by ectopic expression of mature miRNAs or their precursors, such as chemically modified miRNA mimic, adenoviral- or lentiviral-based overexpression and nanopartical-based delivery. Given the fact that multiple DNA damage-responsive miRNAs and miRNAs which regulate DDR genes play a role in the development of neoplasia, modulation of the expression of these miRNAs will be a useful therapeutic tool to overcome chemoresistance when treated with DNA damaging agents.
Concluding Remarks
Taken together, we have shown that expression of ncRNAs is regulated transcriptionally and post-transcriptionally in the DDR. These altered ncRNAs, in turn, contribute to the maintenance and modulation of the DDR. However, biological functions of these ncRNAs are often understudied due to the lack of well-controlled cellular systems and genetically engineered animal models. In particular, as the largest group of ncRNAs, lncRNAs have yet to be fully identified as a whole. There are still many important questions to be addressed in the field of ncRNAs. Further studies on the following aspects might help delineate the mechanisms by which DNA damage signaling is linked to ncRNA expression and functions: (i) the interaction between ncRNA gene promoters and their transcriptional complexes; (ii) tissue and cell specificity of ncRNAs in animals; (iii) differential regulation of ncRNA expression in response to various types of DNA damage; (iv) post-transcriptional modifications of ncRNAs and their roles in regulating ncRNA functions; and (v) biological functions of ncRNA in animal development and diseases. Since many questions remain, it is essential that we continue to identify the molecular and cellular mechanisms involved in ncRNA expression and function. Such research will not only lead to a better understanding on the functional roles of DDR but will also provide new insights into many human diseases with DNA damage-processing defects.
Abbreviations Used
- AGO2
argonaute 2
- AMOs
anti-miRNA oligonucleotides
- ATM
ataxia telangiectasia mutated
- ATR
ataxia telangiectasia and Rad3 related
- BACE1
antisense transcript for β-secretase-1
- BER
base excision repair
- CLL
chronic lymphocytic leukemia
- COX3
cytochrome c oxidase III
- CRM1
exportin 1
- DDR
DNA damage response
- DDRNAs
Dicer- and Drosha-dependent small RNAs
- DDX17
DEAD box RNA helicase p72
- DDX5
DEAD box RNA helicase p68
- DGCR8
DiGeorge Syndrome Critical Region Gene 8
- DSB
double-stranded DNA break
- GSHPX
glutathione peroxidase
- HMOX
heme oxygenase-1
- HR
homologous recombination
- IR
ionizing radiation
- Keap1
Kelch-like ECH-associated protein 1
- KSRP
KH-type splicing regulatory protein
- lincRNAs
large or long intergenic ncRNAs
- LNAs
locked nucleic acids
- lncRNAs
long noncoding RNAs
- MAPK
mitogen-activated protein kinase
- miRNAs
microRNAs
- MMP1
matrix metalloproteinase 1
- MMR
mismatch repair
- ncRNAs
noncoding RNAs
- NCS
neocarzinostatin
- NER
nucleotide excision repair
- NHEJ
nonhomologous end joining
- Nrf2
nulcear factor-erythroid 2-related factor 2
- piRNAs
Piwi-interacting RNAs
- pre-miRNAs
precursor miRNAs
- pri-miRNAs
primary miRNAs
- RISC
RNA-induced silencing complex
- ROS
reactive oxygen species
- rRNAs
ribosomal RNAs
- siRNAs
small interfering RNAs
- snoRNAs
small nucleolar RNAs
- SOD
superoxide dismutatese
- T-ALL
T cell acute lymphoblastic leukaemia
- TGFb1
transforming growth factor beta 1
- TRBP
Tar RNA-binding protein
- tRNAs
transfer RNAs
- TXNRD
thioredoxin reductase
- UTR
untranslated region
- UV
ultraviolet
- XPO5
exportin 5
Acknowledgments
This work is supported by grants to X.L. from National Institutes of Health (CA136549) and American Cancer Society (119135-RSG-10-185-01-TBE), by the Odyssey Program and the Cockrell Foundation Award for Scientific Achievement (to C.H.), and by Harold C. and Mary L. Daily Endowment Fellowship Award (to G.W.) at the University of Texas MD Anderson Cancer Center.
References
- 1.Afanasyeva EA, Mestdagh P, Kumps C, Vandesompele J, Ehemann V, Theissen J, Fischer M, Zapatka M, Brors B, Savelyeva L, Sagulenko V, Speleman F, Schwab M, and Westermann F. MicroRNA miR-885-5p targets CDK2 and MCM5, activates p53 and inhibits proliferation and survival. Cell Death Differ 18: 974–984, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Affymetrix ENCODE Transcriptome Project and Cold Spring Harbor Laboratory ENCODE Transcriptome Project Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature 457: 1028–1032, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aguda BD, Kim Y, Piper-Hunter MG, Friedman A, and Marsh CB. MicroRNA regulation of a cancer network: consequences of the feedback loops involving miR-17-92, E2F, and Myc. Proc Natl Acad Sci U S A 105: 19678–19683, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aguilera A. and Gomez-Gonzalez B. Genome instability: a mechanistic view of its causes and consequences. Nat Rev Genet 9: 204–217, 2008 [DOI] [PubMed] [Google Scholar]
- 5.Alexander RP, Fang G, Rozowsky J, Snyder M, and Gerstein MB. Annotating non-coding regions of the genome. Nat Rev Genet 11: 559–571, 2010 [DOI] [PubMed] [Google Scholar]
- 6.Babiarz JE, Ruby JG, Wang Y, Bartel DP, and Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev 22: 2773–2785, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bai XY, Ma Y, Ding R, Fu B, Shi S, and Chen XM. miR-335 and miR-34a Promote renal senescence by suppressing mitochondrial antioxidative enzymes. J Am Soc Nephrol 22: 1252–1261, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bandiera S, Ruberg S, Girard M, Cagnard N, Hanein S, Chretien D, Munnich A, Lyonnet S, and Henrion-Caude A. Nuclear outsourcing of RNA interference components to human mitochondria. PLoS One 6: e20746, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barrey E, Saint-Auret G, Bonnamy B, Damas D, Boyer O, and Gidrol X. Pre-microRNA and mature microRNA in human mitochondria. PLoS One 6: e20220, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beck D, Ayers S, Wen J, Brandl MB, Pham TD, Webb P, Chang CC, and Zhou X. Integrative analysis of next generation sequencing for small non-coding RNAs and transcriptional regulation in Myelodysplastic Syndromes. BMC Med Genomics 4: 19, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bertozzi D, Iurlaro R, Sordet O, Marinello J, Zaffaroni N, and Capranico G. Characterization of novel antisense HIF-1alpha transcripts in human cancers. Cell Cycle 10: 3189–3197, 2011 [DOI] [PubMed] [Google Scholar]
- 13.Bian Z, Li LM, Tang R, Hou DX, Chen X, Zhang CY, and Zen K. Identification of mouse liver mitochondria-associated miRNAs and their potential biological functions. Cell Res 20: 1076–1078, 2010 [DOI] [PubMed] [Google Scholar]
- 14.Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. . Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bommer GT, Gerin I, Feng Y, Kaczorowski AJ, Kuick R, Love RE, Zhai Y, Giordano TJ, Qin ZS, Moore BB, MacDougald OA, Cho KR, and Fearon ER. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol 17: 1298–1307, 2007 [DOI] [PubMed] [Google Scholar]
- 16.Bonci D, Coppola V, Musumeci M, Addario A, Giuffrida R, Memeo L, D'Urso L, Pagliuca A, Biffoni M, Labbaye C, Bartucci M, Muto G, Peschle C, and De Maria R. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med 14: 1271–1277, 2008 [DOI] [PubMed] [Google Scholar]
- 17.Bottoni A, Piccin D, Tagliati F, Luchin A, Zatelli MC, and degli Uberti EC. miR-15a and miR-16-1 down-regulation in pituitary adenomas. J Cell Physiol 204: 280–285, 2005 [DOI] [PubMed] [Google Scholar]
- 18.Brannan CI, Dees EC, Ingram RS, and Tilghman SM. The product of the H19 gene may function as an RNA. Mol Cell Biol 10: 28–36, 1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Braun CJ, Zhang X, Savelyeva I, Wolff S, Moll UM, Schepeler T, Orntoft TF, Andersen CL, and Dobbelstein M. p53-Responsive micrornas 192 and 215 are capable of inducing cell cycle arrest. Cancer Res 68: 10094–10104, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, and Willard HF. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 349: 38–44, 1991 [DOI] [PubMed] [Google Scholar]
- 21.Bulavin DV, Phillips C, Nannenga B, Timofeev O, Donehower LA, Anderson CW, Appella E, and Fornace AJ, Jr., Inactivation of the Wip1 phosphatase inhibits mammary tumorigenesis through p38 MAPK-mediated activation of the p16(Ink4a)-p19(Arf ) pathway. Nat Genet 36: 343–350, 2004 [DOI] [PubMed] [Google Scholar]
- 22.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, and Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 25: 1915–1927, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, and Croce CM. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A 99: 15524–15529, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, and Croce CM. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A 101: 2999–3004, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Camps C, Buffa FM, Colella S, Moore J, Sotiriou C, Sheldon H, Harris AL, Gleadle JM, and Ragoussis J. hsa-miR-210 Is induced by hypoxia and is an independent prognostic factor in breast cancer. Clin Cancer Res 14: 1340–1348, 2008 [DOI] [PubMed] [Google Scholar]
- 26.Caporali A, Meloni M, Vollenkle C, Bonci D, Sala-Newby GB, Addis R, Spinetti G, Losa S, Masson R, Baker AH, Agami R, le Sage C, Condorelli G, Madeddu P, Martelli F, and Emanueli C. Deregulation of microRNA-503 contributes to diabetes mellitus-induced impairment of endothelial function and reparative angiogenesis after limb ischemia. Circulation 123: 282–291, 2011 [DOI] [PubMed] [Google Scholar]
- 27.Carmell MA, Girard A, van de Kant HJ, Bourc'his D, Bestor TH, de Rooij DG, and Hannon GJ. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell 12: 503–514, 2007 [DOI] [PubMed] [Google Scholar]
- 28.Castanotto D, Lingeman R, Riggs AD, and Rossi JJ. CRM1 mediates nuclear-cytoplasmic shuttling of mature microRNAs. Proc Natl Acad Sci U S A 106: 21655–21659, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Castel SE. and Martienssen RA. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat Rev Genet 14: 100–112, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cerutti H. and Casas-Mollano JA. On the origin and functions of RNA-mediated silencing: from protists to man. Curr Genet 50: 81–99, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, and Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147: 358–369, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chan JA, Krichevsky AM, and Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res 65: 6029–6033, 2005 [DOI] [PubMed] [Google Scholar]
- 33.Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, Arking DE, Beer MA, Maitra A, and Mendell JT. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell 26: 745–752, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen H, Lin YW, Mao YQ, Wu J, Liu YF, Zheng XY, and Xie LP. MicroRNA-449a acts as a tumor suppressor in human bladder cancer through the regulation of pocket proteins. Cancer Lett 320: 40–47, 2012 [DOI] [PubMed] [Google Scholar]
- 35.Chen L, Shen R, Ye Y, Pu XA, Liu X, Duan W, Wen J, Zimmerer J, Wang Y, Liu Y, Lasky LC, Heerema NA, Perrotti D, Ozato K, Kuramochi-Miyagawa S, Nakano T, Yates AJ, Carson WE, 3rd, Lin H, Barsky SH, and Gao JX. Precancerous stem cells have the potential for both benign and malignant differentiation. PLoS One 2: e293, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen Q, Chen X, Zhang M, Fan Q, Luo S, and Cao X. miR-137 is frequently down-regulated in gastric cancer and is a negative regulator of Cdc42. Dig Dis Sci 56: 2009–2016, 2011 [DOI] [PubMed] [Google Scholar]
- 37.Cheng J, Guo JM, Xiao BX, Miao Y, Jiang Z, Zhou H, and Li QN. piRNA, the new non-coding RNA, is aberrantly expressed in human cancer cells. Clin Chim Acta 412: 1621–1625, 2011 [DOI] [PubMed] [Google Scholar]
- 38.Ciafre SA, Galardi S, Mangiola A, Ferracin M, Liu CG, Sabatino G, Negrini M, Maira G, Croce CM, and Farace MG. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun 334: 1351–1358, 2005 [DOI] [PubMed] [Google Scholar]
- 39.Ciccia A. and Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell 40: 179–204, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cichocki F, Lenvik T, Sharma N, Yun G, Anderson SK, and Miller JS. Cutting edge: KIR antisense transcripts are processed into a 28-base PIWI-like RNA in human NK cells. J Immunol 185: 2009–2012, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, and Croce CM. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A 102: 13944–13949, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Congrains A, Kamide K, Katsuya T, Yasuda O, Oguro R, Yamamoto K, Ohishi M, and Rakugi H. CVD-associated non-coding RNA, ANRIL, modulates expression of atherogenic pathways in VSMC. Biochem Biophys Res Commun 419: 612–616, 2012 [DOI] [PubMed] [Google Scholar]
- 43.Costa FF. Non-coding RNAs: Meet thy masters. Bioessays 32: 599–608, 2010 [DOI] [PubMed] [Google Scholar]
- 44.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet 10: 704–714, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Crosby ME, Kulshreshtha R, Ivan M, and Glazer PM. MicroRNA regulation of DNA repair gene expression in hypoxic stress. Cancer Res 69: 1221–1229, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Oliveira PE, Zhang L, Wang Z, and Lazo JS. Hypoxia-mediated regulation of Cdc25A phosphatase by p21 and miR-21. Cell Cycle 8: 3157–3164, 2009 [DOI] [PubMed] [Google Scholar]
- 47.Dent P, Yacoub A, Fisher PB, Hagan MP, and Grant S. MAPK pathways in radiation responses. Oncogene 22: 5885–5896, 2003 [DOI] [PubMed] [Google Scholar]
- 48.Ding DP, Chen ZL, Zhao XH, Wang JW, Sun J, Wang Z, Tan FW, Tan XG, Li BZ, Zhou F, Shao K, Li N, Qiu B, and He J. miR-29c induces cell cycle arrest in esophageal squamous cell carcinoma by modulating cyclin E expression. Carcinogenesis 32: 1025–1032, 2011 [DOI] [PubMed] [Google Scholar]
- 49.Eades G, Yang M, Yao Y, Zhang Y, and Zhou Q. miR-200a regulates Nrf2 activation by targeting Keap1 mRNA in breast cancer cells. J Biol Chem 286: 40725–40733, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Esquela-Kerscher A. and Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer 6: 259–269, 2006 [DOI] [PubMed] [Google Scholar]
- 51.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet 12: 861–874, 2011 [DOI] [PubMed] [Google Scholar]
- 52.Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G, 3rd, Kenny PJ, and Wahlestedt C. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of beta-secretase. Nat Med 14: 723–730, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Feng L, Xie Y, Zhang H, and Wu Y. miR-107 targets cyclin-dependent kinase 6 expression, induces cell cycle G1 arrest and inhibits invasion in gastric cancer cells. Med Oncol 29: 856–863, 2012 [DOI] [PubMed] [Google Scholar]
- 54.Fornari F, Gramantieri L, Ferracin M, Veronese A, Sabbioni S, Calin GA, Grazi GL, Giovannini C, Croce CM, Bolondi L, and Negrini M. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene 27: 5651–5661, 2008 [DOI] [PubMed] [Google Scholar]
- 55.Francia S, Michelini F, Saxena A, Tang D, de Hoon M, Anelli V, Mione M, Carninci P, and d'Adda di Fagagna F. Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature 488: 231–235, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fukuda T, Yamagata K, Fujiyama S, Matsumoto T, Koshida I, Yoshimura K, Mihara M, Naitou M, Endoh H, Nakamura T, Akimoto C, Yamamoto Y, Katagiri T, Foulds C, Takezawa S, Kitagawa H, Takeyama K, O'Malley BW, and Kato S. DEAD-box RNA helicase subunits of the Drosha complex are required for processing of rRNA and a subset of microRNAs. Nat Cell Biol 9: 604–611, 2007 [DOI] [PubMed] [Google Scholar]
- 57.Fulci V, Chiaretti S, Goldoni M, Azzalin G, Carucci N, Tavolaro S, Castellano L, Magrelli A, Citarella F, Messina M, Maggio R, Peragine N, Santangelo S, Mauro FR, Landgraf P, Tuschl T, Weir DB, Chien M, Russo JJ, Ju J, Sheridan R, Sander C, Zavolan M, Guarini A, Foa R, and Macino G. Quantitative technologies establish a novel microRNA profile of chronic lymphocytic leukemia. Blood 109: 4944–4951, 2007 [DOI] [PubMed] [Google Scholar]
- 58.Galasso M, Sana ME, and Volinia S. Non-coding RNAs: a key to future personalized molecular therapy? Genome Med 2: 12, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Galluzzi L, Morselli E, Vitale I, Kepp O, Senovilla L, Criollo A, Servant N, Paccard C, Hupe P, Robert T, Ripoche H, Lazar V, Harel-Bellan A, Dessen P, Barillot E, and Kroemer G. miR-181a and miR-630 regulate cisplatin-induced cancer cell death. Cancer Res 70: 1793–1803, 2010 [DOI] [PubMed] [Google Scholar]
- 60.Garcia AI, Buisson M, Bertrand P, Rimokh R, Rouleau E, Lopez BS, Lidereau R, Mikaelian I, and Mazoyer S. Down-regulation of BRCA1 expression by miR-146a and miR-146b-5p in triple negative sporadic breast cancers. EMBO Mol Med 3: 279–290, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Garzon R, Volinia S, Liu CG, Fernandez-Cymering C, Palumbo T, Pichiorri F, Fabbri M, Coombes K, Alder H, Nakamura T, Flomenberg N, Marcucci G, Calin GA, Kornblau SM, Kantarjian H, Bloomfield CD, Andreeff M, and Croce CM. MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood 111: 3183–3189, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Georges SA, Biery MC, Kim SY, Schelter JM, Guo J, Chang AN, Jackson AL, Carleton MO, Linsley PS, Cleary MA, and Chau BN. Coordinated regulation of cell cycle transcripts by p53-Inducible microRNAs, miR-192 and miR-215. Cancer Res 68: 10105–10112, 2008 [DOI] [PubMed] [Google Scholar]
- 63.Ghildiyal M, Seitz H, Horwich MD, Li C, Du T, Lee S, Xu J, Kittler EL, Zapp ML, Weng Z, and Zamore PD. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science 320: 1077–1081, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gibb EA, Brown CJ, and Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer 10: 38, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gibcus JH, Kroesen BJ, Koster R, Halsema N, de Jong D, de Jong S, Poppema S, Kluiver J, Diepstra A, and van den Berg A. MiR-17/106b seed family regulates p21 in Hodgkin's lymphoma. J Pathol 225: 609–617, 2011 [DOI] [PubMed] [Google Scholar]
- 66.Gillies JK. and Lorimer IA. Regulation of p27Kip1 by miRNA 221/222 in glioblastoma. Cell Cycle 6: 2005–2009, 2007 [DOI] [PubMed] [Google Scholar]
- 67.Girard A, Madani S, Boukortt F, Cherkaoui-Malki M, Belleville J, and Prost J. Fructose-enriched diet modifies antioxidant status and lipid metabolism in spontaneously hypertensive rats. Nutrition 22: 758–766, 2006 [DOI] [PubMed] [Google Scholar]
- 68.Gramantieri L, Ferracin M, Fornari F, Veronese A, Sabbioni S, Liu CG, Calin GA, Giovannini C, Ferrazzi E, Grazi GL, Croce CM, Bolondi L, and Negrini M. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res 67: 6092–6099, 2007 [DOI] [PubMed] [Google Scholar]
- 69.Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, and Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature 432: 235–240, 2004 [DOI] [PubMed] [Google Scholar]
- 70.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, and Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 34: D140–D144, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guan Y, Kuo WL, Stilwell JL, Takano H, Lapuk AV, Fridlyand J, Mao JH, Yu M, Miller MA, Santos JL, Kalloger SE, Carlson JW, Ginzinger DG, Celniker SE, Mills GB, Huntsman DG, and Gray JW. Amplification of PVT1 contributes to the pathophysiology of ovarian and breast cancer. Clin Cancer Res 13: 5745–5755, 2007 [DOI] [PubMed] [Google Scholar]
- 72.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, and Lander ES. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458: 223–227, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, Yang X, Amit I, Meissner A, Regev A, Rinn JL, Root DE, and Lander ES. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477: 295–300, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Guttman M. and Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature 482: 339–346, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hammond SM. MicroRNAs as tumor suppressors. Nat Genet 39: 582–583, 2007 [DOI] [PubMed] [Google Scholar]
- 76.Han C, Wan G, Langley RR, Zhang X, and Lu X. Crosstalk between the DNA damage response pathway and microRNAs. Cell Mol Life Sci 69: 2895–2906, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Han J, Pedersen JS, Kwon SC, Belair CD, Kim YK, Yeom KH, Yang WY, Haussler D, Blelloch R, and Kim VN. Posttranscriptional crossregulation between Drosha and DGCR8. Cell 136: 75–84, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Haque R, Chun E, Howell JC, Sengupta T, Chen D, and Kim H. MicroRNA-30b-mediated regulation of catalase expression in human ARPE-19 cells. PLoS One 7: e42542, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Harries LW. Long non-coding RNAs and human disease. Biochem Soc Trans 40: 902–906, 2012 [DOI] [PubMed] [Google Scholar]
- 80.Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, Yatabe Y, Kawahara K, Sekido Y, and Takahashi T. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res 65: 9628–9632, 2005 [DOI] [PubMed] [Google Scholar]
- 81.He JF, Luo YM, Wan XH, and Jiang D. Biogenesis of MiRNA-195 and its role in biogenesis, the cell cycle, and apoptosis. J Biochem Mol Toxicol 25: 404–408, 2011 [DOI] [PubMed] [Google Scholar]
- 82.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL, Linsley PS, Chen C, Lowe SW, Cleary MA, and Hannon GJ. A microRNA component of the p53 tumour suppressor network. Nature 447: 1130–1134, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hermeking H. MicroRNAs in the p53 network: micromanagement of tumour suppression. Nat Rev Cancer 12: 613–626, 2012 [DOI] [PubMed] [Google Scholar]
- 84.Hong L, Lai M, Chen M, Xie C, Liao R, Kang YJ, Xiao C, Hu WY, Han J, and Sun P. The miR-17-92 cluster of microRNAs confers tumorigenicity by inhibiting oncogene-induced senescence. Cancer Res 70: 8547–8557, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hopper AK. and Phizicky EM. tRNA transfers to the limelight. Genes Dev 17: 162–180, 2003 [DOI] [PubMed] [Google Scholar]
- 86.Hu H, Du L, Nagabayashi G, Seeger RC, and Gatti RA. ATM is down-regulated by N-Myc-regulated microRNA-421. Proc Natl Acad Sci U S A 107: 1506–1511, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hu W, Chan CS, Wu R, Zhang C, Sun Y, Song JS, Tang LH, Levine AJ, and Feng Z. Negative regulation of tumor suppressor p53 by microRNA miR-504. Mol Cell 38: 689–699, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, Attardi LD, Regev A, Lander ES, Jacks T, and Rinn JL. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 142: 409–419, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, Horlings HM, Shah N, Umbricht C, Wang P, Kong B, Langerod A, Borresen-Dale AL, Kim SK, van de Vijver M, Sukumar S, Whitfield ML, Kellis M, Xiong Y, Wong DJ, and Chang HY. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet 43: 621–629, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Huse JT, Brennan C, Hambardzumyan D, Wee B, Pena J, Rouhanifard SH, Sohn-Lee C, le Sage C, Agami R, Tuschl T, and Holland EC. The PTEN-regulating microRNA miR-26a is amplified in high-grade glioma and facilitates gliomagenesis in vivo. Genes Dev 23: 1327–1337, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Iorio MV. and Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med 4: 143–159, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, Menard S, Palazzo JP, Rosenberg A, Musiani P, Volinia S, Nenci I, Calin GA, Querzoli P, Negrini M, and Croce CM. MicroRNA gene expression deregulation in human breast cancer. Cancer Res 65: 7065–7070, 2005 [DOI] [PubMed] [Google Scholar]
- 93.Ivanovska I, Ball AS, Diaz RL, Magnus JF, Kibukawa M, Schelter JM, Kobayashi SV, Lim L, Burchard J, Jackson AL, Linsley PS, and Cleary MA. MicroRNAs in the miR-106b family regulate p21/CDKN1A and promote cell cycle progression. Mol Cell Biol 28: 2167–2174, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jalali S, Bhartiya D, Lalwani MK, Sivasubbu S, and Scaria V. Systematic Transcriptome Wide Analysis of lncRNA-miRNA Interactions. PLoS One 8: e53823, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ji Q, Hao X, Meng Y, Zhang M, Desano J, Fan D, and Xu L. Restoration of tumor suppressor miR-34 inhibits human p53-mutant gastric cancer tumorspheres. BMC Cancer 8: 266, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Juan L, Wang G, Radovich M, Schneider BP, Clare SE, Wang Y, and Liu Y. Potential roles of microRNAs in regulating long intergenic noncoding RNAs. BMC Med Genomics 6Suppl 1: S7, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kalhor HR. and Clarke S. Novel methyltransferase for modified uridine residues at the wobble position of tRNA. Mol Cell Biol 23: 9283–9292, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kang MA, So EY, Simons AL, Spitz DR, and Ouchi T. DNA damage induces reactive oxygen species generation through the H2AX-Nox1/Rac1 pathway. Cell Death Dis 3: e249, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, Suzuki H, Carninci P, Hayashizaki Y, Wells C, Frith M, Ravasi T, Pang KC, Hallinan J, Mattick J, Hume DA, Lipovich L, Batalov S, Engstrom PG, Mizuno Y, Faghihi MA, Sandelin A, Chalk AM, Mottagui-Tabar S, Liang Z, Lenhard B, and Wahlestedt C. Antisense transcription in the mammalian transcriptome. Science 309: 1564–1566, 2005 [DOI] [PubMed] [Google Scholar]
- 100.Kawahara Y. and Mieda-Sato A. TDP-43 promotes microRNA biogenesis as a component of the Drosha and Dicer complexes. Proc Natl Acad Sci U S A 109: 3347–3352, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kawai S. and Amano A. BRCA1 regulates microRNA biogenesis via the DROSHA microprocessor complex. J Cell Biol 197: 201–208, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kazak L, Reyes A, and Holt IJ. Minimizing the damage: repair pathways keep mitochondrial DNA intact. Nat Rev Mol Cell Biol 13: 659–671, 2012 [DOI] [PubMed] [Google Scholar]
- 103.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, Regev A, Lander ES, and Rinn JL. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A 106: 11667–11672, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kim JW, Mori S, and Nevins JR. Myc-induced microRNAs integrate Myc-mediated cell proliferation and cell fate. Cancer Res 70: 4820–4828, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kim VN, Han J, and Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 10: 126–139, 2009 [DOI] [PubMed] [Google Scholar]
- 106.Kirkinezos IG. and Moraes CT. Reactive oxygen species and mitochondrial diseases. Semin Cell Dev Biol 12: 449–457, 2001 [DOI] [PubMed] [Google Scholar]
- 107.Klein U, Lia M, Crespo M, Siegel R, Shen Q, Mo T, Ambesi-Impiombato A, Califano A, Migliazza A, Bhagat G, and Dalla-Favera R. The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell 17: 28–40, 2010 [DOI] [PubMed] [Google Scholar]
- 108.Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, and Xiong Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene 30: 1956–1962, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kozakowska M, Ciesla M, Stefanska A, Skrzypek K, Was H, Jazwa A, Grochot-Przeczek A, Kotlinowski J, Szymula A, Bartelik A, Mazan M, Yagensky O, Florczyk U, Lemke K, Zebzda A, Dyduch G, Nowak W, Szade K, Stepniewski J, Majka M, Derlacz R, Loboda A, Dulak J, and Jozkowicz A. Heme oxygenase-1 inhibits myoblast differentiation by targeting myomirs. Antioxid Redox Signal 16: 113–127, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kren BT, Wong PY, Sarver A, Zhang X, Zeng Y, and Steer CJ. MicroRNAs identified in highly purified liver-derived mitochondria may play a role in apoptosis. RNA Biol 6: 65–72, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kriegel AJ, Fang Y, Liu Y, Tian Z, Mladinov D, Matus IR, Ding X, Greene AS, and Liang M. MicroRNA-target pairs in human renal epithelial cells treated with transforming growth factor beta 1: a novel role of miR-382. Nucleic Acids Res 38: 8338–8347, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kulshreshtha R, Ferracin M, Wojcik SE, Garzon R, Alder H, Agosto-Perez FJ, Davuluri R, Liu CG, Croce CM, Negrini M, Calin GA, and Ivan M. A microRNA signature of hypoxia. Mol Cell Biol 27: 1859–1867, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kumar MS, Lu J, Mercer KL, Golub TR, and Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet 39: 673–677, 2007 [DOI] [PubMed] [Google Scholar]
- 114.Lal A, Pan Y, Navarro F, Dykxhoorn DM, Moreau L, Meire E, Bentwich Z, Lieberman J, and Chowdhury D. miR-24-mediated downregulation of H2AX suppresses DNA repair in terminally differentiated blood cells. Nat Struct Mol Biol 16: 492–498, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Landau DA. and Slack FJ. MicroRNAs in mutagenesis, genomic instability, and DNA repair. Semin Oncol 38: 743–751, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lang N, Liu M, Tang QL, Chen X, Liu Z, and Bi F. Effects of microRNA-29 family members on proliferation and invasion of gastric cancer cell lines. Chin J Cancer 29: 603–610, 2010 [DOI] [PubMed] [Google Scholar]
- 117.Le MT, Shyh-Chang N, Khaw SL, Chin L, Teh C, Tay J, O'Day E, Korzh V, Yang H, Lal A, Lieberman J, Lodish HF, and Lim B. Conserved regulation of p53 network dosage by microRNA-125b occurs through evolving miRNA-target gene pairs. PLoS Genet 7: e1002242, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lee HC, Chang SS, Choudhary S, Aalto AP, Maiti M, Bamford DH, and Liu Y. qiRNA is a new type of small interfering RNA induced by DNA damage. Nature 459: 274–277, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lee JT, Davidow LS, and Warshawsky D. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat Genet 21: 400–404, 1999 [DOI] [PubMed] [Google Scholar]
- 120.Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, Koseki H, Fuchikami T, Abe K, Murray HL, Zucker JP, Yuan B, Bell GW, Herbolsheimer E, Hannett NM, Sun K, Odom DT, Otte AP, Volkert TL, Bartel DP, Melton DA, Gifford DK, Jaenisch R, and Young RA. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125: 301–313, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Leveille N, Elkon R, Davalos V, Manoharan V, Hollingworth D, Oude Vrielink J, le Sage C, Melo CA, Horlings HM, Wesseling J, Ule J, Esteller M, Ramos A, and Agami R. Selective inhibition of microRNA accessibility by RBM38 is required for p53 activity. Nat Commun 2: 513, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Li JH, Xiao X, Zhang YN, Wang YM, Feng LM, Wu YM, and Zhang YX. MicroRNA miR-886-5p inhibits apoptosis by down-regulating Bax expression in human cervical carcinoma cells. Gynecol Oncol 120: 145–151, 2011 [DOI] [PubMed] [Google Scholar]
- 123.Liffers ST, Munding JB, Vogt M, Kuhlmann JD, Verdoodt B, Nambiar S, Maghnouj A, Mirmohammadsadegh A, Hahn SA, and Tannapfel A. MicroRNA-148a is down-regulated in human pancreatic ductal adenocarcinomas and regulates cell survival by targeting CDC25B. Lab Invest 91: 1472–1479, 2011 [DOI] [PubMed] [Google Scholar]
- 124.Liu H, Cao YD, Ye WX, and Sun YY. Effect of microRNA-206 on cytoskeleton remodelling by downregulating Cdc42 in MDA-MB-231 cells. Tumori 96: 751–755, 2010 [DOI] [PubMed] [Google Scholar]
- 125.Liu JJ, Shen R, Chen L, Ye Y, He G, Hua K, Jarjoura D, Nakano T, Ramesh GK, Shapiro CL, Barsky SH, and Gao JX. Piwil2 is expressed in various stages of breast cancers and has the potential to be used as a novel biomarker. Int J Clin Exp Pathol 3: 328–337, 2010 [PMC free article] [PubMed] [Google Scholar]
- 126.Liu M, Lang N, Chen X, Tang Q, Liu S, Huang J, Zheng Y, and Bi F. miR-185 targets RhoA and Cdc42 expression and inhibits the proliferation potential of human colorectal cells. Cancer Lett 301: 151–160, 2011 [DOI] [PubMed] [Google Scholar]
- 127.Liu M, Lang N, Qiu M, Xu F, Li Q, Tang Q, Chen J, Chen X, Zhang S, Liu Z, Zhou J, Zhu Y, Deng Y, Zheng Y, and Bi F. miR-137 targets Cdc42 expression, induces cell cycle G1 arrest and inhibits invasion in colorectal cancer cells. Int J Cancer 128: 1269–1279, 2011 [DOI] [PubMed] [Google Scholar]
- 128.Liu Q, Fu H, Sun F, Zhang H, Tie Y, Zhu J, Xing R, Sun Z, and Zheng X. miR-16 family induces cell cycle arrest by regulating multiple cell cycle genes. Nucleic Acids Res 36: 5391–5404, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Liu X, Li D, Zhang W, Guo M, and Zhan Q. Long non-coding RNA gadd7 interacts with TDP-43 and regulates Cdk6 mRNA decay. EMBO J 31: 4415–4427, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Liu X, Sun Y, Guo J, Ma H, Li J, Dong B, Jin G, Zhang J, Wu J, Meng L, and Shou C. Expression of hiwi gene in human gastric cancer was associated with proliferation of cancer cells. Int J Cancer 118: 1922–1929, 2006 [DOI] [PubMed] [Google Scholar]
- 131.Liu X, Yu J, Jiang L, Wang A, Shi F, Ye H, and Zhou X. MicroRNA-222 regulates cell invasion by targeting matrix metalloproteinase 1 (MMP1) and manganese superoxide dismutase 2 (SOD2) in tongue squamous cell carcinoma cell lines. Cancer Genomics Proteomics 6: 131–139, 2009 [PMC free article] [PubMed] [Google Scholar]
- 132.Liu Y. and Lu X. Non-coding RNAs in DNA damage response. Am J Cancer Res 2: 658–675, 2012 [PMC free article] [PubMed] [Google Scholar]
- 133.Lou JJ, Chua YL, Chew EH, Gao J, Bushell M, and Hagen T. Inhibition of hypoxia-inducible factor-1alpha (HIF-1alpha) protein synthesis by DNA damage inducing agents. PLoS One 5: e10522, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, and Golub TR. MicroRNA expression profiles classify human cancers. Nature 435: 834–838, 2005 [DOI] [PubMed] [Google Scholar]
- 135.Lu X, Nannenga B, and Donehower LA. PPM1D dephosphorylates Chk1 and p53 and abrogates cell cycle checkpoints. Genes Dev 19: 1162–1174, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lu Y, Li C, Zhang K, Sun H, Tao D, Liu Y, Zhang S, and Ma Y. Identification of piRNAs in Hela cells by massive parallel sequencing. BMB Rep 43: 635–641, 2010 [DOI] [PubMed] [Google Scholar]
- 137.Lu Z, Liu M, Stribinskis V, Klinge CM, Ramos KS, Colburn NH, and Li Y. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene 27: 4373–4379, 2008 [DOI] [PubMed] [Google Scholar]
- 138.Lund E, Guttinger S, Calado A, Dahlberg JE, and Kutay U. Nuclear export of microRNA precursors. Science 303: 95–98, 2004 [DOI] [PubMed] [Google Scholar]
- 139.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, and Elledge SJ. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316: 1160–1166, 2007 [DOI] [PubMed] [Google Scholar]
- 140.Mavrakis KJ, Wolfe AL, Oricchio E, Palomero T, de Keersmaecker K, McJunkin K, Zuber J, James T, Khan AA, Leslie CS, Parker JS, Paddison PJ, Tam W, Ferrando A, and Wendel HG. Genome-wide RNA-mediated interference screen identifies miR-19 targets in Notch-induced T-cell acute lymphoblastic leukaemia. Nat Cell Biol 12: 372–379, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Meek DW. Tumour suppression by p53: a role for the DNA damage response? Nat Rev Cancer 9: 714–723, 2009 [DOI] [PubMed] [Google Scholar]
- 142.Mei Y, Yong J, Liu H, Shi Y, Meinkoth J, Dreyfuss G, and Yang X. tRNA binds to cytochrome c and inhibits caspase activation. Mol Cell 37: 668–678, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Melo SA, Moutinho C, Ropero S, Calin GA, Rossi S, Spizzo R, Fernandez AF, Davalos V, Villanueva A, Montoya G, Yamamoto H, Schwartz S, Jr., and Esteller M. A genetic defect in exportin-5 traps precursor microRNAs in the nucleus of cancer cells. Cancer Cell 18: 303–315, 2010 [DOI] [PubMed] [Google Scholar]
- 144.Melo SA, Ropero S, Moutinho C, Aaltonen LA, Yamamoto H, Calin GA, Rossi S, Fernandez AF, Carneiro F, Oliveira C, Ferreira B, Liu CG, Villanueva A, Capella G, Schwartz S, Jr., Shiekhattar R, and Esteller M. A TARBP2 mutation in human cancer impairs microRNA processing and DICER1 function. Nat Genet 41: 365–370, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 145.Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell 133: 217–222, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Meng F, Henson R, Lang M, Wehbe H, Maheshwari S, Mendell JT, Jiang J, Schmittgen TD, and Patel T. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology 130: 2113–2129, 2006 [DOI] [PubMed] [Google Scholar]
- 147.Merritt WM, Lin YG, Han LY, Kamat AA, Spannuth WA, Schmandt R, Urbauer D, Pennacchio LA, Cheng JF, Nick AM, Deavers MT, Mourad-Zeidan A, Wang H, Mueller P, Lenburg ME, Gray JW, Mok S, Birrer MJ, Lopez-Berestein G, Coleman RL, Bar-Eli M, and Sood AK. Dicer, Drosha, and outcomes in patients with ovarian cancer. N Engl J Med 359: 2641–2650, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Michel CI, Holley CL, Scruggs BS, Sidhu R, Brookheart RT, Listenberger LL, Behlke MA, Ory DS, and Schaffer JE. Small nucleolar RNAs U32a, U33, and U35a are critical mediators of metabolic stress. Cell Metab 14: 33–44, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Miki H. and Funato Y. Regulation of intracellular signalling through cysteine oxidation by reactive oxygen species. J Biochem 151: 255–261, 2012 [DOI] [PubMed] [Google Scholar]
- 150.Moskwa P, Buffa FM, Pan Y, Panchakshari R, Gottipati P, Muschel RJ, Beech J, Kulshrestha R, Abdelmohsen K, Weinstock DM, Gorospe M, Harris AL, Helleday T, and Chowdhury D. miR-182-mediated downregulation of BRCA1 impacts DNA repair and sensitivity to PARP inhibitors. Mol Cell 41: 210–220, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Mott JL, Kobayashi S, Bronk SF, and Gores GJ. mir-29 regulates Mcl-1 protein expression and apoptosis. Oncogene 26: 6133–6140, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Nakamachi Y, Kawano S, Takenokuchi M, Nishimura K, Sakai Y, Chin T, Saura R, Kurosaka M, and Kumagai S. MicroRNA-124a is a key regulator of proliferation and monocyte chemoattractant protein 1 secretion in fibroblast-like synoviocytes from patients with rheumatoid arthritis. Arthritis Rheum 60: 1294–1304, 2009 [DOI] [PubMed] [Google Scholar]
- 153.Nakamura Y, Takahashi N, Kakegawa E, Yoshida K, Ito Y, Kayano H, Niitsu N, Jinnai I, and Bessho M. The GAS5 (growth arrest-specific transcript 5) gene fuses to BCL6 as a result of t(1;3)(q25;q27) in a patient with B-cell lymphoma. Cancer Genet Cytogenet 182: 144–149, 2008 [DOI] [PubMed] [Google Scholar]
- 154.Nakashima T, Jinnin M, Etoh T, Fukushima S, Masuguchi S, Maruo K, Inoue Y, Ishihara T, and Ihn H. Down-regulation of mir-424 contributes to the abnormal angiogenesis via MEK1 and cyclin E1 in senile hemangioma: its implications to therapy. PLoS One 5: e14334, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Narasimhan M, Patel D, Vedpathak D, Rathinam M, Henderson G, and Mahimainathan L. Identification of novel microRNAs in post-transcriptional control of Nrf2 expression and redox homeostasis in neuronal, SH-SY5Y cells. PLoS One 7: e51111, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Neijenhuis S, Bajrami I, Miller R, Lord CJ, and Ashworth A. Identification of miRNA modulators to PARP inhibitor response. DNA Repair (Amst), 2013 [DOI] [PubMed] [Google Scholar]
- 157.Ng WL, Yan D, Zhang X, Mo YY, and Wang Y. Over-expression of miR-100 is responsible for the low-expression of ATM in the human glioma cell line: M059J. DNA Repair (Amst) 9: 1170–1175, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 158.Nicoloso MS, Spizzo R, Shimizu M, Rossi S, and Calin GA. MicroRNAs—the micro steering wheel of tumour metastases. Nat Rev Cancer 9: 293–302, 2009 [DOI] [PubMed] [Google Scholar]
- 159.Ofir M, Hacohen D, and Ginsberg D. MiR-15 and miR-16 are direct transcriptional targets of E2F1 that limit E2F-induced proliferation by targeting cyclin E. Mol Cancer Res 9: 440–447, 2011 [DOI] [PubMed] [Google Scholar]
- 160.Park SY, Lee JH, Ha M, Nam JW, and Kim VN. miR-29 miRNAs activate p53 by targeting p85 alpha and CDC42. Nat Struct Mol Biol 16: 23–29, 2009 [DOI] [PubMed] [Google Scholar]
- 161.Paroo Z, Ye X, Chen S, and Liu Q. Phosphorylation of the human microRNA-generating complex mediates MAPK/Erk signaling. Cell 139: 112–122, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Pasmant E, Laurendeau I, Heron D, Vidaud M, Vidaud D, and Bieche I. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res 67: 3963–3969, 2007 [DOI] [PubMed] [Google Scholar]
- 163.Passos JF, Nelson G, Wang C, Richter T, Simillion C, Proctor CJ, Miwa S, Olijslagers S, Hallinan J, Wipat A, Saretzki G, Rudolph KL, Kirkwood TB, and von Zglinicki T. Feedback between p21 and reactive oxygen production is necessary for cell senescence. Mol Syst Biol 6: 347, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Petrelli A, Perra A, Schernhuber K, Cargnelutti M, Salvi A, Migliore C, Ghiso E, Benetti A, Barlati S, Ledda-Columbano GM, Portolani N, De Petro G, Columbano A, and Giordano S. Sequential analysis of multistage hepatocarcinogenesis reveals that miR-100 and PLK1 dysregulation is an early event maintained along tumor progression. Oncogene 31: 4517–4526, 2012 [DOI] [PubMed] [Google Scholar]
- 165.Pichiorri F, Suh SS, Rocci A, De Luca L, Taccioli C, Santhanam R, Zhou W, Benson DM, Jr., Hofmainster C, Alder H, Garofalo M, Di Leva G, Volinia S, Lin HJ, Perrotti D, Kuehl M, Aqeilan RI, Palumbo A, and Croce CM. Downregulation of p53-inducible microRNAs 192, 194, and 215 impairs the p53/MDM2 autoregulatory loop in multiple myeloma development. Cancer Cell 18: 367–381, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 166.Pierson J, Hostager B, Fan R, and Vibhakar R. Regulation of cyclin dependent kinase 6 by microRNA 124 in medulloblastoma. J Neurooncol 90: 1–7, 2008 [DOI] [PubMed] [Google Scholar]
- 167.Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, and Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 465: 1033–1038, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Ponting CP, Oliver PL, and Reik W. Evolution and functions of long noncoding RNAs. Cell 136: 629–641, 2009 [DOI] [PubMed] [Google Scholar]
- 169.Porkka KP, Pfeiffer MJ, Waltering KK, Vessella RL, Tammela TL, and Visakorpi T. MicroRNA expression profiling in prostate cancer. Cancer Res 67: 6130–6135, 2007 [DOI] [PubMed] [Google Scholar]
- 170.Pothof J, Verkaik NS, van IW, Wiemer EA, Ta VT, van der Horst GT, Jaspers NG, van Gent DC, Hoeijmakers JH, and Persengiev SP. MicroRNA-mediated gene silencing modulates the UV-induced DNA-damage response. EMBO J 28: 2090–2099, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Prensner JR. and Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov 1: 391–407, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Qi J, Yu JY, Shcherbata HR, Mathieu J, Wang AJ, Seal S, Zhou W, Stadler BM, Bourgin D, Wang L, Nelson A, Ware C, Raymond C, Lim LP, Magnus J, Ivanovska I, Diaz R, Ball A, Cleary MA, and Ruohola-Baker H. microRNAs regulate human embryonic stem cell division. Cell Cycle 8: 3729–3741, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, and Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129: 1311–1323, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Rosenfeld N, Aharonov R, Meiri E, Rosenwald S, Spector Y, Zepeniuk M, Benjamin H, Shabes N, Tabak S, Levy A, Lebanony D, Goren Y, Silberschein E, Targan N, Ben-Ari A, Gilad S, Sion-Vardy N, Tobar A, Feinmesser M, Kharenko O, Nativ O, Nass D, Perelman M, Yosepovich A, Shalmon B, Polak-Charcon S, Fridman E, Avniel A, Bentwich I, Bentwich Z, Cohen D, Chajut A, and Barshack I. MicroRNAs accurately identify cancer tissue origin. Nat Biotechnol 26: 462–469, 2008 [DOI] [PubMed] [Google Scholar]
- 175.Sachdeva M, Zhu S, Wu F, Wu H, Walia V, Kumar S, Elble R, Watabe K, and Mo YY. p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc Natl Acad Sci U S A 106: 3207–3212, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Saleh AD, Savage JE, Cao L, Soule BP, Ly D, DeGraff W, Harris CC, Mitchell JB, and Simone NL. Cellular stress induced alterations in microRNA let-7a and let-7b expression are dependent on p53. PLoS One 6: e24429, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Sangokoya C, Telen MJ, and Chi JT. microRNA miR-144 modulates oxidative stress tolerance and associates with anemia severity in sickle cell disease. Blood 116: 4338–4348, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Sarkar S, Dey BK, and Dutta A. MiR-322/424 and -503 are induced during muscle differentiation and promote cell cycle quiescence and differentiation by down-regulation of Cdc25A. Mol Biol Cell 21: 2138–2149, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Scheel AH, Beyer U, Agami R, and Dobbelstein M. Immunofluorescence-based screening identifies germ cell associated microRNA 302 as an antagonist to p63 expression. Cell Cycle 8: 1426–1432, 2009 [DOI] [PubMed] [Google Scholar]
- 180.Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, and Sharp PA. Divergent transcription from active promoters. Science 322: 1849–1851, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Sekiya Y, Ogawa T, Iizuka M, Yoshizato K, Ikeda K, and Kawada N. Down-regulation of cyclin E1 expression by microRNA-195 accounts for interferon-beta-induced inhibition of hepatic stellate cell proliferation. J Cell Physiol 226: 2535–2542, 2011 [DOI] [PubMed] [Google Scholar]
- 182.Sevignani C, Calin GA, Siracusa LD, and Croce CM. Mammalian microRNAs: a small world for fine-tuning gene expression. Mamm Genome 17: 189–202, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Sharma AK, Nelson MC, Brandt JE, Wessman M, Mahmud N, Weller KP, and Hoffman R. Human CD34(+) stem cells express the hiwi gene, a human homologue of the Drosophila gene piwi. Blood 97: 426–434, 2001 [DOI] [PubMed] [Google Scholar]
- 184.Shi XB, Xue L, Ma AH, Tepper CG, Kung HJ, and White RW. miR-125b promotes growth of prostate cancer xenograft tumor through targeting pro-apoptotic genes. Prostate 71: 538–549, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Shimada K, Nakamura M, Anai S, De Velasco M, Tanaka M, Tsujikawa K, Ouji Y, and Konishi N. A novel human AlkB homologue, ALKBH8, contributes to human bladder cancer progression. Cancer Res 69: 3157–3164, 2009 [DOI] [PubMed] [Google Scholar]
- 186.Si ML, Zhu S, Wu H, Lu Z, Wu F, and Mo YY. miR-21-mediated tumor growth. Oncogene 26: 2799–2803, 2007 [DOI] [PubMed] [Google Scholar]
- 187.Siomi H. and Siomi MC. Posttranscriptional regulation of microRNA biogenesis in animals. Mol Cell 38: 323–332, 2010 [DOI] [PubMed] [Google Scholar]
- 188.Song L, Lin C, Wu Z, Gong H, Zeng Y, Wu J, Li M, and Li J. miR-18a impairs DNA damage response through downregulation of ataxia telangiectasia mutated (ATM) kinase. PLoS One 6: e25454, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Sotomaru Y, Katsuzawa Y, Hatada I, Obata Y, Sasaki H, and Kono T. Unregulated expression of the imprinted genes H19 and Igf2r in mouse uniparental fetuses. J Biol Chem 277: 12474–12478, 2002 [DOI] [PubMed] [Google Scholar]
- 190.Stachurska A, Ciesla M, Kozakowska M, Wolffram S, Boesch-Saadatmandi C, Rimbach G, Jozkowicz A, Dulak J, and Loboda A. Cross-talk between microRNAs, nuclear factor E2-related factor 2, and heme oxygenase-1 in ochratoxin A-induced toxic effects in renal proximal tubular epithelial cells. Mol Nutr Food Res 57: 504–515, 2013 [DOI] [PubMed] [Google Scholar]
- 191.Su X, Chakravarti D, Cho MS, Liu L, Gi YJ, Lin YL, Leung ML, El-Naggar A, Creighton CJ, Suraokar MB, Wistuba I, and Flores ER. TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nature 467: 986–990, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Suh SO, Chen Y, Zaman MS, Hirata H, Yamamura S, Shahryari V, Liu J, Tabatabai ZL, Kakar S, Deng G, Tanaka Y, and Dahiya R. MicroRNA-145 is regulated by DNA methylation and p53 gene mutation in prostate cancer. Carcinogenesis 32: 772–778, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Sun F, Fu H, Liu Q, Tie Y, Zhu J, Xing R, Sun Z, and Zheng X. Downregulation of CCND1 and CDK6 by miR-34a induces cell cycle arrest. FEBS Lett 582: 1564–1568, 2008 [DOI] [PubMed] [Google Scholar]
- 194.Sun G, Wang Y, Sun L, Luo H, Liu N, Fu Z, and You Y. Clinical significance of Hiwi gene expression in gliomas. Brain Res 1373: 183–188, 2011 [DOI] [PubMed] [Google Scholar]
- 195.Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, and Miyazono K. Modulation of microRNA processing by p53. Nature 460: 529–533, 2009 [DOI] [PubMed] [Google Scholar]
- 196.Taft RJ, Simons C, Nahkuri S, Oey H, Korbie DJ, Mercer TR, Holst J, Ritchie W, Wong JJ, Rasko JE, Rokhsar DS, Degnan BM, and Mattick JS. Nuclear-localized tiny RNAs are associated with transcription initiation and splice sites in metazoans. Nat Struct Mol Biol 17: 1030–1034, 2010 [DOI] [PubMed] [Google Scholar]
- 197.Tagawa H. and Seto M. A microRNA cluster as a target of genomic amplification in malignant lymphoma. Leukemia 19: 2013–2016, 2005 [DOI] [PubMed] [Google Scholar]
- 198.Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, Mitsudomi T, and Takahashi T. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res 64: 3753–3756, 2004 [DOI] [PubMed] [Google Scholar]
- 199.Taubert H, Greither T, Kaushal D, Wurl P, Bache M, Bartel F, Kehlen A, Lautenschlager C, Harris L, Kraemer K, Meye A, Kappler M, Schmidt H, Holzhausen HJ, and Hauptmann S. Expression of the stem cell self-renewal gene Hiwi and risk of tumour-related death in patients with soft-tissue sarcoma. Oncogene 26: 1098–1100, 2007 [DOI] [PubMed] [Google Scholar]
- 200.Tian RQ, Wang XH, Hou LJ, Jia WH, Yang Q, Li YX, Liu M, Li X, and Tang H. MicroRNA-372 is down-regulated and targets cyclin-dependent kinase 2 (CDK2) and cyclin A1 in human cervical cancer, which may contribute to tumorigenesis. J Biol Chem 286: 25556–25563, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Trabucchi M, Briata P, Garcia-Mayoral M, Haase AD, Filipowicz W, Ramos A, Gherzi R, and Rosenfeld MG. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature 459: 1010–1014, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Trompeter HI, Abbad H, Iwaniuk KM, Hafner M, Renwick N, Tuschl T, Schira J, Muller HW, and Wernet P. MicroRNAs MiR-17, MiR-20a, and MiR-106b act in concert to modulate E2F activity on cell cycle arrest during neuronal lineage differentiation of USSC. PLoS One 6: e16138, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Truscott M, Islam AB, Lopez-Bigas N, and Frolov MV. mir-11 limits the proapoptotic function of its host gene, dE2f1. Genes Dev 25: 1820–1834, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Ugalde AP, Ramsay AJ, de la Rosa J, Varela I, Marino G, Cadinanos J, Lu J, Freije JM, and Lopez-Otin C. Aging and chronic DNA damage response activate a regulatory pathway involving miR-29 and p53. EMBO J 30: 2219–2232, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Valeri N, Gasparini P, Fabbri M, Braconi C, Veronese A, Lovat F, Adair B, Vannini I, Fanini F, Bottoni A, Costinean S, Sandhu SK, Nuovo GJ, Alder H, Gafa R, Calore F, Ferracin M, Lanza G, Volinia S, Negrini M, McIlhatton MA, Amadori D, Fishel R, and Croce CM. Modulation of mismatch repair and genomic stability by miR-155. Proc Natl Acad Sci U S A 107: 6982–6987, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.van Attikum H. and Gasser SM. Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol 19: 207–217, 2009 [DOI] [PubMed] [Google Scholar]
- 207.Van Houten B, Woshner V, and Santos JH. Role of mitochondrial DNA in toxic responses to oxidative stress. DNA Repair (Amst) 5: 145–152, 2006 [DOI] [PubMed] [Google Scholar]
- 208.van Jaarsveld MT, Helleman J, Boersma AW, van Kuijk PF, van Ijcken WF, Despierre E, Vergote I, Mathijssen RH, Berns EM, Verweij J, Pothof J, and Wiemer EA. miR-141 regulates KEAP1 and modulates cisplatin sensitivity in ovarian cancer cells. Oncogene, 2012. [Epub ahead of print]; DOI: 10.1038/onc.2012.433 [DOI] [PubMed] [Google Scholar]
- 209.Voorhoeve PM, le Sage C, Schrier M, Gillis AJ, Stoop H, Nagel R, Liu YP, van Duijse J, Drost J, Griekspoor A, Zlotorynski E, Yabuta N, De Vita G, Nojima H, Looijenga LH, and Agami R. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell 124: 1169–1181, 2006 [DOI] [PubMed] [Google Scholar]
- 210.Wagner AE, Boesch-Saadatmandi C, Dose J, Schultheiss G, and Rimbach G. Anti-inflammatory potential of allyl-isothiocyanate—role of Nrf2, NF-(kappa) B and microRNA-155. J Cell Mol Med 16: 836–843, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Wan G, Mathur R, Hu X, Liu Y, Zhang X, Peng G, and Lu X. Long non-coding RNA ANRIL (CDKN2B-AS) is induced by the ATM-E2F1 signaling pathway. Cell Signal 25: 1086–1095, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Wan G, Mathur R, Hu X, Zhang X, and Lu X. miRNA response to DNA damage. Trends Biochem Sci 36: 478–484, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Wang F, Fu XD, Zhou Y, and Zhang Y. Down-regulation of the cyclin E1 oncogene expression by microRNA-16-1 induces cell cycle arrest in human cancer cells. BMB Rep 42: 725–730, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Wang F, Yu J, Yang GH, Wang XS, and Zhang JW. Regulation of erythroid differentiation by miR-376a and its targets. Cell Res 21: 1196–1209, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Wang K, Lin ZQ, Long B, Li JH, Zhou J, and Li PF. Cardiac hypertrophy is positively regulated by MicroRNA miR-23a. J Biol Chem 287: 589–599, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, Wysocka J, Lei M, Dekker J, Helms JA, and Chang HY. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472: 120–124, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Wang P, Zou F, Zhang X, Li H, Dulak A, Tomko RJ, Jr., Lazo JS, Wang Z, Zhang L, and Yu J. microRNA-21 negatively regulates Cdc25A and cell cycle progression in colon cancer cells. Cancer Res 69: 8157–8165, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Wang QE, Han C, Milum K, and Wani AA. Stem cell protein Piwil2 modulates chromatin modifications upon cisplatin treatment. Mutat Res 708: 59–68, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Wang X, Arai S, Song X, Reichart D, Du K, Pascual G, Tempst P, Rosenfeld MG, Glass CK, and Kurokawa R. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature 454: 126–130, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Wang Y, Huang JW, Li M, Cavenee WK, Mitchell PS, Zhou X, Tewari M, Furnari FB, and Taniguchi T. MicroRNA-138 modulates DNA damage response by repressing histone H2AX expression. Mol Cancer Res 9: 1100–1111, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Wang Y, Yu Y, Tsuyada A, Ren X, Wu X, Stubblefield K, Rankin-Gee EK, and Wang SE. Transforming growth factor-beta regulates the sphere-initiating stem cell-like feature in breast cancer through miRNA-181 and ATM. Oncogene 30: 1470–1480, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Wei W, Ba Z, Gao M, Wu Y, Ma Y, Amiard S, White CI, Rendtlew Danielsen JM, Yang YG, and Qi Y. A role for small RNAs in DNA double-strand break repair. Cell 149: 101–112, 2012 [DOI] [PubMed] [Google Scholar]
- 223.Welch C, Chen Y, and Stallings RL. MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene 26: 5017–5022, 2007 [DOI] [PubMed] [Google Scholar]
- 224.Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P, Hogenesch JB, and Schultz PG. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science 309: 1570–1573, 2005 [DOI] [PubMed] [Google Scholar]
- 225.Wouters MD, van Gent DC, Hoeijmakers JH, and Pothof J. MicroRNAs, the DNA damage response and cancer. Mutat Res 717: 54–66, 2011 [DOI] [PubMed] [Google Scholar]
- 226.Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, Henderson JM, Kutok JL, and Rajewsky K. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol 9: 405–414, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Xiao J, Lin H, Luo X, and Wang Z. miR-605 joins p53 network to form a p53:miR-605:Mdm2 positive feedback loop in response to stress. EMBO J 30: 524–532, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Xu D, Takeshita F, Hino Y, Fukunaga S, Kudo Y, Tamaki A, Matsunaga J, Takahashi RU, Takata T, Shimamoto A, Ochiya T, and Tahara H. miR-22 represses cancer progression by inducing cellular senescence. J Cell Biol 193: 409–424, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Xu T, Zhu Y, Xiong Y, Ge YY, Yun JP, and Zhuang SM. MicroRNA-195 suppresses tumorigenicity and regulates G1/S transition of human hepatocellular carcinoma cells. Hepatology 50: 113–121, 2009 [DOI] [PubMed] [Google Scholar]
- 230.Xu Y, Fang F, Zhang J, Josson S, St Clair WH, and St Clair DK. miR-17* suppresses tumorigenicity of prostate cancer by inhibiting mitochondrial antioxidant enzymes. PLoS One 5: e14356, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Yamakuchi M, Ferlito M, and Lowenstein CJ. miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci U S A 105: 13421–13426, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Yan D, Ng WL, Zhang X, Wang P, Zhang Z, Mo YY, Mao H, Hao C, Olson JJ, Curran WJ, and Wang Y. Targeting DNA-PKcs and ATM with miR-101 sensitizes tumors to radiation. PLoS One 5: e11397, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Yan HL, Xue G, Mei Q, Wang YZ, Ding FX, Liu MF, Lu MH, Tang Y, Yu HY, and Sun SH. Repression of the miR-17-92 cluster by p53 has an important function in hypoxia-induced apoptosis. EMBO J 28: 2719–2732, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Yan Z, Hu HY, Jiang X, Maierhofer V, Neb E, He L, Hu Y, Hu H, Li N, Chen W, and Khaitovich P. Widespread expression of piRNA-like molecules in somatic tissues. Nucleic Acids Res 39: 6596–6607, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, Calin GA, Liu CG, Croce CM, and Harris CC. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 9: 189–198, 2006 [DOI] [PubMed] [Google Scholar]
- 236.Yang L, Lin C, Liu W, Zhang J, Ohgi KA, Grinstein JD, Dorrestein PC, and Rosenfeld MG. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell 147: 773–788, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Yang M, Yao Y, Eades G, Zhang Y, and Zhou Q. MiR-28 regulates Nrf2 expression through a Keap1-independent mechanism. Breast Cancer Res Treat 129: 983–991, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Yap KL, Li S, Munoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, Gil J, Walsh MJ, and Zhou MM. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell 38: 662–674, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Yi C, Wang Q, Wang L, Huang Y, Li L, Liu L, Zhou X, Xie G, Kang T, Wang H, Zeng M, Ma J, Zeng Y, and Yun JP. MiR-663, a microRNA targeting p21(WAF1/CIP1), promotes the proliferation and tumorigenesis of nasopharyngeal carcinoma. Oncogene 31: 4421–4433, 2012 [DOI] [PubMed] [Google Scholar]
- 240.Yi R, Qin Y, Macara IG, and Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev 17: 3011–3016, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, Huang Y, Hu X, Su F, Lieberman J, and Song E. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell 131: 1109–1123, 2007 [DOI] [PubMed] [Google Scholar]
- 242.Yu Y, Wang Y, Ren X, Tsuyada A, Li A, Liu LJ, and Wang SE. Context-dependent bidirectional regulation of the MutS homolog 2 by transforming growth factor beta contributes to chemoresistance in breast cancer cells. Mol Cancer Res 8: 1633–1642, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Yuan X, Zhou Y, Casanova E, Chai M, Kiss E, Grone HJ, Schutz G, and Grummt I. Genetic inactivation of the transcription factor TIF-IA leads to nucleolar disruption, cell cycle arrest, and p53-mediated apoptosis. Mol Cell 19: 77–87, 2005 [DOI] [PubMed] [Google Scholar]
- 244.Zeng Y, Sankala H, Zhang X, and Graves PR. Phosphorylation of Argonaute 2 at serine-387 facilitates its localization to processing bodies. Biochem J 413: 429–436, 2008 [DOI] [PubMed] [Google Scholar]
- 245.Zhang H, Ozaki I, Mizuta T, Hamajima H, Yasutake T, Eguchi Y, Ideguchi H, Yamamoto K, and Matsuhashi S. Involvement of programmed cell death 4 in transforming growth factor-beta1-induced apoptosis in human hepatocellular carcinoma. Oncogene 25: 6101–6112, 2006 [DOI] [PubMed] [Google Scholar]
- 246.Zhang J, Sun Q, Zhang Z, Ge S, Han ZG, and Chen WT. Loss of microRNA-143/145 disturbs cellular growth and apoptosis of human epithelial cancers by impairing the MDM2-p53 feedback loop. Oncogene 32: 61–69, 2013 [DOI] [PubMed] [Google Scholar]
- 247.Zhang L, Huang J, Yang N, Greshock J, Megraw MS, Giannakakis A, Liang S, Naylor TL, Barchetti A, Ward MR, Yao G, Medina A, O'Brien-Jenkins A, Katsaros D, Hatzigeorgiou A, Gimotty PA, Weber BL, and Coukos G. microRNAs exhibit high frequency genomic alterations in human cancer. Proc Natl Acad Sci U S A 103: 9136–9141, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Zhang L, Volinia S, Bonome T, Calin GA, Greshock J, Yang N, Liu CG, Giannakakis A, Alexiou P, Hasegawa K, Johnstone CN, Megraw MS, Adams S, Lassus H, Huang J, Kaur S, Liang S, Sethupathy P, Leminen A, Simossis VA, Sandaltzopoulos R, Naomoto Y, Katsaros D, Gimotty PA, DeMichele A, Huang Q, Butzow R, Rustgi AK, Weber BL, Birrer MJ, Hatzigeorgiou AG, Croce CM, and Coukos G. Genomic and epigenetic alterations deregulate microRNA expression in human epithelial ovarian cancer. Proc Natl Acad Sci U S A 105: 7004–7009, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Zhang X, Ng WL, Wang P, Tian L, Werner E, Wang H, Doetsch P, and Wang Y. MicroRNA-21 modulates the levels of reactive oxygen species by targeting SOD3 and TNFalpha. Cancer Res 72: 4707–4713, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 250.Zhang X, Wan G, Berger FG, He X, and Lu X. The ATM kinase induces microRNA biogenesis in the DNA damage response. Mol Cell 41: 371–383, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Zhang X, Wan G, Mlotshwa S, Vance V, Berger FG, Chen H, and Lu X. Oncogenic Wip1 phosphatase is inhibited by miR-16 in the DNA damage signaling pathway. Cancer Res 70: 7176–7186, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Zhu S, Si ML, Wu H, and Mo YY. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1). J Biol Chem 282: 14328–14336, 2007 [DOI] [PubMed] [Google Scholar]
- 253.Zhu S, Wu H, Wu F, Nie D, Sheng S, and Mo YY. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res 18: 350–359, 2008 [DOI] [PubMed] [Google Scholar]