SUMMARY
Increasing evidence suggests that linker histone H1 can influence distinct cellular processes by acting as a gene-specific regulator. However, the mechanistic basis underlying such H1 specificity and whether H1 acts in concert with other chromatin-altering activities remain unclear. Here, we show that one of the H1 subtypes, H1.2, stably interacts with Cul4A E3 ubiquitin ligase and PAF1 elongation complexes, and that such interaction potentiates target gene transcription via induction of H4K31ubiquitylation, H3K4me3 and H3K79me2. H1.2, Cul4A and PAF1 are functionally cooperative, as their individual knockdown results in the loss of the corresponding histone marks and the deficiency of target gene transcription. H1.2 interacts with the serine 2 phosphorylated form of RNAPII and we argue that it recruits the Cul4A and PAF1 complexes to target genes by bridging the interaction between the and the Cul4A and PAF1 complexes. These data define an expanded role for H1 in regulating gene transcription and illustrate its dependence on the elongation competence of RNAPII.
Keywords: Histone, Chromatin, Transcription, Methylation, Ubiquitylation, H1, PAF1, Cul4A
INTRODUCTION
Linker histone H1 is a small basic protein that can bind to the nucleosome and provides the structural and functional flexibility of chromatin. A typical H1 structure consists of a central globular domain flanked by unstructured N-terminal and C-terminal tails (Ponte et al., 2003). In vitro studies revealed that incorporation of H1 into chromatin impairs transcription events by stabilizing the nucleosome, controlling nucleosome spacing, and/or folding nucleosome arrays into 30 nm chromatin fiber (Bustin et al., 2005; Georgel et al., 2003). In contrast to this original view, genetic studies in model organisms suggest that H1 is not a global repressor of transcription but rather plays a more dynamic and gene-targeted role, participating in the up- or down-regulation of small groups of genes (Alami et al., 2003; Brown et al., 1996; Fan et al., 2005; Shen and Gorovsky, 1996). How H1 participates in these specific features of transcriptional responses is still largely unknown, but its functional cooperation with other factors is believed to be part of the underlying mechanism.
Human cells contain at least six somatic histone H1 subtypes, H1.1-H1.5 and H1.0, which exhibit significant sequence divergence in N- and C-terminal tails. While each individual H1 subtype is not essential for cell viability, differential localization of H1 subtypes in the nucleus and variations in their relative concentrations among different cell types allow the assumption that each H1 subtype contributes to the regulation of specific gene transcription (Barra et al., 2000; Jedrusik and Schulze, 2007; Zhang et al., 2012b). Besides distinct patterns of expression and localization in different tissues and cell types, H1 has also been postulated to associate with other regulatory proteins to control their activity. For example, mouse H1b is recruited to the MyoD promoter by Msx1 homeoprotein and cooperates with Msx1 in delaying the differentiation of progenitor cells into muscle (Lee et al., 2004). A single H1 variant exists in Drosophila, and it physically recruits Su(var)3-9 histone methyltransferase to establish heterochromatic gene silencing (Lu et al., 2013). Another striking example is the demonstration made by us that human H1.2 forms a stable complex with a group of proteins and regulates p53-mediated transactivation (Kim et al., 2008). All these results implicate the requirement of extra factors in gene-specific action of H1 subtypes, but the detailed mechanisms have not been elucidated.
Cul4A is the E3 ubiquitin ligase that forms a stable complex with DDB1 and ROC1 to catalyze ubiquitylation of a variety of proteins including core histones. Selective depletion of Cul4A reduces the level of H3 and H4 ubiquitylation but has little effect on H2A and H2B ubiquitylation, indicating that Cul4A is the major ubiquitin ligase activity mediating H3 and H4 ubiquitylation (Wang et al., 2006). While Cul4A shares a high degree of sequence similarity with its homolog Cul4B, the Cul4A −/− lethal phenotype indicates that Cul4A possesses more distinct functions and distinguishes it from the Cul4B E3 ligase (Li et al., 2002; Liu et al., 2009). Ubiquitylation of core histones by Cul4A was originally implicated in cell cycle regulation and cellular responses to DNA damage. However, evidence supporting its involvement in gene regulation comes from studies showing that Cul4A cooperates with other remodeling factors whose activities are closely related to the transcription process (Kotake et al., 2009). Also related to the current study, the PAF1 complex is a well-characterized complex that was originally identified in yeast as an RNAPII-interacting protein complex (Mueller and Jaehning, 2002). The complex is capable of facilitating several histone modifications and domain (CTD) influencing the phosphorylation of the RNAPII carboxy-terminal (CTD), coupling them to transcription elongation through chromatin by RNAPII (Krogan et al., 2003; Ng et al., 2003). As an early step in transcriptional activation in human cells, the PAF1 complex recruits the E3 ubiquitin ligase BRE1 to establish H2B monoubiquitylation on coding regions (Kim et al., 2009). This modification is essential for the recruitment and/or function of specific HMTs that promote H3K4 and K79 methylation events that ultimately result in active transcription. Although these results establish sequential and interdependent modification pathways, H3 methylations at K4 and K79 have also been found to be persistently enriched regardless of neighboring H2BK120 ubiquitylation (Chandrasekharan et al., 2010; Foster and Downs, 2009; Wang et al., 2009). These observations evoke the interesting possibility that an additional mechanism is involved in regulating the methylation reactions.
In this study, we purified factors that interact with each of six human H1 subtypes and determined whether these factors are related to gene-specific functions of H1 subtypes. Our purification identified the selective association of H1.2 with the Cul4A E3 ubiquitin ligase and PAF1 elongation complexes. This association is functional, because H1.2 knockdown severely impaired the ability of the Cul4A and PAF1 complexes to generate active histone marks as well as to facilitate transcriptional elongation. H1.2 interacts physically with the serine 2 phosphorylated form of RNAPII, and so allows the timely recruitment of the Cul4A and PAF1 complexes to target genes at an early elongation stage.
RESULTS
Linker histone H1.2 binds the Cul4A ubiquitin ligase complex and the PAF1 complex
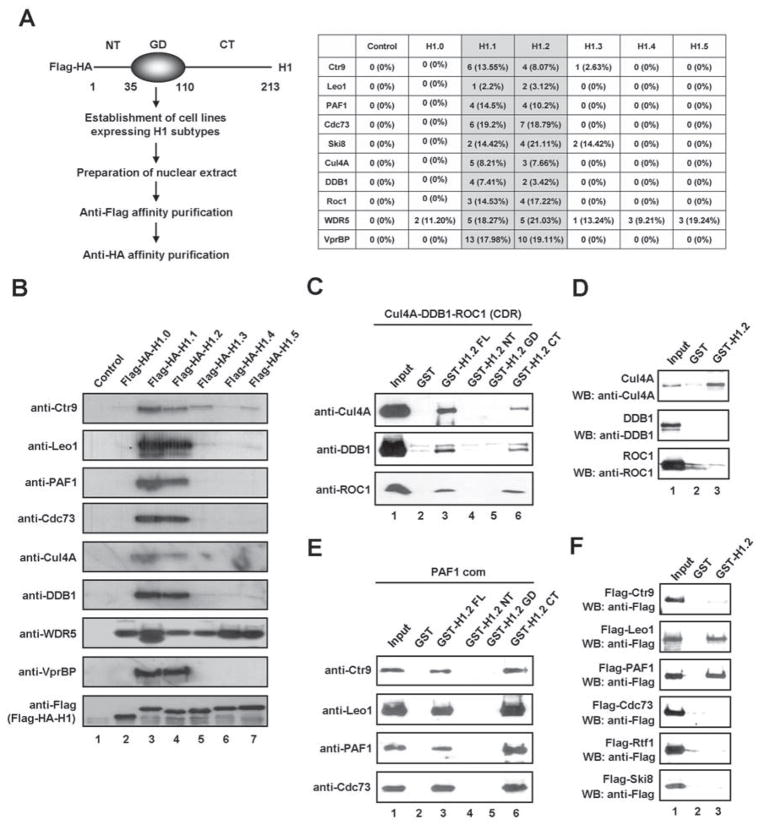
To gain insight into the distinct roles of linker histone H1 subtypes, we generated HeLa S3 cell lines stably expressing six human H1 subtypes fused to Flag and HA epitope-tags. After confirming that the expression levels of the H1 subtypes were comparable (data not shown), ectopic H1 and its associated partners were purified from nuclear extracts derived from the cell lines using sequential immunoprecipitations with anti-Flag and anti-HA antibodies. Recently, H1.2-interacting proteins were purified by utilizing a three-step purification protocol consisting of P11 cation exchange chromatography, anti-Flag affinity chromatography and glycerol gradient centrifugation (Kim et al., 2008). However, in the present study, the rapid two-step purification procedure was employed to ensure optimal protein-protein interactions. Proteins that were co-purified with each of H1 subtypes were subsequently identified by multidimensional protein identification technology (MudPIT). In agreement with our recent study (Kim et al., 2008), the purification of ectopic H1.2 from nuclear extracts detected multiple cofactors and ribosomal proteins as binding partners (data not shown). Most of these proteins were also co-purified with other linker histones (data not shown) Somewhat surprisingly, however, the proteomic analysis and immunoblotting led us to discover that two of the subtypes, H1.1 and H1.2, interact with the Cul4A ubiquitin ligase complex and the PAF1 transcriptional elongation complex (Figures 1A and 1B). Substantial interactions of H1.1 and H1.2 with VprBP, a substrate-specific adaptor for the Cul4 E3 ubiquitin ligase, were also detected. Interestingly, WDR5, another substrate adaptor of the Cul4 E3 ligase, was co-purified with all six linker histone subtypes. These results indicate that the presence of WDR5 in H1-associated complexes does not solely depend on its ability to interact with the Cul4A. Because H1.1 is minimally expressed in HeLa cells (Happel et al., 2009 and Figure S1A), the physiological significance of H1.1 data obtained in our purification could be questioned. For this reason, we decided to focus on the H1.2 interactions with the Cul4A and PAF1 complexes in the present study. Also of note, immunoblot analysis with H1.2 antibody indicated that ectopic H1.2 was expressed at similar levels to endogenous H1.2 and evenly distributed in soluble and insoluble chromatin fractions (Figure S1B).
Figure 1. Identification of the Cul4A and PAF1 complexes as interaction partners of linker histone H1.2.
(A) The six human H1 subtypes and their associated factors were isolated from nuclear extracts of HeLa S3 cells stably expressing Flag-HA-H1 subtypes through sequential immunoaffinity chromatographies. Three independent affinity purifications from HeLa nuclear extracts expressing Flag-HA-H1 subtypes were used for the MudPIT mass spectrometry assay. Among the reproducible and significant (p-value < 0.001) proteins identified in all three analyses, all subunits of both Cul4A and PAF1 complexes were detected by multiple peptides. The table summarizes the peptide count and the amino acid coverage of Cul4A and PAF1 components co-purified with H1 subtypes.
(B) The purified samples shown in (A) were resolved on 4–20% SDS-PAGE, and the presence of the Cul4A and PAF1 complexes was confirmed by immunoblot analysis
(C) The reconstituted Cul4A-DDB1-ROC1 (CDR) E3 ligase complex was incubated with glutathione-Sepharose beads containing GST alone, GST-H1.2 full length (FL), GST-H1.2 N-terminal tail (NT), GST-H1.2 globular domain (GD) and GST-H1.2 C-terminal tail (CT). The bound proteins were analyzed by immunoblotting with the indicated antibodies. Input corresponds to 10% of the CDR complex used in the binding reactions.
(D) GST alone (lane 2) or GST-H1.2 (lane 3), immobilized on glutathione-Sepharose beads, was incubated with recombinant Cul4A, DDB1 and ROC1. After washing with washing buffer, the bound proteins were immunoblotted with anti-Flag antibody. Ten percent of the input proteins were examined by immunoblotting (lane 1).
(E) For the pull-down experiments, the purified PAF1 complex was incubated with GST or the indicated GST-H1.2 fusions and subjected to immunoblotting after extensive washing. Input lane represents 10% of the PAF1 complex used in the binding reactions.
(F) GST pull-down assays were conducted as described in (D), but using Flag-tagged subunits of the PAF1 complex that were individually expressed and purified from Sf9 cells. Binding of each protein was analyzed by immunoblotting. Lane 1 represents 10% of the input. See also Figure S1.
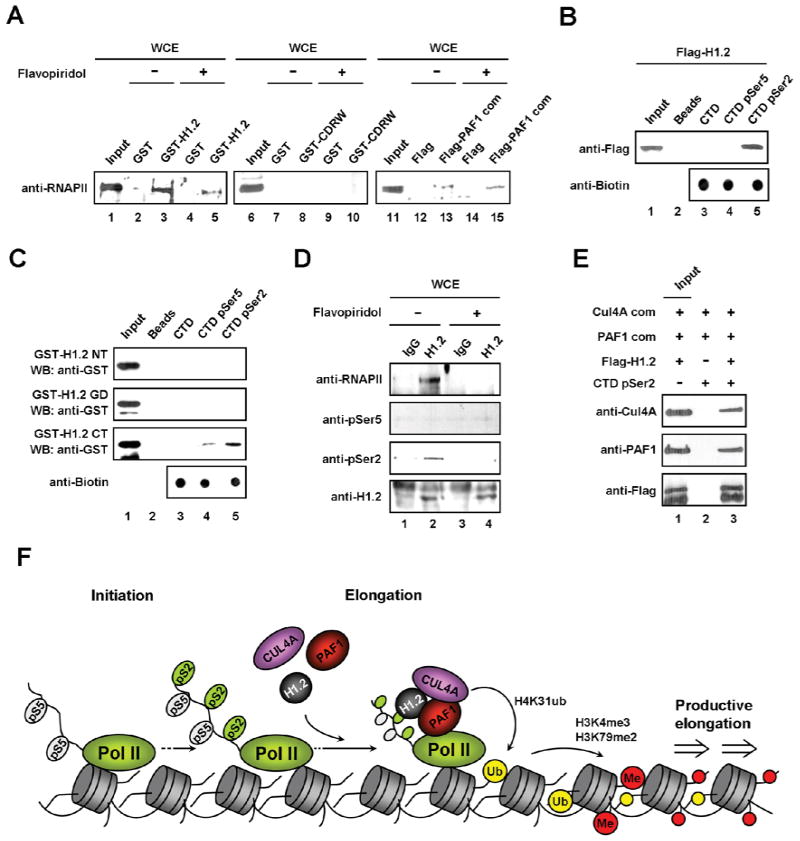
In agreement with our purification results, initial validation experiments involving H1.2 immunoprecipitation confirmed H1.2 interaction with endogenous Cul4A and PAF1 under physiological conditions (Figure S1C). As a more direct approach toward confirming the physical association of H1.2 with the Cul4A and PAF1 complexes, we analyzed the interaction of GST-H1.2 with the highly purified recombinant Cul4A and PAF1 complexes (Figure S1D-1G). Our assays showed that GST-H1.2 protein interacts with both complexes, whereas GST alone does not (Figures 1C and 1E, lanes 2 and 3). To identify the domain of H1.2 required for association with the Cul4A and PAF1 complexes, we performed GST pull-down experiments using three distinct regions of H1.2. The H1.2 C-terminal tail (H1.2 CT) containing amino acids 110-213 interacted with the Cul4A complex, whereas the H1.2 N-terminal tail (H1.2 NT) containing amino acids 1-34 and the H1.2 globular domain (H1.2 GD) containing amino acids 35-109 were unable to show any interaction (Figure 1C). The same sets of H1.2 proteins were also tested for their ability to interact with the PAF1 complex. Similar to the Cul4A complex, the PAF1 complex also retained its affinity only for H1.2 C-terminal tail (Figure 1E, lane 6). To determine the subunits mediating the Cul4A and PAF1 complex interactions with H1.2, GST pull-down experiments were repeated with individual recombinant subunits of the complexes. As shown in Figure 1D, Cul4A was very efficient in binding H1.2, whereas the binding of DDB1 and ROC1 was undetectable or significantly weaker. Additionally, PAF1 and Leo1 bound H1.2 as efficiently as the PAF1 complex (Figure 1F). The observed interactions were specific, because the five other Cullin family proteins (Cul1-3, Cul4B and Cul5) showed no detectable binding to H1.2 (Figure S1H) and two other H1 subtypes (H1.0 and H1.4) are unable to interact with Cul4A and PAF1 (Figure S1I).
The Cul4A requires H1.2 and WDR5 to ubiquitylate histone H4 at K31
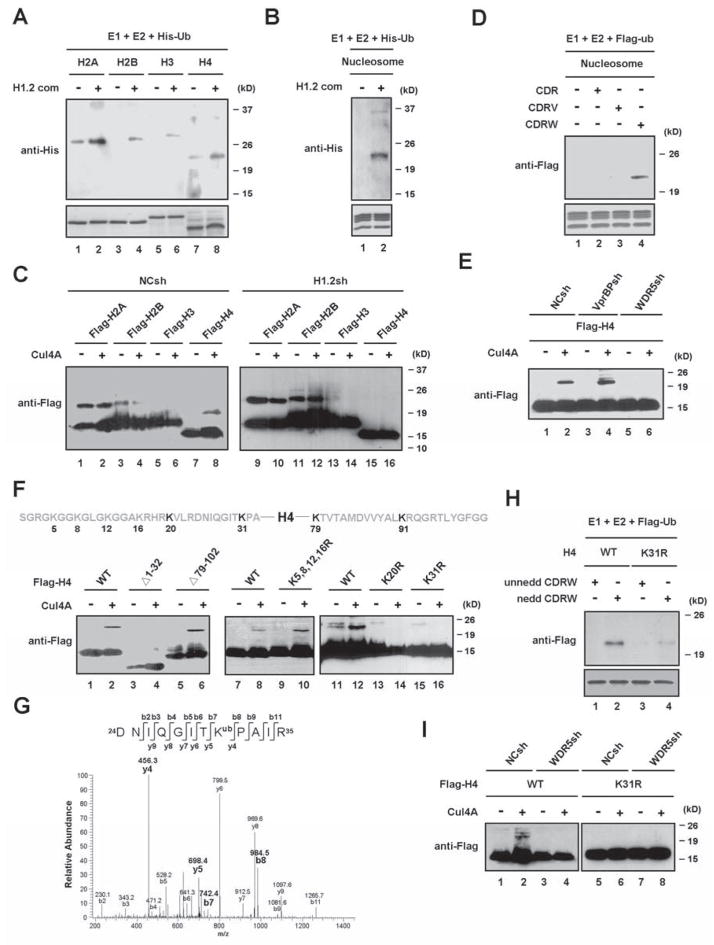
Having demonstrated a direct interaction between H1.2 and the Cul4A complex, we asked whether the Cul4A complex co-purified with H1.2 possesses histone ubiquitin ligase activity. To this end, the purified H1.2-associated factors were incubated with individual histones, histone octamers or nucleosomes in the presence of His-tagged ubiquitin and ubiquitylated histones were differentiated from unmodified forms by immunoblotting with anti-His antibody. Results shown in Figure 2A indicate that the purified factors can mediate ubiquitylation of all individual free histones. Similar assays using histone octamers as substrates revealed that H3 and H4 are preferentially ubiquitylated by the purified factors (Figure S2A). However, when nucleosomes, the more physiological substrates, were used for ubiquitin assays, the H1.2-associated factors showed ubiquitin ligase activity mainly toward H4 present in the nucleosome (Figures 2B and S2B).
Figure 2. Requirements of H1.2 and WDR5 for Cul4A-mediated H4 ubiquitylation.
(A) The purified H1.2-associated factors were assayed for in vitro ubiquitin ligase activity using individual core histones in the presence of E1, E2 and His-ubiquitin. Reactions were separated on 15% SDS-PAGE and analyzed by immunoblotting with anti-His antibody.
(B) In vitro ubiquitylation assays were performed as in (A), but using HeLa H1-depleted oligonucleosomes as substrates.
(C) Mock- or H1.2-depleted 293T cells were transfected with expression vectors for Flag-core histones and/or Cul4A. 48 h post-transfection, whole cell extracts were prepared and immunoprecipitated with anti-Flag antibody. Levels and the monoubiquitylation status of ectopic histones were determined by immunoblotting with anti-Flag antibody. The mobility-shifted bands correspond to monoubiquitylated histones.
(D) The CDR, CDRV and CDRW complexes containing neddylated Cul4A were assayed for ubiquitin ligase activity using HeLa H1-depleted oligonucleosomes as substrates.
(E) Mock-, VprBP-, or WDR5-depleted cells were transfected with expression vectors for Flag-H4 and/or Myc-Cul4A. Flag-H4 proteins were immunoprecipitated and analyzed by immunoblotting with anti-Flag antibody.
(F) Following the expression of Flag-wild type and mutant H4 and/or Cul4A in 293T cells, the ubiquitylation of ectopic H4 proteins was monitored by immunoblotting as in (C).
(G) Cell lysates from 293T cells transfected with Flag-H4 were immunoprecipitated with anti-Flag antibody and resolved in SDS–PAGE. The band corresponding to ubiquitylated H4 was excised and analyzed by LC–MS/MS mass spectrometry. The MS/MS spectrum shows that the Lys 31 residue is ubiquitylated in the peptide 24-DNIQGITKPAIR-35.
(H) The CDRW complex containing un-neddylated or neddylated Cul4A was assayed for ubiquitin ligase activity using wild type or K31-mutated H4.
(I) Mock- or WDR5-depleted cells were transfected with expression vectors for Cul4A and wild type or mutant H4. Ectopic H4 proteins were immunoprecipitated from whole cell lysates, and their ubiquitylation was probed with anti-Flag antibody. See also Figure S2.
To determine whether histones are real physiological substrates for Cul4A, 293T cells expressing HA-tagged ubiquitin were transiently transfected with a plasmid encoding Flag-histone alone or together with a plasmid encoding Myc-Cul4A. Flag-histones were immunopurified from cell lysates, and then subjected to immunoblot analysis with anti-Flag antibody. Equal amounts of Flag-H2A and Flag-H2B were immunopurified from cells transfected with or without Cul4A, and no changes in high molecular mass forms were detected after expression of Cul4A (Figure 2C, lanes 1–4). Similarly, a majority of ectopic H3 immunoprecipitated from Cul4A-transfected cells showed the same migration rates as the unmodified form of H3 in immunoblotting analysis (lanes 5 and 6). In contrast, a slow migrating band was observed in an anti-Flag immunoblot of Flag-H4 purified from Cul4A-transfected cells (lane 8). This shifted band was not detected in mock-transfected control cells (lane 7), demonstrating a direct correlation between Cul4A expression and H4 ubiquitylation.
The finding that H1.2 stably associates with Cul4A in our purification prompted us to check whether Cul4A modifies H4 in an H1.2-dependent manner. To this end, core histones were expressed in 293T cells infected with the lentivirus encoding a control shRNA or H1.2 shRNA (Figure S2C, lanes 1 and 2 and Figure S2D) and then Flag immunoprecipitates were analyzed for the extent of H4 ubiquitylation. Immunoblot analysis showed a significant decrease in H4 ubiquitylation, but no detectable change in H2A, H2B and H3 ubiquitylation, after H1.2 knockdown (Figure 2C, lanes 9–16). Thus, functional and physical interactions between H1.2 and Cul4A are essential for Cul4A to act as the E3 ubiquitin ligase for H4.
The H1.2-associated complex contains two WD40 repeat proteins, VprBP and WDR5, which are known to act as adaptors that recruit Cul4 to specific substrates. To explore their functional participation in Cul4A-mediated H4 ubiquitylation, we prepared the recombinant Cul4A complexes from Sf9 insect cells expressing GST-Cul4A, GST-DDB1 and untagged ROC1 together with His-VprBP or His-WDR5 (Figure S2E). In histone ubiquitylation assays using the reconstituted complexes, the Cul4A-DDB1-ROC1 complex (abbreviated CDR) showed a detectable activity towards H3 and H4 (Figure S2G, lanes 1, 3, 5 and 7), and the presence of VprBP (abbreviated V) influenced the ubiquitylation reactions at varying degrees (lanes 9, 11, 13 and 15). Notably, in the presence of WDR5, the CDR complex showed a higher activity to catalyze H4 ubiquitylation, suggesting that WDR5 (abbreviated W) selectively targets Cul4A activity toward H4 (lanes 17, 19, 21 and 23). Since neddylation is known to facilitate Cul4A E3 ligase activity (Higa and Zhang, 2007), we also prepared neddylated forms of CDR, CDRV and CDRW (Figure S2F) and tested them in our assays. Under our experimental conditions, the neddylation of Cul4A significantly elevated the level of H4 ubiquitylation by the CDRW complex (Figure S2G, compare lane 24 with lane 23), but did not influence H4 ubiquitylation by the CDR and CDRV complexes (lanes 7, 8, 15 and 16). In accordance with these results, similar modification assays using reconstituted nucleosomes confirmed that nucleosomal H4 is specifically ubiquitylated by the neddylated CDRW complex, but not by the neddylated CDRV complex (Figure 2D). After establishing the requirement of WDR5 in Cul4A-mediated H4 ubiquitylation in vitro, we further tested its role in vivo using 293T cells depleted of VprBP or WDR5 (Figure S2C, lanes 5–8). As shown in Figure 2E, H4 ubiquitylation is severely compromised when WDR5 is depleted in cells. In contrast, depletion of VprBP did not cause obvious changes in H4 ubiquitylation. As predicted from these results, VprBP failed to interact with H4, but WDR5 showed a strong interaction with H4 (Figure S2H).
In an attempt to map the primary ubiquitylation site in H4, we deleted the first 32 amino acids or the last 25 amino acids of H4. Co-expression of these deletion mutants with Cul4A revealed that the N-terminally truncated H4 cannot be modified by Cul4A, whereas the C-terminally truncated H4 is still well ubiquitylated (Figure 2F, lanes 1–6). Because the truncated N-terminal region contains six lysine residues, we next mutated them to elucidate which lysines serve as a primary ubiquitylation site(s). Cul4A was able to ubiquitylate H4 mutated at K5, K8, K12 and K16 (lanes 7–10), but mutation of K20 or K31 completely abolished Cul4A-mediated ubiquitylation (lanes 11–16). To investigate further whether H4 is indeed ubiquitylated at these two sites in vivo, Flag-H4 was transiently expressed in cells and immunoprecipitated with anti-Flag antibody. Consistent with previous studies (Danielsen et al., 2011; Yan et al., 2009), our mass spectrometric analysis of the purified Flag-H4 identified ubiquitylations at K31, K79 and K91 (Figures 2G and S2I). Interestingly, although changing K20 to R abolished Cul4A-dependent H4 ubiquitylation, our analysis failed to detect ubiquitylation at this site, leading us to the tentative conclusion that K31 is a physiologically relevant substrate for Cul4A.
Given that Cul4A is the only E3 ubiquitin ligase activity in the purified H1.2-associated factors and is capable of modifying H4K31, we next asked whether Cul4A in the CDRW complex is able to ubiquitylate H4 in a similar way. Mutation of H4K31 led to a marked reduction in the level of H4 ubiquitylation mediated by the reconstituted CDRW complex (Figure 2H). To further confirm this observation in vivo, we tested whether WDR5 is required for the Cul4A-mediated H4K31ubiquitylation in cells. When the level of H4 ubiquitylation was measured by immunoblotting, it was indeed decreased in WDR5 knockdown cells (Figure 2I, lanes 1–4). In contrast, Cul4A had a very limited activity to ubiquitylate H4 carrying K31R mutation, and this basal level of H4K31 ubiquitylation remained unchanged after depletion of WDR5 (lanes 5–8). Collectively, these findings suggest that Cul4A requires WDR5 as a molecular adaptor for efficient ubiquitylation of H4 at K31.
PAF1 is essential for Cul4A-mediated H4 ubiquitylation
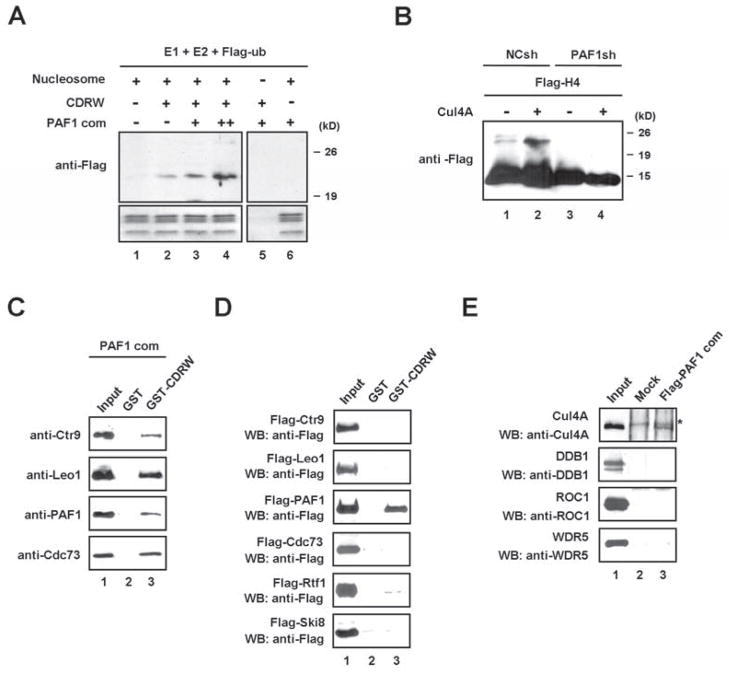
The fact that the PAF1 complex co-purified with the H1.2-associated Cul4A complex raised the question of whether it plays a potential role in regulating the Cul4A E3 ligase activity. To address this question, the PAF1 complex was reconstituted in Sf9 cells and included in the in vitro ubiquitylation reactions. Interestingly, H4 ubiquitylation by the CDRW complex was obviously increased when the PAF1 complex was included in the reaction (Figure 3A). To further evaluate the role of the PAF1 complex in H4K31 ubiquitylation in vivo, 293T cells stably transduced with a control or PAF1 shRNA (Figure S2C, lanes 1 and 4) were transfected with Flag-H4, and then Flag immunoprecipitates were analyzed for the extent of de novo ubiquitylation by immunoblotting with anti-Flag antibody. As expected, silencing endogenous PAF1 almost completely abolished H4 ubiquitylation (Figure 3B). The PAF1 complex had previously been reported to modulate H2B ubiquitylation (Kim et al., 2009; Wood et al., 2003), but to our knowledge this is the first time that it is linked to H4 ubiquitylation.
Figure 3. Regulation of Cul4A activity by PAF1.
(A) HeLa H1-depleted nucleosomes were incubated with neddylated CDRW and/or PAF1 complexes in the presence of E1, E2 and Flag-ubiquitin for in vitro ubiquitylation assays. The ubiquitylated histones were detected by immunoblotting with anti-Flag antibody.
(B) Mock- or PAF1-depleted 293T cells were transfected with expression vectors encoding Flag-H4 and/or Cul4A for 48 h. Flag-H4 was purified and analyzed by immunoblotting with anti-Flag antibody.
(C) Reconstituted PAF1 complex was incubated with GST-CDRW complex or GST control. GST fusion proteins were precipitated and subjected to immunoblot analysis to detect the subunits of the PAF1 complex. Input corresponds to 10% of the materials used in the binding reactions.
(D) GST pull-down assays were performed using components of the PAF1 complex with GST or GST-CDRW complex. After extensive washing, bound proteins were detected by immunoblotting with anti-Flag antibody. Lane 1 represents 10% of the input.
(E) The indicated components of the CDRW complex were individually incubated with Flag-PAF1 complex that was immobilized on M2 agarose. Bound proteins were analyzed by immunoblotting. A non-specific band in the Cul4A immunoblot is indicated by an asterisk. Lane 1 represents 10% of the input. See also Figure S3.
Although several mechanisms could explain the requirement for the PAF1 complex in H4K31 ubiquitylation, one possible explanation is that the PAF1 complex recruits the Cul4A ubiquitin ligase activity to target nucleosomes. In light of this possibility, we assayed for a direct interaction between the PAF1 complex and the CDRW complex. The GST pull-down experiments demonstrated a physical interaction of the purified PAF1 complex with the GST-CDRW complex, but not with GST alone (Figure 3C). To identify the subunits of the PAF1 complex that interact with the CDRW complex, each of the six PAF1 subunits was checked for binding to the GST-CDRW. The PAF1 subunit by itself was able to bind to the CDRW complex, but no binding of other PAF1 subunits was detected (Figure 3D). In parallel binding experiments with an immobilized PAF1 complex, the Cul4A exhibited the binding activity, while three other components of the CDRW complex were incapable of generating detectable interactions (Figures 3E and S3). These results suggest that PAF1 is necessary and is likely to function together with Cul4A for H4K31 ubiquitylation.
H4K31 ubiquitylation by Cul4A is required for H3K4 and H3K79 methylation
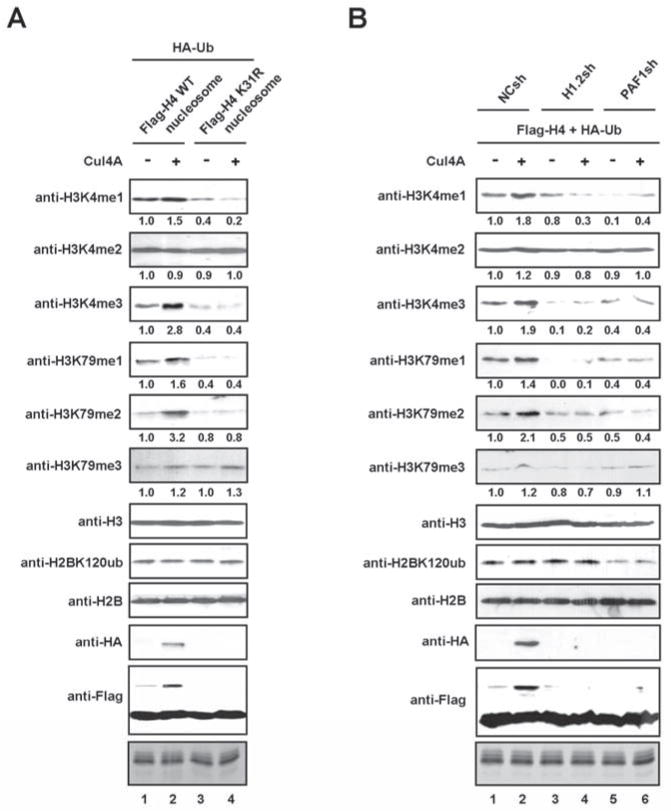
Given the well documented requirement of H2B ubiquitylation for H3K4 and H3K79 methylation (Dover et al., 2002; McGinty et al., 2008; Osley, 2004; Sun and Allis, 2002; Weake and Workman, 2008), we examined whether H4K31 ubiquitylation also affects these two methylation marks. 293T cells were transiently transfected with a plasmid encoding Flag-wild type (WT) or K31-mutated (K31R) H4 together with vectors for HA-ubiquitin and Cul4A. Mononucleosomes were prepared from the transfected cells by extensive sonication of formaldehyde cross-linked chromatin, and ectopic H4-containing nucleosomes were selectively purified with anti-Flag antibody. As confirmed by immunoblotting, similar levels of ectopic H4 proteins were incorporated into the purified mononucleosomes (Figure 4A). Examination of the effect of Cul4A on H3K4 methylation disclosed that H3K4me1 and H3K4me3 were considerably increased after the expression of Cul4A. In contrast, H3K4me2 was not different between Cul4A-transfected cells and control cells. Importantly, Cul4A-induced H3K4me1/me3 exhibited H4K31 ubiquitylation dependence, since the H4K31 mutation markedly reduced these modifications. In the case of H3K79 methylation, H3K79me1 and H3K79me2, but not H3K79me3, reproducibly showed an increase after Cul4A expression. That the observed increase in H3K79me1/me2 was abolished by the H4K31 mutation again indicates these two modifications are dependent on the Cul4A-driven H4K31 ubiquitylation. Also of note, H2BK120 ubiquitylation was not affected by the H4K31 mutation, strongly suggesting that the reduction of H3K4me3/H3K79me2 in Cul4A-depleted cells was independent of the known H2BK120 ubiquitylation pathway. In view of the importance of H1.2 and PAF1 for Cul4A-mediated H4 ubiquitylation, it was also reasonable to assume that H1.2 and PAF1 are necessary to create the H3K4 and H3K79 methylation marks. Indeed, we observed an almost complete disappearance of H3K4me1/me3, H3K79me1/me2 in cells depleted of either H1.2 or PAF1 (Figure 4B). On the contrary, the same depletion resulted in no detectable change in H3K4me2 and H3K79me3 (Figure 4B). Further, as expected from previous studies, H2BK120 ubiquitylation was reduced by PAF1 depletion, but not by H1.2 depletion. Collectively, these results place H3K4me1/me3 and H3K79me1/me2 at a late stage of chromatin remodeling after and dependent of the Cul4A-mediated ubiquitylation of H4K31.
Figure 4. Dependence of H3K4 and K3K79 methylation on H4K31 ubiquitylation.
(A) 293T cells were transfected with plasmids coding for HA-ubiqutin and Flag-wild type or K31-mutated H4 in the presence or absence of Cul4A for 48 h. After formaldehyde-crosslinking, mononucleosomes were prepared and subjected to immunoprecipitations using anti-Flag antibody. The levels of histone modifications in the isolated nucleosomes were analyzed by immunoblotting.
(B) Mock-, H1.2- or PAF1-depleted 293T cells were transfected with expression vectors for HA-ubiquitin and Flag-H4 and/or Cul4A. Mononucleosomes were prepared as in (A), and histone modifications were analyzed by immunoblotting.
H1.2, Cul4A and PAF1 are colocalized and function at shared target genes
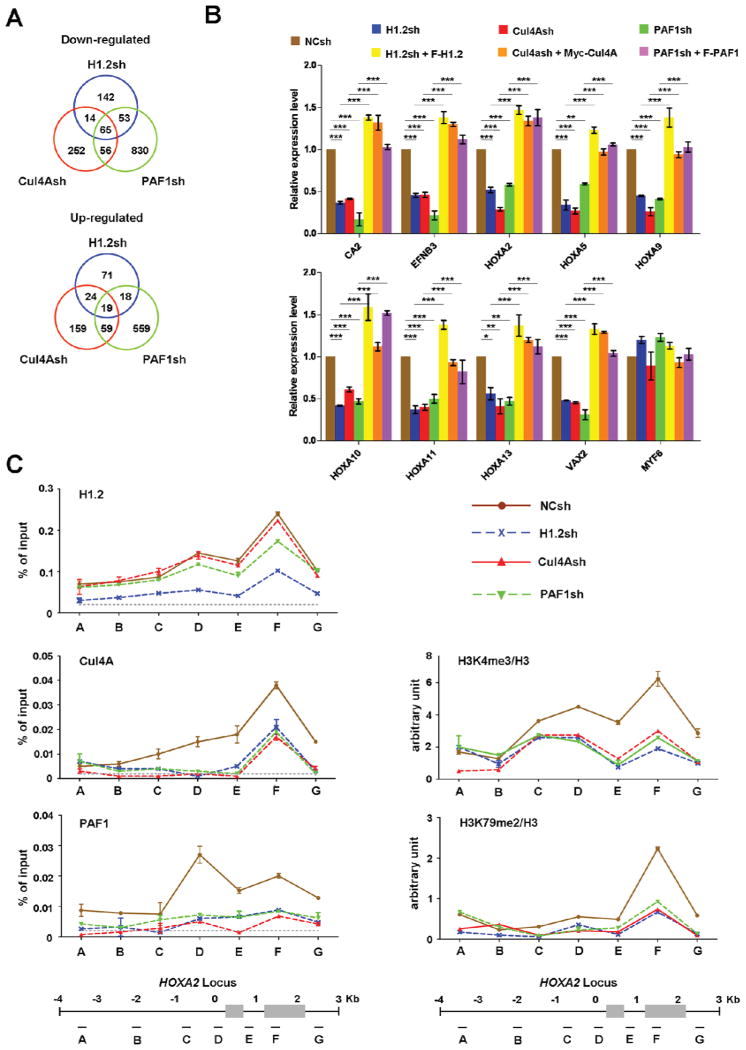
To examine whether the above-described interactions among H1.2, Cul4A and PAF1 reflect functional interactions leading to transcriptional activation, we set out to conduct gene expression microarray analysis with total RNA isolated from 293T cells expressing shRNAs against H1.2, Cul4A or PAF1 (Figure S2C). Using a 1.5-fold cutoff, we detected 1,412 genes down-regulated and 909 genes up-regulated upon knockdown of H1.2, Cul4A or PAF1 (Figure 5A and Table S1). Among the down-regulated genes, the expression of 274, 387 and 1004 genes was found to be reduced by H1.2, Cul4A and PAF1 shRNAs, respectively. When the three gene lists were compared, we detected 65 genes commonly repressed by H1.2, Cul4A and PAF1 shRNAs. Gene ontology analysis showed that genes regulating developmental process and anatomical structural development were overrepresented in the down-regulated genes (Figure S4A). Consistent with the previous publications implicating the PAF1 complex and specific H1 subtypes in the control of Hox gene transcription (Zhang et al., 2012b; Zhu et al., 2005), prominent among the down-regulated genes were the clustered Hox genes. The microarray results were further confirmed by quantitative RT-PCR analyses of 12 down-regulated and 2 unaffected genes (Figures 5B and S4C).
Figure 5. Coordinated actions of H1.2, Cul4A and PAF1 in target gene transcription.
(A) 293T cells were depleted of H1.2, Cul4A or PAF1, and subjected to microarray analysis. Venn diagrams show the overlapping target genes of H1.2, Cul4A and PAF1. See also Table S1.
(B) To validate the gene expression array data, the mRNA levels of the nine down-regulated and one unaffected genes in H1.2/Cul4A/PAF1-depleted cells were quantified by qRT-PCR using primers listed in Supplemental Experimental Procedures. The rescue effects of H1.2/Cul4A/PAF1 expression in H1.2/Cul4A/PAF1-depleted cells were also analyzed as indicated. The values are expressed as fold changes from the mRNA levels in undepleted cells. Results represent the means ± S.D. of three independent experiments (*, P < 0.05; **, P < 0.01; ***, P < 0.001). Statistical tests were performed using one-way ANOVA followed by Bonferroni’s post hoc test.
(C) Chromatin immunoprecipitation (ChIP) experiments were performed in 293T cells depleted of H1.2, Cul4A or PAF1 using the indicated antibodies. Precipitation efficiencies relative to non-enriched input samples were determined for seven locations across the human HoxA2 region by quantitative PCR (qPCR) with primers depicted at the bottom and listed in Supplemental Experimental Procedures. Percentage input is determined as the amount of immunoprecipitated DNA relative to input DNA. Nonspecific background signals obtained from a control rabbit IgG are shown by dotted lines, and error bars represent the standard deviation obtained from three independent experiments. See also Figure S4.
To check weather cooperative functions of H1.2, Cul4A and PAF1 in stimulating gene transcription reflect their targeted recruitments, we next investigated their localization across a large range of HoxA2 gene by ChIP assays. Cross-linked chromatin was isolated from control cells and cells depleted of H1.2, Cul4A or PAF1, and the precipitated DNA was amplified by qPCR using primers specific for seven different regions of the gene. We found that H1.2 binding was enriched in coding regions, especially region F (Figure 5C, H1.2). Additionally, the distribution of Cul4A across the coding regions was similar to that of H1.2, while PAF1 peaked around the transcription start site (region D) and gradually decreased over the coding regions (Cul4A and PAF1). The observed crosstalk among H1.2, Cul4A and PAF1 in mediating HoxA2 gene transcription raised the possibility that they could localize at target genes in a mutually dependent manner. Indeed, depletion of H1.2 reduced the levels of Cul4A and PAF1 at the HoxA2 locus (Cul4A and PAF1). Similarly, individual depletions of Cul4A and PAF1 diminished the localization of both Cul4A and PAF1 (Cul4A and PAF1), but did not lead to any substantial reduction of H1.2 occupancy (H1.2). Consistent with these results, ectopic expression of H1.2 in H1.2-depleted cells increased the levels of Cul4A and PAF1 at HoxA2 locus (Figure S4D) and reactivated target gene transcription (Figures 5B and S4C). Expectedly, however, expressing Cul4A and PAF1 in cells depleted of Cul4A and PAF1 didn’t affect the distribution of H1.2 across HoxA2 gene, but stimulated target gene transcription (Figure S4F). These rescue experiments excluded possible off-target effects of H1.2, Cul4A and PAF1 shRNAs, strengthening our results. In addition, the facts that knockdown of H1.0 or H1.4 has little effect on transcription of H1.2 target genes (Figure S4E) and that H1.2, Cul4A and PAF1 were minimally localized at MYF6 and TMEM55A genes (Figure S4F) strongly suggest that H1.2 is an initial regulator of Cul4A and PAF1 recruitment and function.
Because the PAF1 complex indirectly regulates H3K4me3 and H3K79me2 through its role in H2B ubiquitylation (Kim et al., 2009; Krogan et al., 2003; Muntean et al., 2010), we also checked the levels of these two modifications at the HoxA2 locus. Consistent with our nucleosome purification studies (Figure 4), colocalization of H1.2, Cul4A and PAF1 was accompanied by a marked accumulation of H3K4me3 and H3K79me2 downstream of the transcription start site (Figure 5C, H3K4me3 and H3K79me2). These data raised the question of whether H3K4me3 and H3K79me2 at the HoxA2 gene are dependent on the presence of H1.2, Cul4A and PAF1. In fact, the predicted roles for Cul4A, PAF1 and H1.2 are confirmed by the observation that the levels of H3K4me3 and H3K79me2 were reduced in response to the individual depletion of H1.2, Cul4A and PAF1 (H3K4me3 and H3K79me2). Taken together, these results strongly argue that H1.2, Cul4A and PAF1 function coordinately in up-regulating target genes; their concerted actions are linked to H3K4me3 and H3K79me2.
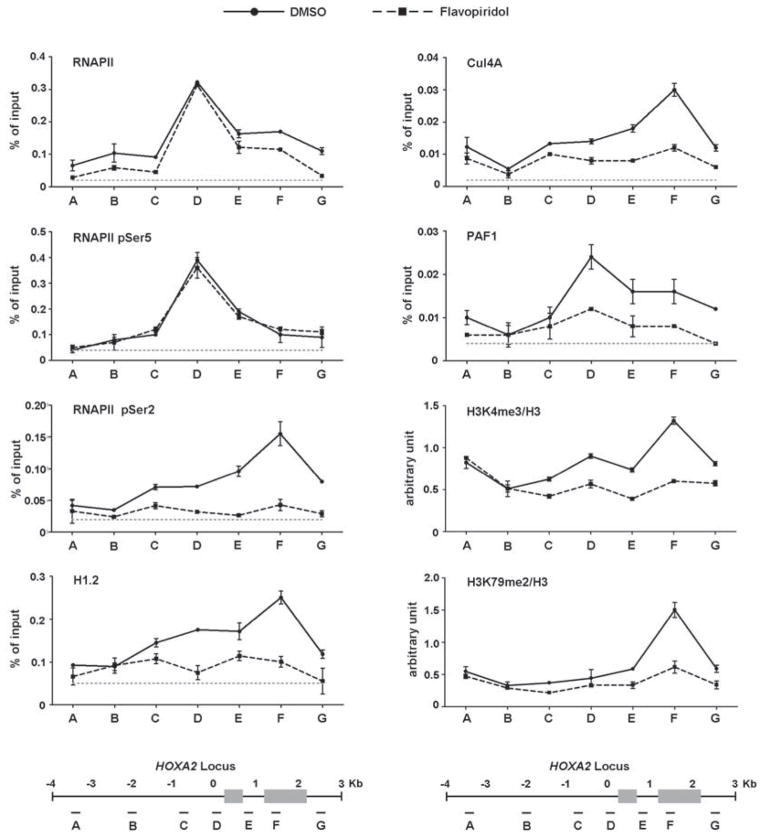
Recruitments of H1.2, Cul4A and PAF1 at target genes are dependent upon RNAPII pSer2
Because PAF1 and H3K4me3/H3K79me2 are known to act in chromatin transcription elongation (Gerber and Shilatifard, 2003; Kim et al., 2010), our results support the view that H1.2, Cul4A and PAF1 could target elongation steps for their activity. Congruent with these ideas, when HoxA2 gene transcription was analyzed by qRT-PCR using primers annealing 3′ ends of mRNA molecules, the levels of 3′ transcripts were significantly decreased after depletion of H1.2, Cul4A or PAF1 (Figure S5A). In contrast, similar levels of 5′ HoxA2 transcripts were detected in cells deficient of H1.2, Cul4A or PAF1 (Figure S5A). These results were further validated by rescue experiments demonstrating that the ectopic expression of H1.2, PAF1 and Cul4A largely overrides the elongation defects arising from depletion of H1.2, Cul4A and PAF1 (Figure S5A). While our approaches measure steady-state mRNA levels, these results suggest that H1.2, Cul4A and PAF1 stimulate Pol II elongation leading a higher level of full-length transcripts. To more directly assess the cooperative roles of H1.2, Cul4A and PAF1 in the elongation stage of transcription, we checked whether the recruitments of H1.2, Cul4A and PAF1 to the HoxA2 gene are dependent on RNAPII and intimately linked to CTD Ser5 and Ser2 phosphorylation which is distinct properties of the initiation and elongation forms of RNAPII. Comparison of overall levels of Ser5 and Ser2 phosphorylation showed a dramatic decrease in Ser2 phosphorylation, but only a modest change in Ser5 phosphorylation in flavopiridol-treated cells relative to control cells (Figure S5B). Beyond global effects, ChIP analysis at the HoxA2 locus also showed a significant drop of Ser2 phosphorylation, but not Ser5 phosphorylation, within the transcribed region after flavopiridol treatment (Figure 6, RNAPII pSer5 and RNAPII pSer2). Flavopiridol treatment also reduced co-occupancy of H1.2, Cul4A and PAF1 around the transcription start site and coding region (H1.2, Cul4A and PAF1). More intriguingly, blocking Ser2 phosphorylation by flavopiridol resulted in a distinct decrease in the levels of H3K4me3 and H3K79me2 around the coding region (H3K4me3 and H3K79me2). These results support the notion that RNAPII is required for the initial recruitment and stable localization of H1.2, Cul4A and PAF1 at the HoxA2 locus and that this process is tightly regulated by CTD Ser2 phosphorylation.
Figure 6. Recruitments of H1.2, Cul4A and PAF1 at HoxA2 locus via RNAPII pSer2.
293T cells were treated with DMSO or flavopiridol (200 nM) for 3 h, and ChIP assays of the HoxA2 locus were performed using indicated antibodies. ChIP-enriched DNA was quantified by qPCR using the primers indicated at the bottom. The dotted lines represent the signal from negative control rabbit IgG. The data are the means of three independent experiments ± S.D. See also Figure S5.
Serine 2 phosphorylation promotes RNAPII association with H1.2
To gain support for the ChIP results above, we next checked whether H1.2, Cul4A and PAF1 can interact with RNAPII to facilitate their occupancy at target genes. In vitro pull-down experiments using whole cell extracts showed that RNAPII bound strongly to H1.2 and moderately to the PAF1 complex, but was not retained by the CDRW complex (Figure 7A, lanes 3, 8 and 13). In line with the requirement of CTD Ser2 phosphorylation for the H1.2-CTD interaction, the observed binding of H1.2 to RNAPII was perturbed in cell extracts prepared from flavopiridol-treated cells (compare lane 5 with lane 3). To further confirm that the interaction between H1.2 and Ser2 phosphorylated CTD is direct, we repeated pull-down experiments using synthetic CTD peptides immobilized on streptavidin-agarose beads. H1.2 can bind Ser2 phosphorylated CTD peptides, but not unmodified or Ser5 phosphorylated CTD (Figure 7B, lane 3–5 and Figure S6). Moreover, the H1.2 interaction with Ser2 phosphorylated CTD was dependent upon its C-terminal tail (Figure 7C). The selective interaction between H1.2 and Ser2 phosphorylated RNAPII is further supported by the failure to detect their interaction in immunoprecipitation of cell extracts prepared from flavopiridol-treated cells (Figure 7D). Moreover, the Cul4A and PAF1 complexes bind to elongating RNAPII in a H1.2-dependent manner (Figure 7E). These data further support our model that H1.2 is critical in regulating the recruitment and function of Cul4A and PAF1 during chromatin transcription elongation.
Figure 7. Selective recognition of RNAPII pSer2 by H1.2.
(A) Cell extracts were prepared from DMSO-treated or flavopiridol-treated cells and incubated with GST (lanes 2, 4, 7 and 8), GST-H1.2 (lanes 3 and 5), GST-CDRW (lanes 8 and 10), Flag peptide (lanes 12 and 14) or Flag-PAF1 complex (lanes 13 and 15) immobilized on beads. After extensive washing, RNAPII binding was analyzed by immunoblot with anti-RNAPII antibody. Lanes 1, 6 and 11 represent 10% of the input.
(B) The biotinylated CTD heptapeptide repeats that were unmodified or phosphorylated at either position Ser2 or Ser5 were immobilized onto streptavidin-agarose beads and incubated with Flag-H1.2. H1.2 binding to the CTD peptides was determined by immunoblotting with anti-Flag antibody. Input corresponds to 10% of H1.2 used in the binding reactions.
(C) Unmodified or phosphorylated CTD peptides were immobilized and incubated with the indicated H1.2 domains, and then pull-down assays were preformed as in (B). Lane 1 represents 10% of H1.2 domains used in the binding reactions.
(D) Cell extracts were prepared from DMSO- or flavopiridol-treated cells as in (A), and immunoprecipitated with anti-H1.2 antibody. The amount and phosphorylation level of RNAPII pulled down were examined by immunoblotting.
(E) The Ser2 phosphorylated CTD peptides were immobilized and incubated with the Cul4A and PAF1 complexes in the presence or absence of H1.2, and then pull-down assays were performed as in (B).
(F) Model for the cooperative role of H1.2, Cul4A and PAF1. Our studies present evidence for H1.2 action targeting post-initiation steps, in which H1.2 selectively recognizes RNAPII CTD Ser2 phosphorylation and brings the Cul4A and PAF1 complexes to target genes. The recruitment and activity of the Cul4A and PAF1 complexes, in turn, stimulate H4K31 ubiquitylation, H3K4me3 and H3K79me2, thereby leading to more productive elongation phase of transcription. Therefore, selective tethering of H1.2 to target loci via CTD Ser2 phosphorylation is essential for the Cul4A and PAF1 complexes to maintain an active state of gene transcription.
DISCUSSION
While linker histone H1 is often regarded as a basic component of chromatin, a growing body of evidence challenges this original idea and suggests that particular H1 subtypes are important in regulating gene expression at a more specific level (Kim et al., 2008; Lee et al., 2004; Studencka et al., 2011; Zhang et al., 2012a). Using a high-confidence proteomic screen, we discovered the stable association of H1.2 with the Cul4A complex and the PAF1 complex and began a systematic investigation of their contribution to chromatin transcription. Our initial results demonstrate that the Cul4A ubiquitin ligase activity mainly targets K31 of histone H4. Another important result was the observation that H1.2 is necessary for the Cul4A activity toward H4K31. This finding is intriguing, because it supports the idea that H1.2 can positively act on the histone modification process, independently of any role related to stabilization of chromatin structure. The observation that H4K31-targeted ubiquitylation by Cul4A is dependent on WDR5 is also important in light of several recent reports suggesting that substrate-specific adaptors are important for cullin E3 ligase activity (Emanuele et al., 2011; Higa et al., 2006; Jackson and Xiong, 2009). WDR5 is an essential functional component of the SET1/MLL complexes, which methylate H3K4 (Muntean et al., 2010; Trievel and Shilatifard, 2009; Wysocka et al., 2005). Thus, it is tempting to speculate that WDR5 on the one hand directly regulates H4K31 ubiquitylation and, on the other hand, acts through H3K4me3 for its stimulation activity. Our results contrast with a recent report (Wang et al., 2006) showing that a complex containing Cul4A, Cul4B, DDB1, DDB2 and ROC1 from HeLa nuclear extracts can modify nucleosomes in the absence of WDR5. These differences could be due to different experimental conditions and approaches. We are also curious whether Cul4B and DDB2 omitted from our complex might have an as yet undescribed role in nucleosome modification.
Our results demonstrating that H4K31 ubiquitylation by Cul4A is dependent on PAF1 fit well with the observed interaction of the PAF1 complex with H1.2 and Cul4A in our initial purification. This is also consistent with the notion that PAF1 actively orchestrates with multiple histone modifying factors in the formation of histone marks linked to transcription (Kim et al., 2009). Of note, although PAF1 can facilitate both H4K31 ubiquitylation and H2BK120 ubiquitylation, what is unanticipated in these results is the H1.2-dependent recruitment and activity of PAF1. These observations argue that the ability of PAF1 to influence transcription-coupled histone modifications is dependent on its interacting factors at target genes. Thus, it will be interesting to learn whether any other chromatin or transcription regulators also affect the distinct role of PAF1 in H4K31 ubiquitylation and H2BK120 ubiquitylation. We also found that Cul4A-mediated H4K31 ubiquitylation influences positively the accumulation of H3K4me3 and H3K79me2. Much lower levels of H3K4me3 and H3K79me2 were detected after H1.2 depletion, again highlighting the critical role of H1.2 within epigenetic regulatory networks and, in particular, its function with PAF1 to establish H3K4me3 and H3K79me2 marks. This striking finding suggests that the presence of H1.2 is critical for H3K4me3 and H3K79me2, linking Cul4A-mediated H4 ubiquitylation to two active methylation marks. Recent studies have indicated that the PAF1 complex is required for H2BK120 ubiquitylation during the transcription process and that H3K4me3 and H3K79me2 are dependent upon prior H2BK120 ubiquitylation (Dover et al., 2002; Kim et al., 2009; McGinty et al., 2008; Sun and Allis, 2002). However, we provided compelling evidence that H4K31 ubiquitylation has a bona fide stimulation effect on H3K4me3 and H3K79me2 and that this activity is independent of H2BK120 ubiquitylation. Therefore, it will be important to determine whether H4K31 ubiquitylation and H2BK120 ubiquitylation are localized differentially and whether they have any distinct roles in generating H3K4me3 and H3K79me2. Interestingly, we also found that blocking H4K31 ubiquitylation by K31R mutation leads to significant decrease in H3K4me1 and H3K79me1. Since H2BK120 ubiquitylation resulted in no changes in H3K4me1 and H3K79me1 (Kim et al., 2009), this finding suggests H4K31 ubiquitylation-dependent H3K4me1 and H3K79me1 and proposes new molecular mechanisms for the crosstalk.
Our microarray analyses also revealed a clear interplay of H1.2, Cul4A and PAF1, and established that these factors play non-redundant, cooperative roles in productive transcription of developmental regulatory genes. This is, to our knowledge, the first demonstration that H1.2 interacts functionally with factors responsible for active transcription. Our ChIP assays on HoxA2 gene whose expression was found to be dependent upon H1.2, Cul4A and PAF1 also show that H1.2, Cul4A and PAF1 are recruited to target loci in a coordinated manner. These findings indicate that the mechanism found in modification processes indeed occurs during gene transcription. To properly regulate their functions at target genes, H1.2, Cul4A and PAF1 appear to have adopted a strategy, wherein PAF1 is positioned at the 5′ end of the gene, more closely to RNAPII, whereas H1.2 and Cul4A are more localized to the coding region. We speculate that the chromatin dynamics and higher order folding render H1.2, Cul4A and PAF1 suitable for their interaction, thereby facilitating H4K31 ubiquitylation and transcription. The requirement for their simultaneous localization in time is dependent upon the physical position of RNAPII near the transcription start site of the gene. Furthermore, we were able to identify an important role for RNAPII Ser2 phosphorylation in regulating H1.2 function based on several lines of evidence. First, H1.2 can interact with the Ser2 phosphorylated CTD, but not with the unmodified or Ser5 phosphorylated CTD. Second, H1.2 is localized at target loci in a manner similar to Ser2 phosphorylated RNAPII. Third, flavopiridol treatment to inhibit P-TEFb and block the CTD Ser2 phosphorylation abolished H1.2 localization to HoxA2 gene. These observations strongly suggest that H1.2 organizes specialized transcription programs at coding regions and thus may participate in reactions that are important for transcription elongation.
Based on our observations, together with previous studies, we propose the following model for how H1.2 activates target gene transcription (Figure 7F). In an initial state, H1.2 is localized at target genes through its interaction with Ser2 phosphorylated RNAPII. Once bound to target genes, H1.2 utilizes its C-terminal tail domain to recruit and/or stabilize the Cul4A and PAF1 complexes. A multistep chromatin remodeling process involving H4K31 ubiquitylation, H3K4me3 and H3K79me2 then takes place to potentiate transcription, especially at the level of elongation. In human cells, Cul4A and PAF1 are free to cooperate with H1.2 through direct protein-protein interactions, such that preventing them from binding to H1.2 could be a novel strategy to control H1.2-mediated transactivation. Furthermore, inactivating Cdk9 can also prevent CTD Ser2 phosphorylation-based localization of H1.2 over the target sites and thus subsequently disrupt concomitant recruitments of Cul4A and PAF1. Future experiments will focus on identification of additional cellular regulators required for gene-specific action of H1.2 and elucidation of the molecular mechanism involved.
EXPERIMENTAL PROCEDURES
Ubiquitylation assays
For in vitro ubiquitylation assays, individual core histones, histone octamers or HeLa H1-depleted oligonucleosomes were incubated with H1.2 complex (100 ng) or reconstituted CDR/CDRV/CDRW complex (100 ng) in the presence of E1 (25 ng, Sigma), E2 (50 ng, UbcH5c, Sigma) and of His-/Flag-ubiquitin (500 ng) in the reaction buffer (50 mM Tris-HCl, pH 7.9, 5 mM MgCl2, 0.4 mM DTT, and 4 mM ATP) for 0.5–1 h at 37 °C. To use the neddylated complex in the assays, CDR/CDRV/CDRW complex was pre-incubated with E1 (APPBP1, Bostone Biochem), E2 (UBCH12, Bostone Biochem) and His-Nedd8 (Bostone Biochem) in the reaction buffer for 2 h at 37 °C. In vivo ubiquitylation assays were performed as described (Yan et al., 2009). In brief, 293T cells stably expressing shRNAs targeting H1.2, VprBP, WDR5 and PAF1 were transfected with expression vectors encoding Flag-histone, HA-ubiquitin and/or Myc-Cul4A. Two days after transfection, cells were washed with PBS and lyzed in immunoprecipitation (IP) buffer (20 mM Tris, pH 7.5, 50 mM NaCl, 2.5% SDS, 2.5% sodium deoxycholate, and 0.5 mM PMSF). After IP with anti-Flag M2 agarose beads, precipitated proteins were eluted with 0.2 mg/ml Flag peptide (Sigma) and analyzed by immunoblotting. To determine H4 ubiquitylation site(s) in vivo, 293T cells were transfected with Flag-H4 for 48 h, and cleared lysates were immunoprecitated with anti-Flag antibody. The precipitated H4 proteins were run on 15% SDS-PAGE, and the ubiquitylated H4 band was cut out from a Coomassie stained gel and subjected to LC-MS/MS analysis.
Details for other experimental procedures can be found in Supplemental Information.
Supplementary Material
HIGHLIGHTS.
H1.2 is a critical regulator of Cul4A-mediated H4K31 ubiquitylation
H3K4me1/me3 and H3K79me1/me2 are dependent of H4K31 ubiquitylation
H1.2 cooperates with Cul4A and PAF1 to activate developmental regulatory genes
H1.2 interaction with PNAPII is required for Cul4A/PAF1 function at target genes
Acknowledgments
We are grateful to O. Rozenblatt-Rosen and M. Meyerson for antibodies against Ctr9, Leo1 and Cdc73. This work was supported by NIH Grant GM84209 (W.A.), ACS Research Scholar Grant DMC-1005001 (W. A.) and NIH Grant CA129325 (R.G.R.).
Footnotes
ACCESSION NUMBERS
The NCBI GEO accession number for microarray data reported in this paper is GSE53414.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alami R, Fan Y, Pack S, Sonbuchner TM, Besse A, Lin Q, Greally JM, Skoultchi AI, Bouhassira EE. Mammalian linker-histone subtypes differentially affect gene expression in vivo. Proc Natl Acad Sci U S A. 2003;100:5920–5925. doi: 10.1073/pnas.0736105100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barra JL, Rhounim L, Rossignol JL, Faugeron G. Histone H1 is dispensable for methylation-associated gene silencing in Ascobolus immersus and essential for long life span. Mol Cell Biol. 2000;20:61–69. doi: 10.1128/mcb.20.1.61-69.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DT, Alexander BT, Sittman DB. Differential effect of H1 variant overexpression on cell cycle progression and gene expression. Nucleic Acids Res. 1996;24:486–493. doi: 10.1093/nar/24.3.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin M, Catez F, Lim JH. The dynamics of histone H1 function in chromatin. Mol Cell. 2005;17:617–620. doi: 10.1016/j.molcel.2005.02.019. [DOI] [PubMed] [Google Scholar]
- Chandrasekharan MB, Huang F, Chen YC, Sun ZW. Histone H2B C-terminal helix mediates trans-histone H3K4 methylation independent of H2B ubiquitination. Mol Cell Biol. 2010;30:3216–3232. doi: 10.1128/MCB.01008-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielsen JM, Sylvestersen KB, Bekker-Jensen S, Szklarczyk D, Poulsen JW, Horn H, Jensen LJ, Mailand N, Nielsen ML. Mass spectrometric analysis of lysine ubiquitylation reveals promiscuity at site level. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.003590. M110 003590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dover J, Schneider J, Tawiah-Boateng MA, Wood A, Dean K, Johnston M, Shilatifard A. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J Biol Chem. 2002;277:28368–28371. doi: 10.1074/jbc.C200348200. [DOI] [PubMed] [Google Scholar]
- Emanuele MJ, Elia AE, Xu Q, Thoma CR, Izhar L, Leng Y, Guo A, Chen YN, Rush J, Hsu PW, et al. Global identification of modular cullin-RING ligase substrates. Cell. 2011;147:459–474. doi: 10.1016/j.cell.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Nikitina T, Zhao J, Fleury TJ, Bhattacharyya R, Bouhassira EE, Stein A, Woodcock CL, Skoultchi AI. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell. 2005;123:1199–1212. doi: 10.1016/j.cell.2005.10.028. [DOI] [PubMed] [Google Scholar]
- Foster ER, Downs JA. Methylation of H3 K4 and K79 is not strictly dependent on H2B K123 ubiquitylation. J Cell Biol. 2009;184:631–638. doi: 10.1083/jcb.200812088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgel PT, Fletcher TM, Hager GL, Hansen JC. Formation of higher-order secondary and tertiary chromatin structures by genomic mouse mammary tumor virus promoters. Genes Dev. 2003;17:1617–1629. doi: 10.1101/gad.1097603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber M, Shilatifard A. Transcriptional elongation by RNA polymerase II and histone methylation. J Biol Chem. 2003;278:26303–26306. doi: 10.1074/jbc.R300014200. [DOI] [PubMed] [Google Scholar]
- Happel N, Warneboldt J, Hanecke K, Haller F, Doenecke D. H1 subtype expression during cell proliferation and growth arrest. Cell Cycle. 2009;8:2226–2232. doi: 10.4161/cc.8.14.8982. [DOI] [PubMed] [Google Scholar]
- Higa LA, Wu M, Ye T, Kobayashi R, Sun H, Zhang H. CUL4-DDB1 ubiquitin ligase interacts with multiple WD40-repeat proteins and regulates histone methylation. Nat Cell Biol. 2006;8:1277–1283. doi: 10.1038/ncb1490. [DOI] [PubMed] [Google Scholar]
- Higa LA, Zhang H. Stealing the spotlight: CUL4-DDB1 ubiquitin ligase docks WD40-repeat proteins to destroy. Cell Div. 2007;2:5. doi: 10.1186/1747-1028-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S, Xiong Y. CRL4s: the CUL4-RING E3 ubiquitin ligases. Trends Biochem Sci. 2009;34:562–570. doi: 10.1016/j.tibs.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedrusik MA, Schulze E. Linker histone HIS-24 (H1.1) cytoplasmic retention promotes germ line development and influences histone H3 methylation in Caenorhabditis elegans. Mol Cell Biol. 2007;27:2229–2239. doi: 10.1128/MCB.01713-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Guermah M, McGinty RK, Lee JS, Tang Z, Milne TA, Shilatifard A, Muir TW, Roeder RG. RAD6-Mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell. 2009;137:459–471. doi: 10.1016/j.cell.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Guermah M, Roeder RG. The human PAF1 complex acts in chromatin transcription elongation both independently and cooperatively with SII/TFIIS. Cell. 2010;140:491–503. doi: 10.1016/j.cell.2009.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Choi J, Heo K, Kim H, Levens D, Kohno K, Johnson EM, Brock HW, An W. Isolation and characterization of a novel H1.2 complex that acts as a repressor of p53-mediated transcription. J Biol Chem. 2008;283:9113–9126. doi: 10.1074/jbc.M708205200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotake Y, Zeng Y, Xiong Y. DDB1-CUL4 and MLL1 mediate oncogene-induced p16INK4a activation. Cancer Res. 2009;69:1809–1814. doi: 10.1158/0008-5472.CAN-08-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Dover J, Wood A, Schneider J, Heidt J, Boateng MA, Dean K, Ryan OW, Golshani A, Johnston M, et al. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol Cell. 2003;11:721–729. doi: 10.1016/s1097-2765(03)00091-1. [DOI] [PubMed] [Google Scholar]
- Lee H, Habas R, Abate-Shen C. MSX1 cooperates with histone H1b for inhibition of transcription and myogenesis. Science. 2004;304:1675–1678. doi: 10.1126/science.1098096. [DOI] [PubMed] [Google Scholar]
- Li B, Ruiz JC, Chun KT. CUL-4A is critical for early embryonic development. Mol Cell Biol. 2002;22:4997–5005. doi: 10.1128/MCB.22.14.4997-5005.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Lee S, Zhang J, Peters SB, Hannah J, Zhang Y, Yin Y, Koff A, Ma L, Zhou P. CUL4A abrogation augments DNA damage response and protection against skin carcinogenesis. Mol Cell. 2009;34:451–460. doi: 10.1016/j.molcel.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Wontakal SN, Kavi H, Kim BJ, Guzzardo PM, Emelyanov AV, Xu N, Hannon GJ, Zavadil J, Fyodorov DV, et al. Drosophila H1 regulates the genetic activity of heterochromatin by recruitment of Su(var)3–9. Science. 2013;340:78–81. doi: 10.1126/science.1234654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinty RK, Kim J, Chatterjee C, Roeder RG, Muir TW. Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature. 2008;453:812–816. doi: 10.1038/nature06906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller CL, Jaehning JA. Ctr9, Rtf1, and Leo1 are components of the Paf1/RNA polymerase II complex. Mol Cell Biol. 2002;22:1971–1980. doi: 10.1128/MCB.22.7.1971-1980.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muntean AG, Tan J, Sitwala K, Huang Y, Bronstein J, Connelly JA, Basrur V, Elenitoba-Johnson KS, Hess JL. The PAF complex synergizes with MLL fusion proteins at HOX loci to promote leukemogenesis. Cancer Cell. 2010;17:609–621. doi: 10.1016/j.ccr.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- Osley MA. H2B ubiquitylation: the end is in sight. Biochim Biophys Acta. 2004;1677:74–78. doi: 10.1016/j.bbaexp.2003.10.013. [DOI] [PubMed] [Google Scholar]
- Ponte I, Vila R, Suau P. Sequence complexity of histone H1 subtypes. Mol Biol Evol. 2003;20:371–380. doi: 10.1093/molbev/msg041. [DOI] [PubMed] [Google Scholar]
- Shen X, Gorovsky MA. Linker histone H1 regulates specific gene expression but not global transcription in vivo. Cell. 1996;86:475–483. doi: 10.1016/s0092-8674(00)80120-8. [DOI] [PubMed] [Google Scholar]
- Studencka M, Konzer A, Moneron G, Wenzel D, Opitz L, Salinas-Riester G, Bedet C, Kruger M, Hell SW, Wisniewski JR, et al. Novel roles of C. elegans heterochromatin protein HP1 and linker histone in the regulation of innate immune gene expression. Mol Cell Biol. 2011 doi: 10.1128/MCB.05229-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- Trievel RC, Shilatifard A. WDR5, a complexed protein. Nat Struct Mol Biol. 2009;16:678–680. doi: 10.1038/nsmb0709-678. [DOI] [PubMed] [Google Scholar]
- Wang H, Zhai L, Xu J, Joo HY, Jackson S, Erdjument-Bromage H, Tempst P, Xiong Y, Zhang Y. Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Mol Cell. 2006;22:383–394. doi: 10.1016/j.molcel.2006.03.035. [DOI] [PubMed] [Google Scholar]
- Wang Z, Cui B, Gorovsky MA. Histone H2B ubiquitylation is not required for histone H3 methylation at lysine 4 in tetrahymena. J Biol Chem. 2009;284:34870–34879. doi: 10.1074/jbc.M109.046250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weake VM, Workman JL. Histone ubiquitination: triggering gene activity. Mol Cell. 2008;29:653–663. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- Wood A, Schneider J, Dover J, Johnston M, Shilatifard A. The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J Biol Chem. 2003;278:34739–34742. doi: 10.1074/jbc.C300269200. [DOI] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, Roeder RG, Brivanlou AH, Allis CD. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- Yan Q, Dutt S, Xu R, Graves K, Juszczynski P, Manis JP, Shipp MA. BBAP monoubiquitylates histone H4 at lysine 91 and selectively modulates the DNA damage response. Mol Cell. 2009;36:110–120. doi: 10.1016/j.molcel.2009.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Cooke M, Panjwani S, Cao K, Krauth B, Ho PY, Medrzycki M, Berhe DT, Pan C, McDevitt TC, et al. Histone h1 depletion impairs embryonic stem cell differentiation. PLoS Genet. 2012a;8:e1002691. doi: 10.1371/journal.pgen.1002691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu Z, Medrzycki M, Cao K, Fan Y. Reduction of Hox gene expression by histone H1 depletion. PLoS One. 2012b;7:e38829. doi: 10.1371/journal.pone.0038829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Zheng Y, Pham AD, Mandal SS, Erdjument-Bromage H, Tempst P, Reinberg D. Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Mol Cell. 2005;20:601–611. doi: 10.1016/j.molcel.2005.09.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.