Abstract
NY-ESO-1 and LAGE-1 represent highly homologous cancer-germline Ags frequently coexpressed by many human cancers, but not by normal tissues, except testis. In contrast to NY-ESO-1, little is known about spontaneous immune responses to LAGE-1. In the current study, we report on spontaneous LAGE-1–specific CD4+ T cells isolated from PBLs of patients with advanced LAGE-1+/ NY-ESO-1+ melanoma and directed against three promiscuous and immunodominant epitopes. Strikingly, although the three LAGE-1–derived epitopes are highly homologous to NY-ESO-1–derived epitopes, LAGE-1–specific CD4+ T cells did not cross-react with NY-ESO-1. LAGE-1–specific CD4+ T cells produced Th1-type and/or Th2-type cytokines and did not exert inhibitory effects on allogenic T cells. We observed that most patients with spontaneous NY-ESO-1–specific responses exhibited spontaneous CD4+ T cell responses to at least one of the three immunodominant LAGE-1 epitopes. Additionally, nearly half of the patients with spontaneous LAGE-1–specific CD4+ T cell responses had circulating LAGE-1–specific Abs that recognized epitopes located in the C-terminal portion of LAGE-1, which is distinct from NY-ESO-1. Collectively, our findings define the hierarchy of immunodominance of spontaneous LAGE-1–specific CD4+ T cell responses in patients with advanced melanoma. These findings demonstrate the capability of LAGE-1 to stimulate integrated cellular and humoral immune responses that do not cross-react with NY-ESO-1. Therefore, they provide a strong rationale for the inclusion of LAGE-1 peptides or protein in vaccine trials for patients with NY-ESO-1+/LAGE-1+ tumors.
LAGE-1 and NY-ESO-1 are cancer-germline Ags expressed by many human tumors but not by normal tissue except testis (1). Because the germ cells in testis do not express MHC molecules (2), cancer-germline Ag-derived epitopes are strictly tumor-specific T cell targets (3). Therefore, they represent interesting candidates for cancer vaccines, as they are less likely to induce tolerance or autoimmunity.
The LAGE-1 gene yields two mRNA transcripts, respectively named LAGE-1a (or LAGE-1S) and LAGE-1b (or LAGE-1L) (4). The primary open reading frame (ORF1) of the genes NY-ESO-1 and LAGE-1a encodes two homologous 180-aa–long proteins, while the LAGE-1b ORF1 encodes a putative 210-aa–long protein. The alternative or nonprimary open reading frames (ORF2) of the genes NY-ESO-1 and LAGE-1 encode two putative proteins that are 58 and 109 aa long, respectively. NY-ESO-1 and LAGE-1 appear to be frequently coexpressed by tumors, although some tumors express only one of these genes (4). LAGE-1a and NY-ESO-1 proteins are highly homologous with ~84% shared identity, while LAGE1b shares ~76.6% homology with NY-ESO-1 in its N-terminal portion (first 141 aa) and then differs in its C-terminal portion. LAGE-1a and LAGE-1b protein sequences are identical for the first 134 aa.
Spontaneous immune responses to NY-ESO-1 in patients with advanced cancers have been extensively studied. NY-ESO-1 appears to be very immunogenic, inducing both spontaneous cellular and humoral responses in patients with NY-ESO-1+ tumors (5, 6). In particular, we and others have identified three immunodominant amino acid sequences (NY-ESO-180–111, NY-ESO-1119–143, and NY-ESO-1157–170) stimulating spontaneous CD4+ T cell responses in patients with advanced NY-ESO-1+ cancers (7–10). In contrast, little is known about spontaneous immune responses to LAGE-1. To date, only one MHC class I epitope (11) and one MHC class II epitope (12) encoded by LAGE-1 but not by NY-ESO-1 have been identified. Whether an overlap exists between the CD4+ T cell responses to LAGE-1 and NY-ESO-1 remains to be determined. Such information will be critical in defining whether NY-ESO-1 peptide- and protein-based vaccines need to be optimized with the inclusion of LAGE-1 epitopes or protein to stimulate broader T cell responses against LAGE-1+/ NY-ESO-1+ tumors. To address this question, we have investigated spontaneous CD4+ T cell responses to LAGE-1 in patients with advanced melanoma with LAGE-1+/NY-ESO-1+ tumors. Our findings showed that spontaneous LAGE-1–specific CD4+ T cells are directed against three promiscuous and immunodominant epitopes and did not cross-react with NY-ESO-1. Additionally, we observed the existence of spontaneous LAGE-1–specific humoral responses. Collectively, our data support the inclusion of LAGE-1 peptides or protein in vaccine trials for patients with LAGE-1+/ NY-ESO-1+ tumors.
Materials and Methods
Cell lines, media
Blood samples were obtained under the University of Pittsburgh Cancer Institute (UPCI) Institutional Review Board-approved protocols 96–099 and 00–079. HLA-DR and HLA-DP genotyping of melanoma patients was performed using commercial typing panels of PCR primers according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA). HLA-DR– transfected cell lines (i.e., L.DR cells) were previously described (9).
Melanoma cell lines UPCI-MEL 285.1 (LAGE-1+) and UPCI-MEL 136.1 (LAGE-1−) were previously described (13). All cell lines were cultured in RPMI 1640 medium (Invitrogen) supplemented with 10% FCS, L-glutamine (2 mM), penicillin (100 U/ml), streptomycin (100 µg/ml) (Gemini Bio-Products, West Sacramento, CA), HEPES (10 mM), and MEM nonessential amino acids solution (0.1 mM) (Invitrogen). CD4+ T cell clones were cultured in Iscove’s medium (Mediatech, Manassas, VA) supplemented with 10% human AB serum (Sigma-Aldrich, St. Louis, MO) as previously reported (9, 10).
Peptide synthesis
The LAGE-1–, NY-ESO-1–, and HIV-derived peptides were synthesized using standard Fmoc chemistry by the University of Pittsburgh Peptide Synthesis Facility (shared resource), were .95% pure as indicated by analytical HPLC, and were validated by mass spectrometry. Lyophilized peptides were dissolved in 100% DMSO at a concentration of 2 mg/ml and stored at −20°C until use.
HLA-DR and HLA-DP peptide binding assays
The binding to the multiple purified HLA-DR and HLA-DP molecules was performed as previously reported (8, 9, 14, 15). Maximal binding was achieved by incubating the biotinylated peptide with the MHC class II molecule in the absence of competitor. Binding specificity for each HLA-DR and HLA-DP molecule was ensured by the choice of the biotinylated peptides. Data are expressed as the concentration of peptide that prevented binding of 50% of the labeled peptide (IC50).
Transfection of COS-7 cells and cell lysates preparation
COS-7 cells were plated in six-well plates (5 × 105 cells/well) 18 h before transfection. The cells were transiently transfected with 4 µg/well pcDNA3-NY-ESO-1 (provided by Drs. Elke Jager and Alexander Knuth, Department of Oncology, Krankenhaus Nordwest, Frankfurt, Germany, and Universita¨tsspital Zu¨rich, Zurich, Switzerland, respectively, and by Dr. Y. T. Chen, Cornell University, New York) or pcDNA3-Lage-1b (provided by Dr. Bernard Lethe, Ludwig Institute for Cancer Research, Brussels, Belgium) using Lipofectamine (Invitrogen). Transfected cells were incubated for 48 h at 37°C and 5% CO2. Transfection efficacy was assessed by flow cytometry. Cells were harvested and lysed in AIM-V medium (Invitrogen) at a concentration of 1 × 107 cells/ml by five rapid freeze-thaw cycles. Cell lysates were centrifuged at 14,000 rpm for 10 min. Supernatants were collected and stored at −20°C until use.
Expansion of CD4+ T cells with peptides
The induction of CD4+ T cells in vitro with dendritic cells (DCs) and LAGE-1–derived peptides was performed as previously reported (8, 9, 16, 17). The T cells were used for in vitro assays 10–12 d following the last stimulation.
IFN-γ ELISPOT assays
The recognition of APCs (L.DR cells) pulsed with LAGE-1–derived peptides by the LAGE-1–specific bulk CD4+ T cells and LAGE-1–specific CD4+ T cell clones expanded in vitro was assessed by ELISPOT assays specific for human IFN-γ as described previously (9). Alternatively, 5 × 103 DCs loaded with recombinant proteins or cell lysates were added to 1 × 103 CD4+ T cell clones/well as previously reported (8). Immature DCs (1 × 106) were incubated with cell lysates from pcDNA3-LAGE-1b or pcDNA-NY-ESO-1– transfected COS-7 cells (3 × 106), or from melanoma cell lines (3 × 106), or with 1 µM recombinant proteins LAGE-1a (GlaxoSmithKline, Rixensart, Belgium) or LAGE-1 ORF2 (13) for 4 h at 37°C in the presence of 1 µg/ml LPS (Sigma-Aldrich). DCs were subsequently washed three times before adding to the CD4+ T cells. Spot numbers and spot sizes were determined with computer-assisted video image analysis (Cellular Technologies, Cleve-land, OH). For statistical evaluation, a t test was used. The p values <0.05 were considered significant.
In vitro sensitization of T cells with peptides and intracellular cytokine staining
PBLs (5 ×106) from melanoma patients and healthy donors were pulsed in 200 µl T cell culture medium with single LAGE-1–derived peptides for 60 min at 37°C in 24-well plates. Culture medium containing 50 IU/ml IL-2 (PeproTech, Rocky Hill, NJ) was then added to a final volume of 2.5 ml/ well. The medium was changed every 2 d. After 10–12 d incubation, intracellular cytokine staining (ICS) for IFN-γ and IL-5 in the presence of the relevant LAGE-1–derived peptides or irrelevant peptide HIVpol711–725 (1 µg/ml) was performed as previously described (18). One million events per sample were acquired during flow cytometric analysis. The frequencies of cytokine-producing LAGE-1–specific CD4+ T cells were calculated by subtracting a mean value obtained for samples restimulated with an irrelevant peptide from values obtained for samples restimulated with LAGE-1–specific peptides.
ELISA
Serum NY-ESO-1– and LAGE-1b–specific Abs were measured with ELISA from sera of patients with NY-ESO-1+/LAGE-1+ or NY-ESO-1−/ LAGE-1− tumors. Briefly, recombinant NY-ESO-1 protein and eight overlapping peptides whose 18-aa–long sequences span the LAGE-1b protein from P132 to P210 (Table IV) were diluted in coating buffer (15 mM Na2CO3, 30 mM NaHCO3 [pH 9.6], with 0.02% NaN3) to the concentration of 1 µg/ml and adsorbed to polypropylene flat-bottom 96-well plates (Corning, Corning, NY) overnight at 4°C. Plates were washed with PBS and saturated overnight at 4°C by addition of 200 µl/well PBS containing 2% BSA. After the plates were washed, serum was diluted in PBS/ 2% BSA buffer and added to the plates (50 µl/well). After 2 h incubation at room temperature, plates were washed, the secondary Ab (goat anti-human IgG-AP; Invitrogen) was added (50 µl/well), and the plates were further incubated for 1 h at room temperature. Following additional washes, 50 µl/well substrate solution (phosphatase substrate system; Kirkegaard & Perry Laboratories, Gaithersburg, MD) was added, and plates were incubated for 25 min at room temperature and read immediately (MRX microplate reader; Dynatech, Chantilly, VA). Sera were tested over a range of 4-fold dilutions as previously described (6).
Table IV.
Assessment of LAGE-1b–specific serum Abs in patients with advanced melanoma
| Peptides |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Patients | LAGE-1b132–149 | LAGE-1b139–156 | LAGE-1b146–163 | LAGE-1b153–170 | LAGE-1b160–177 | LAGE-1b167–184 | LAGE-1b181–198 | LAGE-1b188–210 | NY-ESO-1 |
| MP1 | – | – | – | – | – | – | – | – | +++ |
| MP2 | – | – | – | – | – | – | – | – | +++ |
| MP3 | – | – | – | – | – | – | – | – | +++ |
| MP4 | – | – | – | – | – | – | – | – | +++ |
| MP5 | – | – | – | – | – | ++ | – | ++ | +++ |
| MP6 | – | ++ | – | – | ++ | ++ | – | ++ | ++ |
| MP7 | – | – | – | – | – | ++ | – | ++ | +++ |
| MP8 | – | – | – | – | – | – | – | – | +++ |
| MP9 | – | ++ | – | – | ++ | ++ | – | ++ | +++ |
| MP10 | – | – | – | – | – | – | – | – | +++ |
| MP11 | – | – | – | – | – | – | – | – | +++ |
| MP12 | – | – | – | – | – | – | – | – | +++ |
| MP13 | – | – | – | – | – | – | – | – | +++ |
| MP14 | – | – | – | – | – | – | – | – | +++ |
| MP15 | – | – | – | – | – | – | – | – | +++ |
| MP16 | – | – | – | – | – | – | – | – | ++ |
| MP17 | – | – | – | – | – | – | – | – | – |
| MP18 | – | – | – | – | – | – | – | – | – |
| MP19 | – | – | – | – | – | – | – | – | – |
| MP20 | – | – | – | – | – | – | – | – | – |
| MP21 | – | – | – | – | – | – | – | – | – |
−, Mean of patient sample at 1/400 dilution < (mean [negative control-normal donor] + 3 SD at 1:400 dilution); +, mean of patient sample at 1/400 dilution > (mean [negative control-normal donor] + 3 SD at 1/400 dilution); ++, mean of patient sample at 1/1600 dilution > (mean [negative control-normal donor] + 3 SD at 1/400 dilution); +++, mean of patient sample at 1/6400 dilution > (mean [negative control-normal donor] + 3 SD at 1/400 dilution).
CFSE proliferation assays
Naive CD4+ and CD8+ T cells were purified from human PBMCs obtained from the blood of healthy donors by magnetic cell separation using CD45RA microbeads following the manufacturer’s instructions (Miltenyi Biotec, Auburn, CA) and stained with CFSE (Invitrogen). CFSE-labeled naive T cells (1 × 105) were cultured for 5 d in 96-well flat bottom plates containing 2 × 105 irradiated (5000 rad) autologous CD3-depleted APCs, 0.1 µg/ml anti-CD3 (clone OKT3; eBioscience, San Diego, CA), IL-2 (50 IU/ml) (PeproTech), and different numbers of LAGE-1–specific CD4+ T cell clones. After 5 d, cells were collected and stained with anti–CD4-PE, anti–CD14-ECD, anti–CD19-ECD, and anti– CD8-PE-Cy5 (Beckman Coulter, Fullerton, CA), and proliferation of responder naive cells was analyzed by flow cytometry. As control, CFSE-labeled T cells were also incubated with TRAG-3–specific CD4+ regulatory T cell (Treg) clone 62/3 (19).
Results
LAGE-1 encodes multiple promiscuous HLA-DR–binding peptide sequences
The N-terminal portion (P1–P70) of the LAGE-1a and LAGE-1b proteins (GenBank accession number AJ223093) and the C-terminal portion (P135–P180) of the LAGE-1a protein are almost identical to NY-ESO-1 (NM001327) with only one amino acid difference at P57 and P174, respectively. In contrast, the amino acid sequences spanning from P71 to P134 and from P71 to P210 for LAGE-1a and LAGE-1b, respectively, exhibit less homology to the NY-ESO-1 protein (Fig. 1). To investigate the existence of MHC class II epitopes encoded by LAGE-1 but not by NY-ESO-1, we synthesized 18-aa–long overlapping peptide sequences that span these two amino acid regions (i.e., LAGE-1 P69–P134 and LAGE-1b P69–P210) and evaluated their binding capacities to 10 different HLA-DR and 2 HLA-DP4 molecules, including the 7 molecules encoded by the HLA-DRB1 genes (i.e., HLA-DRB1*0101, HLA-DRB1*0301, HLA-DRB1*0401, HLA-DRB1*0701, HLA-DRB1*1101, HLA-DRB1*1301, and HLA-DRB1*1501), 3 molecules encoded, respectively, by the HLA-DRB3*0101, HLA-DRB4*0101, and HLA-DRB5*0101 genes, and the HLA-DPB1*0401 and HLA-DPB1*0402 molecules (Table I). Binding activity of each peptide was estimated by the IC50 and reported as relative activity to easily compare with high binder peptides.
FIGURE 1.
Alignment of LAGE-1a, LAGE-1b, and NY-ESO-1 amino acid sequences. The identical amino acids are indicated by star symbols. Peptide sequences corresponding to position 97–114 (dotted line), 104–121 (solid line), and 118–135 (dashed line) are underlined. Accession numbers: NY-ESO-1, NM001327; LAGE-1a and LAGE-1b, AJ223093.
Table I.
Binding capacities to HLA-DR and HLA-DP4 molecules of overlapping peptides from LAGE-1
| HLA II Alleles | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Peptides | HLA-DR B1*0101 |
HLA-DR B1*0301 |
HLA-DR B1*0401 |
HLA-DR B1*0701 |
HLA-DR B1*1101 |
HLA-DR B1*1301 |
HLA-DR B1*1501 |
HLA-DR B3*0101 |
HLA-DR B4*0101 |
HLA-DR B5*0101 |
HLA-DP B1*0401 |
HLA-DP B1*0402 |
| Frequency | 17.7 | 20.6 | 10.9 | 26 | 17.6 | 11.6 | 15.4 | 17.6 | 15.2 | 48.2 | 64 | 20.8 |
| LAGE-169–86 | 20,833 | >255 | >4,578 | >12,703 | 105 | >188 | >3,577 | >10,128 | >1,699 | >18,360 | >7,296 | >5,828 |
| LAGE-176–93 | 171 | >255 | 158 | 1,936 | 63 | >188 | 322 | >10,128 | 224 | 6 | >7,296 | >5,828 |
| LAGE-183–100 | 37 | >255 | 16 | 1 | 17 | 3 | 98 | >10,128 | 27 | 181 | 326 | 260 |
| LAGE-190–107 | 9 | >255 | 393 | 37 | 5,701 | >188 | 35 | 3,693 | >1,699 | 977 | 3,266 | 251 |
| LAGE-197–114 | 215 | 50 | 112 | 51 | 1 | 1 | 2 | 2,384 | 12 | 661 | 130 | 23 |
| LAGE-1104–121 | 1 | 102 | 11 | 43 | 32 | 2 | 0.3 | >10,128 | 20 | 3 | >7,296 | >5,828 |
| LAGE-1111–128 | 13,680 | >255 | 9 | 4,472 | 5,292 | >188 | 100 | >10,128 | 181 | 9,454 | >7,296 | >5,828 |
| LAGE-1b118–135 | 58 | 1 | 30 | 1 | 6 | 2 | 3 | 4 | >1,699 | 17 | 43 | 23 |
| LAGE-1b125–142 | 205 | 2 | 100 | 31 | 26 | 2 | >3,577 | >10,128 | >1,699 | 28 | >7,296 | 88 |
| LAGE-1b132–149 | 183 | 19 | 84 | 112 | 47 | 145 | 1,805 | >10,128 | 297 | 0.4 | >7,296 | 215 |
| LAGE-1b139–156 | 4,516 | >255 | >4,578 | 253 | 1,570 | >188 | 26 | >10,128 | 1,534 | 120 | >7,296 | >5,828 |
| LAGE-1b146–163 | 1,389 | >255 | 1,633 | 393 | 1,167 | >188 | 2 | >10,128 | 1,085 | 68 | >7,296 | 833 |
| LAGE-1b153–170 | 19,118 | >255 | >4,578 | >12,703 | >8,367 | >188 | >3,577 | >10,128 | >1,699 | 4,183 | >7,296 | >5,828 |
| LAGE-1b160–177 | 16,667 | >255 | >4,578 | >12,703 | 970 | >188 | >3,577 | >10,128 | >1,699 | 4,610 | >7,296 | >5,828 |
| LAGE-1b167–184 | >39,260 | >255 | >4,578 | >12,703 | >8,367 | >188 | >3,577 | >10,128 | >1,699 | >18,360 | >7,296 | >5,828 |
| LAGE-1b181–198 | >39,260 | >255 | >4,578 | >12,703 | 3 | >188 | 2,375 | >10,128 | >1,699 | >18,360 | >7,296 | >5,828 |
| LAGE-1b188–210 | 3 | >255 | 3 | 15 | 5 | >188 | 260 | >10,128 | >1,699 | 0.3 | 16 | 6 |
Peptides encompassing the sequences of LAGE-1a (P69–P134) and LAGE-1b (P69–P210), which do not overlap with NY-ESO-1, were submitted to binding assays specific for HLA-DR and HLA-DP4 molecules. Reference peptides were used to validate each assay. These peptides are the nonbiotinylated forms of the biotinylated peptides used in the assay and correspond to very good binders. Data are expressed as relative activity (ratio of the IC50 of the peptides to the IC50 of the reference peptide) and are the means of three experiments. Good binders have a relative activity <100 and are in boldface type.
Six peptides encoded by LAGE-1a and/or LAGE-1b (LAGE-183–100, LAGE-197–114, LAGE-1104–121, LAGE-1b118–135, LAGE-1b125–142, and LAGE-1b188–210) exhibited a very large specificity for HLA class II, as they bound to at least seven different HLA class II molecules. In contrast, other peptides located between positions 153 and 184 did not bind to any HLA class II molecules. Except for LAGE-1b188–210, promiscuous MHC class II binder peptides reside in the central part of LAGE-1, especially between positions 83 and 142 (Table I).
We next performed a series of additional experiments to investigate the immunogenicity of the LAGE-1 peptide sequences.
LAGE-1–derived promiscuous epitopes stimulate Th1-type CD4+ T cells isolated from patients with melanoma
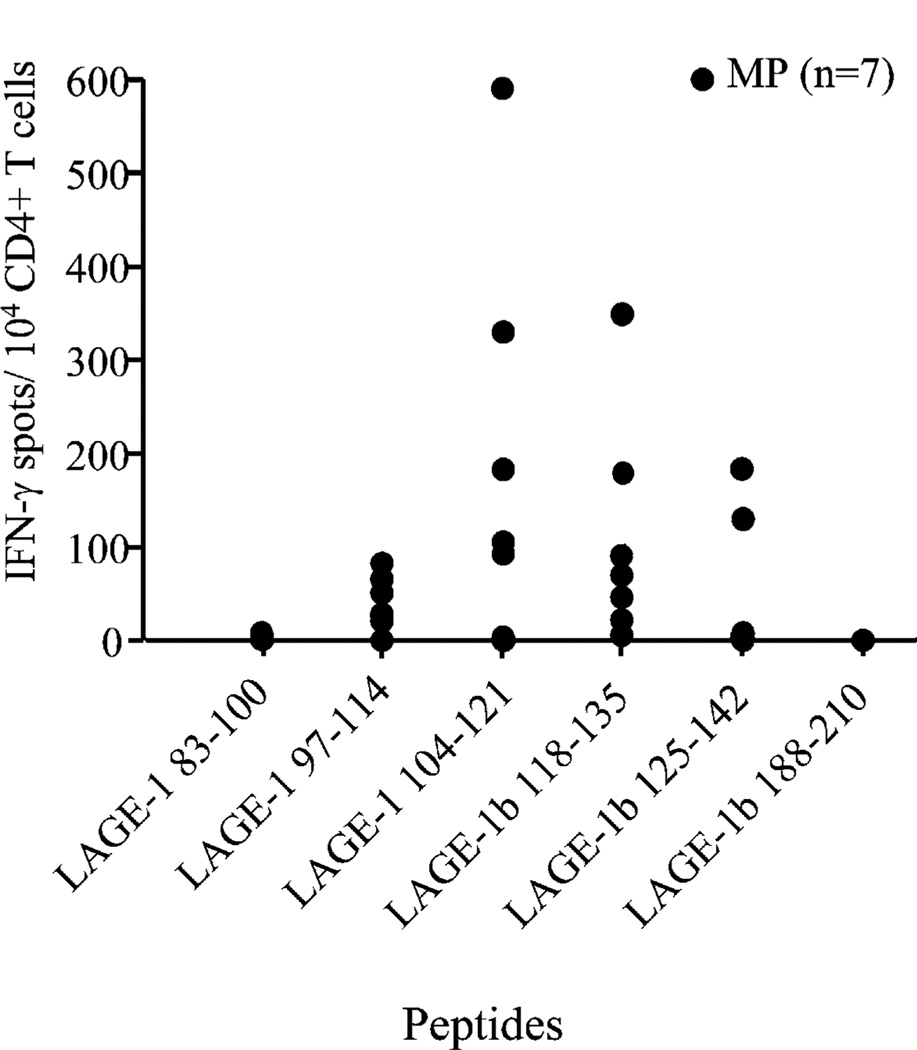
To investigate the immunogenicity of the LAGE-1–derived peptide sequences, we have stimulated CD4+ T cells from seven melanoma patients (MP1–MP7, Table II) with stage IV NY-ESO-1+/ LAGE-1+ tumors against a pool of peptides containing the six promiscuous HLA-DR–binding LAGE-1 peptides. These patients expressed the most common HLA-DR alleles (Table II). Monocyte-derived DCs were incubated with the pool of peptides (10 µg/ml each), irradiated, and used to stimulate autologous CD4+ T cells isolated from PBLs as described in Materials and Methods. After three weekly stimulations with autologous DCs pulsed with the pool of LAGE-1 peptides, we assessed the reactivity of CD4+ T cells against L.DR cells pulsed with each of the six LAGE-1 peptides with the highest binding specificity for HLA class II molecules in IFN-γ ELISPOT assays. Bulk CD4+ T cells produced IFN-γ in the presence of L.DR cells pulsed with peptide LAGE-197–114 in five out of seven of the melanoma patients, with peptide LAGE-1104–121 in five out of seven patients, and with peptide LAGE-1b118–135 in six out of seven melanoma patients (Fig. 2). The CD4+ T cells failed to produce significant amounts of IFN-γ in the presence of APCs pulsed with the pan-DR peptide LAGE-1 ORF285–102, used as an irrelevant peptide (13). Bulk CD4+ T cells from two out of seven melanoma patients recognized L.DR cells pulsed with peptide LAGE-1b125–142. We detected no CD4+ T cell reactivity against peptides LAGE-183–100 and LAGE-1b188–210, although our binding data demonstrated that they were good binders to multiple HLA-DR molecules.
Table II.
HLA typing, stage of disease, NY-ESO-1-Ab, LAGE-1+/NY-ESO-1+ tumor status, LAGE-1-Ab, and response types of the patients and normal donors evaluated in the study
| Th1/Th2 Spontaneous Response Test by ICSa |
||||||||
|---|---|---|---|---|---|---|---|---|
| Donors | HLA-DR | Stage of Disease |
NY-ESO-1 Ab |
NY-ESO-1+/LAGE-1+ (PCR) | LAGE-1 Ab |
LAGE-197–114 | LAGE-1104–121 | LAGE1b118–135 |
| Patients | ||||||||
| MP1 | 1102 | IV | + | + | – | Th1/Th2 (213, 79) | Th1 (551, 0) | |
| MP2 | 0101, 0401 | IV | + | + | – | Th2 (0, 100) | Th2 (0, 110) | |
| MP3 | 0701, 1320 | IV | + | + | – | Th1 (130, 0) | ||
| MP4 | 1501 | IV | + | + | – | Th1 (101, 18) | ||
| MP5 | 1301, 1302 | IV | + | + | + | Th1/Th2 (505, 80) | Th1/Th2 (885, 70) | |
| MP6 | 0101, 0102 | IV | + | + | + | Th1 (110, 20) | Th1/Th2 (351, 185) | Th1/Th2 (141, 55) |
| MP7 | 0301, 1501 | IV | + | + | + | Th2 (0, 435) | Th2 (0, 543) | Th2 (0, 743) |
| MP8 | 1101, 1301 | IV | + | + | – | Th1/Th2 (341, 221) | Th1/Th2 (211, 81) | |
| MP9 | 0101, 1301 | IV | + | + | + | Th1/Th2 (83, 193) | Th1/Th2 (205, 65) | Th1/Th2 (133, 53) |
| MP10 | 0101, 0102 | IV | + | + | – | Th1 (219, 15) | ||
| MP11 | 0301, 0701 | IV | + | + | – | Th1 (480, 0) | ||
| MP12 | NA | IV | + | + | – | Th1 (270, 0) | ||
| MP13 | NA | IV | + | + | – | Th1 (110, 0) | ||
| MP14 | NA | IV | + | + | – | Th1 (400, 0) | ||
| MP15 | NA | IV | + | + | – | Th1/Th2 (60, 530) | ||
| MP16 | NA | IV | + | + | – | Th1 (170, 0) | ||
| MP17 | NA | IV | – | – | – | |||
| MP18 | NA | IV | – | – | – | |||
| MP19 | NA | IV | – | – | – | |||
| MP20 | NA | IV | – | – | – | |||
| MP21 | NA | IV | – | – | – | |||
| Normal donors | ||||||||
| ND1 | 1401, 1501 | – | ||||||
| ND2 | 0301, 1001 | – | ||||||
| ND3 | 0301, 0701 | – | ||||||
| ND4 | NA | – | ||||||
| ND5 | NA | – | ||||||
| ND6 | NA | – | ||||||
| ND7 | NA | – | ||||||
| ND8 | NA | – | ||||||
| ND9 | NA | – | ||||||
| ND10 | NA | – | ||||||
| ND11 | NA | – | ||||||
| ND12 | NA | – | ||||||
| ND13 | NA | – | ||||||
| ND14 | NA | – | ||||||
| ND15 | NA | – | ||||||
| ND16 | NA | – | ||||||
Number of IFN-γ–producing CD4+ T cells per 105 CD4+ T cells, number of IL-5–producing CD4+ T cells per 105 CD4+ T cells. Background has been subtracted. NA, not available; ND, normal donor.
FIGURE 2.
CD4+ T cell responses to immunodominant LAGE-1–de-rived peptides. CD4+ T cells isolated from PBLs of melanoma patients (n = 7) were stimulated weekly with irradiated autologous mature DCs pulsed with pooled LAGE-1 peptides prior to testing reactivity against each of the six LAGE-1 peptides and the irrelevant peptide LAGE-1 ORF285–102 in IFN-γ ELISPOT assays. A positive response is defined as mean of spot numbers (triplicate) obtained for a given LAGE-1 peptide greater than the mean of spot numbers obtained for the irrelevant peptide LAGE-1 ORF285–102 + 3 SD and greater than 20 spots per 104 CD4+ T cells. The means of spot numbers obtained for the irrelevant peptide LAGE-1 ORF285–102 (i.e., background) were always <50/104 CD4+ T cells. Each depicted symbol represents the mean of spot numbers (triplicates) observed in the presence of a given LAGE-1 peptide minus the mean of spot numbers in the presence of the irrelevant peptide LAGE-1 ORF285–102 (i.e., background). Spots were developed and counted by computer-assisted video image analysis. One representative experiment of three performed is depicted.
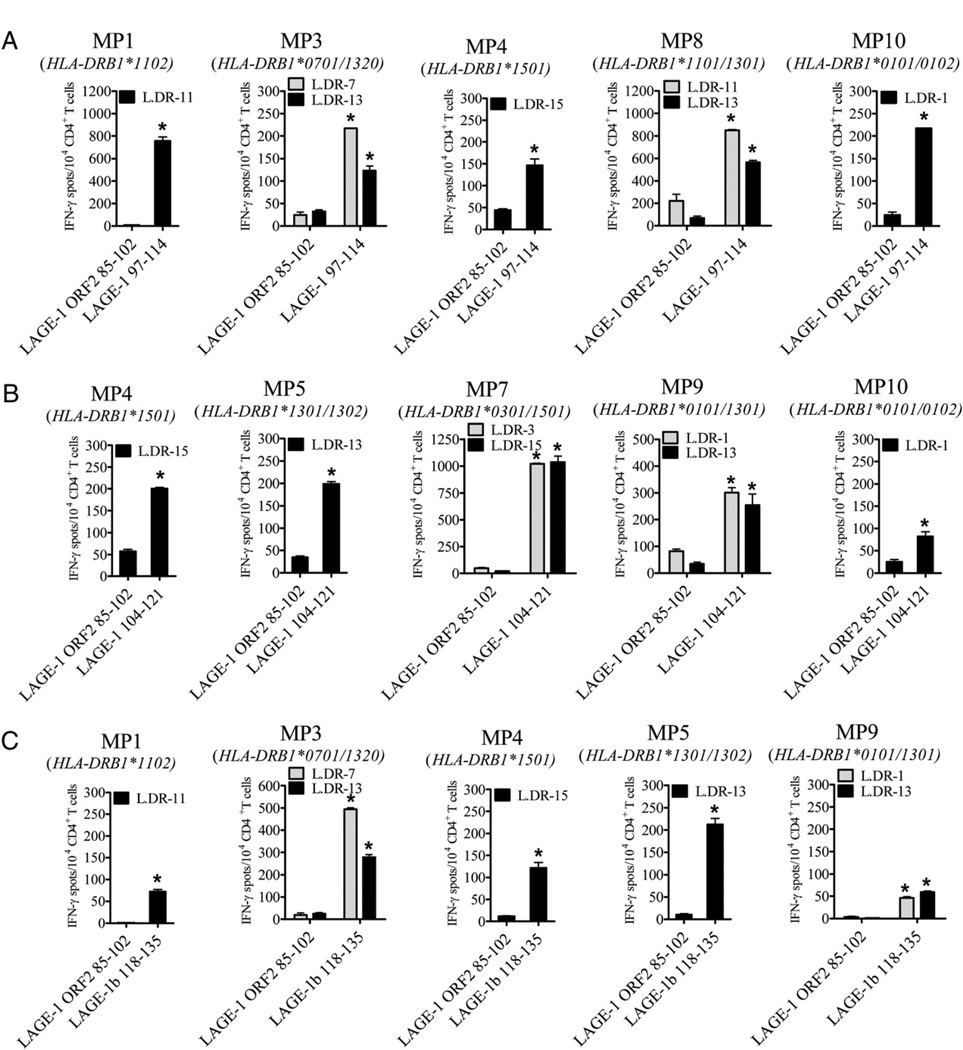
We next stimulated CD4+ T cells isolated from melanoma patients against each of the three LAGE-1–derived peptides, LAGE-197–114, LAGE-1104–121, and LAGE-1b118–135 (Fig. 3, Table III). Bulk CD4+ T cells isolated from MP1, MP3, MP4, MP8, and MP10 and stimulated in vitro with peptide LAGE-197–114 recognized L.DR1, L.DR7, L.DR11, L.DR13, and L.DR15 cells pulsed with relevant peptide but not with an irrelevant peptide (LAGE-1 ORF285–102) in IFN-γ ELISPOT assays (Fig. 3A, Table III). Bulk CD4+ T cells isolated from MP4, MP5, MP7, MP9, and MP10 and stimulated with peptide LAGE-1104–121 peptide specifically recognized L.DR1, L.DR3, L.DR13, and L.DR15 cells pulsed with the relevant peptide but not with an irrelevant peptide (LAGE-1 ORF285–102) in IFN-γ assays (Fig. 3B, Table III). Bulk CD4+ T cells isolated from MP1, MP3, MP4, MP5, or MP9 and stimulated with peptide LAGE-1b118–135 recognized L.DR1, L.DR7, L.DR11, L.DR13, and L.DR15 cells pulsed with relevant peptide but not with an irrelevant peptide (LAGE-1 ORF285–102) in IFN-γ ELISPOT assays (Fig. 3C, Table III).
FIGURE 3.
Peptides LAGE-197–114, LAGE-1104–121, and LAGE-1b118–135 represent promiscuous HLA-DR epitopes recognized by CD4+ T cells isolated from PBLs of melanoma patients. CD4+ T cells were isolated from PBLs of melanoma patients and restimulated four times on a weekly basis with irradiated autologous mature DCs pulsed with individual peptide LAGE-197–114 (A), LAGE-1104–121 (B), or LAGE-1b118–135 (C). CD4+ T cells (1 × 104) were incubated in 48-h IFN-γ ELISPOT assays in the presence of L.DR cells pulsed with relevant LAGE-1–derived peptide or irrelevant peptide LAGE-1 ORF285–102. Spots were developed and counted by computer-assisted video image analysis. Each bar represents the mean spot number of triplicates (±SD) with 1 × 104 CD4+ T cells initially seeded per well. *p < 0.05. One representative experiment of at least three performed is depicted.
Table III.
Numbers of IFN-γ spots produced by bulk CD4+ T cells stimulated with autologous DCs loaded with LAGE-1 peptides
| Bulk Stimulated with LAGE-197–114 |
Bulk Stimulated with LAGE-1104–121 |
Bulk Stimulated with LAGE-1118–135 |
|||||
|---|---|---|---|---|---|---|---|
| Patient No. | Presenting Cell | LAGE-1 ORF285–102 |
LAGE-197–114 | LAGE-1 ORF285–102 |
LAGE-1104–121 | LAGE-1 ORF285–102 |
LAGE-1118–135 |
| 1 | L.DR11 | 7 | 757 | NA | NA | 1 | 72 |
| 3 | L.DR7 | 24 | 217 | NA | NA | 19 | 494 |
| L.DR13 | 33 | 123 | NA | NA | 25 | 279 | |
| 4 | L.DR15 | 45 | 147 | 57 | 201 | 12 | 122 |
| 5 | L.DR13 | NA | NA | 35 | 199 | 10 | 212 |
| 7 | L.DR3 | NA | NA | 48 | 1022 | NA | NA |
| L.DR15 | NA | NA | 23 | 1039 | NA | NA | |
| 8 | L.DR11 | 221 | 848 | NA | NA | NA | NA |
| L.DR13 | 69 | 566 | NA | NA | NA | NA | |
| 9 | L.DR1 | NA | NA | 82 | 301 | 4 | 46 |
| L.DR13 | NA | NA | 34 | 254 | 1 | 59 | |
| 10 | L.DR1 | 24 | 217 | 25 | 82 | NA | NA |
Numbers represent average of triplicate.
NA, not available.
Collectively, our data demonstrate that three LAGE-1–derived peptide sequences out of the six previously defined as good HLA-DR-binders were capable of stimulating IFN-γ–producing CD4+ T cells isolated from PBLs of patients with advanced NY-ESO-1+/ LAGE-1+ melanoma. Each of the eight patients evaluated in this study developed LAGE-1–specific CD4+ T cells against at least one of these three epitopes.
Three immunodominant LAGE-1–derived epitopes stimulate CD4+ T cell responses to LAGE-1 but not NY-ESO-1
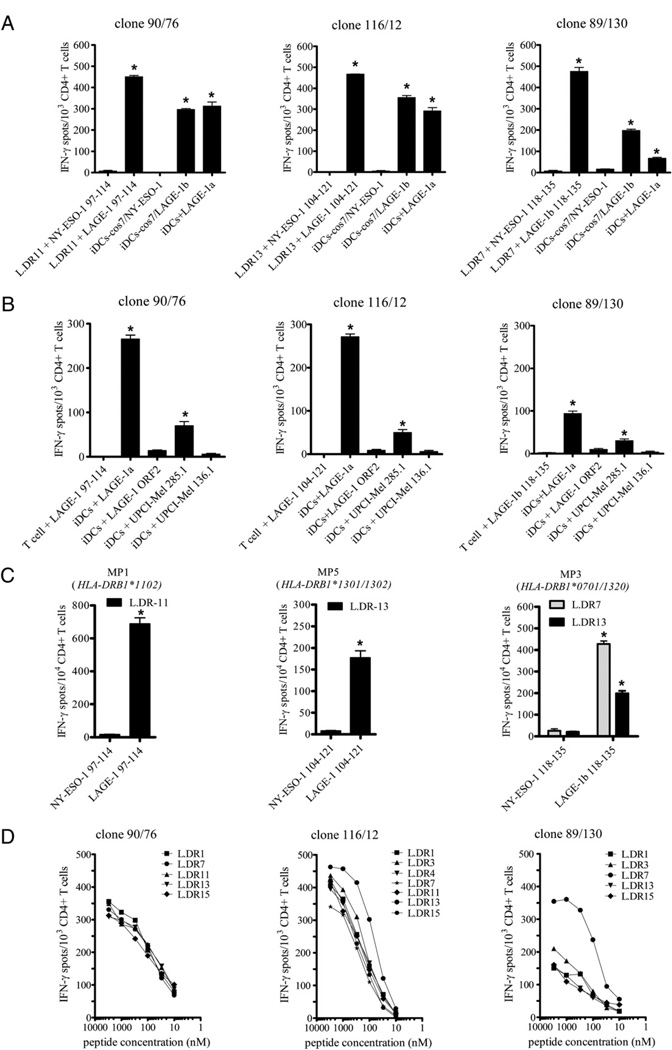
As shown in Fig. 1, the three immunodominant LAGE-1–derived epitopes, LAGE-197–114, LAGE-1104–121, and LAGE-1b118–135, overlapped partially with NY-ESO-1 amino acid sequences. Therefore, we next wanted to investigate whether LAGE-1–specific CD4+ T cells also recognized NY-ESO-1–derived epitopes. Multiple CD4+ T cell clones were obtained by limiting dilution from the LAGE-1–specific CD4+ bulk T cells previously generated.
LAGE-197–114–specific CD4+ T cell clone 90/76 (MP1, Table II), LAGE-1104–121–specific CD4+ T cell clone 116/12 (MP5, Table II), and LAGE-1b118–135–specific CD4+ T cell clone 89/130 (MP11, Table II) were representative of 19, 11, and 21 stable CD4+ T cell clones, respectively. Each clone recognized HLA-matched L.DR cells pulsed with the relevant LAGE-1 peptide and autologous DCs loaded either with the COS-7/LAGE-1b cell lysates, the LAGE-1+ melanoma cell line lysates (UPCI-MEL 285.1), or the LAGE-1a protein in IFN-γ ELISPOT assays (Fig. 4A, 4B). In contrast, LAGE-197–114–specific CD4+ T cell clone 90/ 76, LAGE-1104–121–specific CD4+ T cell clone 116/12, and LAGE-1b118–135–specific CD4+ T cell clone 89/130 did not recognize T cells pulsed with relevant peptides in the absence of APCs or L.DR cells pulsed with peptides NY-ESO-197–114, NY-ESO-1104–121, or NY-ESO-1118–135, respectively. Additionally, LAGE-1–specific CD4+ T cell clones did not recognize autologous DCs fed either with COS-7/NY-ESO-1 cell lysates, with LAGE-1− melanoma cell line lysates (UPCI-MEL 136.1), or with the irrelevant LAGE-1 ORF2 protein (Fig. 4A, 4B). To confirm that LAGE-1–specific CD4+ T cells did not cross-react with NY-ESO-1, we further incubated bulk LAGE-1–specific CD4+ T cells previously generated from PBLs of MP1, MP3, and MP5 in the presence of L.DR cells pulsed with the relevant LAGE-1 peptides (i.e., LAGE-197–114, LAGE-1104–121, or LAGE-1118–135) or NY-ESO-1 peptides overlapping the LAGE-1 peptide sequences (i.e., NY-ESO-197–114, NY-ESO-1104–121, or NY-ESO-1118–135). We observed that LAGE-197–114–specific CD4+ T cells, LAGE-1104–121–specific CD4+ T cells, and LAGE-1b118–135–specific CD4+ T cells recognized L.DR cells pulsed with peptide LAGE-197–114, LAGE-1104–121, and LAGE-1118–135, respectively, but not peptide NY-ESO-197–114, NY-ESO-1104–121, or NY-ESO-1118–135, respectively (Fig. 4C).
FIGURE 4.
LAGE-1–specific CD4+ T cell clones are high-affinity T cells that recognize naturally processed and presented epitopes. A, Each LAGE-1– specific CD4+ T cell clone (1 × 103), 90/76, 116/12, and 89/130, was incubated in 48-h IFN-γ ELISPOT assays in the presence of either L.DR cells pulsed with 3 µM peptide LAGE-197–114 (left), peptide LAGE-1104–121 (middle), or peptide LAGE-1b118–135 (right), respectively, or with autologous immature DCs (iDCs) loaded with cell lysates of COS-7 cells transfected with a plasmid encoding the LAGE-1b protein (pcDNA3-LAGE-1b), or with iDCs loaded with the recombinant LAGE-1a protein. As controls, each clone was tested against NY-ESO-1–derived peptides overlapping the LAGE-1 peptide sequences (i.e., NY-ESO-197–114 [left], NY-ESO-1104–121 [middle], or NY-ESO-1118–135 [right]), or against autologous iDCs loaded with cell lysates from COS-7 cells transfected with a plasmid encoding the NY-ESO-1 protein (pcDNA3-NY-ESO-1). B, Each LAGE-1–specific CD4+ T cell clone was incubated with relevant peptide without APCs or autologous iDCs loaded either with recombinant protein LAGE-1a or with cell lysates from LAGE-1+ melanoma cell line UPCI-MEL 285.1. As controls, CD4+ T cell clones were also incubated with autologous iDCs loaded either with the irrelevant LAGE-1 ORF2 protein or cell lysates from LAGE-1− melanoma cell UPCI-MEL 136.1. For A and B, each bar represents the mean spot number of triplicates ± SD, with 1 × 103 CD4+ T cells initially seeded per well. *p < 0.05. One representative experiment of three performed is depicted. C, Bulk LAGE-1–specific CD4+ T cells (1 × 104) obtained from MP1 (left), MP5 (middle), and MP3 (right), and depicted in Fig. 3, were incubated in 48-h IFN-γ ELISPOT assays in the presence of L.DR cells pulsed with relevant LAGE-1 peptide or NY-ESO-1 peptide. Each bar represents the mean spot number of triplicates ± SD, with 1 × 104 CD4+ T cells initially seeded per well. *p < 0.05. D, L.DR cells were pulsed with titrated doses of peptide LAGE-197–114, LAGE-1104–121, or peptide LAGE-1b118–135 and incubated in the presence of CD4+ T cell clone 90/76 (left), clone 116/12 (middle), or clone 89/130 (right), respectively, in 48-h IFN-γ ELISPOT assays as described in Materials and Methods. Data represent the mean spot number of triplicates, with 1 × 103 CD4+ T cells initially seeded per well. One representative experiment of three performed is depicted.
Notably, LAGE-1104–121–specific CD4+ T cell clone 116/12 also recognized peptide LAGE-1108–120 epitopes, previously reported as the target of tumor Ag (TA)-specific Tregs isolated from tumor-infiltrating lymphocytes (TILs) of melanoma patients (12) (data not shown).
The ability of the LAGE-1–specific CD4+ T cell clones 90/76, 116/12, and 89/130 to produce IFN-γ in the presence of L. DR cells incubated with the relevant LAGE-1 peptide at various concentrations was evaluated to determine the peptide-dose “threshold” for effector T cell recognition. The half-maximal stimulation of CD4+ T cell clones 90/76, 116/12, and 89/130 in the presence of peptide LAGE-197–114, LAGE-1104–121, and LAGE-1118–135, respectively, required peptide “loading” concentrations of ~20, 80, and 30 nM, respectively (Fig. 4D).
Interestingly, clones 90/76 and 116/12 exhibited high recognition levels of peptide LAGE-197–114 and LAGE-1104–121, respectively, not only in the context of the autologous HLA-DR molecule but also in the context of allogenic molecules, suggesting that they represent other examples of cross-reactive TA-specific T cell clones (17).
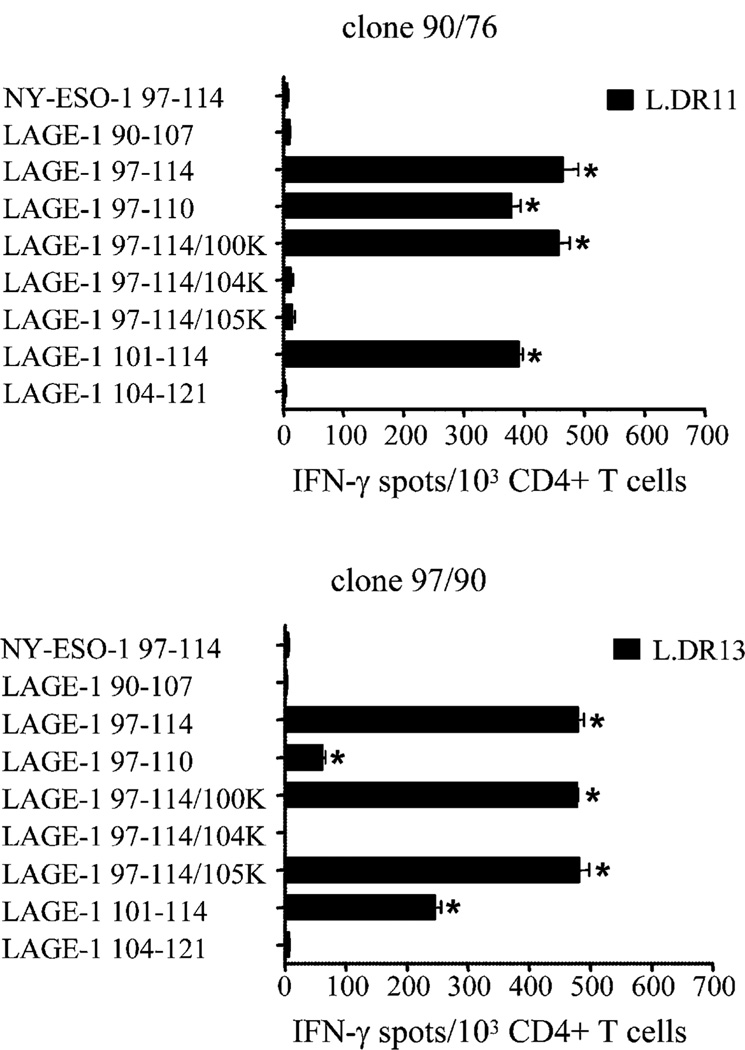
Next, we synthesized a series of peptides from the internal sequence of the immunogenic peptide LAGE-197–114 to delineate the minimal peptide sequence recognized by the CD4+ T cells. The minimum sequence recognized by LAGE-197–114–specific clones 90/76 and 97/90 (MP8, Table II) presented by HLA-DR11 and by HLA-DR13 molecules, respectively, corresponded to peptide LAGE-1101–114 (Fig. 5). Lysine substitution at position 104 abolished recognition by these two clones, suggesting that it corresponds to P1. Additionally, lysine substitution at position 105 abolished T cell recognition only on HLA-DR11 molecules but not on HLA-DR13 molecules, suggesting that peptide LAGE-1101–114 binds to HLA-DR11 according to two distinct peptide binding modes (P1 being either at position 104 or 105).
FIGURE 5.
Epitope core identification of peptide LAGE-197–114. CD4+ T cell clone 90/76 (MP1, Table II) and 97/90 (MP8, Table II) were incubated in 48-h IFN-γ ELISPOT assays in the presence of L.DR cells pulsed either with peptide LAGE-197–114, each of the three single lysine-substituted peptides at positions P100 (LAGE-197–114/100K), P104 (LAGE-197–114/104K), and P105 (LAGE-197–114/105K), peptide LAGE-197–110, peptide LAGE-1101–114, peptide LAGE-190–107, or peptide LAGE-1104–121. As control, each clone was incubated with the peptide NY-ESO-197–114. Each bar represents the mean spot number of triplicates ± SD, with 1 × 103 CD4+ T cells initially seeded per well. *p < 0.05. One representative experiment of three performed is depicted.
Collectively, our data demonstrate that LAGE-1–specific CD4+ T cells obtained from PBLs of patients with advanced melanoma recognized naturally processed and presented epitopes and do not cross-react with NY-ESO-1.
LAGE-1 stimulates TA-specific Th cells
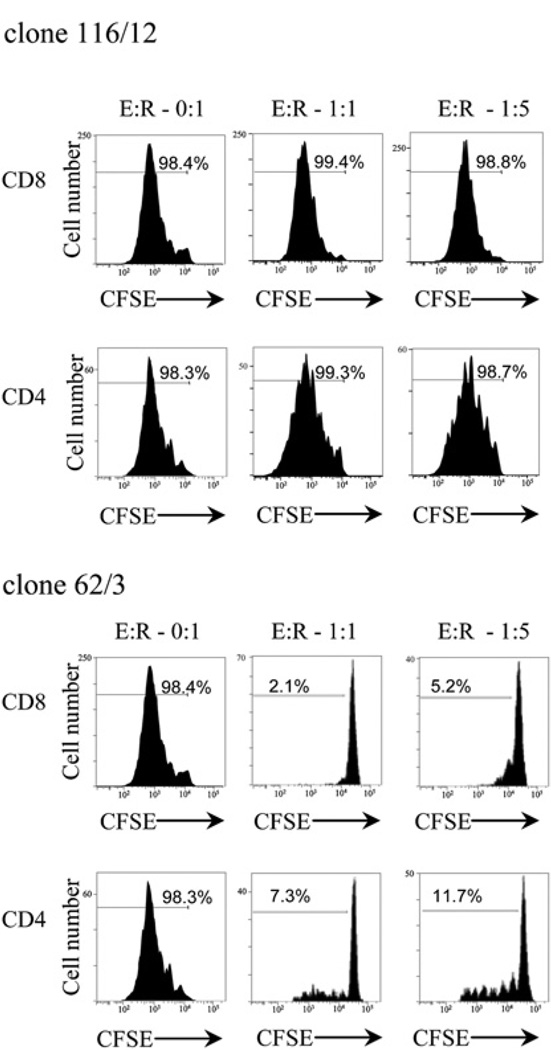
Peptide LAGE-1108–120 has recently been identified as the ligand of TA-specific Tregs isolated from TILs of melanoma patients (12). These LAGE-1–specific Tregs inhibited the proliferation of allogenic T cells and suppressed IL-2 secretion of TA-specific T cells. They produced IFN-γ and IL-10 but not TGF-β. To investigate whether LAGE-1104–121–specific CD4+ T cells isolated from PBLs of patients with melanoma were functionally Th cells or Tregs, we next tested six LAGE-1104–121–specific CD4+ T cell clones isolated from MP5 (Table II) for their capability to suppress the proliferation of allogenic T cells stimulated with anti-CD3 Ab in CFSE-based proliferation assay. The LAGE-1104–121–specific CD4+ T cell clones did not inhibit the proliferation of anti-CD3 Ab-stimulated naive CD8+ and CD4+ T cells as shown for LAGE-1104–121–specific CD4+ T cell clone 116/12 (Fig. 6). As control, TRAG-3–specific CD4+ Treg clone 62/3 inhibited proliferation of allogenic T cells. We performed CFSE-based proliferation assays with the LAGE-197–114–specific CD4+ T cell clone 90/76 and the LAGE-1118–135–specific CD4+ T cell clone 89/130 and observed no inhibition of T cell proliferation, suggesting that both clones were CD4+ Th cells (data not shown).
FIGURE 6.
LAGE-1104–121–specific CD4+ T cell clones are Th cells. Proliferation of allogenic CFSE-labeled naive CD45RA+ T cells obtained from PBLs of healthy donors was assessed after 5 d stimulation with soluble anti-CD3 (0.1 µg/ml) and IL-2 (50 IU/ml) in the presence of autologous non-CD3+ APCsandeithertheLAGE-1104–121–specificCD4+ Tcellclone116/12 or the TRAG-3–specific Treg clone 62/3 as control. Flow cytometry histograms of CFSE dilution by naive T cells are displayed for different ratios of effector CD4+ T cell clones to responder CD4+ and CD8+ naive T cells (E:R).
Collectively, our findings support the capability of LAGE-1 to stimulate TA-specific helper CD4+ T cells isolated from PBLs of patients with advanced melanoma.
Spontaneous CD4+ T cell responses to three immunodominant LAGE-1 epitopes
We next wanted to determine whether spontaneous LAGE-1– specific CD4+ T cell responses exist in multiple patients with advanced LAGE-1+/NY-ESO-1+ tumors. Therefore, we investigated the blood of 16 patients with stage IV LAGE-1+/NY-ESO-1+ melanoma for the presence of CD4+ T cells capable of recognizing the three immunodominant LAGE-1–derived epitopes that we had previously identified. As control, we also included 5 patients with LAGE-1−/NY-ESO-1− melanoma and 16 normal donors (Table II). PBLs were stimulated prior to ICS assays, as described in Materials and Methods.
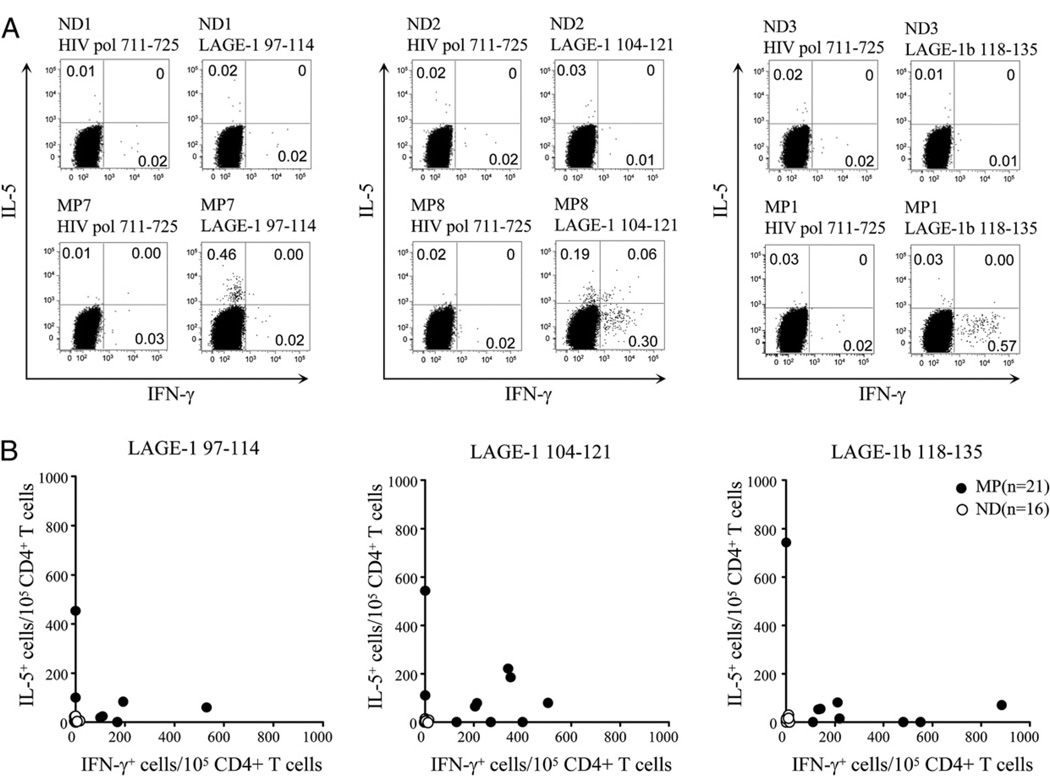
We observed LAGE-197–114–specific CD4+ T cells in 7 out of 16 patients with LAGE-1+/NY-ESO-1+ melanoma (Fig. 7, Table II). These patients either had only Th1-type (three patients), Th2-type (two patients), or both Th1- and Th2-type LAGE-1–specific CD4+ T cells (two patients). We detected LAGE-1104–121–specific CD4+ T cells in 10 out of 16 patients with LAGE-1+/NY-ESO-1+ melanoma (Fig. 7, Table II). These patients either had only Th1-type (three patients), Th2-type (two patients), or both Th1- and Th2-type LAGE-1–specific CD4+ T cells (five patients). We also found LAGE-1118–135–specific CD4+ T cells in 9 out of 16 patients with stage IV LAGE-1+/NY-ESO-1+ melanoma (Fig. 7, Table II). These patients either had only Th1-type (four patients), Th2-type (one patient), or both Th1- and Th2-type LAGE-1–specific CD4+ T cells (four patients). None of the 5 melanoma patients with LAGE-1−/NY-ESO-1− melanoma and none of the 16 normal donors had detectable spontaneous LAGE-1–specific CD4+ T cells.
FIGURE 7.
Th1-type versus Th2-type immunoreactivity of spontaneous LAGE-1–specific CD4+ T cells in patients with melanoma and healthy donors. A and B, Representative dot plots from melanoma patients MP7, MP8, MP1, and healthy donors ND1, ND2, ND3 (A), and summary data for NY-ESO-1+/ LAGE-1+ melanoma patients (n = 16), NY-ESO-1−/LAGE-1− melanoma patients (n = 5), and healthy donors (n = 16) (B) showing the percentages or combined numbers of LAGE-1–specific IFN-γ and/or IL-5–producing CD4+ T cells per 105 CD4+ T cells in 6 h ICS assays performed after 10–12 d IVS with either peptide LAGE-197–114, peptide LAGE-1104–121, or peptide LAGE-1118–135 as described in Materials and Methods. In B, each symbol depicts the number of IFN-γ– and/or IL-5–producing CD4+ T cells per 105 CD4+ T cells in the presence of the relevant LAGE-1 peptide minus the numbers of cytokine producing CD4+ T cells in the presence of the irrelevant peptide HIVpol711–725. A positive response is defined as the number of LAGE-1–specific cytokine-producing cells per 105 CD4+ T cells greater than the mean + 3 SD of the cytokine-producing cell number obtained from all healthy donors and patients against the control peptide HIVpol711–725. The mean values of IFN-γ– and IL-5–producing CD4+ T cells in the presence of the irrelevant peptide HIVpol711–725 were 25 (SD, 10) and 22 (SD, 9), respectively.
Collectively, our data demonstrate the existence of spontaneous LAGE-1–specific CD4+ T cells in PBLs of patients with advanced LAGE-1+/NY-ESO-1+ melanoma that do not cross-react with NY-ESO-1.
Analysis of LAGE-1–specific Ab responses among patients with advanced NY-ESO-1+/LAGE-1+ tumors
We next investigated whether spontaneous LAGE-1b–specific CD4+ Th cells coexist with the presence of LAGE-1b–specific Abs that do not cross-react with NY-ESO-1 in patients with advanced NY-ESO-1+/LAGE-1+ tumors. To this end, serial dilutions of patients’ sera with NY-ESO-1+/LAGE-1+ or NY-ESO-1−/LAGE-1− tumors were tested in ELISA plates coated either with the NY-ESO-1 recombinant protein or each of the 18-aa–long overlapping peptide sequences that span the LAGE-1b protein from P132 to P210.
Among the 16 patients with NY-ESO-1+/LAGE-1+ tumors, 4 had high titers of LAGE-1b Abs recognizing peptide LAGE-1b167–184 and LAGE-1b188–210 (Table IV). These four patients also developed NY-ESO-1–specific Abs. As control, none of the five patients with NY-ESO-1−/LAGE-1− tumors had detectable circulating NY-ESO-1– or LAGE-1–specific Abs.
Collectively, our findings demonstrate that a fraction of melanoma patients with advanced NY-ESO-1+/LAGE-1+ tumors and spontaneous LAGE-1–specific CD4+ T cells develop LAGE-1b– specific Abs targeting linear LAGE-1b–specific epitopes located in the C-terminal portion of the protein.
Discussion
Although immune responses to NY-ESO-1 and LAGE-1 ORF2 have been extensively studied (8, 9, 13), little is known about spontaneous CD4+ T cell and Ab responses to LAGE-1 ORF1. In contrast to LAGE-1 ORF2, LAGE-1 ORF1 is highly homologous to NY-ESO-1 and both are frequently coexpressed by tumors (4). Previous studies have clearly established the hierarchy of immunodominance of spontaneous CD4+ T cell responses to NY-ESO-1 in melanoma patients. Three immunodominant amino acid regions located in the central region of the NY-ESO-1 protein (87–111, 119–143, and 157–170) are recognized by CD4+ T cells isolated from PBLs of patients with advanced melanoma (7–10). Two out of these three amino acid regions (NY-ESO-187–111 and NY-ESO-1119–143) largely overlap with the LAGE-1 amino acid sequence, suggesting the likelihood of CD4+ T cell cross-reactivity between NY-ESO-1 and LAGE-1. In the current study, our findings define the epitope hierarchy of spontaneous CD4+ T cell responses to LAGE-1 directed against three immunodominant epitopes, LAGE-197–114, LAGE-1118–135, and LAGE-1104–121. Strikingly, these three LAGE-1–derived epitopes, which largely overlap with NY-ESO-1 amino acid sequences, stimulate LAGE-1–specific CD4+ T cells that do not cross-react with NY-ESO-1. We did not evaluate the spontaneous immunogenicity of peptide LAGE-1125–142 since LAGE-1125–142–specific CD4+ T cells were detected in a small fraction of melanoma patients, suggesting it does not represent an immunodominant/immunoprevalent epitope. Notably, our findings with a limited number of LAGE-1–specific CD4+ T cell clones do not exclude the existence of cross-reactive CD4+ T cells capable of recognizing both LAGE-1 and NY-ESO-1. These data, together with our previous findings on NY-ESO-1– derived immunodominant epitopes, show that the central protein regions of NY-ESO-1 and LAGE-1 ORF1 are extremely rich in overlapping but distinct promiscuous immunogenic epitopes, supporting the spontaneous immunogenicity of the LAGE-1 and NY-ESO-1 Ags in patients with advanced cancers.
Our data with the minimal peptide and modified sequences from peptide LAGE-197–114 indicate that this sequence likely encompasses at least two LAGE-1–specific but not NY-ESO-1–specific promiscuous HLA-DR–presented sequences. Notably, peptide LAGE-197–114 also encompasses the HLA-A68–restricted LAGE-1103–111 peptide, which belongs to the HLA-A3 supertype (11, 20). Therefore, this peptide represents another example of epitope clustering that may prove useful for stimulating both LAGE-1– specific CD8+ and CD4+ T cells (21).
LAGE-1108–120–specific CD4+ T cells that were previously isolated from TILs of one melanoma patient inhibited the proliferation and suppressed IL-2 production of CD4+ T cells, indicating that they were functionally Tregs (12). LAGE-1 represents the first example of human TA-specific Treg ligand, questioning whether LAGE-1 preferentially stimulates Tregs in patients with advanced LAGE-1–expressing tumors. Our findings address this question and demonstrate that all three immunodominant LAGE-1 epitopes can also stimulate Th1-type CD4+ T cells isolated from PBLs of patients with advanced melanoma. They add to the previous report by Wang et al. (12) of LAGE-1– specific Tregs isolated from TILs of melanoma patients and suggest that LAGE-1 can stimulate both LAGE-1–specific Th cells and Tregs. Notably, we have recently shown that NY-ESO-1 also stimulates both NY-ESO-1–specific CD4+ Th cells and Tregs in PBLs of patients with advanced melanoma (19). Therefore, our findings emphasize the need to develop potent combinatorial immunotherapies to further promote the expansion of Th1-type LAGE-1–specific and NY-ESO-1–specific CD4+ T cells in the context of LAGE-1/NY-ESO-1–based vaccines.
In the current study, spontaneous LAGE-1–specific CD4+ T cell frequencies were <500/105 CD4+ T cells (0.5%) for most melanoma patients. Such frequencies after one round of in vitro stimulation (IVS) are in the range of what we and others have previously reported for other tumor-derived epitopes such as NY-ESO-1 and TRAG-3 (8, 16, 22, 23). Importantly, we did not observe significant frequencies in LAGE-1–specific CD4+ T cells after one round of IVS using PBLs of normal donors or melanoma patients with LAGE-1− tumors, suggesting that we likely expanded the preexisting and low-frequency memory LAGE-1–specific CD4+ T cells in melanoma patients with LAGE-1+ tumors.
In the current study, most patients with spontaneous LAGE-1– specific CD4+ T cells had Th1-type or both Th1- and Th2-type CD4+ T cells, suggesting no Th1/Th2 bias in circulating LAGE-1– specific CD4+ T cell responses in patients with advanced melanoma as previously observed for MAGE-A6–specific CD4+ T cells (24). These data are in line with previous studies of spontaneous circulating NY-ESO-1–specific CD4+ T cells showing no Th1/Th2 bias (8). However, it is important to note that, in this study, we have focused on circulating LAGE-1–specific CD4+ T cells and cannot exclude a Th1/Th2 bias at the tumor sites as previously reported for CEA-specific CD4+ T cells (25).
LAGE-1–specific CD4+ T cell clones recognized the same peptide in the context of autologous but also allogenic HLA-DR molecules. This observation represents other examples of tumor Ag-specific T cell cross-reactivity acting in a peptide-specific, MHC-dependent but unrestricted fashion (17). These findings illustrate the considerable plasticity of TCR recognition of LAGE-1/NY-ESO-1 p-MHC class II complexes and may prove useful for the optimization of adoptive transfer of promiscuous T cells and for the design of TCR gene transfer immunotherapies applicable to most cancer patients independent of their MHC class II phenotype.
An important finding in the current study is the detection of LAGE-1b–specific Abs against linear epitopes located in the C-terminal portion of LAGE-1b, which does not overlap with NY-ESO-1. Taken together with the data on LAGE-1–specific CD4+ T cells, these findings demonstrate the capability of LAGE-1 to stimulate integrated LAGE-1–specific cellular and humoral responses. Although LAGE-1–specific Abs were detectable in nearly half of the patients with spontaneous LAGE-1–specific CD4+ T cells, we cannot exclude that LAGE-1 Abs may be detected more frequently as they might be directed against conformational epitopes. This question will need to be addressed using novel seromic analyses of Ab responses with conformational protein arrays (26).
Collectively, our findings define the epitope hierarchy of LAGE-1–specific CD4+ T cells that do not cross-react with NY-ESO-1, although the LAGE-1 immunodominant epitopes largely overlap with NY-ESO-1 amino acid sequences. They further demonstrate the capability of LAGE-1 to stimulate integrated CD4+ Th cells and Ab responses in patients with advanced melanoma. As LAGE-1 and NY-ESO-1 are commonly coexpressed by tumor cells, our findings strongly support the use of LAGE-1 epitopes/ protein in combination with NY-ESO-1 epitopes/protein in patients with LAGE-1+/NY-ESO-1+ melanoma. Because both LAGE-1 and NY-ESO-1 stimulate Th cells and Tregs, LAGE-1/NY-ESO-1–based vaccines will need to be optimized with combinatorial immunotherapies to promote Th1-type LAGE-1/NY-ESO-1–specific immune responses.
Acknowledgments
This work was supported by National Institutes of Health/National Cancer Institute Grants CA90360 and CA112198 (to H.M.Z.) and a Cancer Research Institute grant (to H.M.Z.).
We thank the patients and their physicians for giving their time and blood samples for the performance of these experiments. We thank Drs. Lloyd Old, Gerd Ritter, and Sacha Gnjatic (Ludwig Institute for Cancer Research, New York, NY) for providing the NY-ESO-1 protein and for sharing information on ELISA for NY-ESO-1–specific Abs. We thank Drs. Elke Jager (Krankenhaus Nordwest, Frankfurt, Germany), Alexander Knuth (Universitätsspital Zürich, Zurich, Switzerland), and Yao T. Chen (Cornell University, New York, NY) for providing the plasmid pcDNA3-NY-ESO-1. We also thank Dr. Bernard Lethe (Ludwig Institute for Cancer Research, Brussels, Belgium) for providing the plasmid pcDNA3-LAGE-1b.
Abbreviations used in this paper
- DC
dendritic cell
- ICS
intracellular cytokine staining
- iDC
immature dendritic cell
- IVS
in vitro stimulation
- MP
melanoma patient
- NA
not available
- ND
normal donor
- ORF
open reading frame
- TA
tumor Ag
- TIL
tumor-infiltrating lymphocyte
- Treg
regulatory T cell
- UPCI
University of Pittsburgh Cancer Institute
Footnotes
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Chen YT, Scanlan MJ, Sahin U, Türeci O, Gure AO, Tsang S, Williamson B, Stockert E, Pfreundschuh M, Old LJ. A testicular antigen aberrantly expressed in human cancers detected by autologous antibody screening. Proc. Natl. Acad. Sci. USA. 1997;94:1914–1918. doi: 10.1073/pnas.94.5.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Uyttenhove C, Godfraind C, Lethé B, Amar-Costesec A, Renauld JC, Gajewski TF, Duffour MT, Warnier G, Boon T, Van den Eynde BJ. The expression of mouse gene P1A in testis does not prevent safe induction of cytolytic T cells against a P1A–encoded tumor antigen. Int. J. Cancer. 1997;70:349–356. doi: 10.1002/(sici)1097-0215(19970127)70:3<349::aid-ijc17>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 3.Boon T, Coulie PG, Van den Eynde BJ, van der Bruggen P. Human T cell responses against melanoma. Annu. Rev. Immunol. 2006;24:175–208. doi: 10.1146/annurev.immunol.24.021605.090733. [DOI] [PubMed] [Google Scholar]
- 4.Lethé B, Lucas S, Michaux L, De Smet C, Godelaine D, Serrano A, De Plaen E, Boon T. LAGE-1, a new gene with tumor specificity. Int. J. Cancer. 1998;76:903–908. doi: 10.1002/(sici)1097-0215(19980610)76:6<903::aid-ijc22>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 5.Jäger E, Gnjatic S, Nagata Y, Stockert E, Jäger D, Karbach J, Neumann A, Rieckenberg J, Chen YT, Ritter G, et al. Induction of primary NY-ESO-1 immunity: CD8+ T lymphocyte and antibody responses in peptide-vaccinated patients with NY-ESO-1+ cancers. Proc. Natl. Acad. Sci. USA. 2000;97:12198–12203. doi: 10.1073/pnas.220413497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stockert E, Jäger E, Chen YT, Scanlan MJ, Gout I, Karbach J, Arand M, Knuth A, Old LJ. A survey of the humoral immune response of cancer patients to a panel of human tumor antigens. [see comments] J. Exp. Med. 1998;187:1349–1354. doi: 10.1084/jem.187.8.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeng G, Wang X, Robbins PF, Rosenberg SA, Wang RF. CD4+ T cell recognition of MHC class II-restricted epitopes from NY-ESO-1 presented by a prevalent HLA DP4 allele: association with NY-ESO-1 antibody production. Proc. Natl. Acad. Sci. USA. 2001;98:3964–3969. doi: 10.1073/pnas.061507398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mandic M, Castelli F, Janjic B, Almunia C, Andrade P, Gillet D, Brusic V, Kirkwood JM, Maillere B, Zarour HM. One NY-ESO-1-derived epitope that promiscuously binds to multiple HLA-DR and HLA-DP4 molecules and stimulates autologous CD4+ T cells from patients with NY-ESO-1-expressing melanoma. J. Immunol. 2005;174:1751–1759. doi: 10.4049/jimmunol.174.3.1751. [DOI] [PubMed] [Google Scholar]
- 9.Zarour HM, Maillere B, Brusic V, Coval K, Williams E, Pouvelle-Moratille S, Castelli F, Land S, Bennouna J, Logan T, Kirkwood JM. NY-ESO-1 119–143 is a promiscuous major histocompatibility complex class II T-helper epitope recognized by Th1- and Th2-type tumor-reactive CD4+ T cells. Cancer Res. 2002;62:213–218. [PubMed] [Google Scholar]
- 10.Zarour HM, Storkus WJ, Brusic V, Williams E, Kirkwood JM. NY-ESO-1 encodes DRB1*0401-restricted epitopes recognized by melanoma-reactive CD4+ T cells. Cancer Res. 2000;60:4946–4952. [PubMed] [Google Scholar]
- 11.Sun Z, Lethé B, Zhang Y, Russo V, Colau D, Stroobant V, Boon T, van der Bruggen P. A new LAGE-1 peptide recognized by cytolytic T lymphocytes on HLA-A68 tumors. Cancer Immunol. Immunother. 2006;55:644–652. doi: 10.1007/s00262-005-0066-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang HY, Lee DA, Peng G, Guo Z, Li Y, Kiniwa Y, Shevach EM, Wang RF. Tumor-specific human CD4+ regulatory T cells and their ligands: implications for immunotherapy. Immunity. 2004;20:107–118. doi: 10.1016/s1074-7613(03)00359-5. [DOI] [PubMed] [Google Scholar]
- 13.Mandic M, Almunia C, Vicel S, Gillet D, Janjic B, Coval K, Maillere B, Kirkwood JM, Zarour HM. The alternative open reading frame of LAGE-1 gives rise to multiple promiscuous HLA-DR-restricted epitopes recognized by T-helper 1-type tumor-reactive CD4+ T cells. Cancer Res. 2003;63:6506–6515. [PubMed] [Google Scholar]
- 14.Texier C, Pouvelle S, Busson M, Hervé M, Charron D, Ménez A, Maillére B. HLA-DR restricted peptide candidates for bee venom immunotherapy. J. Immunol. 2000;164:3177–3184. doi: 10.4049/jimmunol.164.6.3177. [DOI] [PubMed] [Google Scholar]
- 15.Texier C, Pouvelle-Moratille S, Buhot C, Castelli FA, Pecquet C, Ménez A, Leynadier F, Maillère B. Emerging principles for the design of promiscuous HLA-DR-restricted peptides: an example from the major bee venom allergen. Eur. J. Immunol. 2002;32:3699–3707. doi: 10.1002/1521-4141(200212)32:12<3699::AID-IMMU3699>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 16.Janjic B, Andrade P, Wang XF, Fourcade J, Almunia C, Kudela P, Brufsky A, Jacobs S, Friedland D, Stoller R, et al. Spontaneous CD4+ T cell responses against TRAG-3 in patients with melanoma and breast cancers. J. Immunol. 2006;177:2717–2727. doi: 10.4049/jimmunol.177.4.2717. [DOI] [PubMed] [Google Scholar]
- 17.Kudela P, Janjic B, Fourcade J, Castelli F, Andrade P, Kirkwood JM, El-Hefnawy T, Amicosante M, Maillere B, Zarour HM. Cross-reactive CD4+ T cells against one immunodominant tumor-derived epitope in melanoma patients. J. Immunol. 2007;179:7932–7940. doi: 10.4049/jimmunol.179.11.7932. [DOI] [PubMed] [Google Scholar]
- 18.Fourcade J, Kudela P, Sun Z, Shen H, Land SR, Lenzner D, Guillaume P, Luescher IF, Sander C, Ferrone S, et al. PD-1 is a regulator of NY-ESO-1-specific CD8+ T cell expansion in melanoma patients. J. Immunol. 2009;182:5240–5249. doi: 10.4049/jimmunol.0803245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fourcade J, Sun Z, Kudela P, Janjic B, Kirkwood JM, El-Hafnawy T, Zarour HM. Human tumor antigen-specific helper and regulatory T cells share common epitope specificity but exhibit distinct T cell repertoire. J. Immunol. 2010;184:6709–6718. doi: 10.4049/jimmunol.0903612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sidney J, Grey HM, Southwood S, Celis E, Wentworth PA, del Guercio MF, Kubo RT, Chesnut RW, Sette A. Definition of an HLA-A3-like supermotif demonstrates the overlapping peptide-binding repertoires of common HLA molecules. Hum. Immunol. 1996;45:79–93. doi: 10.1016/0198-8859(95)00173-5. [DOI] [PubMed] [Google Scholar]
- 21.Zeng G, Li Y, El-Gamil M, Sidney J, Sette A, Wang RF, Rosenberg SA, Robbins PF. Generation of NY-ESO-1-specific CD4+ and CD8+ T cells by a single peptide with dual MHC class I and class II specificities: a new strategy for vaccine design. Cancer Res. 2002;62:3630–3635. [PMC free article] [PubMed] [Google Scholar]
- 22.Jackson H, Dimopoulos N, Mifsud NA, Tai TY, Chen Q, Svobodova S, Browning J, Luescher I, Stockert L, Old LJ, et al. Striking immuno-dominance hierarchy of naturally occurring CD8+ and CD4+ T cell responses to tumor antigen NY-ESO-1. J. Immunol. 2006;176:5908–5917. doi: 10.4049/jimmunol.176.10.5908. [DOI] [PubMed] [Google Scholar]
- 23.Ayyoub M, Merlo A, Hesdorffer CS, Rimoldi D, Speiser D, Cerottini JC, Chen YT, Old LJ, Stevanovic S, Valmori D. CD4+ T cell responses to SSX-4 in melanoma patients. J. Immunol. 2005;174:5092–5099. doi: 10.4049/jimmunol.174.8.5092. [DOI] [PubMed] [Google Scholar]
- 24.Tatsumi T, Kierstead LS, Ranieri E, Gesualdo L, Schena FP, Finke JH, Bukowski RM, Mueller-Berghaus J, Kirkwood JM, Kwok WW, Storkus WJ. Disease-associated bias in T helper type 1 (Th1)/Th2 CD4+ T cell responses against MAGE-6 in HLA-DRB10401+ patients with renal cell carcinoma or melanoma. J. Exp. Med. 2002;196:619–628. doi: 10.1084/jem.20012142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tassi E, Gavazzi F, Albarello L, Senyukov V, Longhi R, Dellabona P, Doglioni C, Braga M, Di Carlo V, Protti MP. Carcinoembryonic antigen-specific but not antiviral CD4+ T cell immunity is impaired in pancreatic carcinoma patients. J. Immunol. 2008;181:6595–6603. doi: 10.4049/jimmunol.181.9.6595. [DOI] [PubMed] [Google Scholar]
- 26.Gnjatic S, Wheeler C, Ebner M, Ritter E, Murray A, Altorki NK, Ferrara CA, Hepburne-Scott H, Joyce S, Koopman J, et al. Seromic analysis of antibody responses in non-small cell lung cancer patients and healthy donors using conformational protein arrays. J. Immunol. Methods. 2009;341:50–58. doi: 10.1016/j.jim.2008.10.016. [DOI] [PubMed] [Google Scholar]