Abstract
The Musashi (Msi) family of RNA-binding proteins is important in stem and differentiating cells in many species. Here, we present a zebrafish gene/protein trap line gt(msi2b-citrine)ct57a that expresses a Citrine fusion protein with endogenous Musashi-2b (Msi2b). Our results reveal two phases of Msi2b expression: ubiquitous expression in progenitor cells in the early embryo and later, tissue-specific expression in differentiating cells in the olfactory organ, pineal gland and subpopulations of neurons in the central nervous system. Interestingly, this division between early and late phases is paralleled by differential expression of msi2b alternative splicing products. Whereas the full-length and long variant v3 Msi2b predominate at early stages, the later expression of variants in differentiating tissues appears to be tissue-specific. Using the gt(msi2b-citrine)ct57a we characterized tissue-specific expression of Msi2b with cellular resolution in subsets of differentiating cells in the olfactory organ, pineal gland, CNS and ventral neural tube. By performing TALEN-mediated biallelic genome editing or morpholino knockdown of Msi2b in zebrafish, our results show that early inactivation of Msi2b results in severe embryonic defects including hypertrophy of the ventricles and shortening of the body, consistent with an important role in cell proliferation and survival. Moreover, specific inactivation of Msi2b full-length indicates that this species is essential for the early role of Msi2b. This line provides a valuable tool both for live imaging of the endogenous Msi2b at subcellular resolution and manipulation of Msi2b-expressing cells.
Keywords: Musashi, zebrafish, neuronal progenitor, differentiating
INTRODUCTION
The remarkable ability of stem cells to self-renew and differentiate into diverse cell types requires the coordinated regulation of numerous proteins and RNAs involved in homeostasis and cell fate determination. The underlying regulatory mechanisms involve not only gene activation and transcriptional activity, but also post-transcriptional control [1]. RNA-binding proteins (RBPs) are essential for such post-transcriptional regulation and may play a predominant role in controlling protein expression levels [2, 3]. RBPs can associate with specific RNA-binding domains or recognize common mRNA features such as the 5’cap or 3’poly(A) tail within nascent RNA transcripts [4] and control protein levels by modulating transcript longevity, intracellular localization and/or processing/translation rates [5].
Musashi protein was originally described in Drosophila, where it mediates asymmetric division between neuronal and non-neuronal lineages [6, 7]. The Msi family of RNA-binding proteins is evolutionarily conserved across distant taxa, including vertebrates, where its members Msi1 and Msi2 are strongly co-expressed in neural precursor cells, including central nervous system (CNS) stem cells [8, 9]. Vertebrate Msi1 is expressed in proliferating embryonic neural precursors [8, 10, 11] as well as in putative precursors of postnatal and CNS stem cells [12, 13]. In the CNS, Musashi proteins cooperate to maintain the progenitor state [6, 9, 14]. They also function in progenitor cells outside the CNS, such as murine intestinal tissue and human breast stem cells [15, 16].
Msi proteins often function by directly binding to the 3’UTR of many target genes. In Drosophila, dMsi represses translation of ttk69 mRNA in response to Notch signaling during asymmetric cell division and lineage restriction of sensory organ precursors [6]. Likewise, in mice, Msi1 represses mNumb expression by directly interacting with its mRNA 3’UTR [17]. Moreover, Msi1's ability to regulate cell cycle progression has been linked to the binding and translational regulation of p21WAF-1 mRNA [18].
In vertebrate development, Msi2 persists throughout development and is required for self-renewal of embryonic CNS stem cells and mouse ES cells [9, 19]. Msi2 is expressed in non-neuronal cells in mature brains as well as in hematopoietic stem cells [20]. Moreover, dysregulation of Msi2 function is associated with progression of myeloid leukemia from a slow growing chronic to an aggressive phase [20-22]. Taken together, these results suggest that Musashi proteins have important roles in both embryonic and adult tissues.
Here, we present a novel zebrafish gene trap line that reveals expression of endogenous Musashi-2b, the zebrafish co-ortholog of Msi2, at subcellular resolution as a fusion protein with Citrine. This line reveals two phases of Msi2b developmental expression: 1) an initial low-level ubiquitous expression during stages of somitogenesis and 2) higher level tissue-specific expression in the olfactory region, pineal gland and subpopulations of neurons in the CNS. Interestingly, zebrafish Msi2b is expressed as a full-length and two C-terminally truncated splice variants. Not only are these variants differentially expressed, but Msi2b subfunctionalization into early versus late phases also correlates with distinct splice variants. Finally, our results show that zebrafish Msi2b in essential for controlling cell proliferation and cell survival, suggesting a critical developmental role for this protein.
MATERIALS AND METHODS
Zebrafish
Adult fish and embryos were maintained as described [23]. We used the transgenic lines Tg(olig2:DsRed2) [24], Tg(-8.4neurog1:nRFP) [25], Tg(ompk:lyn-mRFP)rw035 [26] and Tg(trpc24.5kb:gab-Venus)rw037 [26].
Cloning of msi2b full-length and splice variants
The trapped gene in the ct57a line was identified by 3’ RACE PCR using citrine-specific primer, as described [27, 28]. For identification of full-length msi2b and splice variants, a 5’RACE library was synthesized from wild-type embryos using oligodT primers and the GeneRacer kit (Invitrogen). Total RNA was extracted using RNAqueous kit (Ambion). PCR amplification of the 5’end was performed using an msi2b specific primer (ct57a_GSP_R, CATCTTGGGTTGAGCGCGTCTTGG). PCR products were gel-purified and cloned in pCRII-TOPO vectors for sequencing. Primers used for cloning full-length splice variants: FT57a_full-length_F, ATGGAGGGAGACGGGAGCCA; FT57a_v1_full-length_R, TCAATGGTATCCGTTTGTAA; FT57a_v2_R, TTATAATAACATTACACACA, and FT57a_v3_R, AAACTCTGCACTCACTACCG.
In situ hybridization
We adapted a whole mount in situ hybridization protocol from [29] that includes high stringent conditions for hybridization and subsequent washes, which were carried out at 70°C in hybridization solution containing 50% formamide; 1.3X SSC; 5mM EDTA pH8.0; 200 μg/ml yeast tRNA; 100μg/ml heparin; 0.2% Tween-20 and 0.5% Chaps. The hybridization signal was detected using NBT/BCIP (Roche, Indianapolis, IN). To determine the distribution of different msi2b variants, we designed probes that differentially target splice variants, with the similarities of msi2b with msi2a and msi1 in the regions targeted by the probes corresponding to 77.7% and 53.6% for v1, 46.6% and 37.9% for v2 and 56.4% and 50.9% for v2, respectively. msi2b v1 probe detects exons 8-13 (873bp); v2 recognizes its 3’UTR, corresponding to intron 7 (765bp), and v3, against the 3’UTR of this variant (or intron 11; 772 bp). Template for probes was PCR-amplified (63812_ish_F: TTGAGAACGAGGATGTGGTG, 63812_ish_R: TTCGGTTTGTTCTTCGTGTG; 174035_ish_F: TTTGTTTTGTGTGCGTTTGC, 174035_ish_R: TCACTCCTCACCTCCCCATA; ct57_v3_ish_F: GATCTGTGTGACTGCTGCTC, ct57_v3_ish_R: AAACTCTGCACTCACTACCG), with forward and reverse primers including SP6 (GATTTAGGTGACACTATAG) or T7 (TAATACGACTCACTATAGGG) extensions, respectively, for in vitro transcription of antisense probe with T7 RNA polymerase. Subcellular distribution of transcripts was resolved by performing tyramide fluorescent in situ hybridization with protocol adapted from [30].
Quantitative analysis of msi2b variants by quantitative PCR
qPCR was performed using cDNA obtained from pools of 10 zebrafish embryos at the stages of 24 and 56 hpf, using the following primers to detect v1 (E12_F: CCAGCCCGGATCAGTATAAG, E13_R: CCGTACAGGTCAGCCACTG), v2 (E7_F: GAAGATGCAATGCTTATGTTTGA, E7’_R: TACACACATGCAAACGCACA) and v3 (i11_F: CTCAAGGCTCTTGCTTGCTT, i11_R: ACCAGTTCCACCGACCATAA). Expression of msi2b variants was normalized using beta actin (beta actin_F: AATCCCAAAGCCAACAGAGA, beta actin_R: ACATACATGGCAGGGGTGTT).
Immunohistochemistry
Immunohistochemistry was performed in whole mount. Fixed embryos were subsequently embedded in 17% gelatin in PBS and fixed in 4% paraformaldehyde in PBS for vibratome-sectioning. The following antibodies were used: rabbit anti-GFP antibody (Invitrogen, 1:300), rabbit anti-GABA (Sigma, 1:300) and anti-rabbit Alexa488 (Invitrogen, 1:1000). Sections were imaged on a Zeiss Pascal confocal microscope.
Western blot
Protein extract was obtained from Gt(Msi2b-Citrine)ct57a homozygote and wild-type zebrafish embryos at 56 hpf. Embryos were de-yolked, lysed in hypotonic buffer (20mM Hepes pH7.9, 1.5mM MgCl2, 10mM KCl, 1mM DTT) with protease inhibitors and centrifuged at maximum speed to obtain cytoplasmic fraction. Nuclear fraction was obtained by lysis in 20 mM Hepes pH7.9, 1.5 mM MgCl2, 0.2 mM EDTA, 20% Glycerol and 420 mM KCl and centrifugation at maximum speed. Detection was performed with rabbit anti-GFP antibody (Torrey Pines, 1:1,000) and mouse anti-tubulin (Sigma, 1:1,000).
Loss-of-function assays
Single-cell embryos were injected with 1.9ng of msi2b translation-blocking morpholino (ATG-MO, 5′-TGGCTTGGCTCCCGTCTCCCTCCAT-3′) or 0.5pg of i7/E8 splice-blocking morpholino (i7/E8-MO, 5’-CGAATCCTGTGAAGAGAGAGAGAGA-3’)(Gene Tools). TALENs were designed to target the splicing donor site of msi2b exon 2 using TALEN targeter [31] and constructed using improved GoldyTalen design [32, 33]. To generate msi2b mutant alleles, embryos were injected with 100pg total E2 TALEN mRNA encoding the targeting sequence for intron 2 (NG_NG_NI_NG_HD_NN_NN_NG_NN_NN_HD_HD_NG_NN_NI_NN_HD_NG_NN_NN) and exon 2 (NN_NI_HD_NI_NN_HD_NN_NI_NI_HD_HD_NI_NG_HD_NN_NN_NG_HD_NG_HD_NG). Mutant alleles were identified by PCR-amplification (I2_F: TAAACACACACCGCTCTGCT, E3_R: TTCGTCGTGGGATCTCTCAT) from genomic DNA extracted from individual embryos, cloning and sequencing.
Comparative analysis of the corresponding regions in msi1 and msi2a sequences was performed to test potential crossed effects of knock-down assays. For the morpholinos, these sequences showed mismatches of 8 bps and 6 bps (ATG) and 10 bps and 6 bps (i7E8) when compared to msi1 and msi2b, respectively. Moreover, these mismatches were distributed throughout the target sequence, such that similarity was not more than 7 contiguous bases, which is less than the 14-15 bps necessary for a morpholino to block a gene transcript [34]. Similarly, the net sequence mismatches for the E2 TALEN was of 12 bps for msi1 and 17 bps for msi2a.
Proliferation and label-retaining assays
To analyze cell proliferation, we used an adapted Click-IT (Invitrogen) protocol [35]. Embryos were injected pericardially with 9nl of EdU (150μM in 2% DMSO, 0.1% phenol red), and incubated at 28°C until the desired stages. For labeling-retaining assays, single-cell embryos were injected with 100 pg of H2B-mCherry mRNA and analyzed by confocal imaging.
RESULTS
Identification of Msi2b line in zebrafish
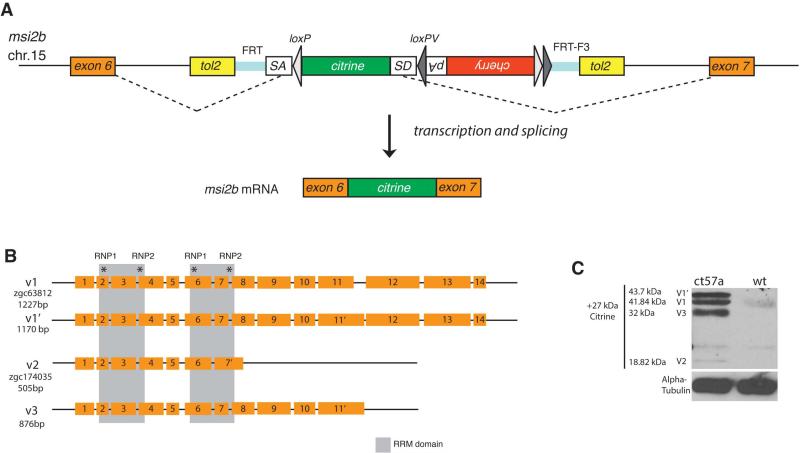
We have identified a novel msi2b zebrafish transgenic line using a Tol2 transposase-based gene/protein trap screen [28]. Integration of the genetrap construct within an intronic region of the trapped locus, results in a Citrine fusion with the endogenous protein. 3’RACE using a citrine-specific primer revealed tagging of musashi-2b (msi2b), a member of the Musashi family of RNA-binding proteins (Fig.1A). This line gt(msi2b-citrine)ct57a has the citrine sequence inserted as an artificial exon between exons 6 and 7. Therefore, the fusion protein contains all the endogenous features of Msi2b, including its 5’ and 3’ UTR. Moreover, Citrine tag does not affect protein function and homozygous fish are viable.
Figure 1. A novel gene trap line depicts expression of Msi2b in zebrafish.
(A) Schematic diagram of the FlipTrap strategy. The FlipTrap construct contains an artificial exon consisting of citrine ORF sequence lacking start and stop codons (green), flanked by rassf8 splice acceptor (SA) and donor (SD) sites. Genomic integration of the construct is mediated by Tol2 transposase. In the gt(msi2b-citrine)ct57a line, the FlipTrap construct has inserted in the chromosome 15, within the intron between exons 6 and 7 of msi2b. The endogenous splicing machinery recognizes the splice acceptor (SA) and splice donor (SD) sites flanking the citrine sequence (dashed lines), resulting in Citrine-fusion with endogenous protein.
(B) Schematic diagram of zebrafish msi2b splice variants. Members of the Msi family of RNA-binding proteins are characterized by the presence of two RNA-binding domains (gray boxes) that include two short highly conserved motifs called RNP-1 and RNP-2 (asterisks). Alternative splicing of msi2b produces 3 splice variants (v1, v2, v3).
(C) Detection of Msi2b protein fused with Citrine-tag reveals Msi2b the presence and relative abundance of variants at the protein level. Expression of Msi2b variants by western blot in gt(msi2b-citrine)ct57a embryos at 56 hpf. Alpha-Tubulin is used as a load control.
Musashi proteins contain two tandem sets of ribonucleoprotein (RNP)-type RNA recognition motifs (RRM) in the N-terminal half of the protein. In zebrafish Msi2b, the RRM are encoded by exons 2-4 and 6-8, with the RNP octomer consensus sequences located in exons 2, 4, 6 and 8 (http://smart.emblheidelberg.de; ExPASy Prosite, http://prosite.expasy.org/). Analysis of endogenous msi2b transcripts by 5’ and 3’RACE revealed 3 distinct splice variants. The full-length (v1) consists of 14 exons (encoding 389 amino acids) and a 3’UTR of 657 bps and alternative splicing of exon 11 produces v1’, in which only 57 nucleotides of intron 11 are included in the coding region of exon 11’, thus producing a protein of 408 amino acids. In v2, splicing of intron 8 does not occur; instead a C-terminally truncated short variant forms by addition of 52bps (14 amino acids) after exon 7, plus a 3’UTR of 1190bps fully overlapping with intron 8. Finally, splice variant v3 spans exons 1-11, plus 83 bps of intron 11, thus producing a 292 amino acids protein. The 3’UTR of v3 corresponds to 856bps of intron 11, with a polyadeylation signal towards the end of the sequence (Fig.1B).
In the gt(msi2b-citrine)ct57a line, endogenous Msi2b is expressed as a fusion protein with Citrine inserted within the intron between exons 6 and 7, without disrupting splice donor and acceptor sites. Hence the Msi2b variants can be detected by western blot using the Citrine-tag (Figure 1C). At 56 hpf, we identified bands that correspond to the expected molecular sizes for V1 (43.7 kDa), V1’ (43.7 kDa) and V3 (32 kDa) as the most abundant forms, while V2 (18.8 kDa) expression is detected at very low levels.
Our results suggest that v2 and v3 variants encode C-terminally truncated forms of the full length Msi2b (v1). In particular, v2 retains only the N-terminal portion of the protein. These truncations may modulate Msi2b protein function, possibly by acting as dominant negative isoforms. Alternative splicing events have been reported for murine Msi1 and Msi2 as well as zebrafish Msi1, although in those cases the variants only differ from the full length proteins by 18 amino acids in the C-terminus, not affecting the RNA-binding domains [8, 19, 36].
Relationship of Msi2b to other Msi family members
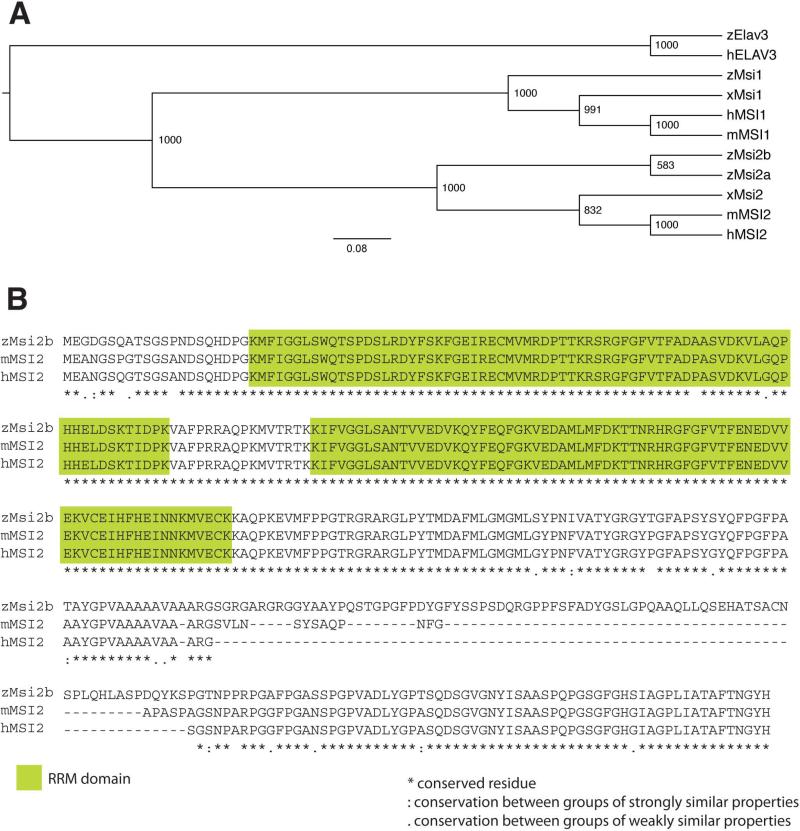
Phylogenetic analysis of Msi RNA-binding proteins in vertebrates using the Neighbor Joining method confirms that Msi2a and Msi2b are co-orthologs of vertebrate MSI2. In addition, it shows that zebrafish Msi2a and Msi2b result from a gene duplication event specific to teleosts (Fig.2A).
Figure 2. Comparison among members of the Msi family of RNA-binding proteins.
(A) Phylogenetic tree of Musashi proteins in vertebrates was generated by ClustalX analysis. Human, hMSI1 and hMSI2; mouse, mMSI1 and mMSI2; Xenopus, xMsi1 and xMsi2 and Zebrafish, zMsi1, zMsi2a, and zMsi2b. Zebrafish and human RNA-binding protein, zElav3 and hELAV3, were included in the analysis as outgroups. In zebrafish, Msi2 has diverged further into zMsi2a and zMsi2b. The numbers on the nodes of the tree represent the bootstrap values from Neighbor joining analysis.
(B) Alignment of zMsi2b protein sequence with mouse and human MSI2. The RNA-binding domains (green boxes) are highly conserved between zMsi2b and mammalian MSI2, with only two amino acids substitutions in the first domain. Sequence divergences consist of an additional sequence of amino acids between positions 263 and 343 of zMsi2b sequence. Full conservation of residues are indicated by asterisk (*); “:” and “.” indicate full conservation of ‘strong’ and ‘weaker’ residues, respectively.
msi2b maps to zebrafish chromosome 15, and Msi2b protein shares 89% homology with Msi2a, encoded on chromosome 10 (ClustalW2, http://www.ebi.ac.uk/Tools/msa/clustalw2/). Msi2a and Msi2b share 73% and 72% similarity, respectively, to zebrafish Msi1. The differences in sequence are mostly confined to the C-terminal half of the proteins. In this region, there was 84% similarity between Msi2a and Msi2b, and 64% identity with zebrafish Msi1 in amino acid sequence. Conversely, the N-terminal portion of Msi2a and Msi2b encompassing the RRM1 and RRM2 regions are highly conserved (94% and 97%, respectively). When compared to Msi1, Msi2a and Msi2b RRM1 are 78% and 79% homologous, respectively, while RRM2 region is more conserved (90% and 93%, respectively).
Zebrafish Msi2b and Msi2a are closer to human MSI2 (93% and 92%, respectively), than to mouse MSI2 (84% and 88%, respectively), with high conservation in the RRM regions. Divergences in sequence consist of an additional stretch of amino acids between positions 263- 343 of Msi2b (Fig.2B).
Two phases of Msi2b expression in zebrafish
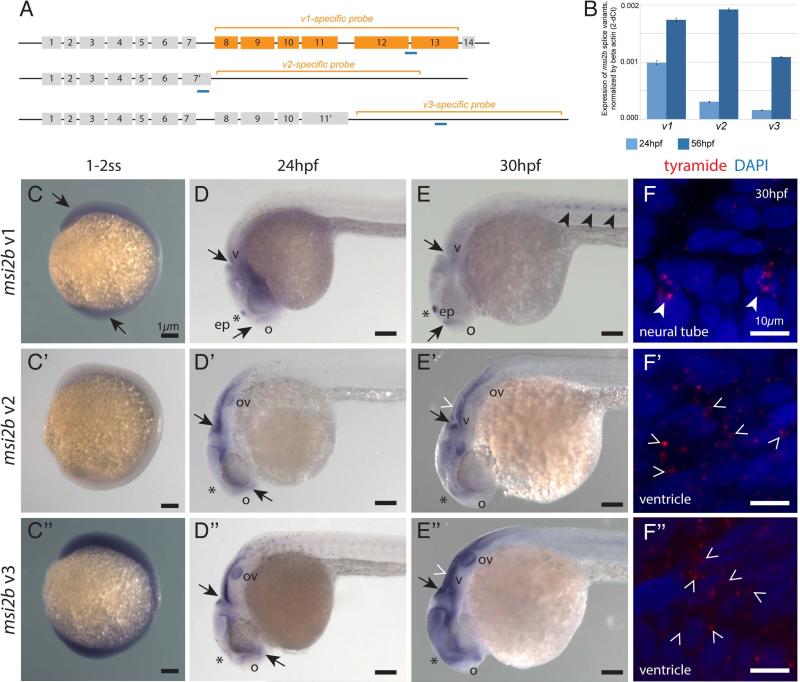
To assess the differential expression of msi2b splice variants through development, we measured relative abundances of each variant by quantitative RT-PCR at two stages of development, using primers that specifically targeted the regions unique to each variant (Fig.3A, B). Furthermore, we characterized spatial and temporal expression patterns of these variants by in situ hybridization (Fig.3C-F”).
Figure 3. msi2b splice variants are differentially expressed in zebrafish.
(A) Schematic diagram of msi2b splice variants. Target regions for anti-sense riboprobes (orange brackets) and qPCR (blue) were designed to specifically detect the distinct msi2b splice variants.
(B) Quantitative analysis of msi2b variants by qPCR. cDNA template from zebrafish embryos at 24 hpf (n=10, light blue) and 56 hpf (n=10, dark blue). Values are expressed in 2-dCt±SD, calculated as a ratio to beta-actin expression.
(C-E”) Expression of msi2b splice variants by whole mount in situ hybridization. Lateral views. Arrows point to areas of msi2b expression in the ventricles (v) and olfactory region (o). Low level, ubiquitous expression of msi2b full-length (v1) and the splice variant v3 at the 1-2 somite-stage (ss) (C,C”). At this stage, expression of v2 is not detected (C’). The full-length msi2b (v1) is expressed at low levels in the head, with tissue-specific expression that corresponds to the expression of Msi2b-Citrine in the epiphysis (ep, *), olfactory placode and CNS at 24 hpf (D), and appearing in the ventral neural tube at 30 hpf (E). msi2b v2 (D’) and v3 (D”) are expressed in the otic vesicle (ov) and lining of the brain ventricles at 24 hpf. At 30 hpf, v2 expression is downregulated in the otic vesicle and localizes to the brain ventricle and CNS (E’), while v3 expression is enhanced in the otic vesicles, ventricles and forebrain (E”). Scale bars: 1 μm.
(F-F”) Subcellular localization of msi2b transcripts by fluorescent in situ hybridization. Transcripts of msi2b variants (red) are localized in the cytoplasm of expressing cells in the neural tube (full arrowheads, E,F) and ventricles (empty arrowheads, E’,F’, and E”,F”), rather than the nuclei counterstained by DAPI (blue). Scale bars: 10 μm.
Using antisense riboprobes specific for each msi2b splice variant (Fig.3A), we identified two phases of msi2b expression. During early somitogenesis, expression of msi2b is characterized by the prevalence of v1 and v3 throughout the embryo (Fig.3C-C”). Interestingly, at later stages, expression of msi2b variants transitions into tissue-specific patterns (Fig.3D-E”). For example, full-length msi2b (v1) is expressed in the epiphysis, olfactory placodes and hindbrain at 24 hpf (Fig.3D), while splice variants v2 and v3 are expressed in the otic vesicle and lining the brain ventricles (Fig.3D’-D”). At 30 hpf, v1 is also detected in the ventral neural tube and ubiquitously in the CNS, at low levels (Fig.3E). In contrast, v2 and v3 remain restricted to the embryonic head region and absent from the ventral neural tube. At this time, v2 is down-regulated in the otic vesicle, though expression is maintained in the forebrain and midbrain (Fig.3E’), whereas v3 is enhanced in the otic vesicle, ventricles and forebrain at 30 hpf (Fig.3E”). Quantitative analysis by qPCR indicates that v1 is the most abundant form expressed at 24 hpf. In the subsequent stages, expression of variants v2 and v3 increases significantly, while v1 levels remain stable by 56 hpf, v2 and v3 levels increase 6 to 7-fold, with full-length v1 level increases by 1.8-fold (Fig.3B).
To differentially detect each msi2b variant by in situ hybridization, we designed riboprobes that target unique regions for each variant. In this strategy, we detect the full-length v1 by targeting the fully spliced region between exons 8 and 13 of msi2b. However, the riboprobes for v2 and v3 could also potentially hybridize with introns 7 and 11 of non-spliced pre-mRNA, respectively. To confirm that the detected transcripts of the splice variants v2 and v3 correspond to mature mRNA, rather than nascent premRNA, we assessed the localization of msi2b transcripts with subcellular resolution by fluorescent tyramide in situ hybridization (Fig.3F-F”). We observed that the transcripts of msi2b variants v2 and v3 are exclusively localized in the cytoplasm and not in the nuclei of expressing cells. Interestingly, our results reveal expression of v1 transcripts as clusters condensed in close association with the nuclei of cells in the ventral neural tube, and even co-localized with the nuclei. This could be an artifact caused by the tight arrangement of these cells, or rather we could be detecting spliced, mature v1 mRNA before it is exported to the cytoplasm.
Live imaging of gt(msi2b)ct57a line depicts expression of endogenous Msi2b in zebrafish
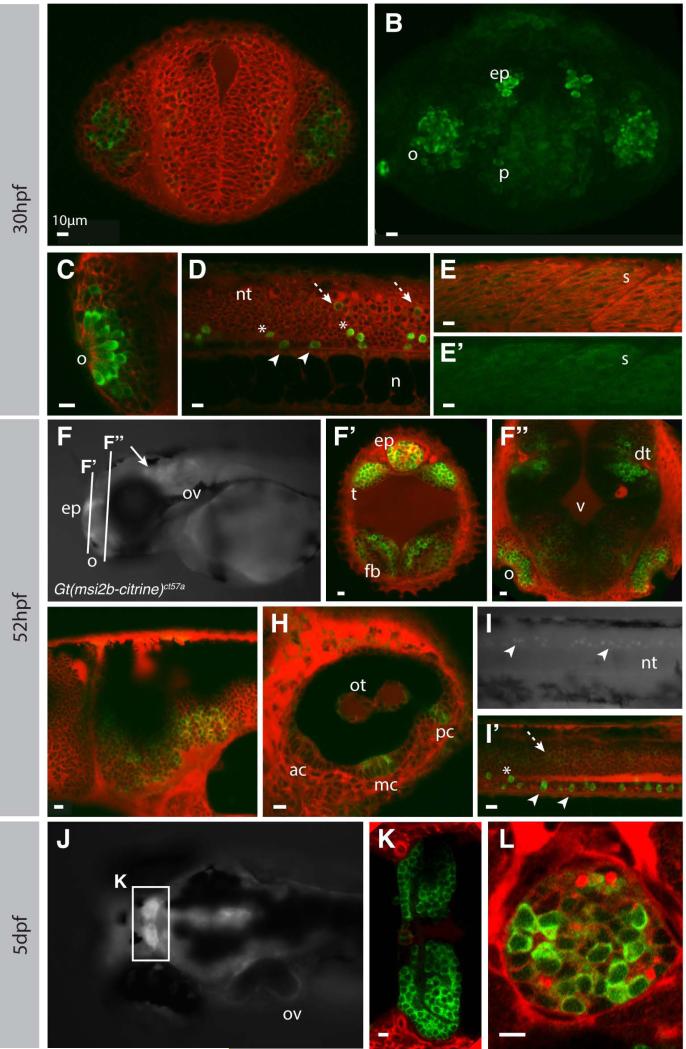
Our 3’RACE results show that all msi2b splice variants contain the citrine insertion in the gt(msi2b-citrine)ct57a embryos. As a consequence, expression of Citrine reflects expression of endogenous Msi2b and all its splice variants, thus providing a valuable tool for assessing the endogenous temporal pattern and sub-cellular distribution of Msi2b fusion proteins. Confocal live imaging reveals cytoplasmic and tissue-specific distribution of Msi2b in the head and trunk of gt(msi2b-citrine)ct57a embryos (Fig.4). At 30 hpf, Msi2b is strongly expressed in the pineal gland and in a subset of cells in the olfactory placode. In the head region at this stage, Msi2b is expressed in the telencephalon, with strong levels of expression in the pallium region (Fig.4A-C). A subset of ventral neural tube neurons strongly expresses Msi2b-Citrine as early as 26 hpf (Fig.4D).
Figure 4. Msi2b is expressed in distinct neuronal populations in zebrafish.
Confocal imaging of live gt(msi2b-citrine)ct57a embryos depicts cytoplasmic expression of Msi2b in zebrafish embryos (green) at 30 hpf (A-E’), 52 hpf (F-I’) and 5 dpf (J-L). Counterstaining by BODIPYTR (red). Olfactory region, o; pallium, p; epiphysis, ep; otic vesicle, ov; neural tube, nt; notochord, n; somite, s; telencephalon, t; forebrain, fb; dorsal telencephalon, dt; midbrain-hindbrain boundary, mhb; otoliths, ot; anterior, median and posterior cristae are respectively ac, mc and pc. Scale bars: 10 μm. (A, B) Frontal view. Single plane imaging (A) and respective confocal projection (B) of Msi2b-Citrine expression in the olfactory placode and pallium, with lower levels of expression in the forebrain. (C-E’) Lateral views. (C) Msi2b is highly expressed in the olfactory placode. (D) Expression of Msi2b in three distinct subpopulations of neurons in the ventral neural tube highly expressing cells within (white arrowhead) and dorsally (asterisks) to the floor plate, and low expression in the periphery of the neural tube (dashed white arrow). (E, E’) Low levels of Mis2b are detected in the somites. (F, G-I’) Lateral views. (F’,G”) Coronal sections. (F) Msi2b is expressed in the epiphysis, olfactory organs, otic vesicles and CNS (white arrow). (F’, F”) Frontal views, optical sections. Expression in the dorsal thalamus and epiphysis as well as in the forebrain and olfactory organs. (G) Msi2b is expressed in the ventral cells of the hindbrain. (H) Expression in the otic vesicle is localized to cells of the otoliths and in the central cells of the cristae of the semicircular canals. (I, I’) Expression of Msi2b in the ventral cells of the neural tube (white arrowhead). Cells dorsal to the floor plate (asterisks) and low expression in the periphery of the neural tube (dashed white arrow). (J) Dorsal view of zebrafish head at 5dpf. Msi2b is expressed in the habenula (K) and is restricted to a subset of neurons in the epiphysis (L).
Our analysis reveals three distinct groups of Msi2b-positive cells in the neural tube: 1) ventral-most cells located within the floor plate expressing strong Citrine, 2) ventral cells dorsal to the floor plate expressing strong Citrine and 3) dorsal-lateral cells expressing lower levels of Msi2b. Interestingly, we also identified low levels of cytoplasmic expression of Msi2b in the muscle fibers of the somites (Fig.4E,E’).
At 52 hpf, Msi2b expression persisted in the head (Fig. 4F), including the pineal gland, dorsal thalamus (Fig.4F’), olfactory organ and forebrain (Fig.4F”). Expression also was observed in the ventral hindbrain (Fig.4G). Msi2b was seen in central cells of the cristae of the semicircular canals and in the otoliths of the otic vesicle (Fig.4H). In the neural tube, Msi2b expression was particularly strong in a subset of ventral neurons (Fig.4I,I’).
In zebrafish larvae at 5dpf, expression of Msi2b persisted in the CNS, with particularly high levels in the telencephalon and habenula (Fig.4J,K). Interestingly, only subsets of cell in the pineal gland maintained strong expression of Msi2b at these later stages (Fig.4L).
Msi2b is expressed in distinct subsets of neural cells in zebrafish
Tissue-specific expression of Msi2b is characterized by distinct expression in the olfactory bulb and pineal gland as well as a subpopulation of ventral neural tube neurons after 24 hpf.
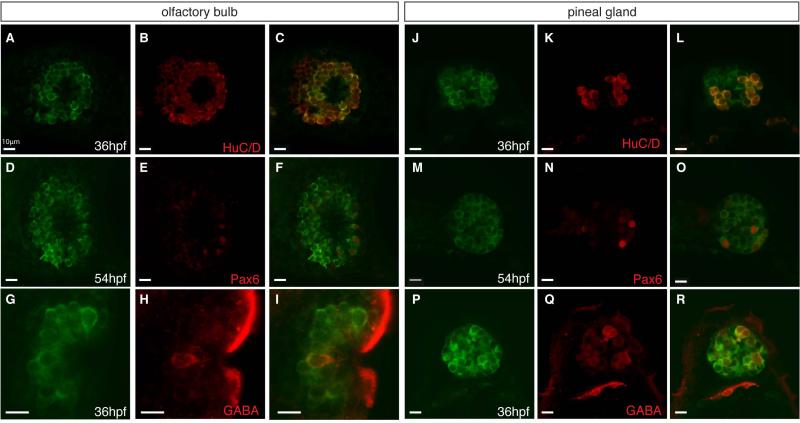
To characterize Msi2b-expressing cells, we examined overlap with RNA-binding proteins HuC/D as a neuronal marker. In the olfactory bulb, Msi2b-positive cells correspond to a mixed population of post-mitotic neurons that express the HuC/D marker (Fig.5A-C). Msi2b-positive olfactory neurons include subpopulations of cells positive for GABA (Fig.5D-F) and Pax6 (Fig.5G-I). In the pineal gland, a subset of Msi2b cells corresponds to post-mitotic neurons expressing HuC/D (Fig.5J-L), with distinct groups expressing GABA (Fig.5M-O) and Pax6 (Fig.5P-R).
Figure 5. Second phase of Msi2b expression is localized in the olfactory organ and pineal gland.
Confocal images of co-expression of neuronal markers (red) with Msi2b-citrine (green) in gt(msi2bcitrine)ct57a line. Scale bars: 10 μm.
(A-C) Msi2b is expressed in the HuC/D-positive neurons (red) in the olfactory bulb at 36 hpf.
(D-F) GABA-positive neurons (red) in the olfactory bulb at 36 hpf. Lateral view.
(G-I) Low levels of Pax6 expression in (red) in the olfactory bulb at 54 hpf.
(J-L) Expression of HuC/D (red) in a subset of Msi2b positive neuron in the pineal gland at 36 hpf.
(M-O) GABA (red) is co-expressed with high levels of Msi2b in the pineal gland at 36 hpf.
(P-R) Strong expression of Pax6 (red) in neurons of the pineal gland at 54 hpf.
Analysis of Msi2b expression in the neural tube revealed high levels of Msi2b-Citrine expression in the ventral neural tube, in cells near the midline (Fig.6A). Interestingly, Msi2b protein expression is complementary to the distribution of post-mitotic cells expressing HuC/D, with little or no expression of HuC/D in the cells expressing high levels of Msi2b-Citrine fusion protein. Conversely, post-mitotic neurons expressing high levels of HuC/D reside in the dorsolateral neural tube and exhibit lower levels of Citrine (Fig.6B,C). These results are consistent with findings in mouse spinal cord, where Msi1 is expressed in mitotically active cells of the ventricular zone, while Hu-positive cells are distributed outside the Msi1-positive area, with a small degree of overlapping expression [8].
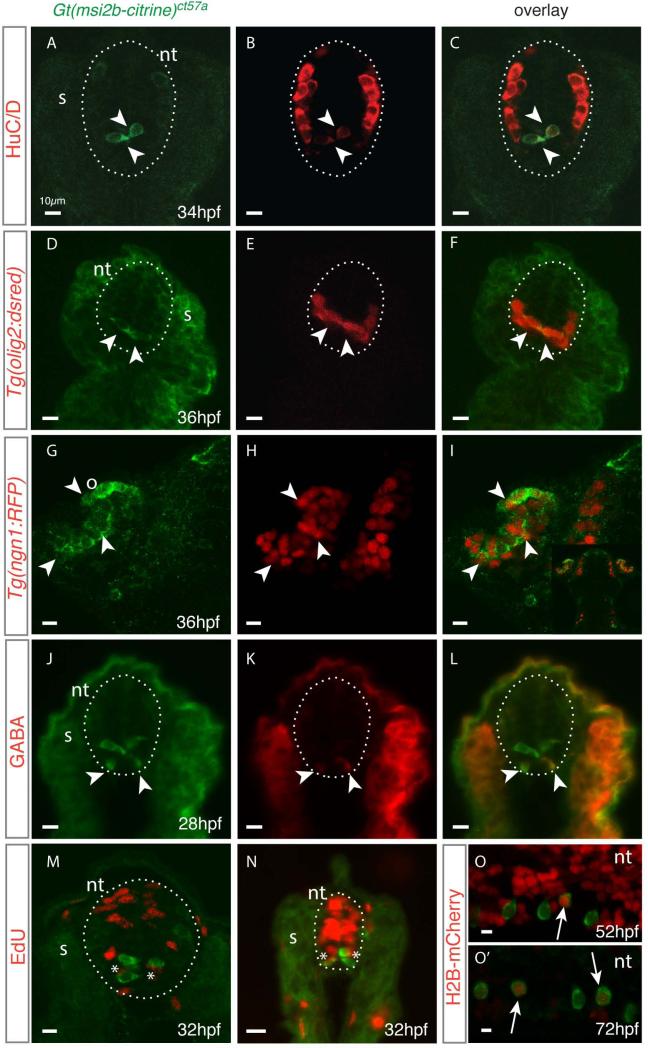
Figure 6. Msi2b expression in the ventral neural tube encompasses distinct cell types in zebrafish.
(A-N) Cross sections of gt(msi2b-citrine)ct57a embryos depict distribution of Msi2b-positive cells in the neural tube (nt, dotted line). Arrowheads point to areas of co-expression of Msi2b-Citrine (green) with the respective neuronal marker (red). Somites, s; olfactory bulb, o. Scale bars: 10 μm.
(A-C) Citrine is highly expressed by a subset of ventral cells closely associated with the midline, while differentiated neurons reside in the dorsal-lateral region of the neural tube, as determined by expression of HuC/D. (D-F) Citrine-positive cells correspond to a subset of olig2-positive cells, as determined by co-expression with dsRed in the Tg(olig2:dsred) transgenic line. (G-I) Crosses with Tg(ngn1:RFP) indicates that Msi2b-positive cells in the olfactory organs correspond to a population of neuronal progenitors. (J-L) Immunodetection reveals that ventral-most Msi2-positive cells express GABA (red). (M,N) Cross-section of neural tube of gt(msi2b-citrine)ct57a embryos treated with EdU (red) at 24 hpf and observed at 32 hpf. Incorporation of EdU (red) is observed in the cells adjacent to Msi2b-positive (green) cells of the dorsal neural tube in the trunk level and in the cells adjacent to the midline in the tail.
(O,O’) Lateral views of the neural tube of gt(msi2b-citrine)ct57a embryos injected with Cherry-H2B mRNA at the 1-cell stage. Scale bars: 10 μm. (O) Expression of Cherry is ubiquitously expressed in the neural tube of embryos at 52 hpf. (O’) At 72 hpf, Cherry-H2B labeling is retained by few cells in the neural tube, including some Msi2b-positive cells (white arrows).
The ventral neural tube contains a population of olig2-expressing pMN precursors with potential to produce motor neurons and oligodendrocyte progenitor cells (OPCs) as well as interneurons and radial glia [37]. By crossing our line with Tg(olig2:DsRed2) fish, we found that a subpopulation of Msi2b-positive cells co-expresses Olig2 (Fig.6D-F) [24, 37]. Moreover, crosses with Tg(-8.4neurog1:nRFP) identify an additional population of progenitor neurons co-expressing Msi2b in the olfactory organ (Fig.6G-I) [25].
To further characterize Msi2b-positive cells, we analyzed Citrine co-localization with neuronal markers. The ventral-most Citrine-positive neural tube cells correspond to GABA-immunoreactive neurons in the spinal cord at 28 hpf (Fig. 6J-L). Their localization in the lateral floor plate suggests they may be GABAergic KA” cells [38]. In zebrafish, a population of GABAergic Kolmer-Agduhr (KA) interneurons designated KA’ and KA” reside in the ventral neural tube and have cell bodies contacting the spinal central canal. No co-expression was observed with Islet1 and Pax2 with Msi2b (data not shown).
We next tested proliferative and label-retaining properties of Msi2b-positive cells as putative progenitors in the neural tube (Fig.6M-O’). Cell proliferation assays reveal extensive cell division associated with the neural tube midline, adjacent to the Msi2b-positive cells. However, no incorporation of EdU was seen in Msi2b-positive cells in embryos treated at 24hpf (Fig.6M,N), raising the possibility that Msi2b-positive cells may be quiescent or slowly-dividing. To explore this possibility, we assessed their label-retaining properties. Single-cell embryos were injected with H2B-Cherry mRNA and distribution of H2B-Cherry signal in the neural tube was analyzed subsequently. Confocal imaging at 52 hpf shows strong Cherry signal ubiquitously distributed in the neural tube, including in Msi2b-positive cells (Fig.6O). In contrast, at 72hpf, Cherry signal was diluted in the neural tube, though we observed a significant number of Citrine-positive cells retain nuclear labeling (Fig.6O’). This suggests that the Msi2b-positive cells in the neural tube are not actively dividing at stages analyzed, either because they represent slow-dividing cells or have already left the cell cycle and differentiated.
Knockdown of Msi2b affects neural development and anterior-posterior extension
Splicing in the gt(msi2b-Citrine)ct57a line results in a fusion protein with Citrine inserted between exons 6/7, the site between the RNP motifs in the second Msi2b RRM. Although this splicing event could potentially disrupt domain spacing, we did not observe an abnormal phenotype after homozygosis of gt(msi2b-Citrine)ct57a, suggesting this was not the case.
To gain insight into Msi2b function, we performed loss-of-function assays using three methods: 1) a translation-blocking morpholino (MO) to knockdown all msi2b splice variants, 2) transcription activator-like effectors nucleases (TALEN) to induce biallelic somatic targeted mutagenesis at the splicing donor junction [39, 40], and 3) a splice-blocking morpholino targeting the junction between intron 7 and exon 8 (i7/E8) to knock-down full-length Msi2b and produce only the v2 C-terminally truncated form (Fig.7A).
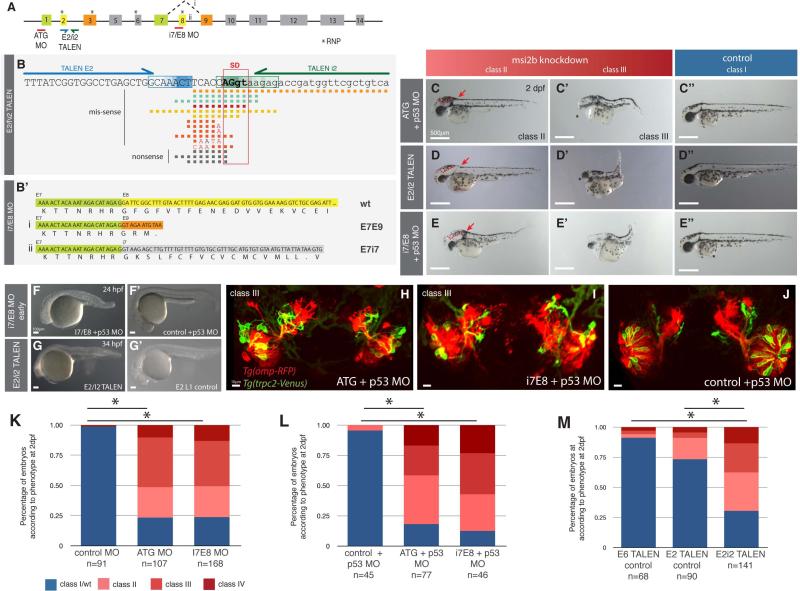
Figure 7. Msi2b loss-of-function by morpholino knockdown and biallelic TALEN-induced somatic mutation.
(A) Schematic diagram of the binding sites for Msi2b translation blocking (ATG) and splice-blocking (i7/E8) morpholinos (MO) as well as the binding sites for E2/i2 TALEN. E2/i2 TALEN was designed to target the splice donor site of Msi2b exon 2. Binding of msi2b i7/E8-MO results in alternative splicing events (i, ii) by disrupting splicing between exons 7 and 8 of Msi2b. Numbered boxes represent exons. Asterisks depict RNP octomer recognition sequences (RNPs).
(B) Schematic diagram of genome editing by E2/i2 TALEN in zebrafish. TALEN E2 (blue arrow) and TALEN i2 (green arrow) respectively bind to regions of exon 2 (E2) and intron 2 (i2) flanking the splice donor (SD) of msi2 E2. Boxed sequences within E2 (blue) and i2 (green) represent the sites of DNA cleavage by FokI nuclease in the TALEN E2/i2 mutant embryos (n=16 clones). The gradient color represents the frequency of DNA cleavage within the region for the mutant embryos analyzed. Nucleotide deletions (squares) and replacements (letters) induced by E2/i2 TALENs are represented under the target sequence. PCR amplification from genomic DNA, cloning and sequencing of multiple clones was obtained for each embryo.
(B’) Schematic diagram of alternative splice events produced by i7/E8 splice-blocking morpholino. Sequences of msi2b exon 7 (E7, green), exon 8 (E8, yellow), exon 9 (E9, orange) and intron 7 (i7, gray) and corresponding translation sequence. Alternative splicing between E7 and E9 (i) or persistence of intron 7 (ii) results in a premature stop signal and truncated of Msi2b protein similar to v2. (C-E”) Inactivation of all Msi2b variants by translation-blocking morphants or E2/i2 TALEN as well as specific truncation of Msi2b full-length (v1) by i7/E8 morpholino in zebrafish is characterized by ventricular edema (red dashed lines), shortening of the body length and atrophic or absent eyes. Scale bars: 500 μm.
(F-G’) Knockdown of msi2b in zebrafish indicates that Msi2b plays an early role in embryonic development. Similar early embryonic phenotype, with truncated body length and tissue degeneration, was observed after specific knockdown of full-length by co-injection of i7/E8 MO and p53 MO (F) and mutagenesis of msi2b by E2/i2 TALEN (G), as compared to embryos co-injected with control MO and p53 MO (F’) or a single TALEN E2 L1 as a control (G’). Scale bars: 100 μm. (H-J) Confocal 3D projection of the olfactory organ in Tg(omp-RFP)/Tg(trpc2-Venus) zebrafish embryos at 52 hpf. Knockdown of msi2b expression by ATG+p53 MO (H) or i7E8+p53 MO (I), as compared to control embryos (J). Scale bars: 10 μm.
(K-M) Quantitative analysis of the distribution of resulting phenotypes after inactivation of msi2b in zebrafish. Asterisks indicate statistical significance, *P<0.001.
The translation blocking morpholino, whose binding encompasses the start site (ATG) of msi2b, resulted in reduced expression of Citrine when injected into gt(msi2b-Citrine)ct57a embryos (not shown). To target the region flanking the splice donor (SD) site of msi2b between exon 2 and intron 2, we injected mRNA encoding for the pair of TALENs E2/i2 (Fig.7B). To assess its efficiency, we analyzed the genomic region encompassing intron 2 to exon 3 of msi2b gene from embryos displaying mild and strong phenotypes (n=6). Because of potential mosaicism resulting from TALEN activity, we cloned and sequenced multiple clones for each mutant, with a total of 22 sequences analyzed. Our analysis shows that DNA cleavage by the FokI nuclease occurred within 6 bp or less from the 3’end of each TALEN in 86.4% of the sequences analyzed (n=19/22)(Fig.7B). In particular, the majority of DNA cleavage sites were clustered in a discrete region, between 4-6 bps and 2-6 bps downstream of the binding regions for TALEN E2 L (91.1%, n=16/17) and TALEN i2 R (75%, n=12/16), respectively. In a single case, the DNA cleavage occurred outside the region between TALENs, producing a 58 bps deletion of intron 2.
Our results show that the combination of E2/i2 TALENs produced efficient mutagenesis of msi2b in zebrafish, with 15 distinct patterns of deletions and nucleotide replacements specifically affecting the target region. Importantly, we found a correlation between higher efficiency mutagenesis and severe phenotypes. For example, all clones analyzed from 4 severely affected embryos showed mutations (n=12), predominantly missense (n=9/12), with non-sense deletions only identified in one embryo. In contrast, mildly affected embryos (n=2) had missense mutations in 54.5% of analyzed clones (n=6/11), 18.2% of non-sense (n=2/11) and 27.3% none. Our results show a high level of genome modification in strongly affected embryos indicating biallelic mutagenesis. Relatively low level of genotypical heterogeneity correlates with early TALEN-mediated gene-editing events. To determine effects of mutations on msi2b splicing, we analyzed the resulting mutagenized sequences for splice sites (http://www.fruitfly.org/seq_tools/splice.html) and performed in silico analysis to predict impact on the resulting protein sequence. We found that mutation of Msi2b protein occurs by premature stop signal sequence resulting mainly from: 1) deletion of SD site and alternative splicing (E1E3), 2) deletion of SD site with persistence of intron region (E2i2 and i2E3) or stop signal at the end of exon 2 (E1E2), and 3) shift of SD site caused by deletion and/or replacement resulting in frameshift (E2E3) (Fig.1S). In all cases, mutagenesis by E2/i2 TALENs produces a truncated Msi2b protein caused by a premature stop signal.
To examine the role of the full-length Msi2b splice variant, we used a splice-blocking MO that targets the junction between intron7-exon8 (i7/E8)(Fig.7B’). This results in truncation of Msi2b protein, with disruption of its second RRM similar to the splice event that produces endogenous variant V2. Efficiency of MO was assessed by reduced expression of Citrine in morphants (not shown).
Msi2b loss-of-function induced by MO-knockdown or targeted mutagenesis by TALENs produced embryos with defects in head development, with a phenotype characterized by distinctive edema of the 4th (hindbrain) ventricle and eye malformation (Fig.7C-E). In severely affected embryos, we also observed a truncated body and atrophy of the tail (Fig.7C’-E’), not seen in controls (Fig.7C”-D”). Importantly, these phenotypes were consistently preceded by a transient phase of increased cell death and tissue degeneration before 36 hpf (Fig.7F-G’), which progressively recovered after 48 hpf. To confirm that cell death was not caused by off-target effects, morpholinos were co-injected with p53 morpholino, which did not alter the observed phenotype. The tissue degeneration during the first phase of development strongly correlated with severe later phenotypes of body truncation. In fact, some of the late embryonic defects, such as small heads and eye malformation, may be secondary to this early cell loss.
To quantify the effects of Msi2b knockdown, we categorized embryos into classes I-IV according to malformations severity at 36hpf (Fig.7K-M). Embryos displaying normal body phenotype, similar to the controls (Fig.7C”-E”), were characterized as class I, although after msi2b knockdown they also often displayed ventricular brain edema. Class II is characterized by defects in tail formation and hydrocephaly (Fig.7C-E). In class III embryos, elongation of the anteroposterior body axis was strongly affected, accompanied by defects in head and eye development as well as cardiac edema and circulatory insufficiency, with blood accumulation over the yolk (Fig.7C’-E’). Finally, class IV embryos had very severe phenotypes comprised of all defects described above (not shown).
Although the general patterning of the organs and tissues in the morphants was not significantly affected, the phenotype at the cellular level was characterized by extensive tissue disorganization, particularly in class III and IV embryos. For example, the assembly of the bundles of ciliated and microvillous neurons in the olfactory organ of class III embryos is perturbed, with apparent reduction in cellularity and signs of tissue degeneration, as determined by expression of the tissue-specific markers Tg(omp:RFP) [26] and Tg(trpc2:Venus) [26] (Fig. 7H-J).
Injection of ATG and i7/E8 morpholinos resulted in a similar distribution of phenotypic severity, with 77% (n=82/107) and 76% (n=128/156) of embryos exhibiting perturbations, respectively (Fig.7K). In these groups, a majority of the embryos showed class III phenotype (41% and 37% of ATG and i7/E8 morphants, respectively), followed by class II (25% and 23%, respectively) and 10% and 13% in class IV, respectively. Interestingly, co-injection of a p53 morpholino along with the msi2b ATG- and i7/E8-MO did not rescue the observed phenotypes; rather 82% (n=63/77) and 88% (n=49/56) of embryos displayed morphant phenotypes, respectively (Fig.7L). This shift corresponded to a more representative population of class II ATG+p53 morphants (40%, n=31/77). For the splice i7/E8 morphants, we observed an increase in the very severe class IV phenotype, from 14% in the i7/E8 morphants to 23% (n=13/56) in i7/E8+p53 MO, at the expense of mildly affected class I embryos (Fig.7L). Similarly, E2/i2 TALEN-mediated mutagenesis of msi2b resulted in 70% of affected embryos (n=98/141), with 32% of the embryos (n=45/141) showing phenotype class II, 24% class III (n=34/141) and 13% class IV (n=19/141)(Fig.7M). The effects of the splice-blocking MO were generally more severe than those observed after using a translation-blocking MO or E2/i2 TALEN. These distinct effects suggest that the balance of variants may refine/modulate Msi2b function.
DISCUSSION
In this study, we characterize Msi2b expression and its role in maintenance of progenitor cell during early zebrafish development. We present a novel zebrafish gene trap line Gt(msi2b-citrine)ct57a that expresses Citrine as a fusion protein with endogenous RNA-binding protein Musashi-2b (Msi2b), thus enabling tissue-specific visualization of endogenous Msi2b expression at cellular resolution in live embryos. Loss-of-function analysis indicates that Msi2b plays an essential role during early embryonic development.
Overlapping functions of Msi1 and Msi2b in zebrafish
We identify two phases of Msi2b expression in zebrafish: early low level distribution throughout the embryo, followed by enriched levels of tissue-specific expression in the olfactory organ and pineal gland; and KV neurons located in the ventral neural tube. These findings are consistent with studies of the dual role of Drosophila Msi in spermatogenesis: it is initially required for maintenance of germ-line stem cells, but later participates in their differentiation [14]. These authors suggest that as murine paralogues Msi1 and Msi2 diverged from the Drosophila Msi protein, Msi1 and Msi2 may differentially account for early versus late Msi functions, suggesting that the ancestral role of dMsi was split in vertebrates to generate a stem cell-specific Msi1 and a role for Msi2 in differentiating tissues.
However, we find early embryonic expression of msi2b, which later switches to tissue-specific localization in various neuronal subtypes in the pineal gland, olfactory bulb and ventral neural tube. The observed severe phenotype after Msi2b loss-of-function is reminiscent of defects in maintenance of the proliferative pool of progenitor cells previously described in Msi1 zebrafish morphants [36]. Thus, Msi2b has also retained overlapping but non-redundant functions with Msi1. This finding is consistent with functional cooperation between murine msi1 and msi2 in proliferation and maintenance of CNS stem cells [9]. This does not exclude a later role for Msi2b in specific tissues, though these may be obscured in our studies by the severity of early defects.
Our characterization of msi2b splice variants shows that transcripts of v2 and v3 are highly expressed in cells lining brain ventricles. Consistent with this, inactivation of Msi2b results in ventricular edema. Similarly, murine msi1 knockdown produces obstructive hydrocephalus resulting from spinal flow blockage and aberrant proliferation of ependymal cells obstructing the Sylvius aqueduct [9], a putative source of postnatal CNS stem cells. Later, Msi2b expression localizes in tissue-specific patterns in the olfactory organ, pineal gland, and KV neurons. These Msi2b-positive cells, however, do not constitute an active population of progenitor cells, but rather subsets of distinct neural cell types.
Tissue-specific expression of Msi2b in zebrafish
The cellular resolution afforded by endogenous Msi2b-Citrine in the ct57a line reveals cell type-specific expression of Msi2b in neural tissue, including subpopulations of cells in the olfactory organ, epiphysis and thalamus, midbrain, hindbrain, ventral neural tube and lining of the ventricles, consistent with reports on later Msi2 expression in mouse and Xenopus [10, 41]. Murine Msi2, for example, is initially strongly co-expressed with Msi1 throughout the telencephalon and spinal cord. Subsequently, expression is detected in the neurons of ventricular and subventricular zones (SVZ) of the telencephalon, spinal cord, ganglionic eminence, and dorsolateral SVZ, known to contain glial precursors for astrocytes, oligodendrocytes [10], and neuronal precursors for the interneurons of the olfactory bulb [42, 43].
In addition, we find that Msi2b is differentially distributed in a subset of spinal cord neurons, in a complementary pattern to neuronal marker HuC/D. As in mouse [10], we observed that high levels of Msi2b coincide with low or absent HuC/D in a subset of ventral cells near the central canal, while postmitotic neurons are localized in the neural tube periphery. These ventrally located Msi2b-positive cells may correspond to pMN precursors identified in the Tg(olig2:dsred) line. pMN cells form motoneurons, interneurons and oligodendrocyte progenitor cells (OPC) [37], but no coexpression was observed with Islet 1, a marker for primary motoneurons [44].
The ventral-most Msi2b-positive cells may correspond to the GABAergic Kolmer-Agduhr (KA) interneurons, which are localized in the lateral floor plate [38, 45] and control spontaneous swimming in zebrafish, representing a major modulator of locomotion in the awake behaving animal [46]. Interestingly, Msi2 expression is also maintained in a subpopulation of fast-spiking PV-containing GABA interneurons, which may enable generation of synchronous oscillations and spatiotemporal information processing within brain regions [10, 47, 48]. It is intriguing to speculate that Msi2b expression in the KA cells may control the local translation or stability of mRNAs for rapid turnover of receptors and channels necessary for their fast activity.
Because Msi2b in the ventral neural tube is found in proximity to the central canal where progenitor neurons are localized [37], we examined their proliferative potential. Our results show that Msi2b-positive cells are not actively dividing at the onset of expression in the neural tube, even though there is considerable proliferation of adjacent spinal cord cells. While some Msi2b-positive cells appear to retain nuclear labeling, these may represent KA interneurons, which by exiting the cell cycle for differentiation would retain high nuclear labeling relative to neighboring actively dividing cells.
Balanced expression of Msi2b splice variants is essential for embryonic development
We identified and characterized zebrafish Msi2b splice variants. Analysis of ESTs suggests that similar short variants with a second truncated RRM domain are present in mouse (AK051269 and AK 031365) (Capra JO369367; Fundulu JW569214, JW570252). Alternative splicing plays a key role in evolution of increased proteomic and functional complexity and is prevalent in the vertebrate nervous system, where it participates in cell-fate determination, axon guidance and synaptogenesis [49-51]. Analysis of alternative splicing in teleosts has suggested a low prevalence in the highly duplicated genome of zebrafish (17% of mapped genes) compared with 43% in the compact genome of pufferfish [52] or 40-60% of human genes.
It has been suggested that Msi roles were differentially subdivided between vertebrate Msi1 and Msi2. However, our results and other reports indicated that Msi2 has retained an early function in progenitor cells as well as a late role in differentiated tissues. Moreover, we identified dynamic changes in expression of msi2b splice variants that parallel the distinct early and late functions of Msi2b, with early ubiquitous and diffuse expression of full-length form being downregulated and/or restricted in the differentiating cells of the epiphysis and spinal cord, while the expression of variants v2 and v3 are distributed in a tissue-specific manner in the differentiating tissues of the ear and CNS.
We speculate that this early-late division may have its roots in alternative splicing. In this scenario, the splice variants could be associated with subfuctionalization events. Rather than between paralogous genes, the division of labor may be between alternative splice variants, with the full-length v1 and the longer variant v3 being expressed early and involved in maintenance of the progenitor state, whereas the C-terminally truncated variant v2 is associated with the late functions. Interestingly, all members of the Msi family share highly conserved RNA-binding motifs, such that their variability lies predominantly in the C-terminus. Thus, this domain possibly contains essential interaction domains, allowing Msi2b full-length protein to perform an early and highly regulated role. In contrast, the v2 variant, which lacks a C-terminus, may perform later tissue-specific functions and control levels of direct targets in those differentiated cells by RNA-binding.
Consistent with this possibility, our analysis shows dynamic tissue-specific expression of msi2b splice variants. However, they all have the same N-terminus, and therefore are likely to have similar cis-regulatory control, as demonstrated by many overlapping expression sites (with the exception of epiphysis and ventral neural tube in case of full-length variant v1). This raises the interesting possibility that those variants are also post-transcriptionally regulated, as they have differing 3’UTRs and hence would differentially respond to either stabilization by RBPs or miRNA-mediated degradation. This is particularly plausible for v2 and v3, which mostly overlap at 24 hpf, whereas later expression is lost in some sites (e.g. the ear for v2) indicating that v3 is differentially retained.
Distinct functions for Msi1 and Msi2
The Musashi family is characterized by the presence of two tandem RNA recognition motifs (RRMs) located at the N-terminus of the protein, each comprised of two highly conserved RNP-1 and RNP-2 motifs [7]. Our results show that zebrafish Msi2b is highly conserved with other vertebrate MSI2. Murine MSI1 inhibits mRNA translation initiation by competing with eIF4G at the polyA binding protein domain (PABP) in the mRNA sequence [53], corresponding to position 190-234 in mouse MSI1. This region is highly conserved in MSI1 across vertebrates, but has only 52% similarity with MSI2. Interestingly, this activity might be specific to MSI1, as the control of translation initiation by the PABP binding activity has not been reported in mouse or Xenopus MSI2 [54].
In addition to cooperating with MSI1 in progenitor cells, MSI2 expression is maintained in a tissue-specific fashion in the CNS. Persistent expression of MSI2 has been reported in GABA interneurons, as well as subpopulations of cholinergic and dopaminergic neurons in regions of the thalamus, hypothalamus and epithalamus, including the medial preoptic and supraoptic nucleus and periventricular nucleus and medial and lateral habenular nuclei [10].
Msi2 is also co-expressed with Msi1 in fetal and adult pancreatic islets of mouse, humans and rats. In alpha and beta pancreatic cells, rather than being a stem cell marker, Musashi is increased in the differentiating tissue. Msi2, in particular, is activated by endoplasmic reticulum (ER) stress [55].
Msi2 expression also participates in normal hematopoiesis: the balance of Msi2b-Numb signaling is a marker of myelogenous leukemia disease progression from a chronic slow growing phase into an aggressive blast crisis [20-22]. Although mice lacking Msi2 are viable, its loss causes severe defects in primitive blood precursors, exacerbated by age [56].
Taken together, these studies show that Msi2 plays an important role in various differentiated tissues, particularly under circumstances induced by stress. Using the gt(msi2b-citrine)ct57a line to characterize the distribution of Msi2b expression with cellular resolution, we identify tissue-specific expression in subsets of differentiating cells in the olfactory organ, pineal gland, CNS and ventral neural tube of zebrafish. Our results support the intriguing possibility that Msi2b expression may be associated with the capacity to respond to stress conditions.
Supplementary Material
Supplemental Figure 1. Knockdown of msi2b by E2/i2 TALEN in zebrafish.
Schematic diagram of genomic region of msi2b targeted by E2/i2 TALENs. SD, splice donor. The squares and letters below the sequence illustrate sites of deletions and replacements, respectively, in the region between exon 2 (upper case) and intron 2 (lower case) of msi2b. Missense mutations are color-coded to indicate resulting splice variant. Gray squares represent non-sense mutations. Exon, E; intron, i.
ACKNOWLEDGEMENTS
We thank Dr. Tatjana Sauka-Spengler for useful discussions, Dr. Marcos Simões-Costa and Dr. Sujata Bhattacharyya for comments on the manuscript, and Dr. Le Trinh and Dr. Scott Fraser for helpful advice. We thank Leigh Ann Fletcher and Kwok Su for fish care, Alice Plein and Ilana Solomon for technical support; and Dr. Bruce Appel for sharing the Tg(olig2:dsred) line; Dr. Uwe Strahle for the Tg(ngn1:nRFP) line and Dr. Yoshihiro Yoshihara, RIKEN BSI and National Bioresource Project of Japan for the Tg(omp:RFG) and Tg(trpc2:Venus) lines. T.H.H. was supported by a Pew Latin American Fellowship in Biomedical Sciences and by California Institute for Regenerative Medicine Training Grant (T2-00006). This work was supported by NIH grants HG004071 and HD037105 (M.E.B).
Footnotes
Authors contributions:
T.H.H. designed and performed experiments, analyzed data and wrote the manuscript. D.E.S.K. and N.M.D. performed experiments, analyzed data and edited the manuscript. M.E.B. supervised the work including helping with experimental design, data, analysis, and manuscript preparation.
REFERENCES
- 1.Sampath P, Pritchard DK, Pabon L, et al. A hierarchical network controls protein translation during murine embryonic stem cell self-renewal and differentiation. Cell Stem Cell. 2008;2:448–60. doi: 10.1016/j.stem.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 2.Schwanhausser B, Busse D, Li N, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–42. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 3.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–45. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lunde BM, Moore C, Varani G. RNA-binding proteins: modular design for efficient function. Nat Rev Mol Cell Biol. 2007;8:479–90. doi: 10.1038/nrm2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dreyfuss G, Kim VN, Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- 6.Okabe M, Imai T, Kurusu M, et al. Translational repression determines a neuronal potential in Drosophila asymmetric cell division. Nature. 2001;411:94–8. doi: 10.1038/35075094. [DOI] [PubMed] [Google Scholar]
- 7.Nakamura M, Okano H, Blendy JA, et al. Musashi, a neural RNA-binding protein required for Drosophila adult external sensory organ development. Neuron. 1994;13:67–81. doi: 10.1016/0896-6273(94)90460-x. [DOI] [PubMed] [Google Scholar]
- 8.Sakakibara S, Imai T, Hamaguchi K, et al. Mouse-Musashi-1, a neural RNA-binding protein highly enriched in the mammalian CNS stem cell. Developmental Biology. 1996;176:230–42. doi: 10.1006/dbio.1996.0130. [DOI] [PubMed] [Google Scholar]
- 9.Sakakibara S, Nakamura Y, Yoshida T, et al. RNA-binding protein Musashi family: roles for CNS stem cells and a subpopulation of ependymal cells revealed by targeted disruption and antisense ablation. PNAS. 2002;99:15194–9. doi: 10.1073/pnas.232087499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sakakibara S, Nakamura Y, Satoh H, et al. Rna-binding protein Musashi2: developmentally regulated expression in neural precursor cells and subpopulations of neurons in mammalian CNS. J Neurosci. 2001;21:8091–107. doi: 10.1523/JNEUROSCI.21-20-08091.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keyoung HM, Roy NS, Benraiss A, et al. High-yield selection and extraction of two promoter-defined phenotypes of neural stem cells from the fetal human brain. Nature Biotechnology. 2001;19:843–50. doi: 10.1038/nbt0901-843. [DOI] [PubMed] [Google Scholar]
- 12.Sakakibara S, Okano H. Expression of neural RNA-binding proteins in the postnatal CNS: implications of their roles in neuronal and glial cell development. J Neurosci. 1997;17:8300–12. doi: 10.1523/JNEUROSCI.17-21-08300.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okano H, Imai T, Okabe M. Musashi: a translational regulator of cell fate. Journal of Cell Science. 2002;115:1355–9. doi: 10.1242/jcs.115.7.1355. [DOI] [PubMed] [Google Scholar]
- 14.Siddall NA, McLaughlin EA, Marriner NL, et al. The RNA-binding protein Musashi is required intrinsically to maintain stem cell identity. PNAS. 2006;103:8402–7. doi: 10.1073/pnas.0600906103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clarke RB, Spence K, Anderson E, et al. A putative human breast stem cell population is enriched for steroid receptor-positive cells. Dev Biol. 2005;277:443–56. doi: 10.1016/j.ydbio.2004.07.044. [DOI] [PubMed] [Google Scholar]
- 16.Potten CS, Booth C, Tudor GL, et al. Identification of a putative intestinal stem cell and early lineage marker; musashi-1. Differentiation. 2003;71:28–41. doi: 10.1046/j.1432-0436.2003.700603.x. [DOI] [PubMed] [Google Scholar]
- 17.Imai T, Tokunaga A, Yoshida T, et al. The neural RNA-binding protein Musashi1 translationally regulates mammalian numb gene expression by interacting with its mRNA. Molecular and Cellular Biology. 2001;21:3888–900. doi: 10.1128/MCB.21.12.3888-3900.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Battelli C, Nikopoulos GN, Mitchell JG, et al. The RNA-binding protein Musashi-1 regulates neural development through the translational repression of p21WAF-1. Mol Cell Neurosci. 2006;31:85–96. doi: 10.1016/j.mcn.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 19.Wuebben EL, Mallanna SK, Cox JL, et al. Musashi2 is required for the self-renewal and pluripotency of embryonic stem cells. PLoS One. 2012;7:e34827. doi: 10.1371/journal.pone.0034827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kharas MG, Lengner CJ, Al-Shahrour F, et al. Musashi-2 regulates normal hematopoiesis and promotes aggressive myeloid leukemia. Nature Medicine. 2010;16:903–8. doi: 10.1038/nm.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barbouti A, Hoglund M, Johansson B, et al. A novel gene, MSI2, encoding a putative RNA- binding protein is recurrently rearranged at disease progression of chronic myeloid leukemia and forms a fusion gene with HOXA9 as a result of the cryptic t(7;17)(p15;q23). Cancer Res. 2003;63:1202–6. [PubMed] [Google Scholar]
- 22.Ito T, Kwon HY, Zimdahl B, et al. Regulation of myeloid leukaemia by the cell-fate determinant Musashi. Nature. 2010;466:765–8. doi: 10.1038/nature09171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Westerfield M. The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Brachydanio rerio) University of Oregon Press; Eugene, OR: 1995. [Google Scholar]
- 24.Kim H, Shin J, Kim S, et al. Notch-regulated oligodendrocyte specification from radial glia in the spinal cord of zebrafish embryos. Dev Dyn. 2008;237:2081–9. doi: 10.1002/dvdy.21620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blader P, Plessy C, Strahle U. Multiple regulatory elements with spatially and temporally distinct activities control neurogenin1 expression in primary neurons of the zebrafish embryo. Mech Dev. 2003;120:211–8. doi: 10.1016/s0925-4773(02)00413-6. [DOI] [PubMed] [Google Scholar]
- 26.Sato Y, Miyasaka N, Yoshihara Y. Mutually exclusive glomerular innervation by two distinct types of olfactory sensory neurons revealed in transgenic zebrafish. J Neurosci. 2005;25:4889–97. doi: 10.1523/JNEUROSCI.0679-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hochgreb-Hägele T, Bronner ME. A novel FoxD3 gene trap line reveals neural crest precursor movement and a role for FoxD3 in their specification. Dev Biol. 2013;374:1–11. doi: 10.1016/j.ydbio.2012.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trinh LA, Hochgreb T, Graham M, et al. A versatile gene trap to visualize and interrogate the function of the vertebrate proteome. Genes & Development. 2011;25:2306–20. doi: 10.1101/gad.174037.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sauka-Spengler T, Meulemans D, Jones M, et al. Ancient evolutionary origin of the neural crest gene regulatory network. Dev Cell. 2007;13:405–20. doi: 10.1016/j.devcel.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 30.Acloque H, Wilkinson DG, Nieto MA. In situ hybridization analysis of chick embryos in whole-mount and tissue sections. Methods in Cell Biology. 2008;87:169–85. doi: 10.1016/S0091-679X(08)00209-4. [DOI] [PubMed] [Google Scholar]
- 31.Doyle EL, Booher NJ, Standage DS, et al. TAL Effector-Nucleotide Targeter (TALE-NT) 2.0: tools for TAL effector design and target prediction. Nucleic Acids Res. 2012;40:W117–22. doi: 10.1093/nar/gks608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carlson DF, Tan W, Lillico SG, et al. Efficient TALEN-mediated gene knockout in livestock. PNAS. 2012;109:17382–7. doi: 10.1073/pnas.1211446109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cermak T, Doyle EL, Christian M, et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Summerton JE. Morpholino, siRNA, and S-DNA compared: impact of structure and mechanism of action on off-target effects and sequence specificity. Curr Top Med Chem. 2007;7:651–60. doi: 10.2174/156802607780487740. [DOI] [PubMed] [Google Scholar]
- 35.Anderson RM, Bosch JA, Goll MG, et al. Loss of Dnmt1 catalytic activity reveals multiple roles for DNA methylation during pancreas development and regeneration. Dev Biol. 2009;334:213–23. doi: 10.1016/j.ydbio.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shibata S, Umei M, Kawahara H, et al. Characterization of the RNA-binding protein Musashi1 in zebrafish. Brain Res. 2012;1462:162–73. doi: 10.1016/j.brainres.2012.01.068. [DOI] [PubMed] [Google Scholar]
- 37.Park HC, Shin J, Appel B. Spatial and temporal regulation of ventral spinal cord precursor specification by Hedgehog signaling. Development. 2004;131:5959–69. doi: 10.1242/dev.01456. [DOI] [PubMed] [Google Scholar]
- 38.Bernhardt RR, Patel CK, Wilson SW, et al. Axonal trajectories and distribution of GABAergic spinal neurons in wildtype and mutant zebrafish lacking floor plate cells. The Journal of Comparative Neurology. 1992;326:263–72. doi: 10.1002/cne.903260208. [DOI] [PubMed] [Google Scholar]
- 39.Bedell VM, Wang Y, Campbell JM, et al. In vivo genome editing using a high-efficiency TALEN system. Nature. 2012;491:114–8. doi: 10.1038/nature11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moore FE, Reyon D, Sander JD, et al. Improved somatic mutagenesis in zebrafish using transcription activator-like effector nucleases (TALENs). PLoS One. 2012;7:e37877. doi: 10.1371/journal.pone.0037877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Good PJ, Rebbert ML, Dawid IB. Three new members of the RNP protein family in Xenopus. Nucleic Acids Res. 1993;21:999–1006. doi: 10.1093/nar/21.4.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lois C, Alvarez-Buylla A. Proliferating subventricular zone cells in the adult mammalian forebrain can differentiate into neurons and glia. PNAS. 1993;90:2074–7. doi: 10.1073/pnas.90.5.2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Levison SW, Goldman JE. Both oligodendrocytes and astrocytes develop from progenitors in the subventricular zone of postnatal rat forebrain. Neuron. 1993;10:201–12. doi: 10.1016/0896-6273(93)90311-e. [DOI] [PubMed] [Google Scholar]
- 44.Lewis KE, Eisen JS. Paraxial mesoderm specifies zebrafish primary motoneuron subtype identity. Development. 2004;131:891–902. doi: 10.1242/dev.00981. [DOI] [PubMed] [Google Scholar]
- 45.Lewis KE, Eisen JS. From cells to circuits: development of the zebrafish spinal cord. Prog Neurobiol. 2003;69:419–49. doi: 10.1016/s0301-0082(03)00052-2. [DOI] [PubMed] [Google Scholar]
- 46.Wyart C, Del Bene F, Warp E, et al. Optogenetic dissection of a behavioural module in the vertebrate spinal cord. Nature. 2009;461:407–10. doi: 10.1038/nature08323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fukuda T, Kosaka T. The dual network of GABAergic interneurons linked by both chemical and electrical synapses: a possible infrastructure of the cerebral cortex. Neurosci Res. 2000;38:123–30. doi: 10.1016/s0168-0102(00)00163-2. [DOI] [PubMed] [Google Scholar]
- 48.Kawaguchi Y, Katsumaru H, Kosaka T, et al. Fast spiking cells in rat hippocampus (CA1 region) contain the calcium-binding protein parvalbumin. Brain Res. 1987;416:369–74. doi: 10.1016/0006-8993(87)90921-8. [DOI] [PubMed] [Google Scholar]
- 49.Li Q, Lee JA, Black DL. Neuronal regulation of alternative pre-mRNA splicing. Nat Rev Neurosci. 2007;8:819–31. doi: 10.1038/nrn2237. [DOI] [PubMed] [Google Scholar]
- 50.Lee CJ, Irizarry K. Alternative splicing in the nervous system: an emerging source of diversity and regulation. Biol Psychiatry. 2003;54:771–6. doi: 10.1016/s0006-3223(03)00375-5. [DOI] [PubMed] [Google Scholar]
- 51.Calarco JA, Superina S, O'Hanlon D, et al. Regulation of vertebrate nervous system alternative splicing and development by an SR-related protein. Cell. 2009;138:898–910. doi: 10.1016/j.cell.2009.06.012. [DOI] [PubMed] [Google Scholar]
- 52.Lu J, Peatman E, Wang W, et al. Alternative splicing in teleost fish genomes: same-species and cross-species analysis and comparisons. Mol Genet Genomics. 2010;283:531–9. doi: 10.1007/s00438-010-0538-3. [DOI] [PubMed] [Google Scholar]
- 53.Kawahara H, Imai T, Imataka H, et al. Neural RNA-binding protein Musashi1 inhibits translation initiation by competing with eIF4G for PABP. The Journal of Cell Biology. 2008;181:639–53. doi: 10.1083/jcb.200708004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gunter KM, McLaughlin EA. Translational control in germ cell development: A role for the RNA-binding proteins Musashi-1 and Musashi-2. IUBMB Life. 2011;63:678–85. doi: 10.1002/iub.499. [DOI] [PubMed] [Google Scholar]
- 55.Szabat M, Kalynyak TB, Lim GE, et al. Musashi expression in beta-cells coordinates insulin expression, apoptosis and proliferation in response to endoplasmic reticulum stress in diabetes. Cell Death Dis. 2011;2:e232. doi: 10.1038/cddis.2011.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.de Andres-Aguayo L, Varas F, Kallin EM, et al. Musashi 2 is a regulator of the HSC compartment identified by a retroviral insertion screen and knockout mice. Blood. 2011;118:554–64. doi: 10.1182/blood-2010-12-322081. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Knockdown of msi2b by E2/i2 TALEN in zebrafish.
Schematic diagram of genomic region of msi2b targeted by E2/i2 TALENs. SD, splice donor. The squares and letters below the sequence illustrate sites of deletions and replacements, respectively, in the region between exon 2 (upper case) and intron 2 (lower case) of msi2b. Missense mutations are color-coded to indicate resulting splice variant. Gray squares represent non-sense mutations. Exon, E; intron, i.