Abstract
Human Monoglyceride Lipase (MGL) is a recently identified lipase and very little is known about its regulation and function in cellular regulatory processes, particularly in context to human malignancy. In this study, we investigated the regulation and function of Monoglyceride Lipase in human cancer(s) and report that MGL expression was either absent or reduced in the majority of primary colorectal cancers. Immunohistochemical studies showed that reduction of MGL expression in the colorectal tumor tissues predominantly occurred in the cancerous epithelial cells. MGL was found to reside in the core surface of a cellular organelle named “lipid body”. Furthermore, it was found to selectively interact with a number of phospholipids including phosphotidic acid and phosphoinositol(3,4,5)P3, phosphoinositol(3,5)P2, phosphoinositol(3,4)P2 and several other phosphoinositides, and among all phosphoinositides analyzed, its interaction with PI(3,4,5)P3 was found to be the strongest. In addition, overexpression of MGL suppressed colony formation in tumor cell lines and knockdown of MGL resulted in increased Akt phosphorylation. Together, our results suggest that MGL plays a negative regulatory role in PI3-K/Akt signaling and tumor cell growth.
Keywords: Monoglyceride Lipase, gene expression, Akt, phosphatidylinositides, colorectal cancer
INTRODUCTION
Colorectal cancer is the most common gastrointestinal (GI) malignancy and the second most common cause of cancer-related deaths (1, 2). Approximately 80–90% of all colorectal cancers are sporadic (3, 4). With equal incidence in males and females, almost 60% of the colon cancers originate in the descending (left) and rectosigmoid colon, and the most common tumor type is adenocarcinoma (4). Recent molecular and genetic studies have revealed that the majority of the sporadic-type colorectal cancers follows the classical adenoma to carcinoma sequence (3, 4). Although significant progress has been made in better understanding the molecular and genetic events underlying the malignant progression of colorectal cancer, the exact mechanism(s) that are responsible for the malignant transformation of colon mucosa need to be further elucidated.
Evidence suggests that dysregulated lipid metabolism appears to contribute to colorectal cancer development (5). For example, secretory group II PLA2 degrades phospholipid to generate lysophospholipids and its overexpression has been found in colorectal adenomas from familial adenomatous polyposis patients (6). Elevated levels of lysophosphatidylcholine and phosphatidylcholine plasmalogen have been found in malignant colorectal tissues when compared to their matching normal tissues (7). Somatic mutations in the p110 subunit of phosphatidylinositol-3 kinase (PI3-K) have been found in a large portion of colon cancers (8); and such mutations increase the kinase’s activity to generate the phosphoinositol-3-phosphate products (such as PI(3,4,5)P3, PI(3,5)P2), which in turn activate its downstream target Akt that plays a very important role in the regulation of cell proliferation and cell survival (9). Evidence suggests that phospholipids are important not only in the formation of the cytoplasmic membrane and membranes of various organelles, but also in the regulation of many cellular processes such as gene transcription, cell signaling, cell survival and proliferation (5). However, the molecular mechanisms underlying the dysregulation of phospholipid metabolism remain largely unclear and need to be further investigated.
In this study, we have investigated the expression and function of Monoglyceride Lipase (MGL) in human cancer. Our results indicate that MGL mRNA and protein expression were significantly reduced or absent in multiple human malignancies, particularly in colon, lung and breast cancers. We also found that MGL overexpression suppressed colony formation in various tumor cell lines. Interestingly, MGL interacted with several phosphatidylinositol derivatives (PIs), particularly with PI(3,4,5)P3 with the highest affinity and knockdown of MGL resulted in increased Akt phosphorylation. Together, our studies suggest for the first time that MGL may play a negative regulatory role in tumor cell growth and cancer development.
RESULTS
Expression of MGL is absent or reduced in multiple human malignancies
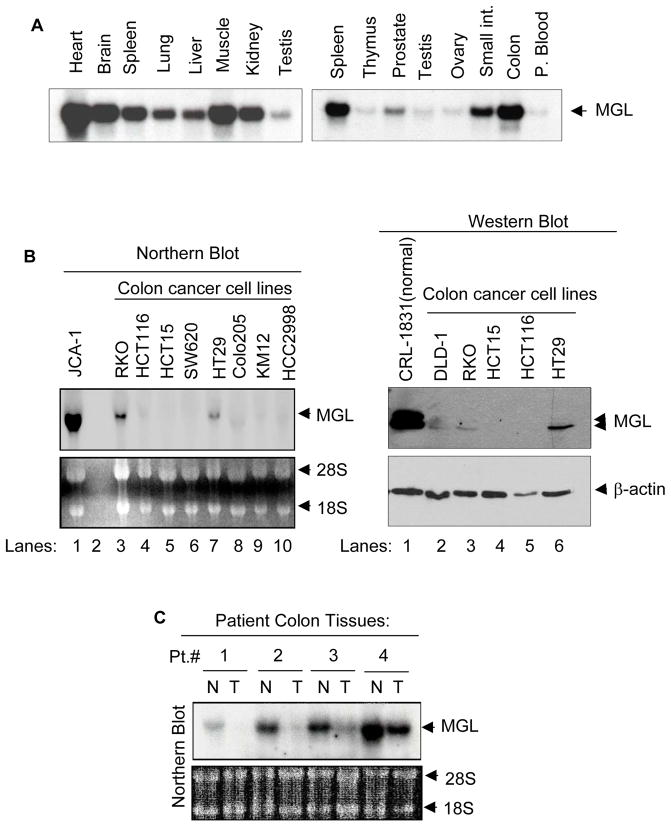
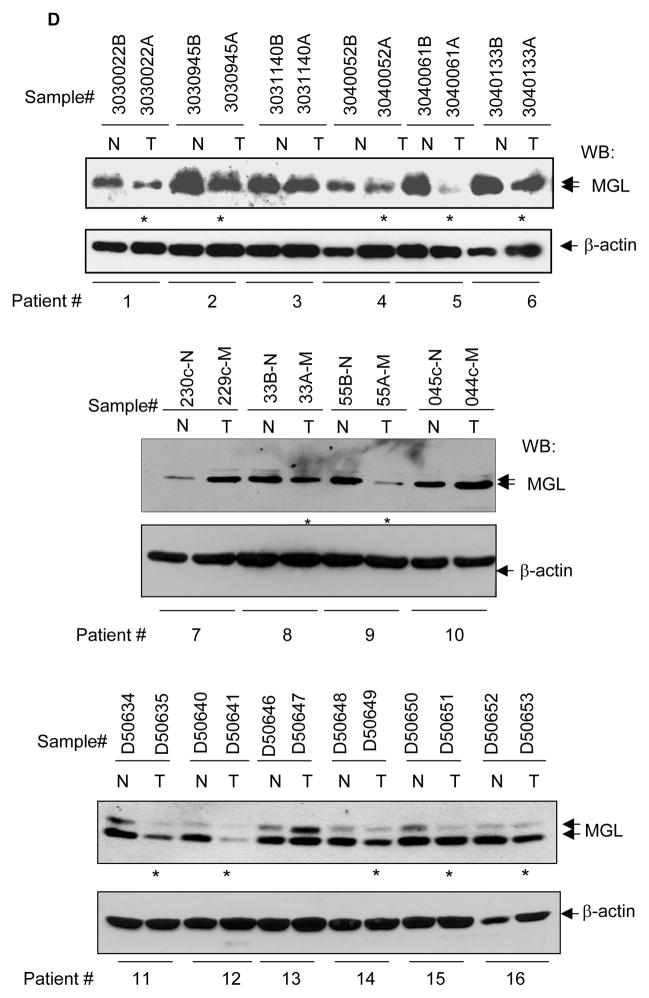
We examined the expression of MGL in multiple normal human tissues. MGL mRNA was detected as an approximately 4.2 kb transcript using a commercially available multi-tissue mRNA membrane. As shown in Fig. 1, higher levels of MGL mRNA expression were seen in the heart, smooth muscle, brain, colon mucosa and spleen, and slightly less in the kidney, lung, liver and small intestine. Testis, prostate, thymus, ovary and peripheral blood showed low levels of MGL expression. To study whether MGL expression was altered in human tumors, we examined MGL expression in various cancer cell lines and primary cancer tissues. In contrast to the high-level expression of MGL seen in the normal colon mucosa (Fig 1A), most of the colon cancer cell lines displayed either undetectable or very low levels of MGL mRNA (Fig. 1B, left panel, lanes 3–10). A high level of MGL mRNA was noted in a bladder cancer cell line JCA-1 (Fig. 1B, lane 1). As shown in Fig. 1B right panel, all colon cancer cell lines (lanes 2–6) examined exhibited a lower or undetectable level of MGL protein when compared to that noted in a normal colon cell line CRL1831 (Fig. 1B right panel, lane 1). We then examined MGL mRNA and protein expression in primary human matching normal and tumor samples. First, we used a commercially available colon tissue RNA membrane carrying samples from matching normal and tumor tissues from 4 patients and noted that MGL mRNA expression was absent or significantly reduced in colon tumor (T) (4 out of 4 patients) compared to their respective matching normal (N) tissues (Fig. 1C). We also sought to investigate MGL expression at the protein level and to that end analyzed matching normal and tumor samples from 61 patients and noted that 33 out of 61 primary colon tumor samples displayed reduced or absent levels of MGL when compared to their matching normal counterparts. Fig. 1D, shows representative results for individuals 1–16 and western blot results for matched normal-tumor tissues of all 61 patients are shown in Supplementary Fig. 1. We also investigated whether MGL expression was only dysregulated in colon cancer or other tumor types were also affected. We found that most of the human breast cancer cell lines examined displayed low levels of MGL mRNA expression (Supplementary Figure 2A). MGL protein expression was also reduced in primary breast cancers when compared to normal tissue samples (Supplementary Figure 2B). Similar dysregulated expression was noted in lung cancer. Supplementary 2C shows an easily detectable level of MGL expression in a normal lung cell line (NL20, lane 1), whereas it was not detectable in 2 out of 4 lung cancer cell lines (Supplementary Figure 2C, lanes 2–5), and MGL expression was also reduced in primary lung cancer when compared to matching normal tissues in 2 out of 3 patients (Supplementary Figure 2D).
Figure 1. MGL expression in human tissues and cell lines.
A. Distribution of MGL in human tissues. Northern blot analysis was performed on multi-tissue blots (2 μg of poly-A RNA per lane, purchased from Clontech) with full-length MGL cDNA as a probe. B. MGL mRNA and protein expression in human colon cancer cell lines. Left panel: Northern blot analysis of MGL mRNA in established human colon cell lines. JCA (human bladder cancer cell line), which expressed high level of MGL was used as a positive control. The membrane was probed with full-length MGL cDNA. The ethidium bromide staining of the membrane in the lower panel serves as loading control. Right panel: Western blot analysis of MGL protein expression in colon normal and cancer cell lines. The membrane was probed with indicated antibodies. It is of note that the normal colon cell line CRL-1831 expressed higher levels of MGL as compared to colon cancer cell lines. C. Northern blot (purchased from Invitrogen) showing MGL expression in primary colon tumor and their matching normal tissues. The membrane was probed with MGL cDNA. The ethidium bromide staining of the membrane in lower panel (provided by the company) serves as the loading control. D. Representative Western blot results showing MGL protein expression in matched human normal colon and primary colon tumor tissues. Membranes were sequentially probed with MGL and β-actin (loading control). The * indicates samples with reduced MGL expression. Representative results of normal and matched tumor samples from 16 patients are shown here and all results of western blot analyses are shown in Supplementary Figure 1.
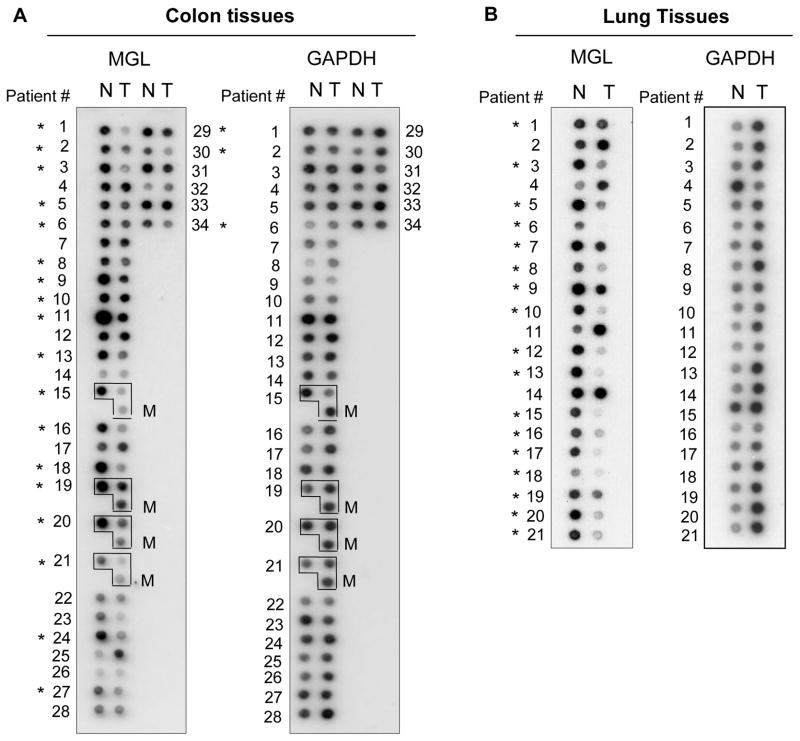
To get a broader view of the MGL expression in human tumors, we used a commercially available Cancer Profiling Array membrane, which contains tumor and matching normal RNA→cDNA samples from 241 individuals representing 13 different tissue types. Figs. 2A and B depict the representative results of Cancer Profiling Array showing that MGL expression was significantly reduced or absent in a large portion of colon and lung tumors (T) compared to their matching normal (N) tissues (left panels). GAPDH probing was also performed to show the loading of all samples (right panels). The overall results of the MGL expression profile in various malignancies are summarized in Table 1. As is shown, when compared to their matching normal tissues, decreased or lost MGL mRNA and protein expression was found in 59.4% (60/101) of colon cancer, 75% (18/24) of lung cancer, 61% (32/52) of breast cancer, 50% (9/18) of rectal cancer and 50% (14/28) of gastric cancer. Interestingly, a decrease in MGL expression was found in a smaller proportion of kidney cancer tissues (35%, 7/20) and endometrium cancer tissues (23.8%, 10/42) and none (0/6) of the thyroid cancer samples was found to have altered MGL expression patterns. In the case of colon cancer, matching normal, tumor and metastatic samples were available from 4 patients and, as shown in Fig 2A, reduced MGL expression was found in both non-metastatic (21/34) and metastatic (4/4) colon tumors when compared to their matching normal counterparts. Together, the presented results indicate that a loss of or reduction in MGL expression occurs commonly in various primary human malignancies, and in the case of colon cancer, also in metastatic tumors.
Figure 2. Cancer Profiling Array showing MGL expression in matching normal and tumor tissues.
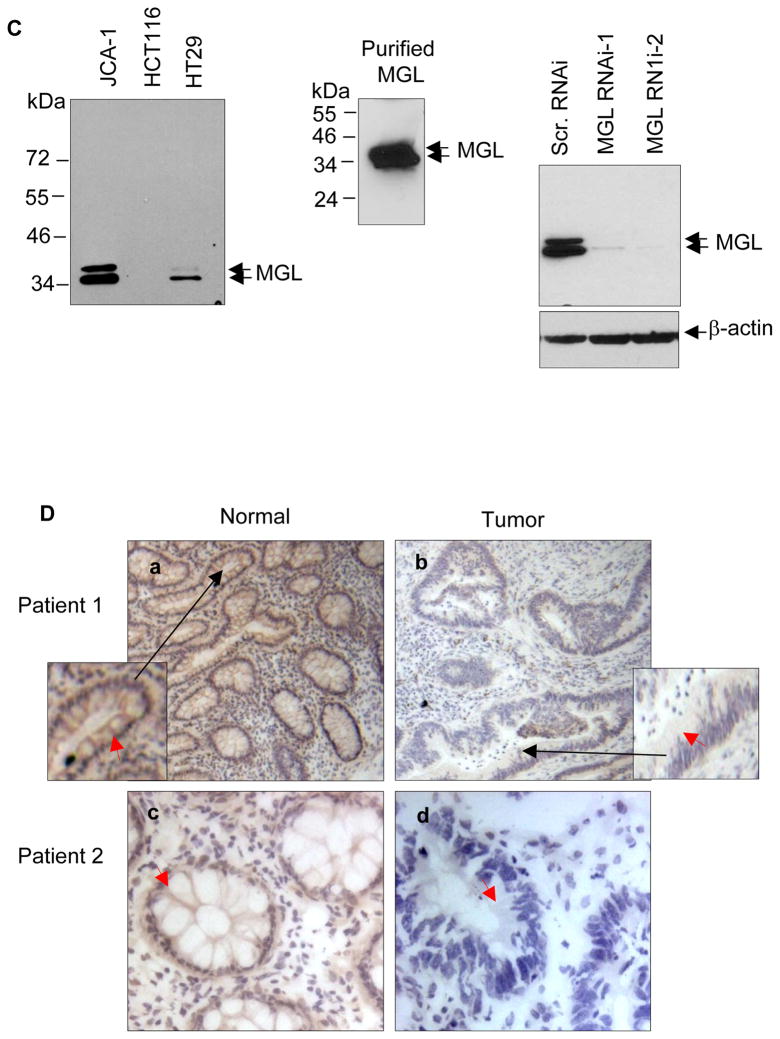
N: normal; T: tumor; M: metastatic tumor. Each number indicates a matching pair of normal and tumor tissues from a single patient. In (A), boxed samples contain normal tissue (upper left), non-metastatic tumor (upper right) and metastatic tumor from same individual. The same membrane was subsequently probed with cDNA of GAPDH as an internal loading control (A & B, right panels). C. Western blot showing the specificity of MGL antibodies against endogenous MGL in human cancer cell lines (left panel), and recombinant purified MGL protein (middle panel). The right panel shows MGL depletion by MGL shRNAs. D. Immunohistochemical staining of MGL protein in patient-matched normal-tumor representing colon tissues. Frozen normal and tumor colon tissues from two individual patients were sectioned and fixed as described in Materials and Methods. The tissue sections were first stained with MGL antibodies and then counterstained with hematoxylin. Brown color indicates the MGL-specific signals. Negative control experiments were also performed using pre-immune serum as primary antibodies followed by the same secondary antibody (data not shown). Red arrows indicate the MGL-specific signal in the epithelial tissues. The small panels within a & b are the enlarged images of the areas indicated by the black arrows.
Table 1.
Summary of MGL expression in patient primary normal and tumor samples*.
| Tissue | Decreased MGL Expression | Tissue | Decreased MGL Expression |
|---|---|---|---|
| Colon | 59.4% (60/101) | Ovary | 50% (7/14) |
| Rectum | 50% (9/18) | Prostate | 50 (2/4) |
| Breast | 61% (32/52) | Kidney | 35% (7/20) |
| Lung | 79% (19/24) | Uterus | 23.8% (10/42) |
| Stomach | 50% (14/28) | Thyroid | 0% (0/6) |
For colon tissues, MGL down or loss expression was detected by Cancer Profiling Array (21/34), Northern blotting (4/4) and Western blotting (33/61), immunohistochemistry (2/2). For breast tissues, information presented here includes data generated by Cancer Profiling Array assay (50 pairs) and Western Blotting (2 pairs). For lung tissues, information was from Cancer Profiling Array (21 pairs) and Western (three pairs). For all other tissues, information was from Cancer Profiling Array only.
Next, we performed immunohistochemistry to investigate MGL expression in matched normal and primary tumor tissues. The specificity of MGL antibodies was demonstrated by Western blot analysis that detected the endogenous and bacterial purified MGL protein, as well as by shRNA knockdown that further confirmed the specific signal for MGL protein (Fig. 2C, left, middle and right panels, respectively). Fig. 2D shows representative results of MGL immunohistochemistry performed on frozen sections of matched normal and tumor tissues from two colon cancer patients. The top and lower left panels show transverse sections of normal colonic mucosa that contains regularly spaced glands of similar size and shape. As is shown, moderate to strong anti-MGL staining is noted in each colonic epithelial cell (red arrows). The fibroblasts in lamina propria were weakly positive in MGL staining. Fig 2D top and lower right panels show sections of colonic adenocarcinoma that contains irregularly sized and shaped tumor glands distributed in desmoplastic stroma. The tumor glands show markedly decreased staining for MGL with focal positivity seen in tumor epithelial cells, capillaries and rarely fibroblasts. It is noteworthy that the positive MGL staining in the connective tissue components of lamina propria was comparable between normal (left panels) and tumor (right panels) tissues which serves as an internal control for MGL staining. These results thus indicate that: 1) MGL is strongly expressed in the normal colorectal epithelial cells and reduced in tumor epithelium, and 2) reduction of MGL expression in the colorectal tumor tissues predominantly occurs in cancerous epithelial cells. Together, results obtained by mRNA, protein and immunohistochemistry analyses all demonstrate that reduction or loss of MGL expression is a common feature in multiple human tumors and decrease or loss of MGL expression may be an important event that favors tumor formation.
MGL localizes to cytoplasmic lipid droplets (CLD)
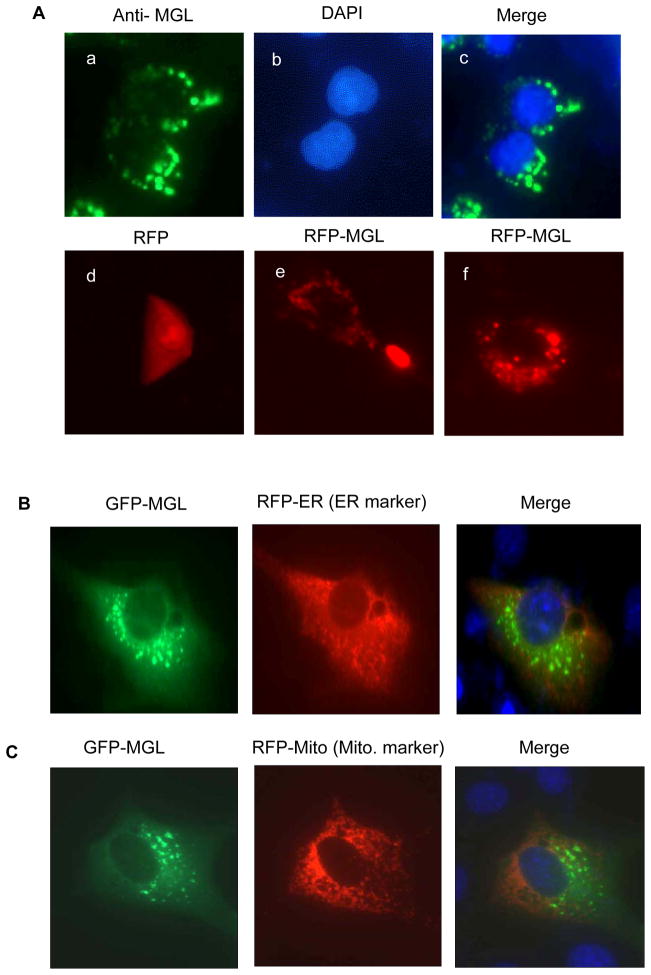
Next, we sought to investigate the cellular localization of MGL protein. Immunofluorescent staining of endogenous MGL protein with an MGL-specific antibody showed MGL was mainly detected in the cytosol with a punctate expression pattern (Fig. 3A, a–c). Exogenously expressed RFP-tagged MGL in NIH3T3 mouse fibroblasts (non-transformed cells) also exhibited similar punctate distribution patterns (Fig. 3A, e & f). Control RFP-only protein distributed evenly throughout whole cells and did not exhibit punctate patterns (Fig. 3A, d). We tried to co-localize MGL with mitochondria, endoplasmic reticulum and lipid droplets (CLDs) (also known as lipid bodies) (10), the cellular organelles that are known to exhibit punctate staining patterns. Recent studies have indicated that lipid bodies not only contain lipid content but also various cellular protein involved in cell signaling (10). Results presented in Figs. 3B & C indicate that GFP-tagged MGL did not predominantly co-localize with the RFP-tagged mitochondrial and ER markers (Fig. 3B). By contrast, RFP-MGL was co-localized with lipid bodies labeled with fluorophore Bodipy (Fig. 3D, a–c), while the staining of RFP-alone (Fig. 3D, d–f) did not exhibit similar patterns. More detailed confocal microscopic analyses revealed that MGL predominantly distributed to the core surface of the CLD and formed “MGL-crescents” around the CLD (Fig. 3E); which was more evident when lipid bodies were enlarged by treatment with oleic acid (Fig. 3E, h–j). Thus, these novel findings indicate that MGL is a CLD-associated protein.
Figure 3. MGL distributes to lipid bodies.
A. a–c: Immunofluorescent staining of endogenous MGL. HT29 colon cancer cells seeded on Lab-Tek chamber slides were fixed and hybridized with MGL-specific antibodies. Cell nuclei were stained with DAPI nuclear dye. d–f: Fluorescent images of RFP (d) or RFP-tagged MGL (e & f). HCT116 cells were transfected with vectors expressing either RFP-only (d) or MGL-RFP fusion protein (e & f). The cells were fixed and analyzed under an Olympus fluorescent microscope. B & C. Cellular localization of GFP-tagged MGL, RFP-ER and RFP-Mito. NIH3T3 cells growing on lab-tek slides were transiently co-transfected with expression vectors expressing GFP-MGL fusion protein (B & C, left panels) or RFP-ER and RFP-Mito fusion proteins (Invitrogen) (B & C, middle panels). It is noteworthy that MGL staining largely did not co-localize with the staining of ER or mitochondrial markers. D. RFP-MGL (red) staining co-localizes with Bodipy staining (green) that is specific to lipid droplets. RKO colon cancer cells were transfected with an RFP-MGL vector (a–c) or a control RFP vector (red) (d–f). Twenty-four hours after transfection, cells were fixed and stained with lipid droplet dye Bodioy (green) and counterstained with nuclear dye DAPI (blue). E. MGL distributes to the core surface of the lipid droplets. RKO (a–g) or HCT116 (h–j) colon cells were transfected with an RFP-MGL expression vector (red). Twenty-four hours after transfection, cells were treated with 300 μM oleic acid overnight (to enlarge the size of the lipid droplets) and then fixed and stained with lipid droplets dye Bodioy (green) followed by counterstaining with DAPI dye (blue). Cells were inspected under either a confocal fluorescent microscope (a–g) or a regular fluorescent microscope (h–j). Image “d” is an enlarged partial image of “c”.
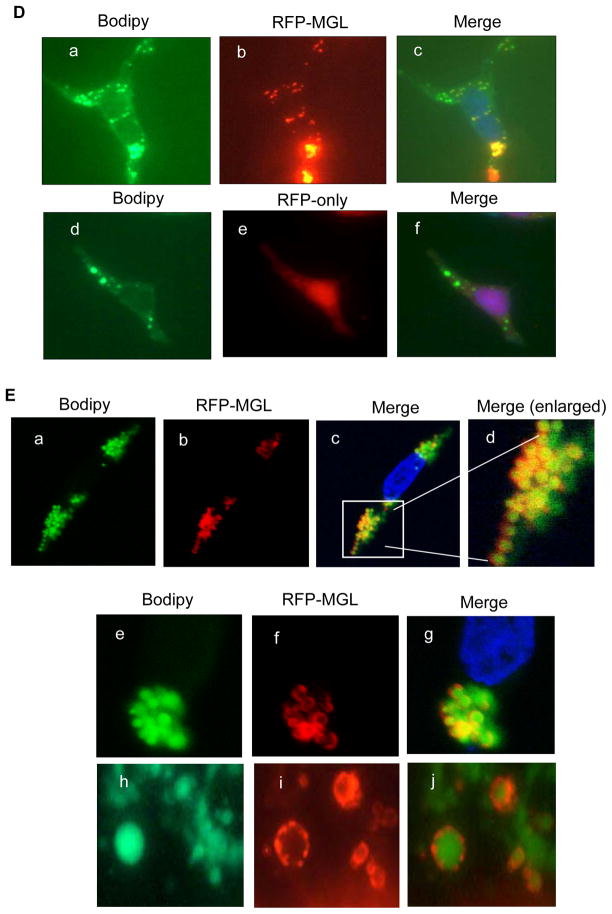
Exogenously expressed MGL suppresses colony formation in colon and lung cancer cells
To determine the effect of MGL on tumor cell growth, we reintroduced exogenous MGL or the control empty vector into tumor cells exhibiting either undetectable or low levels of MGL, and the colony formation of transfected cells was then analyzed. Fig. 4A shows the representative photographs of tissue culture plates depicting colony formation in the presence and absence of exogenous MGL. As is shown, transfection of exogenous MGL significantly reduced colony formation in HCT116 (colon) and H1299 (lung) cancer cell lines. Quantitative results from two independent experiments on HCT116 cells are summarized in Fig. 4B, which indicate that overexpression of MGL suppressed colony formation in HCT116 colon cancer cells by 52–61% when compared to the colonies formed by vector-alone expressing cells. These results suggest that MGL appears to exhibit growth suppression activity in tumor cells.
Figure 4. MGL suppresses colony formation in cancer cells.
A & B. Cells were transfected with control or MGL expression vectors with antibiotic selection marker. Twenty-four hours after transfection, cells were selected using proper antibiotics. Two weeks later, colonies on the tissue culture plates were stained with crystal violet and counted. Representative photographs (A) show tissue culture plates depicting colony formation in the presence and absence of MGL. Quantitative results for two independent colony formation experiments on HCT116 cells are summarized in B.
MGL interacts with phosphatidic acid and phosphoinositol lipids
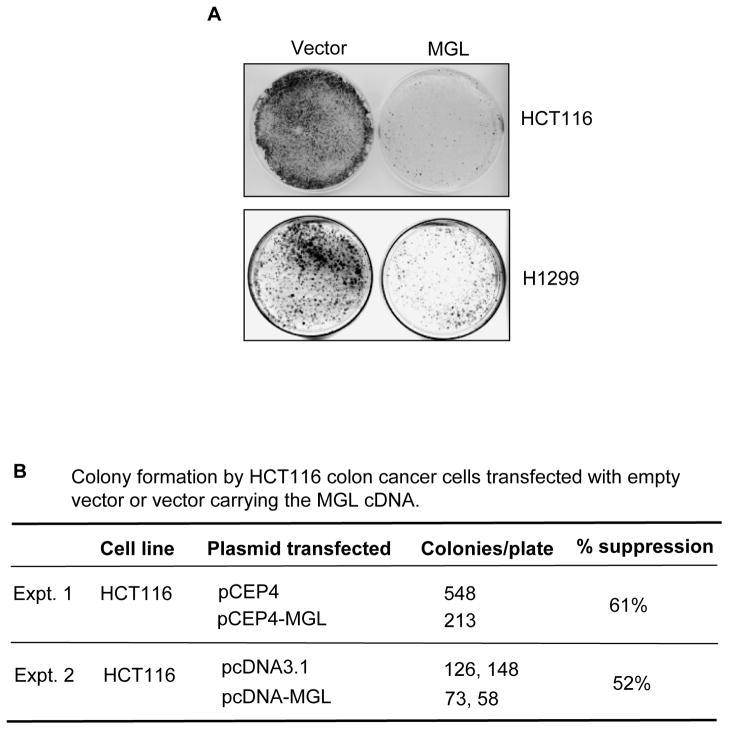
We performed protein-lipid overlay assays to identify MGL-associated lipid(s). As shown in Fig. 5A, eight different phosphoinositol lipids (PtdIns) and phosphatidic acid (PA) showed interaction with MGL on the PIP strips. By contrast, lysophosphatidic acid (LPA), lysophosphocoline (LPC), phosphatidylethanolamine (PE), phosphatidylcholone (PC), sphingosine 1-phosphate (S1P) showed no binding to MGL on the same membrane (Fig. 5A). Phosphatidylserine (PS), on other hand, showed weak binding with MGL (Fig. 5A). We also further analyzed MGL-phosphoinositols binding affinity using the PIP Array membranes that had eight different immobilized phosphoinositides arrayed in amounts ranging from 100 to 1.6 pmol. Fig. 5B shows that MGL displayed highest binding affinity with PtdIns(3,4,5)P3 at 6.25 pmol followed by PtdIns(3,5)P2 and PtdIns(4,5)P2 at 12.5 pmol. MGL showed lower binding affinity with PtdIns(5)P and PtdIns(4)P at 25 pmol and with PtdIns(3)P at 50 pmol. MGL did not show detectable interaction with PtdIns (no phosphate) on the same membrane. These results were reproducible and suggest that MGL interacts with phospholipids, particularly with certain species of phosphoinositides, such as PtdIns(3,4,5)P3 and PtdIns(3,5)P2. Position-3 phosphorylated phosphoinositides are known to be important for activation of Akt, a kinase critical for regulation of cell growth and cell survival.
Figure 5. MGL selectively interacts with phophatidic acid and phosphoinositide derivatives demonstrated by Protein-Lipid Overlay assays.
A. Recombinant purified MGL protein was incubated with commercially available lipid membranes containing various phospholipids as described in Materials and Methods. The lipid-bound MGL was detected as described in Materials and Methods. B. Binding affinity analyses of MGL to various phosphoinositide derivatives. Purified MGL protein was incubated with Phosphoinositide Array membrane, which was spotted with decreased amounts of various phosphoinositide derivatives. MGL-lipid interaction was detected as described in A. Protein-Lipid Overlay assays were performed on the duplicated membranes and similar results were observed.
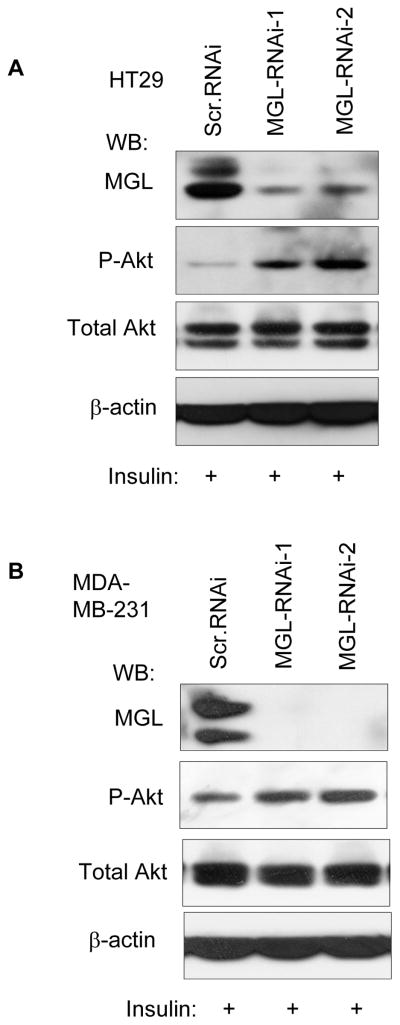
Effect of MGL knockdown on Akt phosphorylation
Next, we sought to determine the effect of MGL on PI3-K/Akt using the lentivirus-mediated MGL knockdown approach and HT29 colon cancer cells and MDA-MB-231 breast cancer cells were used in these experiments. These lines were chosen because that they exhibit easily detectable levels of MGL protein and represent colon and breast cancers in which MGL expression is altered. Cells were infected either with viruses carrying scramble shRNA or two different MGL-specific shRNAs that targeted different regions of MGL mRNA. Akt phosphorylation was then determined in the MGL KD cell or the scramble RNAi control cells. Fig. 6 shows that depletion of MGL significantly enhanced the phosphorylation of Akt but did not affect the protein levels of total Akt protein upon insulin stimulation. The effect of MGL KD on enhancement of Akt phosphorylation was observed in both HT29 colon cancer cells (Fig. 6A) and MDA-MB-231 breast cancer cells (Fig. 6B). Thus, these results indicate that phosphorylation of Akt may be constitutively suppressed by MGL and MGL KD appears to alleviate this suppression, leading to activation of Akt.
Figure 6. MGL knockdown enhances phosphorylation of Akt.
HT29 (A) and MDA-MB-231 (B) colon and breast cancer cells, respectively, were infected with lentivirus carrying MGL shRNA or scramble shRNA. Seven to ten days after infection, cells were treated with 100 μg/ml insulin 30 min prior to being lysed. Western blot analyses were performed sequentially on the same membrane with indicated antibodies.
DISCUSSION
In this study we have investigated MGL expression and function in human cancer. Our results indicate that MGL expression is either absent or reduced in the majority (59.4%, 60/101) of primary colorectal cancer examined. In addition, we also note that reduction in MGL expression also occurred in other types of cancer, including tumors of the lung (75%, 18/24), breast 61% (32/52), stomach (50%, 14/28) and ovary (50% (7/14). Our results also indicate that MGL reduction occurred at both mRNA and protein levels. Decrease in MGL protein expression is likely to be a consequence of reduced mRNA levels in these tumor cells, although we cannot rule out the possibility that MGL alteration may also occur at the protein level. In addition, we also note that MGL expression was increased in a smaller portion [14% (14/101) from the combined results of all our studies] of primary colorectal tumors. Currently, we do not know why a small subset of the colorectal tumors has very different MGL expression pattern. Future studies are needed to determine the mutational status and regulation of MGL in human cancers. Nevertheless, our results presented here have demonstrated that loss or decreased MGL expression is a common feature of multiple human malignancies.
A recent study by Nomura et al. (11) investigated MGL’s role in cancer cells. Their results suggested that elevated MGL lipase activity was associated with more aggressive behavior and metastatic potential of the cancer cells. However, the expression status of endogenous MGL in tumor cells and in primary human tumor tissues was not investigated in their study (11). Interestingly, we found that in 4 out of 4 colon cancer patients, MGL expression was reduced in primary as well as metastatic lesions when compared to matching normal tissues (Fig. 2A). These findings therefore indicate that in these patients, metastasis occurred in the absence of elevated levels of MGL. Further studies are needed to analyze MGL expression in larger cohort of metastatic samples to firmly establish the status of MGL in metastases.
To date, very little is known about MGL in the context of its role in cellular regulatory processes. MGL has been implicated in degrading cannabinoid 2-arachidonoyglycerol (2-AG), which generates free fatty acids (11). However, whether the functional role of MGL is limited to its lipase activity for 2AG is a debatable issue. Our results show that MGL binds to a number of phospholipids including phosphotidic acid (PA) and phosphoinoside derivatives such as PI(3,4,5)P3, PI(3,5)P2, PI(3,4)P2 and other phosphoinositides. The 3-phosphorylated phosphoinositides, products of PI3-kinase, are important signaling molecules that activate Akt kinase, a serine/threonine kinase that is stimulated by the binding of the 3-phosphorylated phosphoinositides (9). Akt phosphorylation/activation plays an important role in regulation of cell proliferation and cell survival (9). In this study, we show that depletion of MGL, via lentivirus-mediated knockdown, increased Akt phosphorylation (Fig. 6). Our results thus suggest that MGL plays a negative role in the regulation of Akt phosphorylation. In addition, MGL may also exhibit functions other than its lipase activity and lipid association. Previous studies have shown that cytoplasmic phospholipases A2 (cPLA2) can exhibit this function by interacting with proteins. For example, it has been shown that cPLA2 interacts with cPLA2-interacting protein (PLIP), the β-unit of BK channel and integrin alphaIIbbeta3 (αIIbβ3) (12, 13, 14). Interactions between cPLA2 and PLIP have been shown to enhance mesangial cells sensitivity to serum withdrawal-induced apoptosis (12) and cPLA2 and αIIbβ3 interaction enhanced each other’s functions during αIIbβ3 signaling (13). The issue of whether MGL could exhibit its growth regulatory function via protein-protein interaction remains to be carefully investigated.
In summary, several lines of evidence presented in our studies do not support an oncogenic role of MGL in human cancers: (1) reduced/lost MGL expression widely occurred in a large portion of human cancers; (2) exogenously expressed MGL in tumor cells lacking MGL suppressed their growth; (3) MGL knockdown activated Akt phosphorylation. Whether MGL has dual roles in cell growth regulation in different cell types is an issue that warrants further investigation. Our data, nevertheless, support the notion that MGL has growth suppressive role and its loss or reduction may favor tumor formation and/or progression in cells lacking this protein. Clearly, further studies are needed to investigate the roles of MGL in human malignancy.
MATERIALS AND METHODS
Antibodies and reagents
The GFP and β-actin antibodies were from Boehringer Mannheim (Indianapolis, IN) and Sigma (St. Louis, MO) respectively; the HA-tag antibody (HA.11) was from Covance (Berkeley, CA); and the p85 and p110α antibodies were from Cell Signaling Technology, Inc. (Beverly, MA) and Santa Cruze Biotechnology Inc (Santa Cruze, CA), respectively. The FITC-conjugated goat anti-rabbit secondary antibody was from Pierce (Rockford, IL), and the peroxidase-conjugated goat anti-rabbit antibody was from Vector Laboratories (Burlingame, CA). The MGL antibody was generated by immunizing rabbits with bacterial purified full-length MGL protein through a commercial source (Pocono Rabbit Farm & Laboratory, Canandensis, PA). The VECTASTAIN ABC kit was from Vector Laboratories (Burlingame, CA). The bodipy lipid probe was from Molecular Probes Inc. (Eugene, OR). The matching human patient normal and tumor samples were from The Cooperative Human Tissue Network (CHTN), an organization sponsored by the National Cancer Institute (NCI).
Cell culture conditions
Human cancer cell lines HT29 (colon), SW480 (colon), JCA-1 (bladder), A549 (lung), H1299 (lung) and MDA-MB-231 (breast) were regularly maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Cellgro, Mediatech, Herndon, VA) supplemented with 10% fetal bovine serum (FBS) (Gemeni Bioproducts Inc., Calabasas, CA). HCT116 cells (colon) were maintained in McCoy’s medium (Cellgro) supplemented with 10% FBS (Gemeni Bioproducts Inc.).
Expression plasmids
RFP-tagged MGL was generated by inserting the full-length MGL cDNA into the pDsRedN1 vector (Clontech, Mountain View, CA). MGL expression vectors used for colony formation assays were generated by inserting the MGL ORF into the multiple cloning site of pCEP4 (with hygromycin selection marker) or pcDNA3.1 (with neomycin selection marker) (Invitrogen Corporation, Carlsbad, CA).
Lentivirus-mediated shRNA silencing
MGL expression was silenced by the lentivirus-mediated shRNA approach. The scramble shRNA construct was purchased from Addgene, Inc. (Cambridge, MA). All other shRNA constructs were purchased from Open Biosystems, Inc. (Huntsville, AL). The two different nucleotide sequences to target the human MGL used in this study were as follows: 5′-ccaatcctgaatctgcaacaa-3′, 5′-caactccgtcttccatgaaat-3′. Virus production and infection were performed per the Addgene’s protocol.
MGL mRNA expression analyses
The human multi-tissue Northern blot membrane (2 μg poly-A mRNA per lane) was purchased from Panomics (Fremont, CA, USA). The Human Cancer Profiling Arrays membrane (contains 241 cancer patients’ matching normal and tumor tissues) was purchased from BD Bioscience/Clontech (Mountain view, CA, USA). The Northern blot membrane containing matching normal and tumor RNAs extracted from 4 colon cancer patients was purchased from Invitrogen Corporation (Carlsbad, CA). Northern blot analyses were performed according to standard procedures as we previously described (15). To detect MGL mRNA expression, full-length MGL cDNA fragment was used as a probe. Full-length GADPH probe was also used for loading control.
Western blot, immunoflorescent staining and immunohistochemistry
Western blot analyses were performed as previously described (16). For immunoflorescent staining of endogenous MGL protein, cells were blocked with 10% goat serum in PBS, and then incubated with MGL antibodies (1:500 diluted in PBS with 1.5% goat serum), followed by FITC-labeled anti-rabbit antibodies (1:500 diluted in PBS with 1.5% goat serum). Cells were counterstained with DAPI nuclear dye and analyzed under an Olympus fluorescent microscope or a fluorescent confocal scanning microscope (Zeiss LSM510).
For immunohistochemistry staining, 6 μm frozen tissue sections were fixed in 4% paraformaldehyde followed by 1% hydrogen peroxide for 10 min. All sections were blocked with 10% goat serum in PBS for 20 minutes before incubation with MGL antibodies followed by peroxidase-labeled anti-rabbit antibodies. The sections were then incubated with Vectastain elite ABC reagent for 30 minutes and then the peroxidase substrate DAB solution was added until the desired stain intensity developed. Finally, all slides were counterstained with hematoxylin (Sigma).
Colony formation assay
Colony formation assays were performed as previously described (16). Cells were transfected with same amounts of control vector or MGL expression vector carrying the antibiotic selection markers via Lipofectamine 2000 as per the protocol of the manufacture (Invitrogen). Cells were subsequently selected with antibiotics (hygromycin 200 μg/ml or G418 500 μg/ml) for 14–20 days and then stained with crystal violet.
Protein-lipid overlay assays
Protein-lipid overlay assays were conducted using commercial phospholipid-containing PIP-Strips or PIs-containing PIP array membrane (Echolon Biosciences, Salt lake city, UT). All membranes were blocked with 3% fatty acid-free BSA in TBST buffer (150 mM NaCl, 10 mM Tris·HCl, pH8.0, and 0.1% Tween-20) for 1 hour at room temperature. Blocked membranes were incubated overnight at 4°C with 0.5 μg/ml purified MGL protein. The membranes were then washed 3X (10 minutes each time) with blocking buffer, followed by incubation with MGL antibodies (1:2000 diluted in TBST with 4% fatty acid free BSA) overnight at 4°C on a shaker. Following three washes over 30 minutes with blocking buffer, the membranes were incubated for 1 hour with peroxidase-conjugated goat anti-rabbit antibodies (1:2000 diluted in blocking buffer) and visualized with Supersignal West Pico chemiluninescent substrate (Pierce, Rockford, IL) according to the manufacturer’s instructions.
Supplementary Material
Acknowledgments
This work was supported by NIH grants DK062136 and CA121850 to YH.
Abbreviations
- MGL
Monoglyceride lipase
- PI3-K
Phosphoinositide 3-Kinase
- PdtIns
Phosphatidylinositol
Footnotes
Conflict of interest: No potential conflict of interest to declare.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, et al. Cancer statistics. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Fearon ER. Molecular genetics of colorectal cancer. Annu Rev Pathol. 2011;6:479–507. doi: 10.1146/annurev-pathol-011110-130235. [DOI] [PubMed] [Google Scholar]
- 3.Rustgi AK. The genetics of hereditary colon cancer. Genes Dev. 2007;21:2525–2538. doi: 10.1101/gad.1593107. [DOI] [PubMed] [Google Scholar]
- 4.Kumar V, Abbas AK, Fausto N, editors. Pathologic basic of disease. 7. Elsevier Inc; Philadelphia, PA: 2005. Robbins and Costran; pp. 856–868. [Google Scholar]
- 5.Duan RD. Phospholipid signals and intestinal carcinogenesis. J Food and Nutrition. 2006;50 (S2):45–53. [Google Scholar]
- 6.Kennedy BP, Soravia C, Moffat J, Xia L, Hiruki T, Collins S, et al. Overexpression of the nonpancreatic secretory group II PLA2 messenger RNA and protein in colorectal adenomas from familial adenomatous polyposis patients. Cancer Res. 1998;58:500–503. [PubMed] [Google Scholar]
- 7.Dueck DA, Chan M, Tran K, Wong JT, Jay FT, Littman C, et al. The modulation of choline phosphoglyceride metabolism in human colon cancer. Mol Cell Biochem. 1996;162:97–103. doi: 10.1007/BF00227535. [DOI] [PubMed] [Google Scholar]
- 8.Karakas B, Bachman KE, Park BH. Mutation of the PIK3CA oncogene in human cancers. Br J Cancer. 2006;94:455–459. doi: 10.1038/sj.bjc.6602970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yuan TL, Cantley LC. PI3K pathway alterations in cancer: variations on a theme. Oncogene. 2008;27:5497–5510. doi: 10.1038/onc.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Accioly MT, Pacheco P, Maya-Monteiro CM, Carrossini N, Robbs BK, Oliveira SS, et al. Lipid bodies are reservoirs of cyclooxygenase-2 and sites of prostaglandin-E2 synthesis in colon cancer cells. Cancer Res. 2008;68:1732–1740. doi: 10.1158/0008-5472.CAN-07-1999. [DOI] [PubMed] [Google Scholar]
- 11.Nomura DK, Long JZ, Niessen S, Hoover HS, Ng SW, Cravatt BF. Monoacylglycerol lipase regulates a fatty acid network that promotes cancer pathogenesis. Cell. 2010;140:49–61. doi: 10.1016/j.cell.2009.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sheridan AM, Force T, Yoon HJ, O’Leary E, Choukroun C, Taheri MR, et al. PLIP, a novel splice variant of Tip60, interacts with group IV cytosolic phospholipase A(2), induces apoptosis, and potentiates prostaglandin production. Mol Cell Biol. 2001;21:4470–4481. doi: 10.1128/MCB.21.14.4470-4481.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li J, Al-Khalili O, Ramosevac S, Eaton DC, Denson DD. Protein-protein interaction between cPLA2 and splice variants of alpha-subunit of BK channels. Am J Physiol Cell Physiol. 2010;298:C251–262. doi: 10.1152/ajpcell.00221.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prévost N, Mitsios JV, Kato H, Burke JE, Dennis EA, Shimizu T, et al. Group IVA cytosolic phospholipase A2 (cPLA2α) and integrin αIIbβ3 reinforce each other’s functions during αIIbβ3 signaling in platelets. Blood. 2009;113:447–457. doi: 10.1182/blood-2008-06-162032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang Y, He Q, Hillman MJ, Rong R, Sheikh MS. Sulindac Sulfide-induced apoptosis involves death receptor 5 and the caspase 8-dependent pathway in human colon and prostate cancer cells. Cancer Res. 2000;61:6918–6924. [PubMed] [Google Scholar]
- 16.Rong R, Montalbano J, Jin W, Zhang J, Garling M, Sheikh MS, et al. Oncogenic Ras-mediated downregulation of Gadd 153/CHOP is required for Ras-induced cellular transformation. Oncogene. 2005;24:4867–4872. doi: 10.1038/sj.onc.1208660. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.